Abstract
ArsR As(III)-responsive transcriptional repressors, members of the ArsR/SmtB family of metalloregulatory proteins, have been characterized biochemically but, to date, no As(III)-bound structure has been solved. Here we report two crystal structures of ArsR repressors from Acidithiobacillus ferrooxidans (AfArsR) and Corynebacterium glutamicum (CgArsR) in the As(III)-bound form. AfArsR crystallized in P21 space group and diffracted up to 1.86Å. CgArsR crystallized in P212121 and diffracted up to 1.6Å. AfArsR showed one As(III) bound in one subunit of the homodimer, while the CgArsR structure showed two As(III) bound with S3 coordination, one in each monomer. Previous studies indicated that in AfArsR As(III) binds to Cys95, Cys96 and Cysl02 from the same monomer, while, in CgArsR, to Cysl5, Cysl6 from one monomer and Cys55 from the other monomer. The dimer interfaces of these structures showed distinct differences from other members of the ArsR/SmtB family of proteins, which potentially renders multiple options for evolving metal(loid) binding sites in this family of proteins. Also, CgArsR presents a new ±2-N binding site, not the previously predicted ±3-N site. Despite differences in the location of the binding cysteines in the primary sequences of these proteins, the two metal binding sites are almost congruent on their structures, an example of convergent evolution. Analyses of the electrostatic surface of the proteins at the DNA binding domain indicate that there two different modes of derepression in the ArsR/SmtB family of metalloregulatory proteins.
Keywords: Crystal structures, ArsR-As(III) complex, arsenic binding, derepression
Introduction:
Arsenic is the most ubiquitous toxic substance in the environment. The United States Agency for Toxic Substances and Disease Registry (ATSDR) has always ranked arsenic first on its priority list of Hazardous Substances (https://www.atsdr.cdc.gov/spl/index.html). It ranks higher than lead, cadmium and mercury, not because it is more toxic than those other metals, but because of its ubiquity, toxicity and potential for human exposure. All life is exposed to arsenic on a daily basis, entering the environment primarily from geochemical sources, with lesser amounts contributed by anthropogenic sources, including herbicides, antimicrobial growth promoters for poultry, wood preservatives and other industrial sources. Arsenic is a Group 1 human carcinogen, associated with various forms such as skin and bladder cancers (Kuo et al., 2017). Arsenic exposure is associated with a number of other arsenic-related diseases, that include cardiovascular and peripheral vascular diseases, neurological disorders, diabetes mellitus and chronic kidney disease. Perhaps, the most serious problems are development problems associated with infant consumption of baby food prepared from arsenic-contaminated rice (Smeester and Fry, 2018).
Prokaryotes evolved in an anoxic environment throughout Earth’s early history. The concentration of arsenic in early oceans was high and primarily in the reduced oxidation state, As(III) (Fru et al., 2015). The first microbes quickly acquired genes for tolerance to As(III) exposure to allow them to thrive and compete with arsenic-sensitive bacteria (Zhu et al., 2014). In nearly every bacterial species arsenic resistance genes are organized in ars operons that are controlled by an ArsR As(III)-responsive transcriptional repressor (San Francisco et al., 1990). These operons encode genes for As(V) reduction (arsC), As(III) efflux (arsB and acr3) (Li et al., 2016), arsenic metallochaperone (arsD) etc . As(III) can be methylated to the more toxic species MAs(III) and DMAs(III) by the ArsM As(III)-S-adenosylmethionine methyltransferase. MAs(III) can be detoxified by C-As bond cleavage (arsl), oxidation to MAs(V) (arsH) or efflux (arsP). ArsR was the first identified member of the large ArsR/SmtB family of metalloregulatory proteins (Morby et al., 1993; San Francisco et al., 1990; Wu and Rosen, 1991). They bind to the promoters of operons for metal tolerance, repressing transcription. Expression of these operons are derepressed by binding of a number of metals including zinc (SmtB, CzrA) (Kuroda et al., 1998; Morby et al., 1993), cadmium (CadC) (Endo and Silver, 1995), cadmium or lead (CmtR)(Zheng et al., 2012), nickel (RcnR) (Iwig et al., 2006) and arsenic. Arsenic responsive ArsR repressors are the best studied member of the ArsR/SmtB family (Busenlehner et al., 2003). The four characterized ArsRs form a heterogeneous group, with the unifying feature that they are regulated by binding of As(III) and/or the organoarsenical methylarsenite (MAs(III)), leading to the hypothesis that the arsenic binding sites evolved by convergent evolution at least three times (Chen and Rosen, 2014) Using ArsR as biosensors for inorganic and organic arsenicals have been explored (Chen and Rosen, 2014), but the lack of crystal structures of these proteins has been an impediment.
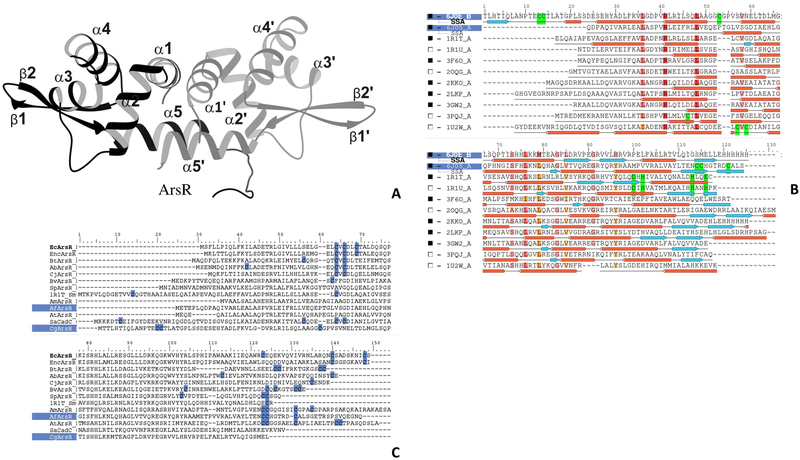
X-ray crystal and NMR structures of a number of proteins in this family of proteins have been solved, but no structure of an As(III)-responsive ArsR has been reported. All members of the ArsR/SmtB family have a HTH winged helix DNA binding domain fold (Figure 1a) and mostly exist as homodimers. Dimerization has been shown to be essential for DNA binding for the Escherichia coli chromosomally-encoded ArsR (Xu and Rosen, 1997). Each monomer of the ArsR/SmtB family of proteins has five helices (α1 to α5) and a two-stranded antiparallel sheet (β1, β2) connected by either a β-turn that form the wing regions located between α4 and α5 or a longer loop. The wing regions and the α4 helices are known to interact with DNA in HTH-DNA complex structures (Gajiwala and Burley, 2000). α4 makes sequence specific DNA interactions and is known as the DNA recognition helix (αR), while the wing regions generally make nonsequence-specific interactions with the binding DNA (Gajiwala and Burley, 2000). The dimer interface is formed by α1 and α5 helices contributed by each monomer, forming an orthogonal bundle of α-helices. To our knowledge, no differences in the orientation of these helices (αl and α5) have been reported. Differences in the dimer interface have implications in forming different types of intra- or inter-molecular metal ion binding sites. To date a total of 13 metal binding sites have been postulated in members of this family (see (Saha et al., 2017)) based on the positions of cysteine or other residues that form the inducer binding site. Figure 1c reveals the diversity in the positions of the cysteine residues in the primary sequences that can potentially form an As(III) binding site.
