Abstract
Sideroflexin 1 (Sfxn1) is a mitochondrial serine transporter involved in one-carbon metabolism in blood and cancer cell lines. The expression of other Sfxn homologs varies across tissues implying that each homolog may have tissue-specific functions. RNA databases suggest that among the Sfxns, Sfxn3 may have a specific function in the brain. Here, we systematically analyzed the level, cellular distribution, and subcellular localization of Sfxn3 protein in the developing and adult rodent brain. We found that, in the cortex and hippocampus, Sfxn3 protein level is low at birth but increases during development and remains at a high level in the mature brains. Similarly, in cultured hippocampal neurons, Sfxn3 protein level is low in young neurons but increases as neurons mature. Sfxn3 protein level is much higher in neurons than in astrocytes. Within neurons, Sfxn3 localizes to mitochondria in all major neuronal compartments. Our results establish that Sfxn3 is a mitochondrial protein enriched in neurons wherein it is developmentally expressed. These findings provide a foundation for future research aimed at understanding the functions of Sfxn3 and one-carbon metabolism in neurons.
Keywords: Sideroflexin, Mitochondria, Brain, Neuron, One-carbon metabolism
Introduction
Previous studies of the Sideroflexin (Sfxn) family of genes and specifically the homolog Sideroflexin 3 (Sfxn3) have been few and vary greatly in their focus. The Sfxn family consists of mitochondrial proteins with five homologs in humans. Sfxn1 and Sfxn3 are the main Sfxn proteins responsible for transporting serine into mitochondria during one-carbon metabolism and show the greatest degree of homology (Kory et al. 2018). One-carbon metabolism includes a series of biochemical reactions that involve the transfer of one-carbon groups and byproducts such as NADP+ and dTMP. The enzymes involved in the pathway and the transfer of carbon play critical roles in nucleotide and amino acid metabolism (Ducker and Rabinowitz 2017). The role of Sfxn3 in serine metabolism may be crucial to cancer growth, because in several types of cancer cells serine metabolism is required for the production of purines, and thus for growth and proliferation (Labuschagne et al. 2014). Serine metabolism also contributes to antioxidant defense, which may play a role in cancer (Yang and Vousden 2016).
Sfxn proteins are found throughout the body, but expression of the different homologs is only partially overlapping (Li et al. 2010). Messenger RNA levels of Sfxn3 and Sfxn5 are relatively high in the brain compared to other parts of the body. Sfxn5 expression is high throughout the adult human brain compared to other tissues (Lockhart et al. 2002). A developmental role for Sfxn3 is suggested from a study showing that Drosophila Sfxn3 mutants exhibit abnormal synaptic morphogenesis at the neuromuscular junction (Amorim et al. 2017). Interestingly, Sfxn3 is a downstream target of α-synuclein, and may be involved in the neurodegenerative process in Parkinson’s disease (Amorim et al. 2017). Sfxn3 may be related to neurodegenerative diseases other than Parkinson’s disease, because in the brain of patients with Alzheimer’s disease, there is lower expression of its close homolog, Sfxn1, which may mediate Tau toxicity (Minjarez et al. 2016). Sfxn3 also may relate to neurological dysfunction in that it alters synaptic morphology, although its exact mechanism is unknown (Labuschagne et al. 2014). The aim of the present research was to characterize Sfxn3 expression and subcellular localization in the developing and adult brain. This basic information will provide a framework for future studies of potential roles for this mitochondrial serine transporter protein in neuroplasticity and neurological disorders.
Results and Discussion
Sfxn3 Protein in the Brain at Different Developmental Stages
We began this study by assessing the level of endogenous Sfxn3 protein in young adult rat brains at postnatal day 30 (p30). We used an Sfxn3 antibody that has been validated for its specificity (Atlas Antibodies; and see “Materials and Methods” section), and also described in a previous study (Amorim et al. 2017). Immunoblotting of brain tissue extracts using the Sfxn3 antibody revealed a major protein band at 31–32 kD, the expected molecular mass for rat Sfxn3 (Fig. 1a). Analysis of several brain regions indicated that Sfxn3 protein levels are higher in the cerebral cortex and hippocampus compared to the cerebellum and olfactory bulb (Fig. 1a).
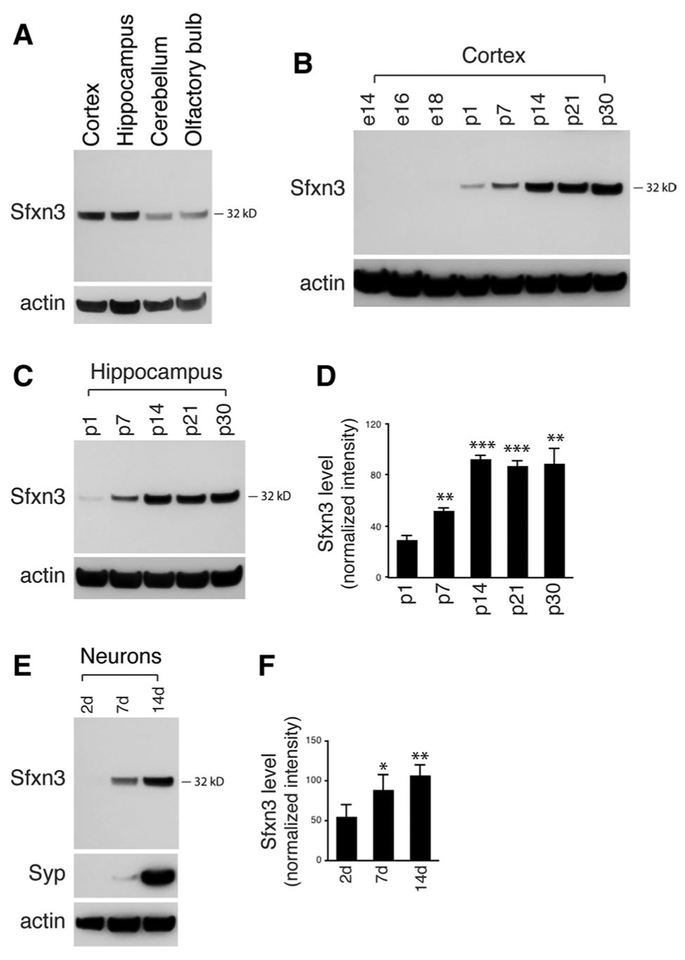
Fig. 1.
Sfxn3 protein level in the brain. a Immunoblot analysis of Sfxn3 protein levels in several areas of the rat brain. The Sfxn3 antibody detects a protein band at 31–32 kDa, the expected molecular mass for rat Sfxn3 protein. The experiment was repeated twice using samples from different rats. b Immunoblot analysis of Sfxn3 protein level in rat cortex from embryonic day 14 (e14) to postnatal day 30 (p30). Sfxn3 protein is undetectable before birth, detectable but low at birth, increases during early postnatal development, and remains at a high level in the young adult brain. The experiment was repeated twice using different sample sets. c, d Immunoblot analysis showing that, similar to cortex, Sfxn3 protein in hippocampus is low at birth, increases postnatally, and is high in the young adult brain. Histogram includes 4 experiments. The values of Sfxn3 protein level represent the Sfxn3 band intensities normalized to the actin band intensities. e, f Immunoblots showing increased Sfxn3 protein level in hippocampal neurons as these neurons mature in cultures. d, days in culture. Histogram includes 3 experiments. In d and f, error bars represent SEM.***p < 0.001, **p < 0.01, *p < 0.05, unpaired t test
We next measured the level of Sfxn3 protein in the cortex at various developmental time points from embryonic day 14 (e14) to p30. We found that Sfxn3 protein was not detectable before birth, was low but detectable at p1, increased rapidly during the first 2 weeks of postnatal development, and remained at a high level in young adult brain (Fig. 1b). Similarly, the hippocampus also exhibited a postnatal surge of Sfxn3 protein expression: its level increased markedly from p1 to p14 and then stayed at a high level (Fig. 1c). Analysis of four biological replicates (number of rats) of hippocampal tissues confirmed that the Sfxn3 protein level at p7 and p14 is significantly higher than at p1 (p7 vs p1, 51.8 ± 2.8 vs 28.8 ± 3.6, p < 0.01; p14 vs p1, 92.5 ± 2.8 vs 28.8 ± 3.6, p < 0.001; Fig. 1d).
We also examined the abundance of Sfxn3 in cultured hippocampal neurons, a well-characterized model system for studies of neuron development (Dotti et al. 1998; Goslin and Banker 1989; Mattson et al. 1988). Similar to the brain tissue extracts, immunoblotting of neuronal cell lysates showed the expected Sfxn3 protein band at 31–32 kD (Fig. 1e). In addition, the Sfxn3 protein levels increased during a 14-day culture period (Fig. 1e, f), suggesting potential roles for Sfxn3 in the process of neurite outgrowth and synaptogenesis that occur during this time period.
Subcellular Localization of Sfxn3 Protein in Embryonic Neurons from Different Brain Regions
We wondered whether our finding of Sfxn3 in hippocampal neurons extended to neurons from other brain regions. We prepared neuronal cultures from three additional brain regions—cortex, midbrain and striatum, and measured Sfxn3 protein level by immunoblots. In neurons of all brain regions examined, we found readily detectable Sfxn3 protein (Fig. 2a).
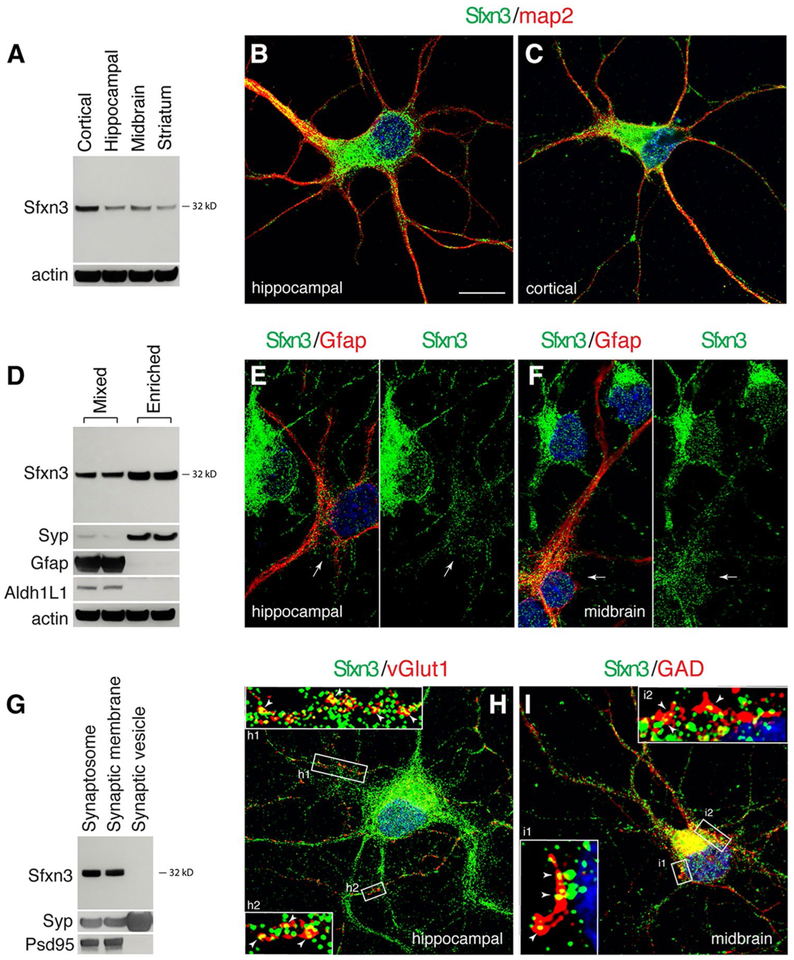
Fig. 2.
Distribution of Sfxn3 protein in neurons. a Immunoblot analysis showing Sfxn3 protein in neurons cultured from several brain regions. b, c, Immunofluorescence images from hippocampal or cortical neuron cultures co-labeled for Sfxn3 (green) and a neuronal marker map2 (red). Sfxn3 labeling is seen throughout neurons. Co-incubating with an Sfxn3 peptide in the immunolabeling nearly completely blocked the Sfxn3 labeling (Supplementary Fig. 1), validating the specificity of the Sfxn3 antibody. d Immunoblots showing higher level of Sfxn3 in neuron-enriched cultures (Enriched) than in neuron-glia mixed cultures (Mixed). Syp, synaptophysin, neuronal synaptic marker; Gfap and Aldh1L1, glial markers. e, f Sample images from hippocampal or midbrain neuron cultures co-labeled for Sfxn3 (green) and the glial marker Gfap (red). Sfxn3 fluorescent intensity is noticeably higher in neurons than in Gfap-labeled glial cells (white arrows). g Immunoblots showing the presence of Sfxn3 protein in synapses but not in synaptic vesicles. Syp, synaptophysin, a synaptic vesicle protein; Psd95, a synaptic protein. h Sample fluorescent image of a hippocampal neuron co-labeled for Sfxn3 (green) and vGlut1 (red). vGlut1 is a marker for excitatory neurons. Zoomed- in views (h1, h2) showing Sfxn3 immunolabeling in close proximity to vGlut1 along neurites. Note that some Sfxn3 green puncta overlap directly with vGlut1 red puncta (white arrowheads); others locate in the shaft of the adjacent neurites. i Sample image of a midbrain neuron co-labeled for Sfxn3 (green) and an inhibitory neuron marker GAD (red). Zoomed-in views in box i1 and i2 showing Sfxn3 immunolabeling in close proximity to GAD labeling on the neuronal soma. Some Sfxn3 green puncta overlap with GAD red puncta (white arrowheads); others are seen in the adjacent region of the soma. Scale in b = 10 μm applies to b, c, e, f, h, i. Additional examples of immunofluorescent images are shown in Supplementary Fig. 2
To visualize distribution patterns of Sfxn3 in neurons, we imaged neurons that were co-labeled with the Sfxn3 antibody and an antibody to a neuronal marker, map2. We observed bright Sfxn3 immunofluorescence labeling in all major compartments of the neurons, as shown in images of a hippocampal neuron (Fig. 2b), a cortical neuron (Fig. 2c), and a midbrain neuron (Supplementary Fig. 2A). Co-incubating a Sfxn3 peptide in the immunolabeling protocol completely eliminated the Sfxn3 labeling (Supplementary Fig. 1), confirming the specificity of the Sfxn3 antibody.
We next asked whether Sfxn3 was preferentially expressed by neurons or glia, taking advantage of the primary neuronal cultures that contain a mixture of neurons and astrocytes. We compared the levels of Sfxn3 in the mixed cultures to the AraC-treated neuron-enriched cultures (see “Materials and Methods” section). The neuron-enriching efficacy of AraC was demonstrated by the robustly increased relative amount of neuronal protein, synaptophysin, and by the disappearance of the astrocytic proteins, Gfap and Aldh1L1 (Fig. 2d; Cahoy et al. 2008). Notably, the Sfxn3 protein level was also increased in the neuron-enriched cultures, although the magnitude of increase was not as great as that for synaptophysin (Fig. 2d). When we immunolabeled the neuron-astrocyte mixed cultures with the Sfxn3 antibody and the Gfap antibody, we found that Sfxn3 immunoreactivity in Gfap-labeled glial cells—while not totally absent— was visibly and consistently lower than in neurons (Fig. 2e, f; Supplementary Fig. 2B).
One of the developmental events taking place in young postnatal brains as well as neurons grown in cultures for 7–14 days is the establishment of synapses. We evaluated the levels of Sfxn3 in the synapses of neurons. Immunoblot analysis of various synaptic fractions prepared from rat brain tissues showed that Sfxn3 protein is not associated with synaptic vesicles but is present in total synaptosomes and in a synaptic membrane fraction (Fig. 2g). We next imaged two distinct subtypes of synapses using an antibody against either the excitatory synapse marker, vesicular glutamate transporter-1 (vGlut1), or the inhibitory synapse marker, glutamate decarboxylase (GAD). We observed that in both types of synapses, Sfxn3-immunolabeled puncta frequently overlapped with puncta of vGlut1 or GAD labeling (Fig. 2h, i; Supplementary Fig. 2C, D). Sfxn3-immunolabeled puncta also were common in the neurites or cell soma adjacent to the vGlut1 or GAD labeling, respectively. These results suggest that Sfxn3 protein is present in most neuronal compartments, including the synapses of the two major subtypes of neurons in the brain.
Sfxn3 is a Mitochondrial Protein in Neurons
In non-neuronal cells, Sfxn1 plays an important role in one-carbon metabolism by transporting serine into mitochondria (Kory et al. 2018). We therefore determined if Sfxn3 is located in mitochondria in neurons. First, we isolated mitochondria from cultured hippocampal neurons and compared Sfxn3 protein in purified mitochondria to mitochondria-depleted cytosol. The purity of the purified mitochondria was validated using an antibody against the mitochondrial protein VDAC (Fig. 3a), and the lack of mitochondria in the mitochondria-depleted cytosol was demonstrated by the absence of VDAC but the presence of the cytosolic protein, Hsp90 (Fig. 3a). Sfxn3 protein was substantially enriched in the purified mitochondria but absent in the mitochondria-depleted cytosol (Fig. 3a).
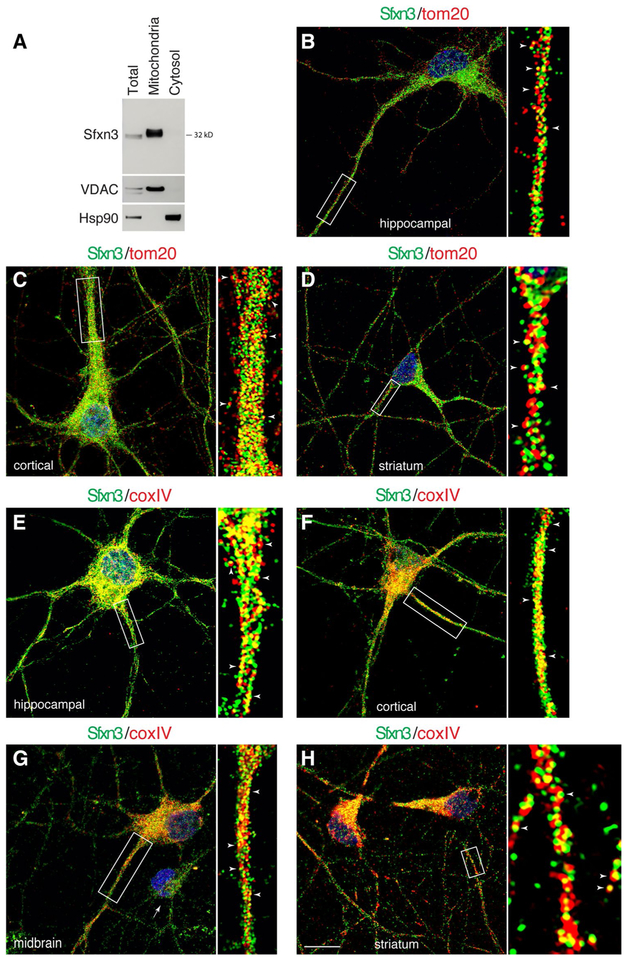
Fig. 3.
Sfxn3 is a mitochondrial protein. a Immunblots show that Sfxn3 protein is exclusively present in mitochondria isolated from neurons (cultured hippocampal neurons) but is absent in mitochondria-depleted cytosol. VDAC, a mitochondrial protein; Hsp90 is a cytosolic protein. b–d Sample images of neurons from different brain regions co-labeled for Sfxn3 (green) and a mitochondrial marker tom20 (red). White arrowheads in the enlarged views depict overlapped Sfxn3 and tom20 labeling. e–h Sample images of neurons from different brain regions co-labeled for Sfxn3 (green) and another mitochondrial marker coxIV (red). White arrowheads in the enlarged views depict overlapped Sfxn3 and coxIV labeling. White arrow in g points to a small low-coxIV-expressing neuron. Scale in h = 10 μm applies to all images. Additional examples of immunofluorescent images are shown in Supplementary Fig. 3
Next, we co-labeled cultured neurons with the Sfxn3 antibody and an antibody against a mitochondrial outer membrane protein, tom20 (Endo and Kohda 2002; Pagliarini et al. 2008). Using confocal microscopy with Airyscan (Zeiss 880, with ~ 120 nm xy and ~ 400 nm z resolution), we observed that Sfxn3-immunolabeled puncta were closely apposed to or colocalized with tom20-labeled mitochondria (Fig. 3b–d; Supplementary Fig. 3A–C). We also immunolabeled the neurons with Sfxn3 and an antibody against an inner mitochondrial membrane protein, cytochrome C oxidase complex IV (coxIV; Pagliarini et al. 2008). We easily observed Sfxn3-immunolabeled puncta colocalized with coxIV-immunolabeled puncta (Fig. 3e–h; Supplementary Fig. 3D,E). We also observed that, unlike tom20, coxIV was not expressed at the same level among all neurons based on coxIV immunofluorescence labeling intensity, and this differential expression was particularly evident among the midbrain neurons (arrow in Fig. 3g; Supplementary Fig. 3E). Nonetheless, Sfxn3 expression and distribution appeared to be similar in neurons expressing low or high levels of coxIV (Fig. 3g; Supplementary Fig. 3E).
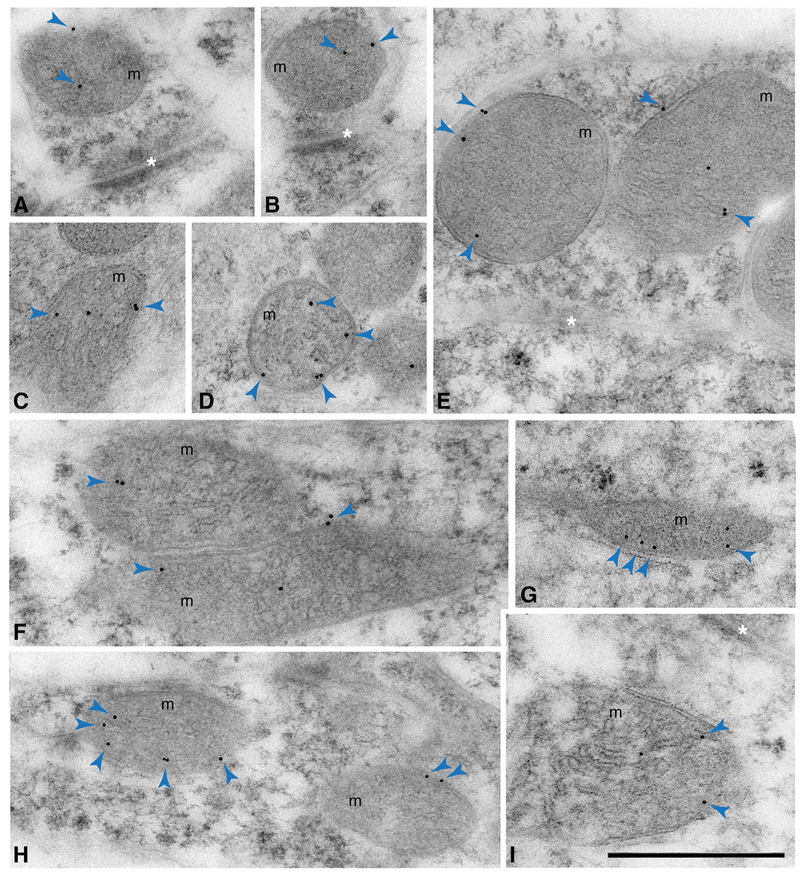
Finally, we further examined the ultrastructural localization of Sfxn3 protein by immunogold electron microscopic analysis of the mouse hippocampus, focusing on neurons in the CA1 and CA3 areas. We found that Sfxn3-associated immunogold particles were almost exclusively located on or in mitochondria (Fig. 4a–i). Taken together, our results demonstrate that Sfxn3 is a mitochondrial protein in neurons.
Fig. 4.
Subcellular localization of Sfxn3 in neurons of the hippocampus. Immunogold labeled-Sfxn3 (arrowheads) is found in or on mitochondria of hippocampal neurons. a–e Presynaptic terminals; f–i postsynaptic dendrites. Examples are from hippocampal CA1 stratum radiatum (a–c, f, g, i) or CA3 stratum lucidum/pyramidale (d, e, h). d An Sfxn3-labeled mitochondrion in a mossy terminal. e Sfxn3 labeled mitochondria in an inhibitory presynaptic terminal on a soma. Asterisks indicate the synaptic clefts of synapses including excitatory synapses with asymmetric synaptic densities (a, b, i) and an inhibitory synapse with symmetric synaptic densities (e). m, mitochondria. Scale bar = 500 nm
Here we characterized a member of the Sfxn family, Sfxn3 protein, in neurons. Sfxn3 protein levels increase during brain development in vivo and as neurons grow axons and dendrites and form synapses in culture. Sfxn3 protein levels are much higher in neurons compared to astrocytes. A previous report by Amorim et al. showed Sfxn3 in mitochondria isolated from mouse brains (Amorim et al. 2017). Our finding is consistent with Amorim et al.’s report—although we specifically demonstrated mitochondrial Sfxn3 in neurons and further visualized the distribution and subcellular localization of the protein by immunofluorescence confocal light microscopy and immunogold electron microscopy. Another member of the Sfxn family, Sfxn1, has been shown to be essential for one-carbon metabolism in human blood and cancer cell lines. Knowledge of the one-carbon metabolism pathway in neurons is sparse. Our study does not answer the questions of whether Sfxn3 protein plays a role in neuronal one-carbon metabolism or whether Sfxn3 plays critical roles in regulating neurite outgrowth and synaptogenesis. Nevertheless, we envision that the characterization of neuronal Sfxn3 described here will inform future studies aimed at elucidating the functions of Sfxn3 in normal brains and its dysfunction in potential involvement in neurological disorders.
Materials and Methods
Animals
All animal procedures were approved by the NIA Animal Care and Use Committee and complied with the NIH Guide for Care and Use of Laboratory Animals. Timed pregnant female Sprague-Dawley rats were used as the source of embryonic brain tissues, and to establish cultures of cortical, hippocampal, midbrain, and striatum neurons. Postnatal rats (postnatal day 1 to p30) of either sex were used as the source of cortical, hippocampal, cerebellar, and olfactory bulb tissues. Hippocampi of 8-week-old male C57BL/6 mice from a previous study (Sun et al. 2011) were used for immunogold labeling.
Antibodies
Commercial antibodies against the following proteins were used: Sfxn3 rabbit polyclonal antibody (Atlas Antibodies, Cat# HPA008028, RRID: AB_1079938; 1:800 immunoblotting, 1:100 immunofluorescence, 1:20 immunogold), Synaptophysin mouse monoclonal antibody (Cat# SVP38, RRID: AB_2315393; 1:2500, immunoblotting), Psd95 mouse monoclonal antibody (Cat# 7E3–1B8; RRID: AB_212825; 1:2000, immunoblotting), Gfap mouse monoclonal antibody (Cat# 3670, RRID: AB_561049; 1:1000 immunoblotting; 1:2000, immunofluorescence), Aldh1L1 rabbit polyclonal antibody (Cat# 278003, RRID: AB_2620010; 1:1000, immunoblotting), map2 mouse monoclonal antibody (Cat# M9942, RRID: AB_477256; 1:2000, immunofluorescence), VDAC rabbit monoclonal antibody (Cat#4661, RRID: AB_10557420; 1:1000 immunoblotting), tom20 mouse monoclonal antibody (Cat# WH0009804M1, RRID: AB_1843992; 1:1000, immunofluorescence), coxIV mouse monoclonal antibody (Cat# 200147; RRID: AB_2722715; 1:1000, immunofluorescence), Hsp90 rabbit monoclonal antibody, (Cat#4877; RRID: AB_2233307, 1:1000 immunoblotting), vGlut1 guinea pig polyclonal (Cat# 135304, RRID: AB_887878; 1:1000, immunofluorescence), GAD2 guinea pig polyclonal (Cat# 198104, RRID: AB_10557995; 1:1000, immunofluorescence). Sfxn3 blocking peptide was from Atlas Antibodies (PrEST Antigen SFXN3, #APrEST71783).
Primary Culture of Rat Hippocampal, Cortical, Midbrain, and Striatum Neurons
Cultures of cortical, hippocampal, midbrain, and striatum neurons were prepared from embryonic day 18 rat brains using the protocol described (Mattson et al. 1989; Kaech and Banker 2006; Yao et al. 2017). Dissociated neurons were seeded at a density of ~ 150 cells/mm2 for cortical or hippocampal neurons, and ~ 300 cells/mm2 for midbrain or striatum neurons. All neurons were grown in Neurobasal medium supplemented with B27 (Invitrogen). For immunoblotting, the neurons were grown in polylysine (0.1 mg/ml)-coated plastic dishes. For immunofluorescence, the neurons were grown on polylysine (1 mg/ml)-coated glass coverslips (no. 1.5). Polylysine was from Sigma (P2636). Neuron-enriched cultures were prepared following a protocol (Tushev et al. 2018) with minor modifications. Briefly, 1 day after initial cell seeding, 2.5 μM of cytosine β-D-arabinofuranoside (AraC) was added. Forty-eight hours later, AraC-containing medium was replaced by the growth medium composed of 50% fresh neurobasal medium with B27, 25% of conditioned medium from cortical neurons, and 25% of conditioned medium from glial cells.
Immunoblotting
Rat brains or cultured neurons were lysed in RIPA buffer (Thermo Scientific, Cat# 89900) containing protease inhibitors (Thermo Scientific, Cat# 78444). The lysed tissues or cells were centrifuged at 10,000×g for 15 min at 4 °C. The supernatant was collected and the amount of total proteins was estimated with a Pierce BCA protein assay kit (Pierce Biotechnology). Protein samples were separated by 4–20% Bis–Tris SDS-PAGE and transferred to nitrocellulose membranes. Following incubation with blocking buffer (5% dry milk and 0.05% Tween20 in PBS), the membranes were incubated overnight at 4 °C in the blocking buffer containing the primary antibody being tested. The membranes were then washed (0.1% Tween20 in PBS) and incubated with appropriate peroxidase-conjugated secondary antibodies. The proteins were visualized using a chemiluminescence kit from Kindle Biosciences. The intensity of protein bands was analyzed using imageJ software.
Mitochondria were isolated using a mitochondria isolation kit for cultured cells (Thermo Scientific, #89874) following the manufacturer’s instructions. Approximately 108 cultured neurons were used for each sample. Mitochondria-depleted cytosol was concentrated 18-fold and then 25-fold using an Amicon centrifugal filter (10 kD). Twenty micrograms of protein from each sample were analyzed by immunoblots.
Immunocytochemistry and Fluorescence Microscopy
Neurons grown on glass coverslips were washed with PBS, fixed with 4% paraformaldehyde and 4% sucrose in PBS, and permeabilized with 0.2% Triton X-100 in PBS. After blocking with 10% bovine serum albumin in PBS, neurons were incubated with primary antibodies followed by species-appropriate secondary antibodies conjugated with Alexa Fluor-488 or −568. Coverslips were mounted on slides using ProLong anti-fade mounting medium.
Images were acquired with an Apochromat 63x/1.4 numerical aperture objective lens on a Zeiss LSM 880 microscope with Airyscan (Carl Zeiss). For each antibody labeling, the image acquisition settings were kept the same between different experiments. The brightness, contrast, and levels of the images were adjusted in Adobe Photoshop, and compiled in Adobe Illustrator. No additional digital image processing was performed. Control cells omitting the primary antibody showed no fluorescence labeling.
Immunogold Electron Microscopy
Postembedding immunogold labeling was performed as described previously (Petralia and Wenthold 1999; Petralia et al. 2010; Sun et al. 2011; Yao et al. 2015; Rivell et al. 2019). Following perfusion with 4% PFA plus 0.5% glutaraldehyde, mouse brain tissue was cryoprotected and frozen in a Leica CPC and then processed for freeze-substitution in Lowicryl HM-20 resin in a Leica AFS. Thin sections from two mice were incubated in 10% normal goat serum, then overnight in primary antibody, and followed by incubation for 1 h with 10-nm immunogold. Sections were stained with uranyl acetate and lead citrate. Control immunogold labeling of sections omitting the primary antibody showed only rare gold particles.
Presentation of Data and Statistics
All graphs were produced using KaleidaGraph (Synergy) software. Statistical comparisons were calculated using the unpaired Student’s t test. All results are expressed as mean ± SEM.
Supplementary Material
Acknowledgements
We thank Dr. Fred E. Indig for assistance in confocal Airyscan imaging. We thank Drs. Lin Lin and Dax Hoffman for mouse tissue for immunogold labeling. This study was supported by the Intramural Research Programs of the National Institutes of Health, National Institute on Aging; and the National Institutes of Health, National Institute on Deafness and Other Communication Disorders. The Advanced Imaging Core code is ZIC DC000081.
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s12017-019-08553-7) contains supplementary material, which is available to authorized users.
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiiations.
References
- Amorim IS, Graham LC, Carter RN, Morton NM, Hammachi F, Kunath T, et al. (2017). Sideroflexin 3 is an α-synuclein-dependent mitochondrial protein that regulates synaptic morphology. Journal of Cell Science, 130, 325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, et al. (2008). A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. Journal of Neuroscience, 28, 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotti CG, Sullivan CA, & Banker GA (1998). The establishment of polarity by hippocampal neurons in culture. Journal of Neuroscience, 8, 1454–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducker GS, & Rabinowitz JD (2017). One-Carbon Metabolism in Health and Disease. Cell Metabolism, 25, 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, & Kohda D (2002). Functions of outer membrane receptors in mitochondrial protein import. Biochimica et Biophysica Acta, 1592, 3–14. [DOI] [PubMed] [Google Scholar]
- Goslin K, & Banker G (1989). Experimental observations on the development of polarity by hippocampal neurons in culture. Journal of Cell Biology, 108, 1507–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S, & Banker G (2006). Culturing hippocampal neurons. Nature Protocols, 1, 2406–2415. [DOI] [PubMed] [Google Scholar]
- Kory N, Wyant GA, Prakash G, de Bos J, Bottanelli F, Pacold ME, et al. (2018). SFXN1 is a mitochondrial serine transporter required for one-carbon metabolism. Science, 362, 6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuschagne CF, van den Broek NJ, Mackay GM, Vousden KH, & Maddocks OD (2014). Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep, 7, 1248–1258. [DOI] [PubMed] [Google Scholar]
- Li X, Han D, Kin Ting Kam R, Guo X, Chen M, Yang Y, et al. (2010). Developmental expression of sideroflexin family genes in Xenopus embryos. Developmental Dynamics, 239, 2742–2747. [DOI] [PubMed] [Google Scholar]
- Lockhart PJ, Holtom B, Lincoln S, Husseym J, Zimprich A, Gasser T, et al. (2002). The human sideroflexin 5 (SFXN5) gene: Sequence, expression analysis and exclusion as a candidate for PARK3. Gene, 285, 229–237. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Dou P, & Kater SB (1988). Outgrowth-regulating actions of glutamate in isolated hippocampal pyramidal neurons. Journal of Neuroscience, 8, 2087–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Murrain M, Guthrie PB, & Kater SB (1989). Fibroblast growth factor and glutamate: Opposing roles in the generation and degeneration of hippocampal neuroarchitecture. Journal of Neuroscience, 9, 3728–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minjarez B, Calderón-González KG, Rustarazo ML, Herrera-Aguirre ME, Labra-Barrios ML, Rincon-Limas DE, et al. (2016). Identification of proteins that are differentially expressed in brains with Alzheimer’s disease using iTRAQ labeling and tandem mass spectrometry. Journal of Proteomics, 139, 103–121. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, et al. (2008). A mitochondrial protein compendium elucidates complex I disease biology. Cell, 134, 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia RS, Wang YX, Hua F, Yi Z, Zhou A, Ge L, et al. (2010). Organization of NMDA receptors at extrasynaptic locations. Neuroscience, 167, 68–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia RS, & Wenthold RJ (1999). Immunocytochemistry of NMDA receptors. Methods in Molecular Biology, 128, 73–92. [DOI] [PubMed] [Google Scholar]
- Rivell A, Petralia RS, Wang YX, Clawson E, Moehl K, Mattson MP, et al. (2019). Sonic hedgehog expression in the postnatal brain. Biology Open. 10.1242/bio.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Maffie JK, Lin L, Petralia RS, Rudy B, & Hoffman DA (2011). DPP6 establishes the A-type K(+) current gradient critical for the regulation of dendritic excitability in CA1 hippocampal neurons. Neuron, 71, 1102–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tushev G, Glock C, Heumüller M, Biever A, Jovanovic M, & Schuman EM (2018). Alternative 3′ UTRs modify the localization, regulatory potential, stability, and plasticity of mrnas in neuronal compartments. Neuron, 98, 495–511. [DOI] [PubMed] [Google Scholar]
- Yang M, & Vousden KH (2016). Serine and one-carbon metabolism in cancer. Nature Reviews Cancer, 16, 650–662. [DOI] [PubMed] [Google Scholar]
- Yao PJ, