Abstract
Background/aim: Chronic myeloid leukaemia (CML) is a myeloproliferative disorder characterized by the presence of breakpoint cluster region–Abelson murine leukemia (BCR–ABL1) gene fusion as a hallmark that is expressed as two major transcripts b2a2 and b3a2. The aim of this study was to compare the BCR–ABL transcripts in the blood cells of patients with CML, and in chemoresistant and chemosensitive CML cell lines to validate their use as a good method to elucidate CML biology. Materials and Methods: Twelve patients with CML and CML cell lines (K562, K562-LUCENA and FEPS) were analyzed by real-time polymerase chain reaction to evaluate gene expression of BCR–ABL transcripts. Results: All patients had the same expression levels of b2a2 and b3a3 transcripts, however, CML cell lines presented only b3a2 expression. There were no significant differences in absolute b3a2 expression between patients and CML cell lines. Conclusion: CML cell lines provide a good in vitro alternative in that they have the same BCR–ABL expression as patients.
Keywords: CML, leukaemia, BCR-ABL, cell lines
Chronic myeloid leukaemia (CML) represents a relevant myeloproliferative disorder with an incidence of 1-2 cases per 100,000 adults, 15% of newly-diagnosed cases of leukaemia (1). Until about a decade ago, chemotherapy of CML was limited to non-specific agents such as hydroxyurea, busulfan and interferon-alpha. However, after the elucidation of the pathogenesis of CML, targeted therapy was developed, dramatically changing the history of the disease. One study showed the improvement of treatment response from 34.7% in a group treated with interferon-alpha plus cytarabine to 87.1% in a group given imatinib mesylate, an inhibitor of breakpoint cluster region (BCR)–Abelson oncogene (ABL)1 (2).
The central pathogenesis of CML is characterized by a reciprocal translocation of chromosome 9 and 22. This translocation results in a shortened chromosome 22 called Philadelphia (Ph+) chromosome, which presents the fusioned gene of BCR–ABL (3). The fusion of BCR located on chromosome 22 and ABL located on chromosome 9 encodes an oncoprotein that has constitutive tyrosine kinase activity, which stimulates proliferation and resistance to cell death (4).
BCR–ABL1 transcript variants depend on the site of cleavage at the BCR gene. Patients with CML have breaks in the major region of the BCR gene, between exon 13 and 15, which fuses with exon 2 of ABL gene, resulting in b2a2 (e13a2) and b3a2 (e14a2) transcripts of BCR–ABL1 gene, which encodes a oncoprotein with 210 kDa (5). Sharma et al. suggested that patients with CML with b2a2 transcripts might have higher response to imatinib mesylate treatment when compared to those with b3a2 transcript variant (6). Therefore, the identification of transcript type for treatment management is indispensable.
The use of cell lines in experimental oncology allows for the study of the development of new antineoplastic drugs, the identification of prognostic markers and their therapeutic efficacy (7). Thus, the detailed characterization of these cell lines is essential for their validation, in addition to facilitating the understanding of the resistance and sensitivity patterns of these chemotherapies, helping in the choice of more efficient therapies for patients (8).
It is worth noting that several leukaemia cell lines have been helpful in the molecular characterization of the disease and biological features, as well as a tool for preclinical screening of new molecules for treatment. As an example, K562, K561-Lucena and FEPS cell lines have been used as important in vitro models for molecular characterization of BCR–ABL1 and for compound screening for chemotherapeutic development to improve CML treatment (9-13).
Therefore, this study aimed to analyse the transcript variant expression of BCR–ABL1 in chemosensitive and chemoresistant CML cell lines in comparison with patients with CML to validate the cell lines as an efficient tool for understanding the biological features of this disease.
Materials and Methods
Cell lines. Leukemia cell lines K-562, K562-Lucena and FEPS were kindly provided by Dr. Vivian Rumjanek (10,11) and were cultured in RPMI 1640 medium supplemented with 10% foetal bovine serum (FBS) at 37˚C and 5% CO2 atmosphere, with regular passages twice a week. In the case of K562-Lucena and FEPS, the medium was supplemented with vincristine sulfate at a final concentration of 60 nM and daunorubicin at a concentration of 466 nM, respectively.
