Abstract
By analyzing the whole-exome sequences of 4,264 schizophrenia cases, 9,343 controls and 1,077 trios, we identified a genome-wide significant association between rare loss-of-function (LoF) variants in SETD1A and risk for schizophrenia (P = 3.3 × 10−9). We found only two heterozygous LoF variants in 45,376 exomes from individuals without a neuropsychiatric diagnosis, indicating that SETD1A is substantially depleted of LoF variants in the general population. Seven of the ten individuals with schizophrenia carrying SETD1A LoF variants also had learning difficulties. We further identified four SETD1A LoF carriers among 4,281 children with severe developmental disorders and two more carriers in an independent sample of 5,720 Finnish exomes, both with notable neuropsychiatric phenotypes. Together, our observations indicate that LoF variants in SETD1A cause a range of neurodevelopmental disorders, including schizophrenia. Combining these data with previous common variant evidence, we suggest that epigenetic dysregulation, specifically in the histone H3K4 methylation pathway, is an important mechanism in the pathogenesis of schizophrenia.
Schizophrenia is a common, debilitating psychiatric disorder that is characterized by positive symptoms (hallucinations, delusions and disorganization) and negative symptoms (impaired motivation, reduced spontaneous speech and social withdrawal). It is associated with cognitive impairment, decreased social and occupational functioning, and increased mortality, with a 12–15-year reduction in lifespan1–3. Schizophrenia has a lifetime risk of ~0.7% and a substantial genetic component, with a sibling recurrence risk ratio of 9.0 and an estimated heritability of up to 81% (refs. 4,5).
The genetic architecture of schizophrenia involves a combination of common, rare and de novo risk variants. At one end of this spectrum, a genome-wide association study of 36,989 cases identified 108 loci containing alleles of individually small effect (median odds ratio = 1.08)6, whereas, at the other, at least 11 rare, recurrent copy number variants (CNVs) (for example, at chromosomes 1q21.1, 15q13.3 and 22q11.2) individually confer substantial risk for schizophrenia (ORs 2–60)7–10. A recent case-control exome sequencing study demonstrated a burden of rare disruptive variants across a set of 2,546 genes selected on the basis of a variety of biological hypotheses about schizophrenia risk and previous genome-wide screens, including GWAS, CNV and de novo mutation studies11. This study did not, however, identify any individual schizophrenia risk genes at a Bonferroni P value of 1.25 × 10−6 (Online Methods). Parent-proband trio studies have sought to increase power by focusing on de novo mutations: the rarity of damaging events makes it possible to observe statistically significant recurrence of mutations in individual genes with smaller sample sizes than would be required in a case-control design. Three such studies in schizophrenia have found suggestive evidence for candidate genes, including EHMT1, DLG2, TAF13 and SETD1A9,12,13. The statistical significance of de novo recurrence is highly dependent on the specification of gene-specific mutation rates, which are difficult to calibrate for indels and CNVs (Online Methods). Because these genes are supported by two de novo events each, of which all but one (in TAF13) are either an indel or CNV, further evidence is needed to firmly establish these as susceptibility genes.
Articles
Two insights have emerged from these early results in schizophrenia. First, genetic risk loci have implicated general biological processes involved in pathogenesis, including histone methylation (common variants)14, transmission at glutamatergic synapses and translational regulation by the fragile X mental retardation protein (rare and de novo variants)11,12. Second, studies of common and rare variation support a highly polygenic architecture involving hundreds of genes, suggesting that very large sample sizes will be required to convincingly identify individual risk genes. This polygenicity is reminiscent of other neuropsychiatric disorders, such as autism spectrum disorder (ASD), which required many thousands of exome sequences and the integration of de novo mutations with case-control burden of rare variants to identify genes at genome-wide significance15,16.
Results
Case-control analysis of schizophrenia exomes
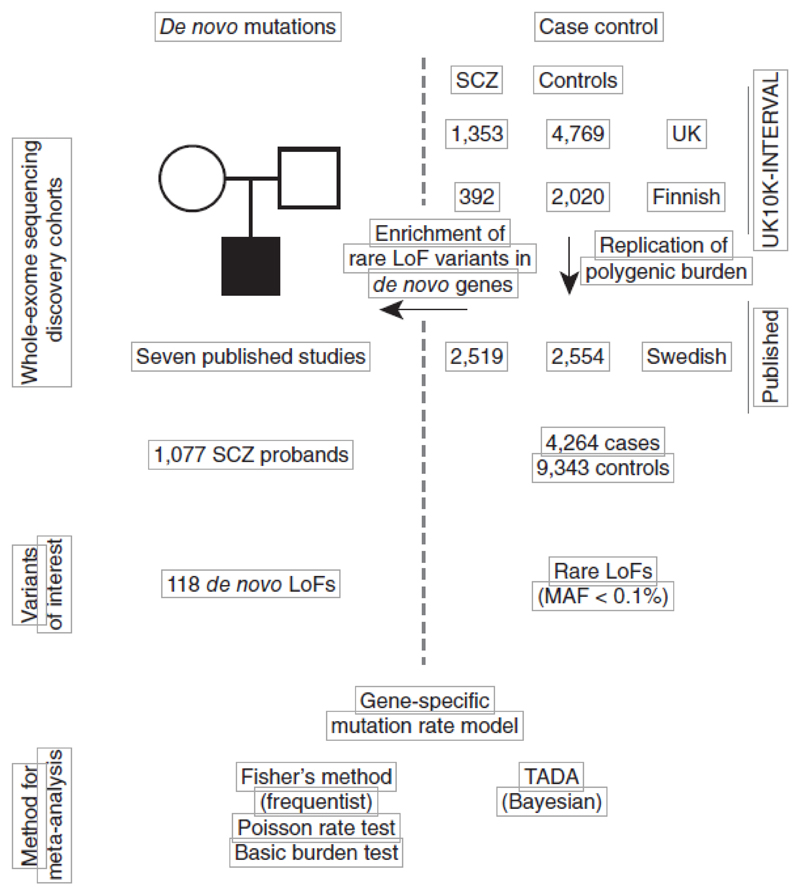
We sequenced the exomes of 1,887 (1,488 UK and 399 Finnish) individuals with schizophrenia and 7,585 (5,469 UK and 2,116 Finnish) individuals without a known neuropsychiatric diagnosis. We jointly called each case set with its nationality-matched controls, but still observed substantial batch effects from the use of different exome capture reagents used at different time points in the experiment (Supplementary Fig. 1). We therefore performed careful quality control (QC) in each set to narrow our analysis to regions with high-quality data in all samples and to remove outlier samples and variants (Online Methods and Supplementary Fig. 2), leaving a total of 1,745 cases and 6,789 controls (Fig. 1). To increase power for gene discovery, we combined our data set with exome sequences of 2,519 Swedish schizophrenia cases and 2,554 controls from a previous study11. The average number of coding SNPs and indels varied among these three sample sets as a result of differences in exome capture technology, QC procedures and sample ancestry, but were closely matched between cases and controls in each set (Supplementary Figs. 3–5). We restricted our analyses to rare variants, stratified by allele frequency (singletons, <0.1%, and <0.5%) and function (LoF and damaging missense variants; Online Methods). In total, this joint discovery set consisted of 357,088 damaging missense and 55,955 LoF variants called in 4,264 cases and 9,343 controls (Fig. 1).
Figure 1.
Study design for the schizophrenia (SCZ) exome meta-analysis. The source of sequencing data, sample sizes, variant classes and analytical methods are described. Details on case-control samples are shown on the right and parent-proband trios are described on the left.
We replicated the enrichment of rare LoF variants in the previously implicated set of 2,456 genes11 in our UK and Finnish schizophrenia data sets (P = 7 × 10−4; Online Methods). Having confirmed that rare disruptive variants spread among many genes are associated with schizophrenia risk, we tested for an excess of disruptive variants in each of 18,271 genes in cases compared with controls (Online Methods). Despite our sample size, the per-gene statistics followed a null distribution in all tests, and we were unable to implicate any gene via case-control burden of disruptive variants (Supplementary Figs. 6 and 7).
LoF variants in SETD1A are associated with schizophrenia To determine whether the integration of de novo mutations with case-control burden might succeed in discovering risk genes in schizophrenia, we aggregated, processed and re-annotated de novo mutations in 1,077 schizophrenia probands from seven published studies, and found 118 LoF and 662 missense variants12,13,17–21 (Supplementary Table 1). 38 genes had two or more de novo nonsynonymous mutations, two of which (SETD1A and TAF13) had been previously suggested as candidate schizophrenia genes12,13. We found that the 754 genes with de novo mutations were significantly enriched in rare LoF variants in cases compared with controls from our main data set. The most significant enrichment across allele frequency thresholds and functional class was for the test of LoF variants with MAF < 0.1% (P = 2.1 × 10−4; OR 1.08, 95% confidence interval (CI) 1.02−1.14), which we focused on for subsequent analysis.
Motivated by this overlap of genes with de novo mutations and excess case-control burden, we metaanalyzed de novo variants in the 1,077 published schizophrenia trios with rare LoF variants (MAF < 0.1%) in 4,264 cases and 9,343 controls. We used two analytical approaches, one based on Fisher’s method to combine de novo and case-control P values, and the other using the transmission and de novo association (TADA) model to integrate de novo, transmitted and case-control variation using a hierarchical Bayesian framework15,22 (Fig. 1). We focused on results that were significant in both analyses and that did not depend on the choice of parameters in TADA (Online Methods and Supplementary Fig. 8). In both methods, loss-of-function mutations in a single gene, SETD1A, were significantly associated with schizophrenia risk (Fisher’s combined P = 3.3 × 10−9; Table 1). We observed three de novo mutations and seven case LoF variants in our discovery cohort and none in our controls (Fig. 2). In one of the seven case carriers, direct genotyping in parents confirmed that the LoF variant (c.5l8-2A>G) was a de novo event, but genotypes were not available for the other parents. We looked for additional SETD1A LoF variants in unpublished whole exomes from 2,435 unrelated schizophrenia cases and 3,685 controls23, but found none (Table 1). Thus, in more than 20,000 exomes, we observed ten case and zero control LoF variants (corrected OR 35.2, 95% CI 4.5–4,528). Although the confidence inter-vals were wide, rare LoF variants in SETD1A conferred substantial risk for schizophrenia. No other gene approached genome-wide significance (Supplementary Table 2 and Supplementary Figs. 9 and 10).
Table 1. Results from statistical tests associating disruptive variants in SETD1A to schizophrenia and developmental disorders.
| Phenotype | Data set | De novo | Case | Control | Test | P value |
|---|---|---|---|---|---|---|
| Schizophrenia | UK10K-INTERVAL | 2 of 1,353 | 0 of 4,769 | |||
| UK10K Finnish | 2 of 392 | 0 of 2,020 | ||||
| Swedish (published) | 3 of 2,519 | 0 of 2,554 | ||||
| All case-control | 7 of 4,264 | 0 of 9,343 | Fisher’s exacta | 0.0003 | ||
| Schizophrenia parent-proband trios | 3 of 1,077 | Poisson exactb | 4.6 × 10−7 | |||
| Case-control + de novo (discovery) | 3 of 1,077 | 7 of 4,264 | 0 of 9,343 | Fisher’s combinedc | 3.3 × 10−9 | |
| Swedish (replication) | 0 of 2,435 | 0 of 3,685 | ||||
| All schizophrenia samples | 3 of 1,077 | 7 of 6,699 | 0 of 13, 028 | Fisher’s combinedc | 5.6 × 10−9 | |
| Other neurodevelopmental phenotypes | DDD study | 2 of 4,281 | 2 of 4,281 | See noted | Fisher’s combinedc | 0.003 |
| ASD trios | 0 of 2,297 | |||||
| ID trios | 0 of 151 | |||||
| Combined | All samples | 5 of 7,806 | 9 of 10,980 | 0 of 13,028 | Fisher’s combinedc | 3.2 × 10−8 |
None of these tests incorporated exomes from the ExAC database. The number of SETD1A LoF variants and the sample size of each data set are indicated in each cell.
The statistical tests were performed as follows:
A one-sided burden test of case-control LoF variants using Fisher’s exact test.
The Poisson probability of observing N de novo variants in SETD1A given as calibrated baseline gene-specific mutation rate.
Meta-analysis of de novo and case-control burden P values using Fisher’s combined probability test.
The INTERVAL data set (n = 4,769) was used as matched controls.
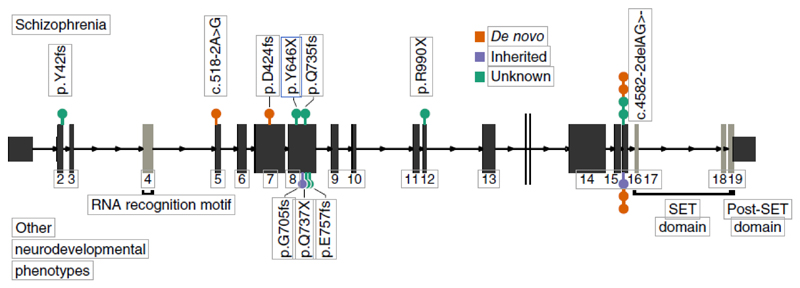
Figure 2.
The genomic position and coding consequences of 16 SETD1A LoF variants observed in the schizophrenia exome meta-analysis, the DDD study and the SiSU project. Variants discovered in patients with schizophrenia are plotted above the gene and those discovered in individuals with other neurodevelopmental disorders (from DDD and SISu) are plotted below. Each variant is colored according to its mode of inheritance. All LoF variants appeared before the conserved SET domain, which is responsible for catalyzing methylation. Seven LoF variants occurred at the same two-base deletion at the exon 16 splice acceptor (c.4582-2delAG>-).
Robustness of the SETD1A association
To validate our observation of the rarity of disruptive variants in SETD1A in unaffected individuals, we examined the exomes of 45,376 individuals without schizophrenia in the Exome Aggregation Consortium (ExAC) database and found only 2 LoF variants24, which represented a substantial depletion compared with chance expectation (Online Methods, expected value 32.5 LoF SNPs, P = 4.4 × 10−8). SETD1A is among the 3% most constrained genes in the human genome24; LoF variants in SETD1A are almost totally absent in the general population. Four of the ten SETD1A carriers with schizophrenia had the same two-base deletion at the exon 16 splice acceptor (c.4582-2delAG>-), at least two of which occurred as de novo mutations (Fig. 2). Given that this variant underpinned the statistical significance of our observation, we investigated it further in several ways. First, to rule out sequencing artifacts, we confirmed a clean call where we had access to the raw sequencing reads (n = 2) and noted that both published de novo mutations at this position had been validated with Sanger sequencing13,20. Second, our model, and therefore the test statistic that we report, is dependent on a gene-specific mutation rate (Online Methods). To address the possibility that the recurrent mutation occurs at a hypermutable site (and thus our model is not well calibrated), we determined that our observations would be exome wide significant (P < 1.25 × 10−6) even if the mutation rate at this position were up to eightfold higher (5.4 × 10−5) than the cumulative LoF rate for all other positions in SETD1A (6.6 × 10−6). If the two-base deletion mutation rate were truly this high (that is, greater than 99.99% of all per-gene LoF mutation rates), we would expect to find 4.9 observations in 45,376 non-schizophrenia exomes in ExAC, but instead we observed only 1 (Fisher’s exact test, P = 0.044). Using a minigene construct, we further found that this two-base deletion resulted in the retention of the upstream intron. This was predicted to lead to the translation of exon 15, the subsequent intron and an out-of-frame translation of exon 16 resulting in a premature stop codon (Supplementary Fig. 11 and Online Methods). Finally, if we ignored the de novo status of variants in our discovery and replication data sets and used ExAC exomes as additional controls (Online Methods and Table 2), LoF variants in SETD1A were significantly associated with schizophrenia using a basic test of case-control burden (P = 2.6 × 10−8, OR 37.6, 95% CI 8.0–353). Taken together, these analyses exclude many possible artifacts and provide confidence in our conclusion that LoF variants in SETD1A confer substantial risk for schizophrenia.