Figure 1: Sequence and structural features of ArsR.
(A) Cartoon representation of a typical ArsR, homo-dimeric protein having HTH-winged helix-like fold. The dimer interface is formed by α1 and α5. (B) Multiple sequence alignment of ArsR/SmtB family of proteins with known structures. Overall secondary structure regions are conserved, the inducer binding site, highlighted in green, is distributed across the total length of the sequence in different proteins. (C) Multiple sequence alignment of ArsRs analyzed in the current study. Cysteine residues in the sequence block and the proteins whose structures have been solved are highlighted.
To date 26 crystal structures of members of ArsR/SmtB family have been deposited in the PDB, including the Zn(II)-responsive repressors cyanobacterial SmtB and S. aureus CzrA. Nineteen of the 26 structures are not arsenical repressors but bind other metal ions such as Ni, Zn or Cd (Supplementary Figure S3). Seven putative ArsR structures include six that either do not have vicinal cysteine residues or are not part of an ars operon, suggesting that those six might not be As(III)-responsive repressors. The seventh structure, 3KTB from B. vulgatus ATCC 8482 has not been published or characterized and, lacks both the winged helix fold and the As(III) binding site, and so is not likely to be a genuine ArsR. Thus, to date, no authentic ArsR structure with or without bound As(III) has been reported, which limits our understanding of the As(III)-induced conformational changes in the DNA binding domain that lead to derepression.
To elucidate the structure of ArsR in complex with arsenic, we undertook structural investigation of several of the ArsRs listed in Figure 1b. Of these, we successfully crystallized and solved the structure of the As(III)-bound form from A. ferrooxidans (AfArsR) (Qin et al., 2007) and C. glutamicum (CgArsR) (Ordóñez et al., 2008). In the AfArsR structure, one trivalent arsenic atom is observed in one subunit (AfArsR-Asl), while two are observed in CgArsR, one in each monomer (CgArsR-As2). In both structures, the dimer interface is formed by two ±l and two ±5 helices, one from each monomer. These adopt different orientations with respect to the DNA recognition helix ±4 of Zn(II)- or Cd(II)-responsive regulators. Also, despite a completely different location of the cysteine residues that form that As(III) binding sites in the primary sequence of AfArsR and CgArsR, their arsenic binding sites are located at identical positions in their respective three dimensional structures. These structures are consistent with our previous proposal of convergent evolution of arsenic binding site in diverse ArsR repressors (Chen and Rosen, 2014; Ordóñez et al., 2008; Qin et al., 2007). Analysis of the electrostatic surfaces of the two proteins indicate that the structural basis of DNA binding/repression could be different in different members of this family of proteins. Elucidation of these ArsR-As(III) structures can lead to more efficient design of effective biosensors for As(III) and MAs(III). More importantly, our data expand our knowledge of the evolution and composition of metal binding sites in regulatory proteins.
Experimental procedures:
Plasmids and constructs:
Construction of plasmids of AfArsR and CgArsR have been described in (Ordóñez et al., 2008; Qin et al., 2007). AfArsR was expressed in cells of E. coli TOP10 induced with 0.4 % arabinose (w/v). CgArsR was expressed in cells of E. coli BL21(DE3) induced with 0.3 mM isopropyl β-D-1-thiogalactopyranoside. Harvested cells were suspended in a buffer containing 50 mM MOPS, pH 7.5, 0.5 M NaCl, 20 mM imidazole and 10 % glycerol (w/v) (buffer A). The cells were lysed using a homogenizer, and insoluble debris and membranes were removed by centrifugation at 105 g. The supernatant solutions containing each His-tagged ArsR were applied to Ni-affinity columns pre-equilibrated with buffer A. The columns were subsequently washed with 6 column volumes of buffer A to remove non-specifically bound proteins, and the His-tagged ArsRs were eluted using a step gradient of imidazole from 20 mM to 200 mM with increments of 20 mM per 5 ml. Both ArsRs eluted with concentrations of imidazole greater than 120 mM. ArsR purity was analyzed by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE) on a 14% acrylamide gel. Fractions with purified ArsR were pooled and exchanged into a buffer containing 50 mM MOPS, pH 7.5 and 0.5 M NaCl and concentrated by centrifugation with 10 kDa cut-off centrifuge filters.
Crystallization and data collection:
Purified ArsRs at concentrations greater than 10 mg/ml were incubated on ice for 30 minutes with dithiothreitol (DTT) and sodium meta arsenite, each at a molar excess over protein, to generate ArsR-As(III) complexes. Unbound As(III) ions were removed using centrifugal filters prior to screening for crystallization using index and PEG-ion commercial screens from Hampton Research, using microbatch under oil method (Chayen et al., 1990). Initial crystals were further optimized using in-house solutions to obtain diffraction-quality crystals. The AfArsR-As(III) complex crystallized in the presence of 1 % (w/v) tryptone and 20 % (w/v) PEG3350 in a buffer containing 50 mM MOPS, 100 mM NaCl at pH 7.5 (buffer B). CgArsR-As(III) complex crystallized in the presence of 200 mM ammonium tartrate and 20 % (w/v) PEG 3350 in buffer B. Initial testing of the diffraction quality of crystals and optimization of cryoprotectants were performed using an in-house rotating anode X-ray diffractometer at the Molecular Biophysics Unit (MBU), Indian Institute of Science (IISc), India. X-ray diffraction data from AfArsR-As(III) complex crystals were collected at MBU-IISc. at 150 mm detector-to-crystal distance, X-ray exposure of 3 min with 1° oscillation per frame. A total of 180 frames were collected. Diffraction data for CgArsR-As(III) complex crystals were collected on beamline 22ID (SER-CAT) at the Advanced Photon Source (APS), Argonne National laboratory equipped with a Microdiffractometer-MD2 and Dectris EIGER hybrid pixel detector (Eiger 16M), which has a two-dimensional array of pin-diodes processed in high sensitivity silicon, connected to an array of readout channels designed in CMOS technology. Multiple-wavelength anomalous diffraction (MAD) data sets for the CgArsR-As(III) complex crystal were collected at wavelengths of 1.04478 Å (peak) and 1.04495 Å (edge). The peak and edge wavelengths values were obtained experimentally by running fluorescence edge scan (As) using a CgArsR-As(III) complex crystal, and only 3% of transmission of beam were used to avoid detector saturation. A total of 960 images (270°) were recorded (fine-slicing) at 180 mm (crystal-to-detector distance) with a 0.25° oscillation angle and an exposure time of 0.25 s for both peak and edge wavelengths. To avoid radiation damage, 6.5% of transmission was used during MAD diffraction experiment.
