EAEC is a remarkable etiologic agent of acute and persistent diarrhea worldwide. The isolates harbor specific subsets of virulence genes and their pathogenesis needs to be better understood. Chemical signaling via histidine kinase sensor QseC has been shown as a potential target to elucidate the orchestration of the regulatory cascade of virulence factors.
KEYWORDS: EAEC, Escherichia coli, O104:H4, QseC, Shiga toxin, VisP, chemical signaling
ABSTRACT
Enteroaggregative Escherichia coli (EAEC) from the O104:H4 specific serotype caused a large outbreak of bloody diarrhea with some complicated cases of hemolytic-uremic syndrome (HUS) in Europe in 2011. The outbreak strain consisted in an EAEC capable to produce the Shiga toxin (Stx) subtype 2a, a characteristic from enterohemorrhagic E. coli. QseBC two-component system detects AI-3/Epi/NE and mediates the chemical signaling between pathogen and mammalian host. This system coordinates a cascade of virulence genes expression in important human enteropathogens. The blocking of QseC of EAEC C227-11 (Stx+) strain by N-phenyl-4-{[(phenylamino) thioxomethyl]amino}-benzenesulfonamide (also known as LED209) in vivo demonstrated a lower efficiency of colonization. The periplasmic protein VisP, which is related to survival mechanisms in a colitis model of infection, bacterial membrane maintenance, and stress resistance, here presented high levels of expression during the initial infection within the host. Under acid stress conditions, visP expression levels were differentiated in an Stx-dependent way. Together, these results emphasize the important role of VisP and the histidine kinase sensor QseC in the C227-11 (Stx+) outbreak strain for the establishment of the infectious niche process in the C57BL/6 mouse model and of LED209 as a promising antivirulence drug strategy against these enteric pathogens.
IMPORTANCE EAEC is a remarkable etiologic agent of acute and persistent diarrhea worldwide. The isolates harbor specific subsets of virulence genes and their pathogenesis needs to be better understood. Chemical signaling via histidine kinase sensor QseC has been shown as a potential target to elucidate the orchestration of the regulatory cascade of virulence factors.
INTRODUCTION
Some strains of Escherichia coli belonging to the O104:H4 serotype may be classified as members of the enteroaggregative E. coli (EAEC) pathovar, which was first described in the mid-1980s and has been recognized as an important cause of diarrheagenic diseases in children and adults in both developed and developing countries (1–4).
In 2011, a large outbreak of foodborne bloody diarrhea and hemolytic-uremic syndrome (HUS) began in Germany and spread throughout other countries; the outbreak was triggered by EAEC serotype O104:H4 lysogenized with Shiga toxin 2a-encoding phage, a common feature related to enterohemorrhagic E. coli (EHEC) or Shiga toxin E. coli. This outbreak emerged with more than 3,800 individuals affected, including primary and secondary cases, leading to 54 reported deaths. Moreover, the HUS cases of this outbreak occurred at a much higher rate than frequently observed for EHEC (5), i.e., approximately 22% of the cases (6, 7).
Previously, E. coli strains O104:H4 (Stx+) have been reported in Europe in sporadic cases of hemorrhagic colitis and HUS (6, 8–11). However, the 2011 outbreak strain is genetically closer to the Stx– O104:H4 strain (55989), which was isolated from an HIV-infected patient in central Africa (12). EAEC was prevalent during a case-control study of children with diarrhea in Salvador-Bahia State, Brazil; among these subjects, an O104:H4 (Stx–) strain was also isolated (13), which is genetically similar to the European isolates.
Bacterial cell-cell chemical signaling may cross over the interspecific and intraspecific differences during communication, in addition to the interkingdom signaling to their respective host cells (14). The two-component system QseBC is closely related to the expression of virulence genes in Enterobacteriaceae. It detects specifically AI-3 signs and adrenergic hormones epinephrine (Epi) and norepinephrine (NE) (15). QseBC and signaling via Epi/NE/AI-3 were described in EHEC (16) and in other pathogens, such as enteropathogenic E. coli (17), Vibrio parahaemolyticus (18), Salmonella enterica serovar Typhimurium (19–22) (among other serovars [23]), Francisella tularensis (24), uropathogenic E. coli (UPEC) (25), Edwardsiella tarda (26), Haemophilus influenzae (27), Aeromonas hydrophila (28, 29), Aggregatibacter actinomycetemcomitans (30, 31), and Legionella pneumophila (32).
QseC sensor kinase in EHEC is a global regulator involved in the expression of >400 genes (33), such as those encoding motility, attaching and effacing (A/E) lesions, and Shiga toxin (33–35), although QseBC’s role in in vivo infections has never been investigated in any EAEC strain that expresses or does not express Shiga toxin.
During characterization studies of histidine kinase sensor QseC in the pathogenesis in vivo of S. enterica serovar Typhimurium, a novel periplasmic protein, VisP, was described that is associated with survival mechanisms, stress response, and the maintenance of membranes (36). VisP is important in the response to heavy metal, osmotic pressure, and acid milieu, all conditions that bacteria may encounter during gastrointestinal (GI) passage and gut colonization. In the periplasmic space of S. Typhimurium, VisP can bind to the sugars of the peptidoglycan layer and also to the internal membrane enzyme LpxO, which is present in Salmonella but absent in E. coli. Directly after binding, VisP blocks the specific modification LpxO-mediated 2-hydroxylation of lipid A (36). Recently, VisP was described as also involved during final O-antigen chain length regulation, together with the Wzz system from S. Typhimurium lipopolysaccharide (LPS) (37).
The VisP role in E. coli was described only in association with bacterial stress, which in turn is related to different pH conditions and stress agents such as hydrogen peroxide and cadmium chloride (38). In S. Typhimurium, VisP is an important player during replication within macrophages in vitro, systemic infection in mice, and response to the same stressors as in E. coli (36). Recently, VisP was described as also involved during final O-antigen chain length regulation, together with the Wzz system from S. Typhimurium LPS (37).
The bacterial stress response in the intestinal environment caused by bacterial pathogens leads to intestinal dysbiosis, which promotes an antagonism between the microbiota and the intruder bacteria in this system, resulting in competition for nutrients, the production of antibiotics, and modulation of the immune system (39, 40). Intestinal bacteria, in order to survive this competitive and stressful environment, must refine mechanisms that, like the pathogens, allow their permanence in the gut. Intestinal pathogens, such as EHEC, have a great ability to survive low stomach pH during GI passage, and their infecting dose is low (>100 CFU); conversely, these characteristics in EAEC were only briefly described (1, 41).
Based on EAEC diversity and its multifactorial properties described in the literature, we investigated further details in this outbreak strain and how the chemical signaling via QseC and auxiliary mechanisms are implicated in the pathogenesis of E. coli O104:H4. Moreover, one of the main aims of this study was to investigate the chemical signaling virulence in EAEC O104:H4 Stx+ and Stx– strains by blocking QseC sensor kinase with LED209 to investigate its role during infection (42). Using in vitro and in vivo assays, we have evaluated the QseC sensor kinase importance in the outbreak O104:H4 (Stx+) compared to another O104:H4 strain lacking Stx production (13). Our data show that QseC participates in the colonization process in an animal model of colonization for EAEC and that LED209 is a potential antivirulence compound for the EAEC O104:H4 Stx+ strain; these findings demonstrate the chemical signaling in vivo proof of principle by blocking QseC sensor kinase. During the QseC studies, compound LED209 was selected from a library of 150,000 molecules and was identified to have QseC inhibitory activity. It has been shown that the blockade of QseC by LED209 leads to the inhibition of important virulence factors in EHEC, S. Typhimurium, and Francisella tularensis (21), in addition to resulting in a higher percentage of survival among infected animals (42). Its activity in the inhibition of biofilm formation in multidrug-resistant (MDR) clinical isolates, such as Pseudomonas aeruginosa, Klebsiella pneumoniae, UPEC, and EAEC, was also reported.
Based on this finding, we sought to assess the chemical signaling virulence in EAEC O104:H4 Stx+ and Stx– strains by blocking the QseC sensor kinase with LED209 to investigate its role during infection. This distinct approach may be considered promising in controlling the spread of pathogenic MDR microorganisms, since LED209 does not interfere with bacterial growth and consequently does not exert strong selective pressure on resistant strains. In addition, it only acts on the bacterial signal transduction pathway, does not demonstrate activity in eukaryotic cell receptors, and is not toxic in animal models. Therefore, LED209 opens perspectives for its possible clinical use in the future.
RESULTS
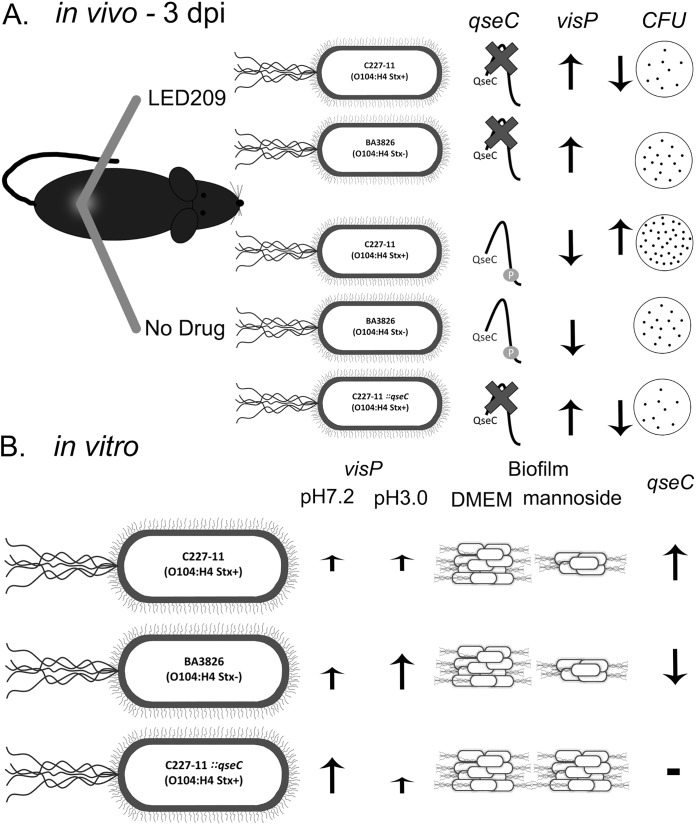
Chemical signaling via QseC plays an important role during O104:H4 infection.
