Abstract
Chronic periodontitis is an inflammatory disease of tooth-supporting tissues associated with Porphyromonas gingivalis. Expansion and invasion of this bacterium into the periodontium is associated with changes in the metabolome of the oral cavity. Materials and Methods: Metabolomics analysis of mouth washout and tongue swab samples based on proton nuclear magnetic resonance (1H-NMR) method was employed to determine metabolic status of the oral cavity in chronic periodontal disease. Results: Mouth washout extracts contained a total of 23 metabolites and tongue swab extracts contained 17. Identified metabolites partially overlap with the content of saliva and gingival crevicular fluid. The colonization of the oral cavity of patients with periodontitis by bacteria was manifested in the change in levels of eight metabolites. Conclusion: NMR-based metabolomics analysis is a potentially useful methodological approach for monitoring the pathological processes observed in the oral cavity in the course of periodontitis.
Keywords: Periodontitis, oral cavity, metabolomics, NMR
Chronic periodontal disease (CPD) is a bacterial-driven inflammatory malady affecting tissues supporting the teeth (1,2). Disease severity ranges from mild and reversible inflammation of soft tissue of the gingiva to chronic destruction of periodontal tissues, including periodontal ligaments and alveolar bone, which leads to formation of periodontal pockets with eventual exfoliation of teeth (3-5). CPD in adults is associated with colonization and proliferation of the gram-negative anaerobic bacterium Porphyromonas gingivalis in periodontal gingival pockets. The presence of P. gingivalis, and several other gram-negative anaerobic bacteria, i.e. Agregatibacter actinomycetemcomitans, Prevotella intermedia, Tannerella forsythia, Fusobacterium nucleatum, Campylobacter rectus, Peptostreptococcus micros, Streptococcus intermedius, Treponema denticola, Eikenella corrodens acting possibly in concert with the absence of beneficial species and the presence of certain immunological deficiencies in the host, appears to play an essential role in disease severity and progression (6,7). The primary site of oral cavity colonization by P. gingivalis includes the supragingival tooth surface and subgingival crevice (the space between the tooth root and the gingiva). P. gingivalis represents a late colonizer of the oral cavity, requiring antecedent organisms to create the necessary environmental conditions. The antecedent bacterial species may facilitate colonization by P. gingivalis via provision of attachment sites for interspecies adherence, supply of nutrients, and reduction of oxygen tension to the low levels required for growth and survival of this obligate anaerobe (7-9).
P. gingivalis does not produce a single potent exotoxin, but is equipped with a set of enzymes, e.g. proteinases, hemolysins, peptidylarginine deiminase and cellular constituents, such as fimbriae and lipopolysaccharides, which along with toxic metabolites, have the potential to impinge upon host tissue integrity thus causing destruction of periodontal supporting tissues (5,7,10-12). The repertoire of enzymes and metabolites that can be detrimental to the host also includes phospholipase A, prostaglandins, alkaline and acid phosphatases, etc. (13-17) and volatile sulfur compounds such as hydrogen sulfide, methylmercaptan, and dimethyl sulfide, which are cytotoxic and can be responsible for protein synthesis inhibition (18,19).
Beyond the primary site of colonization, the oral cavity provides a variety of surfaces to which P. gingivalis can efficiently adhere or invade (13,16). Previous studies have reported detection of periodontopathic bacteria in tongue coating in close association with those in dental plaque (20-22) and periodontal conditions (23-25), indicating that periodontopathic microorganisms in the tongue, equally to dental plaque, are an important factor in the etiopathogenesis of periodontal disease. Therefore, similarly to gingival crevicular fluid (GCF), other oral fluids and surfaces may also constitute a potential reservoir of a variety of inflammatory mediators and tissue-destructive molecules associated with increased host–bacterial interactions compared to individuals with healthy periodontium (25-28).
Metabolomics is a method of choice for diagnosis of different pathological conditions. It allows for comparison of the signature of low molecular weight compounds (MW<1,000-1,500 Da) in biological samples from healthy and disease-affected individuals, mainly by employing nuclear magnetic resonance (NMR) and mass spectrometry (MS) combined with various separation techniques (29-34). Salivary metabolomics studies have been conducted for the detection of many diseases including oral cavity (35). The total composition shown by analysis of salivary gland products should reflect two metabolomes, i.e. first that of the investigated individual, within which all metabolites are associated with the health status, and second, that which represents the oral microbiome. Therefore, the output of different studies may differ due to the contribution of different microbial metabolomes. Among many diseases, metabolomics analysis of oral cavity samples allows distinction between patients with different periodontal diseases and healthy individuals (36-38).
The aim of our study was to demonstrate the feasibility of NMR analysis in determining the metabolic status of the oral cavity in CPD, in easily accessible samples of mouth washout and tongue swab employing an untargeted metabolomics profiling method.
Materials and Methods
Study participants. The study was carried out in accordance with the Declaration of Helsinki and was approved by the Bioethics Committee of the Jagiellonian University, Medical College in Krakow, Poland (KBET/310/B/2012). Adult males and females from Krakow, Malopolska area (Poland) were enrolled in this study. All participants read and signed a written informed consent form prior to inclusion in the study. The study group consisted of 30 donors diagnosed with advanced CPD with a minimum of 20 natural teeth present (aged 37-68 years; 11 males, 19 females). The control group comprised 15 donors in good general health (aged 23-62 years; four males and 11 females). Exclusion criteria included: diagnosis of a medical condition which required pre-medication/pre-treatment prior to dental visits and procedures; five or more decayed untreated teeth at screening (cavities); diagnosis of other diseases of the hard or soft oral tissues; use of antibiotics or antimicrobial drugs within 30 days prior to the first study visit; a history of systemic disease e.g. rheumatoid arthritis, aspiration pneumonia, diabetes mellitus, atherosclerosis or other uncharacterized systemic disease; pregnant and lactating women; tobacco smokers; immune-compromised individuals e.g. diagnosed with HIV, AIDS, taking immuno-suppressive drug therapy, radio- and chemotherapy.
Clinical and periodontal examination. Each participant’s medical, dental, and medication history was recorded by the clinician. The diagnosis of CPD was confirmed by radiographs and peripheral blood samples collected for routine laboratory tests. The following parameters were examined to assess the periodontal status: aproximal periodontal index (API), sulcus bleeding index (SBI), probing pocket depth (PPD) and clinical attachment level (CAL). API was classified as optimal at <25%, moderately good at 26-39%, average at 40-70%, and poor at 71-100% and expressed as the average value per whole oral cavity. SBI was classified as: normal/healthy periodontium at <12%; mild inflammation (slight change in color, little change in texture) at 13-20%; moderate inflammation (redness, edema and hypertrophy) at 21-50%; chronic inflammation (marked redness and edema/hypertrophy, spontaneous bleeding or ulceration) at 51-100% and also expressed as the average value per whole periodontium. PPD and CAL were assessed for five teeth including at least one central incisor and one first molar in the group with periodontitis group and for five random teeth in control group, using the following classification: PPD>7 mm and CAL>5 mm advanced periodontitis; PPD 5-7 mm and CAL 3-4 mm moderate periodontitis; PPD 3-5 mm and CAL 1-2 mm mild periodontitis. All in individuals with periodontal disease included in the study group were classified as having chronic (advanced) periodontitis. In the control group, all assessed parameters were within the reference range.
Sample collection. Participants were asked to refrain from eating or drinking, excluding water, from 11:00 pm of the night preceding sample collection and to brush their teeth and entire mouth in the evening before the visit, but not on the morning of collection. Tongue swabs were obtained before mouth washouts collection. A sterile cotton-tipped swab was used to collect the specimen from the external surface of the tongue, placed in a 1.5 ml sterile polypropylene tube with phosphate-buffered saline (sterile PBS, pH 7.2) and kept for 15 min at 4˚C to dissolve the swab content in the buffer. Next, the samples were frozen in a dry-ice bath and stored at −80˚C until shipping to Wroclaw University of Technology (Poland) for metabolomics analysis. To obtain mouth washout, 0.5 ml of sterile saline (0.9% solution of NaCl) was given to participants to rinse the oral cavity for 20-30 seconds, then the sample was collected in a 2 ml sterile polypropylene tube and immediately frozen in a dry-ice bath until shipping for metabolomics analysis. No preservatives were added to the samples.
Sample preparation for 1H-nuclear magnetic resonance (NMR) measurements. Biological samples were thawed at room temperature and vortexed. Each sample of 120 μl of mouth washout and 120 μl of tongue swab in PBS were transferred to new Eppendorf tubes. The biological samples were then centrifuged for 10 min at 4˚C at 13,845 × g. After centrifugation, 100 μl of supernatant was transferred to a new Eppendorf tube and mixed with 400 μl deuterium oxide (D2O) and 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid sodium salt (TSP) (0.3 mM), and then transferred to an NMR cuvette (5 mm, Type 5SP; ARMAR GmbH, Leipzig, Saxony, Germany).
Proton NMR measurements. The NMR spectra of mouth washout and tongue swab samples were recorded at 300 K using an Avance II spectrometer (Bruker, GmBH Hamburg, Germany) operating at proton frequency of 600.58 MHz. The CPMG pulse sequence with water presaturation (cpmgpr1d in Bruker notation) was used for both types of biological material. Spectra for each sample were collected by 128 subsequent scans with relaxation delay of 3.5 seconds; acquisition time of 2.73 seconds; time domain (TD) of 64k; spectral width (SW) of 20.01 ppm. The 1H-NMR spectra were processed with line broadening of 0.3 Hz and manually phased, baseline-corrected using Mestrelab Research v 11.0, (MestReNova, Santiago de Compostela, Spain). Chemical shift was adjusted to reference signal of TSP at δ=0.00 ppm. The resonance signals were aligned with the use of the correlation optimized warping algorithm (COW) and the icoshift algorithm implemented in Matlab 2014a (Mathworks Inc., Natick, MA, USA) (39,40). Probabilistic quotient normalization (PQN) method was used for spectra normalization (41).
Processing for data analysis. Calculation of the relative integral of any NMR-measured metabolite was obtained as a sum of data points of the non-overlapping resonances (or a cluster of partly overlapping resonances). The metabolite resonances were identified according to assignments published in the literature, the v 8.2 Chenomx software (Chenomx Inc. Edmonton, Alberta, Canada) and on-line databases (Biological Magnetic Resonance Data Bank (www.bmrb.wisc.edu) and Human Metabolome Data Base (www.hmdb.ca). Only those metabolites values that could be calculated based on assumptions of non-overlapping resonances were used for further analysis. A total of 21 mouth washout and 18 tongue swab metabolites were assigned. Data matrix was scaled by unit variance scaling (UV, Autoscaling).
Multivariate data analysis. The Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA) with a 7-fold cross-validation procedure was performed to determine variation between studied groups. Sample order in the data matrix was randomized. The reliability of OPLS-DA models was assessed by cross validation analysis of variance at a significance level α=0.05. The importance of the variables explaining the part of X related to Y were presented by VIPpred plot (Predictive Variable Importance for the Projection). Multivariate analysis calculations were carried out in SIMCA 15 software (Sartorius Stedim Biotech, Göttingen, Lower Saxony, Germany).
Statistical data analysis. Calculated relative integral of metabolites assigned for mouth washouts and tongue swabs were used for univariate statistics. The Shapiro–Wilk test was calculated for each variable at a significance level α=0.05. Depending on the results of normality test, a parametric (Student’s t-test) or a nonparametric (Mann–Whitney–Wilcoxon test) test was used, both at a significance level α=0.05. The changes between relative integrals of metabolites of studied groups were verified by average percentage difference (APD) and median percentage difference (MPD). APD and MPD were calculated in relation to values for the control group, whereby a positive value refers to the value being higher in the patient group and negative one being lower in the patient group than in the control group. The boxplot data visualization and receiver operating characteristics (ROC) curve calculation (classical univariate ROC curve analyses) were prepared with MetaboAnalyst 4.0 platform (42). The cutoff value for area under the ROC curve (AUC-ROC) of 0.750 was implemented for selection of the metabolites most predictive of CPD.
Results
The 1H-NMR measurements allowed assignment of 21 recognized and four unknown metabolites from tongue swabs and mouth washouts as listed in Table I.
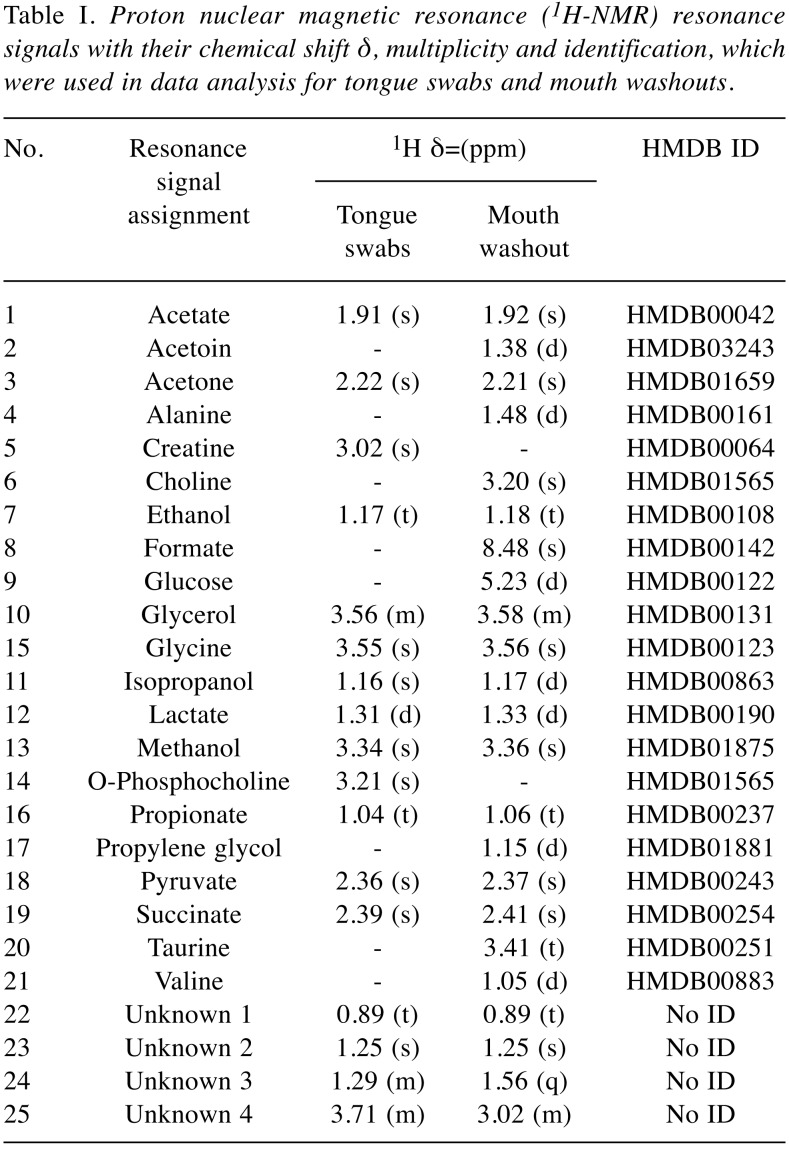
Table I. Proton nuclear magnetic resonance (1H-NMR) resonance signals with their chemical shift δ, multiplicity and identification, which were used in data analysis for tongue swabs and mouth washouts.
