The ubiquitous Pseudomonas species are well adapted to survive in a wide variety of environments. Their success relies on their versatile metabolic, signaling, and transport ability but also on their high intrinsic tolerance to various stress factors. This is why the study of the stress-surviving mechanisms of Pseudomonas species is of utmost importance. The stress tolerance of Pseudomonads is mainly achieved through the high barrier property of their membranes. Here, we present evidence that the TonB-ExbBD-like TonBm-PocAB system is involved in maintaining the membrane homeostasis of Pseudomonas putida, and its deficiency leads to lowered stress tolerance and conditional cell lysis.
KEYWORDS: flagellum localization, growth phase dependence, membrane homeostasis, stress tolerance, swimming motility, whole-cell proteome
ABSTRACT
TonB-ExbB-ExbD-like energy transduction systems are widespread among Gram-negative bacteria. While most species have only one copy of tonB-exbBD genes, the Pseudomonas species possess more TonB-ExbBD homologues. One of them, the TonB3-PocA-PocB complex, was recently shown to be required for polar localization of FlhF and, thus, the flagella in Pseudomonas aeruginosa. Here, we show that the orthologous TonBm-PocA-PocB complex is important for polar localization of FlhF and flagella in Pseudomonas putida as well. Additionally, the system is necessary for maintaining membrane integrity, as the inactivation of the TonBm-PocAB complex results in increased membrane permeability, lowered stress tolerance, and conditional cell lysis. Interestingly, the functionality of TonBm-PocAB complex is more important for stationary than for exponentially growing bacteria. The whole-cell proteome analysis provided a likely explanation for this growth phase dependence, as extensive reprogramming was disclosed in an exponentially growing tonBm deletion strain, while only a few proteomic changes, mostly downregulation of outer membrane proteins, were determined in the stationary-phase ΔtonBm strain. We propose that this response in exponential phase, involving, inter alia, activation of AlgU and ColR regulons, can compensate for TonBm-PocAB’s deficiency, while stationary-phase cells are unable to alleviate the lack of TonBm-PocAB. Our results suggest that mislocalization of flagella does not cause the membrane integrity problems; rather, the impaired membrane intactness of the TonBm-PocAB-deficient strain could be the reason for the random placement of flagella.
IMPORTANCE The ubiquitous Pseudomonas species are well adapted to survive in a wide variety of environments. Their success relies on their versatile metabolic, signaling, and transport ability but also on their high intrinsic tolerance to various stress factors. This is why the study of the stress-surviving mechanisms of Pseudomonas species is of utmost importance. The stress tolerance of Pseudomonads is mainly achieved through the high barrier property of their membranes. Here, we present evidence that the TonB-ExbBD-like TonBm-PocAB system is involved in maintaining the membrane homeostasis of Pseudomonas putida, and its deficiency leads to lowered stress tolerance and conditional cell lysis.
INTRODUCTION
The cell envelope of Gram-negative bacteria contains two membranes separated by a periplasmic space (1). Due to cell envelope architecture, no energy is produced in the outer membrane, which means that transport proteins that require energy need to import it from the cytoplasmic membrane. The energy transfer is carried out by the TonB-ExbB-ExbD complex in the inner membrane that harnesses the proton motive force and directly interacts with the TonB-dependent transporters in the outer membrane (2, 3). Once thought to be restricted only to iron and vitamin B12 transport, the TonB-ExbBD complex promotes the transport of a wide range of substrates that are too large or scarce to enter the cell by diffusion (4).
The TonB-ExbBD system is intensively studied in Escherichia coli, but the exact mechanism of its functioning in energy transduction has remained unclear. TonB and ExbD each have a transmembrane N terminus and are predominantly located in the periplasm, whereas ExbB has three transmembrane domains and a large cytoplasmic domain. ExbB forms a proton channel (5) and provides the structure for the complex (6). Depending on the membrane condition, ExbB proteins form either a pentameric or a hexameric channel with different conductance for protons (7). ExbD is predicted to be the carrier of protons during their translocation from periplasm to cytoplasm (8), but ExbD also interacts with TonB and appears to determine the right conformation of TonB during energy transmission (9, 10). TonB expands across the periplasm and transduces the harvested energy to the outer membrane transporters (11, 12) that have a TonB box domain. This domain is proposed to extend into the periplasm and become available for interaction with TonB after the binding of substrate to the transporter (13).
The TonB-ExbBD complexes are widespread among Gram-negative bacteria. Most species, including E. coli, have one copy of tonB-exbBD genes, but the number of tonB homologues can vary by up to nine per genome (14). The ubiquitous soil and rhizosphere bacterium Pseudomonas putida possesses two TonB homologues encoded by PP_4994 and PP_5308. PP_5308 (tonB) is in an operon with exbB and exbD, and their corresponding proteins appear to have a function similar to that of their E. coli homologs, as the P. putida strains defective in tonB-exbBD genes are impaired in siderophore transport and deficient in iron acquisition (15, 16). Moreover, the exbB of P. putida can complement the lack of exbB in E. coli (15). Besides its importance in iron transport, the TonB-ExbBD complex is required for P. putida for the tolerance of several antibiotics, p‐hydroxybenzoate, and toluene (16, 17) and affects its fitness in colonizing corn seeds and roots (18).
PP_4994 has not been studied in P. putida, but its conserved homologues can be found in most Pseudomonas species (19). The orthologue of PP_4994 in P. aeruginosa, TonB3, likely forms an inner membrane-associated complex with homologues of ExbB and ExbD, named PocA and PocB, respectively (20). Interestingly, P. aeruginosa’s TonB3-PocAB complex does not appear to have a role in promoting iron transport (21). Instead, it is involved in swimming and twitching ability (20, 22). The TonB3-PocAB complex, although not polarly localized itself, is needed for the polar localization of flagella and type IV pili (20). Deleting any one of the tonB3-pocAB genes in P. aeruginosa reduces swimming due to the random localization of flagella, which is caused by the random localization of otherwise polarly localized FlhF protein.
In this study, we investigate the role of PP_4994-encoded TonB and PP_1898-1899-encoded PocAB in P. putida. We show that like its orthologue in P. aeruginosa, the TonB-PocAB complex of P. putida is needed for correct placement of flagella and FlhF. For this shared role in motility, we name the PP_4994-encoded protein TonBm. Interestingly, our results also indicate that the TonBm-PocAB complex has a separate task in maintaining membrane integrity. The TonBm-PocAB-deficient strains have permeable membranes and are sensitive to several chemicals. Curiously, the effects of TonBm-PocAB’s deficiency depend on the physiological state of the inoculum as bacteria originating from stationary phase are significantly more compromised than the exponentially growing TonBm-PocAB knockout mutants. The comparison of whole-cell proteomes of the ΔtonBm strain and wild-type P. putida revealed extensive proteomic changes occurring in the exponentially growing but not in the stationary-phase ΔtonBm strain. We hypothesize that the observed changes compensate for the lack of tonBm in exponentially growing cells.
RESULTS
Inactivation of the TonBm-PocAB complex decreases swimming and results in membrane defects.
The ability of bacteria to bind Congo red (CR) dye has been used as a marker of membrane defects and cell lysis (23). Thus, we screened a transposon mutant library of the P. putida wild-type strain for CR binding mutants to find genes that could be important in membrane homeostasis. In accordance with previous studies (23, 24), the current screen repeatedly detected the CR binding mutants with transposon insertion into the colRS operon. ColR is the response regulator of the ColRS two-component regulatory system that responds to the excess of metals, such as zinc, iron, manganese, and cadmium (25). In the absence of this system, P. putida has problems with maintaining cell membrane integrity not only in the presence of excess metals (25, 26) but also on glucose minimal medium (23, 24). The second most common cause of pink colonies was disruption of PP_4994, which codes for an orthologue of P. aeruginosa’s TonB3. For one pink colony, the transposon had disrupted PP_1898, a probable orthologue of P. aeruginosa’s pocA gene (19). Identifying orthologues of genes that regulate flagellum localization in P. aeruginosa (20) in our membrane stress screen was intriguing, because assuming that PP_4994 and PocA of P. putida have functions similar to those of their orthologues, it is unclear why wrong localization of flagella and decreased motility result in membrane stress (23, 24).
To get further insight into the role(s) of PP_4994 and PocAB in P. putida, the PP_4994 (tonBm), PP_1898 (pocA), and PP_1899 (pocB) single deletion strains as well as the whole TonBm-PocAB complex deletion strain (ΔtonBm ΔpocAB) were constructed. As the decreased swimming ability of P. aeruginosa in the absence of the TonB3-PocAB system is caused by the nonpolar localization of FlhF (20), a protein that determines the place of flagellum formation (27), the flhF (PP_4343) deletion strain was constructed as well.
To test if tonBm, pocA, and pocB are required for motility in P. putida, the swimming ability of all the deletion strains was analyzed. Compared to the ΔflhF strain, which had a severe motility defect, the swimming ability of ΔtonBm, ΔpocA, ΔpocB, and ΔtonBm ΔpocAB strains was less affected but nevertheless clearly inhibited (Fig. 1A). The ΔtonBm ΔflhF double deletion strain displayed swimming defects similar to that of the ΔflhF strain, which is in good correlation with FlhF lying downstream of the TonB3-PocAB system in regulation of flagellum localization in P. aeruginosa (20). Complementation of the ΔtonBm strain with ectopically expressed tonBm restored normal motility (Fig. 1A).
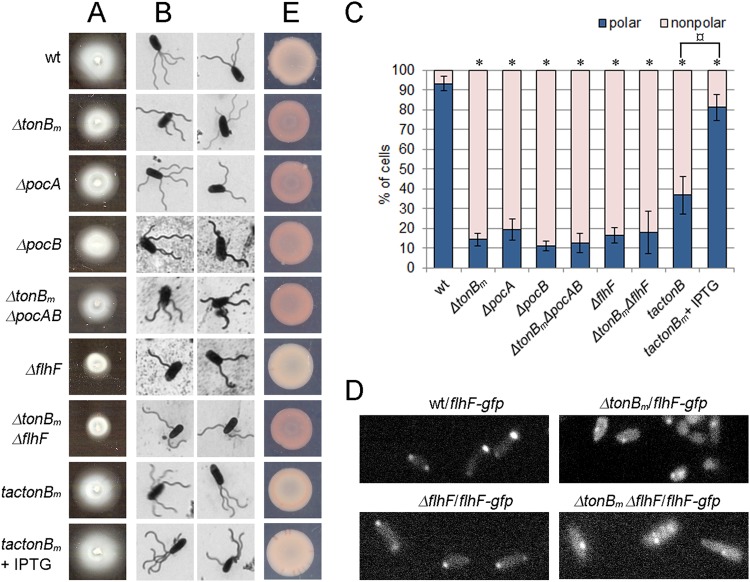
FIG 1.
Effect of TonBm-PocAB system’s deficiency in swimming ability (A), flagellum localization (B and C), FlhF positioning (D), and Congo red binding (E). The cells of P. putida wild type (wt), tonBm, pocA, pocB, and flhF single deletion strains, tonBm pocAB triple deletion strain, tonBm flhF double deletion strain, and tonBm deletion strain complemented with the tonBm gene (tac tonBm) and wild-type, ΔtonBm, ΔflhF, and ΔtonBm ΔpocAB strains carrying the flhF-gfp fusion under the control of the inducible Ptac promoter were grown at 30°C. The expression of TonBm and FlhF-GFP was induced with 0.5 mM IPTG. (A) Swimming ability. The cells were grown on LB medium containing 0.3% agarose for 18 h. (B) Localization of flagella. Exponentially growing cells (OD580 of ∼0.5) were stained with silver and examined using oil immersion light microscopy. (C) Quantification of flagellum localization. Flagellum localization was considered random (nonpolar) if at least one flagellum deviated from polar positioning. Relative proportions of cells with polar and nonpolar flagella (means with 95% confidence intervals from at least four independent experiments) are presented. Two hundred to 800 cells were examined for each strain. Statistically significant differences from the wild type (*, P < 0.001) and difference from the uninduced tac tonBm strain (¤, P = 0.000003) are indicated. (D) Localization of FlhF-GFP. The cells were grown overnight in LB medium supplemented with IPTG and examined using oil immersion light microscopy. (E) Congo red (CR) binding. The cells were grown on glucose minimal medium supplemented with 0.0005% CR for 72 h.