To further investigate the importance of the QseC sensor kinase in O104:H4 infection in vivo, mice were orally infected with 1010 CFU doses and monitored for 14 days. During in vivo assays, we evaluated mice infected with wild-type (WT) C227-11 (Stx+), ::qseC, and BA3826 (Stx–) strains in the presence or absence of LED209. Specifically, we used a C57BL/6J model, described previously for the study of EAEC intestinal colonization (43). Here, we assessed direct intestinal colonization by using CFU counts in feces to provide a numeric readout of the EAEC infection, and mouse weight loss was evaluated to investigate the effect during host regular development to correlate the infection with the disease severity (see Table S1 [https://www2.fcfar.unesp.br/#!/pos-graduacao/biociencias-e-biotecnologias-aplicadas-a-farmacia/docentes2308/corpo-docente/]). The qseC mutant presented lower colonization levels compared to both LED209-treated and nontreated groups; moreover, a remarkable reduction was observed during colonization at day 5 postinfection (p.i.). Clearly, treatment with LED209 led to a more evident effect during the initial days of the infection, especially between days 3 and 5 p.i., that seems to be critical for colonization and qseC expression. The C227-11 (Stx+) strain CFU recovery was highly reduced in LED209 throughout the 14-day p.i. period (Fig. 1A). This was consistent with the observed mouse weight loss for C227-11 mice in the absence of LED209, which points out the clear effect of QseC in the Shiga toxin-producing strain (see Table S1). There was no relevant difference in CFU recovered from feces of the BA3826 (Stx–)-infected mice, even though a reduction trend could be observed in the presence of LED209 (Fig. 1B). LED209 treatment reduced the number of bacteria, indicating that inhibition of QseC prevents high colonization by the C227-11 (Stx+) strain.
FIG 1.
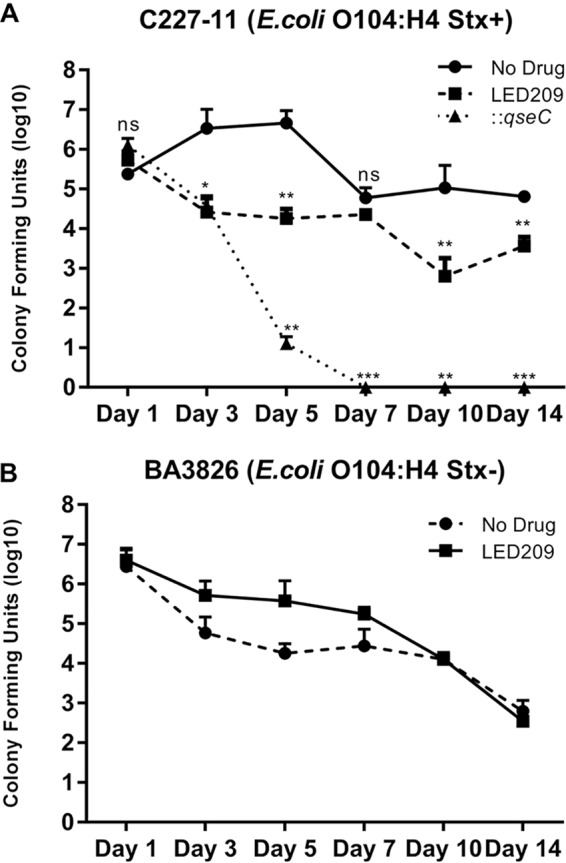
Mouse colonization with 1010 CFU of O104:H4 strains after treatment with LED209. The CFU counts for WT and ::qseC (C227-11) strains (A) or the BA3826 strain (B) recovered days 1 to 14 after the administration of 58.8 mg/kg of LED209 or vehicle were determined. A Student t test was used to determine the statistical significance between treated and nontreated groups. Error bars indicate the SD of the mean (*, P < 0.01; **, P < 0.001; ***, P < 0.0001; ns, not significant).
QseC sensor kinase is critical during the initial GI passage in vivo.
The expression levels were measured via quantitative reverse transcription-PCR (qRT-PCR) to verify qseC gene expression in vitro and during in vivo GI passage based on bacterial RNA extracted direct from feces collected during the mouse assay. We directly assessed here qseC expression during mouse infection under Shiga toxin influence in the C227-11 strain, employing Stx+ and Stx– strains to evaluate QseC possible function in the process. Furthermore, the LED209-treated group displayed significant repression of the qseC expression at the initial days 1 and 3 p.i. of 1.6- and 1.4-fold for C227-11 (Stx+) and 2.0- and 2.4-fold for BA3826 (Stx–) strain in both cases, as expected since LED209 is a QseC blocker, and after 10 days p.i. the compound was degraded, and the differences are surpassed or not effective anymore (Fig. 2A and B). In vitro, the qseC expression levels were very prominent in the C227-11 (Stx+) strain, similar to EHEC 86-24 strain levels, in contrast to the BA3826 (Stx–) strain and the other EAEC prototype strains 042 and 17-2 (Fig. 2C).
FIG 2.
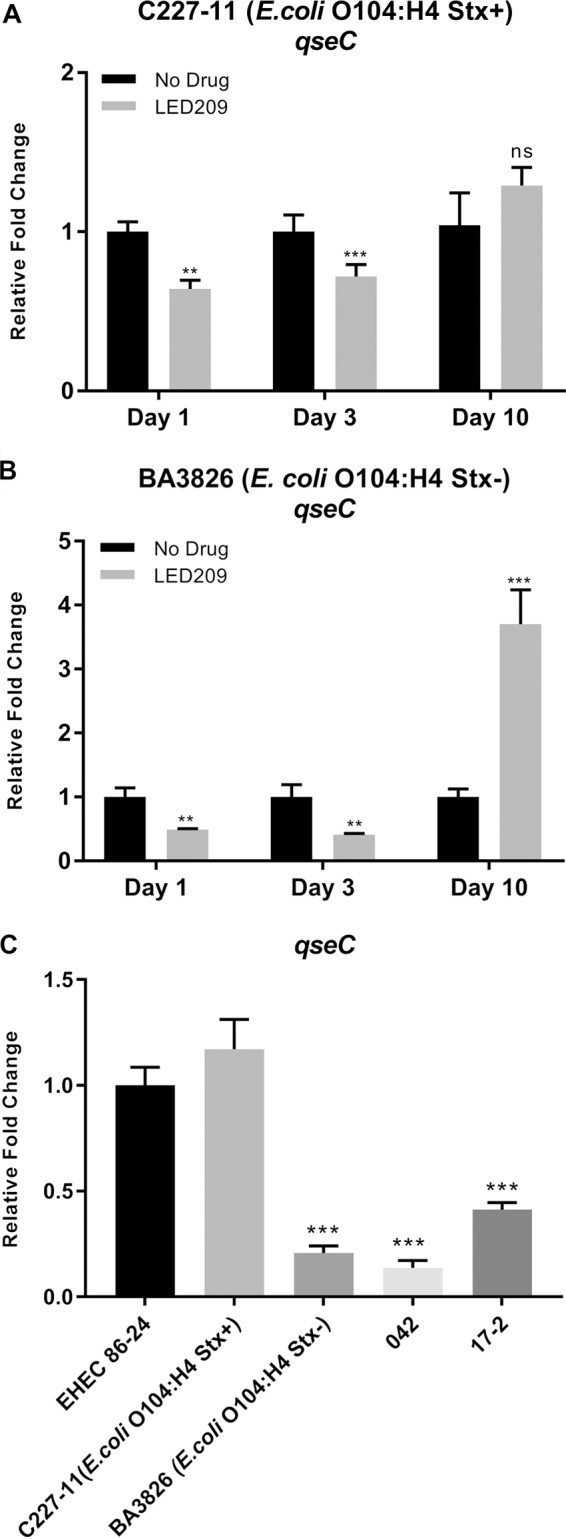
LED209 blocking of QseC during in vivo infection decreases the expression levels of qseC gene in O104:H4 Stx+ and Stx– strains. (A and B) qRT-PCR analyses of qseC from feces collected from days 1 to 10 p.i. of both C227-11 and BA3826 strains. (C) qRT-PCR of qseC with RNA extracted from cultures of EHEC 86-42, C227-11 (Stx+), BA3826 (Stx–), 17-2, and 042 strains grown in LB medium until reaching an OD600 of 1.0. A Student t test was used to determine the statistical significance between treated and nontreated groups, or ANOVA was used for comparisons to EHEC 86-24. Error bars indicate the SD of the mean (**, P < 0.001; ***, P < 0.0001; ns, not significant).
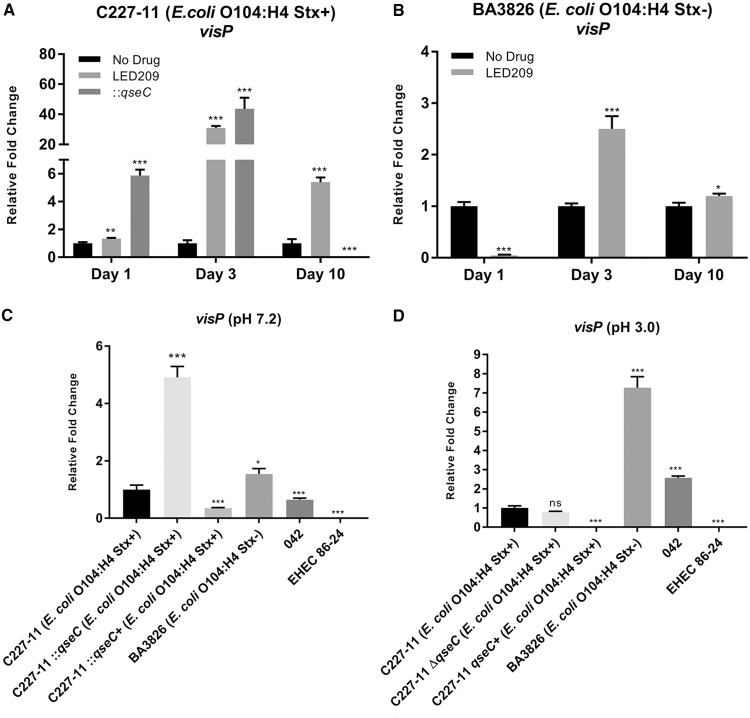
Blocking QseC shows VisP to be an important survival mechanism during mouse infection.