Table 2. Basic burden tests associating disruptive variants in SETD1A to schizophrenia and developmental disorders.
| Phenotype | Data set | Case | Control | Test | P value |
|---|---|---|---|---|---|
| Schizophrenia | All schizophrenia case-control samples (ignoring de novo status) | 10 of 7,776 | 0 of 13,028 | ||
| Non-schizophrenia ExAC exomes | 2 of 45,376 | ||||
| All samples | 10 of 7,776 | 2 of 58,404 | Fisher’s exact | 2.6 × 10−8 | |
| Neurodevelopmental disorders | DDD study | 4 of 4,281 | See notea | Fisher’s exact | 2.9 × 10−4 |
| ASD trios | 0 of 2,297 | ||||
| ID trios | 0 of 151 | ||||
| Combined | All samples | 14 of 14,505 | 2 of 58,404 | Fisher’s exact | 1.2 × 10−8 |
De novo status of variants was ignored was non-schizopherenia exomes from the ExAC database were incorporated as controls. The number of SETD1A LoF variants and the sample size of each data set were indicated in each cell.
The full control data set (n = 58,404) was used to calculated the P value.
SETD1A is associated with severe developmental disorders All heterozygous carriers of SETD1A LoF variants satisfied the full diagnostic criteria for schizophrenia, including classic positive symptoms such as hallucinations, prominent disorganization and paranoid delusions (Table 3). Eight patients had evidence of chronic illness, requiring long-term psychiatric services. Notably, of the seven SETD1A LoF carriers for whom any information on intellectual functioning was available, one was noted to have severe learning difficulties and the other six appeared to have mild to moderate learning difficulties. Four patients were noted to have achieved developmental milestones with clinically salient delays (Table 3). We were unable to confirm whether the three Swedish carriers had any form of cognitive impairment. This is consistent with previous reports that individuals with autism or schizophrenia who have de novo LoF mutations have a higher rate of cognitive impairment12,25.
Table 3. Phenotypes of individuals in the schizophrenia exome meta-analysis who carry LoF variants in SETD1A.
| Variant | Data set | Mode | Clinical features | Intellectual functioning |
|---|---|---|---|---|
| 16:30970178_T/T GATG frameshift | UK10K-Finns | Case | Psychotic episodes with hallucinations and prominent disorganization, requiring psychiatric hospitalization. Chronic illness with deterioration. | Probable mild intellectual disability. Completed compulsory education, but repeated several grades. |
| 16:30974752_A/G splice acceptor | UK10K-Finns | De novo | Disorganized schizophrenia with severe positive and negative symptoms with hallucinations delusions and aggression. Chronic, severe symptoms requiring long psychiatric hospitalization. Early onset at age 10. Has mild facial dysmorphology. | Severe learning difficulties, diagnosed with minimal brain damage, abnormal EEG; mild mental retardation. Unable to complete compulsory education. Development delay. |
| 16:30976334_AC/A frameshift | Takata et al.13 | De novo | Psychotic with persecutory delusions and thought disorder in addition to obsessional thoughts, compulsive behaviors and rituals. Persistent negative symptoms, disorgnized bahavior and delusional thinking. First psychotic break at age 21. As a child (age <10 years), displayed social isolation, excessive fears, inattentiveness, learning difficulties and obsessive-compulsive disorder–like rituals. Moderately deteriorating course. | Learning difficulties noted as a child. Delayed milestones. School performance declined from age 16. Worked as security officer. |
| 16:30977140_C/G stop gained | UK10K | Case | Chronic hallucinations and delusions, partially controlled by depot medication. | Minor problems with memory or understanding. No secondary school diploma. |
| 16:30977405_CAG/C frameshift | Swedish | Case | Two brief admissions, no record of antipsychotic treatment. No immediate family history of psychiatric disorders. | No information on intellectual functioning or educational attainment. |
| 16:30980962_C/T stop gained | Swedish | Case | Multiple hospitalizations, with 8 years of antipsychotic medication. No immediate family history of psychiatric disorders. | No information on intellectual functioning or educational attainment. |
| 16:30992057_CAG/C splice acceptor | UK10K | Case | Breech delivery. Epilepsy with seizures from ages 2 to 18. Socially isolated and dependent on parents till age 40, when presented with bizarre somatic delusions, paranoid delusions and auditory hallucinations including running commentary. Developed negative symptoms alongside ongoing psychotic symptoms and required long-term institutional care. Symptoms were persistent and unresponsive to antipsychotic medication. | Borderline intelligence. Attended mainstream school and left age 17 without a secondary school diploma. Worked as warehouseman. |
| 16:30992057_CAG/C splice acceptor | Swedish | Case | Multiple hospitalizations, with 8 years of antipsychotic medication. No immediate family history of psychiatric disorders. | No information on intellectual functioning or educational attainment. |
| 16:30992057_CAG/C splice acceptor | Takata et al.13 | De novo | Developed schizophrenia aged 18 with delusions, disorganized behavior, poor motivation, flattened affect and social isolation. Compulsive behaviors since 4th grade. Since first episode of psychosis, did not return to previous level of functioning. | Finished high school, but slow learner and inattentive. Delayed developmental milestones. |
| 16:30992057_CAG/C splice acceptor | Guiponni et al.20 | De novo | Undifferentiated schizophrenia. | Developmental delay. |
For each individual, we provide the genomic coordinates of the variant, its mode of inheritance and the study from which each patient was first recruited. ‘Clinical features’ describes notable neuropsychiatric or neurodevelopmental symptoms in each individual and ‘Intellectual functioning’ provides information on reported cognitive phenotypes.
To investigate whether SETD1A might be involved in other neurodevelopmental disorders, we looked for de novo LoF mutations in SETD1A in 3,581 published trios with autism, severe developmental disorders (DD) and/or intellectual disability15,26–28, but found none. We next turned to an additional 3,148 children with diverse, severe, developmental disorders recruited as part of the Deciphering Developmental Disorders (DDD) study, and discovered four probands with LoF variants in SETD1A (Table 4). Three of these were the recur-rent exon 16 splice junction indel described above (two de novo, one maternally inherited) and the fourth was a maternally inherited frameshift insertion (Fig. 2). We validated all four LoF variants using Sanger sequencing. All four probands had developmental delay with additional phenotypes that clustered in the larger DDD study (empirical P = 0.042; Online Methods). A fifth proband was found to have a de novo 650-kb deletion that encompassed SETD1A as well as 29 other genes (Supplementary Fig. 12 and Online Methods). SETD1A did not reach exome-wide significance as a developmental disorder gene in the DDD study alone (P = 3.0 × 10−3), but when we jointly analyzed all samples, the association was clear to both severe developmental disorders and schizophrenia (P = 3.1 × 10−8; Table 1). Because all of the DDD SETD1A carriers were under 12 years of age at recruitment and schizophrenia rarely manifests at this age29, it remains unknown whether these individuals will develop schizophrenia.
Table 4. Phenotypes of individuals in the DDD study and SISu project who carry LoF variants in SETD1A.
| Variant | Data set | Mode | Clinical features | Intellectual functioning |
|---|---|---|---|---|
| 16:30977316_G/GC frameshift | DDD | Maternally inherited | Capillary hemangiomas, abnormality of the eyebrow, broad nasal tip, wide mouth, thick lower lip vermilion, short philtrum, overgrowth, renal duplication. 5.29 years old. | Delayed speech and language development. |
| 16:30992057_CAG/C splice acceptor | DDD | Maternally inherited | Infantile axial hypotonia, delayed gross motor development, midfrontal capillary hemangioma. 0.55 years old. | Not detailed due to age |
| 16:30992057_CAG/C splice acceptor | DDD | De novo | Mild global developmental delay, hypertelorism, wide nasal bridge, hydrocele testis. 3.14 years old. | Aggressive behavior, autoaggression. First words spoken between 2 to 2.5 years of age. |
| 16:30992057_CAG/C splice acceptor | DDD | De novo | Global developmental delay, macrocephaly, nevus flammeus of the forehead, wide and flat nose, mandibular prognathia, hypopigmentation of the skin, wide intermammillary distance, truncal obesity. Has breath-holding attacks and night terrors. 6.09 years old. | Delayed speech and language development. |
| 16:30977411_C/T stop gained | NFID | Case | Short stature, mild facial morphology, EEG abnormalities, delusional disorder, has psychosis. | Mental retardation |
| 16:30977473_G/GC frameshift | NFBC | Case | Epilepsy during childhood (grand mal status epilepticus), diagnosed with personality disorder. | Not detailed |
For each individual, we provide the genomic coordinates of the variant, its mode of inheritance and the study from which each patient was first recruited. ‘Clinical features’ descibes notable neuropsychiatric or neurodevelopmental symptoms in each individual and ‘intellectual functioning’ provides information on reported cognitive phenotypes. NFID, Northern Finnish Intellectual Disability study; NFBC, Northern Finnish Birth Cohort.
