To colonize solid substrates, bacteria often deploy dedicated adhesins that facilitate attachment to surfaces. Caulobacter crescentus initiates surface colonization by secreting a carbohydrate-based adhesin called the holdfast. Because little is known about the chemical makeup of the holdfast, the pathway for its biosynthesis and the physical basis for its unique adhesive properties are poorly understood. This study outlines a method to extract the C. crescentus holdfast and describes the monosaccharide components contained within the adhesive matrix. The composition analysis adds to our understanding of the chemical basis for holdfast attachment and provides missing information needed to characterize enzymes in the biosynthetic pathway.
KEYWORDS: Alphaproteobacteria, adhesion, biofilm, carbohydrate structure, polysaccharide
ABSTRACT
Surface colonization is central to the lifestyles of many bacteria. Exploiting surface niches requires sophisticated systems for sensing and attaching to solid materials. Caulobacter crescentus synthesizes a polysaccharide-based adhesin known as the holdfast at one of its cell poles, which enables tight attachment to exogenous surfaces. The genes required for holdfast biosynthesis have been analyzed in detail, but difficulties in isolating analytical quantities of the adhesin have limited efforts to characterize its chemical structure. In this report, we describe a method to extract the holdfast from C. crescentus cultures and present a survey of its carbohydrate content. Glucose, 3-O-methylglucose, mannose, N-acetylglucosamine, and xylose were detected in our extracts. Our results provide evidence that the holdfast contains a 1,4-linked backbone of glucose, mannose, N-acetylglucosamine, and xylose that is decorated with branches at the C-6 positions of glucose and mannose. By defining the monosaccharide components in the polysaccharide, our work establishes a framework for characterizing enzymes in the holdfast pathway and provides a broader understanding of how polysaccharide adhesins are built.
IMPORTANCE To colonize solid substrates, bacteria often deploy dedicated adhesins that facilitate attachment to surfaces. Caulobacter crescentus initiates surface colonization by secreting a carbohydrate-based adhesin called the holdfast. Because little is known about the chemical makeup of the holdfast, the pathway for its biosynthesis and the physical basis for its unique adhesive properties are poorly understood. This study outlines a method to extract the C. crescentus holdfast and describes the monosaccharide components contained within the adhesive matrix. The composition analysis adds to our understanding of the chemical basis for holdfast attachment and provides missing information needed to characterize enzymes in the biosynthetic pathway.
INTRODUCTION
Bacteria routinely encounter solid objects in their environments that present attractive niches for colonization (1, 2). Attachment to and growth in association with these surfaces are fundamental to survival for many bacteria (3). The diversity of potential colonization substrates ranges from small soil particles, to plant and animal tissues, to the interior walls of pipes and tanks in industrial settings (4). As such, surface associated bacteria present an important obstacle to many clinical, agricultural, and manufacturing processes. Understanding how attached communities form and proliferate has the potential to inform strategies for manipulating surface colonization in a variety of settings.
Surface colonization is a stepwise process (5). Simply contacting a surface requires bacteria to overcome repulsive forces within the hydrodynamic layer that forms at the liquid-solid interface (6). In many cases, active processes such as flagellar motility and pilus oscillation provide the mechanical force needed to achieve direct contact (7–10). Transient interactions between various structures displayed on the cell surface serve to stabilize this initial association, allowing for the deployment of dedicated adhesins that promote permanent attachment (11, 12). Bacterial adhesins are extremely diverse, displaying a range of chemical, functional, and mechanical properties (13). However, irreversible attachment mediated by adhesins is thought to represent the committed step in surface colonization for many organisms (14, 15). It provides cells with the opportunity to grow and divide in association with the target substrate, which can lead to the formation of biofilms.
In the class Alphaproteobacteria, adhesins are often localized to one cell pole, giving each cell a defined geometry for attachment with respect to the surface substrate (16–20). Caulobacter crescentus is a dimorphic organism that has become a model for polar adhesion in this clade (13). During the C. crescentus cell cycle, sessile stalked cells divide asymmetrically to release a motile daughter called a swarmer cell that has type IV pili and a flagellum at one pole (21). Swarmer cells remain motile for a period of time before shedding their pili and flagella and transitioning to stalked cells (22). At developmentally controlled times during the swarmer to stalked transition or in response to a physical encounter with a surface, C. crescentus can produce a specialized adhesin called the holdfast at the old cell pole (Fig. 1A) (23–25). As the swarmer cell transitions, the holdfast remains displayed at the tip of the newly formed stalk, where it is primed for surface attachment (24). The holdfast is one of the strongest adhesives characterized to date (26). It is gelatinous in nature, has elastic characteristics, and can adhere to substrates that have a range of physical and chemical properties, making it an attractive model for developing adhesives to be used in medical and industrial applications (27, 28).
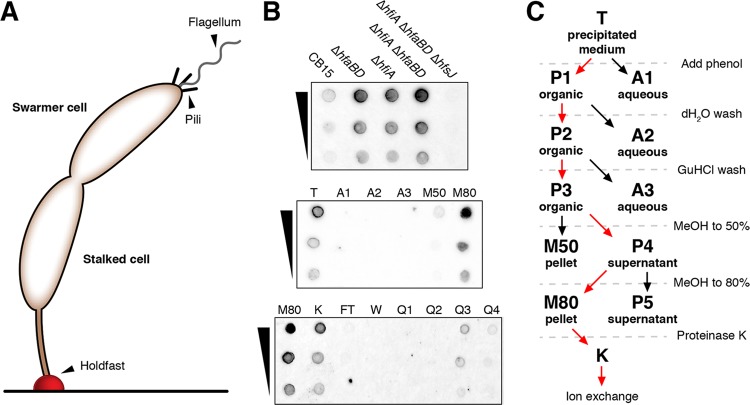
FIG 1.
Extraction of the C. crescentus holdfast. (A) A surface-attached C. crescentus cell at the predivisional stage. The holdfast adhesin (red) is depicted at the tip of the stalked cell, attached to a horizontal surface. Stalked cells divide to release a motile swarmer cell that possesses a flagellum and type IV pili. The released swarmer cell eventually transitions into a stalked cell and secretes a holdfast. (B) Enrichment of the holdfast polysaccharide from C. crescentus culture medium. (Top) Lectin (fWGA)-stained dot blot measuring holdfast released into the spent medium by various mutants. Holdfast was extracted from spent medium of cultures grown in PYE by 60% ethanol precipitation, resuspended in TES buffer (see Materials and Methods) and spotted onto nitrocellulose. (Middle) Phenol extraction of precipitated medium from the ΔhfiA ΔhfaBD strain. The resuspended ethanol precipitation (T) was extracted with phenol. The aqueous phases (A samples) and subsequent methanol precipitates (M samples) are shown. (Bottom) the M80 fraction was treated with proteinase K (K sample) and subjected to anion-exchange chromatography. The flowthrough (FT), wash (W), and step gradient fractions (Q1, Q2, Q3, and Q4) are shown. (C) Schematic of the holdfast enrichment procedure. Fraction names correspond to those shown in panel B.