In addition, the QseBC two-component system in the bacterial inner membrane, the VisP described in S. Typhimurium, participates in this process. Its role is linked to the two-component system in response to stress, general survival mechanisms, lipid A modifications, and membrane maintenance (36, 37). In EHEC, VisP seems to be important in biofilm formation in polystyrene (36). Here, we evaluated the expression levels of the visP gene during the course of infection, and a remarkable upregulation was observed at day 3 p.i. in both WT and ::qseC C227-11 (Stx+) strains of 30.9- and 43.7-fold, respectively. However, the BA3826 (Stx–) strain in the same time period showed only a 2.5-fold difference. Conversely, an increase in visP expression levels was observed in the WT and ::qseC C227-11 (Stx+) strains at day 1 p.i., as well as a decrease for the BA3826 (Stx–) strain. Together, these findings suggest that different stresses are driving VisP fine-tuning upon QseC blockade by LED209 treatment in these strains (Fig. 3A and B). Given the role of VisP in other bacterial pathogens, we further investigated similar stress conditions that O104:H4 usually overcomes during GI passage.
FIG 3.
Stress conditions stimulate overexpression of visP in diarrheagenic E. coli strains. (A and B) qRT-PCR of visP from feces collected from days 1 to 10 p.i. of both C227-11 (Stx+) and BA3826 (Stx–) strains after treatment with LED209. (C and D) qRT-PCR of visP from RNA collected from cultures of WT C227-11 (Stx+), ::qseC, qseC+, BA3826 (Stx–), 042 and EHEC 86-24 strains after 1 h of static incubation at 37°C in pH 7.2 (C) and 3.0 (D). A Student t test was employed to determine statistical significance between treated and nontreated groups or ANOVA comparing to WT C227-11 (Stx+) strain. Error bars indicate the SD of the mean (*, P < 0.01; **, P < 0.001; ***, P < 0.0001; ns, not significant).
VisP role during O104:H4 acid stress conditions.
The periplasmic protein VisP helps bacteria adapt to adverse conditions. Therefore, to verify visP’s importance observed in vivo, visP gene expression in vitro was analyzed in neutral (pH 7.2) and in acidic (pH 3.0) media, and both were compared to the WT C227-11 (Stx+) strain under the same conditions. In neutral pH 7.2, the ::qseC mutant strain showed a 4.9-fold increase, and the BA3826 (Stx–) strain showed a 1.5-fold increase. On the other hand, the prototype 042 and EHEC-86-24 strains were reduced 1.5- and 81.2-fold, respectively (Fig. 3C). Upon assessing the acidic pH (pH 3.0), there was no significant difference for the ::qseC strain, whereas the expression of BA3826 and 042 Stx– strains increased 7.3- and 2.6-fold, and the expression of EHEC 86-24 remained low (83.7-fold) (Fig. 3D).
Taking into consideration this adaptive feature from these EAEC strains, we next evaluated the direct effect in microbiota from mice in vivo upon EAEC infection to better understand whether EAEC plays a role in bacterial prevalence.
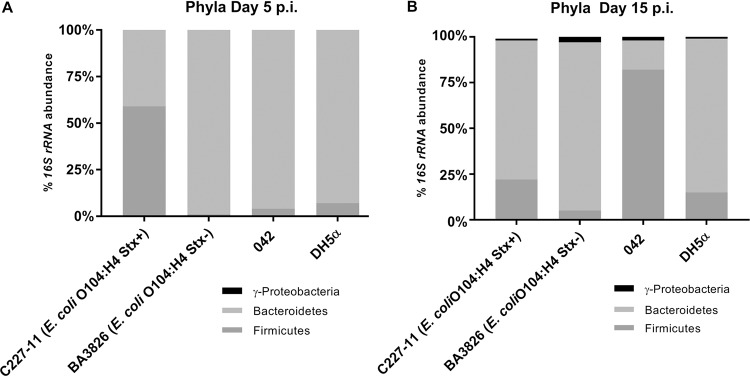
E. coli O104:H4 shifts mouse gut microbiota upon infection.
The intestinal microbiota has been reported to play an important role in resistance to infection in the GI system, and an imbalance in its composition may enhance the ability of pathogens to survive and colonize distinct niches (44). Its composition is unique at the genus and species levels for each individual. However, at higher taxonomic levels, such as phyla, it is more conserved and is predominantly constituted by Bacteroidetes and Firmicutes, followed by Proteobacteria and Actinobacteria (45–47). Therefore, to better evaluate the impact of these EAEC strains on mice with partially depleted microbiota, we analyzed the changes in composition at the phylum level at 5 and 15 days p.i. The prevalence of the Firmicutes phylum (56%) was observed on day 5 p.i. in the group infected with C227-11 (Stx+) strain, while other strains showed a much higher percentage of the Bacteroidetes phylum (Fig. 4A). However, by 15 days p.i., relevant changes occurred, affecting the abundance of the Firmicutes and the reappearance of Proteobacteria members for all strains, especially the EAEC strains. The prevalence of Firmicutes was more evident (83%) for mice infected with the 042 strain compared to other strains and phyla (Fig. 4B). These results indicate the potential intestinal EAEC recolonization as one of the main hallmarks of the infection (1, 4). Other virulence factors of these multifactorial pathogens may help to elucidate their complex regulation.
FIG 4.
Gut microbiota changes in ampicillin mouse model of infection from days 5 (A) and 15 (B) p.i. with various EAEC strains. Proteobacteria, Bacteroidetes, and Firmicutes (more common intestinal phylogenetic groups) abundance analyses by qRT-PCR of 16S rRNA from feces were performed.
Phenotypic behavior in distinct EAEC strains.
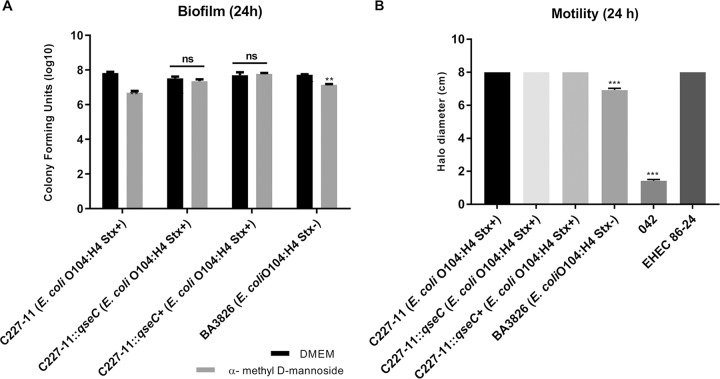
The EAEC multifactorial nature was initially tested concerning its adhesion, commonly known to adhere firmly to epithelial cells and polystyrene surfaces during biofilm formation. Type I fimbriae (TIF) are an important virulence factor in UPEC (48, 49); they participate in intracellular bacterial community formation and bladder colonization in the urinary tract infection (UTI) mouse model (50). Our group has shown that TIF absence in a mutant strain led to a significant reduction in biofilm formation and aggregative adhesion (AA) pattern in EAEC 042 (51). TIF interacts with mannoside residues present in eukaryotic cell receptors (52, 53). To better evaluate its role, we blocked TIF with nonmetabolizable α-methyl-d-mannoside compound during biofilm formation essay. The results showed a significant reduction of 0.5 order of magnitude for both WT C227-11 (Stx+) and BA3826 (Stx–) strains after TIF blocking (Fig. 5A). The adhesion ability in HeLa cells was also evaluated, but no significant difference was demonstrated for the ::qseC mutant (see Fig. S1 [https://www2.fcfar.unesp.br/#!/pos-graduacao/biociencias-e-biotecnologias-aplicadas-a-farmacia/docentes2308/corpo-docente/]).
FIG 5.
General phenotypic differences between EAEC strains during biofilm formation. (A) WT C227-11 (Stx+), ::qseC C227-11 (Stx+), qseC+ C227-11 (Stx+), and BA3826 (Stx–) strains on 96-well polystyrene plate in the presence or absence of 1% α-methyl d-mannoside incubated for 24 h at 37°C. (B) Motility profile between E. coli strains compared to WT C227-11 (Stx+) plated on LB agar 0.3% and incubated at 37°C. Growth halos were measured 24 h after incubation. Statistical analysis was performed using a Student t test between each group or ANOVA. Error bars indicate the SD of the mean (**, P < 0.001; ***, P < 0.0001; ns, not significant).
Generally, flagellum motility helps bacteria (i) to scavenge for nutrients and new colonization niches, (ii) to form biofilms, and (iii) to escape from harmful substances (54). Considered a very relevant virulence factor in several pathogenic bacteria, flagellum motility is involved in host colonization because it mediates previous contact with adhesion surface (55). In particular, type H4 flagella in UPEC are correlated with motility and epithelial cell adhesion and are prevalent in ST131 clonal type strains (56, 57). There are limited data about flagellum importance in EAEC strains.
The motility profile was identical between ::qseC and EHEC 86-24 strains compared to the WT C227-11 (Stx+) isogenic WT strain. However, strain 042 showed a reduction of 82.3%, and strain BA3826 (Stx+) showed a reduction only of 13.5% (Fig. 5B).
Are Shiga toxin and TIF involved in O104:H4 adherence?
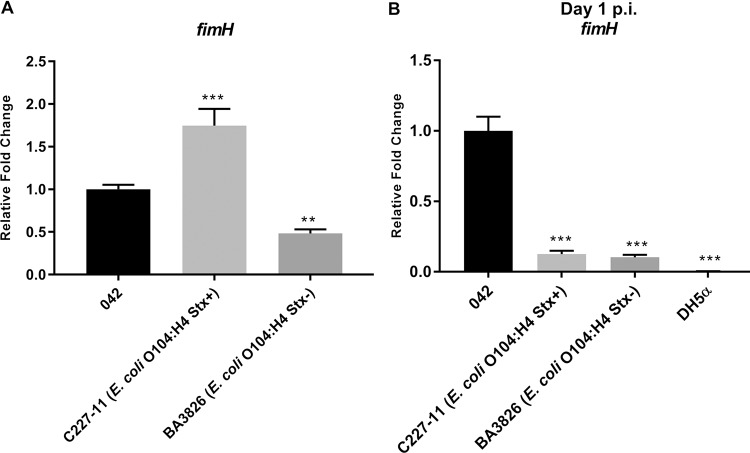
TIF analysis was performed via qRT-PCR through fimH gene expression, which encodes TIF pilin. In vitro, fimH was 1.7-fold higher in the WT C227-11 (Stx+) strain compared to strain 042 and lower in the BA3826 (Stx–) strain (Fig. 6A). In vivo, it was possible to demonstrate the presence of TIF at day 1 p.i. via fimH expression in all EAEC strains. However, the WT C227-11 (Stx+) and BA3826 strains showed significant reductions of 8- and 9.7-fold, respectively, compared to the EAEC prototype strain 042 (Fig. 6B). These results indicate the presence of these fimbriae during adhesion mechanisms of different EAEC strains. Thus, TIF is notably more important for strain 042 during the early stages of infection in the C57BL/6 mouse model, although the C227-11 (Stx+) strain exhibits lower Shiga toxin expression than EHEC 86-24 (Fig. S2) and higher expression of Stx under stress conditions (Fig. S1B), as previously shown (58). The AA pattern mediated by adherence aggregative fimbria enhances its ability to transport the toxin through epithelial cells, as previously reported (59).
FIG 6.
Role of type I fimbriae in O104:H4 Stx+ and Stx– strains during in vitro and in vivo essays. (A) qRT-PCR of fimH of RNA extracted from 042, C227-11 (Stx+), and BA3826 (Stx–) cultivated strains in LB medium until reaching an OD600 of 1.0. (B) fimH expression levels determined via qRT-PCR from RNAs of EAEC and DH5α strains extracted from feces on day 1 p.i. A Student t test or ANOVA was used to determine the statistical significance for each replicate. Error bars indicate the SD of the mean (**, P < 0.001; ***, P < 0.0001).
DISCUSSION
The EAEC pathovar is a highly heterogeneous group of intestinal pathogenic bacteria, whereas each isolate carries an exclusive subgroup of associated virulence genes. Therefore, there is no unique virulence factor associated but rather a set of them linked to its pathogenesis (2, 60–63). The outbreak strain here studied includes a rare combination of classical EAEC features and a recently acquired Stx2a toxin.
The QseC sensor kinase detects environmental cues, such as host stress hormones, and it plays an important role in the expression of relevant virulence genes, as previously reported for important human enteropathogens (64). QseC presents a function similar to that of the G-protein-coupled receptors in the eukaryotic cell membrane, which detect stress hormones and coordinate a regulatory response cascade (33, 65).
The initial study description has shown that LED209 inhibits the autophosphorylation of QseC and directly antagonizes stress hormone epinephrine binding in this histidine kinase sensor. Moreover, in vitro assays with LED209 have affected the transcription of global virulence genes of EHEC, S. Typhimurium, and F. tularensis (21). Very promising results with the interference of QseC by LED209 have reduced the biofilm formation of MDR clinical isolates such as UPEC, Klebsiella, and Pseudomonas aeruginosa, as well as the EAEC O104:H4 Stx+ strain (42). Another recent study showed the importance of LED209 as a therapeutic strategy for colitis-associated bacteria during inflammatory bowel disease caused by adherent-invasive E. coli (66).
LED209 hindered QseC triggering function during mouse infection by the E. coli O104:H4 C227-11 (Stx+) strain; the colonization levels were higher between days 1 and 5 p.i. (Fig. 1A). Thus, LED209 during the initial colonization period, between days 1 and 3, interrupted the QseBC regulatory cascade at a critical initial point of the infection, concomitant with the higher qseC gene expression levels observed without LED209 blocker (Fig. 2A and B). These data corroborate with previous studies in S. Typhimurium- or F. tularensis-infected mice that have shown a significantly higher percentage of survival; in addition, these pathogens presented a lower efficiency during in vivo colonization when LED209 was used, especially during early infection (21, 42). Moreover, the qseC mutant has presented similar results to WT (C227-11) treated with LED209, a finding which concurs with the role of QseC previously reported for EHEC and S. Typhimurium during in vivo infections (19, 21, 67).
Accordingly, the increased qseC expression by day 10 p.i. in both O104:H4 Stx+ and Stx– strains (Fig. 2A and B) coincides with the beginning of colonization decline and likely to less drug availability, since LED209 was used during initial infections in these mice, as previously described (21). The QseC sensor kinase was indirectly measured in vitro via qseC gene expression levels, where both Stx+ strains, EHEC 86-24 and C227-11, showed higher expression levels of qseC gene (Fig. 2C), supporting the importance of the QseC sensor kinase for these Stx+ strains. The data presented here emphasize the relevant role of QseC kinase sensor during the course of disease caused by Shiga toxin-producing strains, such as EHEC O157:H7 infections in mouse, rabbit, and bovine models (15, 68).
VisP has an important role in response to bacterial enteric stress (36, 37), and it seems accentuated upon LED209 treatment during in vivo infection (Fig. 3A and B), which is consistent with a QseC-blocked role and a VisP response to stressors. Similarly, S. Typhimurium and the EHEC 86-24 qseC mutant previously presented upregulation of visP compared to WT levels (36). The acid pH of the stomach (<3) is one of the first barriers to be overcome during the colonization process by enteric bacteria (68, 69). In an acid milieu, such as pH 3.0 (Fig. 3D), the visP expression in the ::qseC mutant did not demonstrate significant change, but its expression was more pronounced, especially in the EAEC Stx– strains. Therefore, this interesting role in acid stress response was observed under certain conditions for different genetic EAEC profiles in specific strains.
Recently, our group also verified that VisP is directly related to the assembly of O antigen, a highly immunogenic polysaccharide chain of LPS in S. Typhimurium, in addition to its important roles in macrophage survival, murine model colonization of infection, and the motility profile (37). LPS is an essential component of the cell envelope of Gram-negative bacteria and is directly related to cellular integrity, as well as in the permeability balance of the outer membrane, preventing the entry of harmful substances (70). However, the VisP role in E. coli LPS assembly remains unclear; the EAEC O104:H4 data here link VisP to the acid response, which may be essential during the whole process of GI tract colonization to survive the low pH of the stomach and possible undescribed LPS changes.
EHEC is a subcategory of Shiga toxin-producing E. coli that, in addition to Stx production, harbors the pathogenicity island LEE (locus of enterocyte effacement). LEE encodes effector proteins and a type III secretion system that are responsible for A/E histopathological lesions in the intestinal mucosa (71, 72). Although the C227-11 (Stx+) strain does not harbor LEE, it presents specialized mechanisms of EAEC adherence, such as aggregative adherence fimbriae (e.g., AAF/I), which are involved in thick biofilm formation and damage to the intestinal mucosa during infection. Thus, the EAEC adherence factors, in association with Stx, result in a very successful combination of pathogenic mechanisms (6, 73, 74).
Bacterial pathogens colonize the GI tract against all host defenses in a complex and intriguing manner that raises another interesting topic regarding how enteric pathogens may change the intestinal microbiota to orchestrate colonization. EAEC has an exclusive adhesion pattern, among other diarrheagenic E. coli, that may present variation within the pathovar (1). Many distinct EAEC animal models such as rabbits, mice, and gnotobiotic piglets have been studied, although these animals do not represent a standard model for colonization and development of the intestinal disease caused by EAEC similar to humans (75), especially the persistent infection observed in EAEC cases (2). A recent study observed an interesting response in C57BL/6 mice upon EAEC O104:H4 (Stx+) infections (43).
The intestinal microbiota plays an important role in human health maintenance, and its imbalance can lead to greater susceptibility to infection (76). All changes caused by pathogenic intruders can lead to future problems; the C57BL/6 mice here studied showed a predominance of Bacteroidetes phylum under all conditions evaluated. The mouse group infected with the C227-11 (Stx+) strain showed a remarkable change by day 5 p.i. (Fig. 4A), characterized by an increase in the Firmicutes phylum, and the level of Bacteroidetes was reestablished in the next few days. These changes probably occurred due to the pathogens’ ability to overcome barriers imposed by microbiota, which led to an exacerbated imbalance in the resident population. Bacteroides spp. are the main representatives of the Bacteroidetes phylum in the GI tract and include about 15 different species (46, 77). Members of this group, such as B. thetaiotaomicron and B. vulgatus, produce enzymes that degrade polysaccharides present in the mucus and make it available to the microbiota, which competes more efficiently for this nutrient compared to invasive pathogens such as EHEC, Shigella, and Salmonella (76). Previously, studies have shown that a significant number of Bacteroidetes strains encode a type VI secretion system (T6SS), and their genes are expressed in an in vivo infection model. Through T6SS, effector proteins may be directly injected into the cytoplasm of target bacteria, such as phospholipases, nucleases, and pore-forming proteins (77–80). Therefore, it is probable that predominance of the Bacteroidetes phylum has a direct or indirect connection to the resistance to the colonization of other enteropathogens that are invading the system, e.g., O104:H4 (Fig. 4).
However, other tested groups in our study did not show significant changes over the course of the study, except for the group infected with EAEC 042 strain on day 15 p.i. (Fig. 4B). An extremely high percentage of Firmicutes, similar to that which occurred at day 5 p.i. with the group infected with C227-11 (Stx+), was observed. Our data suggest that EAEC late colonization is indeed a feature of this in vivo model, similar to EAEC cases of diarrhea that can last for 14 days in humans (2). Together, these data reveal that the relative abundance of microbiota demonstrated a significant difference in the presence of C227-11 (Stx+) versus other EAEC strains; this outbreak strain could damage the microbiota balance and also express Stx during the process. The inflammatory process may lead to persistent disease because of delayed repair of the intestinal mucosa and decreased absorption of nutrients, resulting in deficits in infant growth and development (2, 4, 81).
The EAEC pathology still does not completely elucidate but surely correlates to multifactorial virulence traits and vast strain differences. Therefore, aggregative adhesion is still an important standard manner to gather the strains from this pathovar. Here, we measured both cell and polystyrene adhesion between the O104:H4 strains in the presence or absence of Shiga toxin, and the toxin did not significantly affect the AA pattern (data not shown) and adherent bacterial counts.
