Abstract
Checkpoint inhibitors produce durable responses in numerous metastatic cancers, but immune-related adverse events (irAEs) complicate and limit their benefit. IrAEs can affect organ systems idiosyncratically; presentations range from mild and self-limited to fulminant and fatal. The molecular mechanisms underlying irAEs are poorly understood. Here, we report a fatal case of encephalitis arising during anti-PD-1 therapy. Histologic analyses revealed robust T-cell infiltration and prominent PD-L1 expression. We identified 209 reported cases in global pharmacovigilance databases—across multiple cancer types—of encephalitis associated with checkpoint inhibitor regimens, with a 19% fatality rate. We performed further analyses from the index case and two additional cases to shed light on this recurrent and fulminant irAE. Spatial and multi-omic analyses pinpointed activated memory CD4+ T cells as highly enriched in the inflamed, affected region. We identified a highly oligoclonal T cell receptor (TCR) repertoire, which we localized to activated memory cytotoxic (CD45RO+GZMB+) CD4 cells. We also identified Epstein-Barr-virus-specific T cell receptors and EBV+ lymphocytes in the affected region, which we speculate contributed to neural inflammation in the index case. Collectively, the 3 cases studied here identify CD4+ and CD8+ T cells as culprits of checkpoint inhibitor-associated immune encephalitis.
Main
Monoclonal antibodies blocking key negative regulators of T cell function have transformed the management of numerous cancers1. While immune checkpoint inhibitors (ICI) induce durable responses some patients, aberrant immune-mediated phenomena, termed immune-related adverse events (irAEs) may be unleashed in an unpredictable fashion2. Most irAEs are self-limited or resolve with glucocorticoids3, although highly morbid and even fatal events can occur4–6. Inflamed tissues affected by irAE are characterized by robust infiltration by T lymphocytes and myeloid-lineage cells5–8. Uncommon irAEs affecting the central nervous system (CNS; encephalitis), peripheral nervous system (Guillain-Barre, peripheral sensory neuropathy), and neuromuscular junction (myasthenia gravis) have been described9. The molecular basis of irAEs, including neurologic irAEs, remains poorly understood despite these early findings.
We performed deep molecular profiling of a case of fulminant anti-PD-1-induced encephalitis to provide insight into a severe neurologic irAE. We show that ICI-induced encephalitis occurs across cancer settings and is serious or fatal in many patients. Multi-omic analyses of the index case revealed a clonal CD4+ activated memory cytotoxic T cell population, and robust CNS infiltration by cytotoxic CD4+ and CD8+ T cells.
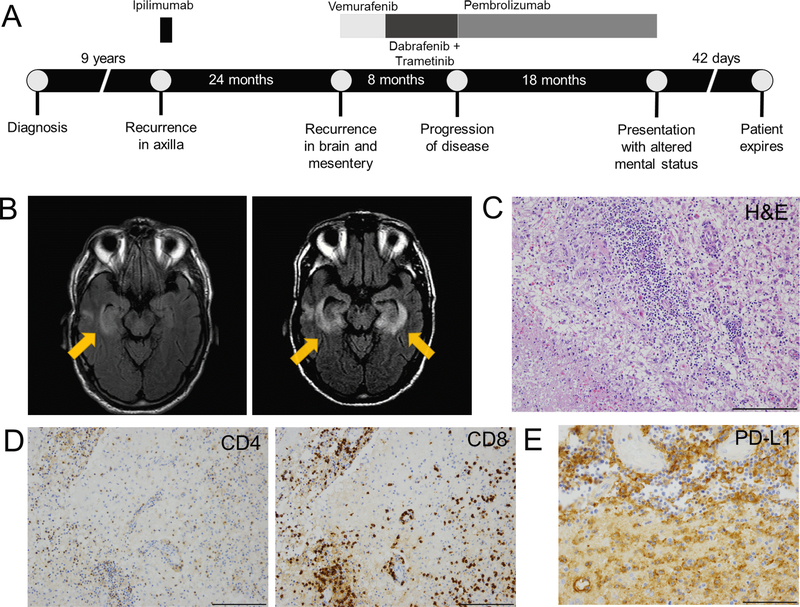
A man in his 70s with metastatic melanoma developed meningoencephalitis while receiving pembrolizumab. His prior history was notable for treatment with ipilimumab (complicated by colitis requiring infliximab), brain metastasis treated with resection and radiation, and BRAF and MEK inhibitor therapy (Figure 1A). Following progression, he received pembrolizumab resulting in near-complete response without toxicity. Approximately 18 months into pembrolizumab therapy, he developed nausea, fevers, confusion, and aphasia progressing over 2 days. Brain MRI showed restricted diffusion and enhancement in the basal ganglia and right temporal lobe consistent with inflammatory or herpes simplex virus (HSV) encephalitis (Figure 1B; Supplementary Table 1). Of note, Epstein-Barr virus (EBV) polymerase chain reaction (PCR) later returned positive with viral load of 1200 copies/mL and was persistently detected in the blood (1888–4321 copies) and across multiple CSF samples. He continued to have neurologic deterioration requiring intubation and mechanical ventilation; MRI showed progressive encephalitis involving both temporal lobes (Figure 1B). Methylprednisolone 1 mg/kg twice daily was initiated five days into the hospitalization, leading to dramatic and rapid clinical improvement.
Figure 1: Clinical course of anti-PD-1-induced encephalitis and histologic findings at autopsy.
A) Timeline of index patient diagnosis, prior therapies and anti-PD-1 therapy. B) (left) Magnetic resonance image (MRI) demonstrating restricted diffusion and hyper-intensity in the right temporal lobe and left basal ganglia from the day of hospital admission. (right) MRI showing progressive bilateral medial temporal encephalitis with bilateral putamen involvement. C) H&E stain shows intense perivascular lymphocytic infiltrate extending to adjacent brain parenchyma, next to a region of infarction (lower left); 20x. D) CD4 and CD8 IHC showing the lymphocytic infiltrate is composed of approximately equal ratios of CD4+ and CD8+ T cells; 20x. E) Diffuse PD-L1 positivity staining in cells with macrophage morphology surrounding the region of infarction; 20x. *Of particular note, tissues collected and assayed in this study anteceded treatment with steroids and anti-TNFα, which could impact the immunologic status of the patient.
Two weeks later, he developed recurrent neurologic symptoms while tapering steroids; MRI showed worsening of encephalitis. Subsequent CSF evaluation showed evolution to a lymphocytic predominance (83 nucleated cells, 8% neutrophils, 91% lymphocytes). He then had brief episodes of clinical improvement with high-dose steroids and intravenous immunoglobulin, followed by progressive deterioration of his mental status and motor strength. He expired 42 days after symptom onset.
To assess the frequency of encephalitis among patients treated with ICI, we queried VigiBase (see Methods). We identified 209 reports of encephalitis; the median age was 61 (range 7−85) and 59% had either melanoma or lung cancer (Table 1). 66% of patients received anti-PD-1 monotherapy; 88% of patients had no other concurrent irAEs reported. The median time to symptom onset was 67 days (range 5−456 days) and 39 patients (19%) died. In contrast to other toxicities (e.g. myocarditis)4, the fatality rate was similar between encephalitis from anti-PD-1 monotherapy vs. combined anti-PD-1 + anti-CTLA-4 (20% vs. 26%, chi-square p=0.50). We next queried 2,501 patients from 4 large academic centers treated with ICI to confirm these findings in clinically-diagnosed cases of encephalitis and identified 22 cases of meningoencephalitis. These findings (Supplementary Table 2), were similar to the pharmacovigilance database, with a median onset of 80 days (range 4−684). The most frequent presenting symptom was confusion (50%), followed by headaches (36%), seizures (23%), and fevers (18%). Temporal or occipital lobe enhancement was the most common imaging finding (22%, n=5). Most patients recovered completely (68%) although 5 (22%) had persistent neurologic symptoms and 2 (9%) died; median hospitalization time was 6.5 days (range 2−120 days). Thus, encephalitis is a rare but recurrent and potentially fatal toxicity of ICI that occurs across cancer types.