To determine if the swimming defect of TonBm-PocAB- and FlhF-deficient strains is caused by incorrect localization of flagella, the position of flagella was ascertained by microscopy. More than 90% of exponentially growing wild-type P. putida cells had all of their flagella (1 to 5 flagella) located at the center of the cell pole (Fig. 1B and C). However, in the TonBm-PocAB single and triple deletion strains, the uniform polar placement of flagella was lost and approximately 80% to 90% of cells had at least one incorrectly placed flagellum (Fig. 1B and C). The ΔflhF and ΔtonBm ΔflhF mutants had a random distribution of flagella similar to that of TonBm-PocAB-deficient strains (Fig. 1C), and their number of flagella per cell was decreased. Already without isopropyl-β-d-thiogalactopyranoside (IPTG) induction, the tonBm complementation strain showed more cells with only polar flagella (35%), which indicates that the tac promoter in the lacIq-Ptac-tonBm cassette was leaky. When the expression of TonBm was induced with 0.5 mM IPTG, about 80% of the cells of the tonBm complementation strain had polar flagella (Fig. 1C). To analyze the localization of FlhF, the flhF-gfp translational fusion was constructed and introduced into the chromosome of P. putida wild-type, ΔtonBm, ΔflhF, and ΔtonBm ΔflhF strains. Fluorescence microscopy showed that FlhF-green fluorescent protein (FlhF-GFP) is polar in wild-type and ΔflhF strains but locates randomly in ΔtonBm and ΔtonBm ΔflhF strains (Fig. 1D). We also observed that the FlhF-GFP foci were brighter in wild-type and ΔflhF bacteria than in the tonBm-deficient bacteria, where more FlhF-GFP seemed to localize in the cytoplasm. Thus, our data indicate that the P. putida TonBm-PocAB system has a role similar to that of TonB3-PocAB in P. aeruginosa.
To investigate whether the motility defect and random localization of flagella correlate with the CR-binding phenotype, the motility-deficient strains were tested on glucose medium supplemented with CR. Interestingly, while strains with the TonBm-PocAB-deficient system stained pink, the ΔflhF strain resembled wild-type P. putida (Fig. 1E). The control experiment with the tonBm complementation strain revealed that the CR binding phenotype of the ΔtonBm strain is suppressed by the overexpression of TonBm. These data show that the motility defect and wrong localization of flagella do not account for the CR binding of the ΔtonBm, ΔpocA, and ΔpocB strains and that in the absence of TonBm-PocAB system, cells must have additional problems not related to the misplacement of flagella.
The absence of TonBm-PocAB complex results in glucose-specific cell lysis.
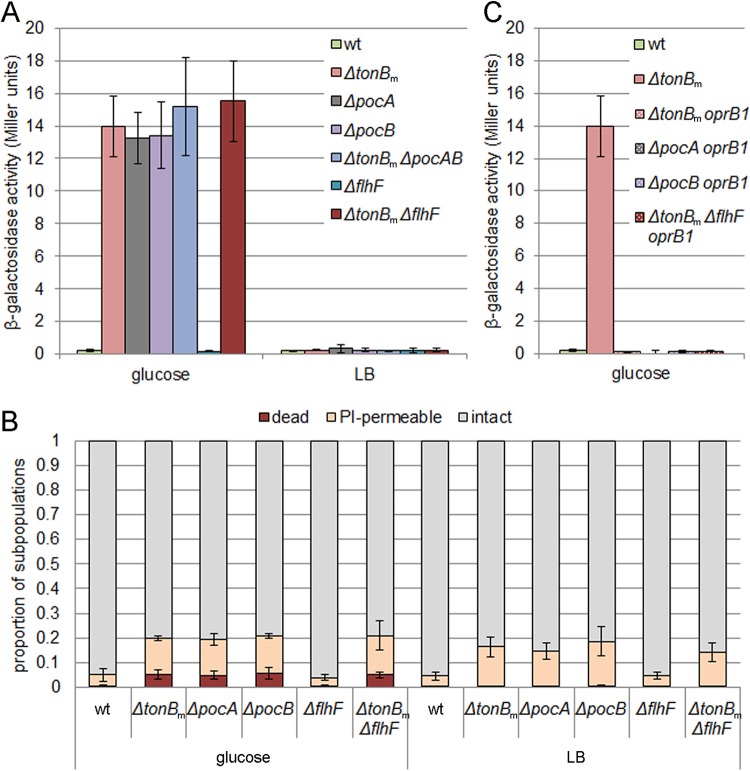
Given that the Congo red binding indicates a severe membrane defect (23, 24), we next tested the motility mutants for membrane leakage. We introduced the β-galactosidase expression plasmid into the wild-type and mutant strains and measured the β-galactosidase activity from the supernatant of glucose-grown bacteria. High extracellular β-galactosidase activity was detected in the case of ΔtonBm, ΔpocA, ΔpocB, ΔtonBm ΔpocAB, and ΔtonBm ΔflhF strains, indicating cell lysis (Fig. 2A). Contrary to this finding, the ΔflhF mutant displayed no β-galactosidase leakage.
FIG 2.
TonBm-PocAB-deficient strains have increased membrane permeability. (A) β-Galactosidase activities measured from the supernatant of glucose- and LB-grown P. putida wt, tonBm, pocA, pocB, and flhF single deletion, tonBm pocAB triple deletion, and tonBm flhF double deletion strains. All strains carry pKTlacZS/C plasmid. Data (means with 95% confidence intervals) from at least three independent experiments are presented. (B) Flow cytometry analysis of glucose- and LB-grown bacteria stained with SYTO9 and propidium iodide (PI). Relative proportions of the subpopulations of intact, PI-permeable, and dead cells (means with 95% confidence intervals) from at least four independent determinations are presented. (C) β-Galactosidase activities measured from the supernatant of glucose-grown P. putida wt, tonBm deletion, oprB1-deficient tonBm, and pocA, pocB, flhF, and tonBm flhF deletion strains. All strains carry pKTlacZS/C plasmid. Data (means with 95% confidence intervals) from at least three independent experiments are presented.
In order to test whether the lysis of the ΔtonBm, ΔpocA, and ΔpocB strains is dependent on the growth medium, the β-galactosidase leakage assay was performed with lysogeny broth (LB)-grown bacteria as well. However, as neither of the LB-grown strains differed from the wild type (Fig. 2A), the cell lysis of ΔtonBm, ΔpocA, and ΔpocB strains seems to be specific to growth on glucose.
Previous results with colR-deficient P. putida have shown that glucose-specific cell lysis is a subpopulation phenotype (24, 28). Therefore, to get insight into the population structure of TonBm-PocAB-deficient strains, flow cytometry analysis of bacteria stained with SYTO9 and propidium iodide (PI) was performed. Three populations could be detected: (i) undamaged cells that stained with SYTO9 only; (ii) cells that stained with both SYTO9 and PI (indicated as PI-permeable); and (iii) cells that stained with PI and SYTO9 but had lower side scatter. The latter subpopulation has been shown to correlate with cell lysis and therefore has been defined as dead cells (28). The single-cell analysis of the glucose solid medium-growing bacteria revealed that colonies of the ΔtonBm, ΔpocA, ΔpocB, and ΔtonBm ΔflhF strains contained more PI-permeable and dead cells than the wild type, whereas the deletion of flhF did not influence either the membrane permeability or death of bacteria (Fig. 2B). Interestingly, analysis of the mutants grown on LB solid medium also showed that the ΔtonBm, ΔpocA, ΔpocB, and ΔtonBm ΔflhF colonies contain more PI-permeable cells, but the amount of dead cells was similar to that of the wild type (Fig. 2B). The LB-grown ΔflhF strain had no difference from the wild type. These results show that while the deficiency in the TonBm-PocAB system increases membrane permeability regardless of the growth medium, the membrane damage is more pronounced on the glucose minimal medium. The unaffected membrane permeability of the ΔflhF strain indicates that wrong placement of flagella does not affect membrane permeability or cause cell lysis.
The inability to tolerate the increased expression of the sugar channel protein OprB1 in the outer membrane has been described as the reason for glucose-specific cell lysis in the colR-deficient mutant (23). To test if the glucose-induced OprB1 could be the reason for cell lysis of glucose-grown ΔtonBm, ΔpocA, ΔpocB, and ΔtonBm ΔflhF cells as well, oprB1-deficient derivatives were constructed from the deletion strains and β-galactosidase leakage was analyzed. Data shown in Fig. 2C indicated that disruption of oprB1 eliminated the glucose-dependent cell lysis of TonBm-PocAB-deficient strains. This indicated that, similar to the colR-deficient mutant, glucose-induced expression of OprB1 was involved in the glucose-specific cell lysis of TonBm-PocAB mutants.
TonBm-PocAB system affects stress tolerance and generation time in a growth phase-dependent manner.
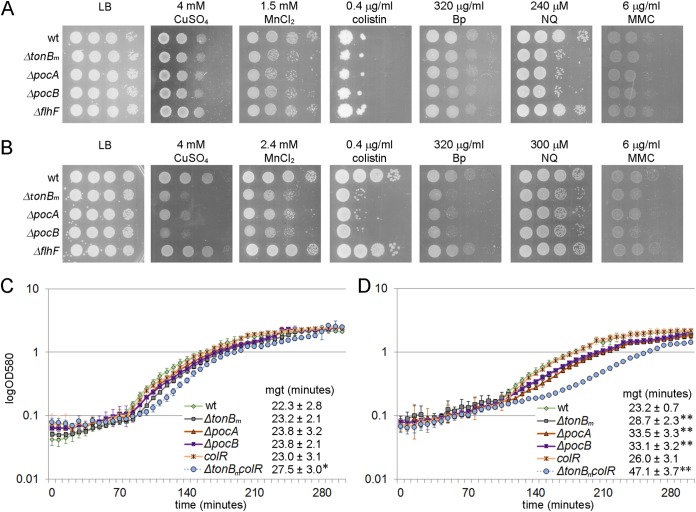
Since the flow cytometry analysis revealed that the TonBm-PocAB system contributes to membrane integrity, we hypothesized that the stress tolerance of these strains is affected. To test this, the growth of the ΔtonBm, ΔpocA, and ΔpocB strains and wild-type P. putida was compared on LB medium supplemented with different chemicals: metal salts (LiCl, NaCl, MnSO4, FeSO4, CoCl2, NiSO4, CuSO4, ZnSO4, RbCl, CdSO4, and K2Cr2O2), antibiotics (colistin, polymyxin B, benzylpenicillin, tetracycline, and rifampin), DNA-damaging chemicals (mitomycin C and 4-nitroquinoline 1-oxide), compounds producing reactive oxygen species (paraquat and H2O2), and EDTA. The ΔflhF strain was used as a control to rule out the effect of mislocalization of flagella on stress tolerance. During the experiments we observed that the results were dependent on the age of the inoculum; thus, the stress tolerance of cells from both stationary and exponential growth phases was analyzed. When the exponential-phase cells were tested, difference in growth could be observed only on media containing MnSO4 or 4-nitroquinoline 1-oxide (nitroquinoline) (Fig. 3A). However, when inoculated cells originated from the stationary growth phase, the TonBm-PocAB deletion strains revealed increased sensitivity to CuSO4, MnSO4, colistin, benzylpenicillin, nitroquinoline, and mitomycin C (Fig. 3B), and their growth was slightly decreased by ZnSO4, NaCl, and EDTA (data not shown). These results indicated that the TonBm-PocAB complex affected P. putida’s stress tolerance, but the magnitude of the effect depended extensively on the growth phase. Given that the ΔflhF strain resembled that of the wild type under all conditions, the increased stress sensitivity could not be caused by mislocalization of flagella. Although the TonBm-PocAB system was not involved in iron tolerance, we also investigated the growth of mutants under iron-limited conditions, but as no differences from the wild-type were detected (data not shown), the TonBm-PocAB complex is likely not involved in iron acquisition.