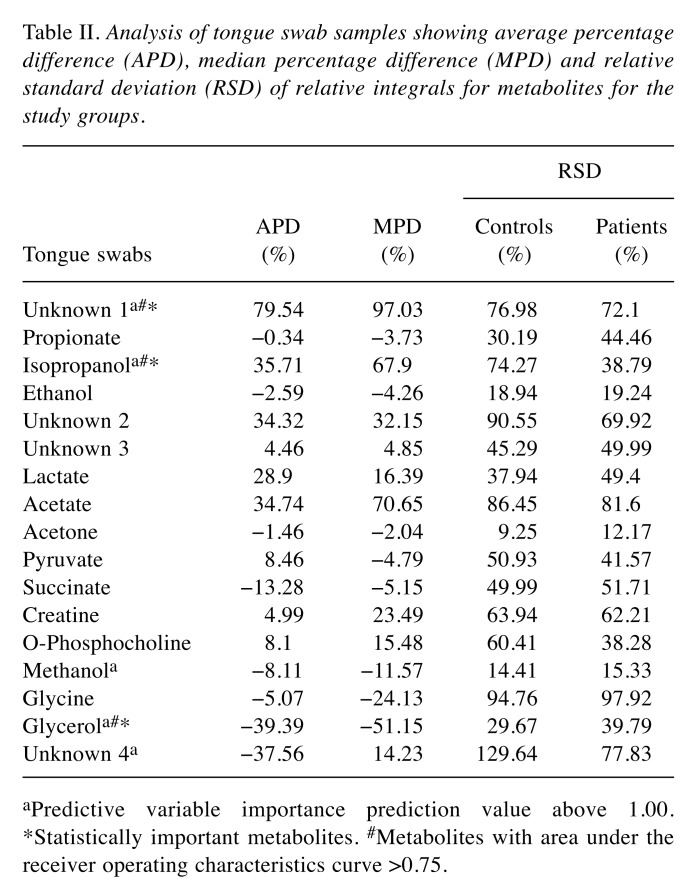
The analysis of the tongue swabs by 1H-NMR method detected 17 metabolites: acetate, acetone, creatine, ethanol, glycerol, glycine, isopropanol, lactate, methanol, O-phosphocholine, propionate, pyruvate, succinate, and four unknown compounds. Among all these metabolites, only three were of statistical significance: Unknown 1, isopropanol, and glycerol (Table II). The first two metabolites were found at a higher level and the third was reduced in the CPD group compared to the control group.
Table II. Analysis of tongue swab samples showing average percentage difference (APD), median percentage difference (MPD) and relative standard deviation (RSD) of relative integrals for metabolites for the study groups.
aPredictive variable importance prediction value above 1.00.*Statistically important metabolites. #Metabolites with area under the receiver operating characteristics curve >0.75.
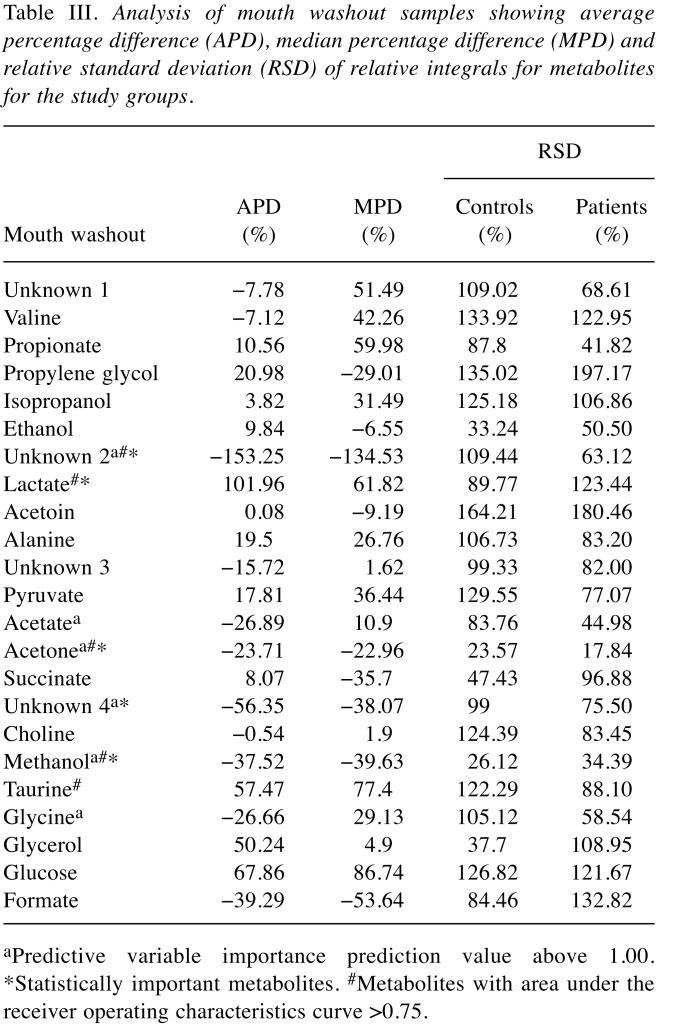
Analysis of the mouth washouts revealed the presence of 23 assigned metabolites, namely: acetate, acetoin, acetone, alanine, choline, ethanol, formate, glucose, glycerol, glycine, isopropanol, lactate, methanol, propionate, propylene glycol, pyruvate, succinate, taurine, valine and unknown compounds 1-4 (Table I). Differences in levels of four metabolites were statistically significant between healthy and study groups: acetone, methanol and unknown 2 were lower, while that of lactate was higher in CPD than in controls (Table III).
Table III. Analysis of mouth washout samples showing average percentage difference (APD), median percentage difference (MPD) and relative standard deviation (RSD) of relative integrals for metabolites for the study groups.
aPredictive variable importance prediction value above 1.00. *Statistically important metabolites. #Metabolites with area under the receiver operating characteristics curve >0.75.
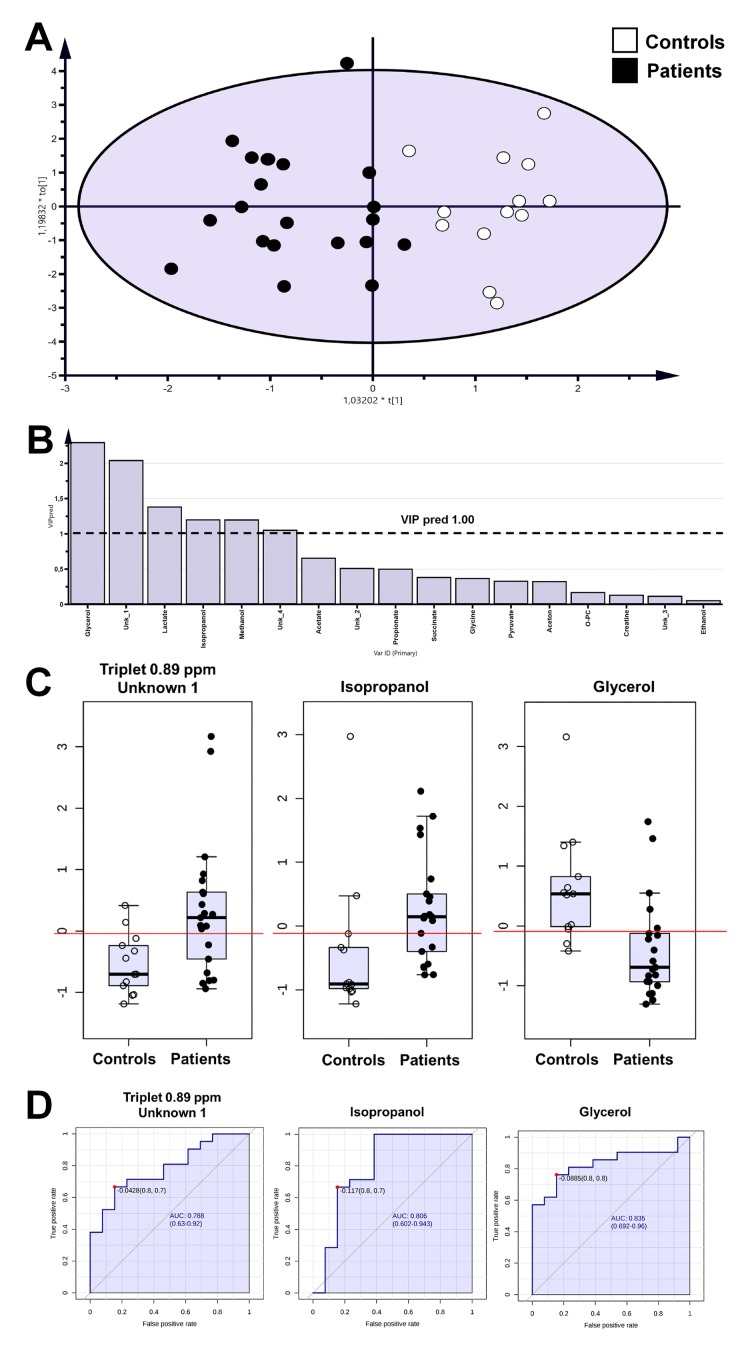
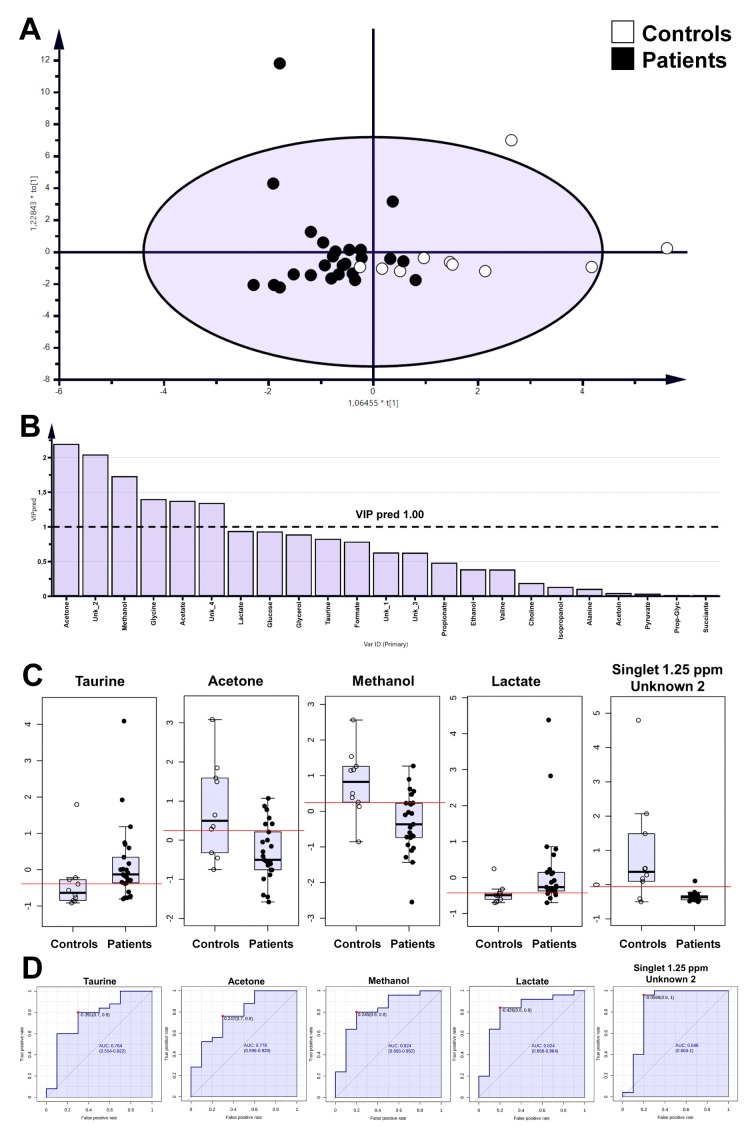
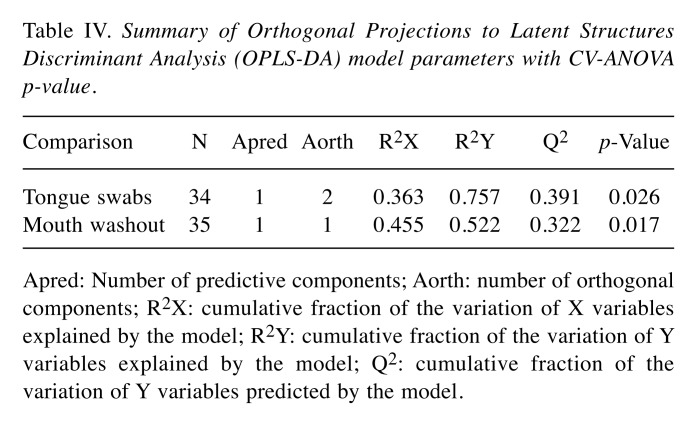
The OPLS-DA analysis showed separation between patient and healthy control groups with satisfactory parameters (Figure 1 and Figure 2; Table IV), where both models passed the validation test at the level of p=0.05.
Figure 1. Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA) models for tongue swab samples (A), predictive variable importance for the projection plot (B), boxplot for statistically important metabolites and these with area under the receiver operating characteristics curve (AUCROC)>0.75 (C), univariate ROC curves for selected metabolites (D).
Figure 2. Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA) models for mouth washout samples (A), predictive variable importance for the projection plot (B), boxplot for statistically important metabolites and/or these with area under the receiver operating characteristics (AUCROC)>0.75 (C), univariate receiver operating characteristics (ROC) curves for selected metabolites (D).
Table IV. Summary of Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA) model parameters with CV-ANOVA p-value.
