Abstract
Background/Aim: Cells suffer from oxidative DNA damage which leads to the accumulation of 8-oxoguanine (8-oxoG) adducts in our genome that can become carcinogenic. The human 8-oxoG DNA glycosylase 1 (hOGG1) plays a central role in repairing these 8-oxoGs via the base excision repair pathway. Mounting evidence has suggested that hOGG1 polymorphisms may affect the activity of hOGG1 and serve as genomic markers for the prediction of personal susceptibility to several cancers. To determine whether the commonly examined hOGG1 rs1052133 (Ser326Cys) polymorphism is associated with the risk of childhood acute lymphoblastic leukemia (ALL) among Taiwanese children, we genotyped the hOGG1 rs1052133 (Ser326Cys) in 266 cases and 266 controls. Results: The distributions of the GG, CG and CC genotypes at the hOGG1 rs1052133 were 49.2, 39.1 and 11.7% in the control group and 48.1, 36.1 and 15.8% in the case group (p=0.3656). The combined genotypes CG+CC were not associated with increased risk of childhood ALL (odds ratio [OR]=1.05, 95% confidence interval [CI]=0.74-1.47, p=0.7947). Conclusion: The hOGG1 rs1052133 polymorphism is not associated with susceptibility to childhood ALL in the Taiwanese population.
Keywords: Childhood leukemia, genotype, hOGG1, polymorphism, Taiwan
Leukemia is the most common cancer among children worldwide, and acute lymphoblastic leukemia (ALL) is a clonal disease of a lymphoblast and the most frequent malignancy in children, accounting for approximately 25% of all pediatric malignancies (1). Studies of developed countries have reported that cancer incidence among children has risen steadily from the 1950s to 2010s, with 38 new cases per million occurring every year (2,3). Like other cancers, mounting evidence has shown that childhood ALL is generally believed to be caused by the interaction of genomic susceptibility factors and environmental factors (2,4-9). It is believed that DNA adducts formed on the genome of hematopoietic precursor cells are essential for the development of leukemia (10,11), and are estimated to occur at a rate of about 10,000 lesions per cell per day (12). In order to maintain genomic stability, several gatekeepers, the DNA repair systems including base excision repair (BER), mismatch repair and nucleotide excision repair have evolved to prevent cells from undergoing carcinogenesis (13-15).
The most abundant oxidative DNA adduct 8-oxoguanine (8-oxoG), mainly produced by reactive oxygen species, causes oxidative damage leading to a transversion from G:C→T:A leading to carcinogenesis (16,17). The human 8-oxoG DNA glycosylase 1 (hOGG1) is the pilot enzyme in the BER pathway used for the detection and recognition of 8-oxoG in our genome (13,18). Functional studies have shown that the genotypes rs1052133 (Ser326Cys) in exon 7 of the hOGG1 gene might determine the activity of the glycosylase (19,20), and may serve as a genomic predictor of an individual’s susceptibility to various types of cancer (21-26). In 2011, Stanczyk and his colleagues reported that the Cys/Cys genotype at hOGG1 rs1052133 increased the risk of childhood ALL in a Polish population (27). Almost at the same time in a Chinese population, Li and his colleagues reported that the combined genotypes Ser/Ser and Ser/Cys at the same hOGG1 rs1052133 polymorphic site were associated with a statistically significant decrease in the risk of childhood ALL (28). There has been no investigation on the hOGG1 rs1052133 polymorphism and the risk of childhood ALL in the Taiwanese population till now. Therefore, in this study, we aimed to determine whether the hOGG1 rs1052133 polymorphism is associated with susceptibility to childhood ALL in a Taiwanese population and its interactions with age and gender.
Materials and Methods
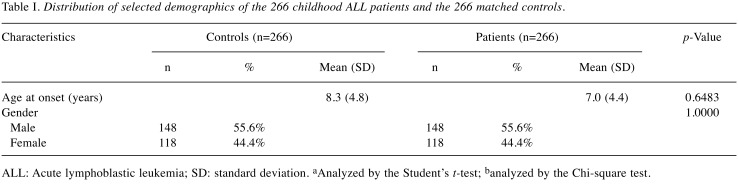
Childhood ALL patients and control subjects. The research design and the detailed procedures of the current study were approved by the Institutional Review Board of China Medical University Hospital (Approval No. DMR103-IRB-153). Written informed consent was obtained from one or both parents of all participants. Briefly, cases with pathologic confirmed childhood leukemia were identified and ascertained into the study by pediatricians, regardless of the patient’s age and stage at diagnosis. In brief, a total of 266 patients who had been diagnosed with childhood ALL were recruited from the general surgery outpatient clinics of the pediatric departments of China Medical University Hospital and the National Taiwan University Hospital, Taiwan, Republic of China during the period of 2005 to 2010. All basic and clinical characteristics of the recruited children, including their histological details, were identified by expert surgeons in the two hospitals. All investigated subjects voluntarily participated in this study, completed a questionnaire form with the help of their parents or guardians, and provided 5 ml of their peripheral blood samples without any uncomfortable feelings. An equal number of age-matched healthy subjects were recruited as the control group in accordance with the method for initial random sampling established by the Health Examination Cohort between 2005 to 2010 as previously described (29-31). Most of the healthy subjects underwent health examinations every 5 to 6 months. A total of 457 volunteers aged under 18 years were recruited in the study and were diagnosed as cancer-free in accordance with the criteria set by the International Classification of Disease (ninth revision, defined by World Health Organization). At last, 266 participants were included in the analysis to match the population structure (number, age, and gender) of our case population. All the participants were Taiwanese and the overall agreement rate in the study exceeded 85%. As shown in Table I, we have provided a concise summary and comparison of the selected recorded characteristics of the case and control groups.
Table I. Distribution of selected demographics of the 266 childhood ALL patients and the 266 matched controls.
ALL: Acute lymphoblastic leukemia; SD: standard deviation. aAnalyzed by the Student’s t-test; banalyzed by the Chi-square test.
Genotyping. Genomic DNA from peripheral blood samples was extracted, aliquoted, and stored. The polymerase chain reaction plus restriction fragment length polymorphism (PCR-RFLP) cycling conditions were: one cycle at 94˚C for 5 min; 35 cycles at 94˚C for 30 sec, 55˚C for 30 sec, and 72˚C for 30 sec, and a final extension at 72˚C for 10 min. The sequences of forward and reverse primers for hOGG1 rs1052133 genotypes were 5’-ACTGTCACTAGTCTCACCAG-3’ and 5’-GGAAGGTGGGAAGGTG-3’, respectively. PCR amplicons were digested by the specific restriction enzyme Fnu4H I for 2 h, and enzyme-digestion products were subjected to 3% DNA gel electrophoresis.
Statistical analysis. The Student’s t-test was used to test the differences in the age between the case and control groups. The Pearson’s χ-square test without Yates’ correction or Fisher’s exact test (when any cell number is less than 5) was used to compare the distribution of hOGG1 rs1052133 genotypes between case and control groups or any other two sub-groups. The associations between the hOGG1 rs1052133 genotypes and the risk of childhood ALL were estimated via computing odds ratios (ORs) and 95% confidence intervals (CIs) through unconditional logistic regression analysis with adjustment for possible confounding factors whenever needed.
Results
Comparison of the basic demographic characteristics of the investigated cases and controls. The basic demographic characteristics of 266 childhood ALL cases and 266 cancer-free controls are presented in Table I. Overall, there was no significant difference between the cases and controls in the distribution of age and gender status (both p>0.05) (Table I).
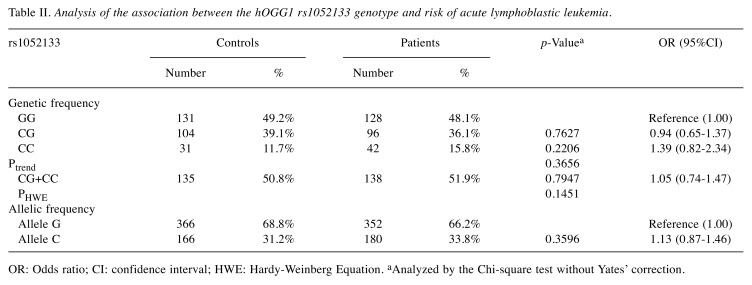
Analysis of the association between hOGG1 rs1052133 genotypes and childhood ALL risk. The observed genotypic and allelic frequencies of hOGG1 rs1052133 among cases and controls and their associations with the risk of childhood ALL are presented in Table II. The hOGG1 rs1052133 genotypes of the control subjects were in agreement with the Hardy–Weinberg equilibrium (p=0.1451). There was no significant difference in the distribution of genotypes of hOGG1 rs1052133 between the control and case groups (p for trend=0.3656). In detail, the hOGG1 rs1052133 CG and CC variant genotypes were present in 39.1 and 11.7%, respectively, in the control group, and in 36.1 and 15.8%, respectively, in the case group (Table II, top part). Multiple logistic regression analysis indicated that the hOGG1 rs1052133 CG and CC variant genotypes were not associated with altered risk of childhood ALL (the OR=0.94 and 1.39, 95%CI=0.65-1.37 and 0.82-2.34, p=0.7627 and 0.2206 for hOGG1 rs1052133 CG heterozygotes and CC homozygotes, respectively) (Table II, top part). We subsequently combined the hOGG1 rs1052133 CG and CC genotypes to construct a dominant genetic model, but there was still no significant association between the combined genotypes and the risk of childhood ALL (Table II, middle part). Allelic frequencies of hOGG1 rs1052133 among cases and controls were also analyzed, but no association was found between the hOGG1 rs1052133 allelic frequency distribution and an altered risk of childhood ALL in Taiwan.
Table II. Analysis of the association between the hOGG1 rs1052133 genotype and risk of acute lymphoblastic leukemia.
OR: Odds ratio; CI: confidence interval; HWE: Hardy-Weinberg Equation. aAnalyzed by the Chi-square test without Yates’ correction.
Discussion
The hOGG1 gene encodes a glycosylase responsible for recognizing the most common oxidative DNA adducts, 8-oxoGs, so as to be removed from our genome by the BER machinery (16,17). In the first step, the hOGG1 enzyme not only recognizes the 8-oxoGs but also cleaves the glycosylic bond between the modified base and the sugar moiety, leaving a basic apurinic/apyrimidinic site for further action of DNA polymerase β and DNA ligase I and III (13). Among leukemia cell lines, the activity of hOGG1 plays an important role in determining their sensitivities to environmental exposures such as radiation and 8-hydroxydeoxyguanosine-induced apoptosis (32,33). The most famous polymorphic site of hOGG1 is hOGG1 rs1052133 (Ser326Cys, C to G) and several functional studies have shown that the glycosylase activity of the “G” variant of the hOGG1 enzyme is more sensitive to inactivation by oxidizing agents than that of the “C” wild-type, and that cells carrying the “G” allele accumulate mutations more readily under oxidative stress (19,34,35).
As mentioned above, two groups have reported that the GG genotype at hOGG1 rs1052133 may be a risk factor for childhood ALL in Polish and Chinese populations (27,28). This is not in accordance with our results which showed no association between the genotypes at the rs1052133 polymorphic site of hOGG1 and childhood ALL in a Taiwanese population. In 2009, two genome-wide association studies also reported that there was no association between hOGG1 rs1052133 polymorphism and childhood ALL risk (36,37). Noticeably, these studies are all valuable since they provide genomic information from different populations. At the same time, it is important to know that the sample sizes of Stanczyk’s, Li’s and ours are at more representative levels than those of Trevino’s and Papaemmanuil’s, who studied a smaller number of children. Additional studies with larger sample sizes and different populations are necessary to validate these results. To the best of our knowledge, this is the first study to show an association between the hOGG1 rs1052133 polymorphism and childhood ALL risk in a Taiwanese population.
As for other solid tumors, the genotypes of CG and/or GG at hOGG1 rs1052133 have been reported to be associated with increased risk of various types of cancer, including oral cancer (26), lung adenocarcinoma (22), breast cancer (38), larynx cancer (39), esophageal cancer (40), colorectal cancer (21), gallbladder cancer (23) and prostate cancer (41,42). At the same time, there are lots of negative findings showing no association between hOGG1 rs1052133 genotypes and specific types of cancer (43,44). Importantly, it has been shown that the G allele may cause reduced glycosylase hOGG1 activity leading to an overall down-regulated BER capacity (19,20). Possible explanations for these inconsistencies may be due to differences in the genetic background between different populations, sampling methodology and small sample size.
In conclusion, our data suggest that the hOGG1 rs1052133 genotypes are not associated with childhood ALL risk among Taiwanese children. More functional examinations will benefit this genotype–phenotype correlation investigation, and a larger sample size with more information regarding exposure to environmental factors for precise stratification analysis will be helpful to reveal the etiology of childhood ALL.
Conflicts of Interest
The Authors have no conflict of interest to declare regarding this study.
Authors’ Contributions
Research Design: Hsu PC, Chen CC and Tzeng HE; Patient and Questionnaire Summarize: Hsu YN, Kuo CC and Pei JS; Experiment Performance: Wang YC and Chang WS; Statistical Analysis: Lin ML and Pei JS; Manuscript Writing: Tsai CW and Bau DT; Reviewing and Revising: Bau DT, Chang WS and Tsai CW.
Acknowledgements
The Authors are grateful to Hsin-Ting Li and Huai-Mei Hsu for their excellent technical assistance. All the participants in this study are appreciated. This study was supported mainly by Taoyuan General Hospital, Ministry of Health and Welfare, Taiwan, ROC to Dr. Hsu (grant number: PTH108019). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Sinnett D, Krajinovic M, Labuda D. Genetic susceptibility to childhood acute lymphoblastic leukemia. Leuk Lymphoma. 2000;38:447–462. doi: 10.3109/10428190009059264. PMID: 10953966. DOI: 10.3109/104281900 09059264. [DOI] [PubMed] [Google Scholar]
- 2.Kaatsch P. Epidemiology of childhood cancer. Cancer Treat Rev. 2010;36:277–285. doi: 10.1016/j.ctrv.2010.02.003. PMID: 20231056. DOI: 10.1016/j.ctrv. 2010.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. PMID: 30207 593. DOI: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Wu KH, Wang CH, Yang YL, Peng CT, Lin WD, Tsai FJ, Lin DT, Bau DT. Significant association of XRCC4 single nucleotide polymorphisms with childhood leukemia in Taiwan. Anticancer Res. 2010;30:529–533. PMID: 20332465. [PubMed] [Google Scholar]
- 5.Wang CH, Lai YL, Chang WS, Wu KH, Lane HY, Chiu CF, Tsai FJ, Lin CC, Bau DT. Significant association of caveolin-1 single nucleotide polymorphisms with childhood leukemia in Taiwan. Cancer Genomics Proteomics. 2013;10:75–79. PMID: 23603343. [PubMed] [Google Scholar]
- 6.Pei JS, Lee YM, Lo HH, Hsu YN, Lin SS, Bau DT. Association of X-ray repair cross-complementing-6 genotypes with childhood leukemia. Anticancer Res. 2013;33:5395–5399. PMID: 24324074. [PubMed] [Google Scholar]
- 7.Pei JS, Hsu CM, Tsai CW, Chang WS, Ji HX, Hsiao CL, Miao CE, Hsu YN, Bau DT. The association of methylenetetrahydrofolate reductase genotypes with the risk of childhood leukemia in Taiwan. PLoS One. 2015;10:e0119776. doi: 10.1371/journal.pone.0119776. PMID: 24324074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pei JS, Chang WS, Hsu PC, Tsai CW, Hsu CM, Ji HX, Hsiao CL, Hsu YN, Bau DT. The association of flap endonuclease 1 genotypes with the risk of childhood leukemia. Cancer Genomics Proteomics. 2016;13:69–74. PMID: 26708601. [PubMed] [Google Scholar]
- 9.Pei JS, Hsu PC, Chou AK, Tsai CW, Chang WS, Hsiao CL, Hsu YN, Cheng SP, Bau DT. Matrix metalloproteinase-1 genotype contributes to the risk of non-solid tumor in childhood leukemia. Anticancer Res. 2016;36:5127–5132. doi: 10.21873/anticanres.11082. PMID: 27798 872. DOI: 10.21873/anticanres.11082. [DOI] [PubMed] [Google Scholar]
- 10.Davies SM, Robison LL, Buckley JD, Radloff GA, Ross JA, Perentesis JP. Glutathione S-transferase polymorphisms in children with myeloid leukemia: A Children’s Cancer Group study. Cancer Epidemiol Biomarkers Prev. 2000;9:563–566. PMID: 10868689. [PubMed] [Google Scholar]
- 11.Nagai F, Hiyoshi Y, Sugimachi K, Tamura HO. Cytochrome P450 (CYP) expression in human myeloblastic and lymphoid cell lines. Biol Pharm Bull. 2002;25:383–385. doi: 10.1248/bpb.25.383. PMID: 11913539. [DOI] [PubMed] [Google Scholar]
- 12.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. PMID: 8469282. DOI: 10. 1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 13.Christmann M, Tomicic MT, Roos WP, Kaina B. Mechanisms of human DNA repair: an update. Toxicology. 2003;193:3–34. doi: 10.1016/s0300-483x(03)00287-7. PMID: 14599765. [DOI] [PubMed] [Google Scholar]
- 14.Dahle J, Brunborg G, Svendsrud DH, Stokke T, Kvam E. Overexpression of human OGG1 in mammalian cells decreases ultraviolet A induced mutagenesis. Cancer Lett. 2008;267:18–25. doi: 10.1016/j.canlet.2008.03.002. PMID: 18406515. DOI: 10.1016/j.canlet.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Hung RJ, Hall J, Brennan P, Boffetta P. Genetic polymorphisms in the base excision repair pathway and cancer risk: A HuGE review. Am J Epidemiol. 2005;162:925–942. doi: 10.1093/aje/kwi318. PMID: 16221808. DOI: 10.1093/aje/kwi318. [DOI] [PubMed] [Google Scholar]
- 16.Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G-T and A-C substitutions. J Biol Chem. 1992;267:166–172. PMID: 1730583. [PubMed] [Google Scholar]
- 17.Luna L, Rolseth V, Hildrestrand GA, Otterlei M, Dantzer F, Bjoras M, Seeberg E. Dynamic relocalization of hOGG1 during the cell cycle is disrupted in cells harbouring the hOGG1-Cys326 polymorphic variant. Nucleic Acids Res. 2005;33:1813–1824. doi: 10.1093/nar/gki325. PMID: 15800211. DOI: 10.1093/nar/gki325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill JW, Evans MK. Dimerization and opposite base-dependent catalytic impairment of polymorphic S326C OGG1 glycosylase. Nucleic Acids Res. 2006;34:1620–1632. doi: 10.1093/nar/gkl060. PMID: 16549874. DOI: 10.1093/nar/gkl060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamane A, Kohno T, Ito K, Sunaga N, Aoki K, Yoshimura K, Murakami H, Nojima Y, Yokota J. Differential ability of polymorphic OGG1 proteins to suppress mutagenesis induced by 8-hydroxyguanine in human cell in vivo. Carcinogenesis. 2004;25:1689–1694. doi: 10.1093/carcin/bgh166. PMID: 15073047. DOI: 10.1093/carcin/ bgh166. [DOI] [PubMed] [Google Scholar]
- 20.Collins AR, Gaivao I. DNA base excision repair as a biomarker in molecular epidemiology studies. Mol Aspects Med. 2007;28:307–322. doi: 10.1016/j.mam.2007.05.005. PMID: 17659329. DOI: 10.1016/j.mam.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Kim JI, Park YJ, Kim KH, Kim JI, Song BJ, Lee MS, Kim CN, Chang SH. hOGG1 Ser326Cys polymorphism modifies the significance of the environmental risk factor for colon cancer. World J Gastroenterol. 2003;9:956–960. doi: 10.3748/wjg.v9.i5.956. PMID: 12717837. DOI: 10.3748/wjg.v9.i5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okasaka T, Matsuo K, Suzuki T, Ito H, Hosono S, Kawase T, Watanabe M, Yatabe Y, Hida T, Mitsudomi T, Tanaka H, Yokoi K, Tajima K. hOGG1 Ser326Cys polymorphism and risk of lung cancer by histological type. J Hum Genet. 2009;54:739–745. doi: 10.1038/jhg.2009.108. PMID: 19881468. DOI: 10.1038/jhg.2009.108. [DOI] [PubMed] [Google Scholar]
- 23.Jiao X, Huang J, Wu S, Lv M, Hu Y, Jianfu, Su X, Luo C, Ce B. hOGG1 Ser326Cys polymorphism and susceptibility to gallbladder cancer in a Chinese population. Int J Cancer. 2007;121:501–505. doi: 10.1002/ijc.22748. PMID: 17417784. DOI: 10.1002/ijc.22748. [DOI] [PubMed] [Google Scholar]
- 24.Weiss JM, Goode EL, Ladiges WC, Ulrich CM. Polymorphic variation in hOGG1 and risk of cancer: a review of the functional and epidemiologic literature. Mol Carcinog. 2005;42:127–141. doi: 10.1002/mc.20067. PMID: 15584022. DOI: 10.1002/mc.20067. [DOI] [PubMed] [Google Scholar]
- 25.Tsai CW, Ho CY, Shih LC, Ying TH, Hsieh YH, Chen YC, Chang WS, Huang CY, Pan SB, Shui HA, Chen CP, Wang PS, Bau DT. The joint effect of hOGG1 genotype and smoking habit on endometriosis in Taiwan. Chin J Physiol. 2013;56:263–268. doi: 10.4077/CJP.2013.BAB142. PMID: 24032711. DOI: 10.4077/CJP.2013.BAB142. [DOI] [PubMed] [Google Scholar]
- 26.Tsai CW, Tsai MH, Tsou YA, Shih LC, Tseng HC, Chang WS, Ho CY, Lee HZ, Bau DT. The joint effect of smoking and hOGG1 genotype on oral cancer in Taiwan. Anticancer Res. 2012;32:3799–3803. PMID: 22993322. [PubMed] [Google Scholar]
- 27.Stanczyk M, Sliwinski T, Cuchra M, Zubowska M, Bielecka Kowalska A, Kowalski M, Szemraj J, Mlynarski W, Majsterek I. The association of polymorphisms in DNA base excision repair genes XRCC1, OGG1 and MUTYH with the risk of childhood acute lymphoblastic leukemia. Mol Biol Rep. 2011;38:445–451. doi: 10.1007/s11033-010-0127-x. PMID: 20364408. DOI: 10.1007/s11033-010-0127-x. [DOI] [PubMed] [Google Scholar]
- 28.Li Q, Huang L, Rong L, Xue Y, Lu Q, Rui Y, Li J, Tong N, Wang M, Zhang Z, Fang Y. hOGG1 Ser326Cys polymorphism and risk of childhood acute lymphoblastic leukemia in a Chinese population. Cancer Sci. 2011;102:1123–1127. doi: 10.1111/j.1349-7006.2011.01928.x. PMID: 21401806. DOI: 10.1111/j.1349-7006.2011.01928.x. [DOI] [PubMed] [Google Scholar]
- 29.Pei JS, Chou AK, Hsu PC, Tsai CW, Chang WS, Wu MF, Wu MH, Hsia TC, Cheng SP, Bau DT. Contribution of matrix metalloproteinase-7 genotypes to the risk of non-solid tumor, childhood leukemia. Anticancer Res. 2017;37:6679–6684. doi: 10.21873/anticanres.12126. PMID: 29187444. DOI: 10.21873/anticanres.12126. [DOI] [PubMed] [Google Scholar]
- 30.Pei JS, Chang WS, Hsu PC, Chen CC, Cheng SP, Wang YC, Tsai CW, Shen TC, Bau DT. The contribution of XRCC3 genotypes to childhood acute lymphoblastic leukemia. Cancer Manag Res. 2018;10:5677–5684. doi: 10.2147/CMAR.S178411. PMID: 30532590. DOI: 10.2147/CMAR.S178411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu PC, Pei JS, Chen CC, Chang WS, Kuo CC, Cheng SP, Tsai CW, Bau DT, Gong CL. Association of matrix metallopeptidase-2 promoter polymorphisms with the risk of childhood leukemia. Anticancer Res. 2019;39:1185–1190. doi: 10.21873/anticanres.13228. PMID: 30842148. DOI: 10.21873/anticanres.13228. [DOI] [PubMed] [Google Scholar]
- 32.Hyun JW, Cheon GJ, Kim HS, Lee YS, Choi EY, Yoon BH, Kim JS, Chung MH. Radiation sensitivity depends on OGG1 activity status in human leukemia cell lines. Free Radic Biol Med. 2002;32:212–220. doi: 10.1016/s0891-5849(01)00793-6. PMID: 11827746. [DOI] [PubMed] [Google Scholar]
- 33.Hyun JW, Jung YC, Kim HS, Choi EY, Kim JE, Yoon BH, Yoon SH, Lee YS, Choi J, You HJ, Chung MH. 8-hydroxydeoxyguanosine causes death of human leukemia cells deficient in 8-oxoguanine glycosylase 1 activity by inducing apoptosis. Mol Cancer Res. 2003;1:290–299. PMID: 12612057. [PubMed] [Google Scholar]
- 34.Bravard A, Vacher M, Moritz E, Vaslin L, Hall J, Epe B, Radicella JP. Oxidation status of human OGG1-S326C polymorphic variant determines cellular DNA repair capacity. Cancer Res. 2009;69:3642–3649. doi: 10.1158/0008-5472.CAN-08-3943. PMID: 19351836. DOI: 10.1158/0008-5472.CAN-08-3943. [DOI] [PubMed] [Google Scholar]
- 35.Zielinska A, Davies OT, Meldrum RA, Hodges NJ. Direct visualization of repair of oxidative damage by OGG1 in the nuclei of live cells. J Biochem Mol Toxicol. 2011;25:1–7. doi: 10.1002/jbt.20346. PMID: 21322094. DOI: 10.1002/jbt.20346. [DOI] [PubMed] [Google Scholar]
- 36.Trevino LR, Yang W, French D, Hunger SP, Carroll WL, Devidas M, Willman C, Neale G, Downing J, Raimondi SC, Pui CH, Evans WE, Relling MV. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1001–1005. doi: 10.1038/ng.432. PMID: 19684603. DOI: 10.1038/ ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, Price A, Olver B, Sheridan E, Kinsey SE, Lightfoot T, Roman E, Irving JA, Allan JM, Tomlinson IP, Taylor M, Greaves M, Houlston RS. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1006–1010. doi: 10.1038/ng.430. PMID: 19684604. DOI: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan W, Xu L, Feng Y, Yang Y, Chen W, Wang J, Pang D, Li D. The hOGG1 Ser326Cys polymorphism and breast cancer risk: a meta-analysis. Breast Cancer Res Treat. 2010;122:835–842. doi: 10.1007/s10549-009-0722-5. PMID: 20058067. DOI: 10.1007/s10549-009-0722-5. [DOI] [PubMed] [Google Scholar]
- 39.Pawlowska E, Janik-Papis K, Rydzanicz M, Zuk K, Kaczmarczyk D, Olszewski J, Szyfter K, Blasiak J, Morawiec-Sztandera A. The Cys326 allele of the 8-oxoguanine DNA N-glycosylase 1 gene as a risk factor in smoking- and drinking-associated larynx cancer. Tohoku J Exp Med. 2009;219:269–275. doi: 10.1620/tjem.219.269. PMID: 19966524. [DOI] [PubMed] [Google Scholar]
- 40.Xing DY, Tan W, Song N, Lin DX. Ser326Cys polymorphism in hOGG1 gene and risk of esophageal cancer in a Chinese population. Int J Cancer. 2001;95:140–143. doi: 10.1002/1097-0215(20010520)95:3<140::aid-ijc1024>3.0.co;2-2. PMID: 11307145. [DOI] [PubMed] [Google Scholar]
- 41.Nam RK, Zhang WW, Jewett MA, Trachtenberg J, Klotz LH, Emami M, Sugar L, Sweet J, Toi A, Narod SA. The use of genetic markers to determine risk for prostate cancer at prostate biopsy. Clin Cancer Res. 2005;11:8391–8397. doi: 10.1158/1078-0432.CCR-05-1226. PMID: 16322300. DOI: 10.1158/1078-0432.CCR-05-1226. [DOI] [PubMed] [Google Scholar]
- 42.Dhillon VS, Yeoh E, Fenech M. DNA repair gene polymorphisms and prostate cancer risk in South Australia--results of a pilot study. Urol Oncol. 2011;29:641–646. doi: 10.1016/j.urolonc.2009.08.013. PMID: 19914098. DOI: 10.1016/j.urolonc.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 43.Ferguson HR, Wild CP, Anderson LA, Murphy SJ, Johnston BT, Murray LJ, Watson RG, McGuigan J, Reynolds JV, Hardie LJ. No association between hOGG1, XRCC1, and XPD polymorphisms and risk of reflux esophagitis, Barrett’s esophagus, or esophageal adenocarcinoma: results from the factors influencing the Barrett’s adenocarcinoma relationship case-control study. Cancer Epidemiol Biomarkers Prev. 2008;17:736–739. doi: 10.1158/1055-9965.EPI-07-2832. PMID: 18349297. DOI: 10.1158/1055-9965.EPI-07-2832. [DOI] [PubMed] [Google Scholar]
- 44.Liu CJ, Hsia TC, Tsai RY, Sun SS, Wang CH, Lin CC, Tsai CW, Huang CY, Hsu CM, Bau DT. The joint effect of hOGG1 single nucleotide polymorphism and smoking habit on lung cancer in Taiwan. Anticancer Res. 2010;30:4141–4145. PMID: 21036733. [PubMed] [Google Scholar]