Table 1:
Characteristics of patients with immune checkpoint inhibitor associated encephalitis (n=209)
| Characteristic | Number (%) |
|---|---|
| Male | 113 (54) |
| Female | 69 (33) |
| Not listed | 27 (13) |
| Age (years; median, range) | 61,16 – 85 |
| Cancer | |
| Melanoma | 44 (21) |
| Lung cancer (NSCLC and other) | 80 (38) |
| Hodgkin’s lymphoma | 14 (7) |
| Non-Hodgkin’s lymphoma | 3 (1.5) |
| Renal | 9 (4.5) |
| Other/not listed | 59 (28) |
| Region reporting | |
| Americas | 117 (56) |
| Europe | 67 (32) |
| Asia | 19 (9) |
| Oceania | 6 (3) |
| Regimen | |
| Anti-PD-1 monotherapy | 137 (66) |
| Combination anti-PD-1 + anti-CTLA-4 | 31 (15) |
| Other combinations* | 14 (7) |
| Anti-PD-L1 monotherapy | 15 (7) |
| Anti-CTLA-4 (ipilimumab) monotherapy | 12 (6) |
| Timing, days (median, range) | 67 (5 – 456) |
| Reporting term | |
| Encephalitis | 145 (69) |
| Encephalitis autoimmune | 36 (17) |
| Limbic encephalitis | 15 (7) |
| Meningoencephalitis | 12 (6) |
| Cerebellitis | 1 (1) |
| Concurrent irAEs | |
| Colitis | 4 (2) |
| Pneumonitis | 6 (3) |
| Thyroiditis | 7 (3) |
| Myocarditis | 2 (1) |
| Vasculitis | 2 (1) |
| Adrenal insufficiency | 4 (2) |
| Dermatitis | 4 (2) |
| Other‡ | 7 (3) |
| Fatal outcome | 39 (19) |
| Reporting year | |
| 2012 – 2014 | 5 (2.5) |
| 2015 | 5 (2.5) |
| 2016 | 36 (17) |
| 2017 | 77 (37) |
| 2018 (through Sep 5) | 86 (41) |
Atezolizumab, chemotherapy (n=2); atezolizumab, bevacizumab (n=2); atezolizumab, cobimetinib (n=1); ipilimumab, crizotinib, nivolumab (n=1); ipilimumab, bevacizumab (n=1); ipilimumab, nivolumab, golimumab (n=1); ipilimumab, interferon (n=1); ipilimumab, nivolumab, cabozantinib (n=1); ipilimumab, aldesleukin (n=1); nivolumab, cabozantinib (n=1); nivolumab, denosumab (n=1); nivolumab, brentuximab (n=1); pembrolizumab, chemotherapy (n=1).
Pancreatitis (n=1), insulin-dependent type 1 diabetes, hypopituitarism (n=2), hypophysitis (n=2), cytokine storm (n=1), autoimmune nephritis (n=1)
Abbreviations: non-small cell lung cancer (NSCLC). We used the following MedDRA 21.0 search term levels for our reaction criteria: encephalitis and/or encephalitis autoimmune and/or limbic encephalitis and/or meningoencephalitis and/or cerebellitis. For our substance criteria, we used pembrolizumab, atezolizumab, nivolumab, durvalumab, avelumab, ipilimumab, tremelimumab, and all combinations thereof.
An autopsy of the index case was performed. No evidence of melanoma was observed in the CNS or elsewhere using pan-melanoma stains S100, SOX10, melanoma antigen recognized by T cells 1 (MelanA/MART-1), and MITF (data not shown). Analysis of T cell receptors (reported subsequently in this letter) also did not identify any known or likely MelanA/MART-1 specific HLA-A*02:01-restricted TCRs. Vigorous inflammation, multiple cerebral infarctions with necrosis, and sporadic hemorrhagic transformation were noted in the bilateral temporal lobes and striatum with surrounding gliosis and numerous macrophages (Figure 1C, Extended Data 1A). Exuberant perivascular inflammation extending to adjacent gray matter characterized by approximately equal ratios of CD4+ and CD8+ T cells was observed (Figure 1D), although we noted that CD4+ cells were present in localized patterns while CD8+ cells were diffuse and consistently present. We also observed frequent CD68+ cells, a macrophage marker which also stains microglial cells (Extended Data 1B). Minimal CD20+ infiltrates or immunoglobulin deposition were observed, suggesting that B cells did not constitute a major component of the inflammatory process (Extended Data 1C). We observed similar patterns of T cell and macrophage infiltration in inflamed meninges and adjacent gray matter, including perivascular spaces (Extended Data 2). By contrast, brain tissue obtained from radiographically normal brain was histologically unremarkable with no inflammatory infiltrate (Extended Data 3).
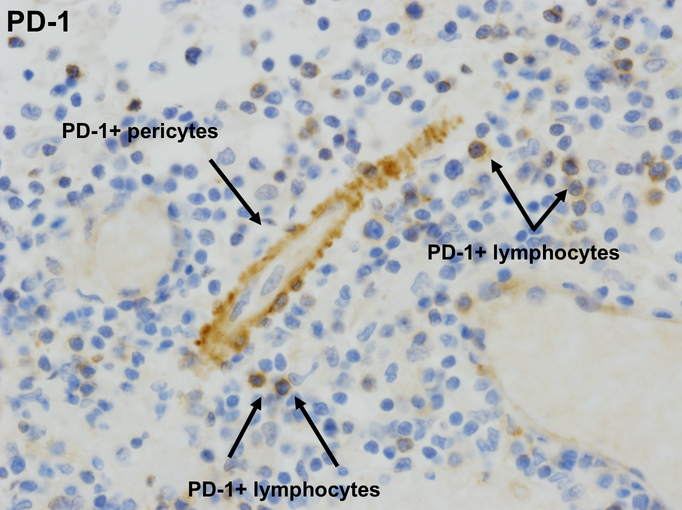
We then profiled the T cell exhaustion markers PD-1 and PD-L1. PD-L1 was diffusely expressed by cells with macrophage morphology (Figure 1E). We observed few-to-no cells expressing PD-L1 in samples from patients with encephalitis of other etiologies (Supplementary Table 3). These samples also showed minimal to no lymphocytic infiltration, suggesting that PD-L1 expression in the brain may represent an attempt to dampen lymphocyte-derived inflammation and injury. PD-1 was expressed at lower levels by infiltrating lymphocytes as well as by pericytes, cells which surround the capillary and venular endothelium and play a critical role in blood-brain barrier maintenance (Extended Data 4)10. This pattern of immune cell infiltration and exhaustion markers were reminiscent of that reported in two cases of fulminant ICI-induced myocarditis5.
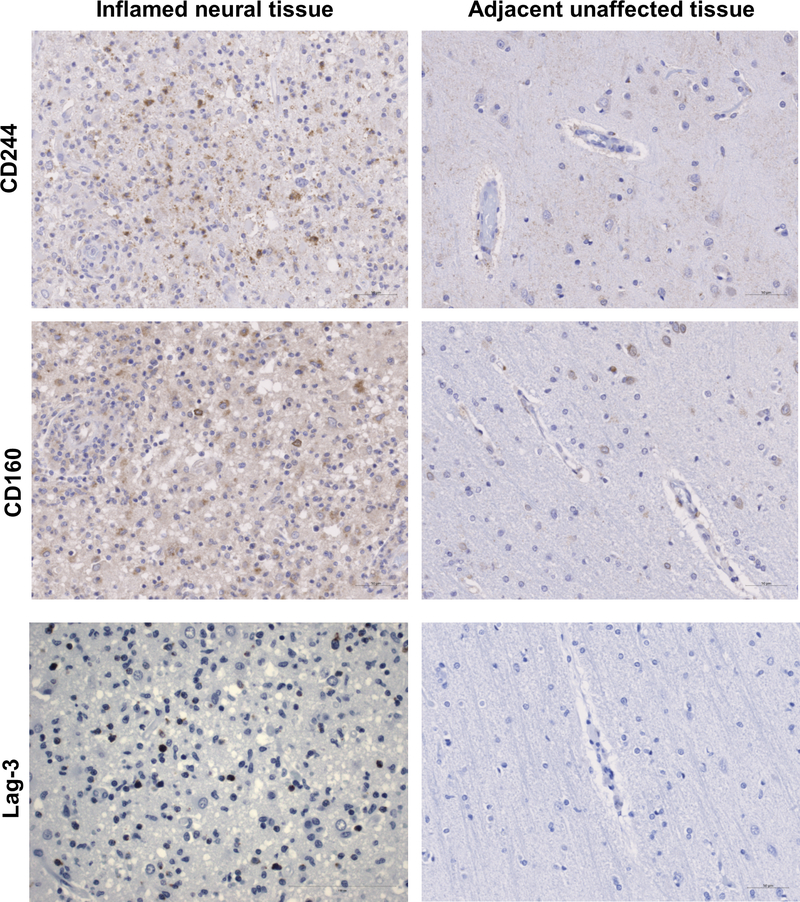
We also profiled other markers of immune exhaustion including CD223/LAG-3 (T cells), CD244/2B4 (NK cells), and CD160 (CD8+ T cells and NK cells) in the inflammatory microenvironment and histologically normal regions (Extended Data 5). LAG3, and to a lesser degree CD244, were highly expressed in the inflamed region but absent in the uninflamed region. We identified CD160+ cells in both tissues, although present at a higher degree in the inflamed region. These results suggested that the infiltrating T cell population likely received negative co-stimulation after repeated antigen exposure.