EAEC has the important clinical features of colonizing various regions of the intestine, forming thick biofilms, and producing a large amounts of mucus (4, 41, 82, 83). Host epithelium cell adherence is an essential step during the bacterial colonization process in the gut. This process in EAEC is facilitated mainly by AAFs, followed by biofilm formation (82, 84, 85). Previous study with T84 cells has shown how AAF/I of the C227-11 O104:H4 (Stx+) strain was sufficient to increase the permeability of the epithelial cell monolayer, thus contributing to an increase in Stx translocation through the epithelium (59). Besides the AAFs, EAEC encodes TIF (mannose-sensitive fimbriae), the most common adhesin found in Enterobacteriaceae and commonly observed in both commensal and pathogenic E. coli isolates. These fimbriae interact with mannose residues presented in eukaryotic cell receptors (86, 87). TIF mediates the colonization of mammalian host tissue, such as intestinal mucosa and the urinary tract. Moreover, this adhesin promotes biofilm formation on abiotic surfaces under static growth conditions. However, α-methyl-d-mannoside inhibits bacterial attachment to both biotic and abiotic surfaces because it is a direct competitive inhibitor of the mannose binding receptor by FimH adhesin of Escherichia coli (48, 52, 53, 88). During biofilm formation, a significant difference (by an order of magnitude of 0.5) was observed with α-methyl-d-mannoside for both strains. Together, these findings highlight the importance of TIF for biofilm formation for both C227-11 (Stx+) and BA3826 (Stx–) strains (Fig. 5A). Based on this, we next evaluated TIF fimH gene expression in vitro and in vivo (Fig. 6) to demonstrate TIF’s important role, especially in the initial adhesion. However, fimH expression in vivo (Fig. 6B) is more evident for EAEC 042 than for O104:H4, consistent with previous data that emphasize the importance of TIF for this strain (51). Gene expression analysis of stx expressed a lower level for C227-11 (Stx+) compared to EHEC 86-24 under the same test conditions, which shows an efficient Stx release by EHEC (Fig. S3). In contrast, EAEC tends to be more efficient when transporting Shiga toxin from intestinal epithelium to the bloodstream due to thick biofilm formation mediated by AAFs and TIF, factors that complicate EAEC pathogenesis. These data are consistent with a study by Zhang et al. (89), who showed that the outbreak strain of patients with hemorrhagic colitis had lost the pAA plasmid during the course of the disease in the European outbreak, which in turn caused a decrease in the ability of the EAEC O104:H4 (Stx+) strain to cause HUS, since these strains become less adherent, showing the genetic plasticity of these bacteria during the course of the outbreak.
Motility has been well described in EHEC, as well as its regulation by QseC (16, 34, 90), similarly to S. Typhimurium mechanisms (19, 22). However, this important virulence feature has been completed neglected with regard to EAEC virulence. In the present study, a pronounced motility phenotype in both EAEC O104:H4 Stx+ and Stx– strains has been observed at high levels similar to EHEC, which may be a novel interesting aspect of EAEC pathogenesis. However, the importance of flagellin and the role of flagella in EAEC pathogenesis still need to be studied more completely. Flagellum motility has been associated in many distinct pathogenic E. coli during the infectious process, such as FliC flagellin H4’s role in ST131 UPEC strain correlated to epithelial cell adhesion, invasion, and increased induction of cytokine interleukin-10 (56).
To summarize, QseC and VisP participate during the initial colonization process in a C57BL/6 mouse model in both O104:H4 Stx+ and Stx– strains. VisP seems more important for the acid response in vitro for the BA3826 (Stx–) strain, during stomach-mimicking conditions, such as low pH, whereas TIF mediates adhesion mechanisms during plastic biofilm formation in EAEC strains, as briefly illustrated and explained in our current model (Fig. 7).
FIG 7.
Proposed model for QseC and VisP role. Blocking of QseC phosphorylation by LED209 or knockout of qseC prevents the recognition of environmental signals and inhibits the transcription cascade of virulence genes in vivo. (A) Interruption of this pathway triggers other survival mechanisms, such as transcription of visP. (B) In vitro, the periplasmic protein VisP plays a role under different pH conditions between O104:H4 Stx+ and Stx– strains, and TIF mediates attachment on the abiotic surface.
Therefore, our study shows that the periplasmic protein VisP is implicated in EAEC stress response and is accentuated upon LED209 treatment to block QseC during in vivo infection. The QseC sensor kinase in EAEC may impact mechanisms of intestinal colonization, bacterial survival, and overexpression of key chemical signaling factors. Finally, we highlight the importance of QseC in C227-11 (Stx+) strain during colonization in an animal model and its role as a promising antivirulence drug target in similar enteric pathogens to prevent future outbreaks of Shiga toxin strain infections.
MATERIALS AND METHODS
Bacterial strains and media.
All of the strains and plasmids used during this study are listed in Table 1 . Strains were aerobically cultivated in Luria-Bertani (LB) broth or LB agar at 37°C. Molecular biology techniques were performed as previously described (91). All of the oligonucleotides used are listed in Table 2 .
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| 86-24 | EHEC O157:H7 LEE and Stx2a (isolated from outbreak of HUS in the USA) | 95 |
| WT C227-11 | WT EAEC O104:H4 pAA AggR AAF/I Stx2a+ Pic SigA SepA (isolated from German outbreak in 2011) | 58 |
| C227-11::qseC | EAEC O104:H4 qseC mutant | This study |
| C227-11::qseC+ | EAEC O104:H4 qseC (pBAD33) complemented strain | This study |
| BA3826 | EAEC O104:H4 pAA, AggR, AAF/III, Pic, EAST-1 (isolated from a case-control study in Brazil) | 13 |
| 042 | EAEC O44:H18 pAA, AggR, AAF/II, Pet, Pic, EAST-1 (isolated from a case of diarrhea in Peru) | 96 |
| 17-2 | EAEC O3:H2 pAA, AggR, AAF/I, EAST-1 (isolated from a case of diarrhea in Chile) | 97 |
| DH5α | E. coli supE44 ΔlacU169(ϕ80lacZΔM15) hsdR17 | Stratagene |
| DH5α(λpir) | DH5α transduced with λpir; Nalr | 98 |
| S17-1(λpir) | Pro Res– mod+ RP4-2 Tc::UM-Km::Tn7 Strr | 99 |
| TOP10 | E. coli F– mcrAΔ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| Plasmids | ||
| pJP5603 | 3.1-kb R6K-based suicide vector; Kmr | 92 |
| pBAD33 | Low-copy-number expression vector; Cmr | 100 |
LEE, locus of enterocyte effacement; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Strr, streptomycin resistance; Nalr, nalidixic acid resistance. AAF/I, pilin subunit of aggregative adherence fimbriae I; SigA, Shigella IgA-like protease homologue; SepA, Shigella extracellular protein A; AAF/III, pilin subunit of aggregative adherence fimbriae III; EAST-1, EAEC heat-stable enterotoxin 1; AAF/II, pilin subunit of aggregative adherence fimbriae II; Pet, protease involved in intestinal colonization.
TABLE 2.
Primers sets used in this study
| Target | Primer set sequence (5′–3′) |
Source or reference | |
|---|---|---|---|
| Forward | Reverse | ||
| qseC | CGCTGAAAGTGCAAACCGAA | CCGCGATAGAGTGAGCAGTT | This study |
| visP | ATGCGTTCAACGATATTGCCG | CGACCGTAGAAAGCGCAAAA | This study |
| fimH | CGGGGTGATGGATTCTCGTT | TCTGGGGATCTCCACCATGT | This study |
| rpoA | GCGCTCATCTTCTTCCGAAT | CGCGGTCGTGGTTATGTG | 22 |
| stx2a | ACCCCACCGGGCAGTT | GGTCAAAACGCGCCTGATA | 21 |
| Universal bacteria, Eub338F/Eub518R | ACTCCTACGGGAGGCAGCAGT | ATTACCGCGGCTGCTGGC | 101 |
| Firmicutes 928F-Firm/1040FirmR | TGAAACTYAAAGGAATTGACG | ACCATGCACCACCTGTC | 102 |
| Bacteroidetes 798cfbF/cfb967R | CRAACAGGATTAGATACCCT | GGTAAGGTTCCTCGCGTAT | 103 |
| Gammaproteobacteria, 1080gF/g1202R | TCGTCAGCTCGTGTYGTGA | CGTAAGGGCCATGATG | 102 |
| qseC-pJP5603 | ATGGTACCCGACGGCAGAATGGTCCTTAA | GAGCTCCCTGAAGGTTATCCAGTGAGTCC | This study |
| qseC-PBAD33 | GCAGAGCTCATGAAATTTACCCAACGTCTTAGTC | GCAGGTACCTTACATCATCACCATCACCACCCAGCTTACCTTCGCCTC | This study |
| M13 | GTAAAACGACGGCCAG | CAGGAAACAGCTATGAC | Invitrogen |
Mutant construction and complementation.
The nonpolar qseC mutant was prepared by using suicide vector pJP5603 (92). An internal portion of the qseC gene of 641 pb was amplified by PCR using qseC-pJP5603 primers. The insert was digested with the restriction enzymes KpnI and SacI (New England Biolabs) and then inserted into the corresponding site in the suicide vector. Recombinant plasmid was transformed into E. coli DH5α(λpir), and the colonies were selected on plates containing kanamycin (50 μg/ml). E. coli S17(λpir) was transformed with selected plasmids or empty suicide vector as a negative control and conjugated to WT C227-11 (Stx+) strain via filter mating on cellulose nitrate membranes. Transconjugant colonies were selected on LB agar plates containing kanamycin (50 μg/ml) and tetracycline (30 μg/ml), and integration of suicide vector pJP5603 in the qseC gene was confirmed by PCR analysis using the primers M13 (reverse) and qseC-pJP5603 (reverse). The qseC gene was cloned into pBAD33 (SacI/KpnI) vector and used to complement the mutant strain. The WT strain with a transformed empty vector was used in all phenotypic and genetic assays as a control for the ::qseC mutant. All PCRs were amplified with Platinum Taq DNA polymerase (Invitrogen) and 1 μM concentrations of each primer (Table 2).
Biofilm formation.
Assays were performed with the O104:H4 strains WT C227-11 (Stx+), ::qseC (C227-11), qseC+ (C227-11), and BA3826 (Stx–) to compare their abilities to form biofilm on polystyrene surface, as previously described (93). Bacteria previously grown in LB broth at 37°C were placed in 96-well plates at 1:100 in Dulbecco modified Eagle medium (DMEM) with or without α-methyl-d-mannoside and incubated for 24 h at 37°C. Finally, the wells were washed three times with sterile PBS, incubated with Tween 0.1% for 10 min, and then plated on LB agar for CFU counting.
Adherence assays.
