Abstract
We investigated gene–environment (G × E) interactions related to childhood antisocial behavior between polymorphisms implicated by recent genome-wide association studies (GWASs) and two key environmental adversities (maltreatment and smoking during pregnancy) in a large population cohort (ALSPAC). We also studied the MAOA candidate gene and addressed comorbid attention-deficit/hyperactivity disorder (ADHD). ALSPAC is a large, prospective, ethnically homogeneous British cohort. Our outcome consisted of mother-rated conduct disorder symptom scores at age 7;9 years. G × E interactions were tested in a sex-stratified way (α = 0.0031) for four GWAS-implicated variants (for males, rs4714329 and rs9471290; for females, rs2764450 and rs11215217), and a length polymorphism near the MAOA-promoter region. We found that males with rs4714329-GG (P = 0.0015) and rs9471290-AA (P = 0.0001) genotypes were significantly more susceptible to effects of smoking during pregnancy in relation to childhood antisocial behavior. Females with the rs11215217-TC genotype (P = 0.0018) were significantly less susceptible to effects of maltreatment, whereas females with the MAOA-HL genotype (P = 0.0002) were more susceptible to maltreatment effects related to antisocial behavior. After adjustment for comorbid ADHD symptomatology, aforementioned G × E’s remained significant, except for rs11215217 × maltreatment, which retained only nominal significance. Genetic variants implicated by recent GWASs of antisocial behavior moderated associations of smoking during pregnancy and maltreatment with childhood antisocial behavior in the general population. While we also found a G × E interaction between the candidate gene MAOA and maltreatment, we were mostly unable to replicate the previous results regarding MAOA–G × E’s. Future studies should, in addition to genome-wide implicated variants, consider polygenic and/or multimarker analyses and take into account potential sex stratification.
Electronic supplementary material
The online version of this article (10.1007/s00406-018-0964-5) contains supplementary material, which is available to authorized users.
Keywords: ALSPAC, Antisocial behavior, Aggression, Gene–environment interaction, Maltreatment, Smoking during pregnancy
Introduction
Conduct disorder (CD) is a pediatric behavioral disorder with a prevalence of approximately 4–16% in boys and about 1–9% in girls [1]. Hallmark of CD are antisocial behaviors, that is, serious violations of basic rights of other people and/or age-appropriate societal norms resulting in severe aggression, deceitfulness, and rule-breaking behavior. The severe nature of these symptoms gives rise to a significant burden for affected patients, families, and societies at large [2]. About 40% of boys and 25% of girls with CD are estimated to display antisocial behaviors persisting into adulthood and develop antisocial personality disorder [3, 4].
CD symptomatology or antisocial behaviors can be considered as continuous traits that are caused by both genetic and environmental risk factors. More specifically, the interplay between genes and environment, also known as gene–environment (G × E) interactions, can provide insight into why some individuals are more susceptible to certain adverse genetic or environmental factors than others. These G × E interactions are assumed to be of great importance in multifactorial traits such as antisocial behavior [5–7].
Recent insights suggest that the classic candidate G × E literature holds important limitations. Most notable, the use of poorly replicated candidate genes, underpowered samples, and inappropriate correction for multiple comparisons are suspected to have resulted in an inflated rate of false-positive findings across studies [8–10]. Instead, hypothesis-free, genome-wide association studies (GWASs) can overcome these issues and thereby provide more robust candidates for both gene discovery and G × E research [8, 11]. Regarding GWAS literature, two large studies identified a number of novel, sex-stratified susceptibility loci for antisocial behavior and antisocial personality disorder recently [12, 13]. Moreover, a G × E interaction was suggested between one of these loci and childhood familial difficulties in males within the general population [13].
So far, one of the most studied G × E interactions in relation to antisocial behavior involves a 30 bp length polymorphic region (LPR) in the monoamine oxidase A (MAOA) gene and exposure to childhood maltreatment. The LPR affects the functionality of the MAOA enzyme resulting in alleles with lower (MAOA-L) and higher (MAOA-H) activity [5, 6, 14]. Since the MAOA gene is located on the X chromosome, males have only one copy of the gene, whereas females have two, supporting sex-stratified analyses. Indeed, meta-analytic evidence has suggested that males with the MAOA-L genotype were more susceptible to effects of maltreatment than males with MAOA-H, while females with the MAOA-H genotype appeared to be more susceptible to maltreatment effects, albeit weakly and less consistent than in males [15]. Similar sex-stratified G × E interaction patterns in relation to antisocial behavior have been reported for MAOA × maternal smoking during pregnancy (i.e., males with MAOA-L were more susceptible to effects of smoking than males with MAOA-H, whereas females with MAOA-H were more susceptible to effects of smoking than females with MAOA-L) [16]. Thus, given location on the X chromosome and (meta-analytic) implication of different functional alleles, more sex-stratified research is needed, taking into account limitations in candidate G × E research.
Regarding sex differences related to antisocial behavior, higher rates of antisocial behavior and crime have been reported in males compared to females [17]. Furthermore, males appear to be over-represented in clinical samples [1]. Considering these observations, sex-stratified investigation of potential risk factors is very much needed. Further reasons for conducting sex-stratified analyses include recent GWAS results pointing to different susceptibility loci for antisocial behavior in males and females [12, 13].
Moreover, another important yet frequently overlooked limitation of a substantial part of the G × E literature arises from a lack of covariate interaction modelling in the G × E analyses [18]. Modelling covariate interactions is important, because both the genetic and environmental factor of interest might be moderated by control variables and the G × E interaction should be adjusted accordingly. Another point of consideration is the inconsistent control for the highly comorbid attention-deficit/hyperactivity disorder (ADHD), which may actually drive part of the associations reported with antisocial behavior [1, 19]. Similarly, further improvements could also be made by adjusting for frequently comorbid internalizing problems [1]. Furthermore, gene–environment correlations (i.e., genetic confounding of the environment) should also be taken into account as a potential driving force behind apparent G × E interactions [20].
In this study, we aimed to address the aforementioned issues concerning the existing G × E literature and investigated G × E interactions in relation to childhood antisocial behavior in the well-powered Avon Longitudinal Study of Parents and Children (ALSPAC). We focused on two key environmental risk factors for antisocial behavior, namely maternal smoking during pregnancy and childhood maltreatment [7, 15, 16, 21], in the interplay with recently identified genetic variants from GWASs of antisocial behavior, while controlling for potential confounding by comorbid ADHD and addressing specific statistical concerns. Furthermore, we aimed to replicate previously reported G × Es for the much studied MAOA candidate gene.
