Salmonella gastroenteritis is one of the most common causes of foodborne disease, possibly affecting millions of people globally each year. Here we characterize the role of a novel small protein, YshB, in mediating Salmonella intracellular survival. This elucidation adds to the body of knowledge regarding how this bacterium achieves intracellular survival.
KEYWORDS: intracellular replication, invasion, Salmonella, small protein, YshB
ABSTRACT
Salmonella virulence requires the initial invasion of host cells, followed by modulation of the intracellular environment for survival and replication. In an effort to characterize the role of small RNAs in Salmonella pathogenesis, we inadvertently identified a 5-kDa protein named YshB that is involved in the intracellular survival of Salmonella. We show here that yshB expression is upregulated upon entry into macrophages. When yshB expression is upregulated before bacterial entry, invasion efficiency is inhibited. Lack of YshB resulted in reduced bacterial survival within the macrophages and led to reduced virulence in a mouse model of infection.
IMPORTANCE Salmonella gastroenteritis is one of the most common causes of foodborne disease, possibly affecting millions of people globally each year. Here we characterize the role of a novel small protein, YshB, in mediating Salmonella intracellular survival. This elucidation adds to the body of knowledge regarding how this bacterium achieves intracellular survival.
INTRODUCTION
Pathogenic bacteria have evolved complex regulatory circuits to regulate their virulence gene expression. Commonly, this is achieved by turning on or off a specific repertoire of genes once the pathogen encounters a different nutritional and host immune environment. This is evidently the case for Salmonella enterica serovar Typhimurium, with active participation in invasion followed by a lengthy intracellular phase of survival and replication within the host cells. The invasion of the nonphagocytic epithelial cells of the intestine is primarily mediated by the effectors of type III secretion system 1 (T3SS-1) (1, 2). The invasive phase is succeeded by multiplication inside the host cells within a specialized compartment called the Salmonella-containing vacuole (SCV). The transformation from the invasive phase to the state in the SCV is mediated by an elaborate network of regulatory proteins regulating the expression of invasion-related genes (3), as well as those required for maintaining bacterial replication inside the SCV (4–9). Many of the effectors of Salmonella pathogenicity island 2 (SPI-2) system are known to be responsible for the formation and biogenesis of the SCV. A few effectors have multifaceted roles in both the invasion and replication phases (10).
The precise control of gene expression related to invasion and subsequent survival is necessary for Salmonella virulence, as gene expression at unwanted times not only is redundant or energy expensive but also may be detrimental. One such example is SipB, a major effector mediating invasion that is also known to induce apoptosis in macrophages (11). Consequently, there is some degree of inverse regulation between the invasive and survival phases. Salmonella is able to sense the cues from its immediate environment as a means to switch the expression of particular sets of genes. For instance, when the bacteria reside within the host macrophage cells, they maneuver the regulatory machinery to repress the genes essential for invasion. The PhoP/PhoQ two-component system, which promotes intracellular survival, also acts as a key genetic switch to mediate repression of invasion-associated genes within macrophages by downregulating the expression of hilA, a master regulator of invasion (5, 12).
Recent studies have identified a large number of noncoding small RNAs (sRNAs), many of which have no known functions (13–15). In an effort to assess the roles of these sRNAs in Salmonella virulence, we inadvertently found a small protein, YshB, and established its role in bacterial virulence. In this report, we show that the small protein YshB is involved in intracellular replication during Salmonella infection.
RESULTS
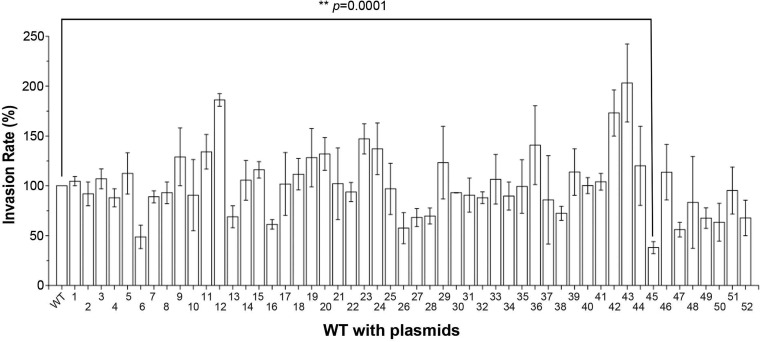
Expression of STnc1450 led to reduced S. Typhimurium invasion.
Regulatory factors often exert their functions when their own expression is upregulated (16–20). In an effort to characterize the roles of noncoding sRNAs in S. Typhimurium SL1344-mediated invasion of epithelial cells, we induced the expression of sRNAs individually from an arabinose-inducible plasmid in the wild-type Salmonella strain background. A total of 52 sRNAs (21) were successfully cloned into the arabinose-inducible vector (pZP1137). The invasion efficiency was then evaluated, using the gentamicin protection assay (22), for each of the strains carrying the sRNA-expressing plasmids, with or without l-arabinose induction. We found that induction of several sRNAs was able to reduce the invasion efficiency, compared to that of the wild-type strain (Fig. 1). Addition of l-arabinose alone did not have any noticeable effect on the invasion efficiency of Salmonella (data not shown), similar to previously reported findings (23). Induction of three sRNAs (STnc840 [P = 0.0005], STnc1420 [P = 0.001], and STnc1430 [P = 0.02]) significantly increased the relative invasion rates. Two sRNAs (STnc520 [P = 0.002] and STnc1450 [P = 0.0001]) significantly decreased invasion. Induction of the sRNA STnc1450 led to the most drastic reduction in Salmonella invasion; therefore, it was chosen for further studies here.
FIG 1.
Expression of STnc1450 led to reduced Salmonella invasion. Relative invasion rates for wild-type Salmonella strains expressing the indicated sRNAs from arabinose-inducible plasmids are shown. Gentamicin protection assays were performed on HeLa cells infected with Salmonella strains with the corresponding sRNAs. The invasion efficiency of the wild-type (WT) strain with an empty vector was normalized as 100. The data are the averages from three independent experiments, with error bars indicating standard deviations. Asterisks indicate a statistically significant difference, compared to the empty vector, using Student's t test, with the P value indicated. The sRNAs were as follows: 1, STnc110-M45; 2, STnc180-M45; 3, STnc190-M45; 4, STnc200-M45; 5, STnc290-M45; 6, STnc520-M45; 7, STnc590-M45; 8, STnc600-M45; 9, STnc740-M45; 10, STnc750-M45; 11, STnc800-M45; 12, STnc840-M45; 13, STnc850-M45; 14, STnc880-M45; 15, STnc1030-M45; 16, STnc1050-M45; 17, STnc1100-M45; 18, STnc1120-M45; 19, STnc1130-M45; 20, STnc1150-M45; 21, STnc1160-M45; 22, STnc1170-M45; 23, STnc1190-M45; 24, STnc1200-M45; 25, STnc1210-M45; 26, STnc1220-M45; 27, STnc1240-M45; 28, STnc1250-M45; 29, STnc1270-M45; 30, STnc1280-M45; 31, STnc1290-M45; 32, STnc1300-M45; 33, STnc1330-M45; 34, STnc1340-M45; 35, STnc1350-M45; 36, STnc1360-M45; 37, STnc1370-M45; 38, STnc1380-M45; 39, STnc1390-M45; 40, STnc1400-M45; 41, STnc1410-M45; 42, STnc1420-M45; 43, STnc1430-M45; 44, STnc1440-M45; 45, STnc1450-M45; 46, STnc1460-M45; 47, STnc1470-M45; 48, STnc1590-M45; 49, STnc1650-M45; 50, STnc2040-M45; 51, STnc2090-M45; and 52, STnc2130-M45.