FIG 3.
TonBm-PocAB system affects stress tolerance and generation time in a growth phase-dependent manner. (A and B) Stress tolerance of exponential-phase (A) and stationary-phase (B) P. putida wt and tonBm, pocA, pocB, and flhF deletion strains. Exponential-phase cells were grown for 3 h (OD580 of ∼1) and stationary-phase cells overnight before inoculating the strains onto LB medium supplemented with different chemicals. The cells were then grown for 20 h at 30°C. Approximately 50,000, 5,000, 500, and 50 cells were inoculated per spot. Bp, benzylpenicillin; NQ, nitroquinoline; MMC, mitomycin C. (C and D) Growth curves and minimal generation times of bacterial cultures inoculated either with exponential-phase (C) or stationary-phase (D) cells. P. putida wt, tonBm, pocA, and pocB deletion strains and colR-deficient and tonBm colR double deficient strains were grown for 3 h (exponential phase) or overnight (stationary phase) before inoculating the cells into LB medium. The strains were grown at 30°C on a microtiter plate. Means from eight parallels of one measurement with 95% confidence intervals are presented. Average minimal generation times (mgt) from three independent measurements for each strain also are shown. Statistically significant differences from the wild type are indicated (*, P = 0.012; **, P < 0.0001).
To further analyze the growth phase effects of TonBm-PocAB-deficient P. putida, the LB medium was inoculated with bacteria of different ages and growth curves were recorded. While the maximum growth rate of the wild type remained the same independent of the inoculum used, the growth rates of TonBm-PocAB-deficient strains were clearly affected by the growth stage of the inoculum. When exponential-phase cells were used for inoculation, the growth curves and minimal generation times of ΔtonBm, ΔpocA, and ΔpocB strains resembled those of the wild type (Fig. 3C). However, when the culture was started with stationary-phase cells, the minimal generation time for the ΔtonBm and ΔpocAB strains was, on average, about 10 min longer (Fig. 3D) and had a statistically significant difference from the wild type. This furthermore demonstrates that the effect of TonBm-PocAB deficiency depends on the age of the P. putida cells.
MexCD-OprJ efflux system is downregulated in ΔtonBm strain in stationary phase.
Considering that the TonBm-PocAB system’s homologue in E. coli, the TonB-ExbBD complex, connects the two membranes by mediating the energy of cytoplasmic membrane to outer membrane transporters (2), we next analyzed different membrane fractions of the TonBm-PocAB-deficient strains from both exponential and stationary growth phases. While the patterns of lipopolysaccharides and periplasmic and cytoplasmic membrane proteins of wild-type and TonBm-PocAB deletion strains were similar (data not shown), the outer membrane protein (OMP) pattern of mutants revealed noticeable differences from the wild type (Fig. 4).
FIG 4.
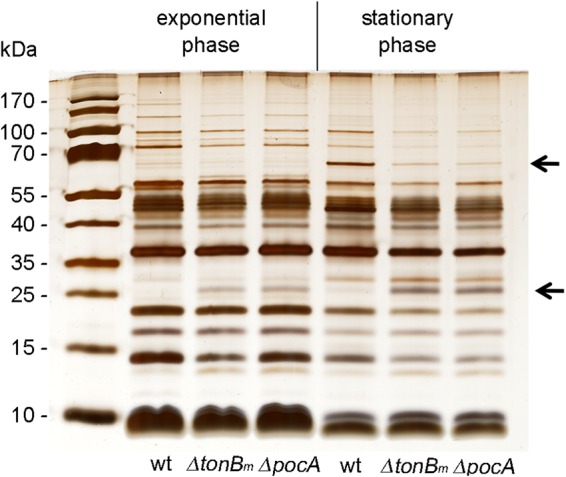
The absence of intact TonBm-PocAB complex causes changes in the outer membrane protein pattern. Outer membrane proteins were prepared from P. putida wt and tonBm and pocA deletion strains, separated by SDS-PAGE, and visualized by silver staining. Cells were grown in LB medium for 3 h (OD580 of ∼1; exponential phase) or 17 h (stationary phase). All lanes contain 1 μg of protein. Major differences between wild-type and tonBm-pocAB-deficient strains are indicated by arrows.
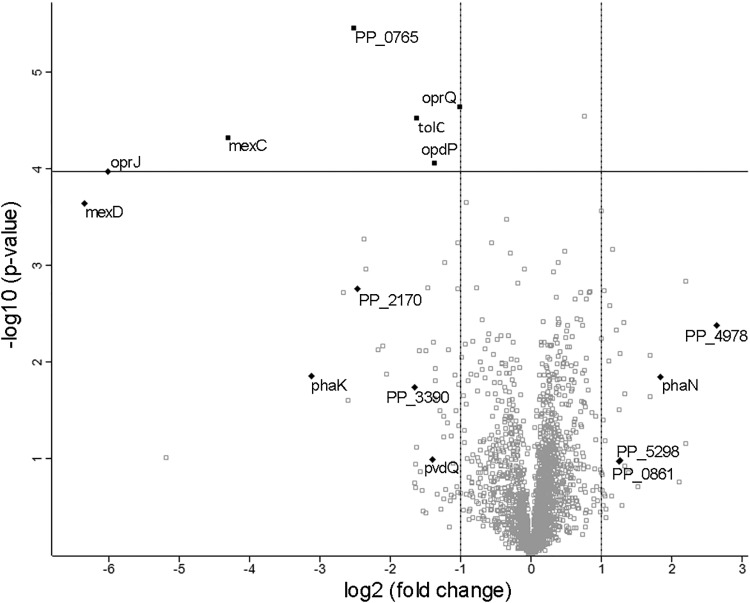
To get more detailed insight into the protein profile of the TonBm-deficient strain, a whole-cell proteome analysis of exponential- and stationary-phase ΔtonBm and wild-type P. putida cells was performed. Extensive growth phase-dependent proteome rearrangements were detected in both strains: 1,426 proteins out of 2,652 in the wild-type strain (see Table S1 in the supplemental material) and 1,326 proteins out of 2,673 in the ΔtonBm strain (Table S2) had a statistically significant and at least 2-fold difference between the growth phases. Interestingly, when the stationary-phase data of the wild type was compared to the corresponding ΔtonBm data, only six proteins were found to be significantly differentially expressed (Fig. 5 and Table S3). Besides these six proteins, we also considered nine differentially expressed proteins that were present in all the samples of one strain but were not detectable in any of the parallels of the other (so-called on-off-regulated proteins) and had at least a 2-fold difference in their expression level when imputed values were used. Notably, 11 of these 15 were membrane proteins, most of them being downregulated in the ΔtonBm strain (Fig. 5 and Table S3). The greatest differences were the 20-, 82-, and 65-fold decreases in the amount of MexC, MexD, and OprJ, respectively, which together form a multidrug efflux system that exports several antimicrobial agents in P. aeruginosa (29). The level of PhaK, an outer membrane channel protein that facilitates the uptake of phenylacetic acid (30), had dropped about 9-fold. This is probably caused by the 4-fold increased level of PhaN, a transcriptional repressor of the phenylacetic acid pathway (30).
FIG 5.
Volcano plot showing differences in protein expression between stationary-phase tonBm-deficient P. putida and the wild type. Filled symbols represent 15 proteins considered differently expressed in the ΔtonBm strain. Diamonds indicate on-off-regulated proteins. Proteins located above the horizontal line have statistically significant changes in their expression values.
In wild-type cells, MexCD-OprJ efflux pump components are upregulated in stationary phase (Table S1), whereas in the ΔtonBm strain, MexC is clearly downregulated and MexD and OprJ are not even detectable in stationary phase (Tables S2 and S3). This remarkable downregulation of MexCD-OprJ efflux pump proteins in the ΔtonBm strain led us to hypothesize that it was the reason for the decreased stress tolerance of the ΔtonBm strain. To test this possibility, we deleted the whole mexCD-oprJ operon from both wild-type and ΔtonBm strains and tested stress tolerance. The deletion of mexCD-oprJ had no effect on the tolerance of CuSO4, MnSO4, nitroquinoline, colistin, benzylpenicillin (Fig. 6), polymyxin B, and mitomycin C (data not shown) in either of the strains, suggesting that MexCD-OprJ did not contribute to the efflux of these compounds and that the downregulation of MexCD-OprJ complex was not the cause of elevated stress sensitivity of the ΔtonBm strain. On the other hand, if the MexCD-OprJ complex was related to the stress tolerance of the ΔtonBm strain, then its overexpression should have alleviated the increased sensitivity.
FIG 6.
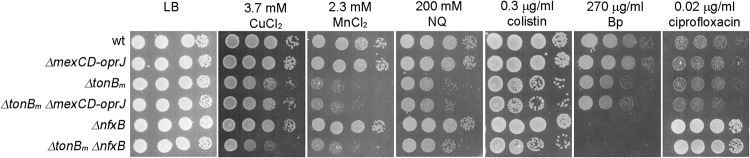
MexCD-OprJ effux pump is not related to the stress susceptibility of tonBm deletion strain. Overnight-grown P. putida wt, mexCD-oprJ, tonBm, and nfxB deletion strains and tonBm mexCD-oprJ and tonBm nfxB double deletion strains were grown on LB medium supplemented with different chemicals for 20 h at 30°C. Approximately 50,000, 5,000, 500, and 50 cells were inoculated per spot.
In P. aeruginosa, the expression of the mexCD-oprJ operon is controlled by two repressors, NfxB (31) and EsrC (32). The loss of NfxB leads to overexpression of mexCD-oprJ genes (33), whereas EsrC is active as a repressor only in the presence of NfxB and its effect on mexCD-oprJ is modest compared to that of NfxB (32). The homologue of NfxB in P. putida is PP_2820, and, expecting its absence to raise the expression of mexCD-oprJ, we constructed ΔnfxB (PP_2820) deletion strains of wild-type and ΔtonBm strains. In P. aeruginosa, the upregulation of MexCD-OprJ due to the loss of NfxB results in increased ciprofloxacin resistance (34). In accordance with that, the ciprofloxacin resistance of the ΔnfxB and ΔtonBm ΔnfxB strains was considerably higher than that of their parent strains (Fig. 6), suggesting that MexCD-OprJ was upregulated in the P. putida ΔnfxB strain. To assess if increased expression of MexCD-OprJ would relieve the reduced stress tolerance of the ΔtonBm strain, we next tested the ΔnfxB strain tolerance to several compounds. The results revealed that while the lack of nfxB decreased the benzylpenicillin tolerance of both wild-type and ΔtonBm strains (Fig. 6), it did not influence the tolerance to other chemicals. These results support the contention that low levels of MexCD-OprJ are not sufficient to explain the decreased stress tolerance of the TonBm-PocAB-deficient strain.
Proteome analysis revealed extensive changes in exponentially growing ΔtonBm cells.