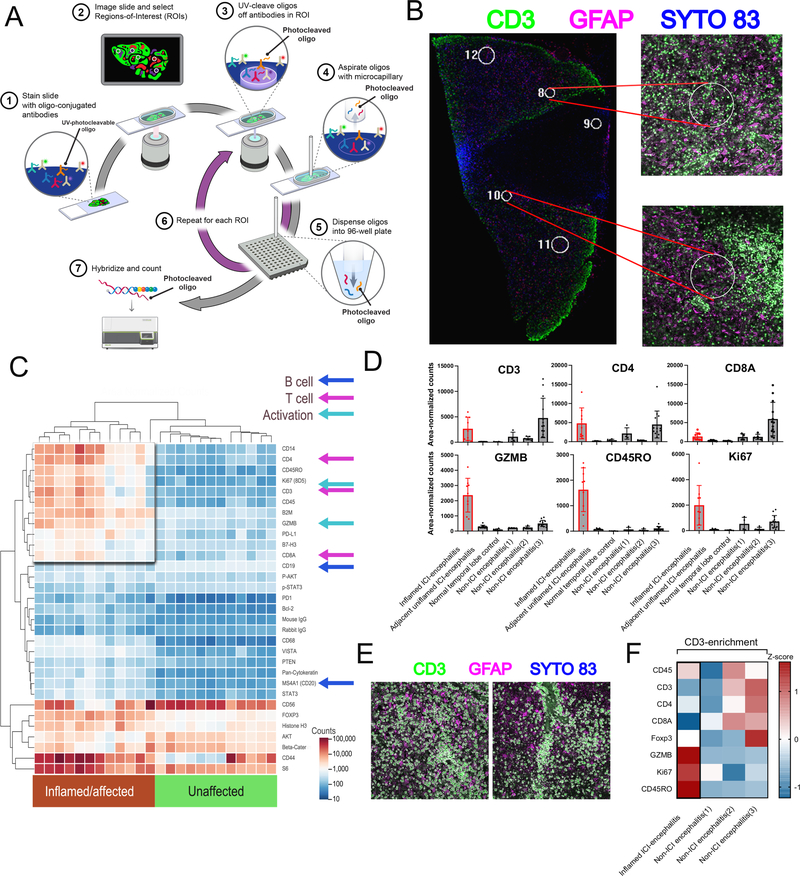
As only formalin-fixed autopsy tissue was available, we used NanoString digital spatial profiling to analyze several regions-of-interest (ROIs) in the immune microenvironment (Figure 2A). We selected 12 ROIs each from inflamed and adjacent unaffected tissue sections (Figure 2B). This analysis demonstrated a high degree of T cell and cytotoxic activation, with large increases in CD4, CD8, CD3, Ki67, PD-L1, GZMB, and CD45RO in the affected region and minimal increase in the B cell markers CD19 and CD20 (Figure 2C). We extended this analysis across additional tissue controls, including a normal temporal lobe specimen and 3 cases of non-ICI-associated encephalitis (Supplementary Table 3). T cell infiltrate (CD3/4/8) was variable but similar across affected tissue in both the present case as well as the non-ICI encephalitic cases; however, there was a preponderance of markers of activated memory phenotypes (expression of GZMB, CD45RO, and Ki67) in the present case (Figure 2D). We next asked what markers were specifically enriched on the infiltrating T cell population, selecting two CD3+ T cell-specific ROIs on the inflamed region of brain, as well as non-ICI encephalitic cases (Figure 2E). While CD3 is expected to be enriched in these ROIs based on its use of fluorescently-labeled selection markers, this analysis further localized Ki67, CD45RO, and GZMB specifically to T cells (Figure 2F). Altogether, we reasoned that activated memory T cells were likely playing an important role in the pathophysiology.
Figure 2: Digital spatial profiling of immune-related protein markers across inflamed and non-inflamed neural tissue.
A) Schematic describing DSP workflow. B). Representative region-of-interest selection under immunofluorescence for analysis. Tissue was stained with anti-GFAP (pink) and anti-CD3 (green) in addition to SYTO 83 nuclear staining for ROI visualization and selection. Circular regions 200 μm in diameter were excited with UV light to release barcodes for collection and analysis. C) Heatmap of area-normalized barcode counts from inflamed and unaffected tissue sections across 12 ROIs. Key antigens representing activation, T cell, and B cell markers are identified by arrows. D) Spatio-regional protein expression patterns across 8–10 ROIs from brain tissues demonstrating enrichment for T cell markers in encephalitis cases, but only memory activated markers in (Ki67HI CD45ROHI GZMBHI) in ICI-encephalitis. E) Representative images of CD3+ T cells gated for barcode collection across 2 spatial ROIs in the inflamed region. F) Area-normalized counts for key antigens demonstrating enrichment of memory activated phenotypes (Ki67HI CD45ROHI GZMBHI) in CD3+ population ROIs from ICI-induced encephalitis versus non-ICI-encephalitis.
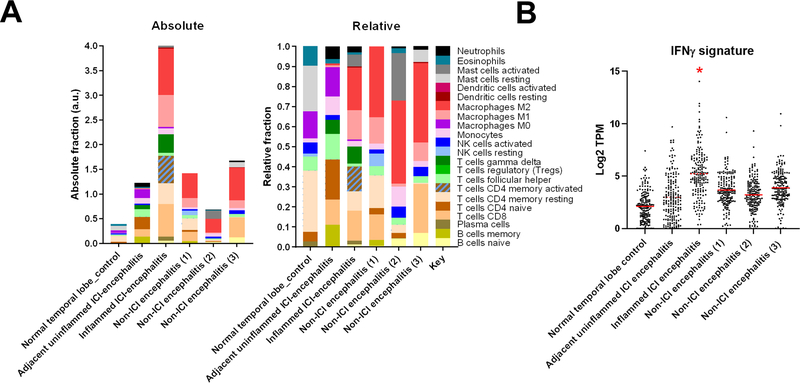
Using RNAseq and CIBERsort analysis (see Methods), we observed several key changes in the inflamed region: 1) conversion of naïve CD4 cells to CD4 memory-activated T cell population in the inflamed tissue 2) conversion from resting to activated mast cells, 3) a loss of memory B cells, and 4) differentiation of M0 to M1/M2 macrophage phenotypes (Extended Data 6A). As response to anti-PD-1 therapy has been linked to type-II interferon responses, we asked whether IFNγ-inducible genes were also over-represented in the inflamed region. Such genes were markedly upregulated (Extended Data 6B; Supplementary Dataset 1).
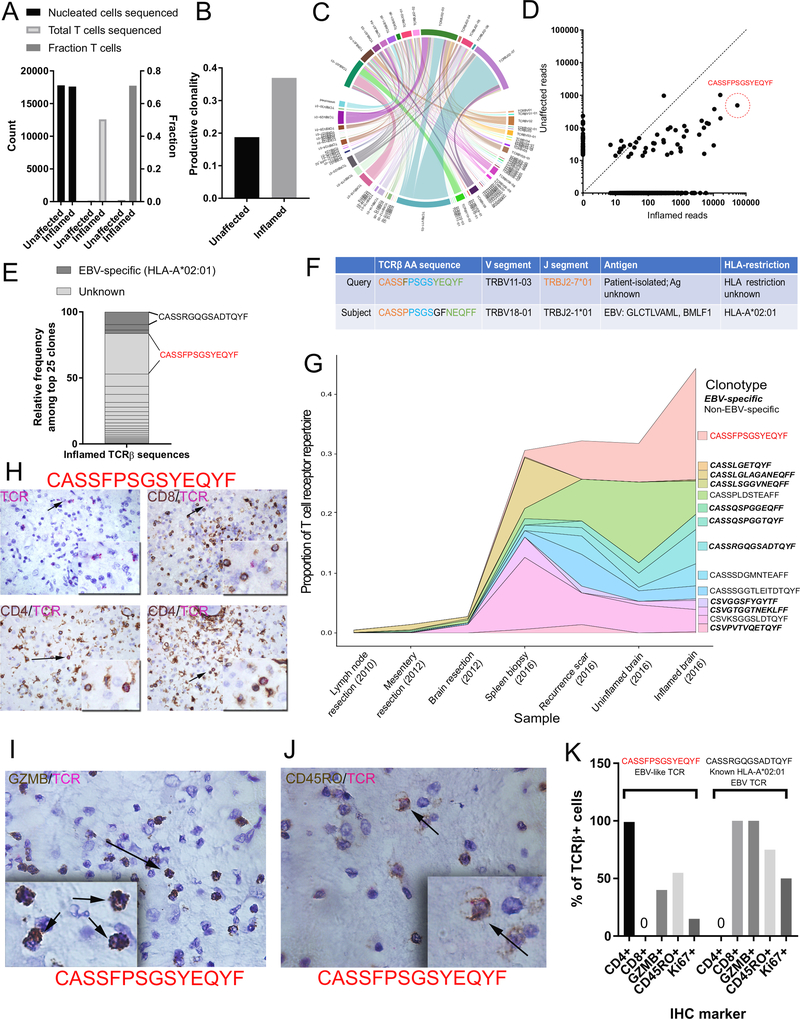
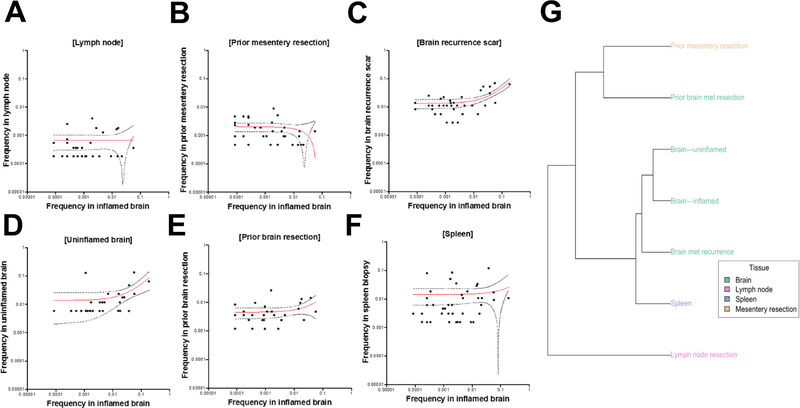
To quantify the clonality of these infiltrating T cells, we performed TCRβ CDR3 sequencing using the immunoSEQ and ArcherDX platforms on: 1) the inflamed and unaffected regions of the brain, 2) prior resections of the tumor-adjacent lymph node, mesentery, and metastasis to the brain, and 3) the recurrence scar and spleen. The inflamed section demonstrated a high T cell fraction (71% of nucleated cells, Figure 3A) and high degree of productive T cell clonality (0.369, Figure 3B and 3C). Notably, a single TCRβ sequence comprised 19.6% of all infiltrating T cells (CDR3 amino acid CASSFPSGSYEQYF, Figure 3D), with two other high-frequency clones comprising 5.98% and 5.91% of infiltrating T cells respectively. For comparison, the most prevalent clone infiltrating the cardiac muscle of two patients with myocarditis5 was approximately 9% (compared to 19.6% in this case of encephalitis). We detected this dominant clone at 1.08% in the spleen and 6.44% in the recurrence scar, though analyses of the TCR repertoire of additional tissues revealed mostly unique repertoires in comparison to the brain (Extended Data 7).