In 5,720 unrelated Finnish individuals exome sequenced as part of the Sequencing Initiative Suomi project (Online Methods), we identified two additional heterozygous LoF variants in SETD1A. One individual with a stop-gain variant was recruited as part of the Northern Finnish Intellectual Disability (NFID) cohort with a diagnosis of mental retardation, short stature, mild facial dysmorphology and EEG abnormalities (Table 4). Notably, this individual was also diagnosed with delusional disorder and unspecified psychosis at 15 years of age. The second SETD1A LoF carrier belonged to the Northern Finnish 1966 Birth Cohort (NFBC), a representative, geographically based population cohort. This individual had epileptic episodes at 7 years of age and was diagnosed with an unspecified personality disorder by a psychiatrist. Thus, in an additional search for SETD1A LoF carriers, only two were found, both in individuals affected by neuropsychiatric disorders.
De novo burden in neurodevelopmental disorders
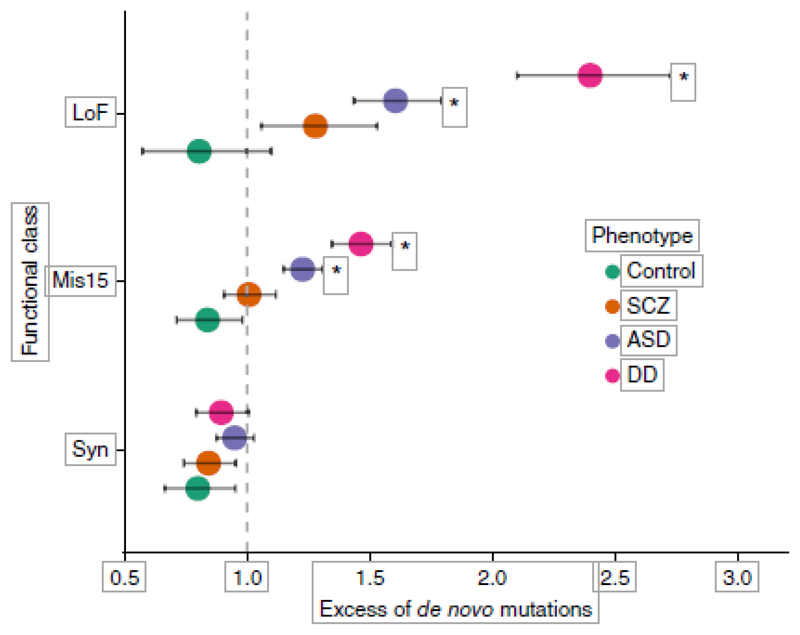
Even though our study had an overall sample size comparable to those of recent ASD and DD studies that identified 7 ASD genes and 32 DD genes15,26, we were only able to implicate a single schizophrenia gene at genome-wide significance. To investigate this further, we aggregated de novo mutations identified in 2,297 ASD, 1,113 DD and 566 control trios with our 1,077 schizophrenia trios and compared the rates of de novo events in each group relative to baseline exome-wide mutation rates (Online Methods). The rates of de novo mutations across damaging missense and LoF variants were significantly higher in DD than in ASD, and higher in ASD than in schizophrenia (Fig. 3). Indeed, the rate of damaging missense variants in schizophrenia was not different from baseline rates (P = 0.45) and only nominally higher than in controls (P = 0.029), and the rates of LoF variants were only slightly elevated (P = 5.7 × 10−3). In ASD, by contrast, missense (P = 9.4 × 10−10) and LoF (P = 3.7 × 10−15) rates were significantly greater than expectation. In developmental disorders, the rates were even higher (missense: P = 2.5 × 10−17; LoF: P = 1.3 × 10−31) (Fig. 3).
Figure 3.
A comparison of genome-wide de novo mutation rates in probands with ASD, DD, schizophrenia (SCZ) and controls. Rates are modeled using calibrated genome-wide mutation rates. Significant excess of de novo mutations when compared to the baseline model, *P < 4 × 10−3 (Bonferroni correction for 12 tests). Nominal significance can be inferred from the error bars (95% CI). Mis15, damaging missense; Syn, synonymous; see Online Methods.
Across all genes in the genome, the rate of disruptive de novo variants differed markedly across these disorders. Because the recurrence of de novo mutations is a particularly powerful way to identify risk genes, the weak excess of de novo variants in schizophrenia provides at least a partial explanation for the limited success of this strategy to date in identifying genes for this disorder.
Discussion
We identified an association between rare LoF variants in SETD1A and risk of schizophrenia and other severe neurodevelopmental phenotypes. A previous report13 suggested SETD1A as a candidate schizophrenia gene on the basis of two of the de novo mutations included in our analysis.
Our study establishes the SETD1A association at a significance exceeding a Bonferroni corrected P value of 1.25 × 10−6 independent of any specification of gene mutation rate. Indeed, in keeping with observations in other neurodevelopmental disorder sequencing studies, even larger meta-analyses of schizophrenia exomes will be required to define the phenotypic spectrum of SETD1A LoF variant carriers, to rule other candidates in or out, and to identify new risk genes.
SETD1A, also known as KMT2F, encodes one of the methyltransferases that catalyze the methylation of lysine residues in histone H3.
Articles
Loss-of-function variants in at least five other genes in this family result in dominant Mendelian disorders characterized by severe developmental phenotypes, including intellectual disability30. These include Wiedemann-Steiner syndrome (KMT2A), Kleefstra syndrome (EHMT1) and Kabuki syndrome (KMT2D) (Supplementary Fig. 13). Moreover, rare de novo LoF mutations and copy number variants in KMT2C, KMT2E, KDM5B and KDM6B have been recently associated with autism risk16. The developmental and cognitive phenotypes of SETD1A carriers are consistent with these other Mendelian conditions of epigenetic machinery; however, among all genes associated with developmental disorders and intellectual disability, SETD1A is the first shown to definitively predispose to schizophrenia, offering insights into the biological differences underlying these conditions26,31. As with other risk genes for severe neurodevelopmental phenotypes, it is possible that an allelic series of LoF variants exists in SETD1A, where different variants increase risk for different clinical features. However, seven of the 16 LoF variant carriers (Fig. 2) have the same two base deletion at the splice acceptor of exon-16 (c.4582-2delAG>-): four in individuals with schizophrenia and three in individuals diagnosed with other developmental disorders. Thus, the same variant is associated with both schizophrenia and developmental disorders.
Detailed phenotypes from the DDD and SISu studies suggest that SETD1A carriers may have distinctive features, including delayed speech and language development, epilepsy, personality disorder and facial dysmorphology (Table 4). Although cognitive and developmental phenotypes in our schizophrenia patients were sparser, four individuals had delayed developmental milestones, one was noted as having mild facial dysmorphology and minimal brain dam-age and another had epileptic seizures during childhood (Table 3). However, impairment of cognitive function is now generally regarded, along with positive and negative symptoms, as an integral feature of schizophrenia rather than a co-morbidity, and our study, as designed, cannot address whether variants in SETD1A are specifically associated with the cognitive features of the disorder. Indeed, it would require a re-sequencing study with detailed cognitive measurements on tens of thousands of patients (Online Methods and Supplementary Fig. 14) to decisively answer this question.
The clinical heterogeneity observed in carriers of SETD1A LoF variants is reminiscent of at least 11 large copy number variant syndromes which cause schizophrenia in addition to many other developmental defects10,32. A canonical example is the 22q11.2 deletion syndrome, which is characterized by schizophrenia in 22.6% of adult carriers33, highly variable intellectual impairment34 and numerous severe neurological and physical defects35. A considerably larger cohort (such as the hundreds of cases of 22q11.2 deletion syndrome studied to date) will be needed to accurately estimate the relative penetrance of SETD1A LoF variants for schizophrenia, developmental disorders and other clinical features.