Structure solution and refinement:
The intensity data were integrated and scaled using AUTOMAR (Bartels and Klein, 2015) for AfArsR-As(III) data, HKL2000 (Otwinowski and Minor, 1997) and SCALEPACK for CgArsR-As(III) data. The AfArsR-As(III) structure was solved by molecular replacement method using PHASER (McCoy et al., 2007) from the CCP4 (Adams et al., 2010) suite. A homology model of AfArsR constructed using the software tool PRIME (Jacobson et al., 2004) from the SCHRODINGER (Schrodinger, v2016-;4) suite. The PDB structure of apo-SmtB (PDP ID: 1R1T) was used as a template. The CgArsR-As(III) structure was solved using experimental phasing using SOLVE/RESOLVE from the PHENIX (Adams et al., 2010) suite. The positions of the anomalous scatterer, arsenic, were identified from the experimental phases, which were further used to solve the structure. The structures were refined by maximum likelihood method using REFMAC (Murshudov et al., 1997) from the CCP4 suite. Electron density map visualization and model fitting were done using COOT (Emsley et al., 2010). The side chain atoms of several residues were modeled with alternate conformations. Occupancy factors of the alternate conformers as well as unstable water molecules were adjusted in accordance to electron density map as well as their thermal atomic displacement parameters.
Bioinformatics analyses:
Multiple sequence alignments were made using ‘clustal omega’ (https://www.ebi.ac.uk/Tools/msa/clustalo/) and plotted using Schrodinger alignment editor (Schrodinger, v2016-4). Phylogeny analysis of ArsR sequences using the maximum-likelihood method was performed using the software tool MEGAvX (Kumar et al., 2016). 500 bootstrap replicates were generated to validate the tree topology. Homology models of the E. coll plasmid R773 ArsR E. coli were constructed using the PRIME module of the SCHRODINGER software suite (Schrodinger, v2016-4). The template structures used were either apo-SmtB (PDB code 1R1T), AfArsR-As1 or CgArsR-As2 structure.
Structure analysis:
Structures were viewed and analysed using molecular visualization tools PyMol (PyMOL: The PyMOL Molecular Graphics System, Schrodinger, LLC), UCSF-Chimera (Pettersen et al., 2004) or VMD (Humphrey et al., 1996) and standard plugins available from their respective official websites. Adaptive Poisson-Boltzman solver electrostatics surface analyses were calculated using the APBS plugin to PyMol (https://pymolwiki.org/index.php/APBS). Root mean square deviation in the positions of common atoms in two protein structures is calculated using the stand alone program LSQMAN of DEJAVU software package (Sierk and Kleywegt, 2004). Angles subtended by interhelical axes were calculated using Chimera. Contact maps were plotted using the software tool CMView (Vehlow et al., 2011).
Accession numbers:
The coordinates and the structure factors of AfArsR-As1 and CgArsR-As2 have been deposited in the protein data bank with accession number 6J05 and 6J0E respectively.
Results:
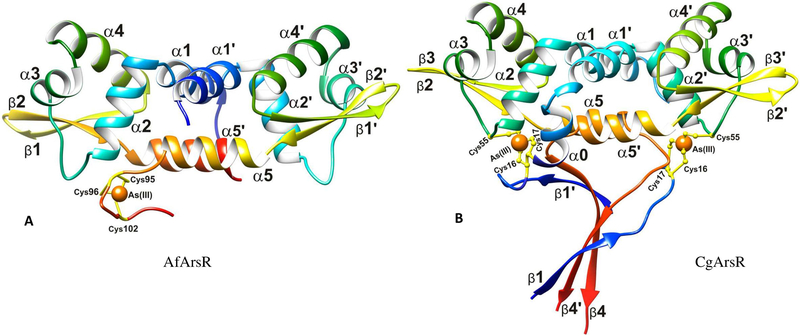
Here we report the crystal structures of A. ferrooxidans and C. glutamicum ArsR in complex with arsenic. AfArsR crystallized in the space group P21 with the cell parameters a= 46.92 Å, b=42.73 µ, c=49.13 µ and β=117.6° and CgArsR in P212121 with the cell parameters a= 43.77 Å, b= 46.39 Å and c= 121.95Å. AfArsR-As(III) diffracted to a maximum resolution of 1.86 Å at the home source diffractometer, CgArsR-As(III) crystals diffracted to 1.6 Å at the synchrotron beamline 22ID, Advanced Photon Source (APS), Argonne National Laboratory, Argonne IL, USA. Crystal data and refinement statistics are given in Table 1. A difference Fourier map of the AfArsR-As(III) complex structure showed one strong peak at > 8 sigma contour and was identified as arsenic. Henceforth, this structure is referred to as AfArsR-Asl. As established from the biochemical studies (Qin et al., 2007), Cys95, Cys96 and Cysl02 form As-S3 coordination in AfArsR-Asl structure (Figure 2a). 100 water molecules were identified from the 2Fo-Fc electron density map in the AfArsR-Asl complex structure. The CgArsR structure was solved using multiple anomalous dispersion data using arsenic as the scatterer. Two arsenic positions were identified from the anomalous data, and the rest of the structure was solved using the heavy atom positions. This structure is henceforth referred to as CgArsR-As2. 236 water and one MPD (2-methyl 2,4 pentane diol) molecules were identified from the difference Fourier map. The residues Cysl5, Cysl6 from one chain and Cys55’ from the other chain formed the arsenic binding site with As-S3 coordination (Figure 2b), as described previously (Ordóñez et al., 2008). The bond length of As-S was constrained to 2.25Å based on the EXAFS studies reported earlier (Ordóñez et al., 2008; Qin et al., 2007). There were a few stray densities in the difference Fourier map near the end of the peptide chains as well as in the solvent region that could not be interpreted. The final R-factors/R-free after refinement to convergence were 16.8%/23.1% for AfArsR-Asl and 17.8%/26.4% for CgArsR-As2. Both structures are dimers with five α-helices and two β strands per monomer forming winged helix-like DNA binding domains. CATH classifies this fold as an orthogonal bundle of α helices as well. CgArsR-As2 shows an additional helix (±O) at the N-terminus of one of the chains and a β strand at the C-termini of both chains. αO is oriented at 87° from ±1 such that Cys15 and Cys16 of one chain is in close proximity to Cys55’ of the other chain. Even in the chain that lacks ±O, Cys15 and Cys16 together with Cys55’ form the As-S3 coordination, but there is a break in the electron density that leaves residues 18 to 20 missing in the electron density map. Both constructs had a C-terminal His-tag, which are not so frequently observed in the electron density maps of crystal structures. In the case of CgArsR-As2, the electron density of the C-terminal His-tag is clearly visible, and the His-tags from both chains form stable mixed β-sheets along with the strands at the N-terminal regions in both chains (Figure 2b). It is to be noted that this additional β-sheet is an artefact induced due to the expression tag at the C-terminal and not part of the CgArsR structure. In the AfArsR-Asl structure, the electron density of the His-tag was not visible.
Table 1: Crystal data and refinement statistics of the two crystals.