Methods
The ALSPAC sample
ALSPAC is an ongoing, prospective, longitudinal birth cohort, which initially recruited 14,541 pregnant women in Avon, UK with expected delivery dates from April 1991 to December 1992 and their subsequently born children. At the time of recruitment, mothers were between age 16 and 45 and represented about 85% of pregnant women in the catchment area. When children reached the age of 7, the initial sample was enriched with eligible cases who had failed to join the study initially. This resulted in an enrollment of 713 additional children. Longitudinally collected data comprise a wide range of phenotypic and environmental measures, as well as biological samples and (epi)genetic data. Further details regarding recruitment, study design, and generalizability have been reported elsewhere [22–24]. Ethical approval for the ALSPAC study was obtained from the ALSPAC Ethics and Law Committee as well as the Local Research Ethics Committees. Details on the ethics committee’s and institutional review boards that approved the study can be found at http://www.bristol.ac.uk/alspac/researchers/research-ethics/. For the present study, we included subjects with data on smoking during pregnancy, maltreatment, childhood antisocial behavior, and genotype data for a specific set of candidate polymorphisms. We only included subjects with a Caucasian ethnicity.
Main outcome: childhood antisocial behavior
As our main outcome, we used childhood antisocial behavior as measured by mother-rated CD symptom scores. Assessments were carried out using the Development and Well-Being Assessment (DAWBA) [25] at the age of 7 years and 9 months. The DAWBA is a psychiatric-diagnostic interview with the parents assessing psychopathology in children and adolescents with good validity [25]. Individual symptoms, derived from the Diagnostic and Statistical Manual of Mental Disorders (DSM) version IV [26], were rated on a three-point scale (0–2). Possible CD symptom scores ranged from 0 to 14. To reduce excess variance and avoid low cell counts given the skewed score distribution, we recoded 32 males with a score > 4 as 4, and 35 females with a score > 3 as 3. Supplementary Table S3 provides frequency tables of childhood antisocial behavior scores.
Genotypes
Details regarding genotyping quality control procedures in the ALSPAC sample are described elsewhere [27]. In summary, genotyping of single-nucleotide polymorphisms (SNPs) was carried out by the Wellcome Trust Sanger Institute, Cambridge, UK, and the Laboratory Corporation of America, Burlington, NC, US, using the Illumina HumanHap550 beadchip array and subsequent quality control filters regarding SNP call rate (0.95), subject call rate (0.97), evidence of Hardy–Weinberg violation (P cut-off 5.00E − 07), minor allele frequency (0.01), and autosomal heterozygosity (outliers were removed). Furthermore, imputation was performed with Impute2 v2.2.2 software, using the 1000 Genomes phase 1 (version 3) reference panel, and subsequent filtering based on Impute Information scores (only SNPs with Info > 0.8 were retained). Our data set obtained from the ALSPAC study included genotype data for 8941 children. Using the PLINK software [28, 29], we extracted four top SNPs from two GWASs. That is, from study one [13], we included rs4714329 (chromosome 6:40273457, G/A, G effect allele) and rs9471290 (chromosome 6:40260515, A/G, A effect allele), which appeared to be mainly a male-driven signal. From study two [12], we included rs2764450 (chromosome 1:180242092, T/C, T effect allele) and rs11215217 (chromosome 11:114689701, T/C, T effect allele), which were identified in female-only analyses. Given, sex-stratified implication, we performed sex-stratified G × E analyses in our sample (i.e., investigating rs4714329 and rs9471290 in males, and rs2764450 and rs11215217 in females). Supplementary Table S1 lists genotype statistics for the included SNPs. Since rs4714329 and rs9471290 were both located within the chromosome 6p21.2 region, we used the web-based application suite LD link (available at https://analysistools.nci.nih.gov/LDlink/) to obtain an estimate of linkage disequilibrium in the European populations. Furthermore, the male-only SNP G × E’s were initially contrasted as an additive model (0/1/2 effect alleles coded as 0/1/2). However, since the data indicated a recessive model (0/1/2 effect alleles coded as 0/0/1) as a better fit, this model was used to contrast the genotypes for the male-only SNPs instead. The extra tests conducted were addressed in the multiple testing corrections applied (see Sect. “Statistical analyses”). Results for male-only SNP G × Es tested as an additive model are provided in Supplementary Table S2. For the female-only SNPs, the cell counts for the T-allele homozygotes were very small (please see Supplementary Table S1 for genotype statistics); therefore, we contrasted these SNPs as a heterozygote model without the T-allele homozygotes.
In addition to GWAS-implicated SNPs, we also investigated a 30 bp length polymorphic region (LPR) near the promoter region of the monoamine oxidase A (MAOA) gene. Therefore, in addition to SNP data, we investigated LPR-genotype data for MAOA, which was available for 9467 subjects. Variants with 2, 3, and 5 repeats were coded as low-activity alleles (MAOA-L), whereas variants with 3.5 and 4 repeats were coded as high-activity alleles (MAOA-H) [30, 31]. As mentioned, MAOA is located on the X chromosome, and therefore, males are hemizygous, while females have two gene copies. As the MAOA-LPR has been implicated in both males and females [15], we investigated its effect in both sexes. Because the extent of X inactivation at the MAOA locus is, however, unclear [32, 33], there remains some debate whether or not to include females with both low- and high-activity alleles [15]. Therefore, we contrasted the MAOA-LPR for males as a hemizygous model for the low-activity allele (MAOA-L) and for females as an additive model for the number of high-activity alleles (MAOA-H) [15].
To investigate population stratification, we merged our genome-wide SNP data with the 1000 Genomes phase 1 reference data set [34], which contains data from 14 different global populations. We then analyzed whether the genetic principal components showed the evidence of population structure by mapping our subjects onto the known populations of the 1000 Genomes data set. In addition, any subjects in our data set scoring less than − 2 or more than + 2 standard deviations on any of the first ten principal components (using only European reference populations from the 1000 Genomes data set) were excluded from the analyses.
Environmental adversities
Maternal smoking during pregnancy was assessed by maternal self-report questionnaires at 18 weeks gestation and was defined as any versus no maternal tobacco smoking during pregnancy. This included the use of cigarettes, cigars, pipes, and other forms of tobacco smoking. Childhood maltreatment consisted of mother reported assessments at multiple time points between birth and 7;9 years of physical (available at 1;6, 1;9, 2;6, 2;9, 3;6, 4;9, 5;1, 5;9, 6;1, 6;9 years), sexual (available at 1;6, 2;6, 3;6, 4;9, 5;9, 6;9 years), or emotional abuse (available at 0;8, 1;9, 2;9, 5;1, 6;1 years) and maladaptive parenting (available at 1;6, 1;9, 2;6, 2;9, 3;6, 3;11, 4;9, 5;9, 6;1, 6;6, 6;9, 7;1 years), the last of which was defined as hitting of, shouting at, or a hostile attitude towards the child. If, at any time point, any type of abuse occurred and affected the child ‘much’ or ‘moderate’, abuse was coded as being present. Hitting and shouting were coded as present at a given time point when they were reported as occurring ‘often’ or ‘sometimes’. Hostility was coded as present at a given time point when the mother responded positively to ‘being often irritated by the child’, ‘having battles of will with the child’ or ‘the child gets on the nerves of mother’. If both at preschool (any time point between 0 and 5 years) and school age (any time point between 5 and 7;9 years), hitting, shouting, or hostility was reported; maladaptive parenting was coded as being present. When abuse, maladaptive parenting, or both were present, maltreatment was coded as present. Otherwise, maltreatment was coded as absent. Definition of maltreatment was considered broadly across multiple time points to obtain a global measure covering the childhood period up to 7;9 years, similar to the construct used by the study of Lereya et al. [35]. Please note that the ALSPAC website contains the details of all the data that are available through a fully searchable data dictionary and variable search tool (http://www.bristol.ac.uk/alspac/researchers/our-data).