Apred: Number of predictive components; Aorth: number of orthogonal components; R2X: cumulative fraction of the variation of X variables explained by the model; R2Y: cumulative fraction of the variation of Y variables explained by the model; Q2: cumulative fraction of the variation of Y variables predicted by the model.
The identified metabolites were verified by the ROC curve by their discrimination potential. The value of 0.75 of AUC-ROC was utilized as the cutoff point for selection of metabolites. According to this limit, metabolites such as unknown 1, isopropanol, and glycerol were important based on tongue swab samples (Table II). Moreover, for mouth washout, acetone, methanol, lactate, unknown 2 and taurine passed the determined critical value for the ROC curve (Table III).
Discussion
To date, metabolomics analysis pertinent to periodontitis has focused only on the metabolome of saliva and GCF (43-47). In this study, we investigated the metabolic status of the oral cavity in CPD using easily accessible, simple to collect and non-invasive sampling of mouth washouts and tongue swabs. The changes in the metabolite levels presented in Tables II and III seem to be related to two phenomena. First, to the activity of the consortium of microorganisms in the oral cavity, which utilize various energy sources during colonization and survival, and second, to the host response to ongoing pathological processes. Eight metabolites altogether can be used as potential markers of periodontitis-associated processes, however, some of them (methanol and isopropanol), which are considered as exogenous compounds, were reported as endogenous metabolites of saliva (43). Moreover, the trends in metabolite level changes in the month washouts and tongue swabs did not always match (Tables II and III). This might have been a result of the methodology of sample collections. Whereas tongue swabs were obtained through physical rubbing on the mucosal surface, the mouth washouts involved gentle rinsing of the oral cavity. A second reason may be associated with the change in solubility or local concentration of the metabolite and gingival response to these stimuli.
Lactic acid was a statistically important metabolite in mouth washout samples. This metabolite is recognized as a side product of oral microbiome activity, e.g. a carbohydrate fermentation (48), due to poor hygiene of the oral cavity. On the other hand, the presence of the Lactobacilli species was associated with their protective function against pathogenic strains (49). Interestingly, in the context of the analysis of variation in trends for metabolites in mouth washout samples from patients with CPD, a similar metabolic profile (formate, propionate) to that found in this study was detected in patients with generalized aggressive periodontitis (G-AgP) (50). The analysis showed significantly increased concentrations of lactic acid, while that of formic acid was lower in comparison to the control group (50) and its concentration was associated with P. gingivalis-positive sites.
Previous studies exploring the levels of antioxidants and oxidative stress in serum, saliva, and GCF of patients with CPD have shown elevated lipid peroxidation and a disturbed antioxidant status (51-53). Although in this study, we did not detect unambiguously any products of protein/lipid peroxidation in CPD-derived mouth washouts, elevated level of taurine was detected. We hypothesize that mobilization of taurine within the oral cavity may constitute a protective effect mediated by its antioxidative properties and regeneration of inflammatory gingival tissue (54).
Acetone originates from breakdown of acetoacetate and α-hydroxybutyrate, accompanying fatty acid degradation, glycolysis, and pyruvate metabolism (55-57). Acetone mouth odor is mainly associated with diabetes and halitosis, however, its higher level in body fluids may be used in clinical diagnostics (56-58). The metabolism of acetone is closely associated with the production of lactic acid, which in this study was increased, therefore can be the product, at least in part of acetone biotransformation (57). Increased level of isopropanol in samples from CPD-affected patients is rather surprising, considering that it is used as a common disinfectant and antiseptic agent. However, the positive correlation between isopropanol and acetone was found, where acetone can also be the product of isopropanol breakdown (56).
In general, glycerol is a metabolite that can originate from glucose, proteins, pyruvate, triacylglycerols, and other metabolic pathways (59). A part of glycerol of biological origin can also be derived from its use as an ingredient of toothpaste. Surprisingly, its level was found to be significantly decreased in CPD, and this phenomenon can be explained by utilization of glycerol as a carbon source or an osmoregulatory agent by bacteria and fungi (59) colonizing the oral cavity during periodontitis. The literature data showed that glycerol-3-phosphate, which is the product of glycerol phosphorylation by glycerol kinase, was found to be elevated in periodontal disease (25,60).
The presence of methanol in the oral cavity remains in conflict with its toxic properties. In general, methanol is considered a toxic agent in mammals. However, it is commonly found as an endogenous metabolite, especially in exhaled breath (61,62). Very recently it was suggested that methanol might regulate mammalian gene activity (63). The role of methanol and its sources in humans are not well known. However, anaerobic fermentation by bacteria in the human gut associated with a pectin-rich plant diet is considered the source of methanol (64). Another process is the transformation of S-adenosyl methionine to methanol (65). Therefore, the presence of methanol in the oral cavity is not surprising and its lowered level in those with CPD than in healthy individuals may be related to its use as a carbon or energy source for bacteria associated with CPD.
All these metabolites might contribute to CPD and determining the pattern of their changes could be helpful to highlight specific biochemical disturbances associated with the transition from oral health to dysbiosis observation in CPD
Conclusion
The current study was performed to recognize the feasibility of using metabolomic analysis in tracing pathological changes in the oral cavity during the transition from health to CPD. Based on the multivariate models, ROC, and statistical analysis, it may be possible to find correlation between specific patterns of metabolite variation and the activity of the microbial consortium in the oral cavity, which utilize various energy sources during colonization.
We conclude that CPD-associated bacterial have a higher metabolic activity as revealed by changes of lactate, acetone, isopropanol, glycerol and methanol levels in comparison to commensal microbiota in a healthy mouth. We suggest that these metabolites are potential candidate biomarkers of CPD and propose NMR-based metabolomics analysis as a potentially useful methodological approach for monitoring the pathological processes observed in the oral cavity in the course of this disease. Further insight is needed to confirm this preliminary observation through a prospective study on a larger cohort of patients with CPD matched by sex and age with healthy individuals.
Ethics Approval and Consent to Participate
The study was carried out in accordance with the Declaration of Helsinki. The Bioethical Committee at the Jagiellonian University (KBET/310/B/2012), Medical College in Krakow approved the protocol, including the clinical/periodontal examination, collection of tongue swabs and mouth washouts.
Consent for Publication
All participants read and signed a written informed consent prior to enrollment in the study.
Availability of Data and Material
The datasets generated during and/or analyzed during the current study are available from the corresponding Author upon a reasonable request.
Conflicts of Interest
The Authors declare that they have no conflict of interests in regard to this study.
Authors’ Contributions
KG: Experiment idea; clinical sample collection; clinical part of the study conduction; manuscript preparation; substantive discussion. WW: Performed the metabolomics experiment; data calculation, article preparation. BQ: Manuscript edition, data preparation. AL: Data preparation. KŁ-B: Clinical part of the study conduction; substantive discussion. PM: Substantive discussion. MC-G: Substantive discussion; JP: Substantive discussion. PM: Experiment idea; manuscript preparation; substantive discussion.
Acknowledgements
The project was financially supported in part by National Science Centre, Poland (UMO-2018/29/B/NZ2/01930 to K.G.) and by Wroclaw University of Technology S40531/Z0303 together with WCB KNOW 2014-2018 grant. JP acknowledges support from Natural Science Centre, Poland (UMO-2018/30/A/NZ5/00650).