The numbers in parentheses are the data for the highest resolution shell.
| Parameter | AfArsR-As1 | CgArsR-As2 (peak) | CgArsR-As2 (edge) |
|---|---|---|---|
| Beamline | Home source | 22ID-APS | 22ID-APS |
| Wavelength (Å) | 1.5414 | 1.04478 | 1.04495 |
| Temperature (K) | 120 | 100 | 100 |
| Detector | MAR2300 | Eiger 16M | Eiger 16M |
| Space group | P21 | P212121 | P212121 |
| Cell dimensions | a=46.92 Å, b=42.73 Å, c=49.13 Å, β=117.6° | a = 43.77 Å, b = 46.39 Å, c = 121.95 Å | a = 43.8 Å, b = 46.5 Å, c = 121.6 Å |
| Resolution (Å) | 1.87 (1.94 – 1.87 ) | 100-1.60 (1.63-1.60) | 100-1.62 (1.65-1.62) |
| Completeness (%) | 97 (83) | 87.0 (78.8) | 86.0 (79.0) |
| Mean I/σ(I) | 6.9 (1.4) | 36.8 (3.7) | 43.8 (3.2) |
| RSym (%) | 7.6 (38.5) | 3.8 (44.8) | 2.8 (39.9) |
| Rpim (%) | 4.42 (21.81) | 1.4 (16.2) | 1.1 (18.1) |
| Multiplicity | 4.04 (3.98) | 8.0 (7.8) | 6.1 (5.0) |
| No. of residues | 180 | 243 | |
| No. of water molecules | 100 | 234 | |
| R/Rfree (%) | 16.5 / 23.1 | 17.8/26.4 | |
| RMSD Bonds (Å) | 0.03 | 0.031 | |
| RMSD Angles (°) | 2.4 | 2.6 | |
| Ramachandran outliers | 0 | 0 | |
Figure 2: X-ray crystal structures of AfArsR-As1 and CgArsR-As2.
(A) Cartoon representation of AfArsR showing one trivalent arsenic ion (orange sphere) bound to C95, C96 and C102. (B) Cartoon representation of CgArsR-As2 showing two arsenic ions (orange spheres) bound to C15, C16 of one chain and C55 from the other chain. The N-terminal residues and the C-terminal his-tag formed a mixed β-sheet.
The dimer interface:
The dimer interface is formed between αl and α5 helices from both monomers in both structures, forming an orthogonal bundle of α-helices. The orientation of a5 with respect to a4 is significantly different in the current structures compared with most other members of the ArsR/SmtB family available in the PDB (Figure 3A). In general, in ArsR/SmtB structures, the helical axes of α4 and α5 subtend an acute angle of about 72° degrees, whereas in the two current structures, an obtuse angle of about 120° is seen. Such a conformation is also seen in the structure 20QG, which, although annotated as an ArsR repressor, is not part of an ars operon, so it is unlikely to be an As(III) regulatory protein. The change in the orientations of the helices at the dimer apparently has not altered the residues involved in the interchain contacts at the dimer interface. Residues 36-40 (LAEFF) of α1 come within proximal contact distance (< 4 Å between the closest atoms) with residues 115-117 (LDHL) in α5 from the other chain in SmtB structures, whereas in CgArsR, residues 31-35 (YADLF) comes into proximity to 106-111 (AELRTV). The physico-chemical properties of the interacting residues are more or less the same in both proteins. Each of the pair of sequences interacting at the dimer interface has at least one acidic residue, either Asp or Glu, one basic residue, either His or Arg, and one Phe. The interface seen in the current structures brings the C-terminal end of α5 (Leu 104) into close proximity to the C-terminal end of the β-sheet (Val104) in the other chain. Interestingly, this also brings the C-terminal end of α5 into close proximity to the arsenic binding site. Thus, binding of arsenic at Cys15/16/55’ will disrupt the hydrophobic interactions at the juncture of α2 and the β-sheet, as well as α5. In AfArsR, arsenic binding at the C-terminal end can result in partial unwinding of the helix α5, which in turn might destabilize the dimer interface. We propose that these interactions play a crucial role in the mechanism of derepression. In SmtB, the N-terminal end of α5 comes into close contact to the a4 and β-turn of the same monomer, whereas in the current structures, the C-terminal end of α5 comes into contact with α4 and β-turn of the other monomer. The schematic given in Figure 3 (Figure 3B, 3C and 3D) explains these differences. This change in α5 orientation is also associated with a change in the location of the metal ion binding sites. The zinc binding sites of SmtB are located across the shorter diameter of the protein at the dimer interface, one each on either side of the N and C-termini of α5. In the CgArsR-As2 and AfArsR-Asl structures, the arsenic binding sites are across the longer axis of the protein away from the plane containing α1/α4 and are closer to the plane of α3/α5. It could be argued that the change in the orientation of the orthogonal bundle of helices is due to the expression tag (6xHis) at the C-terminal of these two proteins. We looked into available winged-helix proteins with orthogonal bundle of α-helices in the PDB and found examples of both possibilities. The structures 20QG, 3F60 have similar orientations as those of the ArsR-As(III) complex structures in the absence of C-terminal His tags. On the other hand, structures 3GW2 and 2KKO, despite having a C-terminal His tag, have their orthogonal bundle of helices formed similar to that of SmtB or other members of ArsR/SmtB family of proteins. These examples indicate that, the addition of the His-tag does not change the orientation; rather the differences may be an inherent aspect of diversity of dimer interface in the HTH winged helix fold.
Figure 3: Unique features in the AfArsR-As1 and CgArsR-As2 structures.
Schematic representation of (A) AfArsR, (B) CgArsR and (C) SmtB in their metal bound forms. Helices are shown as lines, N-terminal end is shown in light cyan, C-terminal in grey or dark. The pair of α5 helices are behind the plane of paper, while α1 & α4 helices are on the upper plane. α2 helices traverse through both the planes, while α3 is located between the planes. The arsenic binding site is along the α5 plane and away from α4. The trivalent arsenic ion is shown as a brown sphere and binding cysteine residues in yellow lines. (D) Superposition of AfArsR-As1 shown in blue cartoon and the zinc regulatory protein, SmtB (grey). The change in the orientations of α5 with respect to α4 is depicted. Here, the pair of α5 helices are shown in the upper plane for clarity. The interhelical angles between α4 and α5 are shown below the superposed structures.
We constructed a contact map of the CgArsR-As2 and SmtB (PDB code 1R22) structures to analyse the common and unique contacts resulting from these two types of α5 orientations. Supplementary figure S1 shows the intra-chain and inter-chain contacts in the dimeric structure of these two proteins, highlighting the contact differences between the two chains of the homodimer. All ArsR repressors form a three-coordinate sulfur binding site involving three cysteine residues. The location of the three cysteines, of which two are usually vicinal, are designed to bring the third cysteine close to the vicinal cysteines. Changes in the fold due to the α4/α5 orientation in ArsR provide more options for evolving metal ion binding sites. Pixels in the quadrants two and four of Figure S1 represent intra-domain contacts. Those in the first and third quadrants are inter-domain contacts. Contacts associated with known metal(loid) binding regions are depicted as indicated. Every pixel in the third quardrant presents a possible inter-domain binding site location.