Although disruptions of SETD1A are very rare events and occur in only a small fraction of schizophrenia cases (0.13% in our meta-analysis, 95% CI 0.062–0.24%), several lines of evidence suggest that histone H3 methylation is more broadly relevant to schizophrenia. The H3K4 methylation gene ontology category (GO:51568) showed the strongest statistical enrichment among 4,939 biological pathways in GWAS data of psychiatric disorders14. This category contains 20 genes, including SETD1A and six others (ASH2L, CXXC1, RBBP5, WDR5, DPY30 and WDR82)36–38 that together form the SET1-COMPASS complex, through which SETD1A regulates transcription by targeted methylation. Indeed, two of the genes in GO:51568 (WDR82 and KMT2E) are near genome-wide significant associations to schizophrenia6. A previous study of de novo CNVs in schizophrenia trios identified one deletion and one duplication overlapping EHMT1, another his-tone methyltransferase9 that has been implicated in developmental delay and a range of congenital abnormalities39. Finally, conserved H3K4me3 peaks identified in prefrontal cortical neurons colocalize with genes related to biological mechanisms in schizophrenia, including glutamatergic and dopaminergic signaling40. Our implication of SETD1A therefore contributes to the growing body of evidence that chromatin modification, specifically histone H3 methylation, is an important mechanism in the pathogenesis of schizophrenia.
Online Methods
Sample collections
Individuals clinically diagnosed with schizophrenia were recruited and exome sequenced as part of eight neurodevelopmental collections (Aberdeen, Collier, Edinburgh, Gurling, Muir, UK-SCZ, Finnish-SCZ and Kuusamo) in the UK10K sequencing project. Matched population controls were selected from non-psychiatric arms of the UK10K project, healthy blood donors from the INTERVAL project, and five Finnish population studies (ENGAGE, Familial dyslipidemia, FINRISK, Health 2000 and METSIM). Additional details on the UK10K data set are described in Supplementary Tables 3 and 4, and the sequence data were deposited into the European Genome-phenome Archive (EGA) under study accession code EGAO00000000079. The Swedish schizophrenia case-control study had been described in an earlier publication11, and we acquired processed VCFs for this data set via dbGaP authorized access (accession code: phs000473.v1.p1). The Deciphering Developmental Disorders study data set included 4,281 children with severe, undiagnosed developmental disorders. Probands and their parents were exome-sequenced in the project in order to identify novel genes associated with developmental disorders. Patient recruitment, sample collection, sequencing production, and initial analysis of the data set were described in detail in a previous publication26. The sequence data were deposited into the EGA under study accession EGAS00001000775.
The Sequencing Initiative Suomi project is an international collaboration generating whole genome and whole exome sequence data from Finnish samples, and consists of a number of prospective and case-control cohorts, including the ENGAGE, FINRISK, Health 2000 and METSIM studies (http://www.sisuproject.fi/content/cohorts). The Northern Finnish 1966 Birth Cohort (NFBC) is a geo-graphically based representative birth cohort including 96% (N = 12,068) of all live births in the two most northern provinces of Finland in 1966. The NFBC began with collection of prenatal information and continued with follow-ups at multiple time points resulting in a rich phenotype database of the study participants that combines information from hospital records, official registers, questionnaires and clinical examinations of the participants. DNA was collected from the study participants during the 31-year follow up and extracted from peripheral blood using standard protocols. All study participants provided a written informed con-sent to participate in the study. The ethical review board of the faculty of medicine, University of Oulu, approved the study. The Northern Finnish Intellectual Disability Cohort (NFID) is an ongoing sample collection of individuals who have been diagnosed with ICD-10 diagnosis of intellectual disability or specific developmental disorder of speech and language of unknown etiology (ICD-10 codes: F70-F79 and F80-F89). The patients were recruited from the Northern Ostrobothnia Hospital District in Finland, including the Oulu University Hospital Policlinic of Medical Genetics and Tahkokangas Care Home for Disabled. Patients were identified through hospital records and during routine visit to the policlinic and were initially contacted by a trained research nurse or by their treating physician. All research subjects and their legal guardians provided a written informed consent to participate in the study. The current sample includes 324 patients and their first-degree family members (N = 631, 92 full trios) with GWAS and WES data available. DNA samples of the participants were extracted primarily from peripheral blood. In few sporadic cases where a blood sample could not be obtained, DNA was extracted from saliva. The ethical committees of the Northern Ostrobothnia Hospital District and the Hospital District of Helsinki and Uusimaa reviewed and approved the study.
Informed consent was obtained for all samples
Further information is avail-able at http://www.uk10k.org/, http://www.ddduk.org, http://www.intervalstudy.org.uk/ and http://www.sisuproject.fi/.
Sequence data production
1–3 μg of DNA was sheared to ~100-400 bp using either a Covaris E210 or LE220 machine (Covaris) and processed using Illumina paired-end DNA library preparation. The DNA was enriched using the Agilent SureSelect Human All Exon v.3 or v.5 kits. All libraries were sequenced on the Illumina HiSeq 2000 with 75 base paired-end reads in multiple batches accord-ing to the manufacturer’s protocol. Sequencing reads that failed quality control (QC) were first removed using the Illumina GA pipeline. Remaining raw reads were mapped to the reference genome (UK10K: GRCh37, INTERVAL: GRCh37_hs37d5) using BWA (v0.5)41 and duplicate fragments were marked using Picard (UK10K: v1.36, INTERVAL: v1.114)42. We used GATK (UK10K: v1.1-5; INTERVAL: v3.2-2) to perform local realignment around indels and recalibrate base qualities in each sample BAM43. All samples were individually called using GATK Haplotype Caller (v3.2), merged into batches of 200 samples using CombineVCFs, and joint-called using GenotypeVCFs, all at default settings44,45. Supplementary Figure 1 showed that the samples enriched using the v.5 kit have lower read depth across the entire exome, but cover a much larger percentage of coding regions than in any previous capture. The samples in the UK10K project are divided into two batches, clearly reflecting a chemistry change that occurred early in the project. The DDD study exomes more closely resembled the UK10K v.3 samples but clear differences in coverage exist between the v.3 and custom v.3 capture. Due to different captures used in the UK10K and INTERVAL data sets, variant calling was performed at the union of the Agilent v.3 and v.5 captures with 100 base pairs of flanking sequence. To harmonize variant calls across all sequencing batches, we limited subsequent QC and analysis to variants covered at 7× or more in at least 80% of samples in each sequencing batch (Supplementary Fig. 1).
Sample-level quality control
Quality control was performed on each population (UK, Finnish and Swedish) separately. We removed samples with a contamination fraction ≥3% estimated using VerifyBamID (v1.0)46 or low coverage (≤75% of the Gencode v.19 coding region covered at ≥10×). Principal components analysis (PCA) was performed using PLINK v1.9 (ref. 47) on a set of high-quality (VQSR tranche 99.0%, missingness < 3% and Hardy-Weinberg P < 10−3), LD-pruned (r2 > 0.2), common (MAF > 5%) SNPs found in our exome capture and in 1000 Genomes Project Phase III data. Ten principal components were estimated using 1000 Genomes samples, onto which we projected all of our cases and controls (Supplementary Fig. 5). We verified whether samples had the same population ancestry (UK, Finnish or Swedish) as reported in the sample manifests and excluded individuals who were of non-European ancestry. We estimated kinship coefficients between each sample pair using KING v1.4 (ref. 48) and excluded one member of any apparent relative pair (kinship ≥ 0.09375). After sample QC, 6,122 UK samples (1,353 cases and 4,769 controls), 2,412 Finnish samples (392 cases and 2,020 controls) and 5,073 Swedish samples (2,519 cases and 2,554 controls) were available for analysis.
Variant-level quality control and annotation
We empirically derived thresholds for site and genotype filters that balanced sensitivity and specificity by training on the following: ExomeChip genotype calls in 295 UK10K cases and doubleton inherited variants (truth sets) and singleton Mendelian inheritance inconsistencies (false set) in 227 trios of the DDD study. We kept SNPs in the VQSR tranche with 99.75% sensitivity and with mean genotype quality (GQ) ≥ 30. Individual genotypes were retained if they had a GQ ≥ 30, alternate allele read depth (DP1) 2, allelic balance (AB) ≥ 0.2, and AB ≤ 0.8. Using these thresholds, we removed 95.63% of Mendelian errors while retaining 98.38% of doubleton inherited variants and 99.62% of heterozygous Exomechip SNPs. We kept indels in the VQSR tranche with 99.50% sensitivity and with mean GQ ≥ 90. Individual genotypes were retained if they had GQ ≥ 90, DP1 ≥ 2, AB ≥ 0.25, and AB ≤ 0.8. Using these thresholds, we removed 92.35% of all indel Mendelian errors and retained 93.60% of all doubleton inherited indels. We further excluded SNPs and indels with missingness > 20%, Hardy-Weinberg equilibrium χ2 P < 10−8, variants within low-complexity regions49 and indels with more than two alternate alleles or within three base pairs of another indel.