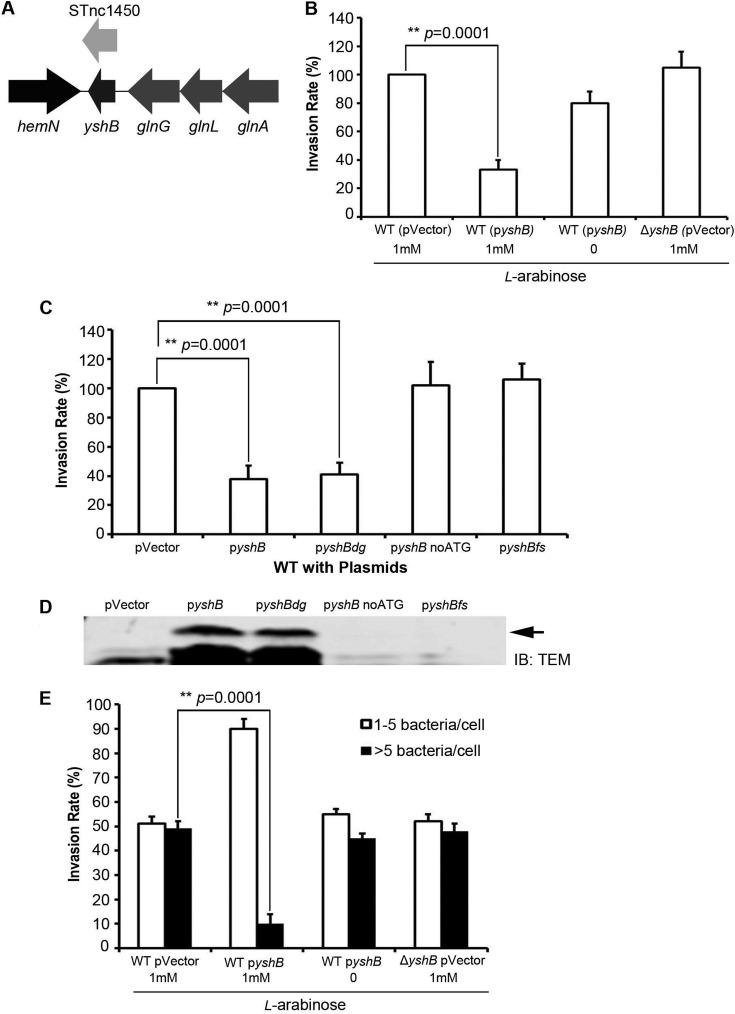
The small protein YshB encoded within the STnc1450 locus is responsible for decreased invasion.
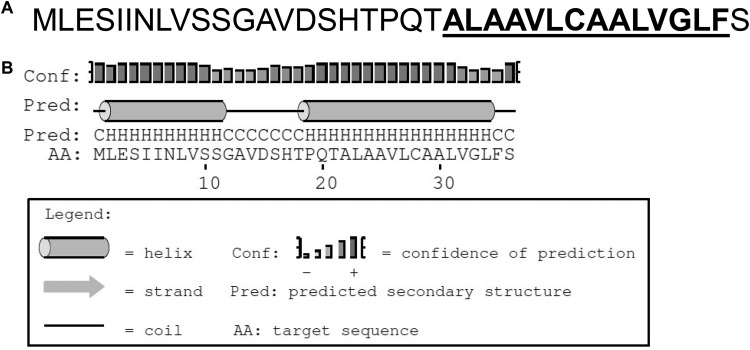
Upon further analysis, we found a putative small open reading frame, named yshB, within the predicted STnc1450 sRNA coding region (Fig. 2A). Given the presence of this putative small protein within the STnc1450 sRNA coding region, the altered invasion phenotype described above might have been due to the induction of either the protein or the sRNA. To differentiate these two possibilities, we constructed plasmids encoding the full-length small protein (pyshB), the full yshB gene without the start codon (pyshBnoATG), the full yshB gene with a frameshift mutation from deletion of the first nucleotide (pyshBfs), and the full yshB gene with a degenerate nucleotide sequence encoding the same 36 amino acids of YshB (pyshBdg). These plasmids were then introduced into wild-type Salmonella, and their invasion efficiency was evaluated using both the gentamicin protection assay and the inside/outside differential fluorescence staining assay (24). The reduced invasion phenotype was conspicuously absent when no ATG or frameshift mutations were present (Fig. 2B and C). To further demonstrate the presence of the small YshB protein, Western blotting was carried out using bacterial lysates expressing tagged YshB-TEM1 in-frame fusion proteins (Fig. 2D). A protein band corresponding to the 35-kDa YshB-TEM1 fusion protein was observed. Interestingly, the singular null yshB mutant did not have any significant change in invasion efficiency (Fig. 2B and E). Furthermore, the growth rate and motility of the bacteria were unaffected, with or without l-arabinose induction. Moreover, the defect in invasion upon YshB induction was not due to any hindrance in the capability of the bacteria to adhere to the host cells (see Fig. S1 in the supplemental material). These results indicated that the reduced invasion phenotype was due to the expression of the small protein YshB, rather than the STnc1450 sRNA. From the amino acid sequence, the secondary structure of this small protein was predicted using the PSIPRED online server (Fig. 3), with the second helix constituting the predicted transmembrane domain (25, 26). The function of YshB remains unknown.
FIG 2.
The small protein YshB within the STnc1450 locus is responsible for the reduced invasion. (A) Genomic location of STnc1450 in S. Typhimurium SL1344, showing the presence of an open reading frame encoding the small protein YshB. (B) Relative invasion rates for Salmonella strains with induced YshB from an arabinose-inducible plasmid (pZP3628) in the wild-type (WT) (SL1344) background, evaluated with the gentamicin protection assay. (C) Relative invasion rates assessed similarly for YshB encoded by degenerate nucleotide sequences (pZP3634), with no start codon (pZP3399), or with frameshifted yshB (pZP3400). (D) Immunoblot showing the expression of YshB from pyshB (pZP3628) and pyshBdg (pZP3634). The bacterial cell lysates were subjected to SDS-PAGE, followed by immunoblotting (IB) with anti-TEM1 antibody. The arrow indicates the 35-kDa YshB-TEM1 fusion protein. (E) Inside/outside differential staining for the wild-type, ΔyshB, and YshB-induced (pZP3628) strains. Percentages of cells with either 1 to 5 or >5 internalized bacteria were enumerated. Asterisks indicate statistically significant differences, with the indicated P values. Three independent experiments were carried out, with the means ± standard deviations (error bars) shown.
FIG 3.
The small protein YshB. (A) Amino acid sequence information for small protein YshB. (B) Predicted secondary structure for YshB using PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred).