Figure 3: TCR sequencing identification of oligoclonal CD4+ cytotoxic T cells in inflamed encephalitic tissue.
A) ImmunoSEQ statistics of total cellular content, composition (fraction of T cells) and total T cells sequenced in inflamed and unaffected neural tissue. B) Enrichment of productive clonality in inflamed region of encephalitic tissue. C) Circos plot depicting the V(D)J rearrangements in TCRβ in the repertoire. D) Biplot demonstrating enrichment of shared TCRs among the unaffected (likely regional presence) and inflamed tissues. The amino acid sequence predicted from the most highly represented DNA sequence is identified (CASSFPSGSYEQYF). E) Representation of top 25 clones according to known EBV-specificity based on patient HLA haplotype. F) Sequence similarity in TCRβ amino acid sequence to a known EBV clone with specificity for the immediate-early HLA-A2-restricted epitopes GLC (BMLF1). G) Clonotype tracking over time and across tissue samples of EBV-specific and non-EBV-specific dominant clones found in the inflamed brain tissue. H) Dual IHC/RNA-ISH analysis for CD4/CD8 and EBV-like CASSFPSGSYEQYF RNA sequence. Upper left: only the TCR probe (pink); upper right: exclusion of TCR probe with CD8+ T cells, lower: Co-expression of CD4 with TCR probe. I) Co-expression of granzyme B with TCR probe J) Co-expression of CD45RO with TCR probe. K) Quantification for Ki67, CD45RO, and granzyme B (GZMB) expressed as percent of all CASSFPSGSYEQYF TCRβ+ cells, or the comparator CASSRGQGSADTQYF TCRβ sequence, a known HLA-A*02:01 restricted EBV clone.
We performed 6-digit HLA haplotyping (see Methods and Supplementary Table 4) on the index case to leverage existing databases of TCR-antigen specificity and provide a contextual understanding of the TCR repertoire. Bioinformatic analyses (see Methods) revealed: 1) a [PSGS] motif expanded in the TCR repertoire in the inflamed brain found exclusively in TCRs recognizing viral proteins and not autoimmune TCRs (Supplementary Table 5), and 2) TCR CDR3β sequences identical to known EBV-specific T cell receptors. Further query of publicly available databases (see Methods) revealed 23 further EBV-specific TCRs, including 7 which exactly matched known HLA-A2-restricted EBV-specific TCRs that were previously described as specific to the epitopes GLC (BMLF1) and YVL (BRLF1)11–17. These sequences represented 10.9% of infiltrating T cells in the infarct, including the second most dominant TCR in the inflamed region of the brain (Figure 3E, 3F). We observed these sequences at frequencies higher than those found in acute-phase infectious mononucleosis patients and in the synovial fluid of rheumatoid arthritis patients, in whom these TCRs were first detected at frequencies of 5% of CD8+ T cells11. While the frequency of these EBV-specific sequences fell to less than 1% of CD8+ T cells over time in acutely infected patients in those studies, we observed a distinct increase in the frequency of EBV-specific sequences in multiple tissues of the index case over time (Figure 3G). These sequences also occupied 16.1% of sequences detected in the spleen biopsy of this patient and 5% of the brain recurrence scar. Notably, we did not detect any previously described HLA-A2-restricted or HLA-restriction-unknown TCRs specific to MelanA epitopes (see Methods).
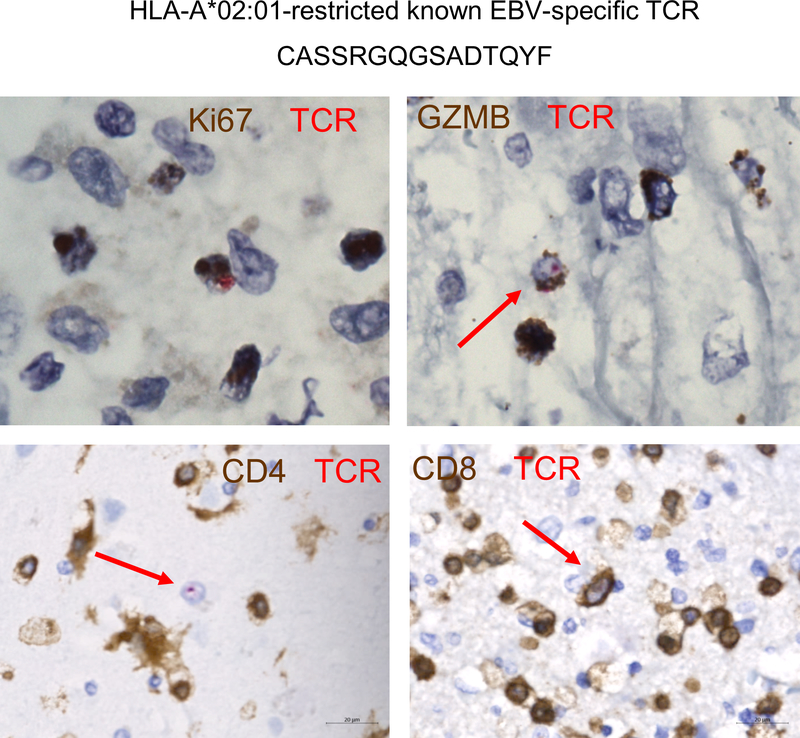
Given consistent findings in our earlier data suggesting a predominance of CD4 memory activated T cells in the inflamed region, we performed dual RNA in situ analysis (using an RNA probe specific to the oligoclonal RNA sequence) and IHC to characterize the cells. This analysis surprisingly yielded a complete localization of CASSFPSGSYEQYF+ cells to CD4+ T cells (Figure 3H). Furthermore, these CD4+ cells were also GZMB+ (40%), CD45RO+ (55%) and Ki67+ (10–15%) (Figure 3I–K). Thus, the oligoclonal cells detected by TCRβ sequencing are likely the same cells identified by digital spatial analysis and CIBERSORT, together corroborating that CD4 memory activated T cells played a key role in this syndrome. Importantly, the second dominant TCR sequence (CASSRGQGSADTQYF), recognizing immediate-early HLA-A2-restricted epitopes GLC (BMLF1) and YVL (BRLF1), was found specifically in CD8+ T cells as expected, and also colocalized with markers of activation (Figure 3J and Extended Data 8).