Following sample and variant QC, the per-sample transition-to-transversion ratio was comparable between all populations (mean ~3.25) (Supplementary Fig. 4). We still observed differences in total variant counts among the UK, Finnish and Swedish collections (Supplementary Fig. 3), likely reflecting differences in sequencing depth, capture reagents, sequencing protocol, read alignment and variant calling. However, variant counts and population genetics metrics were consistent between cases and controls within each population group.
We used the Ensembl Variant Effect Predictor (VEP) version 75 to annotate all variants according to Gencode v.19 coding transcripts50. We grouped frameshift, stop gained, splice acceptor and donor variants as loss-of-function (LoF), and missense or initiator codon variants with a CADD Phred score ≥ 15 as damaging missense51.
Statistical significance and robustness of rare variant association analyses
Previous large sequencing analyses such as the Swedish schizophrenia, DDD and NHLBI myocardial infarction studies11,26,52 have defined genome-wide significance for gene burden tests using a Bonferroni correction for the number of genes and the number of functional and frequency cut-offs tested. For example, P < 1.25 × 10–6 is 0.05 corrected for 20,000 genes tested for nonsynonymous and LoF variants, and a further correction for two frequency thresholds would require the even more stringent cutoff of P < 6.25 × 10−7.
For these thresholds to control false positives, however, the test being used must produce well-calibrated P values. This has been shown to be true for standard approaches in a case-control setting, such as the basic burden test, Fisher’s exact test and the sequence kernel association test (SKAT)53, as long as the cases and controls are well-matched and residual differences are corrected for11,52. On the other hand, parent proband trio studies use a Poisson or Binomial model parameterized by gene-specific mutation rates and the discovery sample size to test for an elevated rate of de novo mutations. While this approach is powerful, it is less robust than the approaches described above. First, de novo test statistics are highly sensitive to the specification of gene-specific mutation rates, which are well established for SNVs but not small indels. Furthermore, the low counts in de novo studies make results sensitive to the size of the discovery data set.
In previous studies of schizophrenia trios, thirty-eight genes had two or more de novo nonsynonymous mutations, two of which (SETD1A, P = 2.4 × 10−6 and TAF13, P = 1 x10−6) were significant enough to be suggested as candidate schizophrenia genes12,13. These two findings illustrate the challenges of interpreting de novo data in small numbers of samples. TAF13 has a coding length of 375 base pairs, making just two observations significant, though no additional evidence has been found in subsequent, much larger studies, including our own. For SETD1A, both mutations are indels, making it hard to accurately calculate Poisson P values (indeed, we have observed one of these de novo three additional times, suggesting it has a high mutation rate). Furthermore, this result is no longer significant when meta-analyzed with the published schizophrenia de novo data sets discussed in the same study17,19, which would be the statistically strongest analysis available at the time. Thus, in keeping with observations in other neurodevelopmental disorder sequencing studies, very large meta-analyses of both case-control and de novo variation from schizophrenia exomes are required to exclude many possible artifacts, rule other candidates in or out, and identify new risk genes.
Case-control analysis
To identify genes with a significant burden of rare, damaging variants, we applied the basic burden test, Fisher’s exact test and the sequence kernel association test (SKAT) as implemented in PLINK/SEQ53,54. For each gene, we tested LoF variants and LoF combined with damaging missense variants. To evaluate significance, we performed 2 million case-control permutations within each population (UK, Finnish and Swedish) to control for ancestry and batch-specific differences. One-sided basic burden and Fisher’s exact tests were applied at three different minor allele frequency (MAF) thresholds (singletons, MAF ≤ 0.1% and MAF ≤ 0.5%). We used default parameters for SKAT (MAF ≤ 5%) and included the first ten principal components as covariates. Consistent with well-matched cases and controls, we observed no genome-wide inflation in either common or rare variant tests (Supplementary Fig. 7).
Gene set enrichment analyses broadly followed the methodology described in ref. 11 and implemented in PLINK/SEQ and the SMP utility. The gene set enrichment statistic was calculated as the sum of single gene burden test-statistics corrected for exome-wide differences between cases and controls. Statistical significance was determined through permutation testing as described above. We adopted the min-P procedure to empirically correct for multiple testing: the same order of phenotypic permutations was applied for all tests, and a joint null distribution of minimal P values was generated to determine the significance of each gene set. The reported odds ratios and confidence intervals from the gene set enrichment analyses were calculated from raw counts without taking into account ancestry and batch-specific differences in cases and controls.
Meta-analysis of de novo mutations and case-control burden
Validated de novo mutations identified in seven published studies of schizophrenia trios were aggregated for analysis with our case-control cohort (Supplementary Table 1). Recurrence of de novo mutations was modeled as the Poisson probability of observing N or more de novo variants in a gene given a baseline gene-specific mutation rate obtained from the method described in ref. 56 modified to produce LoF and damaging missense rates for each canonical Gencode v.19 gene55. The gene-specific mutation rates in our models have been validated as highly reliable in a previous publication56 and subsequently used in the main analyses of large-scale exome sequencing of neurodevelopmental disorders with highly replicable results15,26. A one-sided Fisher’s exact test was used to model the difference in rare LoF (MAF < 0.1%) burden between cases and controls. Subsequently, de novo and case-control burden P-values were meta-analyzed using Fisher’s combined probability method with df = 4 (Supplementary Figs. 8 and 9 and Supplementary Table 2). The odds ratios reported were corrected using penalized maximum likelihood logistic regression model (Firth’s method, implemented in the logistf R package).
We also applied the Transmission and Disequilibrium Association (TADA) method as described in ref. 22 and implemented in ref. 15. Information from the recurrence of de novo mutations was integrated with inherited and case-control burden in a single statistical test. The robustness of results from TADA depends heavily on the specification of its hyperparameters, which are dependent on the (unknown) genetic architecture of the trait. These include the relative risks for de novo and case-control variants (parameterized by γd and γ) and the number of true risk genes in schizophrenia (k). Using estimates from the autism analysis would be incorrect; autism, for instance, has a greater excess of de novo LoF and missense mutations than schizophrenia (Fig. 3). To ensure any results from TADA are robust, we ran the model across a grid of reasonable parameters:
| γ | d ∈{2,4,6,8,10,12,15,20} for LoF variants |
| γ | ∈{1,2,4} for LoF inherited and case-control variants |
| γ | d ∈{1,2,4} for missense variants |
| γ | = 1 for missense inherited and case-control variants |
| k ∈{100,500,1000,2000} | |
We used the default values for the remaining parameters and applied the following restrictions: γd > γ and γ lof > γmis.
After exhaustively generating Bayes factor across a set of reasonable hyper-parameters, the results largely agreed with the results obtained from the Fisher’s combined probability method: only one gene, SETD1A, had reached genome-wide significance (Supplementary Fig. 8). We found that our signal in SETD1A had a q-value < 0.01 as long γd > 4, γ > 4 and k > 100. If we assumed a greater mean relative risk for LoF variants in SETD1A (γd > 8 and γ > 8) as expected for risk alleles in a constrained gene, SETD1A was exome-wide significant for any reasonable specification of k. We found that our signal in SETD1A is robust across frequentist and Bayesian models, under reasonable assumptions about schizophrenia’s genetic architecture (Supplementary Fig. 8). No other gene had a q-value <0.01 under any tested parameterization, including the parameterization used in the previous autism meta-analysis (Supplementary Table 5).
SETD1A LoF variants in the ExAC database
We looked in the ExAC database (v0.3) for the LoF variants in SETD1A. All exomes were joint-called using the GATK v3.2 pipeline, and included other public exome data sets, such as the 1000 Genomes Project and NHLBI-GO Exome Sequencing Project, with additional quality control compared to their original releases. In 60,706 unrelated exomes, we observed seven LoF variants in SETD1A. Since the v0.3 release included the Swedish schizophrenia study, we excluded all samples from this data set, leaving only four LoF variants in 45,376 exomes without a known neuropsychiatric diagnosis. We next applied the same stringent QC metrics we used in our analysis to ExAC data. We found that the 16:30976302-GC/G indel observed in two individuals was located at the same position as a high-quality SNP and occurred at a homopolymer run of cytosines. At the genotype level, both calls had a genotype quality (GQ) phred probability of < 40, far lower than used in our study in which we required indels to have a GQ > 90. In addition, the variant has poor allelic balance (AB < 0.15), and the BAM alignment reflected these low-quality metrics24. Given this evidence, we excluded the putative indel. Two high-quality SETD1A LoF variants in 45,376 unaffected ExAC exomes remained.
