mcr-1 has become an increasing concern due to the fear of rapid transferable resistance to our last resort antibiotics1. While mcr-1 confers resistance to the polymyxin antibiotic colistin, it remains unclear whether this gene also confers resistance to other antimicrobials. Here we report that mcr-1 confers cross-resistance to the cationic host antimicrobial, lysozyme.
mcr-1 positive Escherichia coli isolates, CDF-1 and IHD86_4 (patient isolates from Switzerland2 and Cambodia3, respectively), were cured of their mcr-1 carrying plasmid by serial passage in Lysogeny Broth (LB). Curing of the mcr-1 gene was confirmed using colony PCR comparing parental mcr-1 positive strains vs. mcr-1 cured strains (Fig. 1A). Colony PCR using E. coli specific uspA primers was performed as a positive control (Fig. 1B). When tested by broth microdilution in 25% LB, both cured strains exhibited a four and two-fold increase in susceptibility to colistin and polymyxin B, respectively.
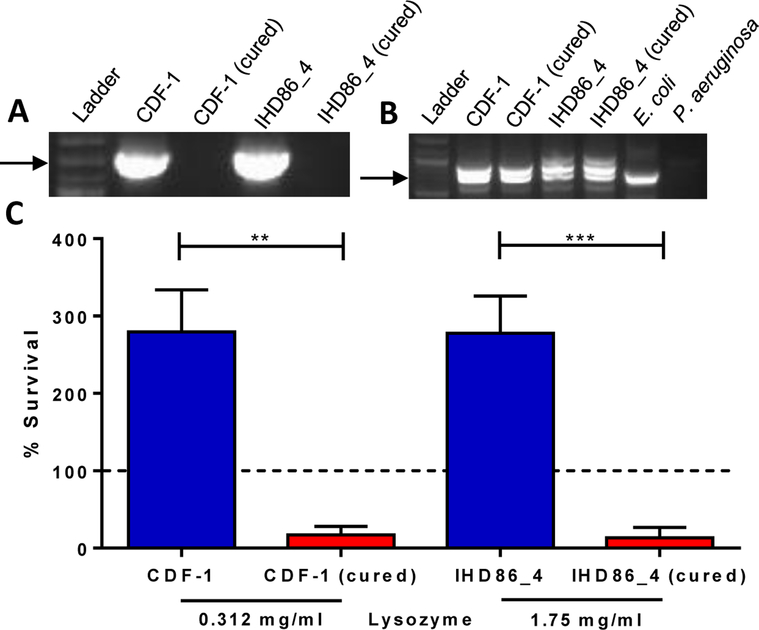
Figure 1. mcr-1 mediates resistance to lysozyme.
A, B, Colony PCR using (A) mcr-1 specific or (B) uspA specific primers on mcr-1 positive isolates CDF-1 and IHD86_4 and their corresponding cured, mcr-1 negative derivatives. A molecular weight ladder was included in each figure, and E. coli strain NCM3722 was used as a positive control and Pseudomonas aeruginosa strain PAO1 was used as a negative control in Figure 1B. C, Percent survival of mcr-1 positive isolates and their corresponding, cured mcr-1 negative derivatives, calculated by dividing the surviving CFUs after three-hour incubation with lysozyme by the initial inoculum (represented by dashed line, 1×106 CFU/ml). Data shown are representative of three biological replicates. Error bars represent mean ± standard deviation, **P=0–0012, ***P=0–0008.
The mcr-1 encoded phosphoethanolamine transferase adds a positively charged phosphoethanolamine moiety to the lipid A portion of the bacterial outer membrane component lipopolysaccharide1. This reduces the overall net negative surface charge of the bacteria, likely leading to repulsion of the cationic antibiotic, colistin. The host’s innate immune system employs multiple positively charged antimicrobials to combat bacterial infection. Therefore, we sought to determine if mcr-1 was capable of providing cross-resistance to the cationic host protein lysozyme. We measured survival rates between mcr-1 positive isolates and cured strains in the presence of lysozyme in 25% LB. Strains lacking mcr-1 were killed in the presence of lysozyme while the parental strains were able to grow (Fig. 1C). Specifically, mcr-1 negative strains were 5 to 20-fold more susceptible to multiple concentrations of lysozyme when comparing percent survival.
To our knowledge, this is the first report of mcr-1 conferring cross-resistance to a host antimicrobial. Resistance to the host’s innate immune defenses through mcr-1 could drive plasmid maintenance in strains carrying mcr-1, mcr-2, or other transferable resistance plasmids leading to propagation of colistin resistance.4,5 Therefore, mammals may currently or in the future serve as reservoirs for bacteria harboring transmissible colistin resistance regardless of polymyxin exposure. More studies are needed to determine the broader effect of mobilized colistin resistance and resistance to cationic antimicrobials within the context of the host.
Acknowledgements
D.S.W. is supported by a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease award, VA Merit Award I01 BX002788, and NIH grant AI098800. D.S.W. had full access to all data presented in the study with final responsibility to submit for publication.
Footnotes
Declaration of Interests
The authors declare no competing financial interests.
References
- 1.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. The Lancet Infectious Diseases. 2015. [DOI] [PubMed] [Google Scholar]
- 2.Nordmann P, Lienhard R, Kieffer N, Clerc O, Poirel L. Plasmid-Mediated Colistin-Resistant Escherichia coli in Bacteremia in Switzerland. Clin Infect Dis. 2016;62(10):1322–3. [DOI] [PubMed] [Google Scholar]
- 3.Stoesser N, Mathers AJ, Moore CE, Day NPJ, Crook DW. Colistin resistance gene mcr-1 and pHNSHP45 plasmid in human isolates of Escherichia coli and Klebsiella pneumoniae. The Lancet Infectious Diseases. 2016;16(3):285–6. [DOI] [PubMed] [Google Scholar]
- 4.Nicolet S, Goldenberger D, Schwede T, Page M, Creus M. Plasmid-mediated colistin resistance in a patient infected with Klebsiella pneumoniae. The Lancet Infectious Diseases. 2016;16(9):998–9. [DOI] [PubMed] [Google Scholar]
- 5.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 2016;21(27). [DOI] [PubMed] [Google Scholar]