Abstract
Background.
Understanding how behaviors are generated by neural circuits requires knowledge of the synaptic connections between the composite neurons. Methods for mapping synaptic connections, such as electron microscopy and paired recordings, are labor intensive and alternative methods are thus desirable.
New Method.
Development of a targeted GFP Reconstitution Across Synaptic Partners(GRASP) method, t-GRASP, for assessing neural connectivity is described.
Results.
Numerous different pre-synaptic and post-synaptic/dendritic proteins were tested for enhancing the specificity of GRASP signal to synaptic regions. Pairing of both targeted pre- and post-t-GRASP constructs resulted in strong preferential GRASP signal in synaptic regions in Drosophila larval sensory neurons, larval neuromuscular junctions, and adult photoreceptor neurons with minimal false-positive signal.
Comparison with Existing Methods.
Activity-independent t-GRASP exhibits an enhancement of GRASP signal specificity for synaptic contact sites as compared to existing Drosophila GRASP methods. Fly strains were developed for expression of both pre- and post-t-GRASP with each of the three Drosophila binary transcription systems, thus enabling GRASP assays to be performed between any two driver pairs of any transcription system in either direction, an option not available for existing Drosophila GRASP methods.
Conclusions.
t-GRASP is a novel targeted GRASP method for assessing synaptic connectivity between Drosophila neurons. Its flexibility of use with all three Drosophila binary transcription systems significantly expands the potential use of GRASP in Drosophila.
Introduction
Understanding how the brain processes information to make behavioral decisions requires identifying the synaptic connections between constituent neurons of a neural circuit. Standard techniques for reliably assessing synaptic connectivity between neurons include electron microscopy (EM) and paired recordings. Unfortunately, due to the labor-intensive nature of these methods they are not easily adapted for high-throughput analysis. Although progress is being made with EM methodology (Eichler et al., 2017; Takemura et al., 2017), it seems unlikely any new comprehensive connectomes of major model genetic organisms will be completed in the near future.
Alternative methods for mapping neuronal connectivity are thus still important for neural circuit mapping and will be for the foreseeable future. One such technique, GFP Reconstitution Across Synaptic Partners (GRASP), involves the use of fluorescence complementation based on split-GFP (GFP1-10 and GFP11). In this technique, each split-GFP fragment is extracellularly expressed in separate neurons and synaptic connections are revealed when these GFP fragments reassemble at synaptic contact sites to produce green fluorescence. This technique was first implemented in C. elegans (Feinberg et al., 2008), but has since also been successfully adapted to Drosophila (Fan et al., 2013; Gordon and Scott, 2009; Macpherson et al., 2015) and mouse (Kim et al., 2011; Yamagata and Sanes, 2012). The original Drosophila GRASP fused the two split-GFP fragments to the extracellular domain of the CD4 transmembrane protein (Gordon and Scott, 2009), hereafter CD4-GRASP, and was not synaptically-targeted, thus potentially leading to false positives at non-synaptic locations. The more recent versions of Drosophila GRASP target the GFP1-10 fragment to pre-synaptic terminals using Neurexin (Fan et al., 2013) (hereafter, Nrx-GRASP) or Neuronal-synaptobrevin (Macpherson et al., 2015) (hereafter, syb-GRASP) and result in an enhancement of synaptic specificity. Nrx-GRASP is activity-independent while Syb-GRASP is activity-dependent. The activity-dependence of syb-GRASP likely contributes to its enhanced synaptic specificity and has the advantage that it can distinguish active from inactive synapses. However, activity-dependence is not without the disadvantage that the strength of the fluorescent signal may not accurately reflect the relative strength of synaptic connectivity. For instance, two weakly connected, highly active neurons may give stronger GRASP signal than two strongly connected, low activity neurons. Moreover, two neurons that have strong synaptic connections, but are only active when exposed to a particular stimulus, could give a false-negative (such as an olfactory sensory neuron and its second-order neuron in the absence of a specific odorant).
Here we describe an alternative, activity-indepdendent, targeted Drosophila GRASP, t-GRASP, in which one of the split-GFP fragments is targeted to pre-synaptic terminals and the other to dendrites. Targeting both the pre- and post-synaptic split-GFP fragments results in enhanced specificity of GRASP signal for synaptic contacts sites as compared to existing Drosophila GRASP alternatives. Fly strains were developed for expressing both the pre-synaptic and dendritically targeted t-GRASP proteins using driver pairs for any of the three Drosophila binary transcription systems, (GAL4, LexA, and Q), thus providing enhanced flexibility and thereby significantly broadening the potential use of GRASP in Drosophila.
Results
GRASP construct design
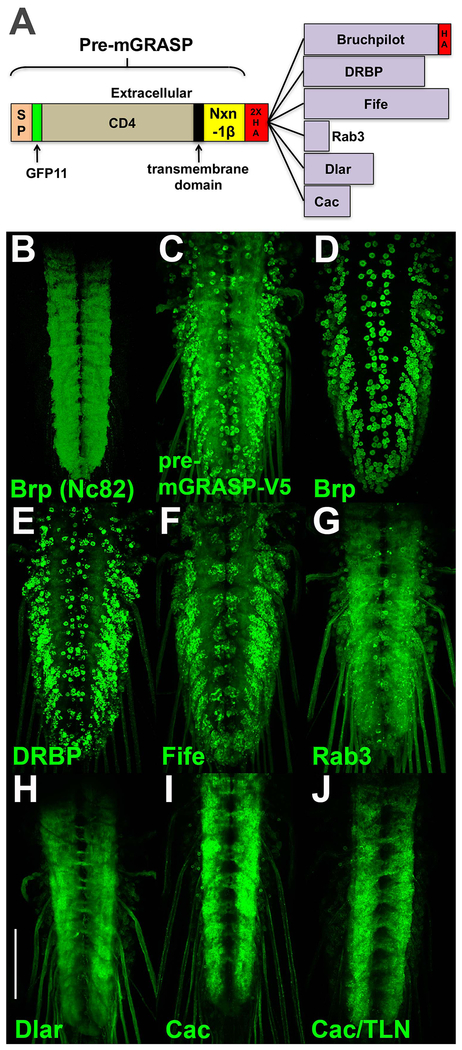
The strategy for generating a Drosophila GRASP with enhanced specificity for synaptic contact sites was to target GFP11 to pre-synaptic terminals and GFP1-10 to dendritic/post-synaptic regions. Initial attempts involved the use of modified versions of the pre- and post-mGRASP proteins that exhibit strong preferential localization to synaptic contact sites in mouse (Kim et al., 2011). An initial attempt involved establishing fly strains containing pre-mGRASP-V5 (a V5 epitope tag was substituted for the cerulean fluorescent protein of the original pre-mGRASP) under QUAS control and post-mGRASPΔT (the tomato fluorescent protein was deleted from the original post-mGRASP) under UAS control. As a stringent test of synapse specificity, both constructs were expressed using the pan-neuronal drivers n-syb-QF2 and n-syb-GAL4 and assessed for specificity of GRASP signal in the neuropil region of third instar larva by direct fluorescence. Pan-neuronal expression of both constructs is a stringent test of synapse specificity because both split-GFP proteins are being expressed in all neurons and thus there is ample opportunity for them to reconstitute outside of synaptic regions both intracellularly and extracellularly. The neuropil region of the third instar larval ventral nerve cord (VNC) where synaptic contact sites are located is visualized in Figure 1B by immunostaining with the nc82 antibody that recognizes the active zone protein Bruchpilot (Brp) (Hofbauer et al., 2009). A synapse-specific GRASP would ideally exhibit reconstituted GFP fluorescence in a similar pattern when pan-neuronally expressed. Unfortunately, pan-neuronal expression of pre-mGRASP-V5 with post-mGRASPΔT did not exhibit the same specificity of fluorescence signal for synapses in Drosophila (Figure 1C) as they do in mouse as significant false-positive signal was observed in the cell bodies outside the neuropil where there are no synaptic contact sites.
Figure 1.
Design and assessment of pre-synaptic GRASP constructs. A) The pre-synaptic GRASP constructs contain at the amino-terminus the pre-mGRASP protein through the Nxn-1β domain and a 2XHA epitope tag fused to the indicated pre-synaptic proteins. The sizes of the pre-synaptic proteins are accurate relative to each other but not to pre-mGRASP. B) nc82 immunostain of a larval ventral nerve cord for the active zone protein Brp reveals the neuropil region where sites of synaptic contact are located. C-J) Direct reconstituted GFP fluorescence imaging of third instar larval ventral nerve cords with both constructs are pan-neuronally expressed. All pre-GRASP constructs are paired with post-mGRASPΔT except J). C) pre-mGRASP-V5 with no carboxy-terminal fusion; D) Brp (1226aa); E) DRBP (1156aa; F) Fife (1315aa); G) Rab3 (219aa); H) Dlar (627aa); I) Cac (418aa). Cac (I) exhibits the most enhanced restriction of GFP signal to the neuropil. J) pre-mGRASP-Cac paired with post-GRASP-TLN. Abbreviations: Brp-Bruchpilot, DRBP-Drosophila Rim binding protein, Dlar-Drosophila liprin-alpha receptor, Cac-Cacophany, SP-signal peptide, Nxn-1β-Neurexin 1-beta. All larval images are direct GFP fluorescence and were acquired and processed identically, except for B) which was immunostained. Scale bar: 100μm.
Figure 1 genotypes.