Different cysteine but same binding site:
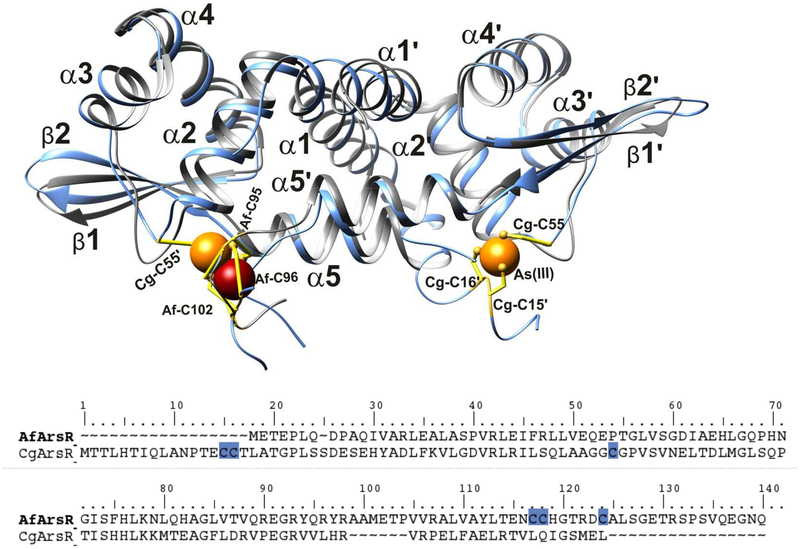
As mentioned above, Cys95, Cys96 and Cysl02 from the same chain in AfArsR-As1 and Cys15A, Cys16 and Cys55’ in CgArsR-As2 form As-S3 coordinations. A pairwise alignment of AfArsR and CgArsR sequences shows 20 % identity and 30 % similarity. A multiple sequence alignment of different ArsRs (Figure 1c) reveals the differences in the location of the cysteine residues in their respective primary structures. It interesting to note that, despite having the arsenic-coordinating cysteine residues at very different locations in the primary sequence, the binding sites on their respective 3D structures are congruent in these two proteins (Figure 4). 13 different modes of arsenic binding sites were proposed previously based on sequence analysis (Saha et al., 2017) that are either intra- or inter-monomer coordination sites. From the current crystal structures, it is apparent that there may be only a few categories of arsenic binding sites, one at the dimer interface, similar to those in the zinc regulatory proteins, a second in the current structures that is close to the dimer interface but at the interface of α2, β-sheet and 鎱5B (as in CgArsR-As2), and a third similar to the E. coli counterpart at the α3 region. These sites may be formed by intra-domain or inter-domain interactions. Thus, there may not be as many structurally different arsenic binding motifs as proposed previously (Saha et al., 2017) among members of ArsR/SmtB family, but there may be a rather limited number of sites, perhaps only three. It should be noted that, to date, no α3 binding motif has been observed experimentally.
Figure 4: Superposition of AfArsR-As1 and CgArsR-As2 structures.
(Top) Superimposition of the AfArsR-As1 (grey) and CgArsR-As2 (light blue) structures reveals the similarity in the metal binding sites in these two proteins. As(III) binding cysteine residues are shown in yellow ball-and-sticks. (Bottom) Pairwise sequence alignment of AfArsR and CgArsR depicting the differences in the locations of the cysteine residues forming the arsenic binding sites. Yet, in 3D structure they are at the same location, shown as red or orange spheres.
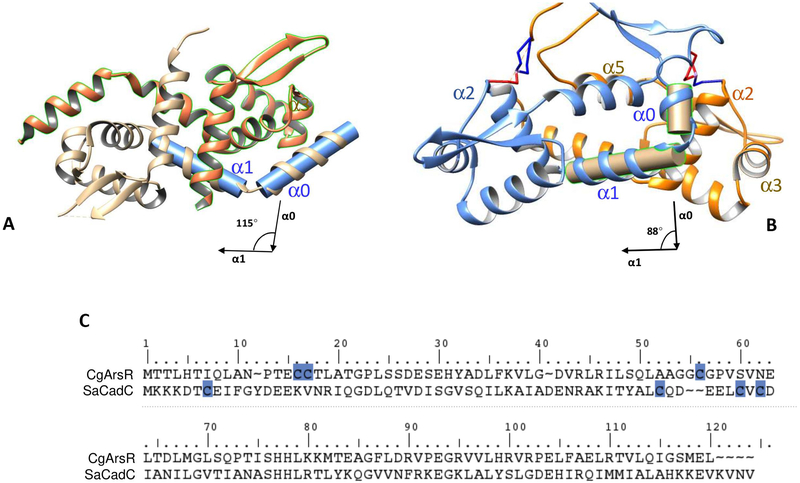
An additional helix α0 in CgArsR:
The sequence alignment in Figure 5C shows CgArsR-As2 having an N-terminal extension that comprises two cysteine residues involved in formation of the metal(loid) binding site. The Cd(II)-responsive CadC repressor from Staphylococcus aureus (SaCadC) also has an additional helix at the N-terminus, similar to CgArsR-As2, which is oriented at an angle close to 115° from α1. This orientation brings the N-terminal vicinal cysteine residues close to Cys58 and Cys60 located at α3 in the other chain, which is predicted to form the cadmium binding site. However, residues Cys7 and Cys11 are not visible in the SaCadC crystal structure (PDB code 1U2W) due to missing electron density. In the CgArsR-As2 structure α0 subtends an angle about 87° with the helical axis of α1 in chain B, which brings Cys15 and Cys16 into close proximity with Cys55’ located at the C-terminal end of α2. This also lies near to the C-terminal end of α5 and the wing region. Thus, despite having similar N-terminal extension residues compared to other members of this family, SaCadC and CgArsR have evolved different metal binding sites. An additional helix α0 is also present in BigR (PDB code 3PQJ), which is a redox-responsive regulator and not a metalloregulatory protein. In spite of responding to different stimuli, the orientation of BigR is similar to that of CgArsR-As2. From sequence alignment it appears that BigR does not have extra residues at the N-terminus, unlike SaCadC or CgArsR (Figure 1b), and its functional cysteine residues are not near the additional helix α0.
Figure 5: Comparison of the α0 helices in CgArsR-As2 and SaCadC.
(A) SaCadC forms an α3-N site, as α1 and α0 subtend 116°. These two helices are shown with a blue cylinder inscribed in them (B) CgArsR forms an α2-N site, characterized by different orientations of α0, wherein α1 and α0 subtend about 87°. The concerned helices are shown with a light brown cylinder inscribed in them. (C) Pairwise sequence alignment of CgArsR and SaCadC. The cysteine residues forming the metal binding site are highlighted.