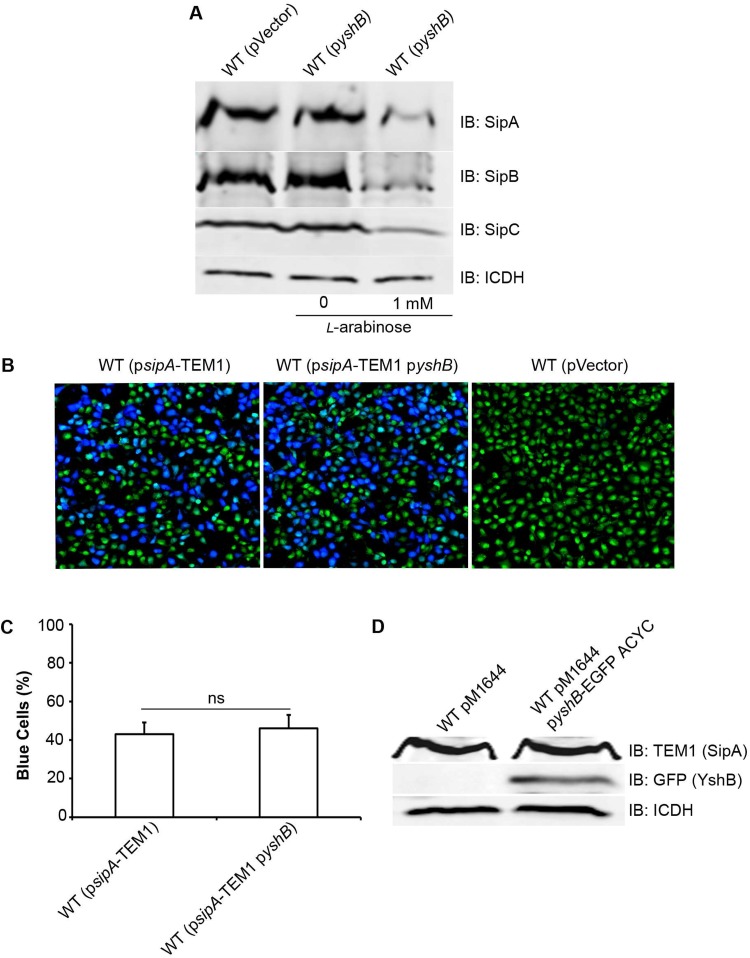
Induction of YshB decreases the expression of invasion-related genes.
SPI-1-encoded T3SS effectors are known to play key roles in modulating host cell actin cytoskeleton rearrangements to promote bacterial invasion (2, 27). It is known that the expression of these T3SS effectors is tightly regulated in response to environmental conditions that the bacteria encounter (28, 29). Thus, it is possible that the YshB-induced reduction in bacterial invasion is mediated by a change in SPI-1 effector expression. To test this possibility, we examined the expression of SipA, SipB, and SipC in bacterial cell lysates when YshB was induced (Fig. 4A). We found that levels of SipA, SipB, and SipC were reduced in strains expressing YshB, compared to the results for the strain carrying the vector only or the results with no YshB induction (Fig. 4). These results are consistent with our notion that the reduced invasion seen for the YshB-induced strain is due to reductions in SipA, SipB, and SipC levels.
FIG 4.
Induction of YshB reduces the expression of invasion genes. (A) Immunoblot (IB) analysis of bacterial whole-cell lysates for the expression of SipA, SipB, and SipC after growth under SPI-1-inducing conditions, with and without YshB induction (pZP3628). Polyclonal rabbit anti-SipA, anti-SipB, and anti-SipC primary antibodies were used, and rabbit anti-ICDH was used to probe the loading control. (B) β-Lactamase-based translocation assay to assess the translocation efficiency of SipA from pM1644 in YshB-induced (pZP3682) cells. Wild-type (WT) Salmonella strains with pM1644 and plasmid pZP2277, in which the SipA from pM1644 was replaced by the RfA cassette of the Gateway vector conversion system, were used as positive and negative controls, respectively. The translocation efficiency was evaluated by enumerating blue versus green cells under a confocal laser scanning microscope. (C) Percentage of blue cells in panel B, to quantitate the SipA-TEM1 translocation efficiency. Three independent experiments were carried out, and the means ± standard deviations (error bars) are shown. (D) Immunoblot analysis of the bacterial culture used in panel B, showing that SipA expression from pM1644 remains unaffected when YshB is overexpressed (pZP3682).
Type III effectors are translocated into nonphagocytic epithelial cells to exert their functions during bacterial invasion (2). Therefore, a change in the translocation efficiency could potentially affect bacterial invasion. To test the translocation efficiency of the SPI-1-encoded T3SS apparatus, we measured the translocation of SipA into HeLa cells using the β-lactamase translocation assay (30). As shown in Fig. 4, the translocation of SipA remained similar with or without the induction of YshB. In addition, we tested the possibility of YshB itself being a T3SS-translocated effector by conducting the same translocation assay (Fig. S2). Our results showed that YshB was not translocated.
YshB is required for efficient intracellular Salmonella replication.
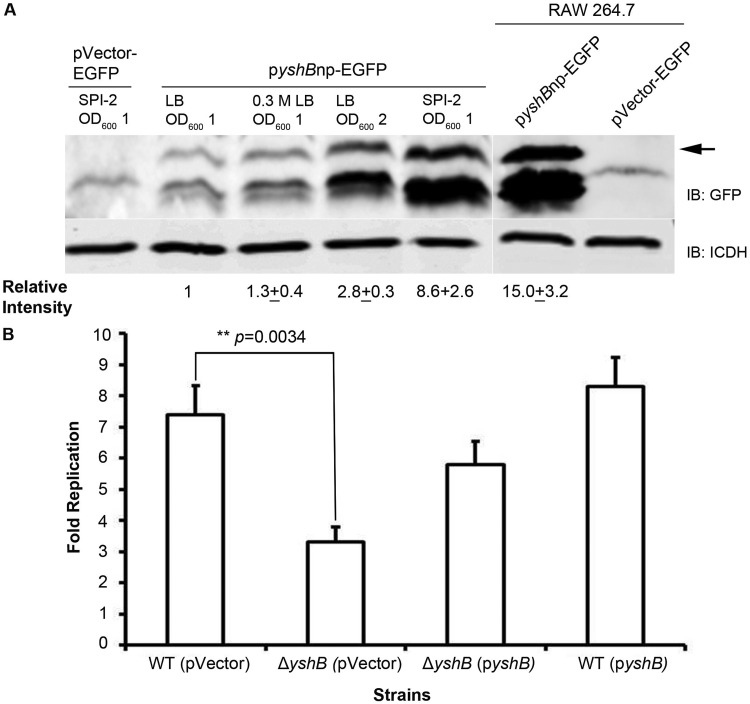
Gene expression is tightly regulated by the immediate environment that the pathogen encounters. Previous studies have suggested that Salmonella responds to the environmental changes by regulating virulence expression to cope with hostile surroundings. To understand how YshB might function, we conducted experiments to assess the expression of YshB from its native promoter under various conditions. For the in vitro conditions, we selected different growth stages grown in Luria-Bertani (LB) broth, with or without 0.3 M NaCl (invasion-inducing medium), or SPI-2 medium (mimicking intracellular conditions). We also examined the YshB expression levels when the bacteria were inside macrophage cells. Our results indicated that YshB expression levels were considerably higher when the bacteria were grown in SPI-2 medium and when the bacteria were inside macrophage cells (intracellular phase of the life cycle) (Fig. 5A).
FIG 5.