Statistical analyses
Calculation of genotype statistics and principal component analyses was done using the PLINK software [28, 29]. Our main analyses were modelled in R [36], using the regression implementation from the ‘MASS’ package [37]. Given the positively skewed and over dispersed outcome data, we used negative binomial regression [38, 39]. Negative binomial regression uses a log-link function, and when regression coefficients are exponentiated, an incidence rate ratio (IRR) is obtained. The IRR gives the ratio of antisocial behavior scores between subjects with the predictor (i.e., risk genotype and risk environment) present versus absent. For example, an IRR of 1.50 indicates that at-risk subjects are predicted to have a 50% higher antisocial behavior score than other subjects. To control the family-wise error rate for multiple comparisons, we divided the nominal significance threshold by the number of tests carried out (for males, we carried out 10 tests, and for females, we carried out 6 tests, totaling 16 tests), resulting in a corrected alpha of 0.05/16 ≈ 0.0031.
Given that our outcome data were measured at the same age for all subjects and we considered sex-stratified G × E models, there was no need to include age or sex as control variables. As preliminary analyses indicated an association with childhood antisocial behavior in our sample, we included the following control variables: presence of a low socioeconomic status (which was determined as the lowest two social classes based on the SOC2000 classification [40]), maternal single-parent status, comorbid ADHD symptom scores (assessment using the DAWBA, possible score ranges 0–36, rated by the mother at age 7;9 years), and comorbid emotional problems (assessment using the Strengths and Difficulties Questionnaire [41], emotional problems subscale, possible score ranges 0–10, rated by mother at age 6;9 years). In addition, we included the first ten genetic principal components. In addition to control variable main effects, control variable interaction terms with both the genetic and environmental factor were included in each G × E model to more robustly control for confounding effects [18]. Furthermore, as G × E interactions may be confounded by gene–environment correlations, we also investigated correlations between our genetic variants and environmental factors. Sensitivity analyses were conducted to investigate potential confounding effects by comorbid ADHD symptoms and comorbid emotional problems.
Results
Descriptive statistics
Table 1 provides sex-stratified descriptive and summary statistics for our sample. Supplementary Table S1 provides the minor allele frequency, Hardy–Weinberg Equilibrium test, call rate, and all genotype frequencies for the included SNPs. Linkage disequilibrium was estimated to be moderate between the two SNPs on chromosome 6 (rs4714329 and rs9471290; D′ = 0.77 and r2 = 0.451). Results from the principal component analysis showed that the ALSPAC subjects constituted a homogeneous sample, both in relation to global and European populations (Fig. 1a, b). Within Europe, ALSPAC appeared to be most proximal to the British and Centre d’Etude du Polymorphisme Humain (CEPH) subclusters of individuals from the 1000 Genomes data set (Fig. 1b).
Table 1.
Descriptive and summary statistics
| Males (N = 2547 max.) N (%) or mean ± SD | Females (N = 2395 max.) N (%) or mean ± SD | |
|---|---|---|
| Age at outcome 7;9 years | 2547 (100%) | 2395 (100%) |
| Caucasian ethnicity (self-report) | 2547 (100%) | 2395 (100%) |
| Childhood antisocial behavior score# | 0.60 ± 1.10 (range 0–10) | 0.46 ± 0.87 (range 0–8) |
| Childhood ADHD score# | 5.83 ± 7.37 (range 0–36) | 3.45 ± 5.32 (range 0–35) |
| Childhood emotional problems score | 1.41 ± 1.64 (range 0–9) | 1.53 ± 1.66 (range 0–10) |
| Smoking during pregnancy | 428 (16.80%) | 394 (16.46%) |
| Maltreatment# | 892 (62.33%) | 712 (54.85%) |
| MAOA-L (males)/HL (females) | 765 (34.23%) | 969 (46.36%) |
| MAOA-HH (females) | 858 (41.05%) | |
| rs4714329 GG | 398 (15.63%) | – |
| rs9471290 AA | 321 (12.60%) | – |
| rs2764450 TC | – | 280 (11.75%) |
| rs11215217 TC | – | 292 (12.32%) |
| Low socioeconomic status | 417 (16.37%) | 375 (15.66%) |
| Single-parent status | 81 (3.18%) | 88 (3.67%) |
MAOA monoamine oxidase A, MAOA-L/H MAOA low-/high-activity allele, ADHD attention-deficit/hyperactivity disorder
#Significant difference between males and females, α = 0.0071 for comparing seven variables (antisocial score, ADHD score, emotional problems score, smoking during pregnancy, maltreatment, low socioeconomic status, and single-parent status) between males and females
Fig. 1.

Genetic principal component analysis. Scatterplots of the first two genetic principal components of ALSPAC merged with the 1000 Genomes phase 1 data set. a Clustering of superpopulations from different continents, whereas b shows clustering of populations within Europe. PC1 principal component (1), PC2 principal component (2), SD standard deviation, CEPH Centre d’Etude du Polymorphisme Humain
Gene–environment correlations
No gene–environment correlations were observed between any of the polymorphisms and the environmental factors (see Supplementary Table S4 for results).
G × E interactions in relation to childhood antisocial behavior in males
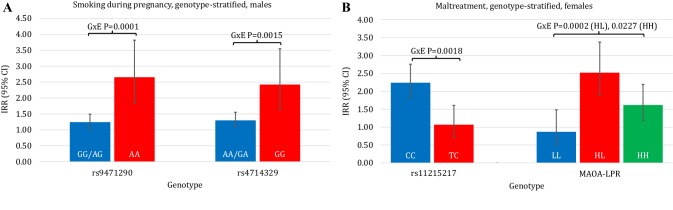
See Table 2 and Fig. 2 for all results (the final number of included subjects for each analysis is also provided in Table 2). Main effects were observed for both smoking during pregnancy [P < 0.0001 (N = 2547)] and childhood maltreatment [P < 0.0001 (N = 1431)], yet none of the genetic polymorphisms showed a main effect. For rs4714329 and rs9471290, effects of smoking during pregnancy were strongest in G- and A-allele homozygotes [significant positive G × E interactions P = 0.0015 (N = 2547) and 0.0001 (N = 2547), respectively]. No interactions between these SNPs and maltreatment were found. For MAOA-LPR, no G × E interactions were seen with smoking during pregnancy or maltreatment. Table S5 provides sex- and genotype-stratified environmental main effects in relation to childhood antisocial behavior.
Table 2.