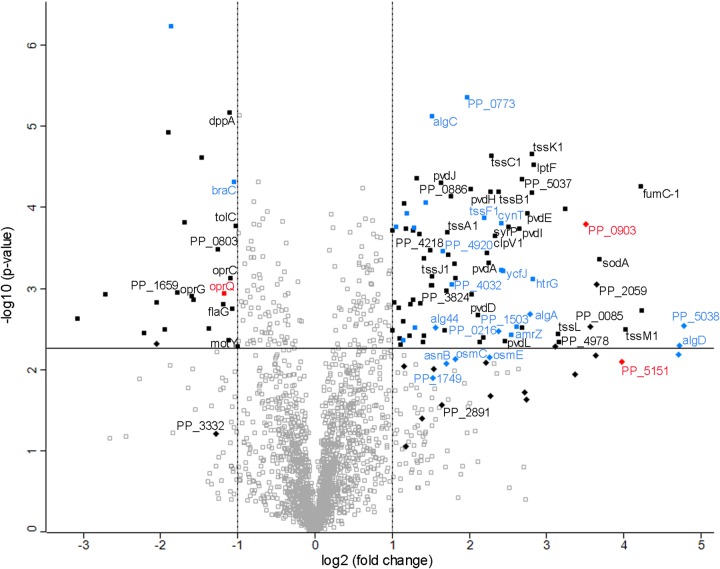
Contrary to the few differences in stationary phase, the proteome analysis of exponential-phase cells showed at least a 2-fold change of 126 proteins in the ΔtonBm strain compared to level for the wild type (Fig. 7 and Table S4). One hundred eight of these changes were statistically significant, but we also included 18 on-off proteins (Table S4). It is remarkable that most of the proteins, 102 out of 126, were upregulated in the ΔtonBm strain (Fig. 7 and Table S4). Only the outer membrane proteins that predominantly belonged to the transport and secretion category were mainly downregulated. About a third of all the changed proteins were associated with either amino acid and protein metabolism (17 proteins) or with general metabolism (22 proteins), suggesting that a considerable metabolic reprogramming was induced in the exponentially growing ΔtonBm strain. Another large group comprised 29 proteins related to transport and secretion. Half of these proteins were downregulated in the ΔtonBm strain and half, including 10 proteins of the type VI secretion system K1 (35), were upregulated (Table S4). It is also noteworthy that 13 proteins associated with stress and defense responses and 10 transcriptional regulators or histidine kinases were responding to TonBm deficiency, and again, most of them were upregulated. Surprisingly, only five proteins (OprQ, OpdP, TolC, PP_0765, and PP_4978) overlapped the differentially expressed proteins of stationary phase.
FIG 7.
Volcano plot showing differences in protein expression between exponentially growing tonBm-deficient P. putida and the wild type. Filled symbols represent 126 proteins considered differently expressed in the ΔtonBm strain. Diamonds indicate on-off proteins. Proteins belonging to the AlgU regulon are shown in blue, and proteins regulated by ColR are in red. A name tag indicates that the protein was also differently expressed in the zinc-exposed colR-deficient strain or is ColR regulated (also in red). Proteins located above the horizontal line have statistically significant changes in their expression values.
AlgU and ColR regulons are activated in ΔtonBm strain.
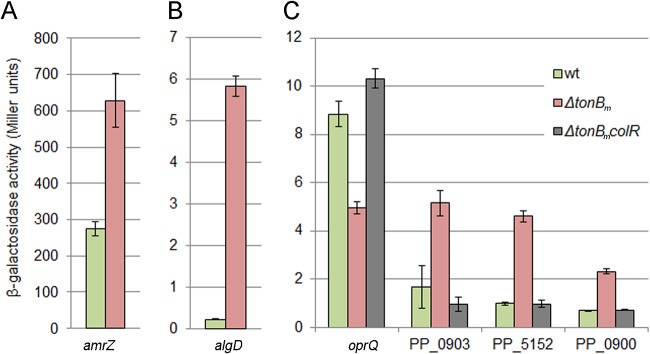
The proteome data of exponential-phase bacteria revealed that several alginate biosynthesis proteins as well as the alginate regulator AmrZ were upregulated in the ΔtonBm strain (Fig. 7 and Table S4). These results were verified by analysis of amrZ-lacZ and algD-lacZ transcriptional fusions in the ΔtonBm strain, showing an increased promoter activity (Fig. 8A and B). Alginate is an exopolysaccharide that contributes to biofilm formation and stress tolerance under water-limiting conditions in P. putida (36). In P. aeruginosa, alginate is produced in response to cell wall stress due to the activation of sigma factor AlgU (37). Analysis of the proteome response of the ΔtonBm strain revealed that 31 out of 126 differentially expressed proteins were orthologues of the P. aeruginosa AlgU regulon (Fig. 7, marked with blue) (37). This suggests that the AlgU regulon is activated in the exponentially growing ΔtonBm strain.
FIG 8.
AlgU and ColR regulon genes respond to tonBm deficiency. (A and B) β-Galactosidase activities measured in P. putida wt and tonBm deletion strains carrying the amrZ or algD transcriptional fusion with lacZ in the plasmid p9TTBlacZ. (C) β-Galactosidase activities measured in P. putida wt, tonBm deletion, and colR-deficient tonBm deletion strains carrying the oprQ, PP_0903, PP_5152, or PP_0900 transcriptional fusion with lacZ. Bacteria were grown in LB medium for 3 h (OD580 of ∼1) at 30°C. Data (means with 95% confidence intervals) from at least four independent experiments are presented.
Besides activation of the AlgU regulon, three proteins of the ColR regulon, OprQ, PP_0903, and PP_5151 (25), responded to TonBm deficiency (Fig. 7, indicated in red) in a direction indicating that the ColRS signaling is active in the ΔtonBm strain. To test this possibility, the ColR-responsive oprQ-lacZ, PP_0903-lacZ, and PP_5152-lacZ transcriptional fusions (PP_5152 is the first gene in the two-gene PP_5152-PP_5151 operon) were analyzed in exponentially grown wild-type, ΔtonBm, and ΔtonBm colR double mutant strains (Fig. 8C). The expression of oprQ is known to be repressed, and the expression of PP_0903 and PP_5152 activated by ColR (25, 38). In accordance with that and verifying the changes seen in the proteome data (Fig. 7), the promoter activity of oprQ was lower and the activities of PP_0903 and PP_5152 were higher in the ΔtonBm than the wild-type strain (Fig. 8C). Given that none of the three promoters responded to the lack of tonBm in the ΔtonBm colR strain (Fig. 8C), ColR is clearly responsible for the altered expression of those genes in the ΔtonBm strain. To further confirm if ColRS signaling is active in TonBm-PocAB-deficient cells, the expression of the ColR-responsive PP_0900, which was not detectable in the proteome, was also analyzed. Corroborating the prior results, the PP_0900-lacZ fusion was induced in TonBm-deficient cells in a ColR-dependent manner (Fig. 8C), confirming the activation of the ColRS system in response to TonBm-PocAB deficiency.
Activation of the ColR regulon is beneficial for the ΔtonBm strain.
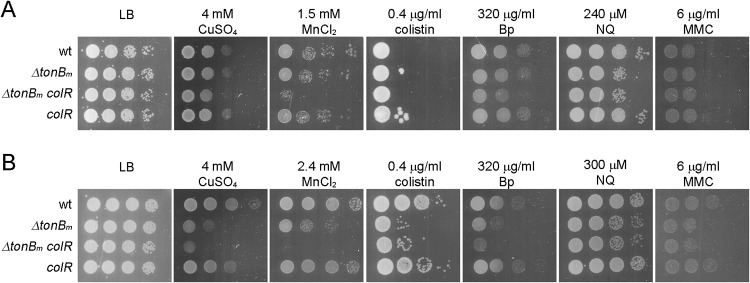
The proteome of colR-deficient P. putida recently has been analyzed (26), and the comparison of these previous data to the current data revealed a remarkable overlap between the proteome responses of exponentially grown ΔtonBm and zinc-stressed colR mutant strains (26). The same 61 proteins, including, for example, several transport and stress response proteins, were differentially expressed in both mutant strains, and all in the same manner (Fig. 7 [the overlapping 61 proteins are indicated with name tags] and Table S4). Considering this partial overlap of the proteome responses and several shared membrane stress-indicating phenotypes (glucose-induced Congo red binding and cell lysis) of ΔtonBm and colR mutant cells (Fig. 1 and 2) (23), it was hypothesized that the activation of ColRS signaling could alleviate the membrane damage of the ΔtonBm mutant. To test this, the growth parameters and stress tolerance of the ΔtonBm colR double mutant was analyzed. The growth curve of the combined mutant in LB medium displayed a prolonged lag phase that was particularly apparent when stationary-phase cells were used as an inoculum (Fig. 3C and D). The minimal generation time of the ΔtonBm colR strain was, on average, about 5 min longer than that of other strains when exponential-phase cells were analyzed (Fig. 3C). The growth rate reduction of the double mutant was even more pronounced when stationary-phase inocula were tested (Fig. 3D). The ΔtonBm colR double mutant was also more compromised in some stress tolerance tests. Exponential-phase ΔtonBm colR cells were less tolerant to MnCl2 than the tonBm single mutant (Fig. 9A), and the stationary-phase ΔtonBm colR double mutant tolerated less CuSO4, MnCl2, and benzylpenicillin than the ΔtonBm strain (Fig. 9B). Given that colR deficiency clearly intensifies the phenotypes of the ΔtonBm strain, the activation of the ColR regulon seems to be beneficial to P. putida lacking TonBm. The accumulation of the effects of tonBm and colR deficiency suggests that the origin of the problems in these strains is different, which implies that although the lack of TonBm-PocAB and ColRS produces similar responses, the two systems operate in separate regulatory pathways.
FIG 9.
Stress tolerance of exponential-phase (A) and stationary-phase (B) P. putida wt, tonBm deletion, colR-deficient tonBm deletion, and colR-deficient strains. Exponential-phase cells were grown for 3 h (OD580 of ∼1) and stationary-phase cells overnight before inoculating the strains onto LB medium supplemented with different chemicals. The cells were then grown for 20 h at 30°C. Approximately 50,000, 5,000, 500, and 50 stationary-phase cells were inoculated per spot.
DISCUSSION
Prior to the current work, the homologue of the TonBm-PocAB complex, TonB3-PocAB, was shown to be required for swimming and twitching in P. aeruginosa (20, 22). In the absence of tonB3, pocA, or pocB, the swimming ability of P. aeruginosa is impaired because the polar localization of flagella is lost due to the random positioning of FlhF (20), which marks the assembly point of new flagella (27). The present study suggests that the TonB-PocAB complex has the same function in other Pseudomonas species, as P. putida tonBm, pocA, and pocB deletion strains have randomly distributed flagella and decreased swimming ability as well. However, our results indicate that besides the role in motility, the TonBm-PocAB complex is needed for the maintenance of membrane integrity. We show that the misplacement of flagella is not responsible for the membrane defects of TonBm-PocAB mutants and rather suggests that the impaired membrane homeostasis is the main cause for the random localization of flagella as well as for other deficiencies.
The subpopulation lysis of TonBm-PocAB mutants on glucose medium, evidenced by the leakage of cytoplasmic β-galactosidase (Fig. 2A), clearly shows that these mutants have a fragile membrane. The finding that glucose-specific cell lysis was abolished by the deletion of oprB1 indicates that TonBm-PocAB-deficient strains cannot tolerate OprB1 porin. OprB1 is a carbohydrate-selective porin that has a relatively high affinity for glucose (39) and is significantly upregulated when glucose becomes limiting (23). In nutrient-rich LB medium, glucose transport is inhibited, as Pseudomonas species prefer to use organic acids and amino acids as a carbon source instead (40, 41). A similar glucose-specific and OprB1-dependent cell lysis has been previously described for a P. putida colR-deficient strain (23, 24). The lysis of the colR mutant was shown to result from the intolerance of the accumulation of OprB1 in the outer membrane, the cell’s normal response to glucose limitation (23). Given that the TonBm-PocAB-deficient strains have similar hypersensitivities to the OprB1 porin, we conclude that the outer membrane of the TonBm-PocAB mutants is compromised. Furthermore, single-cell analysis revealed that, independent of the carbon source, the populations of TonBm-PocAB mutants contain significantly more PI-permeable cells than the wild type (Fig. 2B). As propidium iodide can pass through undamaged outer membrane via porins but cannot cross intact cytoplasmic membrane (42), the increased permeability to PI implies that not only the outer membrane but also the inner membrane of the ΔtonBm mutant is deficient.