Despite significant advances in understanding mechanical properties of the holdfast adhesin, its chemical composition remains poorly characterized. The adhesin binds to the N-acetylglucosamine (GlcNAc)-reactive lectin wheat germ agglutinin (WGA) and is sensitive to the GlcNAc-specific hydrolases lysozyme and chitinase, indicating that it contains GlcNAc (16, 28, 29). Treatment with proteinase K or DNase also alters the mechanical properties, suggesting that protein and DNA may be included in the matrix as well (29). Finally, the genetic determinants of holdfast biosynthesis provide invaluable insight into its chemical makeup. Genes predicted to encode machinery for the production of an extracellular polysaccharide represent the major factors required for holdfast production (Table 1) (30). Among these genes are those encoding four predicted glycosyltransferases (GTs), implying that a complex polysaccharide with a repeating unit of at least four monosaccharides is a critical component of the holdfast matrix (31–33).
TABLE 1.
Genes for holdfast synthesis and assembly
| Gene | Locus | Mutant phenotype | Annotation | Reference(s) |
|---|---|---|---|---|
| hfsA | CC_2431 | No holdfast production (HF–) | Polysaccharide copolymerase Wzz | 30 |
| hfsB | CC_2430 | Few cells with holdfasts; small holdfasts; adhesion defect | Polysaccharide secretion autokinase | 30, 54 |
| hfsC | CC_2429 | Redundant with hfsI; ΔhfsC ΔhfsI mutant is HF– | Polysaccharide polymerase Wzy | 31 |
| hfsD | CC_2432 | No holdfast production (HF–) | Polysaccharide secretin Wza | 30 |
| hfsE | CC_2425 | Redundant with pssY and pssZ; ΔhfsE ΔpssY ΔpssZ mutant is HF– | Hexose phosphate transferase (PHPT) | 31 |
| hfsF | CC_2426 | Smaller holdfasts; adhesion defect | Polysaccharide flippase Wzx | 31, 55 |
| hfsG | CC_2427 | No holdfast production (HF–) | GT2 family glycosyltransferase | 31 |
| hfsH | CC_2428 | Loss of holdfast cohesiveness; adhesion defect | Polysaccharide deacetylase | 31, 50 |
| hfsI | CC_0165 | Redundant with hfsC; ΔhfsC ΔhfsI mutant is HF– | Polysaccharide polymerase Wzy | 31 |
| hfsJ | CC_0095 | No holdfast production (HF–) | WecG/TagA family glycosyltransferase | 32 |
| hfsK | CC_3689 | Loss of holdfast cohesiveness; adhesion defect | CelD family acyltransferase | 51 |
| hfsL | CC_2277 | No holdfast production (HF–) | GT2 family glycosyltransferase | 33 |
| hfaA | CC_2628 | Holdfast anchoring defect; holdfast shedding | CsgA-like curlin protein | 35, 36 |
| hfaB | CC_2629 | Holdfast anchoring defect; holdfast shedding | CsgG family curlin secretion protein | 35, 36 |
| hfaD | CC_2630 | Holdfast anchoring defect; holdfast shedding | Hypothetical protein; signal peptide | 35, 36 |
| hfaE | CC_2639 | Holdfast anchoring defect; holdfast shedding | Hypothetical protein; signal peptide | 33 |
| pssY | CC_0166 | Redundant with hfsE and pssZ; ΔhfsE ΔpssY ΔpssZ mutant is HF– | Hexose phosphate transferase (PHPT) | 31 |
| pssZ | CC_2384 | Redundant with hfsE and pssY; ΔhfsE ΔpssY ΔpssZ mutant is HF– | Hexose phosphate transferase (PHPT) | 31 |
Extracellular polysaccharides are deployed by bacteria for a variety of purposes, including functioning both as adhesins and as components of biofilm matrices (34). The polar polysaccharide adhesins secreted by Alphaproteobacteria are unusual both for their discrete localization within the cell envelope and their exceptional adhesive capabilities (13). Understanding the molecular basis for interactions between polar adhesins and exogenous surfaces requires information about the chemical structure of the adhesive matrix. However, both the insoluble nature of these materials and the limited amounts secreted by producing organisms have hindered efforts to obtain suitable samples for analysis. Here, we describe a method to extract the holdfast from C. crescentus cultures. By analyzing the carbohydrate content of this preparation, we have identified the monosaccharide components of the holdfast and performed a preliminary characterization of their linkage patterns. The results are consistent with a model in which a 1,4-linked backbone of glucose, GlcNAc, mannose, and xylose that is decorated with branches at the C-6 positions of glucose and mannose makes up the holdfast polysaccharide.
RESULTS
Biochemical enrichment of the holdfast.
C. crescentus cells produce only a small patch of holdfast that is tightly associated with the cell envelope, presenting a major challenge to isolating sufficient quantities of material. We took a genetic approach to engineer a strain that would simplify the extraction process. We focused on the holdfast inhibitor A gene (hfiA) that limits holdfast production by directly targeting the glycosyltransferase HfsJ and on the hfa genes that are required for anchoring the holdfast matrix to the cell pole (32, 35, 36). We reasoned that an hfiA hfa double mutant should overproduce holdfast and release it into the spent medium, allowing for the separation of holdfast from cell material by centrifugation. To assess this strategy, we created a series of C. crescentus mutants with deletions at the hfiA and hfa loci. Each strain was grown in complex peptone-yeast extract (PYE) medium to maximize holdfast production and spent medium that had been precipitated with ethanol was probed by dot blotting with fluorescently labeled WGA (fWGA) (Fig. 1B). Wild-type C. crescentus released a small amount of WGA-reactive material, and the signal from ΔhfaBD cultures was higher, consistent with the previously reported shedding phenotype in similar mutants (36). Surprisingly, the ΔhfiA mutant also released a significant amount of WGA-reactive material, suggesting that hyper-holdfast production in this background results in the release of holdfast from the cell surface even when the hfa genes are intact. Finally, the ΔhfiA ΔhfaBD double mutant released more WGA-reactive material than the ΔhfiA or ΔhfaBD single mutants, and the signal was eliminated when the hfsJ gene, encoding a GT required for holdfast biosynthesis, was deleted in the ΔhfiA ΔhfaBD background (32).