G × E interactions in relation to childhood antisocial behavior in males and females
| Contrast | Males | Females | ||||
|---|---|---|---|---|---|---|
| N | IRR (95% CI) | P | N | IRR (95% CI) | P | |
| Smoking during pregnancy | 2547 | 1.43 (1.22–1.68) | < 0.0001* | 2394 | 1.78 (1.51–2.09) | < 0.0001* |
| Maltreatment | 1431 | 1.97 (1.65–2.35) | < 0.0001* | 1298 | 1.99 (1.66–2.40) | < 0.0001* |
| MAOA-L (males)/HL (females) | 2235 | 1.01 (0.87–1.17) | 0.91 | 2090 | 1.03 (0.81–1.31) | 0.80 |
| MAOA-HH (females) | 1.08 (0.85–1.37) | 0.54 | ||||
| rs4714329 GG | 2547 | 0.95 (0.79–1.15) | 0.63 | |||
| rs9471290 AA | 2547 | 1.16 (0.96–1.41) | 0.13 | |||
| rs2764450 TC | 2383 | 1.07 (0.86–1.33) | 0.56 | |||
| rs11215217 TC | 2370 | 0.97 (0.80–1.18) | 0.76 | |||
| MAOA-L (males)/HL (females) × smoking during pregnancy | 2235 | 1.00 (0.70–1.43) | 0.99 | 2089 | 1.15 (0.63–2.10) | 0.64 |
| MAOA-HH (females) × smoking during pregnancy | 1.23 (0.68–2.25) | 0.49 | ||||
| MAOA-L (males)/HL (females) × maltreatment | 1266 | 1.35 (0.90–2.03) | 0.15 | 1135 | 3.27 (1.74–6.14) | 0.0002* |
| MAOA-HH (females) × maltreatment | 2.09 (1.11–3.93) | 0.0227 | ||||
| rs4714329 GG × smoking during pregnancy | 2547 | 1.95 (1.29–2.94) | 0.0015* | |||
| rs4714329 GG × maltreatment | 1431 | 0.76 (0.47–1.25) | 0.28 | |||
| rs9471290 AA × smoking during pregnancy | 2547 | 2.18 (1.47–3.24) | 0.0001* | |||
| rs9471290 AA × maltreatment | 1431 | 1.28 (0.71–2.31) | 0.42 | |||
| rs2764450 TC × smoking during pregnancy | 2382 | 1.22 (0.75–1.97) | 0.43 | |||
| rs2764450 TC × maltreatment | 1291 | 1.61 (0.89–2.91) | 0.11 | |||
| rs11215217 TC × smoking during pregnancy | 2369 | 0.69 (0.40–1.19) | 0.18 | |||
| rs11215217 TC × maltreatment | 1285 | 0.47 (0.29–0.75) | 0.0018* | |||
All analyses were adjusted for socioeconomic status, single-parent status, and the first ten genetic principal components, including covariate interaction terms for the G × E models
Significance values are in bold (P < 0.05)
G × E gene-by-environment interaction, IRR incidence rate ratio, MAOA monoamine oxidase A, MAOA-L/H MAOA low-/high-activity allele
*Significant (i.e., corrected for multiple hypotheses) at α = 0.0031
Fig. 2.
Sex- and genotype-stratified effects of environmental adversities in relation to childhood antisocial behavior. Genotype-moderated effects of smoking during pregnancy in males (a) and maltreatment in females (b). IRR incidence rate ratio, MAOA monoamine oxidase A, LPR length polymorphic region, MAOA-L/H MAOA low-/high-activity allele, Error bars show 95% confidence intervals for the IRR. Gene–environment (G × E) interaction P values are shown on top
G × E interactions in relation to childhood antisocial behavior in females
See Table 2 and Fig. 2 for all results (the final number of included subjects for each analysis is also provided in Table 2). Main effects were observed for both smoking during pregnancy [P < 0.0001 (N = 2394)] and childhood maltreatment [P < 0.0001 (N = 1298)], yet none of the genetic polymorphisms showed a main effect. For rs11215217, we found that the effects of maltreatment were less strong in TC heterozygotes [a significant negative G × E interaction P = 0.0018 (N = 1285)]. No interaction with smoking during pregnancy was observed. For rs2764450, no G × E interactions with smoking during pregnancy or maltreatment were observed. Regarding MAOA-LPR, no G × E interaction was seen with smoking during pregnancy, whereas subjects with both high- and low-activity alleles appeared to be most susceptible to effects of maltreatment [a significant positive G × E interaction, P = 0.0002 (N = 1135)]. Table S5 provides sex- and genotype-stratified environmental main effects in relation to childhood antisocial behavior.
Sensitivity analyses addressing potential confounding by comorbid ADHD symptoms
See Table 3 for all results (the final number of included subjects for each analysis is also provided in Table 3). After adjusting significant G × E interactions for comorbid ADHD symptom scores, we found that the G × Es of rs4714329-GG and rs9471290-AA with smoking during pregnancy (P = 0.0014 and P = 0.0010, respectively, males), as well as the G × E between MAOA-LPR and maltreatment remained significant (P = 0.0013, females). The G × E between rs11215217-TC and maltreatment only reached nominal significance after adjustment for comorbid ADHD symptoms (P = 0.0074, females).
Table 3.
Sensitivity analyses: adjustment for comorbid ADHD symptomatology
| Contrast | N | IRR (95% CI) | P |
|---|---|---|---|
| MAOA-HL (females) × maltreatment | 1129 | 2.74 (1.48–5.08) | 0.0013* |
| MAOA-HH (females) × maltreatment | 1.98 (1.07–3.66) | 0.0298 | |
| rs4714329 GG (males) × smoking during pregnancy | 2529 | 1.85 (1.27–2.70) | 0.0014* |
| rs9471290 AA (males) × smoking during pregnancy | 2529 | 1.87 (1.29–2.71) | 0.0010* |
| rs11215217 TC (females) × maltreatment | 1278 | 0.53 (0.33–0.84) | 0.0074 |
All analyses were adjusted for socioeconomic status, single-parent status, comorbid ADHD symptom scores, and the first ten genetic principal components, including covariate interaction terms for the G × E models. Comorbid ADHD symptom scores were dichotomized closest to the 67th percentile to reduce collinearity with the covariate interaction terms
G × E gene-by-environment interaction, IRR incidence rate ratio, MAOA monoamine oxidase A, MAOA-L/H MAOA low-/high-activity allele, ADHD attention-deficit/hyperactivity disorder
*Significant (i.e., corrected for multiple hypotheses) at α = 0.0031
Sensitivity analyses addressing potential confounding by comorbid emotional problems
See Table 4 for all results (the final number of included subjects for each analysis is also provided in Table 4). After adjusting significant G × E interactions for comorbid emotional problems scores, we found that the G × E’s of rs4714329-GG and rs9471290-AA with smoking during pregnancy (P = 0.0021 and P = 0.0013, respectively, males), as well as the G × E’s of MAOA-LPR and rs11215217-TC with maltreatment remained significant (P = 0.0006 and P = 0.0020, respectively, females).