YshB expression is upregulated following bacterial invasion, and YshB is required for efficient intracellular Salmonella replication. (A) Immunoblot (IB) analysis to compare the levels of endogenous YshB expression from its native promoter (pZP3637) under various in vitro conditions and inside RAW 264.7 macrophage cells. Relative band intensity was determined by dividing the intensity of the YshB-EGFP band by that of the ICDH band, normalized to the value for the LB broth condition (OD600 of 1). The normalized relative intensity is the average value from two independent experiments, with the indicated standard deviation. The arrow indicates the 31-kDa YshB-EGFP. (B) Replication fold of the ΔyshB Salmonella strain, along with the complemented strain (pZP3637) and YshB-induced strain (pZP3637), by the gentamicin protection assay. WT, wild type. Data represent means from three independent experiments, with standard deviations. The asterisks indicate a statistically significant difference, with the P value indicated.
These data showed that YshB expression was upregulated inside the host cells, suggesting that YshB might play a role during the intracellular stage of the bacterial life cycle. Once Salmonella successfully invades host cells, it switches its priorities to modulating the intracellular microenvironment, which consists primarily of a modified phagosome known as the SCV, to promote survival and replication (31). Thus, we tested whether YshB was involved in bacterial survival and replication in macrophages. RAW 264.7 macrophages were infected for 30 min with bacteria opsonized with normal mouse serum, as described in Materials and Methods. The replication fold was determined by dividing the number of intracellular bacteria at 18 h by the number at 2 h. Our data showed that YshB indeed plays a role in the survival of Salmonella inside macrophage cells (Fig. 5B). This finding corroborates the observation that YshB expression is upregulated inside host cells.
YshB is required for full virulence in mice.
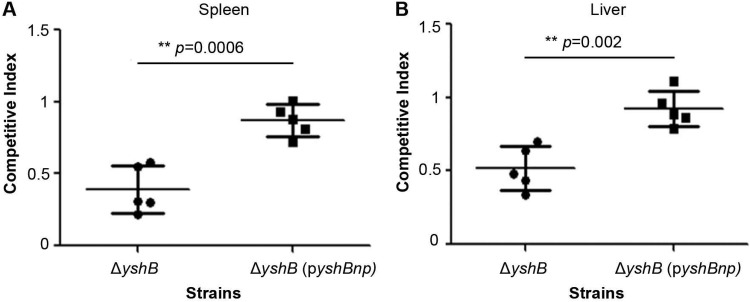
The competitive index assay is frequently used to assess the virulence of bacterial pathogens in a mouse model of infection. To assess whether YshB plays a role in Salmonella virulence in the mouse infection model, we employed mixed infections of the wild-type strain, the ΔyshB mutant, and the ΔyshB strain complemented with a plasmid expressing YshB; this was facilitated by the fact that the ΔyshB strain had a kanamycin cassette replacing the yshB gene, so that it could be selected on double-antibiotic (streptomycin and kanamycin) LB plates, as opposed to the wild-type strain, which is resistant to streptomycin and sensitive to kanamycin. The competitive index for the ΔyshB strain was found to be 0.39 for spleen colonization and 0.52 for liver colonization (Fig. 6). When the small protein was expressed from its native promoter via a plasmid, the virulence was restored almost to the wild-type levels. Our results demonstrated that lack of the yshB gene led to reduced virulence in the mouse model of infection.
FIG 6.
YshB is required for full virulence in mice. The competitive index assay was conducted using mixed bacterial cultures for infection. Mixtures containing wild-type and ΔyshB strains or wild-type and complemented (pZP3637) strains were used to infect BALB/c mice intraperitoneally. The spleen (A) and liver (B) from the animals were harvested 2 days postinfection to determine the competitive index of the strains with respect to the wild-type strain. Asterisks indicate statistical significance, with indicated P values.
DISCUSSION
We initially sought to investigate the roles of Salmonella sRNAs in the invasion of host epithelial cells. Instead, we found a small protein, YshB, encoded within a putative sRNA molecule that was able to affect bacterial invasion and intracellular survival. Wadler and Vanderpool reported that a 43-amino-acid small protein, SgrT, was encoded within a transcript of the sRNA SgrS (32). In recent years, small proteins have been identified as an important group of molecules capable of orchestrating a wide range of cellular processes in bacteria. By convention, small proteins are defined as those with ≤50 amino acids (33). Small proteins mediate a variety of functions in bacteria, including spore formation, transport of nutrients, regulation of membrane-bound enzymes, and signal transduction, among others. Owing to their small sizes, these proteins more commonly act in mechanical ways via direct interaction with their targets (34). One example of such proteins is MciZ, which inhibits the activity of FtsZ, a main player in bacterial cell division, by binding directly to it (35). Similarly, MgrB is a small protein that negatively regulates the PhoP/PhoQ system in Escherichia coli (36). In that study, the authors were able to demonstrate that MgrB binds to and represses the activity of PhoQ, via a negative feedback loop. Sda is another cytosolic small protein, which blocks the interactions between different kinases involved in Bacillus subtilis sporulation (37). Interestingly, the interplay between a small protein and its target can also promote the interactions between domains or proteins. In one example, MgtR, a 30-amino-acid small protein in Salmonella, assists the protease activity of FtsH to degrade the virulence factor MgtC (38). MgtR was shown to directly interact with MgtC, thereby facilitating its degradation by FtsH. This occurred presumably by changing the conformation of MgtC, rendering it sensitive to the protease. Moreover, some small proteins are known to function as chaperones. Smaldone et al. suggested that the small protein FbpB may aid the activity of the sRNA FsrA by acting as its chaperone (39).
YshB is a 36-amino-acid small protein that was first annotated in a screening based on the presence of ribosome binding sites (40) and was later validated as a protein-expressing gene with a predicted transmembrane domain in E. coli (25). This protein is ubiquitously present in Salmonella species, with 100% sequence identity among the common pathogenic serovars S. Typhimurium, S. enterica serovar Typhi, S. enterica serovar Enteritidis, and S. enterica serovar Choleraesuis. Further, among the members of Enterobacteriaceae, the Salmonella YshB sequence is 92% identical (33 of 36 amino acids) to the Escherichia coli (strain K-12), Shigella, and Klebsiella oxytoca protein sequences. Prior to this study, YshB had no known functions or any described roles in bacterial virulence. We reported here that YshB acts as a positive regulator of bacterial intracellular survival. Our data suggest that, when induced within the bacteria, YshB may function to downregulate Salmonella invasion. It is tempting to speculate that the induction of YshB represents the intracellular phase and consequently triggers downregulation of the invasion machinery to facilitate intracellular survival and replication. Although our data demonstrated that the small protein YshB is responsible for coordinating invasion and intracellular survival, we cannot rule out the possibility that the originally annotated sRNA STnc1450 serves an unknown independent function. Such a dual-function example has been described previously (32).
Salmonella encounters various environmental conditions under which the bacteria have to compete to survive before, during, and after infection. In order to ensure a successful transition to a new environment, Salmonella regulates the expression of T3SS-1 genes encoded in SPI-1 in locations where these gene products are needed. Multiple regulatory factors are responsible for regulating this process in response to environmental signals. Once the invasion of the host cell is complete, Salmonella is known to downregulate the expression of T3SS-1 genes and initiates SCV biogenesis, for which T3SS-2 induction is essential (41, 42). A complex network of regulators that are responsible for coordinating their expression has been identified. One example of this is the repression of Salmonella SPI-1 genes as the bacteria reach the proximal sites of the small intestine, where the expression of invasion genes is no longer necessary (43). Furthermore, the cationic peptides produced inside the macrophages are also known to silence the expression of SPI-1 genes when invasion is no longer desired (44).