We precipitated the spent medium from ΔhfiA ΔhfaBD cultures that had been grown in defined M2X medium to minimize contaminants from the medium. The precipitated WGA-reactive signal migrated to the organic phase of a phenol extraction and could be precipitated from the phenol phase by the addition of increasing amounts of methanol. To further enrich the holdfast polysaccharide, we first treated the phenol extracts with proteinase K. WGA staining as detected by dot blotting decreased by ∼50% upon protease treatment. An unknown protein(s) has been shown to contribute to the mechanical properties of the holdfast, and the digestion of these proteins may alter how the material binds to the nitrocellulose membrane or the accessibility of GlcNAc residues within the matrix to WGA (29). Alternatively, some of the digested proteins may themselves be modified with WGA-reactive glycans leading to a staining decrease after treatment. The protease digests were subsequently subjected to anion-exchange chromatography. The WGA-reactive signal bound to Q Sepharose resin and could be eluted with solutions of increasing ionic strength. The peak fraction was dialyzed extensively against H2O and used for subsequent analysis (Fig. 1C).
The UV absorbance spectrum of a typical holdfast preparation is shown in Fig. 2A. No peaks at 260 or 280 nm were present, demonstrating that the preparation is free of significant contamination from protein and nucleic acids. Because the O-antigen from smooth lipopolysaccharide (SLPS) represented an obvious source of potential carbohydrate contamination, we also analyzed the holdfast fraction by SDS-PAGE. The extract did not react with anti-SLPS immune serum (Fig. 2B), and the lack of an O-polysaccharide signature was confirmed by silver staining (Fig. 2C). Interestingly, we did not observe any prominent bands after silver staining. Furthermore, no fWGA reactivity was detected when the holdfast extract was transferred to nitrocellulose after electrophoresis. This finding suggests that the isolated holdfast does not migrate in SDS-PAGE gels either because it is too large to enter the matrix or does not associate with SDS. In support of the former possibility, we were unable to identify size exclusion conditions under which the fWGA reactivity did not elute in the void.
FIG 2.
Characterization of the enriched holdfast extract. (A) A UV-vis absorbance spectrum of a typical holdfast extract lacking peaks in the 260- or 280-nm ranges demonstrated that the sample used for analysis was free of significant protein or nucleic acid contamination. (B) SLPS immunoblot showing the lack of O-antigen signal in the holdfast extract. A cell pellet (cells) and a holdfast extract (hf) from a ΔhfiA ΔhfaBD culture grown in M2X were analyzed. (C) Silver staining analysis of a typical holdfast (hf) extract.
Finally, we examined the extract using light microscopy. Though no phase-contrast signal was detected in the sample, discrete foci were observed when fWGA was added, reminiscent of the staining signal from cell-associated holdfasts (Fig. 3). To confirm that the fluorescent foci were derived from holdfast, we stained an extract prepared from the ΔhfiA ΔhfaBD ΔhfsJ strain. Because this sample was not expected to have a significant signal in the fWGA channel, fluorophore-conjugated beads were added in order to define the focal plane. When viewed with a green fluorescent protein (GFP) filter, the fluorescent beads were evenly dispersed across the slide in the ΔhfiA ΔhfaBD ΔhfsJ extracts, but no fWGA foci were present in the red channel. We concluded that the enrichment yielded discrete WGA reactive particles that could be extracted from cells with an intact holdfast synthesis pathway.
FIG 3.
WGA-reactive particles in holdfast extracts. Extracts prepared from the indicated strains were stained with fWGA in the presence or absence of fluorescently labeled beads. (Top) Red channel (Alexa Fluor 594-WGA); (bottom) green channel (0.2-μm YP microbeads). The scale bars represent 10 μm.
Monosaccharide composition of the holdfast.
To examine the monosaccharide content of the holdfast, we first prepared trimethylsilyl methyl glycosides from acid-hydrolyzed extracts and analyzed them by gas chromatography-mass spectrometry (GC-MS). Because this method can identify neutral monosaccharides, amino sugars, and uronic acids, its broad scope is ideal for exploratory analysis. The preliminary composition profile identified glucose, GlcNAc, mannose, xylose, and a methylated hexose. Guided by these results, we chose to prepare alditol acetates from acid-hydrolyzed holdfast extracts and to analyze both neutral and amino sugar derivatives by GC-MS. Alditol acetate derivatization provides a more sensitive view of monosaccharide composition and can also be used to define the structure of methylated sugars. This analysis confirmed that glucose, GlcNAc, mannose, and xylose were present and defined the methyl-hexose as 3-O-methylglucose (Table 2). Yields from the holdfast extraction procedure were too low to weigh the material accurately, and thus we were unable to determine the percentage of carbohydrate in the samples.
TABLE 2.
Monosaccharide composition of a typical holdfast extracta
| Monosaccharide | Mean composition (mol%) ± SE |
|---|---|
| 3-O-Methylglucose | 12 ± 3 |
| Glucose | 15 ± 4 |
| Mannose | 18 ± 1 |
| N-Acetylglucosamine | 34 ± 4 |
| Xylose | 20 ± 7 |
Values are reported as means from two biological replicates. SE, standard error.
The identification of xylose drew our attention because the cultures used for holdfast extractions had been grown in M2 salts with xylose as the carbon source. Though it seemed unlikely, we wanted to confirm that the growth medium did not contribute to the carbohydrate profile of the holdfast. We repeated the extraction using ΔhfiA ΔhfaBD cultures that had been grown in PYE, a complex medium containing mainly amino acids as the carbon source. The neutral sugar profiles of hydrolyzed holdfast from cells grown in M2X and PYE medium were nearly identical (Fig. 4). We conclude that the holdfast contains glucose, GlcNAc, mannose, xylose, and 3-O-methylglucose and that the growth medium has little effect on this composition profile.
FIG 4.
Comparison of neutral sugar profiles in holdfast extracts from two different growth media. Total ion chromatograms from GC-MS analysis of extracts from cells grown in M2X (A) or PYE (B) medium are shown. Xylose was present in both holdfast extracts, demonstrating that this component is not an artifact derived from cultivation in M2X medium. The similarity in the monosaccharide profiles of cells grown in two distinct media indicates that growth conditions do not significantly influence holdfast composition.