The stress tolerance assays suggested that the fitness effects of TonBm-PocAB’s disruption were more pronounced when stationary-phase bacteria were analyzed (Fig. 3A and B). Considering that the growth characteristics of the ΔtonBm strain also depended on the growth phase of the inoculum under unstressed conditions (Fig. 3C and D), the TonBm-PocAB complex seems to be more important for stationary- than for exponential-phase bacteria. The proteome analysis provided a likely explanation for this growth phase dependence, namely, extensive differences, including upregulation of many metabolism-related stress response and regulatory proteins, were observed in the exponentially growing ΔtonBm strain, while only a few proteomic changes were detected in the stationary-phase ΔtonBm strain. This suggests that exponentially growing cells are actively dealing with the stress caused by the absence of functional TonBm-PocAB, while stationary-phase cells lack this reprogramming, perhaps due to energy limitation, and cannot cope with the effects of TonBm-PocAB deficiency. Thus, we assume that the ability of exponentially growing cells to compensate for the absence of the TonBm-PocAB system allows them to preserve the wild-type-like growth rate in rich medium and start growth in the presence of stress.
Besides growth phase differences, the proteome data confirmed that the TonBm-PocAB complex is necessary for maintaining membrane integrity. First, most of the proteins with changed expression in stationary-phase ΔtonBm cells were membrane proteins that were upregulated in the wild-type but not in the ΔtonBm strain. This hints that the cell envelope of the TonBm-PocAB-deficient strain is sensitive to membrane protein upregulation, somewhat analogously to the sensitivity of OprB1 in glucose medium. Second, as the expression of 31 AlgU regulon proteins was found to be changed in the exponentially growing ΔtonBm strain, AlgU seems to be activated in the ΔtonBm strain. AlgU is an envelope stress response sigma factor that controls the expression of large numbers of genes (37). Under normal growth conditions, AlgU is bound to the anti-sigma factor MucA, which anchors it to the cytoplasmic membrane and prohibits it from regulating gene expression (43). Under cell envelope stress, induced either by cell wall-acting antibiotics or other compounds that disrupt bacterial membranes (37, 44) or by the overexpression of certain outer membrane proteins (43), MucA is degraded and AlgU is released into the cytoplasm (37). Therefore, the activation of the AlgU regulon indicates that the TonBm-PocAB-deficient cells must experience envelope stress. Third, a large part of the differentially expressed proteins in exponentially growing ΔtonBm strain (61 out of 126) overlapped the proteome response previously observed in colR-deficient P. putida treated with ZnSO4 (26). This included many AlgU-regulated and stress-related proteins, the expression of which could be explained by the activation of AlgU in both strains, but also several other proteins, such as pyoverdine synthesis and type VI secretion system proteins. Analogously to AlgU regulon genes, the pyoverdine genes in P. aeruginosa are controlled by extracytoplasmic-function (ECF) sigma factors such as PvdS, FpvI, and SigX (45, 46). Given that activation of ECF sigma factors depends on transmembrane signaling, the upregulation of pyoverdine synthesis proteins may result from membrane stress as well. Since the ΔtonBm strain and the colR mutant possess several common traits indicating their membrane deficiency, the overlapping proteome response can be considered an indicator of a similar type of envelope stress of the two mutants. However, we should emphasize that compared to TonBm-PocAB-deficient strains, the inactivation of ColRS signaling results in significantly milder phenotypes: colR mutant displays a lower level of glucose-dependent lysis and higher stress tolerance (except for metals like zinc, iron, and cadmium) and has no swimming deficiency (Fig. 9 and data not shown).
In addition to the AlgU regulon, the ColR regulon is activated in the ΔtonBm strain as well. This response most likely can somewhat alleviate the envelope stress, because the ΔtonBm colR double mutant displays a stronger growth defect (Fig. 3C and D) and has lower stress tolerance than the strain deficient only in tonBm (Fig. 9A and B). This suggests that the ColRS and TonBm-PocAB systems have some overlap in their roles in membrane homeostasis. The activation of the ColR regulon in LB-growing ΔtonBm cells was somewhat surprising, particularly considering that ColS is a sensor that recognizes the excess of certain metals (25). However, there are two-component systems like PhoP/PhoQ that can sense different stimuli and also detect physical properties of the membrane (47–49). The current study suggests that, besides metals, ColS also senses membrane integrity and ColRS signaling can be triggered by membrane damage.
Both TonBm-PocAB- and FlhF-deficient mutants have randomly placed flagella (Fig. 1B), but only TonBm-PocAB deficiency results in membrane stress, indicating phenotypes like glucose-dependent lysis (Fig. 2A), increased membrane permeability (Fig. 2B), and lowered stress tolerance (Fig. 3). This shows that abnormal placement of flagella per se is not causing any membrane defects. Notably, in P. aeruginosa, the positioning of not only FlhF but also CheA, a chemotaxis histidine kinase with unipolar localization, becomes random in TonB3-PocAB-deficient strains (20), indicating that the TonB3-PocAB complex has a more general role in determining polar localization of different protein complexes. Interestingly, a somewhat similar role in the maintenance of membrane integrity as well as in the polar positioning of certain proteins has been reported for the Tol-Pal complex (50, 51). The transenvelope Tol-Pal complex consists of an outer membrane lipoprotein, Pal, a periplasmic protein, TolB, and inner membrane-situated TolA, TolQ, and TolR that are paralogous to TonBm, PocA, and PocB, respectively. The Tol-Pal complex is part of the cell division machinery. It localizes to the division plane in early predivisional cells, assists proper invagination of the outer membrane, and remains at the new pole until division is completed (51, 52). Besides that, the Tol-Pal complex interacts with chemoreceptors and is required for maintaining the polar positioning of chemoreceptor clusters (50). It is proposed that the Tol-Pal complex physically restricts the departure of the chemoreceptor clusters from the poles after cell division. However, not all polarly localized proteins in E. coli require Tol-Pal for their maintenance in the pole (50). Given that the TonB3-PocAB complex itself is not polarly localized (20), the mechanism of how it determines the polar placement of FlhF and flagella should be indirect and likely does not resemble the mechanism described for the Tol-Pal complex. While the requirements for polar positioning of FlhF are not known, several characteristics of the cell pole can be considered. For example, it has been hypothesized that FlhF detects membrane curvature or recognizes specific proteins or lipids (e.g., cardiolipin and phosphatidylethanolamine) that are enriched at cell poles (53–56). It cannot be ruled out that the TonB-PocAB complex controls the polar placement of FlhF via a specific regulatory mechanism. Still, in light of the current results, we propose that the yet-undetermined polar marker for FlhF is altered due to the membrane defect of the TonBm-PocAB mutant, which then results in the characteristic mislocalization of flagella.
TonB-like proteins connect the inner and outer membrane, as they are situated in the inner membrane and interact with outer membrane proteins. For instance, TonB of E. coli interacts with TonB-dependent outer membrane transporters to mediate siderophore uptake (12, 57). While it is reasonable to assume that the TonBm-PocAB complex has a similar role in bridging the two membranes in P. putida, our attempts to detect the potential interaction partners of TonBm have been unsuccessful so far (data not shown). Hopefully, further studies will reveal the putative interaction partners of TonBm and disclose the true mechanism of how TonBm-PocAB maintains membrane integrity and polar positioning of flagella.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used are listed in Table 1. All strains are derivatives of P. putida PaW85 (58), which is isogenic to the fully sequenced KT2440 (59). Bacteria were grown in lysogeny broth (LB) or on minimal medium (60) containing 0.2% glucose. When selection was necessary, the growth medium was supplemented with benzylpenicillin (800 μg/ml), kanamycin (50 μg/ml), or streptomycin (200 μg/ml) for P. putida and kanamycin (50 μg/ml) or streptomycin (20 μg/ml) for E. coli. E. coli was incubated at 37°C and P. putida at 30°C. Bacteria were electrotransformed according to the protocol of Sharma and Schimke (61).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or characteristics | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli CC118 λpir | Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA1 λpir lysogen | 73 |
| Pseudomonas putida | ||
| PaW85 | Wild type, isogenic to KT2440 | 58 |
| colR strain | PaW85 colR::Km (Kmr) | 74 |
| ΔtonBm strain | PaW85 ΔPP_4994 | This study |
| ΔpocA strain | PaW85 ΔPP_1898 | This study |
| ΔpocB strain | PaW85 ΔPP_1899 | This study |
| ΔtonBm ΔpocAB strain | PaW85 ΔPP_4994 ΔPP_1898-PP_1898 | This study |
| ΔflhF strain | PaW85 ΔPP_4343 | This study |
| ΔtonBm ΔflhF strain | ΔtonBm ΔflhF | This study |
| ΔtonBm tac-tonBm strain | ΔtonBm with genomic lacIq-Ptac-tonBm expression cassette (Smr) | This study |
| wt/flhF-gfp strain | Wild type with genomic lacIq-Ptac-flhF-gfp expression cassette (Gmr) | This study |
| ΔtonBm/flhF-gfp strain | ΔtonBm with genomic lacIq-Ptac-flhF-gfp expression cassette (Gmr) | This study |
| ΔflhF/flhF-gfp strain | ΔflhF with genomic lacIq-Ptac-flhF-gfp expression cassette (Gmr) | This study |
| ΔtonBm ΔflhF/flhF-gfp strain | ΔtonBm ΔflhF with genomic lacIq-Ptac-flhF-gfp cassette (Gmr) | This study |
| ΔtonBm oprB1 strain | ΔtonBm and PP_1019::Sm (Smr) | This study |
| ΔpocA oprB1 strain | ΔpocA and PP_1019::Sm (Smr) | This study |
| ΔpocB oprB1 strain | ΔpocB and PP_1019::Sm (Smr) | This study |
| ΔtonBm ΔflhF-oprB1 strain | ΔtonBm ΔflhF PP_1019::Sm (Smr) | This study |
| ΔmexCD-oprJ strain | PaW85 ΔPP_2817-2819 | This study |
| ΔtonBm ΔmexCD-oprJ strain | ΔtonBm ΔPP_2817-2819 | This study |
| ΔnfxB strain | PaW85 ΔPP_2820 | This study |
| ΔtonBm ΔnfxB strain | ΔtonBm ΔPP_2820 | This study |
| ΔtonBm colR strain | ΔtonBm colR::Km (Kmr) | This study |
| Plasmids | ||
| pUTTn5Sm-lacItac | Delivery plasmid for miniTn5Sm-lacItac (Apr Smr) | 75 |
| pRK2013 | Helper plasmid for conjugal transfer (Kmr) | 64 |
| pKTlacZS/C | Transcriptional fusion of Tn4652 tnpA with lacZ in pKTlacZ | 76 |
| pEMG | Plasmid for homologous recombination, lacZa with two flanking I-SceI sites (Kmr) | 63 |
| pEMG/ΔtonBm | pEMG containing chimeric DNA fragment for deleting PP_4994 (Kmr) | This study |
| pEMG/ΔpocA | pEMG containing chimeric DNA fragment for deleting PP_1898 (Kmr) | This study |
| pEMG/ΔpocB | pEMG containing chimeric DNA fragment for deleting PP_1899 (Kmr) | This study |
| pEMG/ΔpocAB | pEMG containing chimeric DNA fragment for deleting PP_1898 and PP_1899 (Kmr) | This study |
| pEMG/ΔflhF | pEMG containing chimeric DNA fragment for deleting PP_4343 (Kmr) | This study |
| pEMG/ΔmexCD-oprJ | pEMG containing chimeric DNA fragment for deleting ΔPP_2817-2819 operon (Kmr) | This study |
| pEMG/ΔnfxB | pEMG containing chimeric DNA fragment for deleting PP_2820 (Kmr) | This study |
| pSW (I-SceI) | Plasmid coding for I-SceI endonuclease for allelic exchange experiments (Bpr) | 77 |
| p704L/oprB1::Sm | pGP704L with EcoRI fragment of oprB1::Sm from pKS/oprB1::Sm (Apr Smr) | 78 |
| pBRlacItac | Expression vector containing lacIq repressor-controlled Ptac promoter (Ampr) | 79 |
| pBRlacItac/tonBm | pBRlacItac containing tonBm in a 1,044-bp HindIII-XbaI fragment under the Ptac promoter (Ampr) | This study |
| pUCNotKm | Cloning vector (Kmr) | 80 |
| pUCNotKm/tactonBm | pUCNot containing lacIq-Ptac-tonBm expression cassette (Ampr) | This study |
| pBK-miniTn7-ΩSm | pUC19-based delivery plasmid for miniTn7-ΩSm (Ampr Smr) | 81 |
| pminiTn7tac-tonBm | pBK-miniTn7-ΩSm containing lacIq-Ptac-tonBm expression cassette (Ampr Smr) | This study |
| pKTlacItac | Expression vector | 82 |
| pKTlacItac/flhF-gfp | Plasmid for expression of FlhF-GFP fusion protein (Ampr) | This study |
| pGP-mini-Tn7-ΩGm | Delivery plasmid for mini-Tn7-ΩGm (Ampr Gmr) | 83 |
| pGPminiTn7Gm/lacItac-flhF-gfp | Delivery plasmid for miniTn7Gm-lacItac-flhF-gfp (Ampr Gmr) | This study |
| pUXBF13 | Plasmid coding for the Tn7 transposition proteins (Ampr, mob+) | 84 |
| p9TTBlacZ/algD | PP_1288 promoter fused with lacZ in p9TTBlacZ (Ampr Cmr) | 78 |
| p9TTBlacZ/amrZ | PP_4470 promoter fused with lacZ in p9TTBlacZ (Ampr Cmr) | 26 |
| p9TTBlacZ/oprQ | PP_0268 promoter fused with lacZ in p9TTBlacZ (Ampr Cmr) | 78 |
| p9TTBlacZ/903 | PP_0903 promoter fused with lacZ in p9TTBlacZ (Ampr Cmr) | 38 |
| p9TTBlacZ/5152 | PP_5152 promoter fused with lacZ in p9TTBlacZ (Ampr Cmr) | 25 |
| p9TTBlacZ/900 | PP_0900 promoter fused with lacZ in p9TTBlacZ (Ampr Cmr) | 38 |
P. putida transposon mutant library screening for identification of Congo red binding mutants.
Wild-type P. putida was subjected to mutagenesis using a Tn5-based minitransposon, miniTn5Sm-lacItac. Plasmid pUTTn5Sm-lacItac was conjugatively transferred from the E. coli CC118 λpir strain into P. putida PaW85 with the aid of the helper plasmid pRK2013. Transconjugants with random chromosomal insertions of the minitransposon were selected on 0.2% glucose minimal plates supplemented with streptomycin, Congo red (0.0005%), and 2 mM phenol. We searched for pink colonies among the white ones. Pink colonies were analyzed by arbitrary PCR and sequencing. PCR products were generated by two rounds of amplification as described elsewhere (62). In the first round, oligonucleotides prtac, specific for the tac promoter, and the arbitrary Arb6 were used as primers. Second-round PCR was performed with the primers OEint and Arb2. Screening of about 35,000 transposon mutants yielded five independent transposon insertions into the tonBm gene and one into the pocA gene.
Construction of plasmids and strains.
For the generation of deletion strains, the pEMG-based plasmids were constructed according to a protocol described elsewhere (63). The upstream and downstream regions (about 500 bp) of the gene(s) to be deleted were amplified separately and then joined into an approximately 1-kb fragment by overlap extension PCR. Oligonucleotides used in PCR amplifications are listed in Table 1. For construction of the plasmid pEMG/ΔtonBm, the PCR fragment was cut with BamHI and EcoRI. For construction of the pEMG/ΔpocA, pEMG/ΔpocB, pEMG/ΔpocAB, pEMG/ΔflhF, and pEMG/ΔmexCD-oprJ plasmids, the PCR fragments were cut with XbaI and EcoRI. For construction of pEMG/ΔnfxB, EcoRI and SacI were used. The cut fragments were then ligated into the corresponding sites of the plasmid pEMG. The obtained pEMG plasmids were delivered to P. putida PaW85 or its deletion strains by electroporation, and after 2.5 h of growth in LB medium the bacteria were plated onto LB agar supplemented with kanamycin. Kanamycin-resistant cointegrates were selected and electrotransformed with the I-SceI expression plasmid pSW(I-SceI). To resolve the cointegrate, the plasmid-encoded I-SceI was induced with 1.5 mM 3-methylbenzoate overnight. Kanamycin-sensitive colonies were selected and the deletions were verified by PCR. The plasmid pSW(I-SceI) was eliminated from the deletion strains by growing them overnight in LB medium without antibiotics.
For complementation of the ΔtonBm strain, the tonBm gene amplified with the oligonucleotides 4994alg and 4994lopp was first cloned under the control of the tac promoter and lacIq repressor in pBRlacItac. The lacIq-Ptac-tonBm cassette was excised from pBRlacItac/tonBm with BamHI and subcloned into BamHI-opened pUCNotKm, resulting in pUCNotKm/tactonBm. Finally, the TonBm expression cassette was inserted as a NotI fragment into the minitransposon delivery vector pBK-miniTn7-ΩSm. The obtained pminiTn7Sm/tactonBm was introduced to the E. coli CC118 λpir strain and conjugatively transformed into the P. putida ΔtonBm strain with the help of pRK2013. The chromosomal presence of the lacIq-Ptac-tonBm cassette was verified by PCR.
For construction of C-terminal fluorescent fusion to FlhF, the flhF gene was PCR amplified using primers flhFEco and flhFXho. The EcoRI-XhoI-cleaved flhF fragment was then used to replace the colR gene in the colR-gfp translational fusion in plasmid pKTlacItac/colR-gfp (laboratory collection). The lacIq-Ptac-flhF-gfp cassette then was inserted into SmaI-KpnI-opened miniTn7 delivery plasmid pGP-miniTn7-ΩGm. The plasmid pGPminiTn7Gm/lacItac-flhF-gfp was introduced into the P. putida wild-type, ΔtonBm, ΔflhF, and ΔtonBm ΔflhF strains by coelectroporation together with the helper plasmid pUXBF13. The presence of the lacIq-Ptac-flhF-gfp cassette in the attTn7 site was verified by PCR.
For construction of oprB1-deficient strains, p704L/oprB1::Sm plasmid was introduced into the E. coli CC118 λpir strain and conjugatively transformed into P. putida ΔtonBm, ΔpocA, ΔpocB, and ΔtonBm ΔflhF strains with the help of pRK2013 (64). oprB1 deficiency was verified by PCR.
Motility assay.
The motility assay was conducted on petri dishes containing 20 ml of LB medium supplemented with 0.3% agarose. Motility was assessed by inoculating 1 μl of overnight culture into the medium and measuring the diameter of growth after incubating the petri dishes at 30°C for 18 h.
Microscopy analysis of flagellum localization.
Bacteria were grown in 5 ml LB medium at 30°C until the optical density at 580 nm (OD580) was ∼1. The cells were centrifuged at 1,700 × g for 45 s, washed twice with distilled water, and gently resuspended in 1 ml distilled water. Ten-microliter drops of bacterial dilutions (100 cells/μl) were slowly dried on a microscopy slide. The dried drops were covered with stain [0.31% FeCl3, 6.25% tannin, 92 mM AlK(SO4)2] for 4 min, washed with distilled water, and allowed to dry again. The dried drops were then covered with 5% AgNO3 and 0.0125% NH4OH solution for 3 min, washed with distilled water, and allowed to dry. Bacteria were observed using oil immersion light microscopy (×1,000 magnification).
Congo red binding assay.
Bacteria were grown overnight in 5 ml LB and diluted 100-fold, and 5-μl drops were inoculated onto glucose minimal medium plate containing 0.0005% Congo red dye. The plate was incubated at 30°C for 72 h.
β-Galactosidase leakage assay.
Strains containing the pKTlacZS/C plasmid were grown on solid glucose minimal or LB medium at 30°C for 20 h. Cells were suspended in 500 μl M9 buffer and centrifuged for 1 min at 12,000 × g, and the supernatant was used to measure β-galactosidase activity according to a previously described protocol (65).
Flow cytometry analysis.
Bacteria were grown overnight in 5 ml LB and diluted 100-fold, and 5-μl drops were inoculated onto agar plates with glucose minimal medium or LB. After 24 h of growth at 30°C, the cells were scraped off from the plates and suspended in 1 ml M9 buffer. The optical density of the cell suspension was diluted to an OD580 of 0.015. The two components of the LIVE/DEAD BacLight kit (L7012; Invitrogen), 20 mM red fluorescent dye propidium iodide (PI) and 3.34 mM green fluorescent dye SYTO9, were mixed at a 1:1 volume ratio and then diluted 17.6-fold into filter-sterilized M9 buffer. For staining of bacteria, the diluted cell suspension was mixed with the freshly prepared reagent mixture at a 20:1 ratio. Samples were incubated at 30°C in the dark for 30 min, and approximately 10,000 events from every sample were analyzed with a FACSAria flow cytometer (BD Biosciences). Fluorescent dyes were excited at 488 nm. Forward and side scatter of the light and fluorescence emission at 530 and 616 nm were acquired for every event. Populations of intact, PI-permeable, and dead cells were defined as previously described (28). While PI-permeable cells differ from the intact subpopulation only by their PI-permeable membrane, the dead cells are more compromised, containing less DNA than other cells (28).
Stress tolerance plate assay.
To evaluate the stress tolerance, bacteria were grown overnight (stationary phase) or to an OD580 of ∼1 (exponential phase) in 5 ml LB medium. Serial dilutions were spotted as 5-μl drops onto LB agar plates supplemented with different chemicals (specified in Results). Plates were incubated at 30°C for 20 h.
Growth curve and minimal generation time.
To start the assay always with cells in the same physiological state, 50-μl stocks made of overnight-grown bacteria were used. To investigate the growth curve of bacteria starting from stationary phase, cells were inoculated from stock into 5 ml LB medium and grown for 21 h (OD580 of ∼4) at 30°C. For exponential-phase cells, 100 μl of the bacteria grown for 18 h were diluted into fresh LB medium and grown for 3 h (OD580 of ∼1) at 30°C. The optical densities of microbial cultures at 580 nm were measured and the bacteria were diluted in LB medium for the OD580 to be 0.1. Aliquots of 100 μl of the dilutions were transferred into microtiter plate wells, and the cells were grown at 30°C and 400 rpm inside a POLARstar Omega plate reader spectrophotometer. The OD580 was measured every 7 min. Data were collected with the Omega data analysis software.
Minimal generation time was calculated from the slope of the most rapid exponential growth according to the formula G = t/3.3log(b/B), where G marks the generation time, t is the time interval in minutes, and B and b are OD580 at the beginning and the end of the time interval, respectively.
Isolation of outer membrane proteins.
For the isolation of OMPs of stationary-phase cells, bacteria were grown in 30 ml LB medium at 30°C for 17 h. To obtain the OMPs of exponentially growing cells, bacteria were grown in 200 ml LB for 3 h at 30°C to an OD580 of 0.9. Cells were collected by centrifugation at 5,000 × g at 4°C for 10 min, washed with 6 ml of 10 mM HEPES buffer (pH 7.4), and resuspended in 3 ml of the same buffer. Cells were then disrupted by ultrasonication, and cell debris was pelleted by centrifugation at 7,000 × g at 4°C for 15 min. Further OMP isolation was done as described previously (23). One-microgram samples of isolated OMPs were loaded onto 10% SDS-PAGE gel and stained according to a previous protocol (66).
Label-free proteomic analysis.
(i) Growth conditions and nano-LC-MS/MS analysis. Bacteria were pregrown overnight in liquid LB at 30°C, diluted into fresh LB medium (OD580 of ∼0.1), and grown either up to an OD580 of ∼1 (exponential phase) or for 17 h (stationary phase). Cells were harvested at 5,000 × g at 4°C for 10 min, resuspended in lysis buffer (4% SDS, 100 mM Tris, pH 7.5, 10 mM dithiothreitol), heated at 95°C for 5 min, and sonicated. The protein concentration was measured by tryptophan fluorescence, and 30 μg of cellular protein was loaded onto 30-kDa-cutoff Vivacon 500 ultrafiltration spin columns (Sartorius). Three independent samples of exponential- and stationary-phase P. putida wild-type and ΔtonBm cells were digested for 4 h on filter with 1:50 Lys-C (Wako) and then overnight with 1:50 proteomics-grade dimethylated trypsin (Sigma-Aldrich) as described for the filter-aided sample preparation protocol (67). Peptides were desalted with C18 StageTips (68), eluted, dried, and reconstituted in 0.5% trifluoroacetic acid. Nano-liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis was performed as described previously (69) using an Ultimate 3000 RSLCnano system (Dionex) and a Q Exactive mass spectrometer (Thermo Fisher Scientific) operating with top-10 data-dependent acquisition.
(ii) MS raw data processing. Raw data were identified and quantified with the MaxQuant 1.4.0.8 software package (70). Label-free quantification with MaxQuant LFQ algorithm (71) was enabled with default settings. Methionine oxidation and protein N-terminal acetylation were set as variable modifications, while cysteine carbamidomethylation was defined as a fixed modification. A search was performed against the UniProt (www.uniprot.org) P. putida KT2440 database (April 2015 version) using the tryptic digestion rule (cleavage after lysine and arginine without proline restriction). Identifications with minimally one peptide of at least seven amino acids were accepted, and transfer of identifications between runs was enabled. Protein quantification criteria were set to two peptides with a minimum of two consecutive MS1 scans per peptide. Peptide-spectrum match and protein false discovery rate were kept below 1% using a target-decoy approach. All other parameters were kept at default.
(iii) Quantitative protein profiling. Data analysis was performed with the Perseus software (72). The whole data set contained 3,359 different proteins. Parallel samples were grouped together, and groups were compared in pairs: (i) P. putida wild type from exponential (wtE) versus stationary (wtS) phase (2,652 proteins), (ii) ΔtonBm strain from exponential (tonBE) versus stationary (tonBS) phase (2,673 proteins), (iii) wtE versus tonBE (2,406 proteins), and (iv) wtS versus tonBS (2,556 proteins). To be included in the analysis, a protein needed to be detected in all three parallels of one group. Thereafter, missing values were imputed using default settings. Mean protein abundances were compared between two groups using the independent-sample Student t test. The Benjamini-Hochberg multiple-testing correction was applied with the false discovery rate set to 0.05.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sirli Rosendahl and Andres Ainelo for critical reading of the manuscript and Dmitri Lubenets for technical assistance.
This work was supported by the Estonian Research Council grants IUT20-19, IUT2-22, and PUT1351.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00303-19.
REFERENCES
- 1.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. CSH Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Postle K, Larsen RA. 2007. TonB-dependent energy transduction between outer and cytoplasmic membranes. Biometals 20:453–465. doi: 10.1007/s10534-006-9071-6. [DOI] [PubMed] [Google Scholar]
- 3.Wiener MC, Horanyi PS. 2011. How hydrophobic molecules traverse the outer membranes of Gram-negative bacteria. Proc Natl Acad Sci U S A 108:10929–10930. doi: 10.1073/pnas.1106927108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schauer K, Rodionov DA, de Reuse H. 2008. New substrates for TonB-dependent transport: do we only see the”tip of the iceberg”? Trends Biochem Sci 33:330–338. doi: 10.1016/j.tibs.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 5.Celia H, Noinaj N, Zakharov SD, Bordignon E, Botos I, Santamaria M, Barnard TJ, Cramer WA, Lloubes R, Buchanan SK. 2016. Structural insight into the role of the Ton complex in energy transduction. Nature 538:60. doi: 10.1038/nature19757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker KR, Postle K. 2013. Mutations in Escherichia coli ExbB transmembrane domains identify scaffolding and signal transduction functions and exclude participation in a proton pathway. J Bacteriol 195:2898–2911. doi: 10.1128/JB.00017-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maki-Yonekura S, Matsuoka R, Yamashita Y, Shimizu H, Tanaka M, Iwabuki F, Yonekura K. 2018. Hexameric and pentameric complexes of the ExbBD energizer in the Ton system. Elife 7:e35419. doi: 10.7554/eLife.35419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ollis AA, Postle K. 2012. ExbD mutants define initial stages in TonB energization. J Mol Biol 415:237–247. doi: 10.1016/j.jmb.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gresock MG, Kastead KA, Postle K. 2015. From homodimer to heterodimer and back: elucidating the TonB energy transduction cycle. J Bacteriol 197:3433–3445. doi: 10.1128/JB.00484-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ollis AA, Kumar A, Postle K. 2012. The ExbD periplasmic domain contains distinct functional regions for two stages in TonB energization. J Bacteriol 194:3069–3077. doi: 10.1128/JB.00015-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skare JT, Ahmer BMM, Seachord CL, Darveau RP, Postle K. 1993. Energy transduction between membranes–TonB, a cytoplasmic membrane protein, can be chemically cross-linked in-vivo to the outer membrane receptor FepA. J Biol Chem 268:16302–16308. [PubMed] [Google Scholar]
- 12.Ogierman M, Braun V. 2003. Interactions between the outer membrane ferric citrate transporter FecA and TonB: studies of the FecA TonB box. J Bacteriol 185:1870–1885. doi: 10.1128/JB.185.6.1870-1885.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cafiso DS. 2014. Identifying and quantitating conformational exchange in membrane proteins using site-directed spin labeling. Acc Chem Res 47:3102–3109. doi: 10.1021/ar500228s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu BCH, Peacock RS, Vogel HJ. 2007. Bioinformatic analysis of the TonB protein family. Biometals 20:467–483. doi: 10.1007/s10534-006-9049-4. [DOI] [PubMed] [Google Scholar]
- 15.Bitter W, Tommassen J, Weisbeek PJ. 1993. Identification and characterization of the exbB, exbD and tonB genes of Pseudomonas putida WCS358: their involvement in ferric-pseudobactin transport. Mol Microbiol 7:117–130. doi: 10.1111/j.1365-2958.1993.tb01103.x. [DOI] [PubMed] [Google Scholar]
- 16.Godoy P, Ramos-Gonzalez MI, Ramos JL. 2004. Pseudomonas putida mutants in the exbBexbDtonB gene cluster are hypersensitive to environmental and chemical stressors. Environ Microbiol 6:605–610. doi: 10.1111/j.1462-2920.2004.00595.x. [DOI] [PubMed] [Google Scholar]
- 17.Godoy P, Ramos-Gonzalez MI, Ramos JL. 2001. Involvement of the TonB system in tolerance to solvents and drugs in Pseudomonas putida DOT-T1E. J Bacteriol 183:5285–5292. doi: 10.1128/JB.183.18.5285-5292.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molina MA, Godoy P, Ramos-Gonzalez MI, Munoz N, Ramos JL, Espinosa-Urgel M. 2005. Role of iron and the TonB system in colonization of corn seeds and roots by Pseudomonas putida KT2440. Environ Microbiol 7:443–449. doi: 10.1111/j.1462-2920.2005.00720.x. [DOI] [PubMed] [Google Scholar]
- 19.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman F. 2016. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res 44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cowles KN, Moser TS, Siryaporn A, Nyakudarika N, Dixon W, Turner JJ, Gitai Z. 2013. The putative Poc complex controls two distinct Pseudomonas aeruginosa polar motility mechanisms. Mol Microbiol 90:923–938. doi: 10.1111/mmi.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shirley M, Lamont IL. 2009. Role of TonB1 in pyoverdine-mediated signaling in Pseudomonas aeruginosa. J Bacteriol 191:5634–5640. doi: 10.1128/JB.00742-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang BX, Ru K, Yuan Z, Whitchurch CB, Mattick JS. 2004. tonB3 is required for normal twitching motility and extracellular assembly of type IV pili. J Bacteriol 186:4387–4389. doi: 10.1128/JB.186.13.4387-4389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Putrinš M, Ainelo A, Ilves H, Hõrak R. 2011. The ColRS system is essential for the hunger response of glucose-growing Pseudomonas putida. BMC Microbiol 11:170. doi: 10.1186/1471-2180-11-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Putrinš M, Ilves H, Kivisaar M, Hõrak R. 2008. ColRS two-component system prevents lysis of subpopulation of glucose-grown Pseudomonas putida. Environ Microbiol 10:2886–2893. doi: 10.1111/j.1462-2920.2008.01705.x. [DOI] [PubMed] [Google Scholar]
- 25.Ainsaar K, Mumm K, Ilves H, Hõrak R. 2014. The ColRS signal transduction system responds to the excess of external zinc, iron, manganese, and cadmium. BMC Microbiol 14:162. doi: 10.1186/1471-2180-14-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mumm K, Ainsaar K, Kasvandik S, Tenson T, Hõrak R. 2016. Responses of Pseudomonas putida to zinc excess determined at the proteome level: pathways dependent and independent of CoIRS. J Proteome Res 15:4349–4368. doi: 10.1021/acs.jproteome.6b00420. [DOI] [PubMed] [Google Scholar]
- 27.Pandza S, Baetens M, Park CH, Au T, Keyhan M, Matin A. 2000. The G-protein FlhF has a role in polar flagellar placement and general stress response induction in Pseudomonas putida. Mol Microbiol 36:414–423. doi: 10.1046/j.1365-2958.2000.01859.x. [DOI] [PubMed] [Google Scholar]
- 28.Putrinš M, Ilves H, Lilje L, Kivisaar M, Hõrak R. 2010. The impact of ColRS two-component system and TtgABC efflux pump on phenol tolerance of Pseudomonas putida becomes evident only in growing bacteria. BMC Microbiol 10:110. doi: 10.1186/1471-2180-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother 44:3322–3327. doi: 10.1128/AAC.44.12.3322-3327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olivera ER, Minambres B, Garcia B, Muniz C, Moreno MA, Ferrandez A, Diaz E, Garcia JL, Luengo JM. 1998. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc Natl Acad Sci U S A 95:6419–6424. doi: 10.1073/pnas.95.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purssell A, Poole K. 2013. Functional characterization of the NfxB repressor of the mexCD-oprJ multidrug efflux operon of Pseudomonas aeruginosa. Microbiology 159:2058–2073. doi: 10.1099/mic.0.069286-0. [DOI] [PubMed] [Google Scholar]