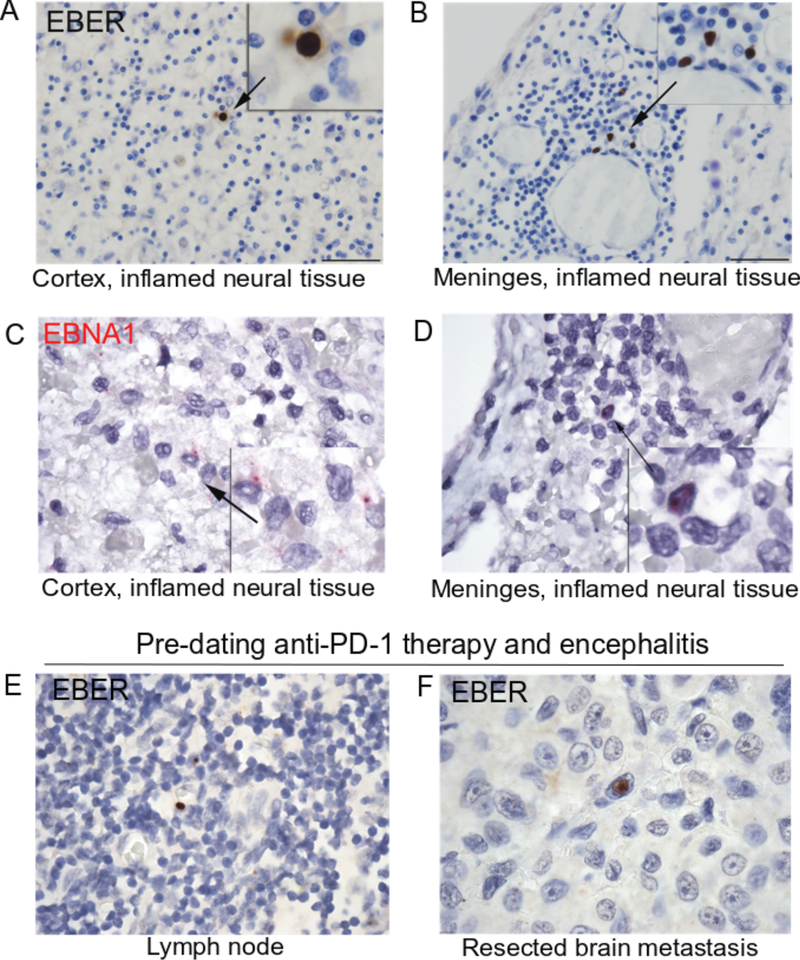
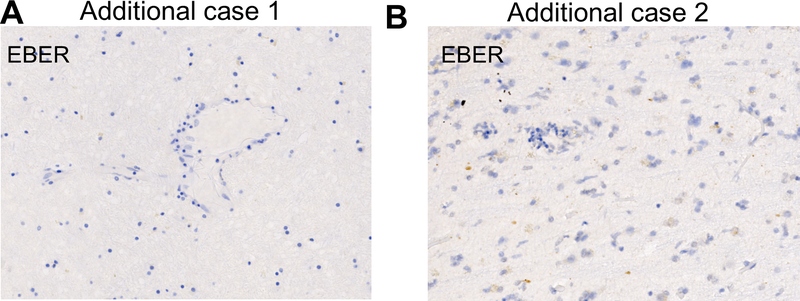
Given the persistently positive EBV PCR from multiple CSF samples and EBV-specific TCRβ sequences (reported below) identified in the brain, we performed a diagnostic RNA in situ hybridization (RNA-ISH) stain against EBV-encoded RNAs (EBER1/2). We identified rare but replicable EBER+ lymphocytes in the cortex and meninges (Extended Data 9A and B, respectively) of the affected region of the brain. EBNA1-specific RNA-ISH confirmed the presence of EBV-infected cells (Extended Data 9C and D). Staining of a lymph node dissection anteceding pembrolizumab demonstrated EBER+ cells, suggesting that the encephalitis did not arise from acute infection (Extended Data 9E). Surprisingly, EBER staining of the patient’s resected brain metastasis, also predating immunotherapy and symptoms, identified rare melanoma cells with EBER reactivity (Extended Data 9F). In contrast, EBNA1 staining of 9 additional non-immunotherapy related encephalitis cases demonstrated complete absence of EBER+ lymphocytes in all cases, suggesting that EBV+ lymphocyte presence is an uncommon finding in cases of encephalitis caused by other etiologies (Supplementary Table 3). Notably, archival neural tissue from two other cases of ICI-encephalitis, which were characterized by substantially less robust inflammation, had negative EBER staining (Extended Data 10).
The molecular basis of ICI toxicities is unclear and difficult to analyze as such cases seem to arise stochastically. While these toxicities can affect nearly all organ systems, encephalitis is the most common neurological toxicity associated with ICI therapy18. Here, we report for the first time the clinical and molecular features of a fulminant and fatal case of anti-PD-1-induced encephalitis. Treatment with steroids and anti-TNFα anteceded collection of tissues in this study, potentially affecting the immunologic status of the patient. Analyses of affected brain tissue demonstrated robust and clonal infiltration of cytotoxic CD4+ activated memory T cells, PD-L1 expression, and evidence of concurrent EBV infection. To our knowledge, this is the first molecular description of immune-therapy induced neurotoxicity and the first suggestion of interplay between viral infection and irAEs.
The cellular mechanisms of ICI toxicity remain poorly characterized. We and others have suggested that causes or correlations could include shared antigens present in tumor and affected tissue5, pre-existing subclinical autoimmune responses19,20, early B cell changes21, and specific intestinal microbiota22. We observed an extremely clonal cytotoxic CD4+ T cell population in this sample. Such cells have central roles in autoimmunity23, antitumor immunity24,25, and antiviral defense26,27. We cannot exclude the possibility that the clone we describe above is restricted by HLA-A2, as both CD4+ T cells restricted by HLA class I and CD8+ T cells restricted by HLA class II have been described in numerous healthy and disease states28–31. The frequency and phenotype of the CD4+ clone described above may suggest a novel role for cytotoxic memory CD4+ T cells if the clone is tumor-reactive, with respect to tumor control and dangerous autoreactivity as shown by others24,25. Indeed, select TCRs are restricted by peptides presented on both HLA class I and class II, implying that such TCRs possess multiple specificities32. The presence and frequency of both CD4+ and CD8+ cytotoxic T cell populations in the brain and tumor microenvironment merit further study in additional cancer types and in autoimmune models, particularly given that the additional cases we examined both showed marked inflammation-associated T cell infiltration of the CNS.
In conclusion, this is the first molecular analysis of neurotoxicity due to immune checkpoint inhibitors to our knowledge. In addition, we believe this is the first evidence linking concurrent viral infection with irAEs, and the first detailed identification and phenotyping of culprit immune cell populations. Further studies are needed to determine the best interventions for prevention and management of these rare but fulminant toxicities. [a little redundant with first paragraph of conclusions, may need to discuss further]
Methods
Index patient and autopsy data
The index patient was consented to protocols that permitted de-identified research use of biospecimens, genetic testing results, and clinical data (Vanderbilt IRB numbers 100178, 181685, and 030220) and his family consented to a post-mortem histopathologic evaluation; cases from outside institutions were approved with institution-specific protocols. Clinical data was obtained through chart review. During the autopsy, evaluation for melanoma using standard melanoma stains (MITF, MelanA, SOX10, S100), and histopathologic evaluation of infiltrating immune cell subsets using CD3, CD20, and CD68 was performed in accordance with institutional protocols.
Additional cases of checkpoint inhibitor-associated encephalitis
Additional case 1 was seen at the University Hospital, Zurich, Switzerland and has been previously described33. Briefly, she was a 60-year-old (2012) patient with an isolated lesion in the right gyrus frontalis medius as well as bilateral pulmonary metastases. After neurosurgical resection followed by adjuvant radiotherapy (with a total of 30 Gray (Gy)) the patient was treated with ipilimumab (3 mg/kg IV) for a total of 4 doses. Three months after the last dose, the patient reported generalized tiredness, weakness, and loss of appetite. However, serum endocrinological work-up revealed a non-significant cortisol shift without osmolality changes, which was not specific for hypopituitarism or adrenal insufficiency. No new metastases were found in the MRI of the brain. A new PET-CT scan showed bilateral pulmonary progression. Subsequent therapy with pembrolizumab was initiated (1 infusion). The patient passed away without any antecedent evidence of altered physical status within 2 weeks. At autopsy, gross inspection revealed a 3 cm defect in the right frontal lobe. No other lesions were seen on further sectioning. Microscopic examination was remarkable for scattered perivascular and parenchymal collections of mature lymphocytes, mostly CD8+ T cells, in the hemispheres and brainstem. In addition, sections of brainstem showed CD68+ microglial cells, occasionally organized in loose nodules. IHC preparations for cytomegalovirus, herpes simplex virus I and II, simian virus 40, toxoplasmosis and varicella zoster virus were negative. There was evidence of melanoma metastasis, even after additional sampling. A diagnosis of encephalitis was rendered based on the presence of intraparenchymal and perivascular CD8-positive lymphocytes accompanied by microglial proliferation.
Additional case 2 was seen at Dana-Farber Cancer Institute in Boston, MA and initially presented in 2013 with a T1a melanoma of the left lower extremity. He was followed until 2015 when he noted increased dyspnea on exertion and a CT scan showed numerous enlarged lymph nodes throughout the chest, abdomen and pelvis. The largest of these was a 7.4 X 5.3 cm left pelvic sidewall node with numerous other large nodes noted. He underwent an ultrasound guided biopsy which demonstrated metastatic melanoma. He was started on single agent pembrolizumab and after the first dose started reporting increased tremors and leg weakness. Due to the subacute nature of these symptoms he received two more treatments with pembrolizumab with progressively worsening symptoms. He developed increased weakness and memory issues and the pembrolizumab was held and he was admitted for work-up of this. Lumbar puncture X3, MRI of the brain and spine, neurologic evaluation and extensive work-up for other causes of his symptoms was performed without an etiology. He received a short course of high dose steroids followed by IVIG and a longer steroid taper without benefit. His functional status continued to decline without a clear etiology despite these interventions and brain biopsy was performed which showed T-cell inflammatory infiltrate and gliosis suspicious for pembrolizumab-related encephalopathy. He subsequently died from progressive neurologic decline.
Additional cases of encephalitis of other etiologies
Additional cases of encephalitis not associated with checkpoint inhibitor use were collected under an IRB-approved protocol to study cellular heterogeneity in normal brain and brain neoplasms (IRB#180238). Cases were stained with EBNA1 RNA-ISH as detailed below, and with anti-PD-L1 IHC as detailed below. Details of the cases are listed in Supplementary Table 3. Positive and negative control probes were used in all cases to verify specificity and presence of intact RNA for analysis.