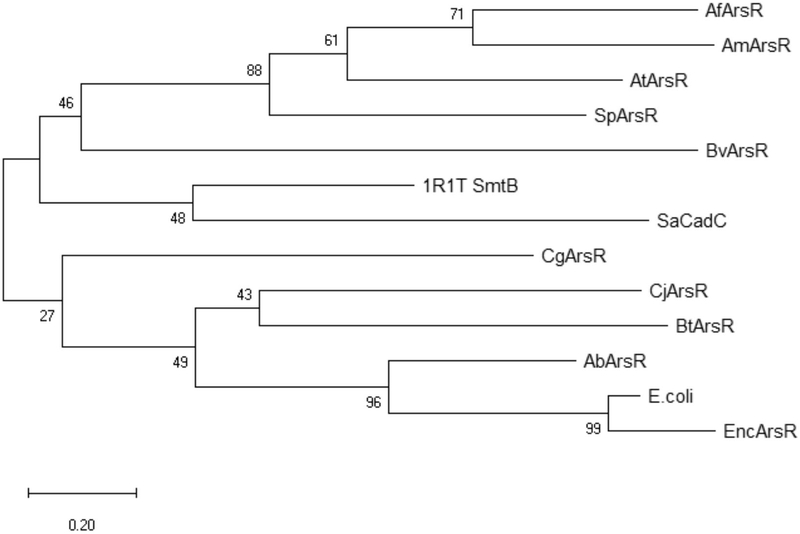
Phylogenetic analysis (Figure 6) places CgArsR and EcArsR in the same clade, although CgArsR is divergent from other members of its clade albeit with lower bootstrap value. ArsR repressors that have intra-domain binding sites near the C-terminus, close to the dimer interface as in AfArsR, form the second major phylogenetic clade. Both EcArsR and SaCadC have α3 binding sites, but are in different clades. However, SmtB, which has an α5 binding site, and SaCadC are in this clade. It is to be noted that cysteine residues are present at the α3 region of SmtB but do not form functional metal binding sites. SaCadC also has an α5 zinc binding site similar to SmtB that is not involved in metal sensing. We propose that metal(loid) binding sites arose independently in the ancestral repressor in response to environmental challenges so that there is no linear sequence of events that connect the evolutionary pathway of the repressors , but rather multiple events from multiple starting points. (Qin et al., 2007; Ordóñez et al., 2008).
Figure 6: Evolutionary relationships of members of the ArsR/SmtB family/SpArsR with ArsRs from members of other bacterial species.
Evolutionary relatedness was inferred using the Maximum Likelihood method and JTT matrix-based model (Jones et al., 1992) . The tree with the highest log likelihood (−2135.41) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 13 amino acid sequences. All positions with less than 95% site coverage were eliminated, i.e., fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position (partial deletion option). There were a total of 77 positions in the final dataset. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018).
Structural analyses of ArsR/SmtB family members currently available in the PDB show that the wing region, which comprises the two stranded anti-parallel β-sheet with a β-turn, is not perfectly symmetrical between monomers. Mostly type II or type IV β turns comprising three residues are observed at the wing region. Two distinct modes of DNA binding have been demonstrated in HTH winged helix DNA binding proteins (Gajiwala and Burley, 2000). We calculated electrostatic surfaces using Adaptive Poisson-Boltzman Solver electrostatic surfaces with a PyMol plugin (https://pymolwiki.org/index.php/APBS)) and found two different types of electrostatic surfaces. Type-I surfaces have a continuous of stretch of positive electrostatic surface across the long axis of the protein, and Type-II have two positive surfaces separated by a zone of positive surface. In CgArsR-As2, Type-I electrostatic charge distribution covers the longer diameter, running through both monomers as a single stretch, whereas AfArsR-As1 shows Type-II distributions. SaCadC also shows a similar electrostatic surface as CgArsR. In the apo form of zinc regulatory protein SmtB (PDB code 1R1T), Type-II electrostatic distributions span the shorter diameter of the protein, one on either monomer that includes the α4 and wing region, separated by a distinctly negatively charged region (Supplementary figure S2). This indicates a possible difference in the mode of DNA binding compared with either SmtB or AfArsR. It is to be noted that the electrostatic surface charge distribution did not change drastically in the zinc-bound form of the same protein (PDB code 1R22). A homology model of EcArsR constructed using AfArsR-Asl as template shows an electrostatic surface similar to CgArsR or SaCadC, but not AfArsR (data not shown). The DNA-bound forms of these repressors must be determined to elucidate the structural basis of repression/derepression.
Discussion:
ArsR is the patriarch of the ArsR/SmtB family of metal(loid)-responsive transcriptional repressors. The first member of family to be identified was the ArsR repressor of E. coli plasmid R773 (San Francisco et al., 1990) . In the absence of As(III), the ArsR homodimer binds to its promoter, repressing its own expression and the downstream ars genes. When As(III) enters the cell, usually through the aquaglyceroporin GlpF, it binds to a cysteine triad, Cys32, Cys34 and Cys37 in ArsR. This is proposed to induce a conformational change that results in dissociation of ArsR from the operator DNA and subsequent expression of ars genes. In an effort to understand how metal ion binding results in derepression, the 1.9 Å X-ray crystal structure of the SaCadC Cd(II)/Pb(II)/Zn(II)-responsive homodimeric repressor was determined (Ye et al., 2005). CadC has a Cd(II)/Pb(II)/Zn(II)-binding site composed of four cysteine resides, Cys7, Cys11, Cys58 and Cys60, in the putative DNA binding domain. SaCadC also has a structural Zn(II) binding site. There are at least 25 structures deposited in the PDB annotated as ArsR. Detailed analysis of their gene structures indicates that most repressors annotated as ArsR are not, in fact, authentic As(III)-responsive regulatory proteins, but, rather, are members of the ArsR/SmtB family that bind other divalent cations such Zn(II), Cd(II) or Ni(II).
The goal of this study was to determine the structure of one or more genuine ArsR repressors. This was complicated by the fact that there are at least three different types of ArsR repressors with different As(III) binding sites. The chromosomally-encoded EcArsR of E. coli is a close ortholog of R773 ArsR and has the same Cys32, Cys34, Cys37 As(III) binding site. Two other ArsR repressors with As(III) binding sites different from R733 ArsR and EcArsR were identified in the ars operons of A. ferrooxidans (AfArsR) and C. glutamicum (CgArsR). The As(III) binding site in AfArsR is composed of Cys95, Cys96 and Cys102, and the As(III) binding site in CgArsR is composed of Cys15, Cys16 from one subunit and Cys55 from the other subunit.
From biochemical analysis, AfArsR is predicted to have an intradomain β5-C binding site, while CgArsR is predicted to have an α3-N type binding site. The crystal structures presented in this study confirm the binding predictions for AfArsR, while As(III) binds to CgArsR in a novel interdomain α2-N site. Despite the fact that the location of the arsenic binding cysteine residues is drastically different in the primary structure of these two proteins, arsenic binding sites in theirs structures are nearly congruent (Figure 5). A least squares superposition of the common atoms in the dimer gives an RMSD of 3.6 Å with the two As(III) ions at a distance of 3.8 Å. In the AfArsR structure, only a single As(III) is visible, whereas in CgArsR, two arsenic atoms are visible, one in each monomer. In the CgArsR-As2 homodimer the electron density map for several residues next to the arsenic binding site in one of the monomers is not visible, while, in the other monomer, an additional helix α0 is observed. It is possible that arsenic binding is cooperative, i.e., one of the monomers has higher affinity than the other for As(III). It is not clear whether binding of As(III) to only one site is sufficient to produce derepression. Interestingly, the structures of both AfArsR-As1 and CgArsR-As2 show a very characteristic tail formed by about 10 amino acid residues from either the N- or C-terminii. In spite of their different locations in the primary structure, they are co-located in the two 3D structures. The sequence of the two tails are not similar to each other, and the lack of conserved basic or cysteine residues suggests that they may not be involved in either DNA or As(III) binding. Further biochemical analysis is required to understand if they have a role to play in function of these proteins.