Patients. The blood samples evaluated in this study were obtained from 12 patients previously diagnosed with CML at the Walter Cantídio University Hospital (Fortaleza, Ceará) from October 2017 to October 2018, who are part of the biological bank of samples from the Laboratory of Pharmacogenetics of the Federal University of Ceará. All patients’ samples were collected before any therapeutic intervention.
Ethics. The blood sample collections were performed after the approval of the Ethics and Research Committee under protocol number 2.439.294. Assurance was provided that the use of biological materials and participation in the study would not cause any harm to nor have any negative influence on patient treatment.
RNA extraction. RNA from patients’ blood samples and from K-562, K562-Lucena and FEPS cell lines were extracted using TRIzol® (Life Technologies, Carlsbad, CA, USA) according to manufacturer’s instructions and reversely transcribed to cDNA using High Capacity cDNA Reverse Transcriptase (Life Technologies) in a Verity® PCR System thermal cycler (Applied Biosystems®, Foster City, CA, USA).
Real-time quantitative reverse transcription (RT-qPCR). The genes selected for expression analysis in patients and in cell lines were BCR–ABL1 transcripts: b2a2 (Hs03024541_ft), and b3a2 (Hs03024541_ft), with ABL (Hs 99999002_mH) being used as internal control. Such genes are commercially available as TaqMan® Gene expression assays (Life Technologies). RT-qPCR was performed using QuantStudio5 Real-Time PCR system (Applied Biosystems®). For each sample was used concentrations following: 3 μl of cDNA, 1 μl of each primer/probe, 12.5 μl of TaqMan® Gene Expression Master Mix (Life Technologiess) and 8.5 μI of Ultra-pure water. Each assay was performed at least three times according to Minimum Information for Publication of Quantitative Real-Time PCR Experiments Guidelines (14). The gene-expression levels were based on absolute and relative analyses and calculated using the 2−ΔΔCT (delta-delta threshold cycle) method, the expression level of the gene of interest is reported relative to the reference gene for each sample (15).
Statistical analysis. All groups of samples were tested for normality using Kolmogorov–Smirnov test assuming 95% of confidence intervals, indicating that the samples were normally distributed. Results are graphically shown as the mean±standard deviation (SD) for patients and cell line samples. The differences between groups were compared using t-test or analysis of variance (ANOVA) followed by Bonferroni post-test, assuming 95% of confidence intervals.
Results and Discussion
BCR–ABL1 fusion gene is a hallmark of CML and a crucial driver of leukaemogenesis due to its constitutive tyrosine kinase activity, which contributes to cell proliferation, resistance to cell death and deregulation of cell differentiation (16). Therefore, advances in the understanding of the molecular basis of CML, including the BCR–ABL1 pathway, are invaluable in improving the treatment and management of CML (17).
Regarding BCR–ABL1 studies, leukaemia cell lines are important tools in this research area, helping to understand the pathways in microenvironments. However, it is still necessary to evaluate whether BCR–ABL1 expression in CML cell lines can be extrapolated to patients with CML.
The patients’ samples were submitted to analysis of BCR–ABL1 transcript expression. Among 34 suspected samples, only 12 (35.3%) patients were diagnosed with CML by the presence of BCR–ABL1 gene fusion. Seven out of the 12 patients were female. The median age was 40.5 years (ranging from 24 to 74 years). However, the worldwide incidence of CML shows a median age at diagnosis of 55 to 60 years, occurring with a slight predominance in males (18,19). Other studies in Brazil have shown the median age at diagnosis to be at least 10 years lower than that found in the international literature (20,21). Data from the present study corroborate a study by Bortolheiro and Chiattone (22), where patients with disease refractory to interferon-alpha had a median age at diagnosis of 40 years.