B) yw; C) yw; 10XQUAS-pre-mGRASPV5/20XUAS-post-mGRASPΔT; n-syb-GAL4, n-sybQF2/+; D) yw; 20XUAS-post-mGRASPΔT /+; n-syb-GAL4, n-sybQF2/10XQUAS-pre-mGRASP-Brp; E) yw; 20XUAS-post-mGRASPΔT /+; n-syb-GAL4, n-sybQF2/10XQUAS-pre-mGRASP-DRBP; F) yw; 20XUAS-post-mGRASPΔT /+; n-syb-GAL4, n-sybQF2/10XQUAS-pre-mGRASP-Fife; G) yw; 20XUAS-post-mGRASPΔT /+; n-syb-GAL4, n-sybQF2/10XQUAS-pre-mGRASP-Rab3; H) yw; 20XUAS-post-mGRASPΔT /+; n-syb-GAL4, n-sybQF2/10XQUAS-pre-mGRASP-Dlar; I) yw; 20XUAS-post-mGRASPΔT /+; n-syb-GAL4, n-sybQF2/10XQUAS-pre-mGRASP-Cac; J) yw; n-syb-GAL4, n-sybQF2/10XQUAS-pre-mGRASP-Cac, 20XUAS-post-mGRASP-TLN.
In an attempt to enhance the specificity of the GRASP signal for synaptic contact sites, all or part of the six pre-synaptic Drosophila proteins Brp (Kittel et al., 2006; Wagh et al., 2006), Drosophila Rim-Binding Protein (DBRP) (Liu et al., 2011), Fife (Bruckner et al., 2012), Rab3 (DiAntonio et al., 1993; Graf et al., 2009), Drosophila liprin-alpha receptor (Dlar) (Krueger et al., 1996), and Cacophony (Cac) (Kawasaki et al., 2000) were separately fused to the carboxy-terminus of the pre-mGRASP protein just after a 2XHA epitope tag (for tracking the protein) located immediately downstream of the Nxn-1β domain (Figure 1A). Transgenic fly strains for each of these constructs under QUAS control were established, pan-neuronally expressed in partnership with post-mGRASPΔT, and assessed for specificity of the signal to the neuropil. The results are shown in Figures 1D–I in increasing order of specificity. Surprisingly, pre-mGRASP fusions to the active zone proteins Brp (Figure 1D), DRBP (Figure 1E), and Fife (Figure 1F) resulted in decreased or, at best, similar synapse specificity as pre-mGRASP-V5. Fusions to the synaptic vesicle protein Rab3 (Figure 1G), the intracellular tail of Dlar (Figure 1H), and the intracellular tail of Cac (Figure 1I) enhanced neuropil specificity with Cac exhibiting the highest degree of enhancement.
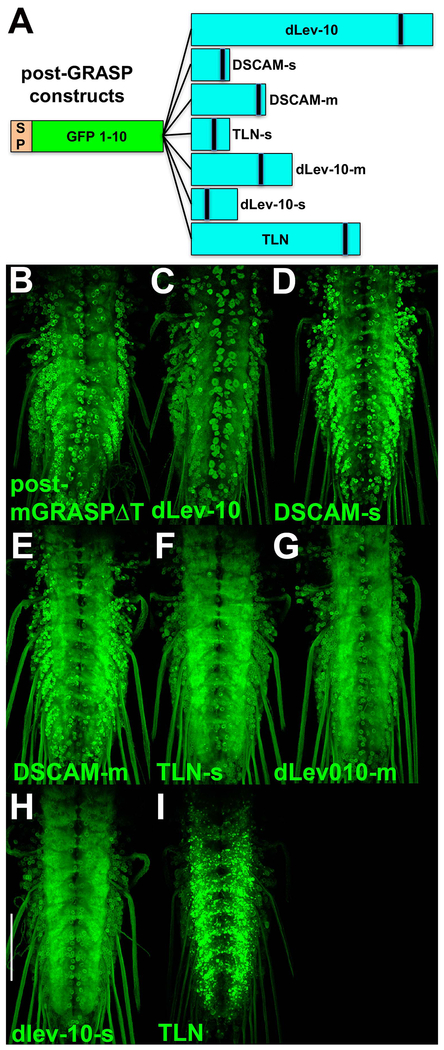
In an attempt to enhance the synaptic specificity of the post-GRASP construct, GFP1-10 was separately fused to the amino terminus of varying portions of the extracellular domains of three transmembrane proteins known to preferentially localize to dendrites or post-synaptic regions, Drosophila homolog (dLev-10/CG42613) of C.elegans Lev-10 (Gally et al., 2004), hereafter dLev-10, Drosophila Down Syndrome Cell Adhesion Molecule (DSCAM) (Shi et al., 2007), and mouse Telencephalin (TLN/ICAM5) (Nicolai et al., 2010). Fly strains containing each of these under UAS control were established, pan-neuronally expressed in partnership with pre-mGRASP-V5, and assessed for specificity of the signal to the neuropil. The results are shown in Figures 2C–I in order of increasing specificity. Fusions to dLev-10 (Figure 2C), DSCAM-s (s for short) (Figure 2D), and DSCAM-m (m for mid-length) (Figure 2E) exhibited similar or reduced specificity as compared to post-mGRASPΔT (Figure 2B). Fusions to TLN-s (Figure 2F), dLev-10-m (Figure 2G), dLev-10-s (Figure 2H), and TLN (Figure 2I) exhibited enhanced neuropil specificity with TLN showing the most significant enhancement. Pairing of pre-GRASP-Cac with post-GRASP-TLN (Figure 1J) exhibits a dramatic enhancement of synaptic specificity as compared to the pre-mGRASP-V5/post-mGRASPΔT starting point (Figures 1C, 2B) and approaches optimal synaptic specificity (Figure 1B). Hereafter, pre-GRASP-Cac is referred to as pre-t-GRASP and post-GRASP-TLN as post-t-GRASP.
Figure 2.
Design and assessment of the post-synaptic GRASP constructs. A) The post-synaptic GRASP constructs contain the GFP1-10 fragment at the amino-terminus fused to the indicated dendritic/post-synaptic proteins. The sizes of the post-synaptic proteins are accurate relative to each other, but not to GFP1-10. B-I) Direct reconstituted GFP fluorescence imaging of third instar larval ventral nerve cords with both constructs are pan-neuronally expressed. All post-GRASP constructs are paired with pre-mGRASP-V5. B) post-mGRASPΔT; C) dLev-10 (1154aa); D) DSCAM-s (184aa); E) DSCAM-m (356aa); F) TLN-s (181aa); G) dlev-10-m (490aa); H) dLev-10-s (219aa); I) TLN (806aa). TLN (I) exhibits the strongest preference for the neuropil. Scale bar: Abbreviations: dLev-10-Drosophila Lev-10, DSCAM-Drosophila cell adhesion molecule, TLN-Telencephalin/ICAM5, SP-signal peptide. All larval images are direct GFP fluorescence and were acquired and processed identically. Scale bar: 100μm.
Figure 2 genotypes. B) yw; 10XQUAS-pre-mGRASPV5/20XUAS-post-mGRASPΔT; n-syb-GAL4, n-sybQF2/+; C) yw; 10XQUAS-pre-mGRASPV5/+; n-syb-GAL4, n-sybQF2/20XUAS-post-GRASP-dLev-10; D) yw; 10XQUAS-pre-mGRASPV5/+; n-syb-GAL4, n-sybQF2/20XUAS-post-GRASP-DSCAM-short; E) yw; 10XQUAS-pre-mGRASPV5/+; n-syb-GAL4, n-sybQF2, 20XUAS-post-GRASP-DSCAM-mid; F) yw; 10XQUAS-pre-mGRASPV5/+; n-syb-GAL4, n-sybQF2/20XUAS-post-GRASP-TLN-short; G) yw; 10XQUAS-pre-mGRASPV5/+; n-syb-GAL4, n-sybQF2/20XUAS-post-GRASP-dLev-10-mid; H) yw; 10XQUAS-pre-mGRASPV5/+; n-syb-GAL4, n-sybQF2/20XUAS-post-GRASP-dLev-10-short; I) yw; 10XQUAS-pre-mGRASPV5/+; n-syb-GAL4, n-sybQF2/20XUAS-post-GRASP-TLN.
Specificity and signal strength comparison to existing Drosophila GRASP methods
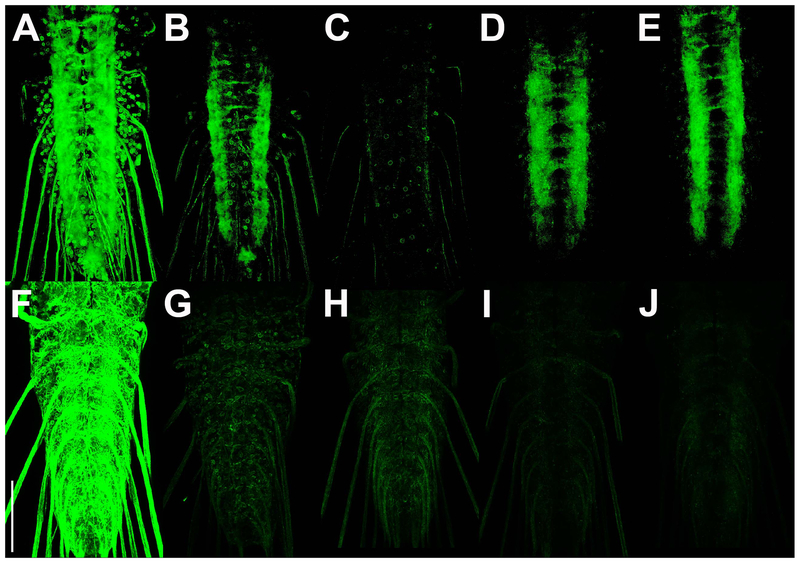
To directly compare the specificity and signal strength of the existing Drosophila GRASP methods with t-GRASP, both split-GFP fragments of each method were expressed pan-neuronally in the third instar larval VNC using the same pan-neuronal drivers. CD4-GRASP exhibits substantial non-specific GRASP signal outside the neuropil in the cell bodies and in the nerves extending from the VNC (Figure 3A). Nrx-GRASP and syb-GRASP both show a major reduction in non-specific GRASP signal outside the neuropil as compared to CD4-GRASP, although some non-specific signal is still apparent (Figs 3B,C, respectively). t-GRASP in both directions exhibits a further reduction in non-specific GRASP signal to a level that is near the limit of detection (Figs 3D,E).