Determination of monosaccharide linkages.
To characterize the linkage patterns of monosaccharides in the holdfast extract, we prepared partially methylated alditol acetates (PMAAs) and analyzed them by GC-MS. Because neutral monosaccharides and amino sugars cannot be separated efficiently with the same GC method, we analyzed the two sets of PMAAs on separate columns. We merged the two data sets by normalizing peak area percentages to the signal for 4-linked glucose, which was present in both chromatograms (Table 3). The analysis identified 4-linked forms of glucose, GlcNAc, mannose, and xylose as the major residues. Terminal galactose, terminal glucose, terminal GlcNAc, terminal mannose, 2-linked xylose, 4,6-linked mannose, and 4,6-linked glucose were detected as minor components.
TABLE 3.
Analysis of monosaccharide linkages from a typical holdfast extract using CH3I derivatization
| Residue | Normalized peak (%) |
|---|---|
| Terminal galactopyranosyl | 1 |
| Terminal glucopyranosyl | 13 |
| Terminal mannopyranosyl | 2 |
| Terminal N-acetylglucosaminopyranosyl | 2 |
| Terminal xylopyranosyl | 1 |
| 2-Linked xylopyranosyl | 4 |
| 4-Linked glucopyranosyl | 27 |
| 4-Linked mannopyranosyl | 18 |
| 4-Linked N-acetylglucosaminopyranosyl | 22 |
| 4-Linked xylopyranosyl | 7 |
| 4,6-Linked glucopyranosyl | ∼1 |
| 4,6-Linked mannopyranosyl | 1 |
To perform linkage analysis, the hydroxyls in an intact polysaccharide are methylated, the methylated polysaccharide is hydrolyzed, and the resulting partially methylated monosaccharides are reduced, acetylated, and analyzed by GC-MS. Both glucose and 3-O-methylglucose are present in the holdfast, and methylation of the polysaccharide renders PMAAs derived from glucose and those derived from 3-O-methylglucose indistinguishable. To define the linkage state of 3-O-methylglucose, we repeated the linkage analysis by treating the holdfast polysaccharide with CD3I instead of CH3I, which is normally used to prepare PMAAs. This allowed us to use mass spectrometry to differentiate methyl groups in the native polysaccharide from those added during derivatization. The neutral sugar profile obtained with CD3I methylation was similar to that obtained using CH3I (Tables 3 and 4). 4-Linked glucose, 4-linked mannose, and 4-linked xylose were the major components. Terminal 3-O-methylglucose, terminal xylose, 4,6-linked glucose, and 4,6-linked mannose were minor components. We identified terminal 3-O-methylglucose by the presence of 121, 165, 167, and 211 m/z ions (Fig. 5), demonstrating that the methyl groups in the holdfast polysaccharide are attached to C-3 of terminal glucose residues.
TABLE 4.
Analysis of neutral monosaccharide linkages from a typical holdfast extract using CD3I derivatization
| Residue | Peak (%) |
|---|---|
| Terminal 3-O-methylglucopyranosyl | 9 |
| Terminal xylopyranosyl | 12 |
| 4-Linked glucopyranosyl | 21 |
| 4-Linked mannopyranosyl | 27 |
| 4-Linked xylopyranosyl | 23 |
| 4,6-Linked glucopyranosyl | 4 |
| 4,6-Linked mannopyranosyl | 5 |
FIG 5.

Mass spectrum of PMAA derived from terminal-3-O-methylglucopyranose. The predicted fragmentation pattern of a CD3I-treated derivative of terminal 3-O-methylglucopyranose is shown in the top right corner. Characteristic ions used to define the position of the polysaccharide-derived methyl group are shown in red.
DISCUSSION
Permanent attachment is a critical stage in the process of surface colonization by bacteria (37). As the first irreversible step, it represents the point at which an organism commits to colonizing a substrate (38). A unipolar adhesin called the holdfast is the primary determinant of surface colonization in the model alphaproteobacterium Caulobacter crescentus (33). Biophysical studies of the C. crescentus holdfast have confirmed the material’s high versatility and adhesive strength, while also providing evidence for a layered substructure within the matrix (28, 29). Detailed genetic analyses have shown that the production of an extracellular polysaccharide is the key determinant of holdfast synthesis (30). Binding of the GlcNAc-specific lectin WGA to the holdfast matrix and its sensitivity to lysozyme and chitinase further support the notion that it is a polysaccharide-based material (16).
Linking the mechanical properties of the holdfast to specific chemical features has been limited by a lack of information about its composition. To understand the molecular basis for holdfast-based adhesion and to begin reconstituting the biosynthetic pathway, chemical analysis of the polysaccharide is needed. Despite decades of interest, such experiments have been precluded by an inability to isolate analytical quantities of holdfast material. We took a genetic approach to streamline the extraction process by deleting the genes for the holdfast inhibitor HfiA and components of the holdfast anchor. The resulting strain produced more holdfast than did the wild type and released it from the cell surface, allowing for the enrichment of holdfast material directly from spent medium (Fig. 1). This approach yielded a sample free of protein and nucleic acid that could be used for carbohydrate analysis (Fig. 2). More broadly, the extraction we describe here provides a framework for isolating unipolar adhesins that can be optimized and scaled to facilitate more detailed structural analyses.
Glucose, GlcNAc, mannose, and xylose were the major monosaccharides in our preparations. These residues were predominantly 4-linked, leading us to propose that they constitute a 1,4-linked backbone within the holdfast polysaccharide. Determining the order of residues in this chain will require additional approaches, including nuclear magnetic resonance analysis, which can be undertaken once sufficient material has been obtained. We also identified 3-O-methylglucose as a minor, terminally linked component of the holdfast. Methylation of glucose at the nonreducing end of the polysaccharide could serve as a termination signal for polymerization as it does for certain ABC transporter-dependent synthesis pathways (39, 40). However, the relative abundance of 3-O-methylglucose suggests that it is more likely a substoichiometric component of the repeating unit and not present in a single copy exclusively at the chain terminus. We propose that 3-O-methylglucose sits at the termini of branches within the repeating unit. Indeed, we consistently detected 4,6-linked glucose and 4,6-linked mannose in our samples, indicating that glucose and mannose residues within the 1,4-linked backbone are substituted at C-6. The identification of glucose and mannose in both the 4- and 4,6-linked forms suggests that C-6 substitutions at these residues are nonstoichiometric and that branching is heterogeneous.