Table 4.
Sensitivity analyses: adjustment for comorbid emotional problems
| Contrast | N | IRR (95% CI) | P |
|---|---|---|---|
| MAOA-HL (females) × maltreatment | 1095 | 2.97 (1.59–5.56) | 0.0006* |
| MAOA-HH (females) × maltreatment | 1.94 (1.02–3.67) | 0.0427 | |
| rs4714329 GG (males) × smoking during pregnancy | 2358 | 1.92 (1.27–2.91) | 0.0021* |
| rs9471290 AA (males) × smoking during pregnancy | 2358 | 1.97 (1.30–2.96) | 0.0013* |
| rs11215217 TC (females) × maltreatment | 1239 | 0.45 (0.27–0.74) | 0.0020* |
All analyses were adjusted for socioeconomic status, single-parent status, comorbid emotional problems scores, and the first ten genetic principal components, including covariate interaction terms for the G × E models. Comorbid emotional problem scores were dichotomized closest to the 67th percentile to reduce collinearity with the covariate interaction terms
G × E gene-by-environment interaction, IRR incidence rate ratio, MAOA monoamine oxidase A, MAOA-L/H MAOA low-/high-activity allele, ADHD attention-deficit/hyperactivity disorder
*Significant (i.e., corrected for multiple hypotheses) at α = 0.0031
Discussion
In this study, we performed sex-stratified analyses of G × E interactions in relation to childhood antisocial behavior in a large population cohort for recent GWAS-implicated SNPs and MAOA with two well-known environmental risk factors, namely maternal smoking during pregnancy and childhood maltreatment. Regarding males, our most important findings are that G-allele homozygotes of the rs4714329 SNP and A-allele homozygotes of the rs9471290 SNP appeared to be more susceptible to effects of smoking during pregnancy in relation to antisocial behavior. Regarding females, we found that heterozygotes of the rs11215217 SNP appeared to be less susceptible, and carriers of both low- and high-activity allele of the MAOA-LPR appeared to be more susceptible to effects of childhood maltreatment in relation to antisocial behavior.
In males, the related SNPs rs4714329 and rs9471290 appeared to moderate the relation between smoking during pregnancy and antisocial behavior in such a way that risk allele homozygotes appeared to be more vulnerable to effects of maternal smoking than the other genotypes. More specifically, in risk allele homozygotes, antisocial behavior scores were more than twice as high in smoking-exposed subjects compared to unexposed subjects. By using the open-access GTEx database (available at https://www.gtexportal.org/home/), the SNP rs4714329 was linked to the expression of nearby genes LINC00951 and LRFN2 in the brain [13]. LRFN2 encodes a protein suggested to be involved in neural developmental processes such as neurite outgrowth and synaptic plasticity [42]. LRFN2 is part of a larger protein class characterized by a leucine-rich repeat domain. Many leucine-rich repeats containing transmembrane proteins are thought to be involved in nervous system development and neurodevelopmental disorders [43, 44]. LRFN2 regulates the post-synaptic PSD-95 complex, and has also been implicated in erythropoiesis, working memory, and autistic features [42, 45–48]. LINC00951 is an intergenic, long non-protein coding RNA gene, which is also expressed in the brain [13]. While many of these RNAs remain to be characterized, in general, they are assumed to be involved in gene expression regulation at both epigenetic and (post) transcriptional levels as well as other processes such as genomic imprinting [49]. In addition, these RNAs may play an important role in neurodevelopmental disorders [50].
Smoking during pregnancy has been one of the more strongly associated prenatal risk factors in relation to CD [7, 51], although this may, in part, be due to genetic and/or familial confounding [20, 52, 53]. Tobacco smoke consists of a mixture of many chemicals including nicotine, carbon monoxide, polycyclic aromatic hydrocarbons, and heavy metals, all of which may affect the developing fetus by various mechanisms [54–57]. A number of recent studies investigating gene expression patterns in relation to smoking reported the gene LRRN3 among their top hits of smoking-related differentially expressed genes [58–60]. Similar to LRFN2, LRRN3 is a leucine-rich repeat domain containing transmembrane protein expressed in the brain, and suggested to play a role in the development and maintenance of the nervous system [61]. Functionally, LRRN3 has been implicated in autism, antidepressant action, and cortical thickness (alterations of which are associated with conduct and psychopathic features) [62–64]. Although the before mentioned studies of smoking did not specifically target effects of smoking during pregnancy and gene expression alterations might be reversible, the reported results suggest that smoking might exert effects on pathways that are also affected by genetic risk factors related to antisocial behavior. Conversely, G × E interplay might be expected, i.e., moderation effects among genotype and environment such as observed in the present study.
Furthermore, as mentioned before, the use of smoking during pregnancy as an exclusively and independent environmental factor has been a point of discussion. As confounding by both genetic and socio-environmental factors has been suggested [20, 52, 53], this could indicate that the observed G × E with smoking during pregnancy may at least in part be a proxy for a gene–gene interaction and/or G × E interaction with the other environmental factors. However, as we did not observe gene–environment correlations between the selected genetic variants and smoking during pregnancy and controlled our analyses for covariate interactions, we at least addressed part of these confounding issues.
Therefore, although the exact nature of the identified G × E interaction with smoking during pregnancy is not clear, both the genetic and environmental factors in this G × E may affect brain development through effects on leucine-rich repeat protein interaction networks thought to be involved in functions such as synapse and neural circuit formation, and thereby predispose offspring for antisocial behavior [43, 44]. This also implies that future studies should also take into account related neural leucine-rich repeat protein (regulatory) genes when attempting to replicate or extent present findings.
A G × E interaction between the SNP rs11215217 and childhood maltreatment was observed in relation to offspring antisocial behavior in females. The nearest gene to this SNP is a non-coding, uncharacterized RNA gene (LOC105369506). As before mentioned, multiple (regulatory) functions of non-coding RNA genes have been described and their role in neurodevelopmental disorders highlighted [49, 50]. Of note, when adjusted for comorbid ADHD symptoms, the interaction became only nominally significant, which might indicate that the effect could be partially driven by comorbid ADHD.
Furthermore, the GWAS in relation to antisocial personality disorder by Rautiainen et al. [13] suggested a male-specific interaction between the SNP rs4714329 and childhood familial difficulties (severe conflicts and/or economic difficulties) in the general population [13]. Since we did not find any (male) G × E interactions between maltreatment and rs4714329 (or the related SNP rs9471290), we conclude that this suggested interaction does not appear to extend to childhood maltreatment in relation to pediatric antisocial behavior.