References
- 1.Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. PMID: 23134607. DOI: 10.1111/ j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wright CJ, Burns LH, Jack AA, Back CR, Dutton LC, Nobbs AH, Lamont RJ, Jenkinson HF. Microbial interactions in building of communities. Mol Oral Microbiol. 2013;28:83–101. doi: 10.1111/omi.12012. PMID: 23253299. DOI: 10.1111/omi.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. PMID: 9841671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eke PI, Dye BA, Wei L, Thornton Evans GO, Genco RJ. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res. 2012;91:914–920. doi: 10.1177/0022034512457373. PMID: 22935673. DOI: 10.1177/0022034512457373. [DOI] [PubMed] [Google Scholar]
- 5.Demmer RT, Papapanou PN. Epidemiologic patterns of chronic and aggressive periodontitis. Periodontol 2000. 2010;53:28–44. doi: 10.1111/j.1600-0757.2009.00326.x. PMID: 20403103. DOI: 10.1111/j.1600-0757.2009. 00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haffajee AD, Socransky SS. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994;5:78–111. doi: 10.1111/j.1600-0757.1994.tb00020.x. PMID: 963164. DOI: 10.1111/j.1600-0757.1994.tb00020.x. [DOI] [PubMed] [Google Scholar]
- 7.Socransky SS, Haffajee AD. The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol. 1992;63:322–331. doi: 10.1902/jop.1992.63.4s.322. PMID: 1573546. DOI: 10.1902/jop.1992. 63.4s.322. [DOI] [PubMed] [Google Scholar]
- 8.Bostanci N, Belibasakis GN. Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiol Lett. 2012;333:1–9. doi: 10.1111/j.1574-6968.2012.02579.x. PMID: 22530835. DOI: 10.1111/j.1574-6968.2012.02579.x. [DOI] [PubMed] [Google Scholar]
- 9.Theilade E. Factors controlling the microflora of the healthy mouth, In Hill MJ, Marsh PD. (Eds). Human Microbial Ecology CRC Press. 1990:1–56. [Google Scholar]
- 10.McGraw WT, Potempa J, Farley D, Travis J. Purification, characterization, and sequence analysis of a potential virulence factor from Porphyromonas gingivalis, peptidylarginine deiminase. Infect Immun. 1999;67:3248–3256. doi: 10.1128/iai.67.7.3248-3256.1999. PMID: 10377098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pyrc K, Milewska A, Kantyka T, Sroka A, Maresz K, Kozieł J, Nguyen KA, Enghild JJ, Knudsen AD, Potempa J. Inactivation of epidermal growth factor by Porphyromonas gingivalis as a potential mechanism for periodontal tissue damage. Infect Immun. 2013;81:55–64. doi: 10.1128/IAI.00830-12. PMID: 23090954. DOI: 10.1128/IAI.00830-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gawron K, Bereta G, Nowakowska Z, Lazarz–Bartyzel K, Lazarz M, Szmigielski B, Mizgalska D, Buda A, Koziel J, Oruba Z, Chomyszyn Gajewska M, Potempa J. Peptidylarginine deiminase from Porphyromonas gingivalis contributes to infection of gingival fibroblasts and induction of prostaglandin E2-signaling pathway. Mol Oral Microbiol. 2014;29:321–332. doi: 10.1111/omi.12081. PMID: 25176110. DOI: 10.1111/omi.12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank RM, Voegel JC. Bacterial bone resorption in advanced cases of human periodontitis. J Periodontal Res. 1978;13:251–261. doi: 10.1111/j.1600-0765.1978.tb00177.x. PMID: 149195. DOI: 10.1111/j.1600-0765.1978.tb00177.x. [DOI] [PubMed] [Google Scholar]
- 14.Slots J. Enzymatic characterization of some oral and nonoral gram-negative bacteria with the API ZYM system. J Clin Microbiol. 1981;14:288–294. doi: 10.1128/jcm.14.3.288-294.1981. PMID: 7026598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Offenbacher S, Odle BM, Van Dyke TE. The use of crevicular fluid prostaglandin E2 levels as a predictor of periodontal attachment loss. J Periodontal Res. 1986;21:101–112. doi: 10.1111/j.1600-0765.1986.tb01443.x. PMID: 2937899. DOI: 10.1111/j.1600-0765.1986.tb01443.x. [DOI] [PubMed] [Google Scholar]
- 16.Offenbacher S, Heasman PA, Collins JG. Modulation of host PGE2 secretion as a determinant of periodontal disease expression. J Periodontol. 1993;64:432–444. doi: 10.1902/jop.1993.64.5s.432. PMID: 8315565. DOI: 10.1902/jop.1993.64.5s.432. [DOI] [PubMed] [Google Scholar]
- 17.Preshaw PM, Heasman PA. Prostaglandin E2 concentrations in gingival crevicular fluid: observations in untreated chronic periodontitis. J Clin Periodontol. 2002;29:15–20. doi: 10.1034/j.1600-051x.2002.290103.x. PMID: 11846844. DOI: 10.1034/j.1600-051x.2002.290103.x. [DOI] [PubMed] [Google Scholar]
- 18.Lamont RJ, Jenkinson HF. Life Below the Gum Line: Pathogenic Mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. PMID: 9841671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tonzetich J, McBride BC. Characterization of volatile sulphur production by pathogenic and non-pathogenic strains of oral Bacteroides. Arch Oral Biol. 1981;26:963–969. doi: 10.1016/0003-9969(81)90104-7. DOI: 10.1016/0003-9969(81)90104-7. [DOI] [PubMed] [Google Scholar]
- 20.Mager DL, Ximenez Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol. 2003;30:644–654. doi: 10.1034/j.1600-051x.2003.00376.x. PMID: 12834503. DOI: 10.1034/j.1600-051X.2003.00376.x. [DOI] [PubMed] [Google Scholar]
- 21.Tanner AC, Paster BJ, Lu SC, Kanasi E, Kent R Jr, Van Dyke T, Sonis ST. Subgingival and tongue microbiota during early periodontitis. J Dent Res. 2006;85:318–323. doi: 10.1177/154405910608500407. PMID: 16567551. DOI: 10.1177/154405910608500407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kishi M, Ohara Nemoto Y, Takahashi M, Kishi K, Kimura S, Aizawa F, Yonemitsu M. Prediction of periodontopathic bacteria in dental plaque of periodontal healthy subjects by measurement of volatile sulfur compounds in mouth air. Arch Oral Biol. 2013;58:324–330. doi: 10.1016/j.archoralbio.2012.09.019. PMID: 23107050. DOI: 10.1016/ j.archoralbio.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 23.Cortelli JR, Aquino DR, Cortelli SC, Fernandes CB, Filho de Carvalho J, Franco GC, Costa FO, Kawai T. Etiological analysis of initial colonization of periodontal pathogens in oral cavity. J Clin Microbiol. 2008;46:1322–1329. doi: 10.1128/JCM.02051-07. PMID: 18287326. DOI: 10.1128/JCM.02051-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuboniwa M, Amano A, Kimura KR, Sekine S, Kato S, Yamamoto Y, Okahashi N, Iida T, Shizukuishi S. Quantitative detection of periodontal pathogens using real-time polymerase chain reaction with TaqMan probes. Oral Microbiol Immunol. 2004;19:168–176. doi: 10.1111/j.0902-0055.2004.00135.x. PMID: 15107068. DOI: 10.1111/ j.0902-0055.2004.00135.x. [DOI] [PubMed] [Google Scholar]