Interestingly, even the SPI-2 system has been known to regulate the expression of SPI-1 genes involved in bacterial invasion (45–47). PhoP is a response regulator and is known to repress the expression of SPI-1 genes (48), while activating the expression of pag genes, including mgtCB, pmrB, and SPI-2 genes, inside macrophages. The fact that pag genes are transcriptionally activated when the bacteria are inside macrophages was observed when as many as eight proteins were absent in a phoP mutant, compared to the wild-type strain, following macrophage uptake (49). This differential regulation corresponds to the spatial localization of the bacteria in the macrophages, where pag genes are needed, as opposed to being in the small intestine, where most SPI-1 genes are needed (12, 46, 50–52). Furthermore, the PhoP/PhoQ two-component regulatory system needs to remain downregulated for the induction of SPI-1 genes to promote bacterial invasion (12, 53). A seemingly narrow cation concentration range is responsible for the activation of most pag genes, with optimal expression at concentrations of <100 μM; at concentrations of ≥2 mM, expression is completely turned off (51). This corresponds well with the respective sites in the host, with the cation concentrations being relatively higher in the intestinal environment, compared to conditions when the bacteria are inside the phagosomes (52). The PhoP/PhoQ system represents an exquisite example of spatiotemporal control of gene expression in Salmonella. Our data showed that YshB was upregulated when the bacteria were inside the macrophages. Further study is required to elucidate whether YshB interacts with the PhoP/PhoQ system to regulate the transition from the extracellular phase to that inside the host cells.
Our data showed that, under the conditions in which YshB expression is induced, such as inside macrophages, the invasion efficiency is markedly reduced, and this is important for intracellular survival as well. It is known that the expression of unnecessary genes results in energy expenditure and sometimes even is detrimental. In a study to examine the temporal expression of invasion genes, Boddicker and Jones found that downregulation of invasion gene expression was partly mediated by the Lon protease and this downregulation was essential for the intracellular survival of Salmonella (54). Uncalled expression of SPI-1 genes inside the macrophage results in excessive induction of apoptosis, which is countered by the Lon protease (54, 55). This was supported by the previous findings by Takaya et al. that transcription of SPI-1 genes was enhanced in a Δlon mutant (56, 57).
We attempted to identify Salmonella proteins that may interact with YshB by using genetic bacterial two-hybrid screening and biochemical coimmunoprecipitation assays, and we did not find any valuable targets. It is possible that we failed to identify the potential target of YshB in these screenings. Alternatively, the target for this small protein might be nonprotein, such as a sRNA. Hence, further analysis of how YshB modulates bacterial virulence is needed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and mammalian cell lines.
Salmonella and E. coli strains were routinely grown in LB broth. For the induction of SPI-1 machinery, Salmonella strains were grown in LB broth with 0.3 M NaCl (final concentration). N-salts minimal medium with a low Mg2+ concentration (SPI-2 medium) was used for the induction of SPI-2 components (46). Where applicable, antibiotics were used at the following concentrations: streptomycin, 150 μg ml−1; ampicillin, 120 μg ml−1; kanamycin, 40 μg ml−1; tetracycline, 15 μg ml−1.
Isogenic derivatives of the virulent wild-type SL1344 strain of Salmonella Typhimurium were used in this study (58). In-frame chromosomal deletions of genes and chromosomal-flag-tagged constructs in Salmonella strains were generated by the one-step gene disruption strategy using the λ-Red recombination system (59, 60). HeLa (CCL-2) and RAW 264.7 cell lines were purchased from ATCC (Manassas, VA) and maintained in Dulbecco's modified Eagle's medium (DMEM) (VWR) supplemented with 10% fetal bovine serum (FBS) (Gibco).
Primers used in this study are listed in Table S1 in the supplemental material. For inducible expression of various YshB constructs, PCR-generated DNA fragments were cloned into the EcoRI and XmaI sites of an arabinose-inducible plasmid (pZP1137) so that it was in frame with the M45 tag at its C terminus. To facilitate the visualization of YshB by Western blotting, a translational fusion with a double M45-TEM1 tag derived from plasmid pM1644 (31) was constructed at the C-terminal end of YshB in pZP1137 to generate plasmid pyshB (pZP3628). The resulting YshB-M45-TEM1 fusion protein expressed from this plasmid had a molecular size of approximately 35 kDa. The same M45-TEM1 tag was also cloned into the empty pZP1137 vector (pVector-TEM1 [pZP3635]) and used as a control. Another control, pVector-enhanced green fluorescent protein (EGFP) plasmid (pZP3636), was constructed by subcloning the 759-nucleotide fragment generated from the XmaI/XbaI digest of pEGFP-N2 into the same sites of pZP1137. To construct a plasmid in which yshB would be expressed from its native promoter (pyshBnp [pZP3637]), a 254-nucleotide fragment was amplified from the Salmonella genome, including 130 nucleotides upstream of yshB. This fragment was cloned into the XmaI site of the aforementioned vector control plasmid (pVector-EGFP [pZP3636]). The resulting plasmid expressed a YshB-EGFP fusion protein with a molecular size of 31 kDa. To construct the plasmid pyshBnoATG (pZP3399), the forward primers were designed so that the ATG for yshB was omitted. The PCR product was amplified and cloned into the EcoRI and XmaI sites of pZP1137. Plasmid pyshBfs (pZP3400) was constructed by designing a primer in which the first adenosine nucleotide for yshB was omitted, which resulted in a shift for the rest of the reading frame. Moreover, plasmid pyshBdg (pZP3634) was constructed by annealing degenerate oligonucleotides coding for the same 36 amino acids of YshB into the EcoRI and XmaI sites of pVector-TEM1. Briefly, degenerate nucleotide sequences from the wild-type sequence of the yshB gene were designed so that they had EcoRI and XmaI overhangs. The overlapping oligonucleotides were resuspended in annealing buffer (10 mM Tris [pH 7.5 to 8.0], 50 mM NaCl, 1 mM EDTA) and mixed in equimolar concentrations. The annealing step was performed by heating the mixture to 100°C for 5 min and slowly equilibrating it to room temperature (61). The insert was then cloned into the EcoRI and XmaI sites of pVector-TEM1.
For expression within macrophages, yshB was expressed from its native promoter (pZP3637). Furthermore, a plasmid used for cotransformation with pM1644 was constructed by subcloning yshB-EGFP into the XhoI and NotI sites of pACYC-184SK to generate pyshB-EGFP-ACYC (pZP3682). The sipA gene in pM1644 was replaced by yshB by subcloning to generate plasmid pyshB-TEM1 (pZP3639), which was used to assess the translocation of YshB into host cells. All plasmid constructs were verified by restriction digestion and sequencing.
Growth rate and motility assays.
For growth rate measurements, overnight LB broth cultures of bacteria were subcultured at a 1:100 dilution in 10 ml LB broth and grown at 37°C in a shaker (200 rpm). Serial dilutions of the culture were then plated every 40 min, and bacteria were enumerated. A swarm plate assay was performed to assess bacterial motility (62). Briefly, semisolid LB plates (0.25% agar) with or without 1 mM l-arabinose were prepared. From the overnight cultures of bacteria, 100 μl was inoculated into 2.5 ml LB broth and allowed to reach mid-log phase (optical density at 600 nm [OD600] of ∼0.5 to 0.6). From this culture, 1 μl was spotted on a semisolid agar plate and incubated at 37°C. The size of the halo around the site of inoculation was measured every 30 min, to evaluate the bacterial swarm/motility.