Figure 3.
Comparison of non-specific GRASP signal between existing Drosophila GRASP methods and t-GRASP in neurons and glia. A-E) Pan-neuronal expression. A) spGFP11::CD4 and spGFP1-10::CD4; B) spGFP11::CD4 and spGFP1-10::Nrx; C) spGFP11::CD4 and syb::spGFP1-10; D) pre-t-GRASP and post-t-GRASP (LexAop2-pre and UAS-post); E) pre-t-GRASP and post-t-GRASP (UAS-pre and LexAop2-post). F) pan-glial expression of spGFP1-10::CD4 with pan-neuronal expression of spGFP11::CD4; G) pan-neuronal expression of spGFP11::CD4 with pan-glial expression of spGFP1-10::Nrx; H) pan-neuronal expression of spGFP11::CD4 with pan-glial expression of syb::spGFP1-10; I) pan-glial expression of post-t-GRASP with pan-neuronal expression of pre-t-GRASP; J) pan-glial expression of pre-t-GRASP with pan-neuronal expression of post-t-GRASP; All images are of direct GFP fluorescence. Collection of images and post-imaging processing was identical for A-E, and distinct, but identical for F-J. Scale bar: 100μm.
Figure 3 genotypes. A) yw; n-syb-LexA/+; n-syb-GAL4/LexAop-spGFP11::CD4, UAS-spGFP1-10::CD4; B) w; n-syb-LexA/+; n-syb-GAL4/LexAop-spGFP11::CD4, UAS-spGFP1-10::Nrx; C) w; n-syb-LexA/LexAop-spGFP11::CD4, UAS-syb::spGFP1-10; n-syb-GAL4/+; D) yw; n-syb-LexA/+; n-syb-GAL4/13XLexAop2-pre-t-GRASP, 20XUAS-post-t-GRASP/+; E) yw; n-syb-LexA/+; n-syb-GAL4/13XLexAop2-post-t-GRASP, 20XUAS-pre-t-GRASP/+. F) yw; n-syb-LexA/+; repo-GAL4/LexAop-CD4-GFP11, UAS-CD4-GFP1-10; G) w; n-syb-LexA/+; repo-GAL4/LexAop-spGFP11::CD4, UAS-spGFP1-10::Nrx; H) w; n-syb-LexA/LexAop-spGFP11::CD4, UAS-syb::spGFP1-10; repo-GAL4/+; I) yw; n-syb-LexA/+; repo-GAL4/13XLexAop2-pre-t-GRASP, 20XUAS-post-t-GRASP; J) yw; n-syb-LexA/+; repo-GAL4/13XLexAop2-post-t-GRASP, 20XUAS-pre-t-GRASP.
Differences in GRASP signal intensity were also observed among the GRASP methods. CD4-GRASP exhibited the strongest GRASP signal (Fig 3A) while Nrx-GRASP (Fig 3B) and t-GRASP in both directions (Figs 3D,E) exhibited only slightly reduced signal intensity, with syb-GRASP showing the weakest GRASP signal (Fig 3C). The enhanced synaptic specificity of t-GRASP was thus achieved without a major reduction in GRASP signal strength.
In a separate experiment, the extent of non-specific GRASP signal between neurons and glia was compared between the different GRASP methods using the same pan-neuronal and pan-glial drivers. Since neurons and glia do not form synaptic contacts, any GRASP signal between them should be false-positive. In this experiment one of the split-GFP fragments was expressed pan-neuronally while the other split-GFP fragment was expressed pan-glially. CD4-GRASP exhibited strong non-specific reconstituted GFP fluorescence throughout the ventral nerve cord (Fig 3F). Nrx-GRASP and Syb-GRASP showed greatly reduced signal (Figs 3G,H, respectively), while t-GRASP in both directions showed a noticeable further reduction to near the limits of detection (Figs 3I,J).
Specificity for synaptic contacts in larval sensory neurons
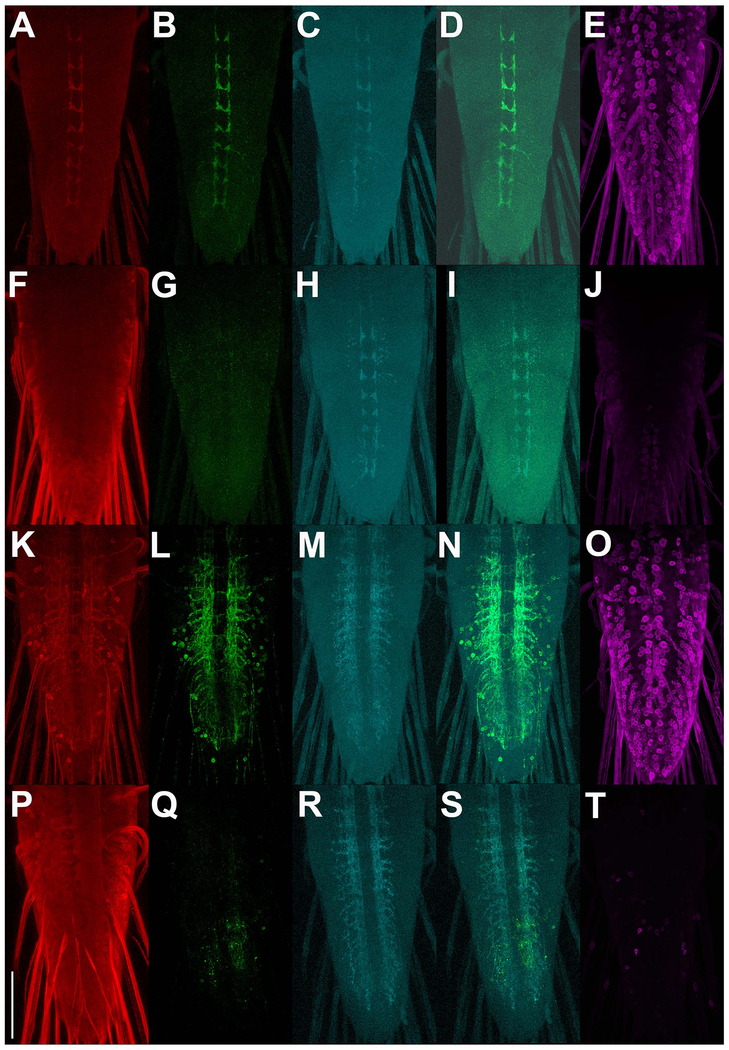
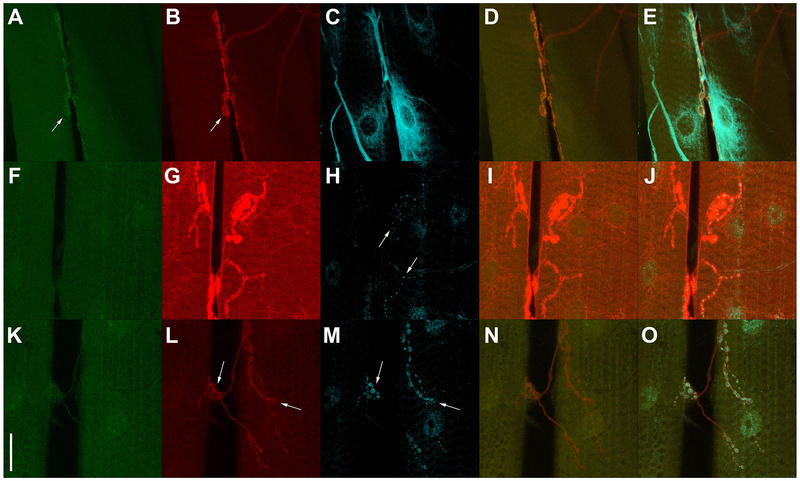
Although t-GRASP exhibits reconstituted GFP signal preferentially in the synaptic neuropil as demonstrated above, this does not guarantee that the signal within the neuropil is restricted to neurons that are synaptically connected as opposed to neurons that are just in close proximity. To assess the specificity of t-GRASP for legitimate synaptic contact sites, the STaR method (Chen et al., 2014), a conditionally-expressible, V5-tagged version of the active zone marker Brp was included in t-GRASP experiments identifying synaptic contact sites of larval sensory neuron types III and IV. The cell bodies and dendrites of these neurons are peripherally located just under the larval cuticle and their axons project to the neuropil region of the larval ventral nerve cord where their synaptic contact sites are located (Prokop and Meinertzhagen, 2006). Ideally, in these experiments GRASP signal will be restricted to regions of overlap with conditionally-expressed Brp-V5. In these reciprocal experiments, either pre- or post-t-GRASP is expressed in larval sensory neurons while the complementary t-GRASP is pan-neuronally expressed. Again, pan-neuronal t-GRASP expression makes this experiment a stringent test of synapse specificity for reasons previously mentioned. These experiments enhance the GRASP signal using an anti-GFP antibody that specifically recognizes reconstituted GFP, but not pre-t-GRASP or post-t-GRASP (Figure S1). Expression of pre-t-GRASP in type IV sensory neurons (Fig. 4A) in combination with pan-neuronal expression of post-t-GRASP (Fig 4E), results in GRASP signal (Fig. 4B) coincident with type IV sensory neuron active zones (Figs 4C, D) even at high magnification (Fig S3). The reciprocal experiment in which pre-t-GRASP was pan-neuronally expressed (Fig. 4F) and post-t-GRASP was expressed in type IV sensory neurons (Fig. 4J) exhibits very little GRASP signal (Fig. 4G) in type IV sensory neuron active zones (Figs. 4H,I), thus demonstrating the effectiveness of the pre-synaptic and dendritic targeting of pre- and post-t-GRASP, respectively.