Following the approach in ref. 56, we determined the significance of the depletion of SETD1A LoF variants in ExAC using a signed Z-score of the chi-squared deviation between observed and expected counts. We scaled the expected LoF counts provided by ExAC (43 in 60,706) to 45,376 exomes (expected value 32.5), and calculated the one-tailed P-value of the signed Z-score assuming two observed LoF variants. The degree of constraint relative to other coding genes was based on the pLI score24.
If we disregarded de novo status of our variants, our combined schizophrenia data set was composed of 7,776 cases and 13,028 controls. After including unaffected ExAC exomes as additional controls, we observed ten LoF variants in 7,776 cases and two LoF variants in 58,404 controls, which was significantly different by a Fisher’s exact test. This result was driven by ten very rare variants in our schizophrenia cases: six observed in only one individual each and the seventh observed in four individuals. Two of these four were de novo and the other two were found in unrelated individuals of different ancestry (one from Sweden and one from the UK). Similarly, of the two LoF variants in ExAC, one was observed in only one individual and the other was the recurrent indel in an individual of African ancestry. Thus, our burden test of very rare variants in SETD1A would not be confounded by systematic differences between subpopulations in the ExAC exomes and our data set.
Validation of SETD1A variants
We designed primers using Primer3 to produce products between 400 and 600 bp in length centered on the site of interest. Using genomic DNA from all trio members as templates, PCR reactions were carried out using Thermo-Start Taq DNA Polymerase (Thermo Scientific), following the manufacturer’s protocol, and successful PCR products were capillary sequenced. Traces from all trio members were aligned, viewed, and scored for the presence or absence of the variant.
Functional consequence of the exon 16 splice acceptor deletion
To assess the impact of the exon 16 splice acceptor site variant, we created a custom minigene construct. We cloned the entire 696-bp genomic region encompassing exons 15, 16, 17 and intervening introns of human SETD1A, fused inframe to a C-terminal GFP. The entire cassette was flanked by a strong upstream promoter and a down-stream polyadenylation sequence. Plasmids containing either reference or deletion-containing forms were transfected into HELA cells, which were then grown for 2 d under standard conditions. RNA was extracted (RNEasy, Qiagen) from the transfected cells and used to synthesize cDNA (SuperscriptIII, Invitrogen). We designed minigenespecific primers to avoid amplification of endogenous HELA derived transcripts. The first pair of primers spanned all three exons, thus allowing us to detect overall splicing changes (Pair 1, Forward 2: TCGAAG AGTCATAAACACTGCCATG, Reverse 9: GTGAACAGCTCCTCGCCCTTG). We also designed pairs of exonic, intron-spanning primers to distinguish splic-ing events upstream (Pair 2, Forward 1: TTTGCAGGATCCCATCGAAGAG TC, exon 16 reverse: CACTGTCCATGATGGCGGAGGTA) and downstream (Pair 3, exon16 forward: CTGCTGAGCGCCATCGGTAC, exon17 reverse: CTGAACTTGTGGCCGTTTACGTC) of exon 16. PCRs were performed on cDNA from two transfection replicates of each sample. Agarose gels identified PCR product size differences (DNA ladder: 2-log ladder, New England BioLabs), which were further analyzed by capillary sequencing.
As expected, strong GFP expression was detected from the reference sequence construct. This suggested correct splicing between exons, leading to in-frame GFP translation. The mutant form displayed dramatically weaker GFP expression. mRNA was extracted from the transfected cells, and PCRs spanning all three exons revealed an increased transcript size in the mutant form compared to refer-ence (Supplementary Fig. 11a). A PCR spanning just the first 2 exons (15/16) revealed a similar shift in size, suggesting that the splice site deletion/mutation was causing intron retention between exons 15 and 16 (Supplementary Fig. 11b). Sanger sequencing of the PCR products confirmed this aberrant splicing outcome (Supplementary Fig. 11c). The predicted translation product would therefore include translation of exon 15, the subsequent intron and out-of-frame translation of exon 16, resulting in a premature stop within this exon. The downstream splicing event to exon 17 was not affected. These data indicate that in human cells, the recurrent indel we observe in probands results in a premature stop codon and a truncated SETD1A protein.
De novo CNV deleting a single copy of SETD1A found in the DDD study
We observed a de novo CNV deleting 650 kilobases around SETD1A (chr16:30,964,376-31,614,891, Supplementary Fig. 12) in a DDD proband. CNV calling and quality control in the DDD study was described in a previous publication26, and the call was supported by signal from 156 probes. The proband had global developmental delay, absent speech, motor delay, sleep disturbance, developmental regression, feeding difficulties in infancy and generalized myoclonic seizures.
Phenotype clustering in DDD probands
Clinical geneticists systematically recorded phenotypes of DDD probands using the Human Phenotype Ontology (HPO)57. These terms were used to assess the probability that the probands shared more similar clinical phenotypes than expected by chance. Similarity testing used the Human Phenotype Ontology version 2013-11-30. For each pair of terms we determined the information content (defined as the negative logarithm of the probability of the terms’ usage within the DDD cohort of 4,295 probands) for the most informative common ancestor. The similarity of HPO terms between two individuals was estimated as the maximum information content (maxIC) from pairwise comparisons of the HPO terms for the two individuals. The phenotype similarity for a set of N probands was estimated as the sum of all the pairwise maxIC scores. A null distribution of similarity scores was simulated from randomly sampled sets of N probands. The P-value was calculated as the proportion of scores greater than or equal to the observed score.
Comparison of de novo mutation rates
This analysis aggregated and analyzed de novo mutations from four different studies: 1,113 probands with develop-mental disorders26, 2,297 ASD probands15 and 566 control probands25,58. De novo mutations (xd) in each neurodevelopmental condition was modeled as xd ~ Pois(2NtμG), where Nt is the number of trios, μG is the genome-wide mutation rate for a particular functional class and xd is the observed number of de novo mutations in Nt trios. The genome-wide mutation rate of each variant class was calculated as the sum of all gene-specific mutation rates in Samocha et al.56 (μsyn = 0.137, μdamaging mis = 0.165, μLoF = 0.043). We modeled de novo mutations in control trios to ensure that the genome-wide mutation rates were well calibrated. We report the probability of observing xd or more mutations in Nt trios given the genome-wide mutation rate. We used the Poisson exact test to determine if pairwise differences in de novo rates existed between control, SCZ, ASD and DD trios, and reported the two-sided P-values and rate ratios. Bonferroni correction was used to adjust for multiple testing.
Power calculations to show co-morbid cognitive impairment in schizophrenia SETD1A carriers
We estimated the sample size required to show that LoF variants in SETD1A specifically give rise to decreased cognitive function beyond their effect on schizophrenia risk. We assumed that pre-morbid IQ in individuals diagnosed with schizophrenia followed a Gaussian distribution with mean μ0 and s.d. σ. We further assumed that the distribution of pre-morbid IQ in carriers of SETD1A LoF variants was also Gaussian, shared the same s.d. σ, but has a shifted mean μ1. To calculate the sample size needed to show that μ0 and μ1 were statistically different, we performed power calculations using a one-sided t-test of means across a range of parameters for the effect size and frequency of SETD1A LoF variants. We define the following:
N = sample size (individuals diagnosed with schizophrenia)
or the effect size (in s.d. units) of SETD1A LoF variants on premorbid IQ
α = 0.05, type I error probability
p = frequency of LoF variants in SETD1A in schizophrenia cases
Supplementary Figure 14 shows power to detect this effect across the following parameter combinations:
N ∈ {5000,10000,…100000}
d ∈ {0.5,1}, or μ1 = μ0–σ×d
p ∈ {1×10−4, 5×10−4, 1×10−3}
Assuming a modest effect on cognition (d = 0.5) and that only one in 10,000 schizophrenia patients carries a SETD1A LoF variant, a sample size of over 100,000 individuals would be required for 50% power to detect the effect. If the effect on cognition was greater (d = 1) and the true frequency was similar to the 0.1% observed in our study, a sample size of over 10,000 individuals would have >50% power.