Invasion efficiency.
The invasion efficiencies were assessed with the classic gentamicin protection assay (63). In brief, overnight LB broth cultures of Salmonella were subcultured at 37°C after a 1:30 dilution in 0.3 M NaCl LB broth to an OD600 of 1.0. HeLa cells were then infected for 15 min in a 24-well plate at a multiplicity of infection (MOI) of 10 and incubated at 37°C in a 5% CO2 incubator. After infection, cells were washed three times with phosphate-buffered saline (PBS), followed by the addition of DMEM containing 100 μg ml−1 of gentamicin and further incubation for 1 h to kill extracellular bacteria. Cells were again washed three times with PBS and were lysed with 0.1% sodium deoxycholate. Cell lysates were then serially diluted and plated on LB plates for colony enumeration.
The ability of bacteria to invade host cells was also evaluated with the inside/outside differential staining assay, as described previously (24). Briefly, HeLa cells were infected for 15 min with Salmonella at an MOI of 5. The bacteria that remained outside the infected cells were identified using rabbit anti-Salmonella O-antigen and were visualized with anti-rabbit IgG conjugated with Alexa Fluor 488 (Molecular Probes). This was followed by permeabilization of the infected cells with 0.2% Triton X-100 and counterstaining of both intracellular and extracellular bacteria with Texas Red (Molecular Probes). Host cell nuclei and bacterial DNA were stained with 4,6-diamidino-2-phenylindole (DAPI) (Molecular Probes). The bacteria outside would appear green or yellow and the bacteria inside would be red when visualized under a double filter for both Alexa Fluor 488 and Texas Red. The invasion efficiency was determined by enumerating the percentages of cells with either 1 to 5 or >5 internalized bacteria per host cell. The experiment was performed in triplicate, with approximately 300 cells counted for each sample.
Intracellular survival in macrophages.
Bacterial infection of RAW 264.7 macrophages and intracellular survival assays were carried out by the gentamicin protection assay, as described previously (47). Briefly, Salmonella strains were grown to early stationary phase and diluted to an OD600 of 0.1. The bacteria were opsonized for 20 min at 37°C in DMEM containing 10% normal mouse serum (Gemini Bio-Products, Woodland, CA). Opsonized bacteria were added to RAW 264.7 macrophages at an MOI of 10 in 24-well plates and were incubated for 30 min at 37°C in 5% CO2 for infection. This was followed by washing the cells three times with PBS and further incubating them for 1 h in DMEM with 10% FBS and 100 μg ml−1 of gentamicin, to kill extracellular bacteria. Infected cells were then washed again three times with PBS and incubated for the indicated times in DMEM containing 16 μg ml−1 of gentamicin. At 2 and 18 h postinfection, the infected macrophages were washed three times with PBS and lysed with 0.1% sodium deoxycholate. Cell lysates were then serially diluted and plated onto LB plates for bacterial enumeration. The replication fold was determined by dividing the number of intracellular bacteria at 18 h by the number at 2 h.
Competitive index assay in a mouse model of infection.
Bacteria were grown overnight at 37°C in LB medium. The bacteria were then harvested by centrifugation and resuspended in sterile PBS. Indicated combinations of Salmonella strains were mixed in a 1:1 ratio. Serial dilutions were prepared for each bacterial mixture and plated onto LB plus streptomycin plates and LB plus streptomycin plus kanamycin plates to calculate the input inocula. Both wild-type and mutant strains would grow on the LB plus streptomycin plates, whereas LB plus streptomycin plus kanamycin plates would act as selective medium for only the mutant strains used in this study. Around 104 CFU of bacteria in 200 μl were injected intraperitoneally into 6- to 8-week-old female BALB/c mice (5 mice for each mixture sample). The mice were sacrificed 2 days postinfection by CO2 euthanasia followed by cervical dislocation. The spleen and liver from each mouse were harvested and homogenized aseptically in sterile PBS. Serial dilutions were carried out and plated onto LB plus streptomycin plates and LB plus streptomycin plus kanamycin plates for output enumeration. The competitive index was calculated as the ratio between the mutant strain and the wild-type strain in the output divided by the ratio of the two strains in the input (64). All animal experiments were conducted in strict accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, following a protocol approved by the Purdue Animal Care and Use Committee (protocol 1111000320).
Adherence and protein translocation assays.
To measure bacterial adherence to mammalian cells, HeLa cells were treated with cytochalasin D (Sigma), an inhibitor of actin filamentation, at a final concentration of 1 μg ml−1 for 30 min before infection (65) to stall bacterial internalization. Salmonella strains were added to the treated cells at an MOI of 10 and incubated for 15 min to facilitate adherence to the host cells. The cells were then washed three times with PBS to remove the unattached bacteria, followed by plating for bacterial enumeration.
To measure translocation of bacterial effectors into the host cells, HeLa cells were infected with Salmonella strains expressing the corresponding β-lactamase fusions (derived from pM1644) in a 96-well plate at an MOI of 10. CCF4-AM (Invitrogen, Carlsbad, CA) was then added to the wells 15 min postinfection, and the cells were incubated for 2 h at room temperature. CCF4, which emits green fluorescence, is a β-lactamase substrate and emits blue fluorescence upon cleavage. Infected cells were then examined under the fluorescence microscope to quantify the numbers of green and blue cells. Experiments were performed in triplicate, and approximately 300 cells were counted in each sample.
SDS-PAGE and immunoblotting.
To prepare bacterial samples for SDS-PAGE and immunoblotting, bacterial strains were grown overnight in LB broth, followed by subculturing with 1:30 dilutions in the respective medium and incubation under the indicated conditions. The bacterial cultures were then centrifuged at 10,000 × g for 5 min, and the cell pellets were lysed with 2× Laemmli sample buffer and boiled for 10 min. For intracellular infections, a stationary-phase LB culture was used to infect macrophages at an MOI of 10 for 30 min. Infected macrophages were lysed with 0.1% sodium deoxycholate 12 h postinfection. The lysate was centrifuged at 500 × g for 5 min to remove cell debris. The supernatant was then centrifuged at 10,000 × g for 5 min to collect the intracellular bacteria. The pellet was subsequently lysed with 2× Laemmli sample buffer and boiled for 10 min. Samples were run on a 10% SDS-polyacrylamide gel and immunoblotted using the respective antibodies. The SipA, SipB, SipC, and GFP primary antibodies were rabbit polyclonal antibodies, whereas the anti-TEM1 was a mouse monoclonal antibody. Rabbit anti-isocitrate dehydrogenase (anti-ICDH) was used to probe ICDH protein as a loading control. The anti-TEM1 mouse monoclonal antibody and rabbit anti-ICDH polyclonal antibody were gifts from Zhao-qing Luo (Purdue University).
Supplementary Material
ACKNOWLEDGMENTS
We thank Jay Hinton for providing the list of sRNAs.
This work was funded by NIH grant R21AI128134 to D.Z.
R.B., M.Z., and D.Z. conceptualized the project, R.B., M.Z., and D.Z. designed the experiments, R.B. and M.Z. performed the experiments, R.B. and M.Z. wrote the original manuscript, R.B., M.Z., and D.Z. reviewed and edited the manuscript, and D.Z. supervised the project.
We declare no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00314-19.