Figure 4.
Synaptic specificity assessment of t-GRASP in larval sensory neurons. A-E) trpA1-QF drives expression of pre-t-GRASP in type IV sensory neurons and n-syb-GAL4 drives pan-neuronal expression of post-t-GRASP. F-J) Opposite polarity experiment of A-E where trpA1-QF drives post-t-GRASP in type IV sensory neurons and n-syb-GAL4 drives pan-neuronal expression of pre-t-GRASP . K-O) nompC-QF drives expression of pre-t-GRASP in type III sensory neurons and n-syb-GAL4 drives pan-neuronal expression of post-t-GRASP. P-T) Opposite polarity experiment of K-O where nompC-QF drives expression of post-t-GRASP and n-syb-GAL4 drives pan-neuronal expression of pre-t-GRASP. pre-t-GRASP expression (red) (A, F, K, P) was visualized with an anti-HA antibody; reconstituted GFP fluorescence (green) (B, G, L, Q) was visualized with anti-GFP Tag Abfinity mAb; active zone marker Brp-V5 conditional expression (cyan) (C, H, M, R) was visualized with an anti-V5 antibody; overlay of reconstituted GFP fluorescence with Brp-V5 (D, I, N, S); post-t-GRASP expression (magenta) (E, J, O, T) was visualized with an anti-TLN antibody. pre-t-GRASP, reconstituted GFP fluorescence, and conditional Brp-V5 expression are from the same animal. Post-t-GRASP expression is from a different animal of the same genotype as the others on the same row. All images in the same column were acquired and processed identically. Scale bar: 100μm.
Figure 4 genotypes. A-E) yw; brp-FRT-STOP-FRT-V5, QUAS-FLP/+; trpA1-QF, n-syb-GAL4/10XQUAS-pre-t-GRASP, 20XUAS-post-t-GRASP; F-J) yw; brp-FRT-STOP-V5, QUAS-FLP/+; trpA1-QF, n-syb-GAL4/20XUAS-pre-t-GRASP, 10XQUAS-post-t-GRASP; K-O) yw; brp-FRT-STOP-FRT-V5, QUAS-FLP/+; nompC-QF, n-syb-GAL4/10XQUAS-pre-t-GRASP, 20XUAS-post-t-GRASP; P-T) yw; brp-FRT-STOP-V5, 10XQUAS-FLP/+; nompC-QF, n-syb-GAL4/20XUAS-pre-t-GRASP, 10XQUAS-post-t-GRASP.
Similar experiments were performed for type III larval sensory neurons. Expression of pre-t-GRASP in type III larval sensory neurons (Fig. 4K) and pan-neuronal expression of post-t-GRASP (Fig. 4O) resulted in GRASP signal (Fig. 4L) that mostly overlapped with type III larval neuron active zones (Figs. 4M,N), although some GRASP signal in cell bodies was also apparent. This result indicates t-GRASP can yield false-positive signal dependent on the particular driver pair, most likely due to strong drivers that result in high level expression of pre- or post-t-GRASP. The reciprocal experiment in which post-t-GRASP was pan-neuronally expressed (Fig. 4P) and pre-t-GRASP was expressed in type III larval sensory neurons (Fig. 4T) exhibited a slight GRASP signal (Fig 4Q) that mostly coincides with type III larval neuron active zones (Figs. 4R,S). As this GRASP signal is only a small fraction of that observed in the previous reciprocal experiment, it again demonstrates the effectiveness, albeit not 100%, of the pre-synaptic and dendritic targeting of pre- and post-t-GRASP.
Since pre-t-GRASP and post-t-GRASP are both being expressed in type IV or type III sensory neurons in both directions of the above experiments, and GRASP signal is much stronger in the experiments done in the initial direction (Figs 4B and L) than in the reciprocal direction (Figs 4G and Q), these results indicate the vast majority of the GRASP signal in the initial direction is due to extracellular trans GFP reconstitution and not cis GFP reconstitution.
Trans vs cis GFP reconstitution at the larval NMJ
To further address trans vs cis GFP reconstitution with the t-GRASP method, reciprocal experiments were performed at the third instar larval NMJ in which pre-t-GRASP and post-t-GRASP were expressed in motor neurons and muscles. In the first experiment, pre-t-GRASP was expressed in motor neurons and post-t-GRASP in muscles. In this experiment, GRASP signal was detected at the NMJ (Fig 5A,D,E) coincident with localization of pre-t-GRASP (Fig 5B,D,E) and post-t-GRASP (Fig 5C,E). In the reciprocal experiment, pre-t-GRASP was expressed in muscles and post-t-GRASP was expressed in motor neurons. Surprisingly, in this experiment pre-t-GRASP showed significant accumulation at post-synaptic regions of the muscle (Fig 5G, I, J) and post-t-GRASP showed detectable accumulation at presynaptic terminals (Fig 5H, J). Nevertheless, GRASP signal was not detected in the reciprocal experiment (Fig 5F, I, J).
Figure 5.
Assessment of trans vs cis GFP reconstitution at the third instar larval NMJ. A-E) vGlut-LexA drives expression of pre-t-GRASP in motor neurons and 24B-GAL4 drives expression of post-t-GRASP in muscles. F-J) 24B-GAL4 drives expression of pre-t-GRASP in muscle and vGlut-LexA drives expression of post-t-GRASP in motor neurons. K-O) vGlut-LexA drives simultaneous expression of both pre-t-GRASP and post-t-GRASP in motor neurons. GRASP signal (green) is observed when pre-t-GRASP (red) is expressed in motor neurons and post-t-GRASP (cyan) is expressed in muscle (arrow A), but not when post-t-GRASP is expressed in motor neurons and pre-t-GRASP is expressed in muscle (F) or when both pre-t-GRASP and post-t-GRASP are expressed in motor neurons (K). pre-t-GRASP accumulates in the pre-synaptic terminals of motor neurons (arrows B, L) and at the post-synaptic region of muscles (G). post-t-GRASP accumulates outside the nucleus and at the post-synaptic region of muscles (C) and in the pre-synaptic terminals of motor neurons (arrows H, M). Overlay of GRASP signal with pre-t-GRASP (D, I, N) and with pre-t-GRASP and post-t-GRASP (E, J, O). The observation that GRASP signal is only observed when pre-t-GRASP is expressed in motor neurons and post-t-GRASP is expressed in muscles, but not in the reciprocal experiment or when both pre-t-GRASP and post-t-GRASP is expressed in motor neurons reveals the t-GRASP method exhibits a significant preference for extracellular trans over intracellular cis GFP reconstitution. Reconstituted GFP, pre-t-GRASP, and post-t-GRASP were visualized with anti-GFP Tag Abfinity mAb, anti-HA, and anti-TLN antibodies, respectively. Scale bar 30μm.
Figure 5 genotypes. A-E) yw; vGlut-LexA/+ ;24B-GAL4/13XLexAop2-pre-t-GRASP, 20XUAS-post-t-GRASP; F-J) yw; vGlut-LexA/+; 24B-GAL4/20XUAS-pre-t-GRASP/13XLexAop2-post-t-GRASP; K-O) yw; vGlut-LexA/+; 13XLexAop2-pre-t-GRASP, 20XUAS-post-t-GRASP.
A third experiment was also performed in which both pre-t-GRASP and post-t-GRASP were simultaneously expressed in motor neurons. Despite the observation that pre-t-GRASP (Fig 5L, N, O) and post-t-GRASP (Fig 5M,O) accumulate in close proximity in motor neuron pre-synaptic terminals, no GRASP signal is observed (Fig 5K, N, O). The continuous outline of the NMJ by pre-t-GRASP as compared to the more punctate distribution of post-t-GRASP suggests that within the pre-synaptic terminal the two constructs are being differentially localized.
For each of these three experiments, animals of the same genotypes were alternatively immunostained such that endogenous Brp was substituted for post-t-GRASP (Figure S2). GRASP signal (Fig S2A,D,E) outlines the motor neuron pre-synaptic nerve terminal as indicated by the active zone marker Brp (Fig S2C,D,E). Taken together, the results of these NMJ experiments demonstrate t-GRASP exhibits a significant preference for trans over cis GFP reconstitution.
Specificity for synaptic contacts in adult photoreceptors
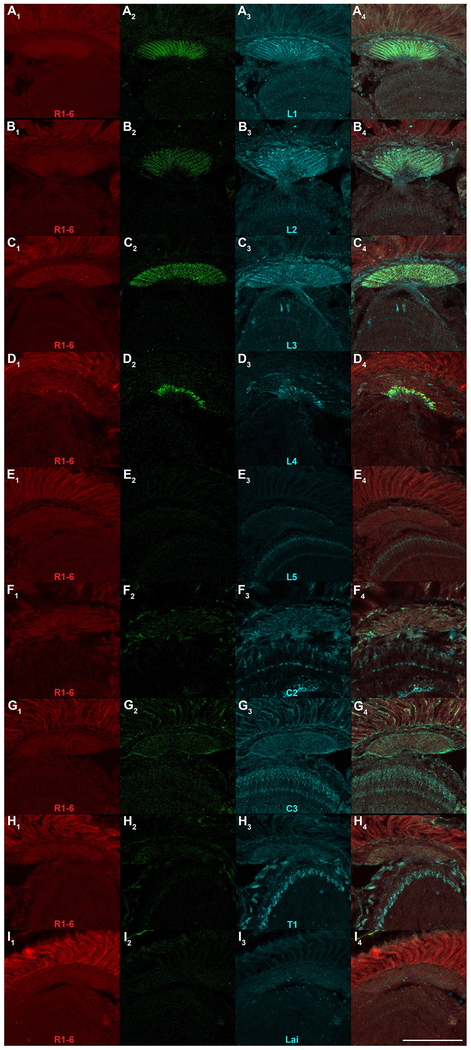
To further assess the specificity of t-GRASP for legitimate synaptic contact sites, pre-t-GRASP was expressed in R1-6 photoreceptors using Rh1-QF2 and evaluated for GRASP signal in combination with expression of post-t-GRASP in each of nine different lamina neuron subtypes. The R1-6 photoreceptor/lamina synapses were chosen for assessment because the synaptic connections are known from EM studies (Meinertzhagen and O’Neil, 1991) and split-Gal4 drivers specific for each lamina subtype are available (Tuthill et al., 2013). Since the pre-synaptic region of R1-6 photoreceptors and all nine distinct lamina subtypes are tightly packed into the lamina region of the adult optic lobe, this will provide a rigorous test of synapse-specificity as the dendrites of six of the nine lamina subtypes are in close proximity to R1-6 pre-synaptic terminals but are not synaptically connected. R1-6 photoreceptors form direct synaptic connections onto lamina subtypes L1, L2, and L3, but not the other six lamina subtypes (Meinertzhagen and O’Neil, 1991), and thus GRASP signal is only expected in lamina subtypes L1, L2, and L3.
The results of the R1-6 photoreceptor/lamina experiments are shown in Figure 5. R1-6 photoreceptor expression of pre-t-GRASP is presented in the leftmost column (Figs 5A1–I1), lamina subtype expression of post-t-GRASP in the third column (Figs. 5A3–I3), GRASP signal in the second column (Figs. 5A2–I2), and the overlay in the rightmost column (Figs 5A4–I4). As expected from EM studies, significant GRASP signal is observed for lamina subtypes L1 (Fig. 5A2), L2 (Fig. 5B2), and L3 (Fig 5C2) thus demonstrating t-GRASP is sensitive enough to detect all known synaptic connections between R1-6 photoreceptors and their post-synaptic lamina neuron subtypes. It should also be noted that since there is no overlap in neuronal expression of pre-t-GRASP and post-t-GRASP in these experiments, the GRASP signal observed must be due to trans, and not cis, GFP reconstitution. As expected from EM studies, no GRASP signal was detected for L5 (Fig 5E2), C2 (Fig 6F2), C3 (Fig 5G2), T1 (Fig 5H2), or Lai (Fig 5I2), thus demonstrating false-positives were not occurring due to non-synaptic trans GFP reconstitution.
Figure 6.
Synaptic specificity assessment of t-GRASP in lamina region of adult optic lobe. Rh1-QF2 was used to drive expression of pre-t-GRASP in R1-6 photoreceptors in all genotypes and split-GAL4 drivers specific for each of the nine columnar lamina subtype neurons were used to drive expression of post-t-GRASP as indicated. A) L1; B) L2; C) L3; D) L4; E) L5; F) C2; G) C3; H) T1; I) Lai. Subscripts are as follows: (1) pre-t-GRASP (red) immuno-labeled with anti-HA; (2) reconstituted GRASP (green) immuno-labeled with anti-GFP Tag Abfinity mAb; (3) post-t-GRASP (cyan) immuno-labeled with anti-TLN; (4) triple overlay. L1, L2, and L3 exhibit GRASP signal throughout the lamina while L4 shows GRASP signal in the medial lamina. No GRASP signal is observed in L5, C2, C3, T1, and Lai. All images in the same column were acquired and processed identically. Scale bar: 100μm.
Figure 6 genotypes. A) yw; Rh1-QF2 cn bw/R48A08AD cn bw; R66A01DBD/10XQUAS-pre-t-GRASP, 20XUAS-post-t-GRASP; B) yw; Rh1-QF2 cn bw/R53G02AD cn bw; R29G11DBD/10XQUAS-pre-t-GRASP, 20XUAS-post-t-GRASP; C) yw; Rh1-QF2 cn bw/R64B03AD cn bw; R14B07DBD/10XQUAS-pre-t-GRASP, 20XUAS-post-t-GRASP; D) yw; Rh1-QF2 cn bw/cn bw; R31C06AD, R34G07DBD/10XQUAS-pre-t-GRASP, 20XUAS-post-t-GRASP; E) yw; Rh1-QF2 cn bw/R21A05AD cn bw; R31H09DBD/10XQUAS-pre-t-GRASP, 20XUAS-post-t-GRASP; F) yw; Rh1-QF2 cn bw/R25B02AD cn bw; R48D11DBD/10XQUAS-pre-t-GRASP, 20XUAS-post-t-GRASP; G) yw; Rh1-QF2 cn bw/R26H02AD cn bw; R29G11DBD/10XQUAS-pre-t-GRASP, 20XUAS-post-t-GRASP; H) yw; Rh1-QF2 cn bw/R31F10AD cn bw; R30F10DBD/10XQUAS-pre-t-GRASP, 20XUAS-post-t-GRASP; I) yw; Rh1-QF2 cn bw/R92A10AD cn bw; R66A02DBD/10XQUAS-pre-t-GRASP, 20XUAS-post-t-GRASP.
The sole exception from expectations based on EM analysis was L4 (Fig. 5D2), where GRASP signal is present in a thin strip of the most proximal portion of the lamina, and this result must therefore be considered a false-positive. Interestingly, although R1-6 photoreceptors do not synapse onto L4 lamina neurons, L4 lamina neurons do synapse onto R1-6 photoreceptors (Meinertzhagen and O’Neil, 1991) as part of an apparent feedback loop. Thus, the signal observed for lamina subtype L4 is a bona fide synaptic contact site detected by t-GRASP even though based on the polarity of this synapse its detection was unexpected. This result indicates the strength of expression of this particular driver pair was sufficient to overcome the effectiveness of the targeting of pre- and post-t-GRASP. Nevertheless, in eight of nine specificity tests the expected results were observed with only a small portion of the lamina exhibiting aberrant, yet synapse-specific, signal in the single case of L4.
Discussion
The development and characterization of t-GRASP, a new Drosophila genetic tool for assessing neuronal connectivity has been described. t-GRASP was developed by fusing a number of pre-synaptic and post-synaptic/dendritic proteins to either of the two split-GFP fragments and selecting the one from each side of the synapse exhibiting the most significant enhancement of reconstituted GFP fluorescence signal to synaptic regions. The effect of targeting both split-GFP constructs enhances GRASP signal specificity for synaptic regions as compared to existing Drosophila GRASP alternatives. t-GRASP should thus be a useful alternative GRASP option to Nrx-GRASP or syb-GRASP, especially for experiments involving low activity neurons where the activity-dependence of syb-GRASP may make it sub-optimal.
In the majority of driver pairs tested in larval sensory neurons, NMJs, and adult photoreceptors, GRASP signal was observed coincident with active zones and likely represents synaptic contact sites. For a small minority of the driver pairs tested some false positives were, however, observed, presumably due to one or both drivers being sufficiently strong to express the t-GRASP constructs to levels beyond the capacity of the neurons to localize them to their targeted pre-synaptic or post-synaptic subcellular locations. Thus, t-GRASP is not infallible and positive results with t-GRASP alone should not be interpreted as definitive without corroboration from other assays such as optogenetic activation of pre-synaptic neurons coupled with functional calcium imaging or electrophysiological recordings in post-synaptic neurons.
Various strategies can be incorporated into t-GRASP experiments for recognizing and/or minimizing false-positives. One is to use t-GRASP in combination with the STaR method, as demonstrated above (Figure 4), where false-positive GRASP signal can be recognized when it is not coincident with conditional Brp-V5 active zone labeling. Another strategy possible with the Gal4 and Q systems is to reduce driver strength by including repressors whose activities can be controlled in a dose-dependent manner via the addition of exogenously added compounds (Potter et al., 2010; Sethi and Wang, 2017). A third strategy is to use driver pairs with expression patterns as sparse a possible to minimize the chances of producing false-positive signal.
Fly strains for expressing both pre-t-GRASP and post-t-GRASP with all three Drosophila transcription systems (GAL4, LexA, and Q) have been developed and characterized. With t-GRASP it is possible to perform GRASP experiments using any two driver pairs among the three Drosophila binary transcriptions in either direction. This flexibility significantly broadens the potential application of GRASP in Drosophila for neural circuit mapping and thus adds another useful tool to the already expansive Drosophila genetic toolbox.
Experimental procedures
Plasmid construction
All entry and expression clones were assembled using Gateway MultiSite cloning as previously described (Petersen and Stowers, 2011). The components of the Gateway MultiSite LR reactions are shown in Table 2. The destination vector for all LR reactions was pDESTsvaw (Shearin et al., 2013). The following entry clones were previously described: L1-20XUAS-L4 and L3-Rab3-L2 (Petersen and Stowers, 2011); L1-13XLexAop2-L4 (Shearin et al., 2013). New entry clones and templates were as follows: pENTR L1-10XQUAS-L4>pENTR L1-10XQUAS-L5 (Shearin et al., 2013); pENTR L5-QF2-L2>n-syb-QF2 (Riabinina et al., 2015), pENTR L1-Rh1-R5>Drosophila genomic DNA; L5-pre-mGRASP-V5-L2>pre-mGRASP (Kim et al., 2011); L5-post-mGRASP-ΔT-L2>post-mGRASP (Kim et al., 2011); R4-pre-mGRASP-2XHA-R3> pre-mGRASP (Kim et al., 2011); R4-GFP1-10-R3>post-mGRASP (Kim et al., 2011); L3-cac-L2>UAS-cac (Kawasaki et al., 2002); L3-Brp-L2>BrpD1-3 (Fouquet et al., 2009), L3-DRBP-L2>AT04807 DRBP cDNA (Drosophila Genomics Resource Center); L3-Fife-L2>Fife cDNA (Bruckner et al., 2012); L3-Dlar-L2>Dlar cDNA (Krueger et al., 1996) ; L3-dLev-10-s-L2 and L3-dLev-10-m-L2>GH21941 cDNA (Drosophila Genomics Resource Center) ; L3-dLev-10-L2>dLev-10 full-length cDNA generated by RT-PCR and hybrid PCR with GH21941; L3-DSCAM-s-L2 and L3-DSCAM-L-L2>DSCAM 17.1 cDNA (Shi et al., 2007); L2-TLN-L2>TLN cDNA (Nicolai et al., 2010). The destination vector pDESTsvaw has been previously described (Shearin et al., 2013). A single C nucleotide deletion mutation at base pair 1272 (ΔC1272) was initially overlooked in L3-cac-L2. This mutation results in omission of the C-terminal 93aa of cac. Subsequently, this mutation was corrected and flies containing pre-mGRASP-2XHA fused to the wildtype cac tail were compared to the cac ΔC1272 for specificity of localization of GFP fluorescence to the neuropil (Figure S4). Since the GFP fluorescence signal from cac ΔC1272 was more tightly localized to the neuropil than wildtype cac, the cac ΔC1272 mutant was used in all fly strains described in the text. Annotated protein-coding sequences of each pre- and post-GRASP construct are shown in Fig S5. Sequences of entry clones are available upon request.
Table 2.
Expression clones and component entry clones of Gateway MultiSite LR reactions.
| Expression clones | Component entry clones | ||
|---|---|---|---|
| 10XQUAS-pre-mGRASP-V5 | L1-10XQUAS-R5 | L5-pre-mGRASP-V5-L2 | |
| 20XUAS-post-mGRASPΔT | L1-20XUAS-R5 | L5-post-mGRASPΔT-L2 | |
| 10XQUAS-pre-GRASP-Brp | L1-10XQUAS-L4 | R4-pre-mGRASP-2XHA-R3 | L3-Brp-L2 |
| 10XQUAS-pre-GRASP-DRBP | L1-10XQUAS-L4 | R4-pre-mGRASP-2XHA-R3 | L3-DRBP-L2 |
| 10XQUAS-pre-GRASP-Fife | L1-10XQUAS-L4 | R4-pre-mGRASP-2XHA-R3 | L3-Fife-L2 |
| 10XQUAS-pre-GRASP-Rab3 | L1-10XQUAS-L4 | R4-pre-mGRASP-2XHA-R3 | L3-Rab3-L2 |
| 10XQUAS-pre-GRASP-Dlar | L1-10XQUAS-L4 | R4-pre-mGRASP-2XHA-R3 | L3-Dlar-L2 |
| 10XQUAS-pre-GRASP-Cac | L1-10XQUAS-L4 | R4-pre-mGRASP-2XHA-R3 | L3-Cac-L2 |
| 10XQUAS-pre-GRASP-CacWT | L1-10XQUAS-L4 | R4-pre-mGRASP-2XHA-R3 | L3-CacWT-L2 |
| 20XUAS-pre-GRASP-Cac | L1-20XUAS-L4 | R4-pre-mGRASP-2XHA-R3 | L3-Cac-L2 |
| 13XLexAop-pre-GRASP-Cac | L1-13XLexAop-L4 | R4-pre-mGRASP-2XHA-R3 | L3-Cac-L2 |
| 20XUAS-post-GRASP-DLev10-s | L1-20XUAS-L4 | R4-GFP1-10-R3 | L3-DLev10-s-L2 |
| 20XUAS-post-GRASP-DLev10-m | L1-20XUAS-L4 | R4-GFP1-10-R3 | L3-DLev10-m-L2 |
| 20XUAS-post-GRASP-DLev10 | L1-20XUAS-L4 | R4-GFP1-10-R3 | L3-DLev10-L2 |
| 20XUAS-post-GRASP-DSCAM-s | L1-20XUAS-L4 | R4-GFP1-10-R3 | L3-DSCAM-s-L2 |
| 20XUAS-post-GRASP-DSCAM-m | L1-20XUAS-L4 | R4-GFP1-10-R3 | L3-DSCAM-m-L2 |
| 20XUAS-post-GRASP-TLN-s | L1-20XUAS-L4 | R4-GFP1-10-R3 | L3-TLN-s-L2 |
| 20XUAS-post-GRASP-TLN | L1-20XUAS-L4 | R4-GFP1-10-R3 | L3-TLN-L2 |
| 10XQUAS-post-GRASP-TLN | L1-10XQUAS-L4 | R4-GFP1-10-R3 | L3-TLN-L2 |
| 13XLexAop-post-GRASP-TLN | L1-13XLexAop-L4 | R4-GFP1-10-R3 | L3-TLN-L2 |
| Rh1-QF2 | L1-Rh1-R5 | L5-QF2-L2 | |
Drosophila stocks
Injections of transgenes for fly strains reported in this study were performed by Bestgene, Inc. Other fly stocks: repo-GAL4 (Sepp et al., 2001) (BDSC #7415), n-syb-GAL4 (Bushey et al., 2009), n-syb-QF2 (Riabinina et al., 2015), n-syb-LexA (Shearin et al., 2013), TrpA1-QF, and nompC-QF (Petersen and Stowers, 2011), QUAS-FLP (Potter et al., 2010), Brp-FRT-STOP-FRT-V5 (Chen et al., 2014), LexAop-GFP11 and UAS-CD4-GFP1-10 (Gordon and Scott, 2009), UAS-spGFP1-10::Nrx (Fan et al., 2013), UAS-syb::spGFP1-10 (Macpherson et al., 2015), vGlut-LexA (Baek et al., 2013), 24B-GAL4 (Brand and Perrimon, 1993).
Immunostaining.
Larval immuno-labeling was performed as previously described (Shearin et al., 2013). Heads for cryostat sectioning were prepared as follows. After removal of the proboscis and internal air sacs, heads were dehydrated in 5% sucrose 10 minutes, 10% sucrose 10 minutes, 25% sucrose 1 hour, fixed 1 hour in 4% paraformaldehyde, followed by dehydration being repeated for 10 minutes each step. Heads were then embedded and frozen in O.C.T. (Tissue-Tek 4583) on dry ice before cryostat sectioning. After blocking, 10 µm orthogonal head sections were incubated 1 hour at 37°C in primary antibodies, washed in PBS-T (0.5% Triton), and incubated 4 hours at room temperature in secondary antibodies. After washing, slides were mounted in a 10% Mowiol 4-88 (Sigma-Aldrich 81381)/2.5% Dabco 33-LV (Sigma-Aldrich 290734) mixture before imaging on a Leica SP5 confocal microscope.
Primary antibodies and dilutions: nc82 (E. Buchner, Developmental Studies Hybridoma Bank), rabbit anti-GFP Tag Abfinity (ThermoFisher G10362), mouse anti-HA.11/16B12 (Covance MMS-101P-500), rat anti-HA (3F10, Roche 11 867 431 001), rat anti-V5 (Biorbyt orb256445), mouse anti-V5 (Biorbyt orb256432), and goat anti-TLN/ICAM-5 (Bio-techne AF1173). All antibodies were diluted 1:300 for larva and 1:100 for cryostat sections (except nc82 which was used at 1:50) in PBS with 0.5% Triton X-100 and 5% BSA. Secondary antibodies: F’Ab2 donkey anti-rabbit AF488 (Jackson ImmunoResearch 711-546-152), F’Ab2 donkey anti-rat-Cy3 (Jackson ImmunoResearch 712-166-153), F’Ab2 donkey anti-rat AL647 (712-606-153), F’ab2 donkey anti-mouse Cy3 (Jackson ImmunoResearch 715-166-151), F’Ab2 donkey anti-mouse 647 (Jackson ImmunoResearch 715-606-151), and donkey anti-goat AL647 (ThermoFisher A-21447). Secondary antibodies were diluted 1:300.