Our results showed no significant differences of BCR–ABL1 expression between male and female patients (p=0.8329), nor between patients aged 40 years or more and those less than 40 years old (p=0.7430), consistent with previous results obtained which also did not find significant differences (23,24).
BCR–ABL1-negative CML occurs commonly in older patients (69 years) and men (57%) (23). Given the divergent data in the literature, age and gender would not be considered important factors in BCR–ABL1 expression analysis, but this needs to be confirmed by a study with a high representative number of patients. Nevertheless, men and patients with a median age of 55 years tended to be more adherent to treatment with BCR–ABL1 inhibitors than did women and younger patients (25).
Another finding of our study is that patients with CML did not present the b2a2 breakpoint variant, while six patients (50%) presented both b2a2 and b3a2 transcripts, and six patients (50%) presented only b3a2 transcript expression. Moreover, grouping patients into b2a2 and b3a2 groups, the absolute expression levels were not significantly different between the two groups (p=0.3940).
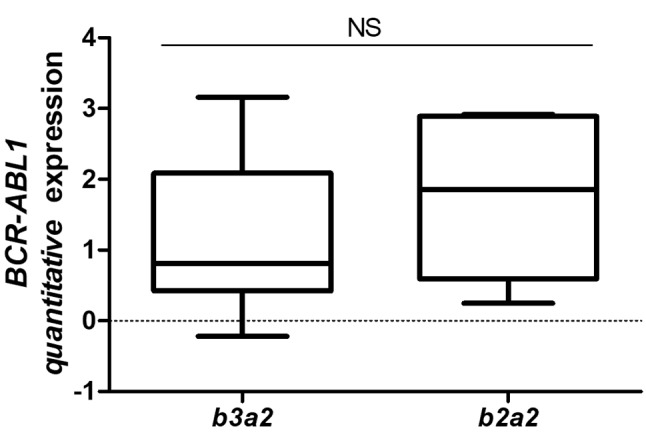
The b3a2 and b2a2 transcripts essentially had the same level of expression in the patients (Figure 1). As mentioned, several studies have shown the importance of BCR–ABL1 transcripts in treatment response, and contradictory results have been demonstrated in different studies that analysed expression of BCR–ABL1 transcript variants, showing an advantage for patients with b3a2. Patients with b3a2 are more likely to achieve favourable molecular major response (MMR) because they may have lower tyrosine kinase activity than those with b2a2, and thus be more sensitive to imatinib treatment (26-30). In an apparent contradiction, de Lemos et al. reported that b2a2 patients had better MMR at 6 months (31), whereas another study showed no significant differences between patients with b3a2 and b2a2 in MMR at 6 months (32).
Figure 1. Absolute quantitative transcript levels of breakpoint cluster region (BCR)–Abelson murine leukemia (ABL1) b3a2 and b2a2 in 12 patients with chronic myeloid leukaemia. Absolute expression data are represented as the mean±SD and groups were statistically analyzed by t-test. NS: Not significant.
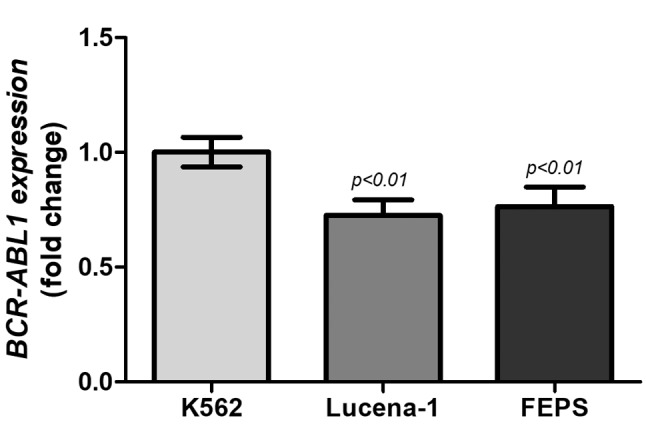
Despite this contradiction, we also tested the expression of BCR–ABL1 transcripts in leukaemia cell lines to show how representative they are for preclinical trials or BCR–ABL1 expression analysis. The presence of b2a2 transcript was not detected in chemosensitive and chemoresistant CML cell lines, only b3a2 transcript expression. However, as shown in Figure 2, the relative expression indicated that chemoresistant cell lines had low transcript level compared to the parental K562 cell line (p<0.01), which confirmed BCR–ABL1-independent resistance that often occurs in patients with CML, such as hyperexpression of membrane transporters (17). Indeed, Moreira et al. have already demonstrated that the resistance mechanism of K562-Lucena1 and FEPS were mediated by membrane transporter genes ATP-binding cassette sub-family B member 1 (ABCB1) and ATP-binding cassette sub-family C member 1 (ABCC1) (33).