REFERENCES
- 1.Galan JE, Curtiss R III.. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci U S A 86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galan JE, Zhou D. 2000. Striking a balance: modulation of the actin cytoskeleton by Salmonella. Proc Natl Acad Sci U S A 97:8754–8761. doi: 10.1073/pnas.97.16.8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golubeva YA, Sadik AY, Ellermeier JR, Slauch JM. 2012. Integrating global regulatory input into the Salmonella pathogenicity island 1 type III secretion system. Genetics 190:79–90. doi: 10.1534/genetics.111.132779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller SI, Kukral AM, Mekalanos JJ. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci U S A 86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behlau I, Miller SI. 1993. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J Bacteriol 175:4475–4484. doi: 10.1128/jb.175.14.4475-4484.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tu X, Latifi T, Bougdour A, Gottesman S, Groisman EA. 2006. The PhoP/PhoQ two-component system stabilizes the alternative sigma factor RpoS in Salmonella enterica. Proc Natl Acad Sci U S A 103:13503–13508. doi: 10.1073/pnas.0606026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakowski MA, Braun V, Brumell JH. 2008. Salmonella-containing vacuoles: directing traffic and nesting to grow. Traffic 9:2022–2031. doi: 10.1111/j.1600-0854.2008.00827.x. [DOI] [PubMed] [Google Scholar]
- 8.Schroeder N, Mota LJ, Meresse S. 2011. Salmonella-induced tubular networks. Trends Microbiol 19:268–277. doi: 10.1016/j.tim.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Galan JE. 2001. Salmonella interactions with host cells: type III secretion at work. Annu Rev Cell Dev Biol 17:53–86. doi: 10.1146/annurev.cellbio.17.1.53. [DOI] [PubMed] [Google Scholar]
- 10.Agbor TA, McCormick BA. 2011. Salmonella effectors: important players modulating host cell function during infection. Cell Microbiol 13:1858–1869. doi: 10.1111/j.1462-5822.2011.01701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hersh D, Monack DM, Smith MR, Ghori N, Falkow S, Zychlinsky A. 1999. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc Natl Acad Sci U S A 96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bajaj V, Lucas RL, Hwang C, Lee CA. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol 22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- 13.Sittka A, Pfeiffer V, Tedin K, Vogel J. 2007. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol 63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong H, Vu GP, Bai Y, Chan E, Wu R, Yang E, Liu F, Lu S. 2011. A Salmonella small non-coding RNA facilitates bacterial invasion and intracellular replication by modulating the expression of virulence factors. PLoS Pathog 7:e1002120. doi: 10.1371/journal.ppat.1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hebrard M, Kroger C, Srikumar S, Colgan A, Handler K, Hinton JC. 2012. sRNAs and the virulence of Salmonella enterica serovar Typhimurium. RNA Biol 9:437–445. doi: 10.4161/rna.20480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, Morozov I, Baker CS, Georgellis D, Babitzke P, Romeo T. 2003. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol Microbiol 48:657–670. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- 17.Pessi G, Williams F, Hindle Z, Heurlier K, Holden MT, Camara M, Haas D, Williams P. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J Bacteriol 183:6676–6683. doi: 10.1128/JB.183.22.6676-6683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sledjeski D, Gottesman S. 1995. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc Natl Acad Sci U S A 92:2003–2007. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fettes PS, Forsbach-Birk V, Lynch D, Marre R. 2001. Overexpresssion of a Legionella pneumophila homologue of the E. coli regulator csrA affects cell size, flagellation, and pigmentation. Int J Med Microbiol 291:353–360. doi: 10.1078/1438-4221-00141. [DOI] [PubMed] [Google Scholar]
- 20.Nishino K, Yamaguchi A. 2001. Overexpression of the response regulator evgA of the two-component signal transduction system modulates multidrug resistance conferred by multidrug resistance transporters. J Bacteriol 183:1455–1458. doi: 10.1128/JB.183.4.1455-1458.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kroger C, Dillon SC, Cameron AD, Papenfort K, Sivasankaran SK, Hokamp K, Chao Y, Sittka A, Hebrard M, Handler K, Colgan A, Leekitcharoenphon P, Langridge GC, Lohan AJ, Loftus B, Lucchini S, Ussery DW, Dorman CJ, Thomson NR, Vogel J, Hinton JC. 2012. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci U S A 109:E1277–E1286. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Cai X, Wu S, Bomjan R, Nakayasu ES, Handler K, Hinton JCD, Zhou D. 2017. InvS coordinates expression of PrgH and FimZ and is required for invasion of epithelial cells by Salmonella enterica serovar Typhimurium. J Bacteriol 199:e00824-16. doi: 10.1128/JB.00824-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López-Garrido J, Puerta-Fernández E, Cota I, Casadesús J. 2015. Virulence gene regulation by l-arabinose in Salmonella enterica. Genetics 200:807–819. doi: 10.1534/genetics.115.178103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li D, Wang X, Wang L, Zhou D. 2013. The actin-polymerizing activity of SipA is not essential for Salmonella enterica serovar Typhimurium-induced mucosal inflammation. Infect Immun 81:1541–1549. doi: 10.1128/IAI.00337-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hemm MR, Paul BJ, Schneider TD, Storz G, Rudd KE. 2008. Small membrane proteins found by comparative genomics and ribosome binding site models. Mol Microbiol 70:1487–1501. doi: 10.1111/j.1365-2958.2008.06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hemm MR, Paul BJ, Miranda-Rios J, Zhang A, Soltanzad N, Storz G. 2010. Small stress response proteins in Escherichia coli: proteins missed by classical proteomic studies. J Bacteriol 192:46–58. doi: 10.1128/JB.00872-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou D. 2006. Bacterial invasion into non-phagocytic cells, p 247–273. In Nickerson CA, Schurr MJ (ed), Molecular paradigms of infectious disease: a bacterial perspective. Springer Science+Business Media, New York, NY. [Google Scholar]
- 28.Akbar S, Schechter LM, Lostroh CP, Lee CA. 2003. AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella typhimurium. Mol Microbiol 47:715–728. doi: 10.1046/j.1365-2958.2003.03322.x. [DOI] [PubMed] [Google Scholar]
- 29.Rakeman JL, Bonifield HR, Miller SI. 1999. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J Bacteriol 181:3096–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlumberger MC, Kappeli R, Wetter M, Muller AJ, Misselwitz B, Dilling S, Kremer M, Hardt WD. 2007. Two newly identified SipA domains (F1, F2) steer effector protein localization and contribute to Salmonella host cell manipulation. Mol Microbiol 65:741–760. doi: 10.1111/j.1365-2958.2007.05823.x. [DOI] [PubMed] [Google Scholar]
- 31.Malik-Kale P, Jolly CE, Lathrop S, Winfree S, Luterbach C, Steele-Mortimer O. 2011. Salmonella: at home in the host cell. Front Microbiol 2:125. doi: 10.3389/fmicb.2011.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wadler CS, Vanderpool CK. 2007. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc Natl Acad Sci U S A 104:20454–20459. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramamurthi KS, Storz G. 2014. The small protein floodgates are opening; now the functional analysis begins. BMC Biol 12:96. doi: 10.1186/s12915-014-0096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Storz G, Wolf YI, Ramamurthi KS. 2014. Small proteins can no longer be ignored. Annu Rev Biochem 83:753–777. doi: 10.1146/annurev-biochem-070611-102400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Handler AA, Lim JE, Losick R. 2008. Peptide inhibitor of cytokinesis during sporulation in Bacillus subtilis. Mol Microbiol 68:588–599. doi: 10.1111/j.1365-2958.2008.06173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salazar ME, Podgornaia AI, Laub MT. 2016. The small membrane protein MgrB regulates PhoQ bifunctionality to control PhoP target gene expression dynamics. Mol Microbiol 102:430–445. doi: 10.1111/mmi.13471. [DOI] [PubMed] [Google Scholar]
- 37.Jacques DA, Streamer M, Rowland SL, King GF, Guss JM, Trewhella J, Langley DB. 2009. Structure of the sporulation histidine kinase inhibitor Sda from Bacillus subtilis and insights into its solution state. Acta Crystallogr D Biol Crystallogr 65:574–581. doi: 10.1107/S090744490901169X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alix E, Blanc-Potard AB. 2008. Peptide-assisted degradation of the Salmonella MgtC virulence factor. EMBO J 27:546–557. doi: 10.1038/sj.emboj.7601983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smaldone GT, Antelmann H, Gaballa A, Helmann JD. 2012. The FsrA sRNA and FbpB protein mediate the iron-dependent induction of the Bacillus subtilis lutABC iron-sulfur-containing oxidases. J Bacteriol 194:2586–2593. doi: 10.1128/JB.05567-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosuge T, Abe T, Okido T, Tanaka N, Hirahata M, Maruyama Y, Mashima J, Tomiki A, Kurokawa M, Himeno R, Fukuchi S, Miyazaki S, Gojobori T, Tateno Y, Sugawara H. 2006. Exploration and grading of possible genes from 183 bacterial strains by a common protocol to identification of new genes: Gene Trek in Prokaryote Space (GTPS). DNA Res 13:245–254. doi: 10.1093/dnares/dsl014. [DOI] [PubMed] [Google Scholar]
- 41.Cirillo DM, Valdivia RH, Monack DM, Falkow S. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol 30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 42.Hensel M. 2000. Salmonella pathogenicity island 2. Mol Microbiol 36:1015–1023. doi: 10.1046/j.1365-2958.2000.01935.x. [DOI] [PubMed] [Google Scholar]
- 43.Prouty AM, Gunn JS. 2000. Salmonella enterica serovar Typhimurium invasion is repressed in the presence of bile. Infect Immun 68:6763–6769. doi: 10.1128/iai.68.12.6763-6769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bader MW, Navarre WW, Shiau W, Nikaido H, Frye JG, McClelland M, Fang FC, Miller SI. 2003. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol Microbiol 50:219–230. doi: 10.1046/j.1365-2958.2003.03675.x. [DOI] [PubMed] [Google Scholar]
- 45.Deiwick J, Nikolaus T, Shea JE, Gleeson C, Holden DW, Hensel M. 1998. Mutations in Salmonella pathogenicity island 2 (SPI2) genes affecting transcription of SPI1 genes and resistance to antimicrobial agents. J Bacteriol 180:4775–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deiwick J, Nikolaus T, Erdogan S, Hensel M. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol 31:1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x. [DOI] [PubMed] [Google Scholar]
- 47.Hensel M, Shea JE, Waterman SR, Mundy R, Nikolaus T, Banks G, Vazquez-Torres A, Gleeson C, Fang FC, Holden DW. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol 30:163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- 48.Miller SI, Mekalanos JJ. 1990. Constitutive expression of the Phop regulon attenuates Salmonella virulence and survival within macrophages. J Bacteriol 172:2485–2490. doi: 10.1128/jb.172.5.2485-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buchmeier NA, Heffron F. 1990. Induction of Salmonella stress proteins upon infection of macrophages. Science 248:730–732. doi: 10.1126/science.1970672. [DOI] [PubMed] [Google Scholar]
- 50.Pegues DA, Hantman MJ, Behlau I, Miller SI. 1995. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol Microbiol 17:169–181. doi: 10.1111/j.1365-2958.1995.mmi_17010169.x. [DOI] [PubMed] [Google Scholar]
- 51.Vescovi EG, Soncini FC, Groisman EA. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165–174. doi: 10.1016/S0092-8674(00)81003-X. [DOI] [PubMed] [Google Scholar]
- 52.Lucas RL, Lee CA. 2000. Unravelling the mysteries of virulence gene regulation in Salmonella typhimurium. Mol Microbiol 36:1024–1033. doi: 10.1046/j.1365-2958.2000.01961.x. [DOI] [PubMed] [Google Scholar]
- 53.Lucas RL, Lostroh CP, DiRusso CC, Spector MP, Wanner BL, Lee CA. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J Bacteriol 182:1872–1882. doi: 10.1128/jb.182.7.1872-1882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boddicker JD, Jones BD. 2004. Lon protease activity causes down-regulation of Salmonella pathogenicity island 1 invasion gene expression after infection of epithelial cells. Infect Immun 72:2002–2013. doi: 10.1128/iai.72.4.2002-2013.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takaya A, Suzuki A, Kikuchi Y, Eguchi M, Isogai E, Tomoyasu T, Yamamoto T. 2005. Derepression of Salmonella pathogenicity island 1 genes within macrophages leads to rapid apoptosis via caspase-1- and caspase-3-dependent pathways. Cell Microbiol 7:79–90. doi: 10.1111/j.1462-5822.2004.00435.x. [DOI] [PubMed] [Google Scholar]
- 56.Takaya A, Tomoyasu T, Tokumitsu A, Morioka M, Yamamoto T. 2002. The ATP-dependent Lon protease of Salmonella enterica serovar Typhimurium regulates invasion and expression of genes carried on Salmonella pathogenicity island 1. J Bacteriol 184:224–232. doi: 10.1128/jb.184.1.224-232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takaya A, Suzuki M, Matsui H, Tomoyasu T, Sashinami H, Nakane A, Yamamoto T. 2003. Lon, a stress-induced ATP-dependent protease, is critically important for systemic Salmonella enterica serovar Typhimurium infection of mice. Infect Immun 71:690–696. doi: 10.1128/iai.71.2.690-696.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoiseth SK, Stocker B. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 59.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc Natl Acad Sci U S A 98:15264–15269. doi: 10.1073/pnas.261348198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McIntyre GJ, Fanning GC. 2006. Design and cloning strategies for constructing shRNA expression vectors. BMC Biotechnol 6:1. doi: 10.1186/1472-6750-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ren D, Sims JJ, Wood TK. 2001. Inhibition of biofilm formation and swarming of Escherichia coli by (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone. Environ Microbiol 3:731–736. doi: 10.1046/j.1462-2920.2001.00249.x. [DOI] [PubMed] [Google Scholar]
- 63.Vaudaux P, Waldvogel FA. 1979. Gentamicin antibacterial activity in the presence of human polymorphonuclear leukocytes. Antimicrob Agents Chemother 16:743–749. doi: 10.1128/aac.16.6.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beuzon CR, Holden DW. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect 3:1345–1352. doi: 10.1016/S1286-4579(01)01496-4. [DOI] [PubMed] [Google Scholar]
- 65.Finlay BB, Falkow S. 1988. Comparison of the invasion strategies used by Salmonella cholerae-suis, Shigella flexneri and Yersinia enterocolitica to enter cultured animal cells: endosome acidification is not required for bacterial invasion or intracellular replication. Biochimie 70:1089–1099. doi: 10.1016/0300-9084(88)90271-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.