Supplementary Material
Figure S1. Specificity assessment of rabbit anti-GFP Tag Abfinity mAb. A-C) pan-neuronal expression of both pre-t-GRASP and post-t-GRASP. A) pre-t-GRASP; B) GFP; C) post-t-GRASP. D-F) pan-neuronal expression of pre-t-GRASP only. D) pre-t-GRASP; E) GFP; F) post-t-GRASP. G-I) pan-neuronal expression of post-t-GRASP only. G) pre-t-GRASP; H) GFP; I) post-t-GRASP. pre-t-GRASP immuno-labeling-anti-HA (red), GFP immuno-labeling-rabbit anti-GFP Tag Abfinity mAb (green), post-t-GRASP immuno-labeling-anti-TLN (cyan). All images in the same column were acquired and processed identically. Scale bar: 100μm.
Figure S1 genotypes. A-C) yw; n-syb-GAL4/+; n-syb-QF2/10XQUAS-pre-t-GRASP/20XUAS-post-t-GRASP. D-F) yw; n-syb-QF2/10XQUAS-pre-t-GRASP/20XUAS-post-t-GRASP. G-I) yw; n-syb-GAL4/+; +/10XQUAS-pre-t-GRASP/20XUAS-post-t-GRASP.
Figure S2. Further assessment of trans vs cis GFP reconstitution at the third instar larval NMJ. A-E) vGlut-LexA drives expression of pre-t-GRASP in motor neurons and 24B-GAL4 drives expression of post-t-GRASP in muscles. F-J) 24B-GAL4 drives expression of pre-t-GRASP in muscle and vGlut-LexA drives expression of post-t-GRASP in motor neurons. K-O) vGlut-LexA drives simultaneous expression of both pre-t-GRASP and post-t-GRASP in motor neurons. GRASP signal is observed when pre-t-GRASP is expressed in motor neurons and post-t-GRASP is expressed in muscle (A), but not when post-t-GRASP is expressed in motor neurons and pre-t-GRASP is expressed in muscle (F) or when both pre-t-GRASP and post-t-GRASP are expressed in motor neurons (K). pre-t-GRASP accumulates in the pre-synaptic terminals of motor neurons (B, L) and at post-synaptic sites of muscles (G). Endogenous Brp accumulates in discrete puncta in the pre-synaptic terminals of motor neurons (C, H, M). Overlay of GRASP signal with Brp (D, I, N) and with pre-t-GRASP and Brp (E, J, O). GRASP signal extends throughout the motor neuron pre-synaptic nerve terminal indicated by the active zone marker Brp. Reconstituted GFP, pre-t-GRASP, and Brp were visualized with anti-GFP Tag Abfinity mAb, anti-HA, and anti-Brp antibodies, respectively. Scale bar 30μm.
Figure S2 genotypes. A-E) yw; vGlut-LexA/+ ;24B-GAL4/13XLexAop2-pre-t-GRASP, 20XUAS-post-t-GRASP; F-J) yw; vGlut-LexA/+; 24B-GAL4/20XUAS-pre-t-GRASP/13XLexAop2-post-t-GRASP; K-O) yw; vGlut-LexA/+; 13XLexAop2-pre-t-GRASP, 20XUAS-post-t-GRASP.
Figure S3. High magnification images of GRASP signal at larval type IV sensory neuron synapses. A-C) trpA1-QF drives expression of pre-t-GRASP in type IV sensory neurons and n-syb-GAL4 drives pan-neuronal expression of post-t-GRASP. pre-t-GRASP expression (red) (A,E) was visualized with an anti-HA antibody; reconstituted GFP fluorescence (green) (B,F) was visualized with anti-GFP Tag Abfinity mAb; active zone marker Brp-V5 conditional expression (cyan) (C,G) was visualized with an anti-V5 antibody; overlay of reconstituted GFP fluorescence with Brp-V5 (D,H). GRASP signal is coincident with the active zone marker Brp-V5. Scale bars: A) 30μm; E) 10μm.
Figure S3 genotype. A-C) yw; brp-FRT-STOP-FRT-V5, QUAS-FLP/+; trpA1-QF, n-syb-GAL4/10XQUAS-pre-t-GRASP, 20XUAS-post-t-GRASP.
Figure S4. Neuropil specificity assessment of cac ΔC1272 and cac wildtype. Direct fluorescence imaging of pan-neuronal expression of post-mGRASPΔT with A) cac ΔC1272; B) cac WT. GFP fluorescence is more restricted to the neuropil in cac ΔC1272. Scale bar: 100μm.
Figure S4 genotypes. A) yw; 20XUAS-post-mGRASPΔT/+; n-syb-GAL4, n-syb-QF2/10XQUAS-pre-cac ΔC1272; B) yw; 20XUAS-post-mGRASPΔT/+; n-syb-GAL4, n-syb-QF2/2110XQUAS-pre-cac wildtype.
Table 1.
Transgenes and insertion sites.
| Transgene | Insertion Site |
|---|---|
| 10XQUAS-pre-mGRASP-V5 | VK27 |
| 20XUAS-post-mGRASPΔT | attP2 |
| 10XQUAS-pre-GRASP-Brp | VK27 |
| 10XQUAS-pre-GRASP-DRBP | VK27 |
| 10XQUAS-pre-GRASP-Fife | VK27 |
| 10XQUAS-pre-GRASP-Rab3 | VK27 |
| 10XQUAS-pre-GRASP-Dlar | VK27 |
| 10XQUAS-pre-GRASP-Cac | VK27 |
| 10XQUAS-pre-GRASP-CacWT | VK27 |
| 20XQUAS-pre-GRASP-Cac | VK27 |
| 13XLexAop2-pre-GRASP-Cac | VK27 |
| 20XUAS-post-GRASP-dLev-10-s | attP2 |
| 20XUAS-post-GRASP-dLev-10-m | attP2 |
| 20XUAS-post-GRASP-dLev-10 | attP2 |
| 20XUAS-post-GRASP-DSCAM-s | attP2 |
| 20XUAS-post-GRASP-SDCAM-m | attP2 |
| 20XUAS-post-GRASP-TLN-s | attP2 |
| 20XUAS-post-GRASP-TLN | attP2 |
| 20XUAS-post-GRASP-TLN | attP2 |
| 20XUAS-post-GRASP-TLN | attP2 |
| Rh1-QF2 | attP40 |
Acknowledgements
This work was supported by NIH grant R21NS082922. The nc82 monoclonal antibody developed by E. Buchner was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. We thank the Drosophila Genomics Resource center supported by NIH grant 2P40OD010949 for cDNAs. We thank Hannah McKinney for helpful comments on the manuscript.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- Baek M, Enriquez J, and Mann RS (2013). Dual role for Hox genes and Hox co-factors in conferring leg motoneuron survival and identity in Drosophila. Development 140, 2027–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, and Perrimon N (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Bruckner JJ, Gratz SJ, Slind JK, Geske RR, Cummings AM, Galindo SE, Donohue LK, and O’Connor-Giles KM (2012). Fife, a Drosophila Piccolo-RIM homolog, promotes active zone organization and neurotransmitter release. J Neurosci 32, 17048–17058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey D, Tononi G, and Cirelli C (2009). The Drosophila fragile X mental retardation gene regulates sleep need. J Neurosci 29, 1948–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Akin O, Nern A, Tsui CY, Pecot MY, and Zipursky SL (2014). Cell-type-specific labeling of synapses in vivo through synaptic tagging with recombination. Neuron 81, 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiAntonio A, Burgess RW, Chin AC, Deitcher DL, Scheller RH, and Schwarz TL (1993). Identification and characterization of Drosophila genes for synaptic vesicle proteins. J Neurosci 13, 4924–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler K, Li F, Litwin-Kumar A, Park Y, Andrade I, Schneider-Mizell CM, Saumweber T, Huser A, Eschbach C, Gerber B, et al. (2017). The complete connectome of a learning and memory centre in an insect brain. Nature 548, 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan P, Manoli DS, Ahmed OM, Chen Y, Agarwal N, Kwong S, Cai AG, Neitz J, Renslo A, Baker BS, et al. (2013). Genetic and neural mechanisms that inhibit Drosophila from mating with other species. Cell 154, 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg EH, Vanhoven MK, Bendesky A, Wang G, Fetter RD, Shen K, and Bargmann CI (2008). GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron 57, 353–363. [DOI] [PubMed] [Google Scholar]
- Fouquet W, Owald D, Wichmann C, Mertel S, Depner H, Dyba M, Hallermann S, Kittel RJ, Eimer S, and Sigrist SJ (2009). Maturation of active zone assembly by Drosophila Bruchpilot. J Cell Biol 186, 129–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gally C, Eimer S, Richmond JE, and Bessereau JL (2004). A transmembrane protein required for acetylcholine receptor clustering in Caenorhabditis elegans. Nature 431, 578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon MD, and Scott K (2009). Motor control in a Drosophila taste circuit. Neuron 61, 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf ER, Daniels RW, Burgess RW, Schwarz TL, and DiAntonio A (2009). Rab3 dynamically controls protein composition at active zones. Neuron 64, 663–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofbauer A, Ebel T, Waltenspiel B, Oswald P, Chen YC, Halder P, Biskup S, Lewandrowski U, Winkler C, Sickmann A, et al. (2009). The Wuerzburg hybridoma library against Drosophila brain. J Neurogenet 23, 78–91. [DOI] [PubMed] [Google Scholar]