- 25.Barnes VM, Teles R, Trivedi HM, Devizio W, Xu T, Mitchell MW, Milburn MV, Guo L. Acceleration of purine degradation by periodontal diseases. J Dent Res. 2009;88:851–855. doi: 10.1177/0022034509341967. PMID: 19767584. DOI: 10.1177/0022034509341967. [DOI] [PubMed] [Google Scholar]
- 26.Akalin FA, Baltacioglu E, Alver A, Karabulut E. Lipid peroxidation levels and total oxidant status in serum, saliva and gingival crevicular fluid in patients with chronic periodontitis. J Clin Periodontol. 2007;34:558–565. doi: 10.1111/j.1600-051X.2007.01091.x. PMID: 17555410. DOI: 10.1111/j.1600-051X.2007.01091.x. [DOI] [PubMed] [Google Scholar]
- 27.Pradeep AR, Manjunath SG, Swati PP, Shikha C, Sujatha PB. Gingival crevicular fluid levels of leukotriene B4 in periodontal health and disease. J Periodontol. 2007;78:2325–2330. doi: 10.1902/jop.2007.070135. PMID: 18052705. DOI: 10.1902/jop.2007.070135. [DOI] [PubMed] [Google Scholar]
- 28.Alpagot T, Bell C, Lundergan W, Chambers DW, Rudin R. Longitudinal evaluation of GCF MMP-3 and TIMP-1 levels as prognostic factors for progression of periodontitis. J Clin Periodontol. 2001;28:353–359. doi: 10.1034/j.1600-051x.2001.028004353.x. PMID: 11314892. DOI: 10.1034/j.1600-051x.2001.028004353.x. [DOI] [PubMed] [Google Scholar]
- 29.Dawiskiba T, Deja S, Mulak A, Ząbek A, Jawień E, Pawełka D, Banasik M, Mastalerz Migas A, Balcerzak W, Kaliszewski K, Skóra J, Barć P, Korta K, Pormańczuk K, Szyber P, Litarski A, Młynarz P. Serum and urine metabolomic fingerprinting in diagnostics of inflammatory bowel diseases. World J Gastroenterol. 2014;20:163–174. doi: 10.3748/wjg.v20.i1.163. PMID: 24415869 DOI: 10.3748/wjg.v20.i1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rathnayake N, Akerman S, Klinge B, Lundegren N, Jansson H, Tryselius Y, Sorsa T, Gustafsson A. Salivary biomarkers for detection of systemic diseases. PLoS ONE. 2013;8:e61356. doi: 10.1371/journal.pone.0061356. PMID: 23637817. DOI: 10.1371/journal.pone.0061356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deja S, Porebska I, Kowal A, Zabek A, Barg W, Pawelczyk K, Stanimirova I6, Daszykowski M, Korzeniewska A, Jankowska R, Mlynarz P. Metabolomics provide new insights on lung cancer staging and discrimination from chronic obstructive pulmonary disease. J Pharm Biomed Anal. 2014;100:369–380. doi: 10.1016/j.jpba.2014.08.020. PMID: 25213261. DOI: 10.1016/j.jpba.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 32.Bessonneau V, Bojko B, Pawliszyn J. Analysis of human saliva metabolome by direct immersion solid-phase microextraction LC and benchtop orbitrap MS. Bioanalysis. 2013;5:783–792. doi: 10.4155/bio.13.35. PMID: 23534423. DOI: 10.4155/bio.13.35. [DOI] [PubMed] [Google Scholar]
- 33.Zhang A, Sun H, Wang X. Saliva metabolomics opens door to biomarker discovery, disease diagnosis, and treatment. Appl Biochem Biotechnol. 2012;168:1718–1727. doi: 10.1007/s12010-012-9891-5. PMID: 22971835. DOI: 10.1007/s12010-012-9891-5. [DOI] [PubMed] [Google Scholar]
- 34.Rathnayake N, Akerman S, Klinge B, Lundegren N, Jansson H, Tryselius Y, Sorsa T, Gustafsson A. Salivary biomarkers of oral health: a cross-sectional study. J Clin Periodontol. 2013;40:140–147. doi: 10.1111/jcpe.12038. PMID: 23174014. DOI: 10.1111/jcpe.12038. [DOI] [PubMed] [Google Scholar]
- 35.Spielmann N, Wong DT. Saliva: diagnostics and therapeutic perspectives. Oral Dis. 2011;17:345–354. doi: 10.1111/j.1601-0825.2010.01773.x. PMID: 21122035. DOI: 10.1111/j.1601-0825.2010.01773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aimetti M, Cacciatore S, Graziano A, Tenori L. Metabolomics analysis of saliva reveals generalized chronic periodontitis signature. Metabolomics. 2012;8:465–474. DOI: 10.1007/s11306-011-0331-2. [Google Scholar]
- 37.Barnes VM, Ciancio SG, Shibly O, Xu T, Devizio W, Trivedi HM, Guo L, Jönsson TJ. Metabolomics revels elevated macromolecular degradation in periodontal diseases. J Dent Res. 2011;90:1293–1297. doi: 10.1177/0022034511416240. PMID: 21856966. DOI: 10.1177/0022034 511416240. [DOI] [PubMed] [Google Scholar]
- 38.Huang Y, Zhu M, Li Z, Sa R, Chu Q, Zhang Q, Zhang H, Tang W, Zhang M, Yin H. Mass spectrometry-based metabolomic profiling identifies alterations in salivary redox status and fatty acid metabolism in response to inflammation and oxidative stress in periodontal disease. Free Radic Biol Med. 2014;70:223–232. doi: 10.1016/j.freeradbiomed.2014.02.024. PMID: 24607715. DOI: 10.1016/j.freeradbiomed. 2014.02.024. [DOI] [PubMed] [Google Scholar]
- 39.Tomasi G, van den Berg F, Andersson C. Correlation optimized warping and dynamic time warping as preprocessing methods for chromatographic data. J Chemometrics. 2004;18:231–241. DOI: 10.1002/cem.859. [Google Scholar]
- 40.Savorani F, Tomasi G, Engelsen SB. Icoshift A versatile tool for the rapid alignment of 1D NMR spectra. J Magn Reson. 2010;202:190–202. doi: 10.1016/j.jmr.2009.11.012. PMID: 20004603. DOI: 10.1016/j.jmr. 2009.11.012. [DOI] [PubMed] [Google Scholar]
- 41.Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem. 2006;78:4281–4290. doi: 10.1021/ac051632c. PMID: 16808434. DOI: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- 42.Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, Wishart DS, Xia J. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486–W494. doi: 10.1093/nar/gky310. PMID: 29762782. DOI: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dame ZT, Aziat F, Mandal R, Krishnamurthy R, Bouatra S, Borzouie S, Guo AC, Sajed T, Deng L, Lin H, Liu P, Dong E, Wishart DS. The human saliva metabolome. Metabolomics. 2015;11:1864–1883. DOI: 10.1007/s11306-015-0840-5. [Google Scholar]