Adherence assays were performed to complement the adhesion profile (see the supplemental material [https://www2.fcfar.unesp.br/#!/pos-graduacao/biociencias-e-biotecnologias-aplicadas-a-farmacia/docentes2308/corpo-docente/]) with confluent HeLa cells as previously described (94). Cells were cultivated in DMEM (Gibco-BRL, Gaithersburg, MD) containing 10% of fetal bovine serum. Assays were performed in 24-well plates with or without α-methyl-d-mannoside (a nonmetabolizable analogue of mannose) to investigate the role of TIF in the AA pattern. Strains were grown in LB broth overnight and then used to infect HeLa cells at a multiplicity of infection of 100:1. The strains were incubated in a CO2 incubator at 37°C for 3 or 6 h. The wells were then washed three times with sterile PBS, incubated with Triton X-100 (1%) for 10 min, and then plated on LB agar for CFU counting.
Stress response assays.
For acid response assays using qRT-PCR, bacteria were statically incubated in both LB broth (pH 3.0) and PBS (pH 7.2) at 37°C for 1 h, and RNA was extracted. The acidic pH was adjusted using HCl (36).
Motility assays.
The motility assays were performed using LB broth containing 0.3% agar, as described previously (14, 22). Overnight cultures grown with shaking at 37°C were inoculated at the center of each plate, followed by incubation at 37°C for 24 h, and halo diameters were measured.
qRT-PCR.
RNA extraction was performed with bacteria aerobically grown in LB until reaching an optical density at 600 nm (OD600) of 1.0 or fecal pellets collected from days 0 to 15 after C57BL/6J mouse infections. Then, 1 ml of TRIzol (Life Technologies) was used per 100 mg of samples (bacterial or fecal pellets). RNA was extracted by using a RiboPure bacterial isolation kit (Ambion) according to the instructions of the manufacturer. qRT-PCR was carried out with primers to detect virulence genes and predesigned RNA 16S of bacterial phyla (Table 1). QuantStudio3 (Thermo Fisher Scientific) was used for one-step qRT-PCR. Data were normalized with rpoA (RNA polymerase subunit A) as an endogenous control, virulence genes, and EUB (universal bacteria) for the microbiota phylum. In order to verify the relative expression of each taxon, results were normalized within day 0 (before infection). Final results were analyzed by determining the comparative critical threshold (ΔΔCT) method, as described previously (35), and statistical significance tests were performed using a Student t test.
Mice.
Mice were acquired from CEMIB/UNICAMP and maintained at our Animal Facility in the Biological Sciences Department at School of Pharmaceutical Sciences/UNESP. The experiment with animals was previously approved by the Animal Ethics Committee (CEUA/FCF/Car 23/2016). Animals were 3- to 5-week-old female C57BL/6J UNIB mice, weighing between 12 and 15 g. They were pretreated with 20 mg/kg of ampicillin via oral gavage, 24 h before infection, for the depletion of microbiota. Assays were divided into four experimental groups of animals with five mice per group; one group was inoculated with E. coli K-12 DH5a (nonpathogenic) as a negative control. The other three groups were infected, respectively, with EAEC C227-11 (Stx+), EAEC BA3826 (Stx–), and 042 strains. All experiments were repeated at least twice to ensure the results presented here.
Strains were cultivated for 16 to 18 h, centrifuged, and resuspended in PBS. Animals were infected with 1010 bacteria via oral gavage. All experiments were performed as previously described by Zangari et al. (43), except for minor modifications already mentioned. Feces were collected from mice from days 1 to 14 p.i. to recover CFU and determine gene expression by qRT-PCR. Euthanasia was performed on day 14. Mice were monitored for weight change over 14 days. Weight loss was considered for animals with a decrease of at least 5% of total body mass (43).
LED209.
A total of 24 female C57BL/6J UNIB mice (3 to 5 weeks old, 12 to 15 g), received 20 mg/kg of ampicillin via oral gavage for microbiota depletion by 24 h before infection. LED209 (58.8 mg/kg) or vehicle (5% dimethyl sulfoxide, 23% polyethylene glycol 400, 70% sodium bicarbonate [pH 9], 2% Tween 80) only were administered 3 h before, at the time of, and 3 h after infection. Mice were infected with 1010 CFU of O104:H4 C227-11 (Stx+) and O104:H4 BA3826 (Stx–) strains. Feces were collected from mice from days 1 to 14 p.i. to recover CFU and determine gene expression by qRT-PCR. Euthanasia was performed on day 14.
Statistical analysis.
The statistical significance was evaluate by one-way analysis-of-variance (ANOVA) and Student t test using GraphPad Prism (v7). Experiments were repeated at least three times independently. The error bars indicate the standard deviations (SD) of the mean.
ACKNOWLEDGMENTS
We acknowledge financial support from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; universal project 441884/2014-8) and the Fundação de Amparo à Pesquisa do Estado de São Paulo (Young Investigator Project 2014/06779-2), as well as Graduate Program funding from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior.
REFERENCES
- 1.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 2.Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin Microbiol Rev 11:142–201. doi: 10.1128/CMR.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang DB, Mohanty A, DuPont HL, Okhuysen PC, Chiang T. 2006. A review of an emerging enteric pathogen: enteroaggregative Escherichia coli. J Med Microbiol 55:1303–1311. doi: 10.1099/jmm.0.46674-0. [DOI] [PubMed] [Google Scholar]
- 4.Nataro JP, Steiner T, Guerrant RL. 1998. Enteroaggregative Escherichia coli. Emerg Infect Dis 4:251–261. doi: 10.3201/eid0402.980212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gould LH, Demma L, Jones TF, Hurd S, Vugia DJ, Smith K, Shiferaw B, Segler S, Palmer A, Zansky S, Griffin PM. 2009. Hemolytic uremic syndrome and death in persons with Escherichia coli O157:H7 infection, foodborne diseases active surveillance network sites, 2000-2006. Clin Infect Dis 49:1480–1485. doi: 10.1086/644621. [DOI] [PubMed] [Google Scholar]
- 6.Bielaszewska M, Mellmann A, Zhang W, Köck R, Fruth A, Bauwens A, Peters G, Karch H. 2011. Characterization of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect Dis 11:671–676. doi: 10.1016/S1473-3099(11)70165-7. [DOI] [PubMed] [Google Scholar]
- 7.Frank C, Werber D, Cramer JP, Askar M, Faber M, An der Heiden M, Bernard H, Fruth A, Prager R, Spode A, Wadl M, Zoufaly A, Jordan S, Kemper MJ, Follin P, Muller L, King LA, Rosner B, Buchholz U, Stark K, Krause G. 2011. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med 365:1771–1780. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- 8.Mellmann A, Bielaszewska M, Kock R, Friedrich AW, Fruth A, Middendorf B, Harmsen D, Schmidt MA, Karch H. 2008. Analysis of collection of hemolytic uremic syndrome-associated enterohemorrhagic Escherichia coli. Emerg Infect Dis 14:1287–1290. doi: 10.3201/eid1408.071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scavia G, Morabito S, Tozzoli R, Michelacci V, Marziano ML, Minelli F, Ferreri C, Paglialonga F, Edefonti A, Caprioli A. 2011. Similarity of Shiga toxin-producing Escherichia coli O104:H4 strains from Italy and Germany. Emerg Infect Dis 17:1957–1958. doi: 10.3201/eid1710.111072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmed SA, Awosika J, Baldwin C, Bishop-Lilly KA, Biswas B, Broomall S, Chain PS, Chertkov O, Chokoshvili O, Coyne S, Davenport K, Detter JC, Dorman W, Erkkila TH, Folster JP, Frey KG, George M, Gleasner C, Henry M, Hill KK, Hubbard K, Insalaco J, Johnson S, Kitzmiller A, Krepps M, Lo CC, Luu T, McNew LA, Minogue T, Munk CA, Osborne B, Patel M, Reitenga KG, Rosenzweig CN, Shea A, Shen X, Strockbine N, Tarr C, Teshima H, van Gieson E, Verratti K, Wolcott M, Xie G, Sozhamannan S, Gibbons HS. 2012. Genomic comparison of Escherichia coli O104:H4 isolates from 2009 and 2011 reveals plasmid, and prophage heterogeneity, including Shiga toxin encoding phage Stx2. PLoS One 7:e48228. doi: 10.1371/journal.pone.0048228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guy L, Jernberg C, Arven Norling J, Ivarsson S, Hedenstrom I, Melefors O, Liljedahl U, Engstrand L, Andersson SG. 2013. Adaptive mutations and replacements of virulence traits in the Escherichia coli O104:H4 outbreak population. PLoS One 8:e63027. doi: 10.1371/journal.pone.0063027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernier C, Gounon P, Le Bouguénec C. 2002. Identification of an aggregative adhesion fimbria (AAF) type III-encoding operon in enteroaggregative Escherichia coli as a sensitive probe for detecting the AAF-encoding operon family. Infect Immun 70:4302–4311. doi: 10.1128/iai.70.8.4302-4311.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bueris V, Sircili MP, Taddei CR, dos Santos MF, Franzolin MR, Martinez MB, Ferrer SR, Barreto ML, Trabulsi LR. 2007. Detection of diarrheagenic Escherichia coli from children with and without diarrhea in Salvador, Bahia, Brazil. Mem Inst Oswaldo Cruz 102:839–844. doi: 10.1590/S0074-02762007005000116. [DOI] [PubMed] [Google Scholar]
- 14.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. 2003. Bacterium-host communication: the language of hormones. Proc Natl Acad Sci U S A 100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. 2006. The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci U S A 103:10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sperandio V, Torres AG, Kaper JB. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in Escherichia coli. Mol Microbiol 43:809–821. doi: 10.1046/j.1365-2958.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- 17.Sircili MP, Walters M, Trabulsi LR, Sperandio V. 2004. Modulation of enteropathogenic Escherichia coli virulence by quorum sensing. Infect Immun 72:2329–2337. doi: 10.1128/iai.72.4.2329-2337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakano M, Takahashi A, Sakai Y, Nakaya Y. 2007. Modulation of pathogenicity with norepinephrine related to the type III secretion system of Vibrio parahaemolyticus. J Infect Dis 195:1353–1360. doi: 10.1086/513275. [DOI] [PubMed] [Google Scholar]
- 19.Bearson BL, Bearson SM. 2008. The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb Pathog 44:271–278. doi: 10.1016/j.micpath.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Merighi M, Septer AN, Carroll-Portillo A, Bhatiya A, Porwollik S, McClelland M, Gunn JS. 2009. Genome-wide analysis of the PreA/PreB (QseB/QseC) regulon of Salmonella enterica serovar Typhimurium. BMC Microbiol 9:42. doi: 10.1186/1471-2180-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasko DA, Moreira CG, Li DR, Reading NC, Ritchie JM, Waldor MK, Williams N, Taussig R, Wei S, Roth M, Hughes DT, Huntley JF, Fina MW, Falck JR, Sperandio V. 2008. Targeting QseC signaling and virulence for antibiotic development. Science 321:1078–1080. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreira CG, Weinshenker D, Sperandio V. 2010. QseC mediates Salmonella enterica serovar Typhimurium virulence in vitro and in vivo. Infect Immun 78:914–926. doi: 10.1128/IAI.01038-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pullinger GD, van Diemen PM, Dziva F, Stevens MP. 2010. Role of two-component sensory systems of Salmonella enterica serovar Dublin in the pathogenesis of systemic salmonellosis in cattle. Microbiology 156:3108–3122. doi: 10.1099/mic.0.041830-0. [DOI] [PubMed] [Google Scholar]
- 24.Mokrievich AN, Kondakova AN, Valade E, Platonov ME, Vakhrameeva GM, Shaikhutdinova RZ, Mironova RI, Blaha D, Bakhteeva IV, Titareva GM, Kravchenko TB, Kombarova TI, Vidal D, Pavlov VM, Lindner B, Dyatlov IA, Knirel YA. 2010. Biological properties and structure of the lipopolysaccharide of a vaccine strain of Francisella tularensis generated by inactivation of a quorum sensing system gene qseC. Biochemistry 75:443–451. [DOI] [PubMed] [Google Scholar]
- 25.Hadjifrangiskou M, Kostakioti M, Chen SL, Henderson JP, Greene SE, Hultgren SJ. 2011. A central metabolic circuit controlled by QseC in pathogenic Escherichia coli. Mol Microbiol 80:1516–1529. doi: 10.1111/j.1365-2958.2011.07660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Wang Q, Yang M, Xiao J, Liu Q, Wu H, Zhang Y. 2011. QseBC controls flagellar motility, fimbrial hemagglutination, and intracellular virulence in fish pathogen Edwardsiella tarda. Fish Shellfish Immunol 30:944–953. doi: 10.1016/j.fsi.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 27.Unal CM, Singh B, Fleury C, Singh K, Chávez de Paz L, Svensäter G, Riesbeck K. 2012. QseC controls biofilm formation of non-typeable Haemophilus influenzae in addition to an AI-2-dependent mechanism. Int J Med Microbiol 302:261–269. doi: 10.1016/j.ijmm.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 28.Khajanchi BK, Kozlova EV, Sha J, Popov VL, Chopra AK. 2012. The two-component QseBC signaling system regulates in vitro and in vivo virulence of Aeromonas hydrophila. Microbiology 158:259–271. doi: 10.1099/mic.0.051805-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozlova EV, Khajanchi BK, Popov VL, Wen J, Chopra AK. 2012. Impact of QseBC system in c-di-GMP-dependent quorum sensing regulatory network in a clinical isolate SSU of Aeromonas hydrophila. Microb Pathog 53:115–124. doi: 10.1016/j.micpath.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Juarez-Rodriguez MD, Torres-Escobar A, Demuth DR. 2013. ygiW and qseBC are coexpressed in Aggregatibacter actinomycetemcomitans and regulate biofilm growth. Microbiology 159:989–1001. doi: 10.1099/mic.0.066183-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weigel WA, Demuth DR, Torres-Escobar A, Juárez-Rodríguez MD. 2015. Aggregatibacter actinomycetemcomitans QseBC is activated by catecholamines and iron and regulates genes encoding proteins associated with anaerobic respiration and metabolism. Mol Oral Microbiol 30:384–398. doi: 10.1111/omi.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrison CF, Kicka S, Kranjc A, Finsel I, Chiriano G, Ouertatani-Sakouhi H, Soldati T, Scapozza L, Hilbi H. 2015. Adrenergic antagonists restrict replication of Legionella. Microbiology 161:1392–1406. doi: 10.1099/mic.0.000094. [DOI] [PubMed] [Google Scholar]
- 33.Hughes DT, Clarke MB, Yamamoto K, Rasko DA, Sperandio V. 2009. The QseC adrenergic signaling cascade in enterohemorrhagic Escherichia coli. PLoS Pathog 5:e1000553. doi: 10.1371/journal.ppat.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clarke MB, Sperandio V. 2005. Transcriptional autoregulation by quorum sensing E. coli regulators B and C (QseBC) in enterohemorrhagic Escherichia coli. Mol Microbiol 58:441–455. doi: 10.1111/j.1365-2958.2005.04819.x. [DOI] [PubMed] [Google Scholar]
- 35.Walters M, Sperandio V. 2006. Autoinducer 3 and epinephrine signaling in the kinetics of locus of enterocyte effacement gene expression in enterohemorrhagic Escherichia coli. Infect Immun 74:5445–5455. doi: 10.1128/IAI.00099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreira CG, Herrera CM, Needham BD, Parker CT, Libby SJ, Fang FC, Trent MS, Sperandio V. 2013. Virulence and stress-related periplasmic protein (VisP) in bacterial/host associations. Proc Natl Acad Sci U S A 110:1470–1475. doi: 10.1073/pnas.1215416110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.da Silva P, Manieri FZ, Herrera CM, Trent MS, Moreira CG. 2018. Novel role of VisP and the Wzz system during O-antigen assembly in Salmonella enterica serovar Typhimurium pathogenesis. Infect Immun 86:e00319-18. doi: 10.1128/IAI.00319-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J, Hiibel SR, Reardon KF, Wood TK. 2010. Identification of stress-related proteins in Escherichia coli using the pollutant cis-dichloroethylene. J Appl Microbiol 108:2088–2102. doi: 10.1111/j.1365-2672.2009.04611.x. [DOI] [PubMed] [Google Scholar]
- 39.Kendall MM, Sperandio V. 2016. What a dinner party! mechanisms and functions of interkingdom signaling in host-pathogen associations. mBio 7:e01748. doi: 10.1128/mBio.01748-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sekirov I, Finlay BB. 2009. The role of the intestinal microbiota in enteric infection. J Physiol 587:4159–4167. doi: 10.1113/jphysiol.2009.172742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nataro JP, Deng Y, Cookson S, Cravioto A, Savarino SJ, Guers LD, Levine MM, Tacket CO. 1995. Heterogeneity of enteroaggregative Escherichia coli virulence demonstrated in volunteers. J Infect Dis 171:465–468. doi: 10.1093/infdis/171.2.465. [DOI] [PubMed] [Google Scholar]
- 42.Curtis MM, Russell R, Moreira CG, Adebesin AM, Wang C, Williams NS, Taussig R, Stewart D, Zimmern P, Lu B, Prasad RN, Zhu C, Rasko DA, Huntley JF, Falck JR, Sperandio V. 2014. QseC inhibitors as an antivirulence approach for Gram-negative pathogens. mBio 5:e02165. doi: 10.1128/mBio.02165-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zangari T, Melton-Celsa AR, Panda A, Boisen N, Smith MA, Tatarov I, De Tolla LJ, Nataro JP, O’Brien AD. 2013. Virulence of the Shiga toxin type 2-expressing Escherichia coli O104:H4 German outbreak isolate in two animal models. Infect Immun 81:1562–1574. doi: 10.1128/IAI.01310-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yurist-Doutsch S, Arrieta M-C, Vogt SL, Finlay BB. 2014. Gastrointestinal microbiota mediated control of enteric pathogens. Annu Rev Genet 48:361–382. doi: 10.1146/annurev-genet-120213-092421. [DOI] [PubMed] [Google Scholar]
- 45.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Human Microbiome Project Consortium. 2012. Structure, function, and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baumler AJ, Sperandio V. 2016. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pratt LA, Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis, and type I pili. Mol Microbiol 30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 49.Johnson JR. 1991. Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev 4:80–128. doi: 10.1128/cmr.4.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Totsika M, Kostakioti M, Hannan TJ, Upton M, Beatson SA, Janetka JW, Hultgren SJ, Schembri MA. 2013. A FimH inhibitor prevents acute bladder infection and treats chronic cystitis caused by multidrug-resistant uropathogenic Escherichia coli ST131. J Infect Dis 208:921–928. doi: 10.1093/infdis/jit245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moreira CG, Carneiro SM, Nataro JP, Trabulsi LR, Elias WP. 2003. Role of type I fimbriae in the aggregative adhesion pattern of enteroaggregative Escherichia coli. FEMS Microbiol Lett 226:79–85. doi: 10.1016/S0378-1097(03)00561-5. [DOI] [PubMed] [Google Scholar]
- 52.Yamamoto T, Fujita K, Yokota T. 1990. Adherence characteristics to human small intestinal mucosa of Escherichia coli isolated from patients with diarrhea or urinary tract infections. J Infect Dis 162:896–908. doi: 10.1093/infdis/162.4.896. [DOI] [PubMed] [Google Scholar]
- 53.Bloch CA, Stocker BA, Orndorff PE. 1992. A key role for type 1 pili in enterobacterial communicability. Mol Microbiol 6:697–701. doi: 10.1111/j.1365-2958.1992.tb01518.x. [DOI] [PubMed] [Google Scholar]
- 54.Park SY, Pontes MH, Groisman EA. 2015. Flagellum-independent surface motility in Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci U S A 112:1850–1855. doi: 10.1073/pnas.1422938112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Houdt R, Michiels CW. 2005. Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res Microbiol 156:626–633. doi: 10.1016/j.resmic.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 56.Kakkanat A, Totsika M, Schaale K, Duell BL, Lo AW, Phan MD, Moriel DG, Beatson SA, Sweet MJ, Ulett GC, Schembri MA. 2015. The role of H4 flagella in Escherichia coli ST131 virulence. Sci Rep 5. doi: 10.1038/srep16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riley LW. 2014. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin Microbiol Infect 20:380–390. doi: 10.1111/1469-0691.12646. [DOI] [PubMed] [Google Scholar]
- 58.Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, Paxinos EE, Sebra R, Chin CS, Iliopoulos D, Klammer A, Peluso P, Lee L, Kislyuk AO, Bullard J, Kasarskis A, Wang S, Eid J, Rank D, Redman JC, Steyert SR, Frimodt-Moller J, Struve C, Petersen AM, Krogfelt KA, Nataro JP, Schadt EE, Waldor MK. 2011. Origins of the Escherichia coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med 365:709–717. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boisen N, Hansen AM, Melton-Celsa AR, Zangari T, Mortensen NP, Kaper JB, O’Brien AD, Nataro JP. 2014. The presence of the pAA plasmid in the German O104:H4 Shiga toxin type 2a (Stx2a)-producing enteroaggregative Escherichia coli strain promotes the translocation of Stx2a across an epithelial cell monolayer. J Infect Dis 210:1909–1919. doi: 10.1093/infdis/jiu399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okeke IN, Lamikanra A, Steinruck H, Kaper JB. 2000. Characterization of Escherichia coli strains from cases of childhood diarrhea in provincial southwestern Nigeria. J Clin Microbiol 38:7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Czeczulin JR, Whittam TS, Henderson IR, Navarro-Garcia F, Nataro JP. 1999. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect Immun 67:2692–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boisen N, Scheutz F, Rasko DA, Redman JC, Persson S, Simon J, Kotloff KL, Levine MM, Sow S, Tamboura B, Toure A, Malle D, Panchalingam S, Krogfelt KA, Nataro JP. 2012. Genomic characterization of enteroaggregative Escherichia coli from children in Mali. J Infect Dis 205:431–444. doi: 10.1093/infdis/jir757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Estrada-Garcia T, Navarro-Garcia F. 2012. Enteroaggregative Escherichia coli pathotype: a genetically heterogeneous emerging foodborne enteropathogen. FEMS Immunol Med Microbiol 66:281–298. doi: 10.1111/j.1574-695X.2012.01008.x. [DOI] [PubMed] [Google Scholar]
- 64.Sperandio V, Torres AG, Giron JA, Kaper JB. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J Bacteriol 183:5187–5197. doi: 10.1128/jb.183.17.5187-5197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gilman AG. 1987. G proteins: transducers of receptor-generated signals. Annu Rev Biochem 56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 66.Rooks MG, Veiga P, Reeves AZ, Lavoie S, Yasuda K, Asano Y, Yoshihara K, Michaud M, Wardwell-Scott L, Gallini CA, Glickman JN, Sudo N, Huttenhower C, Lesser CF, Garrett WS. 2017. QseC inhibition as an antivirulence approach for colitis-associated bacteria. Proc Natl Acad Sci U S A 114:142–147. doi: 10.1073/pnas.1612836114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moreira CG, Russell R, Mishra AA, Narayanan S, Ritchie JM, Waldor MK, Curtis MM, Winter SE, Weinshenker D, Sperandio V. 2016. Bacterial adrenergic sensors regulate virulence of enteric pathogens in the gut. mBio 7:e00826-16. doi: 10.1128/mBio.00826-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arnold KW, Kaspar CW. 1995. Starvation- and stationary-phase-induced acid tolerance in Escherichia coli O157:H7. Appl Environ Microbiol 61:2037–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gorden J, Small PL. 1993. Acid resistance in enteric bacteria. Infect Immun 61:364–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu Rev Biochem 71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A 92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elliott SJ, Wainwright LA, McDaniel TK, Jarvis KG, Deng YK, Lai LC, McNamara BP, Donnenberg MS, Kaper JB. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol Microbiol 28:1–4. [DOI] [PubMed] [Google Scholar]
- 73.Boisen N, Melton-Celsa AR, Scheutz F, O’Brien AD, Nataro JP. 2015. Shiga toxin 2a and Enteroaggregative Escherichia coli: a deadly combination. Gut Microbes 6:272–278. doi: 10.1080/19490976.2015.1054591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boisen N, Struve C, Scheutz F, Krogfelt KA, Nataro JP. 2008. New adhesin of enteroaggregative Escherichia coli related to the Afa/Dr/AAF family. Infect Immun 76:3281–3292. doi: 10.1128/IAI.01646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Philipson CW, Bassaganya-Riera J, Hontecillas R. 2013. Animal models of enteroaggregative Escherichia coli infection. Gut Microbes 4:281–291. doi: 10.4161/gmic.24826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McKenney ES, Kendall MM. 2016. Microbiota and pathogen “pas de deux”: setting up and breaking down barriers to intestinal infection. Pathog Dis 74:ftw051. doi: 10.1093/femspd/ftw051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Coyne MJ, Roelofs KG, Comstock LE. 2016. Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics 17:58. doi: 10.1186/s12864-016-2377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Coyne MJ, Zitomersky NL, McGuire AM, Earl AM, Comstock LE. 2014. Evidence of extensive DNA transfer between bacteroidales species within the human gut. mBio 5:e01305. doi: 10.1128/mBio.01305-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB, Chou S, Gonen T, Goodlett DR, Goodman AL, Mougous JD. 2014. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16:227–236. doi: 10.1016/j.chom.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wilson MM, Anderson DE, Bernstein HD. 2015. Analysis of the outer membrane proteome and secretome of Bacteroides fragilis reveals a multiplicity of secretion mechanisms. PLoS One 10:e0117732. doi: 10.1371/journal.pone.0117732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Steiner TS, Lima AA, Nataro JP, Guerrant RL. 1998. Enteroaggregative Escherichia coli produce intestinal inflammation and growth impairment and cause interleukin-8 release from intestinal epithelial cells. J Infect Dis 177:88–96. doi: 10.1086/513809. [DOI] [PubMed] [Google Scholar]
- 82.Hicks S, Candy DC, Phillips AD. 1996. Adhesion of enteroaggregative Escherichia coli to pediatric intestinal mucosa in vitro. Infect Immun 64:4751–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nataro JP, Hicks S, Phillips AD, Vial PA, Sears CL. 1996. T84 cells in culture as a model for enteroaggregative Escherichia coli pathogenesis. Infect Immun 64:4761–4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang DB, Okhuysen PC, Jiang ZD, DuPont HL. 2004. Enteroaggregative Escherichia coli: an emerging enteric pathogen. Am J Gastroenterol 99:383–389. doi: 10.1111/j.1572-0241.2004.04041.x. [DOI] [PubMed] [Google Scholar]
- 85.Czeczulin JR, Balepur S, Hicks S, Phillips A, Hall R, Kothary MH, Navarro-Garcia F, Nataro JP. 1997. Aggregative adherence fimbria II, a second fimbrial antigen mediating aggregative adherence in enteroaggregative Escherichia coli. Infect Immun 65:4135–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ofek I, Doyle RJ. 1994. Bacterial adhesion to cells and tissues, p 239–320. In Ofek I, Doyle RJ (ed), Bacterial adhesion to cells and tissues, 2nd ed, vol 735 Chapman & Hall, New York, NY. [Google Scholar]
- 87.Hernandes RT, Silva RM, Carneiro SM, Salvador FA, Fernandes MC, Padovan AC, Yamamoto D, Mortara RA, Elias WP, da Silva Briones MR, Gomes TA. 2008. The localized adherence pattern of an atypical enteropathogenic Escherichia coli is mediated by intimin omicron and unexpectedly promotes HeLa cell invasion. Cell Microbiol 10:415–425. doi: 10.1111/j.1462-5822.2007.01054.x. [DOI] [PubMed] [Google Scholar]
- 88.Aronson M, Medalia O, Schori L, Mirelman D, Sharon N, Ofek I. 1979. Prevention of colonization of the urinary tract of mice with Escherichia coli by blocking of bacterial adherence with methyl α-d-mannopyranoside. J Infect Dis 139:329–332. doi: 10.1093/infdis/139.3.329. [DOI] [PubMed] [Google Scholar]
- 89.Zhang W, Bielaszewska M, Kunsmann L, Mellmann A, Bauwens A, Kock R, Kossow A, Anders A, Gatermann S, Karch H. 2013. Lability of the pAA virulence plasmid in Escherichia coli O104:H4: implications for virulence in humans. PLoS One 8:e66717. doi: 10.1371/journal.pone.0066717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Njoroge J, Sperandio V. 2012. Enterohemorrhagic Escherichia coli virulence regulation by two bacterial adrenergic kinases, QseC and QseE. Infect Immun 80:688–703. doi: 10.1128/IAI.05921-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidmam JG, Smith JA, Struhl K. 1992. Short protocols in molecular biology , 3rd ed Greene Publishing Associates/John Wiley Sons, Inc., New York, NY. [Google Scholar]
- 92.Penfold RJ, Pemberton JM. 1992. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118:145–146. doi: 10.1016/0378-1119(92)90263-O. [DOI] [PubMed] [Google Scholar]
- 93.Sheikh J, Hicks S, Dall’Agnol M, Phillips AD, Nataro JP. 2001. Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol Microbiol 41:983–997. [DOI] [PubMed] [Google Scholar]
- 94.Cravioto A, Gross RJ, Scotland SM, Rowe B. 1979. An adhesive factor found in strains of Escherichia coli belonging to the traditional infantile enteropathogenic serotypes. Curr Microbiol 3:95–99. doi: 10.1007/BF02602439. [DOI] [Google Scholar]
- 95.Griffin PM, Ostroff SM, Tauxe RV, Greene KD, Wells JG, Lewis JH, Blake PA. 1988. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann Intern Med 109:705–712. [DOI] [PubMed] [Google Scholar]
- 96.Nataro JP, Baldini MM, Kaper JB, Black RE, Bravo N, Levine MM. 1985. Detection of an adherence factor of enteropathogenic Escherichia coli with a DNA probe. J Infect Dis 152:560–565. [DOI] [PubMed] [Google Scholar]
- 97.Vial PA, Mathewson JJ, DuPont HL, Guers L, Levine MM. 1990. Comparison of two assay methods for patterns of adherence to HEp-2 cells of Escherichia coli from patients with diarrhea. J Clin Microbiol 28:882–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Elliot SJ, Kaper JB. 1997. Role of type 1 fimbriae in EPEC infections. Microb Pathog 23:113–118. [DOI] [PubMed] [Google Scholar]
- 99.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784–791. [Google Scholar]
- 100.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fierer N, Jackson JA, Vilgalys R, Jackson RB. 2005. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl Environ Microbiol 71:4117–4120. doi: 10.1128/aem.71.7.4117-4120.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bacchetti De Gregorisa T, Aldred N, Clare AS, Burgess JG. 2011. Improvement of phylum- and class-specific primers for real-time PCR quantification of bacterial taxa. J Microbiol Methods 86:351–356. doi: 10.1016/j.mimet.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 103.Guo X, Xia X, Tang R, Zhou J, Zhao H, Wang K. 2008. Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett Appl Microbiol 47:367–373. doi: 10.1111/j.1472-765X.2008.02408.x. [DOI] [PubMed] [Google Scholar]