The diversity of the ArsR repressors is an example of convergent evolution from a common ancestral DNA binding protein. It is likely that the As(III)-binding site in R773/EcArsR, CgArsR and AfArsR arose independently by introduction of cysteine triads at different positions in each protein. Almost all known ArsRs have HTH-winged helix-like DNA binding fold with an orthogonal bundle of α-helices at the dimer interface, but their inducer binding sites are significantly different in their respective primary sequences. Structurally the arsenic binding sites may be located at either the dimer interface or at the extremities along the longer axis of the protein. Their DNA binding may also exhibit at least two modes, as inferred from the electrostatic surface analysis. The electrostatic surfaces of AfArsR-As1 and CgArsR-As2 structures show different distributions of charged residues, yet the As(III) binding sites are congruent both. Thus, the DNA binding mechanism may differ in these two proteins, yet the mechanism of derepression may be similar. It is interesting to note that the As(III) binding sites evolved and converged to one of the two possible sites (α3* or α5*). Similarly, the DNA binding/repressing mechanism has also evolved and converged to one of two possible mechanisms. The availability of α3 binding site structures may also be of value for engineering a repressor with both α3 and α5 binding sites that could sense/trap arsenic for use in bioremediation of this toxic metalloid.
Conclusion:
X-ray crystal structures of arsenic responsive repressor ArsR from A. ferrooxidans and C. glutamicum have been solved in complex with trivalent arsenic ions. The arsenic binding cysteine residues are located at extremely different positions in the primary sequences of these proteins but structurally they are at identical locations. C. glutamicum ArsR and S. aureus CadC have similar binding cysteine residues forming an inter-domain metal binding site in their protein sequence, but the structures indicate their binding site locations are different. Thus, the structures present instances of two proteins with different sequences having a similar metal binding site, as well as those with similar sequences but different metal binding sites. Also, the dimer interface of the helix-tum-helix winged helix proteins present a unique orthogonal bundle of α-helices which to the best of our knowledge have not been reported earlier. The structural information provided in the current study will expand our understanding of the evolution of regulatory metal binding sites and may also be useful for the design of biosensors for detecting environmental arsenic.
Supplementary Material
Supplementary Figure SI: Contact map of SmtB and CgArsR. Black dots in the lower triangle indicate common contacts. Green dots are unique to CgArsR, magenta ones are unique to SmtB. The upper triangle depicts the same as a heat map. Blue regions are common to both, and red regions are those with highest differences. Gaps in sequence alignment between SmtB and CgArsrR are blank stripes. II and IV quadrants represent in the intra domain contacts, I and III represent inter-domain contacts. Maroon rhombus points to intra-domain α3 site in EcArsR like proteins, blue square points to α5 site, as in AfArsR. The blue star points to α2-N’ site as in CgArsR, and the circle points to α5-α5’ inter-domain site as in SmtB. The same symbols in quadrant 1 are equivalent to sites formed in the second monomer.
Supplementary Figure S2: The electrostatic surface of CgArsR (A, B) and AfArsR (C,D), calculated using Adaptive Poisson-Boltzman Solver plugin to PyMol, shows two major modes of charge distribution. (A) The electrostatic surface of the CgArsR-As(III) complex shows a continuous positive surface track (blue region) on the protein. (B) shows the same surface rotated 180° about the longer axis, showing a predominantly negative surface (red region). (C) The electrostatic surface of AfArsR-As(III) complex shows two distinct positive regions separated by a negative surface . (D) shows the AfArsR electrostatic surface rotated 180° about longer axis.
Supplementary Figure S3: Structure analysis of ars operons. (A) Representative operon structures of the ArsR/SmtB family of proteins were obtained from the NCBI database. Most of the putative ArsR structures deposited in the PDB are not part of ars operons. (B) Operon structures of proteins described in the current study demonstrate that they are included in ars operons.
Highlights.
Two different crystal structures of ArsR As(III)-responsive transcriptional regulators in complex with trivalent arsenic solved from two microbial species
Despite differences in the locations of the cysteine residues in the primary sequence, the As(III)-S3 binding sites are located at identical positions in both proteins
Differences in the orientations of the orthogonal bundle of alpha helices are observed for the first time.
Binding of arsenic at Cys15/16/55’ in CgArsR can potentially disrupt the hydrophobic interactions at the juncture of α2 and the β-sheet, as well as α5.
In AfArsR, arsenic binding at the C-terminal end can result in partial unwinding of the helix α5, which in turn might destabilize the dimer interface.