Immunohistochemistry
Archival tissue from the autopsy was obtained and IHC evaluation was performed. Antibodies utilized were anti-CD45RO (Thermo Scientific, cat# MA5–11532, 1:1,600), anti-GZMB (Granzyme B, Biocare cat# ACI 3202 AA, prediluted), anti-Ki67 (Dako, cat# M7240, 1:200), anti-CD4 (StatLab, cat# RM27–10, prediluted), anti-CD8 (StatLab, cat# MM39–10, prediluted), anti-PD-L1 (Cat.#PA5–28115 ThermoFisher, Grand Island, NY; 1:7500), anti-CD68 (PA0191, Leica, Buffalo Grove, IL; prediluted), anti-PD-1 (HPA035981, Sigma-Aldrich Co., St. Louis, MO; 1:75), anti-CD20 (PA0906, Leica, Buffalo Grove, IL, prediluted), anti-LAG3 (Cell Signaling, Catalog# 15372, dilution 1:200), CD244 (Proteintech, Catalog# 16677–1-AP, dilution 1:200), CD160 (Abcam, Catalog# ab202845, dilution 1:600). The Bond Polymer Refine detection system or Envision system was used for visualization. Slides were then dehydrated, cleared, and coverslipped.
RNA in situ hybridation (RNA-ISH) analysis
For RNAscope RNA-ISH (Advanced Cell Diagnostics) analysis of EBNA1, standard RNAscope manufacturer’s protocols were followed using the RNAscope H2O2 and Protease pretreatment kit, (ACD, reference# 322381), RNAscope Target retrieval buffer (ACD, reference# 322000), and appropriate positive and negative RNA probes for controls. For custom BaseScope RNA-ISH (Advanced Cell Diagnostics) TCRβ analysis (ACD, reference# 712111), standard BaseScope manufacturer’s protocols were followed using BaseScope™ Detection Reagents– RED (ACD, reference# 322910). Dual ISH-IHC was performed using the custom TCRβ target probe(s) followed by incubation with primary antibodies (CD4, CD8, Ki67, CD45RO, and GZMB) as described above, using the Envision (Dako) system, DAB (Dako), and hematoxylin counterstain. For EBER staining, slides were placed on the Leica Bond Max IHC stainer, and all steps, besides dehydration, clearing and coverslipping, were performed on the Leica Bond Max. Slides were deparaffinized, and enzyme retrieval was performed using Proteinase K (Ref# S3020, Dako, Santa Clara, CA) for 5 minutes. Slides were put through ISH Hybridization with the Ready-To-Use EBER probe (Ref# ISH5687-A, Leica, Buffalo Grove, IL) for two hours. Slides were placed in an anti-Fluorescein antibody (Ref# AR0222, Leica, Buffalo Grove, IL) for 15 minutes. The Bond Polymer Refine detection system was used for visualization. Slides were then dehydrated, cleared and coverslipped. For dual staining, data were quantified by colocalization of the signal as a fraction of the TCR+ cells, by a licensed pathologist (PIE-G).
RNA sequencing
Total RNA quality was assessed using the 2200 TapeStation (Agilent). At least 20ng of DNase-treated total RNA having at least 30% of the RNA fragments with a size >200 nt (DV200) was used to generate RNA Access libraries (Illumina) following manufacturer’s recommendations. Library quality was assessed using the 2100 Bioanalyzer (Agilent) and libraries were quantitated using KAPA Library Quantification Kits (KAPA Biosystems). Pooled libraries were subjected to 75 bp paired-end sequencing according to the manufacturer’s protocol (Illumina HiSeq3000). Bcl2fastq2 Conversion Software (Illumina) was used to generate de-multiplexed FASTQ files.
QC for the paired-end raw sequencing reads of all samples were performed using FastQC34 for the analysis of sequence quality, GC content, adapter content, over-represented k-mers and duplicated reads. Sequencing reads were mapped to human reference genome GRCh38 (Release-85, Ensembl) using STAR 2.2.1 with 2-pass mapping35. QC for read alignment and mapping was evaluated with RSeQC36 for sequencing saturation, mapped reads clipping profile, mapped read distribution, and coverage uniformity. Transcript-per-million (TPM) values were calculated using RSEM37 and used to assess the global quality and replicability of the RNA-seq dataset and exported for downstream data analyses.
CIBERSORT and gene set analysis
CIBERSORT analysis was performed on RNAseq TPM level data generated above, using both the ‘relative’ and ‘absolute’ methods available on the analysis website (https://cibersort.stanford.edu/). The CIBERSORT method used is explained in full detail elsewhere38. To complete this analysis, we used the LM22 dataset which was carefully developed and validated in the manuscript. The genes utilized, along with computational methods and development/validation approach are provided as supplemental data in the original report38 and also freely available on the CIBERSORT website above. Analysis was performed as previously described38. For interferon gene set analysis, the HALLMARK_INTERFERON_GAMMA_RESPONSE (M5913; 200 genes, 177 genes in overlap/matching) signature was downloaded from the Molecular Signatures Database (http://software.broadinstitute.org/gsea/msigdb). For each gene, the log2 fold change was calculated between the inflamed and unaffected region, or across additional tissues as indicated.
T cell receptor (TCR) sequencing
TCR sequencing and clonality quantification was assessed in neuropathologist-selected (author BM) FFPE samples of highly inflamed and non-inflamed brain parenchyma using survey level immunoSEQ™ and the Immunoverse™ assay, as previously described (Adaptive Biotechnologies, ArcherDX)39,40. Sequencing results were evaluated using the immunoSEQ analyzer version 3.0. Shannon entropy, a measure of sample diversity, was calculated on the clonal abundance of all productive TCR sequences in the data set. Shannon entropy was normalized by dividing Shannon entropy by the logarithm of the number of unique productive TCR sequences. This normalized entropy value was then inverted (1 − normalized entropy) to produce the clonality metric.
TCR repertoire analysis: data processing
We processed raw immunoSEQ data for downstream analysis using VDJTools41. We excluded clonotypes if a TRBV or TRBJ gene was not assigned or if they were non-productive. We then recalculated the frequency of each remaining productive sequence and collapsed the resulting TCR repertoire dataset by CDR3 amino acid sequence (i.e. the counts for two clonotypes with different nucleotide sequences encoding the same CDR3 AA sequence would be summed).
TCR repertoire analysis: clonotype annotation and known antigen assignment
We first retrieved continuous and discontinuous CDR3β motifs significantly enriched at least 3-fold above a naïve repertoire of 200,000 unselected CD4 and CD8 T cells at a resampling depth of 10,000 using the GLIPH algorithm17. We then annotated each TCR repertoire for known and possible antigen-specificity and pathogenic associations using the greedy VDJMatch algorithm42 in VDJDB against each CDR3β and significant motifs from GLIPH17, and a Levenshtein distance of 1 for sequences stored in McPAS-TCR43. We also searched 2,460 HLA-A2 restricted previously published44 melanoma-specific TCRs for matches and homology to our TCR repertoire data.
TCR repertoire analysis: in silico epitope prediction
Finally, we annotated each clonotype sequence for potential recognition of 38 viral and 5 cancer T cell epitopes using CDR3β sequence, TRBV gene, and TRBJ gene as input to the random forest TCRex model45 at a false positive rate of 1 in 10,000 (0.01%).
Pharmacovigilance database analysis
We queried VigiBase, the WHO database of individual case safety reports containing more than 18 million ICSRs submitted by national pharmacovigilance centers since 1967, on January 23, 2018. These reports originate from sources including healthcare professionals, patients, and pharmaceutical companies and are generally reported post-marketing. Of >18,000,000 individual reports, 47,240 were from patients receiving nivolumab, pembrolizumab, ipilimumab, atezolizumab, durvalumab, avelumab, or tremelimumab. We used the following Medical Dictionary for Regulatory Activities (MedDRA) preferred terms for encephalitis associated with ICI: encephalitis, encephalitis autoimmune, limbic encephalitis, meningoencephalitis, and cerebellitis for the following drugs: nivolumab, pembrolizumab, ipilimumab, atezolizumab, durvalumab, avelumab, and tremelimumab. We collected date of report, therapy, indication (cancer type), age, gender, concurrent adverse events, time of onset, and outcome for all patients when available. We used descriptive statistics to quantify medians and range for continuous variables and percentages for categorical variables. We used the R statistical language (3.5.1) to perform the χ2 test for contingency tables.
Digital spatial profiling (DSP) of protein
Single FFPE slides from inflamed and non-inflamed brain tissue were selected, as well as from non-inflammatory ‘healthy’ brain (epileptic) and 3 additional cases of non-ICI-induced encephalitis. Ten regions of interest (ROIs) within each slide were selected by pathologists (authors RA and BM). Multiplexed protein profiles were generated with the NanoString® GeoMX™ digital spatial profiling platform (RUO – Research Use Only). Formalin-fixed paraffin-embedded tissue sections of 5um thickness were subjected to antigen retrieval (citrate buffer pH6) and stained with a cocktail of antibodies labeled with photocleavable DNA-indexing oligos (to generate quantitative protein profiles), fluorescent anti-CD3 and anti-GFAP (visualization markers) and SYTO 83 nuclear dye (Thermo Fisher, S11364). Tissues were imaged by fluorescence microscopy on the GeoMX platform and regions of interest (ROI) were chosen for molecular profiling. ROIs were selected as circles 200 μm in diameter from inflamed or heathy tissue or by custom masking using the fluorescent CD3 signal to specifically identify T cells and a digital micromirror device to precisely control the pattern of UV illumination. ROIs were exposed to UV light (365 nm) to release oligos which were captured via microfluidics and stored in individual wells of a microtiter plate. Following collection from all ROIs, oligos were hybridized to unique 4-color, 6-spot optical barcodes and enumerated on the nCounter® platform. Data were normalized to ERCC-sequence specific probes to control for technical variation in hybridization efficiency, followed by area normalization to control for ROI size and control IgG (rabbit and mouse) to normalize for background. Data were visualized by unsupervised hierarchical clustering or grouped dot plots.