Figure 2. Relative breakpoint cluster region (BCR)–Abelson murine leukemia (ABL1) b3a2 transcript expression in the chemosensitive (K562) and chemoresistant (K562 Lucena-1 and FEPS) cell lines. Relative expression data are represented as mean±SD and groups were statistically analysed by ANOVA followed by Bonferroni post-test.
In order to compare the b3a2 transcript level between patients and leukaemia cell lines, absolute quantitative expression was performed. There were no significant differences of b3a2 transcript level between patients and chemosensitive and chemoresistant CML cell lines (Figure 3).
Figure 3. Comparison of breakpoint cluster region (BCR)–Abelson murine leukemia (ABL1) b3a2 transcript expression between patients and chemosensitive (K562) and chemoresistant (K562 Lucena-1 and FEPS) leukaemia cell lines. Absolute expression data are represented as the mean±SD and groups were statistically analysed by ANOVA followed by Bonferroni post-test. NS: Not significant.
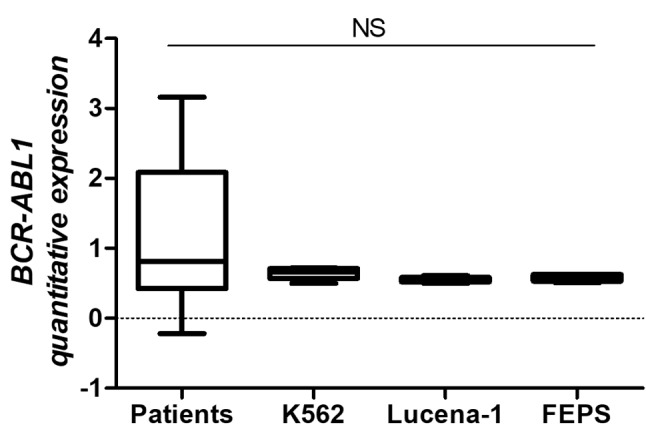
As described before, many studies in the literature have not yet elucidated the clinical relevance of b2a2 and b3a2 transcripts in patients with CML and their influence on response to treatment, thus, the b3a2 transcript expression data presented both in patients and in the cell lines analyzed in this study, demonstrate that the similarity between the models tested may be useful in drug screening regardless of the transcript expressed by the patient.
In agreement with the present study, work performed by Ferreira et al. (7) and Jaeger et al. (34), emphasize the importance of the use of cell lines as an in vitro model to study the different biological aspects of cancer. They also help in the identification of biomarkers, which play a key role in the early stages of drug screening, thus reducing costs with clinical trials.
In addition, although cell lines remain an excellent model for the study of mechanisms of resistance/sensitivity of chemotherapeutics already in use in the treatment of cancer, a study recently published by Ben-David et al. showed that established strains, considered clonal and genetically stable, are in fact highly heterogeneous (35). Cell characterization and standardization of culture conditions between laboratories to minimize the variability of results is essential.
Conclusion
The chemosensitive and resistant CML cell lines evaluated in this study, despite presenting only the BCR–ABL1 b3a2 transcript, showed no difference in expression of this transcript when compared to patients. In summary, the data from this study validate the use of CML cell lines as an indispensable tool for validation of molecular studies, for understanding of biological mechanisms, as well as for the elucidation of mechanisms of resistance to chemotherapy and for screening of new anticancer drugs, mimicking the molecular characteristics that give rise to the tumour model of the disease in vivo.
Conflicts of Interest
The Authors declare no conflicts of interest regarding this study.
Authors’ Contributions
Moreira-Nunes CA, Mesquita FP, Portilho AJS and Montenegro RC performed the study design; Sales LO and Portilho AJS performed the cell culture analysis; Sales LO, Mesquita FP, Portilho AJS and Moreira-Nunes CA performed the molecular analysis; Mesquita FP, Moraes-Filho MO, Moraes MEA, Montenegro RC and Moreira-Nunes CA wrote the article. All Authors read and approved the final article.
Acknowledgements
This study was supported by Brazilian funding agencies National Counsel of Technological and Scientific Development (CNPq; to CAMN, RCM, MEAM, MOMF and FPM) and Coordination for the Improvement of Higher Education Personnel (CAPES; to CAMN and AJSP).
References
- 1.Jabbour E, Kantarjian H. Chronic myeloid leukemia: 2014 update on diagnosis, monitoring, and management. Annu Clin Upd Hematol Malign. 2014;89:547–556. doi: 10.1002/ajh.23691. PMID: 24727196. DOI: 10.1002/ajh.23691. [DOI] [PubMed] [Google Scholar]
- 2.O’Brien SG, Guilhot F, Larson RA, Gathmann I, Baccarani M, Cervantes F, Cornelissen JJ, Fischer T, Hochhaus A, Hughes T, Lechner K, Nielsen JL, Rousselot P, Reiffers J, Saglio G, Shepherd J, Simonsson B, Gratwohl A, Goldman JM, Kantarjian H, Taylor K, Verhoef G, Bolton AE, Capdeville R, Druker BJ. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2003;348:994–1004. doi: 10.1056/NEJMoa022457. PMID: 12637609. DOI: 10.1056/NEJMoa022457. [DOI] [PubMed] [Google Scholar]
- 3.Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. PMID: 4126434. DOI: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 4.Pophali PA, Patnaik MM. The role of new tyrosine kinase inhibitors in chronic myeloid leukemia. Cancer Jl. 2016;22:40–50. doi: 10.1097/PPO.0000000000000165. PMID: 26841016. DOI: 10.1097/PPO.0000000000000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood. 2000;96:3343–3356. PMID: 11071626. [PubMed] [Google Scholar]
- 6.Sharma P, Kumar L, Mohanty S, Kochupillai V. Response to Imatinib mesylate in chronic myeloid leukemia patients with variant BCR–ABL fusion transcripts. Ann Hematol. 2009;89(3):241–247. doi: 10.1007/s00277-009-0822-7. PMID: 19714331. DOI: 10.1007/s00277-009-0822-7. [DOI] [PubMed] [Google Scholar]
- 7.Ferreira D, Adega F, Chaves R. The importance of cancer cell lines as in vitro models in cancer methylome analysis and anticancer drugs testing. Oncogenomics and cancer proteomicsnovel approaches in biomarkers discovery and therapeutic targets in cancer. First ed. InTechOpen. 2013 DOI: 10.55772/ 53110. [Google Scholar]
- 8.Sharma SV, Haber DA, Settelman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer. 2010;10(4):241–253. doi: 10.1038/nrc2820. PMID: 20300105. DOI: 10.1038/nrc2820. [DOI] [PubMed] [Google Scholar]
- 9.Uphoff CC, Habig S, Fombonne S, Matsuo Y, Drexler HG. ABL-BCR expression in the BCR–ABL-positive human leukemia cell lines. Leuk Res. 1999;23:1055–1060. doi: 10.1016/s0145-2126(99)00131-9. PMID: 10576511. DOI: 10.1016/S0145-2126(99)00131-9. [DOI] [PubMed] [Google Scholar]
- 10.Rumjanek VM, Trindade GS, Wagner Souza K, de-Oliverira MC, Marques Santos LF, Maia RC. Multidrug resistance in tumour cells: Characterisation of the multidrug resistant cell line K562-Lucena 1. Anais Acad Brasil Ciências. 2001;73(1):57–69. doi: 10.1590/s0001-37652001000100007. PMID: 11246270. DOI: 10.1590/S0001-37652001000100007. [DOI] [PubMed] [Google Scholar]
- 11.Rumjanek VM, Vidal RS, Maia RC. Multidrug resistance in chronic myeloid leukaemia: How much can we learn from MDR–CML cell lines. Biosci Rep. 2013;33(6):875–888. doi: 10.1042/BSR20130067. PMID: 24070327. DOI: 10.1042/BSR20130067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma L, Roderick J, Kelliher MA, Green MR. High-throughput screening of tyrosine kinase inhibitor resistant genes in CML. Meth Mol Biol. 2016;1465:159–173. doi: 10.1007/978-1-4939-4011-0_14. PMID: 27581147. DOI: 10.1007/978-1-4939-4011-0_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daflon Yunes N, Pinto Silva FE, Vidal RS, Novis BF, Berguetti T, Lopes RR, Polycarpo C, Rumjanek VM. Characterization of a multidrug-resistant chronic myeloid leukemia cell line presenting multiple resistance mechanisms. Mol Cell Biochem. 2013;383(1-2):123–135. doi: 10.1007/s11010-013-1761-0. PMID: 23877223. DOI: 10.1007/ s11010-013-1761-0. [DOI] [PubMed] [Google Scholar]
- 14.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pffaffl MW, Shipley GL, Vandemsompele J, Wittwer CT. The MIQE Guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. PMID: 23570709. DOI: 10.1373/clinchem.2013.206375. [DOI] [PubMed] [Google Scholar]
- 15.Schmittgen TD, Livak K. Analyzing real-time PCR data by the comparative Ct method. Nat Prot. 2008;3(6):1101–1107. doi: 10.1038/nprot.2008.73. PMID: 18546601. DOI: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 16.Quintás-Cardana A, Cortes J. Molecular biology of BCR–ABL1-positive chronic myeloid leukemia. Blood. 2009;113(8):1619–1630. doi: 10.1182/blood-2008-03-144790. PMID: 18827185. DOI: 10.1182/blood-2008-03-144790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang K, Fu LW. Mechanisms of resistance to BCR–ABL TKIs and the therapeutic strategies: A review. Crit Rev Oncol Hematol. 2015;93(3):277–292. doi: 10.1016/j.critrevonc.2014.11.001. PMID: 25500000. DOI: 10.1016/ j.critrevonc.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Baccarani M, Deininger MW, Rosti G, Hochhaus A, Soverini S, Apperley JF, Cervantes F, Clarck RE, Cortes JE, Guilhot F, Hjorth Hansen H, Hughes TP, Kantarjian HM, Kim DW, Larson RA, Lipton JH, Mahon FX, Martinelli G, Mayer J, Müller MC, Nierderwieser D, Pane F, Radich JP, Rousselot P, Saglio G, SauBele S, Schiffer C, Silver R, Simonsson B, Steegmann JL, Goldman JM, Hehlmann R. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood. 2013;(122)6:872–884. doi: 10.1182/blood-2013-05-501569. PMID: 23803709. DOI: 10.1182/blood-2013-05-501569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann VS, Baccarani M, Hasford J, Lindoerfer D, Burgstaller S, Sertic D, Costeas P, Mayer J, Indrak K, Everaus H, Koskenvesa P, Guilhot J, Schubert-Fritschle G, Castagnetti F, DiRaimondo F, Lejniece S, Griskevicius L, Thielen N, Sacha T, Hellmann A, Turkina AG, Zaritskey A, Bogdanovic A, Sniska Z, Zupan I, Steegmann JL, Simonsson B, Clark RE, Covelli A, Guidi G, Hehlmann R. The EUTOS population-based registry: incidence and clinical characteristics of 2904 CML patients in 20 European Countries. Leukemia. 2015;29(6):1336–1343. doi: 10.1038/leu.2015.73. PMID: 25783795. DOI: 10.1038/leu.2015.73. [DOI] [PubMed] [Google Scholar]
- 20.Chauffaille MLLF. Neoplasias mieloproliferativas: revisão dos critérios diagnósticos e dos aspectos clínicos. Rev Brasil Hematol Hemoter. 2010;32(4):308–316. DOI: 10.1590/S1516-84842010005000091. [Google Scholar]
- 21.Di Felice E, Rocanglia F, Venturelli F, Mangone L, Luminari S, Cirilli C, Caroozzi G, Rossi PG. The impact of introducing tyrosine kinase inhibitors on chronic myeloid leukemia survival: A population-based study. BMC Cancer. 2018;18(1):1069–1069. doi: 10.1186/s12885-018-4984-3. PMID: 30400842. DOI: 10.1186/s12885-018-4984-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bortolheiro TC, Chiattone CS. Leucemia Mielóide Crônica: história natural e classificação. Rev Brasil Hematol Hemoter. 2008;30(1):3–7. DOI: 10.1590/S1516-84842008000700003. [Google Scholar]
- 23.Balatzenko G, Vundinti BR, Margarita G. Correlation between the type of BCR–ABL transcripts and blood cell counts in chronic myeloid leukemia – a possible influence of MRD1 gene expression. Hematol Rep. 2011;3:5–9. doi: 10.4081/hr.2011.e3. PMID: 22184525. DOI: 10.4081/hr.2011.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin HX, Sjaarda J, Dyck J, Stringer R, Hillis C, Harvey M, Carter R, Ainsworth P, Leber B, Pare G, Sadikovic B. Gender and BCR–ABL transcript type are correlated with molecular response to imatinib treatment in patients with chronic myeloid leukemia. Eur J Haematol. 2015;96:360–366. doi: 10.1111/ejh.12597. PMID: 26059983. DOI: 10.1111/ejh.12597. [DOI] [PubMed] [Google Scholar]
- 25.Giri S, Pathak R, Martin MG, Bhatt VR. Characteristics and survival of BCR/ABL-negative chronic myeloid leukemia: A retrospective analysis of the Surveillance, Epidemiology and End Results database. Therapeut Adv Hematol. 2015;6(6):308–312. doi: 10.1177/2040620715607416. PMID: 2662299. DOI: 10.1177/2040620715607416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geissler J, Sharf G, Bombaci F, Daban M, De Jong J, Gavin T, Pelouchova J, Dziwinski E, Hasford J, Hoffmann VS. Factors influencing adherence in CML and ways to improvement: Results of a patient-driven survey of 2546 patients in 63 countries. J Cancer Res Clin Oncol. 2017;143(7):1167–1176. doi: 10.1007/s00432-017-2372-z. PMID: 28289895. DOI: 10.1007/s00432-017-2372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lucas CM, Harris RJ, Giannoudis A, Davies A, Knight K, Watmough SJ, Wang L, Clark RE. Chronic myeloid leukemia patients with the e13a2 BCR–ABL fusion transcript have inferior responses to imatinib compared to patients with the e14a2 transcript. Haematologica. 2009;94(10):1362–1367. doi: 10.3324/haematol.2009.009134. PMID: 19713230. DOI: 10.3324/haematol.2009.009134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanfstein B, Lauseker M, Hehlmann R, Sausselle S, Erben P, Dietz C, Fabarius A, Proetel U, Schnittger S, Haferlach C, Krause SW, Schubert J, Einsele H, Hanel M, Dengler J, Falge C, Kanz L, Neubauer A, Kneba M, Stegelmann F, Pfreundschuh M, Waller CF, Spiekermann K, Baerlocher GM, Pfirmann M, Hasford J, Hofmann WK, Hochauss A, Muller MC, SAKK and German CML Study Group Distinct characteristics of e13a2 versus e14a2 BCR–ABL1 driven chronic myeloid leukemia under first-line therapy with imatinib. Haematologica. 2014;99:1441–1447. doi: 10.3324/haematol.2013.096537. PMID: 24837466. DOI: 10.3324/haematol.2013. 096537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deb P, Chakrabarti P, Chakrabarty S, Aich R, Nath U Ray SS, Chaudhuri U. Incidence of BCR–ABL transcript variants in patients with chronic myeloid leukemia: Their correlation with presenting features, risk scores and response to treatment with imatinib mesylate. Ind J Med Paed Oncol. 2014;35(1):26–30. doi: 10.4103/0971-5851.133707. PMID: 25006280. DOI: 10.4103/0971-5851.133707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castagnetti F, Gugliotta G, Breccia M, Iurlo A, Levato L, Albano F, Vigneri P, Abruzzese E, Rossi G, Rupoli S, Cavazzini F, Martino B, Orlandi E, Pregno P, Annunziata M, Usala E, Tiribelli M, Sica S, Bonifacio M, Fava C, Gherlinzoni F, Bocchia M, Soverini S, Bochicchio MT, Cavo M, Giovanni M, Saglio G, Pane F, Baccarani M, Rosti G, GIMEMA CML Working Party The BCR–ABL1 transcript type influences response and outcome in Philadelphia chromosome-positive chronic myeloid leukemia patients treated frontline with imatinib. Am J Hematol. 2017;92(8):797–805. doi: 10.1002/ajh.24774. PMID: 28466557. DOI: 10.1002/ajh.24774. [DOI] [PubMed] [Google Scholar]
- 31.de Lemos JA, de Oliveira CM, Scerni AC, Bentes AQ, Beltrao AC, Bentes IR, Azevedo TC, Maradei Pereira LM. Differential molecular response of the transcripts b2a2 and b3a2 to imatinib mesylate in chronic myeloid leukemia. Genet Mol Res. 2005;4:803–811. PMID: 16475128. [PubMed] [Google Scholar]
- 32.Polampalli S, Choughule A, Negi N, Shinde S, Baisane C, Amre P, Subramanian PG, Gujral S, Prabhash K, Parikh P. Analysis and comparison of clinicohematological parameters and molecular and cytogenetic response of two BCR/ABL fusion transcripts. Genet Mol Res. 2008;7:1138–1149. doi: 10.4238/vol7-4gmr485. PMID: 19048492. [DOI] [PubMed] [Google Scholar]
- 33.Moreira MA, Bagni C, de Pinho MB, Mac-Cormick TM, Mota dos Santos M, Pinto-Silva FE, Daflon-Yunes N, Rumjanek VM. Changes in gene expression profile in two multidrug-resistant cell lines derived from a same drug sensitive cell line. Leuk Res. 2014;38(8):983–987. doi: 10.1016/j.leukres.2014.06.001. PMID: 24996974. DOI: 10.1016/j.leukres.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Jaeger S, Duran-Frigola M, Aloy P. Drug sensitivity in cancer cell lines is not tissue-specific. Mol Cancer. 2015;14(1):40–40. doi: 10.1186/s12943-015-0312-6. PMID: 25881072. DOI: 10.1186/s12943-015-0312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ben-David U, Siranosian B, Ha G, Tang H, Oren Y, Hinohara K, Strathdee CA, Dempster J, Lyons NJ, Burns R, Nag A, Kugener G, Cimini B, Tsvetkov P, Maruvka YE, O’Rourke R, Garrity A, Tubelli AA, Bandopahayay P, Tsherniak A, Vazquez F, Wong B, Birger C, Ghandi M, Thorner AR, Bittker JA, Meyerson M, Getz G, Beroukhim R, Golub TR. Genetic and transcriptional evolution alters cancer cell line drug response. Nature. 2018;560(7718):325–358. doi: 10.1038/s41586-018-0409-3. PMID: 30089904. DOI: 10.1038/ s41586-018-0409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]