- Kawasaki F, Collins SC, and Ordway RW (2002). Synaptic calcium-channel function in Drosophila: analysis and transformation rescue of temperature-sensitive paralytic and lethal mutations of cacophony. J Neurosci 22, 5856–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki F, Felling R, and Ordway RW (2000). A temperature-sensitive paralytic mutant defines a primary synaptic calcium channel in Drosophila. J Neurosci 20, 4885–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Zhao T, Petralia RS, Yu Y, Peng H, Myers E, and Magee JC (2011). mGRASP enables mapping mammalian synaptic connectivity with light microscopy. Nat Methods 9, 96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittel RJ, Wichmann C, Rasse TM, Fouquet W, Schmidt M, Schmid A, Wagh DA, Pawlu C, Kellner RR, Willig KI, et al. (2006). Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science 312, 1051–1054. [DOI] [PubMed] [Google Scholar]
- Krueger NX, Van Vactor D, Wan HI, Gelbart WM, Goodman CS, and Saito H (1996). The transmembrane tyrosine phosphatase DLAR controls motor axon guidance in Drosophila. Cell 84, 611–622. [DOI] [PubMed] [Google Scholar]
- Liu KS, Siebert M, Mertel S, Knoche E, Wegener S, Wichmann C, Matkovic T, Muhammad K, Depner H, Mettke C, et al. (2011). RIM-binding protein, a central part of the active zone, is essential for neurotransmitter release. Science 334, 1565–1569. [DOI] [PubMed] [Google Scholar]
- Macpherson LJ, Zaharieva EE, Kearney PJ, Alpert MH, Lin TY, Turan Z, Lee CH, and Gallio M (2015). Dynamic labelling of neural connections in multiple colours by trans-synaptic fluorescence complementation. Nat Commun 6, 10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinertzhagen IA, and O’Neil SD (1991). Synaptic organization of columnar elements in the lamina of the wild type in Drosophila melanogaster. J Comp Neurol 305, 232–263. [DOI] [PubMed] [Google Scholar]
- Nicolai LJ, Ramaekers A, Raemaekers T, Drozdzecki A, Mauss AS, Yan J, Landgraf M, Annaert W, and Hassan BA (2010). Genetically encoded dendritic marker sheds light on neuronal connectivity in Drosophila. Proc Natl Acad Sci U S A 107, 20553–20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen LK, and Stowers RS (2011). A Gateway MultiSite recombination cloning toolkit. PLoS One 6, e24531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Tasic B, Russler EV, Liang L, and Luo L (2010). The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell 141, 536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokop A, and Meinertzhagen IA (2006). Development and structure of synaptic contacts in Drosophila. Semin Cell Dev Biol 17, 20–30. [DOI] [PubMed] [Google Scholar]
- Riabinina O, Luginbuhl D, Marr E, Liu S, Wu MN, Luo L, and Potter CJ (2015). Improved and expanded Q-system reagents for genetic manipulations. Nat Methods 12, 219–222, 215 p following 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepp KJ, Schulte J, and Auld VJ (2001). Peripheral glia direct axon guidance across the CNS/PNS transition zone. Dev Biol 238, 47–63. [DOI] [PubMed] [Google Scholar]
- Sethi S, and Wang JW (2017). A versatile genetic tool for post-translational control of gene expression in Drosophila melanogaster. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearin HK, Dvarishkis AR, Kozeluh CD, and Stowers RS (2013). Expansion of the gateway multisite recombination cloning toolkit. PLoS One 8, e77724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Yu HH, Yang JS, and Lee T (2007). Specific Drosophila Dscam juxtamembrane variants control dendritic elaboration and axonal arborization. J Neurosci 27, 6723–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura SY, Aso Y, Hige T, Wong A, Lu Z, Xu CS, Rivlin PK, Hess H, Zhao T, Parag T, et al. (2017). A connectome of a learning and memory center in the adult Drosophila brain. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuthill JC, Nern A, Holtz SL, Rubin GM, and Reiser MB (2013). Contributions of the 12 neuron classes in the fly lamina to motion vision. Neuron 79, 128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Durrbeck H, Buchner S, Dabauvalle MC, Schmidt M, Qin G, et al. (2006). Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron 49, 833–844. [DOI] [PubMed] [Google Scholar]
- Yamagata M, and Sanes JR (2012). Transgenic strategy for identifying synaptic connections in mice by fluorescence complementation (GRASP). Front Mol Neurosci 5, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Specificity assessment of rabbit anti-GFP Tag Abfinity mAb. A-C) pan-neuronal expression of both pre-t-GRASP and post-t-GRASP. A) pre-t-GRASP; B) GFP; C) post-t-GRASP. D-F) pan-neuronal expression of pre-t-GRASP only. D) pre-t-GRASP; E) GFP; F) post-t-GRASP. G-I) pan-neuronal expression of post-t-GRASP only. G) pre-t-GRASP; H) GFP; I) post-t-GRASP. pre-t-GRASP immuno-labeling-anti-HA (red), GFP immuno-labeling-rabbit anti-GFP Tag Abfinity mAb (green), post-t-GRASP immuno-labeling-anti-TLN (cyan). All images in the same column were acquired and processed identically. Scale bar: 100μm.
Figure S1 genotypes. A-C) yw; n-syb-GAL4/+; n-syb-QF2/10XQUAS-pre-t-GRASP/20XUAS-post-t-GRASP. D-F) yw; n-syb-QF2/10XQUAS-pre-t-GRASP/20XUAS-post-t-GRASP. G-I) yw; n-syb-GAL4/+; +/10XQUAS-pre-t-GRASP/20XUAS-post-t-GRASP.
Figure S2. Further assessment of trans vs cis GFP reconstitution at the third instar larval NMJ. A-E) vGlut-LexA drives expression of pre-t-GRASP in motor neurons and 24B-GAL4 drives expression of post-t-GRASP in muscles. F-J) 24B-GAL4 drives expression of pre-t-GRASP in muscle and vGlut-LexA drives expression of post-t-GRASP in motor neurons. K-O) vGlut-LexA drives simultaneous expression of both pre-t-GRASP and post-t-GRASP in motor neurons. GRASP signal is observed when pre-t-GRASP is expressed in motor neurons and post-t-GRASP is expressed in muscle (A), but not when post-t-GRASP is expressed in motor neurons and pre-t-GRASP is expressed in muscle (F) or when both pre-t-GRASP and post-t-GRASP are expressed in motor neurons (K). pre-t-GRASP accumulates in the pre-synaptic terminals of motor neurons (B, L) and at post-synaptic sites of muscles (G). Endogenous Brp accumulates in discrete puncta in the pre-synaptic terminals of motor neurons (C, H, M). Overlay of GRASP signal with Brp (D, I, N) and with pre-t-GRASP and Brp (E, J, O). GRASP signal extends throughout the motor neuron pre-synaptic nerve terminal indicated by the active zone marker Brp. Reconstituted GFP, pre-t-GRASP, and Brp were visualized with anti-GFP Tag Abfinity mAb, anti-HA, and anti-Brp antibodies, respectively. Scale bar 30μm.
Figure S2 genotypes. A-E) yw; vGlut-LexA/+ ;24B-GAL4/13XLexAop2-pre-t-GRASP, 20XUAS-post-t-GRASP; F-J) yw; vGlut-LexA/+; 24B-GAL4/20XUAS-pre-t-GRASP/13XLexAop2-post-t-GRASP; K-O) yw; vGlut-LexA/+; 13XLexAop2-pre-t-GRASP, 20XUAS-post-t-GRASP.
Figure S3. High magnification images of GRASP signal at larval type IV sensory neuron synapses. A-C) trpA1-QF drives expression of pre-t-GRASP in type IV sensory neurons and n-syb-GAL4 drives pan-neuronal expression of post-t-GRASP. pre-t-GRASP expression (red) (A,E) was visualized with an anti-HA antibody; reconstituted GFP fluorescence (green) (B,F) was visualized with anti-GFP Tag Abfinity mAb; active zone marker Brp-V5 conditional expression (cyan) (C,G) was visualized with an anti-V5 antibody; overlay of reconstituted GFP fluorescence with Brp-V5 (D,H). GRASP signal is coincident with the active zone marker Brp-V5. Scale bars: A) 30μm; E) 10μm.
Figure S3 genotype. A-C) yw; brp-FRT-STOP-FRT-V5, QUAS-FLP/+; trpA1-QF, n-syb-GAL4/10XQUAS-pre-t-GRASP, 20XUAS-post-t-GRASP.
Figure S4. Neuropil specificity assessment of cac ΔC1272 and cac wildtype. Direct fluorescence imaging of pan-neuronal expression of post-mGRASPΔT with A) cac ΔC1272; B) cac WT. GFP fluorescence is more restricted to the neuropil in cac ΔC1272. Scale bar: 100μm.
Figure S4 genotypes. A) yw; 20XUAS-post-mGRASPΔT/+; n-syb-GAL4, n-syb-QF2/10XQUAS-pre-cac ΔC1272; B) yw; 20XUAS-post-mGRASPΔT/+; n-syb-GAL4, n-syb-QF2/2110XQUAS-pre-cac wildtype.