Extended Data
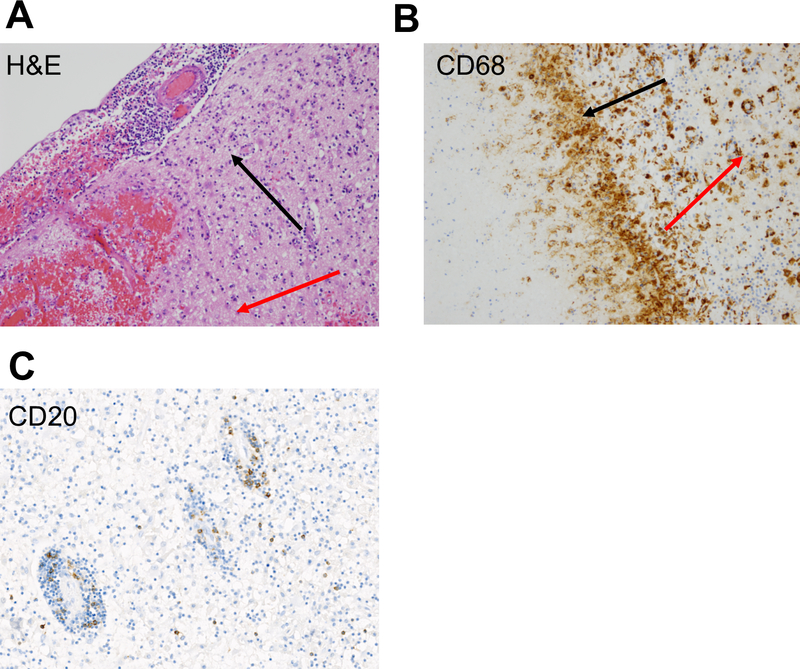
Extended Data 1. Inflammatory and myeloid/microglial infiltrate in infarct regions of brain.
A) H&E stain shows a dense chronic inflammatory infiltrate in the meninges (upper left; black arrow), focal hemorrhage in the underlining brain parenchyma (lower left; red arrow), and gliotic gray matter; 20x. B) CD68 positive cells at the interface of the necrotic area (black arrow) and the adjacent brain parenchyma (red arrow) 20x. C) Limited expression of CD20 staining in perivascular and parenchymal regions of brain; 20x.
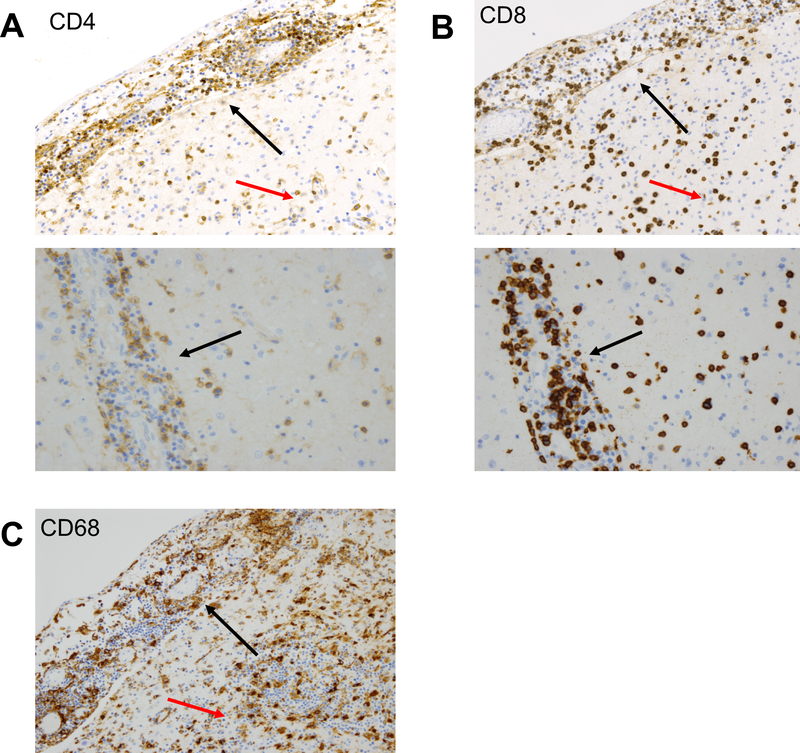
Extended Data 2. Lymphocytic and myeloid infiltrate in meninges, perivascular, and parenchymal regions.
A) The lymphocytic infiltrate involving the meninges (upper image; upper left of image, black arrow), parenchyma and brain perivascular regions (upper image; lower right of image, red arrow and lower image) includes CD4 and (B) CD8 positive T cells; 20x. C) CD68 positive cells accompanying lymphocytic infiltrate in meninges (upper left of image, black arrow) and brain parenchyma (lower right, red arrow); 20x.
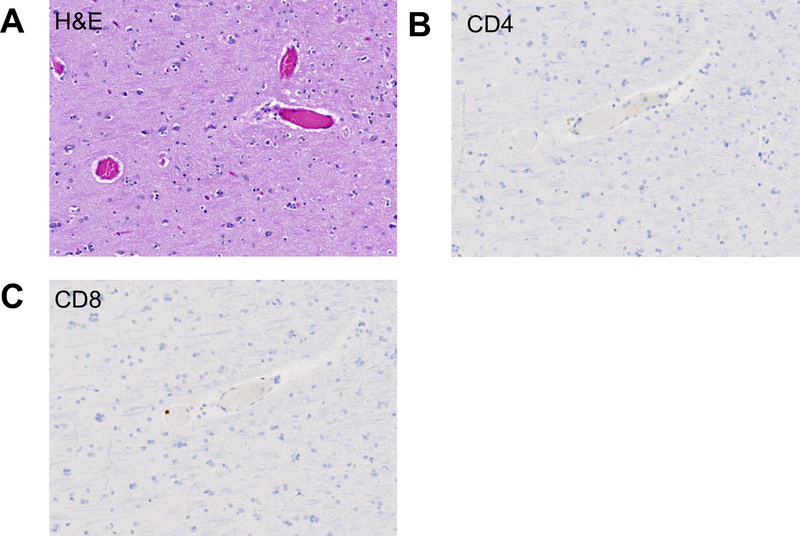
Extended Data 3. Absence of substantial inflammatory infiltrate in radiologically and macroscopically non-affected area.
A) H&E corresponding to radiologically and macroscopically non-affected area with preserved architecture and no apparent inflammatory infiltrate; 20x. B) CD4 and (C) CD8 stains demonstrating sparse presence or absence of T cell infiltrates in radiologically and macroscopically non-affected area; 20x.
Extended Data 4. PD-1 expression in inflamed region of brain.
PD-1 positive perivascular lymphocytes and pericytes; 20x.
Extended Data 5. Expression of T cell and NK cell markers of immune cell exhaustion.
Inflamed and non-inflamed adjacent regions of neural tissue were immunostained for CD244, CD160, and LAG-3; 20x.
Extended Data 6. RNAseq analysis of encephalitic and unaffected tissue.
A) Absolute and relative quantification of immune subsets by CIBERSORT. B) Interferon-γ-inducible genes (HALLMARK_INTERFERON_GAMMA_RESPONSE; M5913; 177 overlapping genes) quantified in inflamed and unaffected regions by RNAseq, as well as additional cases identified in Supplementary Table 3. *adjusted P value <0.0001 versus all other samples via ANOVA with Tukey’s post-hoc test.
Extended Data 7. Analysis of repertoire overlap between inflamed brain and other resected and biopsied tissues.
In all panels, we display the frequency of TCRs detected in both the inflamed brain and the listed tissue using the ArcherDX Immunoverse platform, as well as the linear regression and its 95% confidence intervals on a log10 scale. A) Lymph node biopsy. B) Prior mesentery resection. C) Brain recurrence scar; the TCR repertoire of the the brain recurrence scar and the D) uninflamed brain overlapped significantly with that of the inflamed brain (r2 of 0.46 and 0.17, p < 0.0001 and p = 0.02 respectively). EBV-specific clones in both the inflamed brain and the spleen (E and F) were observed at high frequency, though the shared TCR repertoires detected in the brain and spleen were poorly correlated. G) Hierarchical clustering on the F2 distance metric was used to evaluate the similarity of the TCR repertoires from each of the tissue sites sampled from the patient. As also observed above, we the brain and spleen samples from 2016 and the time of death were most similar to each other, and that the brain-resident TCR repertoire appeared to be somewhat stable over time. The TCR repertoire of the lymph node nearest to the original tumor was highly distinct in comparison to the other samples.
Extended Data 8. Overlap of HLA-A*02:01-restricted known EBV-specific TCR with CD8+ Ki67+ and GZMB+ phenotypes.
Representative images of the TCRβ RNAish probe overlaid with IHC markers.
Extended Data 9. Evidence of latent EBV infection at the site of encephalitic inflammation.
A) EBER(1/2) staining of lymphocytes by RNA in situ hybridization in the cortex and meninges (B) of the encephalitic infarct. C) EBNA1 staining of lymphocytes by RNA in situ hybridization in the cortex and meninges (D) of the encephalitic infarct. E) Positive EBER staining of rare lymphocytes in lymph node resection pre-dating anti-PD-1 therapy, suggesting historic EBV infection. F) Positive EBER staining of rare tumor cells in brain metastasis resection pre-dating anti-PD-1 therapy, also suggesting historic EBV infection.
Extended Data 10. Lack of detection of EBER+ cells in 2 additional cases of checkpoint-inhibitor encephalitis.
A) EBER stain of neural tissue from additional case 1 and additional case 2 (details of case in methods).
Supplementary Material
Acknowledgements
The supplied data from VigiBase come from a variety of sources. The likelihood of a causal relationship is not the same in all reports. The information does not represent the opinion of the World Health Organization (WHO).
We thank the patients and their families for participating in this study. WJM was supported by NHBLI grant T32HL069765, NIDDK grants R01DK112262 and R56DK108352, and NHLBI grant K12HL143956. SAM was supported by NIAID grant P30AI110527. DBJ is supported by NIH/NCI K23 CA204726 and the James C. Bradford Jr. Melanoma Fund. JES was supported by the Cancer ITMO of the French National Alliance for Life and Health Sciences (AVIESAN): “Plan Cancer 2014–2019”. JMB was supported by Department of Defense Era of Hope Award BC170037 and NIH/NCI R00CA181491. In addition, we acknowledge the Translational Pathology Shared Resource supported by NCI/NIH Cancer Center Support Grant 5P30 CA68485-19 and the Vanderbilt Mouse Metabolic Phenotyping Center Grant 2 U24 DK059637-16.
Footnotes
Competing interests
Kristi Barker, Joseph Beechem, Yan Liang, and Sarah Warren are employees of NanoString and receive compensation as such. Douglas Johnson serves on advisory boards for Array, Bristol Myers Squibb, Genoptix, Incyte, Merck, and research funding from Bristol Myers Squibb and Incyte. Justin Balko receives consulting fees from Novartis and research support from Genentech and Incyte. Javid Moslehi serves as a consultant or in an advisory role for BMS, Daiichi Sankyo, Novartis, Pfizer, Regeneron, Takeda, Myokardia, Deciphera, and Ipsen and has received research funding from BMS and Pfizer. Cody Chastain receives grant/research funding from Gilead Sciences, Inc. (related to hepatitis C virus).
Data availability
Source data for RNAseq analyses (IFNγ genes) are included as Supplementary Dataset 1. All other data, including processed TCR sequencing data, are available on request from the corresponding authors.
References
- 1.Darvin P, Toor SM, Sasidharan Nair V & Elkord E Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp. Mol. Med. 50, 165 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson DB, Chandra S & Sosman JA Immune Checkpoint Inhibitor Toxicity in 2018. JAMA 320, 1702–1703 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Postow MA, Sidlow R & Hellmann MD Immune-Related Adverse Events Associated with Immune Checkpoint Blockade. N. Engl. J. Med. 378, 158–168 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Moslehi JJ, Salem J-E, Sosman JA, Lebrun-Vignes B & Johnson DB Increased reporting of fatal immune checkpoint inhibitor-associated myocarditis. Lancet 391, 933 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson DB et al. Fulminant Myocarditis with Combination Immune Checkpoint Blockade. N. Engl. J. Med. 375, 1749–1755 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naidoo J et al. Pneumonitis in Patients Treated With Anti-Programmed Death-1/Programmed Death Ligand 1 Therapy. J. Clin. Oncol. 35, 709–717 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez RS et al. PD-1 inhibitor gastroenterocolitis: case series and appraisal of ‘immunomodulatory gastroenterocolitis’. Histopathology 70, 558–567 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Verschuren EC et al. Clinical, Endoscopic, and Histologic Characteristics of Ipilimumab-Associated Colitis. Clin. Gastroenterol. Hepatol. 14, 836–842 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Larkin J et al. Neurologic Serious Adverse Events Associated with Nivolumab Plus Ipilimumab or Nivolumab Alone in Advanced Melanoma, Including a Case Series of Encephalitis. Oncologist 22, 709–718 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rustenhoven J, Jansson D, Smyth LC & Dragunow M Brain Pericytes As Mediators of Neuroinflammation. Trends Pharmacol. Sci. 38, 291–304 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Annels NE, Callan MF, Tan L & Rickinson AB Changing patterns of dominant TCR usage with maturation of an EBV-specific cytotoxic T cell response. J. Immunol. 165, 4831–4841 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Lim A et al. Frequent contribution of T cell clonotypes with public TCR features to the chronic response against a dominant EBV-derived epitope: application to direct detection of their molecular imprint on the human peripheral T cell repertoire. J. Immunol. 165, 2001–2011 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Cohen GB et al. Clonotype Tracking of TCR Repertoires during Chronic Virus Infections. Virology 304, 474–484 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Koning D et al. In vitro expansion of antigen-specific CD8(+) T cells distorts the T-cell repertoire. J. Immunol. Methods 405, 199–203 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Grant EJ et al. Lack of Heterologous Cross-reactivity toward HLA-A*02:01 Restricted Viral Epitopes Is Underpinned by Distinct αβT Cell Receptor Signatures. J. Biol. Chem. 291, 24335–24351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dash P et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature 547, 89–93 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glanville J et al. Identifying specificity groups in the T cell receptor repertoire. Nature 547, 94–98 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang DY et al. Fatal Toxic Effects Associated With Immune Checkpoint Inhibitors: A Systematic Review and Meta-analysis. JAMA Oncol 4, 1721–1728 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwama S et al. Pituitary expression of CTLA-4 mediates hypophysitis secondary to administration of CTLA-4 blocking antibody. Sci Transl Med 6, 230ra45 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Osorio JC et al. Antibody-mediated thyroid dysfunction during T-cell checkpoint blockade in patients with non-small-cell lung cancer. Ann. Oncol. 28, 583–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das R et al. Early B cell changes predict autoimmunity following combination immune checkpoint blockade. J. Clin. Invest. 128, 715–720 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubin K et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun 7, 10391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peeters LM et al. Cytotoxic CD4+ T Cells Drive Multiple Sclerosis Progression. Front Immunol 8, 1160 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Curran MA et al. Systemic 4–1BB activation induces a novel T cell phenotype driven by high expression of Eomesodermin. J. Exp. Med. 210, 743–755 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirschhorn-Cymerman D et al. Induction of tumoricidal function in CD4+ T cells is associated with concomitant memory and terminally differentiated phenotype. J. Exp. Med. 209, 2113–2126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Penaloza-MacMaster P et al. Vaccine-elicited CD4 T cells induce immunopathology after chronic LCMV infection. Science 347, 278–282 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeuchi A & Saito T CD4 CTL, a Cytotoxic Subset of CD4+ T Cells, Their Differentiation and Function. Front Immunol 8, 194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ranasinghe S et al. Antiviral CD8+ T Cells Restricted by Human Leukocyte Antigen Class II Exist during Natural HIV Infection and Exhibit Clonal Expansion. Immunity 45, 917–930 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyle LH, Goodall JC & Gaston JSH Major histocompatibility complex class I-restricted alloreactive CD4+ T cells. Immunology 112, 54–63 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang M et al. High-affinity human leucocyte antigen class I binding variola-derived peptides induce CD4+ T cell responses more than 30 years post-vaccinia virus vaccination. Clin. Exp. Immunol. 155, 441–446 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Legoux F et al. Characterization of the human CD4(+) T-cell repertoire specific for major histocompatibility class I-restricted antigens. Eur. J. Immunol. 43, 3244–3253 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Heemskerk MH et al. Dual HLA class I and class II restricted recognition of alloreactive T lymphocytes mediated by a single T cell receptor complex. Proc. Natl. Acad. Sci. U.S.A. 98, 6806–6811 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 33.Bossart S et al. Case Report: Encephalitis, with Brainstem Involvement, Following Checkpoint Inhibitor Therapy in Metastatic Melanoma. Oncologist 22, 749–753 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.S. A FastQC: a quality control tool for high throughput sequence data. (2010). [Google Scholar]
- 35.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L, Wang S & Li W RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Li B & Dewey CN RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newman AM et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tumeh PC et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathew JM et al. Generation and Characterization of Alloantigen-Specific Regulatory T Cells For Clinical Transplant Tolerance. Sci Rep 8, 1136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shugay M et al. VDJtools: Unifying Post-analysis of T Cell Receptor Repertoires. PLoS Comput. Biol. 11, e1004503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shugay M et al. VDJdb: a curated database of T-cell receptor sequences with known antigen specificity. Nucleic Acids Res. 46, D419–D427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tickotsky N, Sagiv T, Prilusky J, Shifrut E & Friedman N McPAS-TCR: a manually curated catalogue of pathology-associated T cell receptor sequences. Bioinformatics 33, 2924–2929 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Chen G et al. Sequence and Structural Analyses Reveal Distinct and Highly Diverse Human CD8(+) TCR Repertoires to Immunodominant Viral Antigens. Cell Rep 19, 569–583 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gielis S et al. TCRex: a webtool for the prediction of T-cell receptor sequence epitope specificity. bioRxiv (2018). doi: 10.1101/373472 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.