We observed some variability among the less abundant signals in our extracts. Glucose, GlcNAc, mannose, and xylose, though primarily 4-linked, were also detected in terminally linked forms at low levels, and the presence of these signals was inconsistent (Tables 3 and 4). It seems unlikely that small amounts of each monosaccharide are present at branch termini. Instead these signals likely reflect nonspecific hydrolysis of the 1,4-linked backbone during either the culture growth or the extraction processes. Trace amounts of galactose were also detected only in some extracts, and we identified both 2-linked xylose and terminal galactose in samples containing galactose. This suggests that some branches may contain xylose substituted at C-2 with galactose. Variability in the linkage analysis is likely due in part to the difficulty of quantitatively permethylating and hydrolyzing regions with branched residues, particularly with limited quantities of sample. Determining the exact nature of the C-6 substitutions on glucose and mannose will require optimizing the extraction described here to increase yields.
The genes required for holdfast production encode a predicted wzy-type polysaccharide assembly pathway (30, 33). In the first stage of this process, GT enzymes add monosaccharides to a lipid carrier, producing a glycolipid repeating unit. Enzymatic machinery located in the cell envelope then polymerizes the repeating unit and secretes the resulting polysaccharide to the cell surface (41, 42). Although the polymerization and export machinery is conserved among bacteria, each pathway uses a characteristic set of GTs with various specificities to assemble a repeating unit, explaining the enormous diversity of wzy-dependent carbohydrates (43–45). Extensive genetic analysis using saturating mutagenesis has shown that four GTs are required for holdfast production (31–33), but our results suggest that repeating units with up to six or even seven sugars may be present in the polysaccharide. GTs that perform multiple monosaccharide additions using a single active site have been described (46–48), and such activities may explain how four enzymes synthesize a repeating unit with greater than four sugars. For instance, a bifunctional GT might both elongate the 1,4-linked backbone and introduce a branch by sequentially adding monosaccharides to C-4 and C-6 of the terminal sugar in a glycolipid intermediate. Alternatively, GTs required for C-6 branching may not be essential for holdfast production. Mutating these genes could disrupt branching without affecting the adhesive properties of the holdfast, making them difficult to identify using forward genetics. In vitro studies with the holdfast GTs should help to clarify this point.
Though polysaccharide production is the main genetic determinant of holdfast-based adhesion, there is evidence that other macromolecules are associated with the matrix (29). Proteinase K treatment alters the elastic properties of surface-attached holdfasts, and our biochemical fractionation supports the presence of protein (29). Not only did treating holdfast extracts with proteinase K reduce their WGA reactivity, but the material also generally partitioned like a protein; it migrated to the organic phase of a phenol extraction and precipitated in phenol with high concentrations of added methanol. Nucleic acid, on the other hand, was not detected in the final holdfast fraction despite the lack of a nuclease treatment in the isolation process. This suggests that, despite the holdfast’s reactivity toward nucleic acid dyes and mild sensitivity to DNase, nucleic acids likely represent a loosely associated component that is lost easily during the extraction process (29, 49).
Imaging of isolated holdfast showed that the matrix retained some degree of structural integrity despite the extensive enrichment process (Fig. 3). This persistent insolubility agrees with atomic force microscopy studies of surface attached holdfasts that proposed the presence of a feature promoting association between polysaccharide strands in the matrix (28). Two holdfast biosynthesis genes, hfsH and hfsK, are predicted to encode a polysaccharide N-deacetylase and an N-acetyltransferase, respectively, and deleting either gene disrupts the cohesiveness of the holdfast (50, 51). Both enzymes would be predicted to target amino groups from the 4-linked GlcNAc residues detected in our samples. GlcNAc modification could be used to cross-link adjacent strands or incorporate proteins into the matrix and likely contributes significantly to the polysaccharide’s mechanical and adhesive properties. Unfortunately, such modifications would have been masked by the acid hydrolysis and acetylation sequence that is used to perform carbohydrate composition and linkage analysis. Determining the nature of GlcNAc decorations, the order of monosaccharides in the 1,4-linked backbone, and the specifics of the polysaccharide’s branching pattern will require more nuanced analytical techniques. The results presented here establish the foundation for characterizing these features using the isolated polysaccharide and also define the substrate pool needed to characterize the biosynthetic pathway in vitro with purified enzymes. Given the challenge of obtaining large quantities of holdfast, the convergence of analytical and enzymatic approaches will likely be required to completely define the structure and biosynthesis of the holdfast.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and genetic manipulations.
The strains used in this study are shown in Table 5. C. crescentus was grown at 30°C in PYE or M2 xylose (M2X) medium supplemented with 3% (wt/vol) sucrose, 25 μg/ml kanamycin (solid medium), or 5 μg/ml kanamycin (liquid medium) when necessary. E. coli was grown at 37°C in LB medium supplemented with 50 μg/ml kanamycin when appropriate. The plasmid for hfaBD deletion (pDH108) was created using the primers dhfaB a (5′-GCTACGTAATACGACTCACTAGTGTCCAGCGCCCTTCCCAG-3′), dhfaB b (5′-CGTCTGGCTAGTAGACCATCATTTCGGAGTACCGCCC-3′), dhfaB c (5′-CCGAAATGATGGTCTACTAGCCAGACGACGACGCC-3′), and dhfaB d (5′-CCAGATATCCTGCAGAGAAGCTTGGTGAGCCGCCACGTCG-3′), with which the first and last six nucleotides of the CC_2629 open reading frame were used to link the fragments upstream and downstream of the gene. The inserts were fused into pNTPS138 using Gibson assembly. Deletion plasmids for hfiA and hfsJ have been described previously (32). Plasmids were isolated using the GeneJet plasmid miniprep kit (Thermo Fisher) and introduced into C. crescentus by electroporation. Unmarked, in-frame deletions were created using a standard two-step procedure with sacB-based counterselection. Cultures for holdfast extraction were prepared by inoculating either M2 salts containing 0.15% (wt/vol) xylose or PYE with an overnight starter culture of the appropriate strain grown in PYE.
TABLE 5.
Caulobacter strains used in this study
| Strain | Genotype | Description | Source or reference |
|---|---|---|---|
| FC19 | CB15 | Wild type | ATCC 19089 |
| FC1365 | ΔhfiA | In-frame deletion of CC_0817 | 32 |
| FC3343 | ΔhfaBD | Deletion of CC_2629; removes 77 nucleotides of CC_2630 (hfaD) | This study |
| FC3344 | ΔhfiA ΔhfaBD | Deletion of CC_2629 in FC1365 background; removes 77 nucleotides of hfaD | This study |
| FC3345 | ΔhfiA ΔhfaBD ΔhfsJ | In-frame deletion of CC_0095 in FC3344 background | This study |
Wheat germ agglutinin blotting.