Supplementary Material
Acknowledgments
We thank the thousands of patients who participated in these studies. We thank H. Firth and D. FitzPatrick for discussions. The UK10K project was funded by Wellcome Trust grant WT091310. The DDD Study is funded by HICF-1009-003. The DDD and the INTERVAL sequencing studies are funded by Wellcome Trust grant WT098051. T.S. is supported by the Williams College Dr. Herchel Smith Fellowship. P.F.S. is supported by NIH R01 MH077139. A.P. is supported by Academy of Finland grants 251704 and 286500, NIMH U01MH105666 and the Sigrid Juselius Foundation. The work at Cardiff University was funded by Medical Research Council (MRC) Centre (G0801418) and Program Grants (G0800509). The key groups of the Sequencing Initiative Suomi (SISu) project are from the Universities of Eastern Finland, Oulu and Helsinki and The Institute for Health and Welfare, Finland, Lund University, The Wellcome Trust Sanger Institute, University of Oxford, The Broad Institute, University of Michigan, Washington University in St. Louis and University of California, Los Angeles (UCLA). The SiSu project is coordinated in the Institute for Molecular Medicine Finland at the University of Helsinki. Participants in INTERVAL were recruited with the active collaboration of NHS Blood and Transplant England, which has supported fieldwork and other elements of the trial. DNA extraction and genotyping was funded by the National Institute of Health Research (NIHR RP-PG-0310-1004), the NIHR BioResource and the NIHR Cambridge Biomedical Research Centre. The academic coordinating center for INTERVAL was supported by core funding from NIHR Blood and Transplant Research Unit in Donor Health and Genomics, UK Medical Research Council (G0800270) and British Heart Foundation (SP/09/002). M.I.K. was supported by Instrumentarium Science Foundation, Finland; Finnish Foundation for Cardiovascular Research; Orion Research Foundation and the University of Eastern Finland, Saastamoinen Foundation.
Footnotes
Author Contributions
T.S., S.S.G., E.L.C., D.G., M.E.H., M.C.O’D. A.P., M.J.O. and J.C.B. conceived and designed the experiments. S.S.G., E.L.C. and E.P. performed the experiments. T.S., D. Curtis, S.M.P., L.C., J.M. and H.C. performed the statistical analysis. T.S., M.I.K., S.M.P., L.C., J.M., H.C., G.B. and E.R. analyzed the data. M.I.K., S.M.P., J. Suvisaari, D.B., G.B., O.P., D. Collier, M.J.D., J.D., N.B.F., M.J., G.K., J.K., O.K., P.H., C.M.H., M.M., S.A.M., P.M., A.M.M., A.M., J.S.M., C.M., W.O., T.P., D.R., J. Sambrook, P.S., D.S.C., J.V., J.T.R.W., H.W. and P.F.S. contributed reagents, materials and analysis tools. T.S., P.F.S., M.E.H., M.J.O. and J.C.B. wrote the paper. J. Suvisaari, D.B., M.A., M.B., T.C., D. Collier, N.C., M.D., A.F., S.J., C.I., J.L., R.M.M., R.N.-E., T.P. and D.S.C. recruited patients.
References
- 1.Perälä J, et al. Lifetime prevalence of psychotic and bipolar I disorders in a general population. Arch Gen Psychiatry. 2007;64:19–28. doi: 10.1001/archpsyc.64.1.19. [DOI] [PubMed] [Google Scholar]
- 2.van Os J, Kapur S. Schizophrenia. Lancet. 2009;374:635–645. doi: 10.1016/S0140-6736(09)60995-8. [DOI] [PubMed] [Google Scholar]
- 3.Saha S, Chant D, McGrath J. A systematic review of mortality in schizophrenia: is the differential mortality gap worsening over time? Arch Gen Psychiatry. 2007;64:1123–1131. doi: 10.1001/archpsyc.64.10.1123. [DOI] [PubMed] [Google Scholar]
- 4.Lichtenstein P, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 6.Ripke S, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirov G, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signaling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rees E, et al. Analysis of copy number variations at 15 schizophrenia-associated loci. Br J Psychiatry. 2014;204:108–114. doi: 10.1192/bjp.bp.113.131052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purcell SM, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fromer M, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takata A, et al. Loss-of-function variants in schizophrenia risk and SETD1A as a candidate susceptibility gene. Neuron. 2014;82:773–780. doi: 10.1016/j.neuron.2014.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The Network and Pathway Analysis Subgroup of the Psychiatric Genomics Consortium. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci. 2015;18:199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Rubeis S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanders S, et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Girard SL, et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- 18.Xu B, et al. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulsuner S, et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guipponi M, et al. Exome sequencing in 53 sporadic cases of schizophrenia identifies 18 putative candidate genes. PLoS One. 2014;9:e112745. doi: 10.1371/journal.pone.0112745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy SE, et al. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry. 2014;19:652–658. doi: 10.1038/mp.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He X, et al. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 2013;9:e1003671. doi: 10.1371/journal.pgen.1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Genovese G, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. 2015 doi: 10.1038/nature19057. Preprint at http://www.biorxiv.org/content/early/2015/10/30/030338. [DOI] [PMC free article] [PubMed]
- 25.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rauch A, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 28.de Ligt J, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 29.Rajji TK, Ismail Z, Mulsant BH. Age at onset and cognition in schizophrenia: meta-analysis. Br J Psychiatry. 2009;195:286–293. doi: 10.1192/bjp.bp.108.060723. [DOI] [PubMed] [Google Scholar]
- 30.Fahrner JA, Bjornsson HT. Mendelian disorders of the epigenetic machinery: tipping the balance of chromatin states. Annu Rev Genomics Hum Genet. 2014;15:269–293. doi: 10.1146/annurev-genom-090613-094245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Firth HV, et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am J Hum Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirov G, et al. The penetrance of copy number variations for schizophrenia and developmental delay. Biol Psychiatry. 2014;75:378–385. doi: 10.1016/j.biopsych.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bassett AS, et al. Clinical features of 78 adults with 22q11 deletion syndrome. Am J Med Genet. 2005;138:307–313. doi: 10.1002/ajmg.a.30984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Butcher NJ, et al. Functional outcomes of adults with 22q11.2 deletion syndrome. Genet Med. 2012;14:836–843. doi: 10.1038/gim.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryan AK, et al. Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: a European collaborative study. J Med Genet. 1997;34:798–804. doi: 10.1136/jmg.34.10.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J, Tate CM, You J, Skalnik DG. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J Biol Chem. 2007;282:13419–13428. doi: 10.1074/jbc.M609809200. [DOI] [PubMed] [Google Scholar]
- 37.Lee J, Skalnik DG. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol. 2008;28:609–618. doi: 10.1128/MCB.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.The Uniprot Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2014;43:204–212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kleefstra T, et al. Further clinical and molecular delineation of the 9q subtelomeric deletion syndrome supports a major contribution of EHMT1 haploinsufficiency to the core phenotype. J Med Genet. 2009;46:598–606. doi: 10.1136/jmg.2008.062950. [DOI] [PubMed] [Google Scholar]
- 40.Dincer A, et al. Deciphering H3K4me3 broad domains associated with gene-regulatory networks and conserved epigenomic landscapes in the human brain. Transl Psychiatry. 2015;5:e679. doi: 10.1038/tp.2015.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Picard. [accessed 1 March 2011]; http://broadinstitute.github.io/picard/
- 43.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;11:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jun G, et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am J Hum Genet. 2012;91:839–848. doi: 10.1016/j.ajhg.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thornton T, et al. Estimating kinship in admixed populations. Am J Hum Genet. 2012;91:122–138. doi: 10.1016/j.ajhg.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30:2843–2851. doi: 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McLaren W, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Do R, et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2014;518:102–106. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu MC, et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.PLINK/SEQ version 0.09. [accessed 1 February 2014]; http://atgu.mgh.harvard.edu/plinkseq/
- 55.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Samocha KE, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Köhler S, et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acis Res. 2014;42:D966–D974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moore C, et al. The INTERVAL trial to determine whether intervals between blood donations can be safely and acceptably decreased to optimise blood supply: study protocol for a randomized controlled trial. Trials. 2014;15:363. doi: 10.1186/1745-6215-15-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.