- 44.Syrjänen S, Piironen P, Markkanen H. Free amino-acid content of wax-stimulated human whole saliva as related to periodontal disease. Arch Oral Biol. 1987;32:607–610. doi: 10.1016/0003-9969(87)90032-x. PMID: 3481959. DOI: 10.1016/0003-9969(87)90032-X. [DOI] [PubMed] [Google Scholar]
- 45.Qiqiang L, Huanxin M, Xuejun G. Longitudinal study of volatile fatty acids in the gingival crevicular fluid of patients with periodontitis before and after nonsurgical therapy. J Periodontal Res. 2012;47:740–749. doi: 10.1111/j.1600-0765.2012.01489.x. DOI: 10.1111/j.1600-0765.2012.01489.x. [DOI] [PubMed] [Google Scholar]
- 46.Lu RF, Meng HX, Gao XJ, Feng L, Xu L. Analysis of short chain fatty acids in gingival crevicular fluid of patients with aggressive periodontitis. Zhonghua Kou Qiang Yi Xue Za Zhi. 2008;43:664–667. PMID: 15938880. [PubMed] [Google Scholar]
- 47.Syrjänen S, Piironen P, Markkanen H. Free amino-acid composition of wax-stimulated whole saliva in human subjects with healthy periodontium, severe chronic periodontitis and post-juvenile periodontitis. Arch Oral Biol. 1984;29:735–738. doi: 10.1016/0003-9969(84)90181-x. DOI: 10.1016/0003-9969(84)90181-X. [DOI] [PubMed] [Google Scholar]
- 48.Nobuhiro T. Microbial ecosystem in the oral cavity, Metabolic diversity in an ecological niche and its relationship with oral diseases. International Congress Series. 2005;1284:103–112. DOI: 10.1016/j.ics.2005.06.071. [Google Scholar]
- 49.Sookkhee S, Chulasiri M, Prachyabrued W. Lactic acid bacteria from healthy oral cavity of Thai volunteers: inhibition of oral pathogens. J Appl Microbiol. 2001;90:172–179. doi: 10.1046/j.1365-2672.2001.01229.x. PMID: 11168719. DOI: 10.1046/j.1365-2672.2001.01229.x. [DOI] [PubMed] [Google Scholar]
- 50.Lu R, Meng H, Gao X, Xu L, Feng X. Effect of non-surgical periodontal treatment on short chain fatty acid levels in gingival crevicular fluid of patients with generalized aggressive periodontitis. J Periodontal Res. 2014;49:574–583. doi: 10.1111/jre.12137. PMID: 25340203. DOI: 10.1111/jre.12137. [DOI] [PubMed] [Google Scholar]
- 51.Canakci CF, Cicek Y, Yildirim A, Sezer U, Canakci V. Increased levels of 8-hydroxydeoxyguanosine and malondialde-hyde and its relationship with antioxidant enzymes in saliva of periodontitis patients. Eur J Dent. 2009;3:100–106. PMID: 19421389. [PMC free article] [PubMed] [Google Scholar]
- 52.Chapple IL, Brock G, Eftimiadi C, Matthews JB. Glutathione in gingival crevicular fluid and its relation to local antioxidant capacity in periodontal health and disease. Mol Pathol. 2002;55:367–373. doi: 10.1136/mp.55.6.367. PMID: 12456773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moore S, Calder KA, Miller NJ, Rice Evans CA. Antioxidant activity of saliva and periodontal disease. Free Radic Res. 1994;21:417–425. doi: 10.3109/10715769409056594. PMID: 7834056. [DOI] [PubMed] [Google Scholar]
- 54.Chaudhry S, Tandon B, Gupta A, Gupta S. Taurine: A potential mediator for periodontal therapy. Indian J Dent Res. 2018;29:808–811. doi: 10.4103/ijdr.IJDR_123_17. DOI: 10.4103/ijdr.IJDR_123_17. [DOI] [PubMed] [Google Scholar]
- 55.Synthesis and degradation of ketone bodies – Homo sapiens (human) https://www.genome.jp/kegg-bin/show_pathway?org_name=map&mapno=00072&mapscale=&show_description=hide %5D]. [Google Scholar]
- 56.Ruzsányi V, Péter Kalapos M. Breath acetone as a potential marker in clinical practice. J Breath Res. 2017;11:024002–024002. doi: 10.1088/1752-7163/aa66d3. PMID: 28294105. DOI: 10.1088/1752-7163/aa66d3. [DOI] [PubMed] [Google Scholar]
- 57.Kalapos MP. On the mammalian acetone metabolism: from chemistry to clinical implications. Biochim Biophys Acta. 2003;1621:122–139. doi: 10.1016/s0304-4165(03)00051-5. PMID: 12726989. DOI: https://doi.org/ 10.1016/S0304-4165(03)00051-5. [DOI] [PubMed] [Google Scholar]
- 58.Aylikci BU, Colak H. Halitosis: From diagnosis to management. J Nat Sci Biol Med. 2013;4:14–23. doi: 10.4103/0976-9668.107255. PMID: 23633830. DOI: 10.4103/0976-9668.107255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brisson D, Vohl MC, St-Pierre J, Hudson TJ, Gaudet D. Glycerol: a neglected variable in metabolic processes. Bioessays. 2001;23:534–542. doi: 10.1002/bies.1073. PMID: 11385633. DOI: 10.1002/bies.1073. [DOI] [PubMed] [Google Scholar]
- 60.Liu B, Faller LL, Klitgord N, Mazumdar V, Ghodsi M, Sommer DD, Gibbons TR, Treangen TJ, Chang Y, Li S, Stine OC, Hasturk H, Kasif S, Segrè D, Pop M, Amar S. Deep Sequencing of the Oral Microbiome Reveals Signatures of Periodontal Disease. PLoS ONE. 2012;7:e37919. doi: 10.1371/journal.pone.0037919. PMID: 22675498. DOI: 10.1371/ journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang T, Pysanenko A, Dryahina K, Španel P, Smith D. Analysis of breath, exhaled via the mouth and nose, and the air in the oral cavity. J Breath Res. 2008;2:037013–037013. doi: 10.1088/1752-7155/2/3/037013. PMID: 21386174. DOI: 10.1088/1752-7155/2/3/037013. [DOI] [PubMed] [Google Scholar]
- 62.Fisher JW, Dorman DC, Medinsky MA, Welsch F, Conolly RB. Analysis of respiratory exchange of methanol in the lung of the monkey using a physiological model. Toxicol Sci. 2000;52:185–193. doi: 10.1093/toxsci/53.2.185. PMID: 10696766. DOI: 10.1093/toxsci/53.2.185. [DOI] [PubMed] [Google Scholar]
- 63.Komarova TV, Petrunia IV, Shindyapina AV, Silachev DN, Sheshukova EV, Kiryanov GI, Dorokhov YL. Endogenous methanol regulates mammalian gene activity. PLoS ONE. 2014;9:e90239. doi: 10.1371/journal.pone.0090239. PMID: 24587296. DOI: 10. 1371/journal.pone. 0090239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dorokhov YL, Shindyapina AV, Sheshukova EV, Komarova TV. Metabolic methanol: molecular pathways and physiological roles. Physiol Rev. 2015;95:603–644. doi: 10.1152/physrev.00034.2014. PMID: 25834233. DOI: 10.1152/physrev.00034.2014. [DOI] [PubMed] [Google Scholar]
- 65.Axelrod J, Daly J. Pituitary gland: enzymic formation of methanol from S- adenosylmethionine. Science. 1965;150:892–893. doi: 10.1126/science.150.3698.892. PMID: 5835789. DOI: 10.1126/science.150.3698.892. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding Author upon a reasonable request.