In addition, while interactions between the near-promoter LPR in MAOA and childhood maltreatment in relation to antisocial behavior have been reported for both sexes previously [15], we only observed a G × E interaction in females. More specifically, we observed a disadvantage mostly for maltreatment-exposed females with both low- and high-activity alleles (showing antisocial behavior scores more than twice as high compared to unexposed females), which is slightly different from the (additive) H-allele effect suggested in a previous meta-analysis [15]. Furthermore, in males with a low-activity allele, we did not observe any interaction with maltreatment. While this null finding does not replicate previous meta-analytic results [15], the largest study in the aforementioned meta-analysis also failed to find any interaction between MAOA and stressful life events in relation to conduct problems, both in males and females [65]. This study was also conducted within ALSPAC; however, important differences with the current study include the use of childhood life event scores instead of a specific measure of maltreatment, and the use of more general behavioral questionnaire data rather than diagnostic assessments of antisocial behavior. In addition to emphasizing our null finding in males, these differences may also explain the different female G × E results compared to the current study. Regarding smoking during pregnancy, we also failed to replicate the previous G × E findings for MAOA [16] in both sexes. Therefore, to conclude, while we reported a G × E between MAOA-HL and maltreatment in females, we consider our other negative results regarding MAOA as a sign to be slightly cautious when interpreting the earlier candidate gene-based G × E studies in this area [8, 10, 18].
Strengths and limitations
Strengths of the current study have been the use of well-powered GWAS-implicated variants as novel targets for G × E research, the use of a large, ethnically homogeneous population sample with prospective measurements of smoking during pregnancy and childhood maltreatment, and more robust confounding control through modelling of covariates in interaction with both the genetic and environmental factors. Another strength has been the use of diagnostic interview data to measure childhood behavior. Moreover, we also performed adjustments for comorbid ADHD and internalizing problems, which is frequently lacking in both G × E and main effect studies. While we did not find main effects of the genetic variants (which may be due to methodological and/or clinical differences with the original studies), we did observe clear G × E interactions, which points to the importance of this field of study and implies that G × E’s (as part of the broad sense heritability model) might be able to explain part of the so-called ‘missing heritability’ [66, 67]. Of note, ALSPAC is one of the samples used in the GWAS meta-analysis of antisocial behavior by Tielbeek et al. [12]. However, since we failed to replicate the genetic main effect of the female-only SNPs implicated by that study, the meta-analytic genome-wide signals for these SNPs may be driven by the other cohorts in that study. While, on average, antisocial behavior levels were low (as expected in a population cohort), we observed relative effect sizes of moderate-to-large magnitude resulting from common genetic variants and environmental exposures, emphasizing the clinical relevance of these results.
Nevertheless, we need to acknowledge limitations of the present study. First, the use of singular genetic variants does not necessarily provide a comprehensive picture of G × E interactions as the genetic architecture of antisocial behavior is expected to be of a complex nature [6, 12]. Alternative approaches to address this issue include the use of polygenic risk scores, gene-set (for example combining all genetic variants of a specific pathway), or gene-based (i.e., combining all variants related to a gene) analyses rather than singular variants. Nevertheless, we were able to identify different genetic loci that are likely to be of relevance given their implication as GWAS top hits. Furthermore, the top SNPs identified by the Rautiainen et al. GWAS were located only about 8 Mb distance (6p21.2) from the major histocompatibility complex (MHC) region at chromosome 6. The MHC region is highly polymorphic, displays extended LD structures and numerous disease associations have been reported for this region [68]. However, as reported by Rautiainen et al., there was no LD between the identified top SNPs at 6p21.2 and SNPs showing up at the MHC region [13]. Finally, maternal self-report measures of smoking during pregnancy and maltreatment, although measured prospectively may be subject to underreporting due to social desirability bias, which may affect the accuracy of effect estimates.
Conclusions
We studied sex-stratified G × E interactions in relation to antisocial behavior in a large population cohort and found interactions between recently (GWAS-)implicated variants and well-known environmental adversities. In males, G × E interactions with smoking during pregnancy were observed, which may be related to specific leucine-rich repeat protein networks involved in neurodevelopment. In females, G × E interactions with childhood maltreatment were found for one GWAS top SNP and MAOA. We were, however, unable to replicate other previously reported G × E interactions involving the MAOA gene. We conclude on a more general level that G × E studies do, indeed, contribute valuable information about the multifactorial nature of antisocial behavior, and we support the notion that well-powered GWASs provide more robust variants for G × E studies than classical candidate genes. Future studies should, in addition to GWAS top hits, incorporate polygenic, multimarker approaches, while addressing statistical robustness and potential sex differences when studying G × E interactions related to antisocial behavior.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses. The UK Medical Research Council and Wellcome (Grant ref: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. A comprehensive list of Grants funding is available on the ALSPAC website (http://www.bristol.ac.uk/alspac/external/documents/grant-acknowledgements.pdf). Genotype data were generated by Sample Logistics and Genotyping Facilities at Wellcome Sanger Institute and LabCorp (Laboratory Corporation of America) using support from 23andMe. This publication is the work of the authors and this research is supported by the European Community’s Seventh Framework Programme (FP7/2007–2013) under Grant agreement no. 603016 (MATRICS).
Compliance with ethical standards
Conflict of interest
I. Hyun Ruisch, Pieter J. Hoekstra, and Andrea Dietrich reported no financial interests or potential conflicts of interest. Jan K. Buitelaar was a consultant to/member of advisory board of/and/or speaker for Janssen-Cilag BV, Eli Lilly, Shire, Novartis, Roche, and Servier. Jeffrey C. Glennon has, in the past 3 years, been a consultant to Boehringer Ingelheim GmbH. Neither Jan K. Buitelaar nor Jeffrey C. Glennon is employees of any of these companies, and neither are stock shareholders of any of these companies.
Contributor Information
I. Hyun Ruisch, Phone: +31 50 361 61 61, Email: i.h.ruisch@umcg.nl.
Andrea Dietrich, Email: a.dietrich@accare.nl.
Jeffrey C. Glennon, Email: j.glennon@donders.ru.nl
Jan K. Buitelaar, Email: jan.buitelaar@radboudumc.nl
Pieter J. Hoekstra, Email: p.hoekstra@accare.nl
References
- 1.Olsson M. DSM diagnosis of conduct disorder (CD)—a review. Nord J Psychiatry. 2009;63:102–112. doi: 10.1080/08039480802626939. [DOI] [PubMed] [Google Scholar]
- 2.American Psychiatric Association . Diagnostic and statistical manual of mental disorders. 5. Washington, DC: American Psychiatric Association; 2013. [Google Scholar]
- 3.Black D. The natural history of antisocial personality disorder. Can J Psychiatr. 2015;60:309–314. doi: 10.1177/070674371506000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robins L. Epidemiology of antisocial personality disorder. In: Michels R, Cavenar JO, editors. Psychiatry. Philadelphia: Lippincott; 1987. pp. 1–14. [Google Scholar]
- 5.Waltes R, Chiocchetti AG, Freitag CM. The neurobiological basis of human aggression: a review on genetic and epigenetic mechanisms. Am J Med Genet Part B Neuropsychiatr Genet. 2016;171:650–675. doi: 10.1002/ajmg.b.32388. [DOI] [PubMed] [Google Scholar]
- 6.Holz NE, Zohsel K, Laucht M, et al. Gene×environment interactions in conduct disorder: implications for future treatments. Neurosci Biobehav Rev. 2016 doi: 10.1016/j.neubiorev.2016.08.017. [DOI] [PubMed] [Google Scholar]
- 7.Ruisch IH, Dietrich A, Glennon JC, et al. Maternal substance use during pregnancy and offspring conduct problems: a meta-analysis. Neurosci Biobehav Rev. 2018;84:325–336. doi: 10.1016/j.neubiorev.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Dick DM, Agrawal A, Keller MC, et al. Candidate gene–environment interaction research. Perspect Psychol Sci. 2015;10:37–59. doi: 10.1177/1745691614556682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vassos E, Collier DA, Fazel S. Systematic meta-analyses and field synopsis of genetic association studies of violence and aggression. Mol Psychiatry. 2014;19:471–477. doi: 10.1038/mp.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kendler KS. What psychiatric genetics has taught us about the nature of psychiatric illness and what is left to learn. Mol Psychiatry. 2013;18:1058–1066. doi: 10.1038/mp.2013.50. [DOI] [PubMed] [Google Scholar]
- 11.Psychiatric GWAS Consortium Steering Committee A framework for interpreting genome-wide association studies of psychiatric disorders. Mol Psychiatry. 2009;14:10–17. doi: 10.1038/mp.2008.126. [DOI] [PubMed] [Google Scholar]
- 12.Tielbeek JJ, Johansson A, Polderman TJC, et al. Genome-wide association studies of a broad spectrum of antisocial behavior. JAMA Psychiatry. 2017 doi: 10.1001/jamapsychiatry.2017.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rautiainen M, Paunio T, Repo-Tiihonen E, et al. Genome-wide association study of antisocial personality disorder. Transl Psychiatry. 2016;6:e883. doi: 10.1038/tp.2016.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buckholtz JW, Meyer-Lindenberg A. MAOA and the neurogenetic architecture of human aggression. Trends Neurosci. 2008;31:120–129. doi: 10.1016/j.tins.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 15.Byrd AL, Manuck SB. MAOA, childhood maltreatment, and antisocial behavior: meta-analysis of a gene–environment interaction. Biol Psychiatry. 2014;75:9–17. doi: 10.1016/j.biopsych.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wakschlag LS, Kistner EO, Pine DS, et al. Interaction of prenatal exposure to cigarettes and MAOA genotype in pathways to youth antisocial behavior. Mol Psychiatry. 2010;15:928–937. doi: 10.1038/mp.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stephenson Z, Woodhams J, Cooke C. Sex differences in predictors of violent and non-violent juvenile offending. Aggress Behav. 2014;40:165–177. doi: 10.1002/ab.21506. [DOI] [PubMed] [Google Scholar]
- 18.Keller M. Gene-by-environment interaction studies have not properly controlled for potential confounders: the problem and the (simple) solution. Biol Psychiatry. 2014;75:18–24. doi: 10.1016/j.biopsych.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mannuzza S, Klein RG, Abikoff H, Moulton JL. Significance of childhood conduct problems to later development of conduct disorder among children with ADHD: a prospective follow-up study. J Abnorm Child Psychol. 2004;32:565–573. doi: 10.1023/B:JACP.0000037784.80885.1a. [DOI] [PubMed] [Google Scholar]
- 20.Rice F, Harold GT, Boivin J, et al. Disentangling prenatal and inherited influences in humans with an experimental design. Proc Natl Acad Sci USA. 2009;106:2464–2467. doi: 10.1073/pnas.0808798106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keiley MK, Howe TR, Dodge K, et al. The timing of child physical maltreatment: a cross-domain growth analysis of impact on adolescent externalizing and internalizing problems. Dev Psychopathol. 2001;13:891–912. [PMC free article] [PubMed] [Google Scholar]
- 22.Boyd A, Golding J, Macleod J, et al. Cohort profile: the’children of the 90s’—the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2013;42:111–127. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fraser A, Macdonald-wallis C, Tilling K, et al. Cohort profile: the avon longitudinal study of parents and children: ALSPAC mothers cohort. Int J Epidemiol. 2013;42:97–110. doi: 10.1093/ije/dys066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golding J, Pembrey M, Jones R, ALSPAC study team ALSPAC–the Avon Longitudinal Study of Parents and Children. I. Study methodology. Paediatr Perinat Epidemiol. 2001;15:74–87. doi: 10.1046/j.1365-3016.2001.00325.x. [DOI] [PubMed] [Google Scholar]
- 25.Goodman R, Ford T, Richards H, et al. The development and well-being assessment: description and initial validation of an integrated assessment of child and adolescent psychopathology. J Child Psychol Psychiatry Allied Discip. 2000;41:645–655. doi: 10.1111/j.1469-7610.2000.tb02345.x. [DOI] [PubMed] [Google Scholar]
- 26.American Psychiatric Association . Diagnostic and statistical manual of mental disorders. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- 27.Riglin L, Collishaw S, Richards A, et al. Schizophrenia risk alleles and neurodevelopmental outcomes in childhood: a population-based cohort study. Lancet Psychiatry. 2017;1:57–62. doi: 10.1016/S2215-0366(16)30406-0. [DOI] [PubMed] [Google Scholar]
- 28.Purcell S, Chang C (2017) PLINK software, version 1.9. https://www.cog-genomics.org/plink/1.9/
- 29.Chang C, Chow C, Telier L, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sabol S, Hu S, Hamer D. A functional polymorphism in the monoamine oxidase A gene promoter. Hum Genet. 1998;103:273–279. doi: 10.1007/s004390050816. [DOI] [PubMed] [Google Scholar]
- 31.Deckert J, Catalano M, Syagailo Y, et al. Excess of high activity monoamine oxidase A gene promoter alleles in female patients with panic disorder. Hum Mol Genet. 1999;8:621–624. doi: 10.1093/hmg/8.4.621. [DOI] [PubMed] [Google Scholar]
- 32.Benjamin D, Van Bakel I, Craig I. A novel expression based approach for assessing the inactivation status of human X-linked genes. Eur J Hum Genet. 2000;8:103–108. doi: 10.1038/sj.ejhg.5200427. [DOI] [PubMed] [Google Scholar]
- 33.Carrel L, Willard H. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 34.Auton A, Abecasis GR, Altshuler DM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lereya ST, Copeland WE, Costello EJ, Wolke D. Adult mental health consequences of peer bullying and maltreatment in childhood: two cohorts in two countries. Lancet Psychiatry. 2015;2:524–531. doi: 10.1016/S2215-0366(15)00165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.R Core Team (2018) R: a language and environment for statistical computing, version 3.4.3. R Foundation for Statistical Computing. Vienna, Austria. https://www.R-project.org
- 37.Venables W, Ripley B. Modern applied statistics with S. 4. New York: Springer; 2002. [Google Scholar]
- 38.Karazsia BT, van Dulmen MHM. Regression models for count data: illustrations using longitudinal predictors of childhood injury. J Pediatr Psychol. 2008;33:1076–1084. doi: 10.1093/jpepsy/jsn055. [DOI] [PubMed] [Google Scholar]
- 39.Zuur A, Ieno E, Walker N, et al. Mixed effects models and extensions in ecology with R. New York: Springer; 2009. Zero-truncated and zero-inflated models for count data; pp. 261–293. [Google Scholar]
- 40.U.K. Office of National Statistics (2000) Standard Occupational Classification 2000, vol 1. Structure and descriptions of unit groups. The Stationary Office, London (ISBN: 0 11 621388 4)
- 41.Goodman RPD. Psychometric properties of the strengths and difficulties questionnaire. J Am Acad Child Adolesc Psychiatry. 2001;40:1337–1345. doi: 10.1097/00004583-200111000-00015. [DOI] [PubMed] [Google Scholar]
- 42.Morimura N, Inoue T, Katayama K, Aruga J. Comparative analysis of structure, expression and PSD95-binding capacity of Lrfn, a novel family of neuronal transmembrane proteins. Gene. 2006;380:72–83. doi: 10.1016/j.gene.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 43.de Wit J, Ghosh A. Control of neural circuit formation by leucine-rich repeat proteins. Trends Neurosci. 2014;37:539–550. doi: 10.1016/j.tins.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frei JA, Stoeckli ET. SynCAMs—from axon guidance to neurodevelopmental disorders. Mol Cell Neurosci. 2017;81:41–48. doi: 10.1016/j.mcn.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 45.Castellanos A, Lang G, Frampton J, Weston K. Regulation of erythropoiesis by the neuronal transmembrane protein Lrfn2. Exp Hematol. 2007;35:724–734. doi: 10.1016/j.exphem.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 46.Thevenon J, Souchay C, Seabold GK, et al. Heterozygous deletion of the LRFN2 gene is associated with working memory deficits. Eur J Hum Genet. 2016;24:911–918. doi: 10.1038/ejhg.2015.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morimura N, Yasuda H, Yamaguchi K, et al. Autism-like behaviours and enhanced memory formation and synaptic plasticity in Lrfn2/SALM1-deficient mice. Nat Commun. 2017 doi: 10.1038/ncomms15800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Kim R, Cho YS, et al. Lrfn2-mutant mice display suppressed synaptic plasticity and inhibitory synapse development and abnormal social communication and startle response. J Neurosci. 2018 doi: 10.1523/JNEUROSCI.3321-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hung T, Chang HY. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol. 2010;7:582–585. doi: 10.4161/rna.7.5.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van deVondervoort IIGM, Gordebeke PM, Khoshab N, et al. Long non-coding RNAs in neurodevelopmental disorders. Front Mol Neurosci. 2013;6:1–9. doi: 10.3389/fnmol.2013.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruisch IH, Buitelaar JK, Glennon JC, et al. Pregnancy risk factors in relation to oppositional-defiant and conduct disorder symptoms in the Avon Longitudinal Study of Parents and Children. J Psychiatr Res. 2018;101:63–71. doi: 10.1016/j.jpsychires.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 52.D’Onofrio BM, Van Hulle CA, Waldman ID, et al. Smoking during pregnancy and offspring externalizing problems: an exploration of genetic and environmental confounds. Dev Psychopathol. 2008;20:139–164. doi: 10.1017/S0954579408000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gilman SE, Gardener H, Buka SL. Maternal smoking during pregnancy and children’s cognitive and physical development: a causal risk factor? Am J Epidemiol. 2008;168:522–531. doi: 10.1093/aje/kwn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lammer EJ, Shaw GM, Iovannisci DM, et al. Maternal smoking and the risk of orofacial clefts: susceptibility with NAT1 and NAT2 polymorphisms. Epidemiology. 2004;15:150–156. doi: 10.1097/01.ede.0000112214.33432.cc. [DOI] [PubMed] [Google Scholar]
- 55.Munger RG, Sauberlich HE, Corcoran C, et al. Maternal vitamin B-6 and folate status and risk of oral cleft birth defects in the Philippines. Birth Defects Res Part A Clin Mol Teratol. 2004;70:464–471. doi: 10.1002/bdra.20037. [DOI] [PubMed] [Google Scholar]
- 56.Ziaei S, Nouri K, Kazemnejad A. Effects of carbon monoxide air pollution in pregnancy on neonatal nucleated red blood cells. Paediatr Perinat Epidemiol. 2005;19:27–30. doi: 10.1111/j.1365-3016.2004.00619.x. [DOI] [PubMed] [Google Scholar]
- 57.Mochizuki M, Maruo T, Masuko K, Ohtsu T. Effects of smoking on fetoplacental-maternal system during pregnancy. Am J Obstet Gynecol. 1984;149:413–420. doi: 10.1016/0002-9378(84)90156-X. [DOI] [PubMed] [Google Scholar]
- 58.Vink JM, Jansen R, Brooks A, et al. Differential gene expression patterns between smokers and non-smokers: cause or consequence? Addict Biol. 2017;22:550–560. doi: 10.1111/adb.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huan T, Joehanes R, Schurmann C, et al. A whole-blood transcriptome meta-analysis identifies gene expression signatures of cigarette smoking. Hum Mol Genet. 2016;25:4611–4623. doi: 10.1093/hmg/ddw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin F, Talikka M, Hoeng J, Peitsch MC. Identification of gene expression signature for cigarette smoke exposure response—from man to mouse. Hum Exp Toxicol. 2015;34:1200–1211. doi: 10.1177/0960327115600364. [DOI] [PubMed] [Google Scholar]
- 61.Haines BP, Gupta R, Jones CM, et al. The NLRR gene family and mouse development: modified differential display PCR identifies NLRR-1 as a gene expressed in early somitic myoblasts. Dev Biol. 2005;281:145–159. doi: 10.1016/j.ydbio.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 62.Sousa I, Clark TG, Holt R, et al. Polymorphisms in leucine-rich repeat genes are associated with autism spectrum disorder susceptibility in populations of European ancestry. Mol Autism. 2010;1:1–7. doi: 10.1186/2040-2392-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsapakis EM, Fernandes C, Moran-Gates T, et al. Effects of antidepressant drug exposure on gene expression in the developing cerebral cortex. Synapse. 2014;68:209–220. doi: 10.1002/syn.21732. [DOI] [PubMed] [Google Scholar]
- 64.Kochunov P, Charlesworth J, Winkler A, et al. Transcriptomics of cortical gray matter thickness decline during normal aging. Neuroimage. 2013;82:273–283. doi: 10.1016/j.neuroimage.2013.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Enoch MA, Steer CD, Newman TK, et al. Early life stress, MAOA, and gene–environment interactions predict behavioral disinhibition in children. Genes Brain Behav. 2010;9:65–74. doi: 10.1111/j.1601-183X.2009.00535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2012;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kennedy AE, Ozbek U, Dorak MT. What has GWAS done for HLA and disease associations? Int J Immunogenet. 2017;44:195–211. doi: 10.1111/iji.12332. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.