Acknowledgements:
This work was supported by DST-SERB, Government of India, sanction no EMR/2014/000299 to ST and NIH grant GM55425 to BPR. An institutional grant to IBAB from the department of IT, BT, science and technology, the Government of Karnataka is acknowledged. IBAB is also supported by DST-FIST vide sanction no SR/FST/LSI-536/2012. SER-CAT is supported by its member institutions (www.ser-cat.org/members.html), and equipment grants (S10_RR25528 and S10_ RR028976) from the National Institutes of Health. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. The home source diffractometer facility at the Molecular Biophysics Unit, Indian Institute of Science is acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Adams PD, Afonine PV, Bunkñczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH, 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels K, Klein C, 2015. automar Data Processing Suite. [Google Scholar]
- Busenlehner LS, Pennella MA, Giedroc DP, 2003. The SmtB/ArsR family of metalloregulatory transcriptional repressors: Structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27, 131–43. [DOI] [PubMed] [Google Scholar]
- Chayen NE, Shaw Stewart PD, Maeder DL, Blow DM, IUCr, 1990. An automated system for micro-batch protein crystallization and screening. J. Appl. Crystallogr. 23, 297–302. 10.1107/S0021889890003260 [DOI] [Google Scholar]
- Chen J, Rosen B, 2014. Biosensors for Inorganic and Organic Arsenicals. Biosensors 4, 494–512. 10.3390/bios4040494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K, 2010. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo G, Silver S, 1995. CadC, the transcriptional regulatory protein of the cadmium resistance system of Staphylococcus aureus plasmid pI258. J. Bacteriol. 177, 4437–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fru EC, Arvestal E, Callac N, El Albani A, Kilias S, Argyraki A, Jakobsson M, 2015. Arsenic stress after the Proterozoic glaciations. Sci. Rep. 5, 17789 10.1038/srepl7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajiwala KS, Burley SK, 2000. Winged helix proteins. Curr. Opin. Struct. Biol. 10, 110–6. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K, 1996. VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Iwig JS, Rowe JL, Chivers PT, 2006. Nickel homeostasis in Escherichia coli ? the rcnR-rcnA efflux pathway and its linkage to NikR function. Mol. Microbiol. 62, 252–262. 10.1111/j.1365-2958.2006.05369.x [DOI] [PubMed] [Google Scholar]
- Jacobson MP, Pincus DL, Rapp CS, Day TJF, Honig B, Shaw DE, Friesner RA, 2004. A hierarchical approach to all-atom protein loop prediction. Proteins Struct. Funct. Bioinforma. 55, 351–367. 10.1002/prot.10613 [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM, 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–82. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K, 2018. MEGAX: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K, 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C-C, Moon KA, Wang S-L, Silbergeld E, Navas-Acien A, 2017. The Association of Arsenic Metabolism with Cancer, Cardiovascular Disease, and Diabetes: A Systematic Review of the Epidemiological Evidence. Environ. Health Perspect. 125, 087001 10.1289/EHP577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M, Bhattacharjee H, Rosen BP, 1998. Arsenical pumps in prokaryotes and eukaryotes. Methods Enzymol. 292, 82–97. [DOI] [PubMed] [Google Scholar]
- Li J, Pawitwar SS, Rosen BP, 2016. The organoarsenical biocycle and the primordial antibiotic methylarsenite. Metallomics 8, 1047–1055. 10.1039/c6mt00168h [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ, IUCr, 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674. 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morby AP, Turner JS, Huckle JW, Robinson NJ, 1993. SmtB is a metal-dependent repressor of the cyanobacterial metallothionein gene smtA: identification of a Zn inhibited DNA-protein complex. Nucleic Acids Res. 21, 921–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ, 1997. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr. Sect. D Biol. Crystallogr. 53, 240–255. 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- Ordóñez E, Thiyagarajan S, Cook JD, Stemmler TL, Gil J. a, Mateos LM, Rosen BP, 2008. Evolution of metal(loid) binding sites in transcriptional regulators. J. Biol. Chem. 283, 25706–14. 10.1074/jbc.M803209200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W, 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–26. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, 2004. UCSF Chimera: A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- PyMOL: The PyMOL Molecular Graphics System, n.d.
- Qin J, Fu H-L, Ye J, Bencze KZ, Stemmler TL, Rawlings DE, Rosen BP, 2007. Convergent evolution of a new arsenic binding site in the ArsR/SmtB family of metalloregulators. J. Biol. Chem. 282, 34346–55. 10.1074/jbc.M706565200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha RP, Samanta S, Patra S, Sarkar D, Saha A, Singh MK, 2017. Metal homeostasis in bacteria: the role of ArsR-SmtB family of transcriptional repressors in combating varying metal concentrations in the environment. BioMetals 30, 459–503. 10.1007/s10534-017-0020-3 [DOI] [PubMed] [Google Scholar]
- San Francisco MJ, Hope CL, Owolabi JB, Tisa LS, Rosen BP, 1990. Identification of the metalloregulatory element of the plasmid-encoded arsenical resistance operon. Nucleic Acids Res. 18, 619–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger, L., n.d. Small-Molecule Drug Discovery Suite. [Google Scholar]
- Sierk ML, Kleywegt GJ, 2004. Deja Vu All Over Again. Structure 12, 2103–2111. 10.1016/j.str.2004.09.016 [DOI] [PubMed] [Google Scholar]
- Smeester L, Fry RC, 2018. Long-Term Health Effects and Underlying Biological Mechanisms of Developmental Exposure to Arsenic. Curr. Environ. Heal. Reports 5, 134–144. 10.1007/s40572-018-0184-1 [DOI] [PubMed] [Google Scholar]
- Vehlow C, Stehr H, Winkelmann M, Duarte JM, Petzold L, Dinse J, Lappe M, 2011. CMView: Interactive contact map visualization and analysis. Bioinformatics 27, 1573–1574. 10.1093/bioinformatics/btr163 [DOI] [PubMed] [Google Scholar]
- Wu J, Rosen BP, 1991. The ArsR protein is a trans-acting regulatory protein. Mol. Microbiol. 5, 1331–6. [DOI] [PubMed] [Google Scholar]
- Xu C, Rosen BP, 1997. Dimerization is essential for DNA binding and repression by the ArsR metalloregulatory protein of Escherichia coli. J. Biol. Chem. 272, 15734–8. [DOI] [PubMed] [Google Scholar]
- Ye J, Kandegedara A, Martin P, Rosen BP, 2005. Crystal Structure of the Staphylococcus aureus pI258 CadC Cd(II)/Pb(II)/Zn(II)-Responsive Repressor. J. Bacteriol. 187, 4214–4221. 10.1128/JB.187.12.4214-4221.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Li Y, Nie L, Qian L, Cai L, Liu J, 2012. Transcriptional and functional studies of a Cd(II)/Pb(II)-responsive transcriptional regulator(CmtR) from Acidithiobacillus ferrooxidans ATCC 23270. Curr. Microbiol. 65, 117–21. 10.1007/s00284-012-0117-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y-G, Yoshinaga M, Zhao F-J, Rosen BP, 2014. Earth Abides Arsenic Biotransformations. Annu. Rev. Earth Planet. Sci. 42, 443–467. 10.1146/annurev-earth-060313-054942 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure SI: Contact map of SmtB and CgArsR. Black dots in the lower triangle indicate common contacts. Green dots are unique to CgArsR, magenta ones are unique to SmtB. The upper triangle depicts the same as a heat map. Blue regions are common to both, and red regions are those with highest differences. Gaps in sequence alignment between SmtB and CgArsrR are blank stripes. II and IV quadrants represent in the intra domain contacts, I and III represent inter-domain contacts. Maroon rhombus points to intra-domain α3 site in EcArsR like proteins, blue square points to α5 site, as in AfArsR. The blue star points to α2-N’ site as in CgArsR, and the circle points to α5-α5’ inter-domain site as in SmtB. The same symbols in quadrant 1 are equivalent to sites formed in the second monomer.
Supplementary Figure S2: The electrostatic surface of CgArsR (A, B) and AfArsR (C,D), calculated using Adaptive Poisson-Boltzman Solver plugin to PyMol, shows two major modes of charge distribution. (A) The electrostatic surface of the CgArsR-As(III) complex shows a continuous positive surface track (blue region) on the protein. (B) shows the same surface rotated 180° about the longer axis, showing a predominantly negative surface (red region). (C) The electrostatic surface of AfArsR-As(III) complex shows two distinct positive regions separated by a negative surface . (D) shows the AfArsR electrostatic surface rotated 180° about longer axis.
Supplementary Figure S3: Structure analysis of ars operons. (A) Representative operon structures of the ArsR/SmtB family of proteins were obtained from the NCBI database. Most of the putative ArsR structures deposited in the PDB are not part of ars operons. (B) Operon structures of proteins described in the current study demonstrate that they are included in ars operons.