- 32.Purssell A, Fruci M, Mikalauskas A, Gilmour C, Poole K. 2015. EsrC, an envelope stress-regulated repressor of the mexCD-oprJ multidrug efflux operon in Pseudomonas aeruginosa. Environ Microbiol 17:186–198. doi: 10.1111/1462-2920.12602. [DOI] [PubMed] [Google Scholar]
- 33.Poole K, Gotoh N, Tsujimoto H, Zhao QX, Wada A, Yamasaki T, Neshat S, Yamagishi JI, Li XZ, Nishino T. 1996. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol 21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 34.Jeannot K, Elsen S, Köhler T, Attree I, van Delden C, Plesiat P. 2008. Resistance and virulence of Pseudomonas aeruginosa clinical strains overproducing the MexCD-OprJ efflux pump. Antimicrob Agents Chemother 52:2455–2462. doi: 10.1128/AAC.01107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernal P, Allsopp LP, Filloux A, Llamas MA. 2017. The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J 11:972–987. doi: 10.1038/ismej.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang WS, van de Mortel M, Nielsen L, de Guzman GN, Li XH, Halverson LJ. 2007. Alginate production by Pseudomonas putida creates a hydrated microenvironment and contributes to biofilm architecture and stress tolerance under water-limiting conditions. J Bacteriol 189:8290–8299. doi: 10.1128/JB.00727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wood LF, Ohman DE. 2009. Use of cell wall stress to characterize sigma(22) (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol Microbiol 72:183–201. doi: 10.1111/j.1365-2958.2009.06635.x. [DOI] [PubMed] [Google Scholar]
- 38.Kivistik PA, Kivi R, Kivisaar M, Hõrak R. 2009. Identification of CoIR binding consensus and prediction of regulon of CoIRS two-component system. BMC Mol Biol 10:46. doi: 10.1186/1471-2199-10-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wylie JL, Worobec EA. 1995. The OprB porin plays a central role in carbohydrate uptake in Pseudomonas aeruginosa. J Bacteriol 177:3021–3026. doi: 10.1128/jb.177.11.3021-3026.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno R, Martinez-Gomariz M, Yuste L, Gil C, Rojo F. 2009. The Pseudomonas putida Crc global regulator controls the hierarchical assimilation of amino acids in a complete medium: evidence from proteomic and genomic analyses. Proteomics 9:2910–2928. doi: 10.1002/pmic.200800918. [DOI] [PubMed] [Google Scholar]
- 41.Rojo F. 2010. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol Rev 34:658–684. doi: 10.1111/j.1574-6976.2010.00218.x. [DOI] [PubMed] [Google Scholar]
- 42.Gh MS, Wilhelm MJ, Sheffield JB, Dai HL. 2015. Living E. coli is permeable to propidium iodide: a study by time-resolved second-harmonic scattering and fluorescence microscopy. Biophys J 108:148A–149A. doi: 10.1016/j.bpj.2014.11.819. [DOI] [Google Scholar]
- 43.Qiu DR, Eisinger VM, Rowen DW, Yu H. 2007. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 104:8107–8112. doi: 10.1073/pnas.0702660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wood LF, Leech AJ, Ohman DE. 2006. Cell wall inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: roles of sigma(22) (AlgT) and the AlgW and Prc proteases. Mol Microbiol 62:412–426. doi: 10.1111/j.1365-2958.2006.05390.x. [DOI] [PubMed] [Google Scholar]
- 45.Chevalier S, Bouffartigues E, Bazire A, Tahrioui A, Duchesne R, Tortuel D, Maillot O, Clamens T, Orange N, Feuilloley MGJ. 2018. Extracytoplasmic function sigma factors in Pseudomonas aeruginosa. Biochim Biophys Acta Gene Regul Mech 1862:706–721. doi: 10.1016/j.bbagrm.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 46.Llamas MA, Imperi F, Visca P, Lamont IL. 2014. Cell-surface signaling in Pseudomonas: stress responses, iron transport, and pathogenicity. FEMS Microbiol Rev 38:569–597. doi: 10.1111/1574-6976.12078. [DOI] [PubMed] [Google Scholar]
- 47.Vescovi EG, Soncini FC, Groisman EA. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165–174. doi: 10.1016/S0092-8674(00)81003-X. [DOI] [PubMed] [Google Scholar]
- 48.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho US, Xu WQ, Klevit RE, Le Moual H, Miller S. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 49.Yuan J, Jin F, Glatter T, Sourjik V. 2017. Osmosensing by the bacterial PhoQ/PhoP two-component system. Proc Natl Acad Sci U S A 114:E10792–E10798. doi: 10.1073/pnas.1717272114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santos TMA, Lin TY, Rajendran M, Anderson SM, Weibel DB. 2014. Polar localization of Escherichia coli chemoreceptors requires an intact Tol-Pal complex. Mol Microbiol 92:985–1004. doi: 10.1111/mmi.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeh YC, Comolli LR, Downing KH, Shapiro L, McAdams HH. 2010. The Caulobacter Tol-Pal complex is essential for outer membrane integrity and the positioning of a polar localization factor. J Bacteriol 192:4847–4858. doi: 10.1128/JB.00607-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer P. 2007. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer membrane invagination during cell constriction in E. coli. Mol Microbiol 63:1008–1025. doi: 10.1111/j.1365-2958.2006.05571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao T, Shi MM, Ju LL, Gao HC. 2015. Investigation into FlhFG reveals distinct features of FlhF in regulating flagellum polarity in Shewanella oneidensis. Mol Microbiol 98:571–585. doi: 10.1111/mmi.13141. [DOI] [PubMed] [Google Scholar]
- 54.Green JCD, Kahramanoglou C, Rahman A, Pender AMC, Charbonnel N, Fraser GM. 2009. Recruitment of the earliest component of the bacterial flagellum to the old cell division pole by a membrane-associated signal recognition particle family GTP-binding protein. J Mol Biol 391:679–690. doi: 10.1016/j.jmb.2009.05.075. [DOI] [PubMed] [Google Scholar]
- 55.Kondo S, Imura Y, Mizuno A, Homma M, Kojima S. 2018. Biochemical analysis of GTPase FIhF which controls the number and position of flagellar formation in marine Vibrio. Sci Rep 8:12115. doi: 10.1038/s41598-018-30531-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kusumoto A, Shinohara A, Terashima H, Kojima S, Yakushi T, Homma M. 2008. Collaboration of FlhF and FlhG to regulate polar-flagella number and localization in Vibrio alginolyticus. Microbiology 154:1390–1399. doi: 10.1099/mic.0.2007/012641-0. [DOI] [PubMed] [Google Scholar]
- 57.Higgs PI, Larsen RA, Postle K. 2002. Quantification of known components of the Escherichia coli TonB energy transduction system: TonB, ExbB, ExbD and FepA. Mol Microbiol 44:271–281. doi: 10.1046/j.1365-2958.2002.02880.x. [DOI] [PubMed] [Google Scholar]
- 58.Bayley SA, Duggleby CJ, Worsey MJ, Williams PA, Hardy KG, Broda P. 1977. Two modes of loss of the Tol function from Pseudomonas putida mt-2. Mol Gen Genet 154:203–204. doi: 10.1007/BF00330838. [DOI] [PubMed] [Google Scholar]
- 59.Regenhardt D, Heuer H, Heim S, Fernandez DU, Strompl C, Moore ERB, Timmis KN. 2002. Pedigree and taxonomic credentials of Pseudomonas putida strain KT2440. Environ Microbiol 4:912–915. doi: 10.1046/j.1462-2920.2002.00368.x. [DOI] [PubMed] [Google Scholar]
- 60.Adams MH. 1959. Bacteriophages. Interscience Publishers Inc., New York, NY. [Google Scholar]
- 61.Sharma RC, Schimke RT. 1996. Preparation of electro-competent E. coli using salt-free growth medium. Biotechniques 20:42–44. doi: 10.2144/96201bm08. [DOI] [PubMed] [Google Scholar]
- 62.O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 63.Martinez-Garcia E, de Lorenzo V. 2011. Engineering multiple genomic deletions in Gram-negative bacteria: analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ Microbiol 13:2702–2716. doi: 10.1111/j.1462-2920.2011.02538.x. [DOI] [PubMed] [Google Scholar]
- 64.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- 66.Chevallet M, Luche S, Rabilloud T. 2006. Silver staining of proteins in polyacrylamide gels. Nat Protoc 1:1852–1858. doi: 10.1038/nprot.2006.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wisniewski JR, Zougman A, Nagaraj N, Mann M. 2009. Universal sample preparation method for proteome analysis. Nat Methods 6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 68.Rappsilber J, Mann M, Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 69.Kasvandik S, Samuel K, Peters M, Eimre M, Peet N, Roost AM, Padrik L, Paju K, Peil L, Salumets A. 2016. Deep quantitative proteomics reveals extensive metabolic reprogramming and cancer-like changes of ectopic endometriotic stromal cells. J Proteome Res 15:572–584. doi: 10.1021/acs.jproteome.5b00965. [DOI] [PubMed] [Google Scholar]
- 70.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 71.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tyanova S, Temu T, Cox J. 2016. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 73.Herrero M, de Lorenzo V, Timmis KN. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol 172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hõrak R, Ilves H, Pruunsild P, Kuljus M, Kivisaar M. 2004. The ColR-ColS two-component signal transduction system is involved in regulation of Tn4652 transposition in Pseudomonas putida under starvation conditions. Mol Microbiol 54:795–807. doi: 10.1111/j.1365-2958.2004.04311.x. [DOI] [PubMed] [Google Scholar]
- 75.Ainelo A, Tamman H, Leppik M, Remme J, Hõrak R. 2016. The toxin GraT inhibits ribosome biogenesis. Mol Microbiol 100:719–734. doi: 10.1111/mmi.13344. [DOI] [PubMed] [Google Scholar]
- 76.Hõrak R, Kivisaar M. 1998. Expression of the transposase gene tnpA of Tn4652 is positively affected by integration host factor. J Bacteriol 180:2822–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wong SM, Mekalanos JJ. 2000. Genetic footprinting with mariner-based transposition in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 97:10191–10196. doi: 10.1073/pnas.97.18.10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kivistik PA, Putrinš M, Püvi K, Ilves H, Kivisaar M, Hõrak R. 2006. The ColRS two-component system regulates membrane functions and protects Pseudomonas putida against phenol. J Bacteriol 188:8109–8117. doi: 10.1128/JB.01262-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ojangu EL, Tover A, Teras R, Kivisaar M. 2000. Effects of combination of different-10 hexamers and downstream sequences on stationary-phase-specific sigma factor sigma(S)-dependent transcription in Pseudomonas putida. J Bacteriol 182:6707–6713. doi: 10.1128/JB.182.23.6707-6713.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Juurik T, Ilves H, Teras R, Ilmjärv T, Tavita K, Ukkivi K, Teppo A, Mikkel K, Kivisaar M. 2012. Mutation frequency and spectrum of mutations vary at different chromosomal positions of Pseudomonas putida. PLoS One 7:e48511. doi: 10.1371/journal.pone.0048511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Koch B, Jensen LE, Nybroe O. 2001. A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into Gram-negative bacteria at a neutral chromosomal site. J Microbiol Methods 45:187–195. doi: 10.1016/S0167-7012(01)00246-9. [DOI] [PubMed] [Google Scholar]
- 82.Tamman H, Ainelo A, Ainsaar K, Hõrak R. 2014. A moderate toxin, GraT, modulates growth rate and stress tolerance of Pseudomonas putida. J Bacteriol 196:157–169. doi: 10.1128/JB.00851-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jakovleva J, Teppo A, Velts A, Saumaa S, Moor H, Kivisaar M, Teras R. 2012. Fis regulates the competitiveness of Pseudomonas putida on barley roots by inducing biofilm formation. Microbiology 158:708–720. doi: 10.1099/mic.0.053355-0. [DOI] [PubMed] [Google Scholar]
- 84.Bao Y, Lies DP, Fu H, Roberts GP. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167–168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.