Portions (2.75 μl) of each sample to be analyzed, along with two 2-fold serial dilutions prepared in TU buffer (15 mM Tris-HCl [pH 8.8] and 4 M urea), were spotted onto a nitrocellulose membrane. The membrane was allowed to dry for 30 m and then blocked for a minimum of 1 h using TBST buffer (20 mM Tris-HCl [pH 8.0], 137 mM NaCl, 2.7 mM KCl, and 0.1% Tween 20) containing 5% (wt/vol) bovine serum albumin (BSA). The membrane was then stained for a minimum of 1 h using 1.5 μg/ml Alexa Fluor 594-WGA (Thermo Fisher) dissolved in TBST containing 1.25% BSA. After three washes with TBST, the membrane was imaged using a Bio-Rad Gel Doc Imager on the Alexa Fluor 647 setting.
Phenol extraction of shed holdfast.
The ΔhfiA ΔhfaBD strain was grown for 24 h in 1 liter of M2X or PYE medium. The cell material was removed by centrifuging the culture (4°C) for 1 h at 17,000 × g. Then, 750 ml of the cell-free spent medium was brought to a concentration of 60% (vol/vol) ethanol, followed by incubation overnight at 4°C. The resulting precipitate was isolated by centrifugation (4°C) for 1 h at 17,000 × g, resuspended in 110 ml of TES buffer (10 mM Tris-HCl [pH 7.4], 2% SDS, and 1 mM EDTA), and sonicated for 30 s. An equal volume of unbuffered aqueous phenol (Fisher Scientific) was added, and the solution was incubated at 68°C for 15 m. After the phases were allowed to separate, the upper aqueous phase was removed and discarded. Next, 80 ml of dH2O was added to the remaining phenol phase, the solution was again incubated at 68°C for 15 min, and the aqueous phase was again discarded. Then, 12 ml of TEG buffer (10 mM Tris-HCl [pH 7.4], 4 M guanidinium-HCl, and 1 mM EDTA) was added to the remaining phenol phase. The upper phase was discarded, and the remaining phenol phase was washed a final time with 6 ml of dH2O. An equal volume of methanol was added to the phenol phase, and the resulting solution was incubated for 1 h at room temperature. The 50% methanol solution was centrifuged (4°C) for 40 m at 3,000 × g, and the insoluble material was discarded. The supernatant was brought to a concentration of 80% methanol and incubated overnight at –20°C. The 80% methanol solution was then centrifuged (4°C) for 45 min at 17,000 × g. The pellet was washed with ice-cold 70% ethanol and recentrifuged, leaving a holdfast-enriched pellet.
Proteinase K digestion.
The precipitated phenol phase (above) was resuspended in 5 ml of TU buffer and sonicated for 30 s. Proteinase K was added to 50 μg/ml. The digest was incubated for 2 h at 37°C, followed by an additional 2 h of incubation at 60°C.
Ion exchange chromatography.
Proteinase K digested material was loaded on a 1.5-ml Fast Flow Q (Amersham Biosciences) column that had been equilibrated in TU buffer. The column was then washed with 5 ml of TU buffer and eluted sequentially with a step gradient of 15 mM Tris-HCl (pH 8.5) containing 30 mM NaCl, 30 mM guanidinium-HCl, 300 mM guanidinium-HCl, and 4 M guanidinium-HCl in 3.5-ml fractions. The 300 mM guanidinium-HCl fraction, which contained the bulk of the WGA-reactive signal, was added to 3.5-kDa cutoff dialysis tubing and dialyzed four times against dH2O for 24 h. The dialyzed sample was added to a glass tube, frozen at –80°C, and lyophilized to dryness.
Fluorescence microscopy.
Solutions of 10 μg/ml Alexa Fluor 594-WGA and 0.005% Fluoresbrite 0.1-μm YG microspheres (Polysciences, Inc.) were prepared in PYE and used to stain holdfast extracts. Then, 0.5 μl of fWGA solution and 1 μl of fluorescent bead solution (or 1 μl of PYE blank) was added to 2.5 μl of H2O dialyzed holdfast extract. Imaging was performed using a Leica DM500 microscope equipped with a HCX PL APO 63×/1.4 NA Ph3 objective. fWGA staining was visualized using an RFP fluorescence filter (Chroma set 41043), and the fluorescent beads were visualized with a GFP fluorescence filter (Chroma set 41017).
SDS-PAGE.
A cell pellet from 0.75 ml of a ΔhfiA ΔhfaBD culture grown to early log phase in M2X medium was collected by centrifugation and resuspended in 50 μl of SDS loading dye. Next, 1 ml of dialyzed holdfast extract (above) was lyophilized to dryness and resuspended in 50 μl of SDS loading dye. Samples were heated for 5 min at 95°C, and 3 μl was separated in a 12% Tris-glycine gel. Gels were silver stained by the method of Kittelberger et al. (52), with an extended oxidation step as described by Walker et al. (53).
Immunoblotting.
Electrophoresed samples were transferred to nitrocellulose and blocked in TBST containing 5% milk powder. The membrane was then probed sequentially with rabbit serum raised to C. crescentus SLPS (53) dissolved at a 1:20,000 dilution in TBST containing 5% milk and horseradish peroxidase-conjugated goat anti-rabbit antibody (Thermo Fisher) dissolved in TBST with 5% milk. The blot was visualized using chemiluminescent peroxidase substrate.
Monosaccharide composition analysis.
Before hydrolysis, 20 μg of myoinositol was added to the holdfast samples as an internal standard. Samples were hydrolyzed using 2 M trifluoroacetic acid (400 μl, 2 h, 121°C), reduced with NaBD4 (10 mg/ml in 1 M NH4OH, room temperature, overnight), and acetylated using acetic anhydride-pyridine (250 μl plus 250 μl, 1 h, 100°C). The resulting alditol acetates were analyzed on an Agilent 7890A GC interfaced to a 5975C MSD (mass selective detector, electron impact ionization mode). Separation was performed on a 30-m Supelco SP-2331 bonded-phase fused silica capillary column (neutral sugars) and on a Supelco Equity-1 fused silica capillary column (30 m by 0.25 mm [inner diameter]) (amino sugars). Samples were analyzed both at a 100:1 split and a 10:1 split in SP-2331 and splitless in Equity-1. The molar percentages for each component were calculated based on previously determined response factors for each monosaccharide relative to the inositol standard.
Linkage analysis of partially methylated alditol acetates.
Holdfast samples were preacetylated, permethylated, hydrolyzed, reduced, and reacetylated. The resultant partially methylated alditol acetates (PMAAs) were analyzed by GC-MS. Before permethylation, the samples were acetylated with acetic anhydride-pyridine (250 μl plus 250 μl, 1 h, 100°C). Permethylation was performed by using two rounds of treatment with sodium hydroxide (15 min) and methyl iodide or deuteromethyl iodide (45 min). Subsequently, the permethylated material was hydrolyzed with 2 M trifluoroacetic acid (400 μl, 2 h, 121°C), reduced with NaBD4 (10 mg/ml in 1 M NH4OH, room temperature, overnight), and acetylated using acetic anhydride-pyridine (250 μl plus 250 μl, 1 h, 100°C). The resulting PMAAs were analyzed on an Agilent 7890A GC interfaced to a 5975C MSD (mass selective detector, electron impact ionization mode). Separation was performed on a 30-m Supelco SP-2331 bonded-phase fused silica capillary column (neutral sugars) and on a Supelco Equity-1 fused silica capillary column (30 m by 0.25 mm [inner diameter]) (amino sugars).
ACKNOWLEDGMENTS
We thank members of the Crosson lab for helpful discussions.
This study was supported by National Institutes of Health (NIH) grant R01GM087353 to S.C. and by a Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, U.S. Department of Energy, grant (DE-SC0015662) to P.A. at the Complex Carbohydrate Research Center. D.M.H. is supported by the Helen Hay Whitney Foundation.
We declare that we have no conflicts of interest with the contents of this article.
REFERENCES
- 1.Henrici AT. 1933. Studies of freshwater bacteria. I. A direct microscopic technique. J Bacteriol 25:277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zobell CE. 1943. The effect of solid surfaces upon bacterial activity. J Bacteriol 46:39–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davey ME, O’Toole GA. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 5.O’Toole G, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu Rev Microbiol 54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 6.Donlan RM. 2002. Biofilms: microbial life on surfaces. Emerg Infect Dis 8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pratt LA, Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis, and type I pili. Mol Microbiol 30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 8.O’Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 9.Watnick PI, Kolter R. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol Microbiol 34:586–595. doi: 10.1046/j.1365-2958.1999.01624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bodenmiller D, Toh E, Brun YV. 2004. Development of surface adhesion in Caulobacter crescentus. J Bacteriol 186:1438–1447. doi: 10.1128/jb.186.5.1438-1447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marshall KC, Stout R, Mitchell R. 1971. Mechanism of the initial events in the sorption of marine bacteria to surfaces. J Gen Microbiol 68:337–348. doi: 10.1099/00221287-68-3-337. [DOI] [Google Scholar]
- 12.Hinsa SM, Espinosa-Urgel M, Ramos JL, O’Toole GA. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol 49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- 13.Berne C, Ducret A, Brun YV, Hardy GG. 2015. Adhesins involved in attachment to abiotic surfaces by Gram-negative bacteria. Microbiol Spectr 3:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caiazza NC, O’Toole GA. 2004. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J Bacteriol 186:4476–4485. doi: 10.1128/JB.186.14.4476-4485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffman MD, Zucker LI, Brown PJB, Kysela DT, Brun YV, Jacobson SC. 2015. Timescales and frequencies of reversible and irreversible adhesion events of single bacterial cells. Anal Chem 87:12032–12039. doi: 10.1021/acs.analchem.5b02087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merker RI, Smit J. 1988. Characterization of the adhesive holdfast of marine and freshwater caulobacters. Appl Environ Microbiol 54:2078–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams A, Wilkinson A, Krehenbrink M, Russo DM, Zorreguieta A, Downie JA. 2008. Glucomannan-mediated attachment of Rhizobium leguminosarum to pea root hairs is required for competitive nodule infection. J Bacteriol 190:4706–4715. doi: 10.1128/JB.01694-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J, Kim J, Danhorn T, Merritt PM, Fuqua C. 2012. Phosphorus limitation increases attachment in Agrobacterium tumefaciens and reveals a conditional functional redundancy in adhesin biosynthesis. Res Microbiol 163:674–684. doi: 10.1016/j.resmic.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segev E, Tellez A, Vlamakis H, Kolter R. 2015. Morphological heterogeneity and attachment of Phaeobacter inhibens. PLoS One 10:e0141300. doi: 10.1371/journal.pone.0141300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fritts RK, LaSarre B, Stoner AM, Posto AL, McKinlay JB. 2017. A Rhizobiales-specific unipolar polysaccharide adhesin contributes to Rhodopseudomonas palustris biofilm formation across diverse photoheterotrophic conditions. Appl Environ Microbiol 83:e03035-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poindexter JS. 1964. Biological properties and classification of the Caulobacter group. Bacteriol Rev 28:231–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shapiro L, Agabian-Keshishian N, Bendis I. 1971. Bacterial differentiation. Science 173:884–892. doi: 10.1126/science.173.4000.884. [DOI] [PubMed] [Google Scholar]
- 23.Poindexter JS. 1981. The caulobacters: ubiquitous unusual bacteria. Microbiol Rev 45:123–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levi A, Jenal U. 2006. Holdfast formation in motile swarmer cells optimizes surface attachment during Caulobacter crescentus development. J Bacteriol 188:5315–5318. doi: 10.1128/JB.01725-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li G, Brown PJB, Tang JX, Xu J, Quardokus EM, Fuqua C, Brun YV. 2012. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol Microbiol 83:41–51. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsang PH, Li G, Brun YV, Freund LB, Tang JX. 2006. Adhesion of single bacterial cells in the micronewton range. Proc Natl Acad Sci U S A 103:5764–5768. doi: 10.1073/pnas.0601705103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li G, Smith CS, Brun YV, Tang JX. 2005. The elastic properties of the Caulobacter crescentus adhesive holdfast are dependent on oligomers of N-acetylglucosamine. J Bacteriol 187:257–265. doi: 10.1128/JB.187.1.257-265.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berne C, Ma X, Licata NA, Neves BRA, Setayeshgar S, Brun YV, Dragnea B. 2013. Physiochemical properties of Caulobacter crescentus holdfast: a localized bacterial adhesive. J Phys Chem B 117:10492–10503. doi: 10.1021/jp405802e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernando-Pérez M, Setayeshgar S, Hou Y, Temam R, Brun YV, Dragnea B, Berne C. 2018. Layered structure and complex mechanochemistry underlie strength and versatility in a bacterial adhesive. mBio 9:e02359-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith CS, Hinz A, Bodenmiller D, Larson DE, Brun YV. 2003. Identification of genes required for synthesis of the adhesive holdfast in Caulobacter crescentus. J Bacteriol 185:1432–1442. doi: 10.1128/jb.185.4.1432-1442.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toh E, Kurtz HD, Brun YV. 2008. Characterization of the Caulobacter crescentus holdfast polysaccharide biosynthesis pathway reveals significant redundancy in the initiating glycosyltransferase and polymerase steps. J Bacteriol 190:7219–7231. doi: 10.1128/JB.01003-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fiebig A, Herrou J, Fumeaux C, Radhakrishnan SK, Viollier PH, Crosson S. 2014. A cell cycle and nutritional checkpoint controlling bacterial surface adhesion. PLoS Genet 10:e1004101. doi: 10.1371/journal.pgen.1004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hershey DM, Fiebig A, Crosson S. 2019. A genome-wide analysis of adhesion in Caulobacter crescentus identifies new regulatory and biosynthetic components for holdfast assembly. mBio 10:e02273-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flemming H-C, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 35.Ong CJ, Wong ML, Smit J. 1990. Attachment of the adhesive holdfast organelle to the cellular stalk of Caulobacter crescentus. J Bacteriol 172:1448–1456. doi: 10.1128/jb.172.3.1448-1456.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardy GG, Allen RC, Toh E, Long M, Brown PJB, Cole-Tobian JL, Brun YV. 2010. A localized multimeric anchor attaches the Caulobacter holdfast to the cell pole. Mol Microbiol 76:409–427. doi: 10.1111/j.1365-2958.2010.07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berne C, Ellison CK, Ducret A, Brun YV. 2018. Bacterial adhesion at the single-cell level. Nat Rev Microbiol 16:616–627. doi: 10.1038/s41579-018-0057-5. [DOI] [PubMed] [Google Scholar]
- 38.Petrova OE, Sauer K. 2012. Sticky situations: key components that control bacterial surface attachment. J Bacteriol 194:2413–2425. doi: 10.1128/JB.00003-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greenfield LK, Whitfield C. 2012. Synthesis of lipopolysaccharide O-antigens by ABC transporter-dependent pathways. Carbohydr Res 356:12–24. doi: 10.1016/j.carres.2012.02.027. [DOI] [PubMed] [Google Scholar]
- 40.Heiss C, Burtnick MN, Black I, Azadi P, Brett PJ. 2012. Detailed structural analysis of the O-polysaccharide expressed by Burkholderia thailandensis E264. Carbohydr Res 363:23–28. doi: 10.1016/j.carres.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem 75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 42.Islam ST, Lam JS. 2014. Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can J Microbiol 60:697–716. doi: 10.1139/cjm-2014-0595. [DOI] [PubMed] [Google Scholar]
- 43.Stenutz R, Weintraub A, Widmalm G. 2006. The structures of Escherichia coli O-polysaccharide antigens. FEMS Microbiol Rev 30:382–403. doi: 10.1111/j.1574-6976.2006.00016.x. [DOI] [PubMed] [Google Scholar]
- 44.Hong Y, Reeves PR. 2014. Diversity of O-antigen repeat unit structures can account for the substantial sequence variation of Wzx translocases. J Bacteriol 196:1713–1722. doi: 10.1128/JB.01323-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geno KA, Gilbert GL, Song JY, Skovsted IC, Klugman KP, Jones C, Konradsen HB, Nahm MH. 2015. Pneumococcal capsules and their types: past, present, and future. Clin Microbiol Rev 28:871–899. doi: 10.1128/CMR.00024-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luzhetskyy A, Fedoryshyn M, Dürr C, Taguchi T, Novikov V, Bechthold A. 2005. Iteratively acting glycosyltransferases involved in the hexasaccharide biosynthesis of landomycin A. Chem Biol 12:725–729. doi: 10.1016/j.chembiol.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 47.O’Reilly MK, Zhang G, Imperiali B. 2006. In vitro evidence for the dual function of Alg2 and Alg11: essential mannosyltransferases in N-linked glycoprotein biosynthesis. Biochemistry 45:9593–9603. doi: 10.1021/bi060878o. [DOI] [PubMed] [Google Scholar]
- 48.Troutman JM, Imperiali B. 2009. Campylobacter jejuni PglH is a single active site processive polymerase that utilizes product inhibition to limit sequential glycosyl transfer reactions. Biochemistry 48:2807–2816. doi: 10.1021/bi802284d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berne C, Kysela DT, Brun YV. 2010. A bacterial extracellular DNA inhibits settling of motile progeny cells within a biofilm. Mol Microbiol 77:815–829. doi: 10.1111/j.1365-2958.2010.07267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wan Z, Brown PJB, Elliott EN, Brun YV. 2013. The adhesive and cohesive properties of a bacterial polysaccharide adhesin are modulated by a deacetylase. Mol Microbiol 88:486–500. doi: 10.1111/mmi.12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sprecher KS, Hug I, Nesper J, Potthoff E, Mahi M-A, Sangermani M, Kaever V, Schwede T, Vorholt J, Jenal U. 2017. Cohesive properties of the Caulobacter crescentus holdfast adhesin are regulated by a novel c-di-GMP effector protein. mBio 8:e00294-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kittelberger R, Hilbink F. 1993. Sensitive silver-staining detection of bacterial lipopolysaccharides in polyacrylamide gels. J Biochem Biophys Methods 26:81–86. doi: 10.1016/0165-022X(93)90024-I. [DOI] [PubMed] [Google Scholar]
- 53.Walker SG, Karunaratne DN, Ravenscroft N, Smit J. 1994. Characterization of mutants of Caulobacter crescentus defective in surface attachment of the paracrystalline surface layer. J Bacteriol 176:6312–6323. doi: 10.1128/jb.176.20.6312-6323.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Javens J, Wan Z, Hardy GG, Brun YV. 2013. Bypassing the need for subcellular localization of a polysaccharide export-anchor complex by overexpressing its protein subunits. Mol Microbiol 89:350–371. doi: 10.1111/mmi.12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hardy GG, Toh E, Berne C, Brun YV. 2018. Mutations in sugar-nucleotide synthesis genes restore holdfast polysaccharide anchoring to Caulobacter crescentus holdfast anchor mutants. J Bacteriol 200:e00597-17. [DOI] [PMC free article] [PubMed] [Google Scholar]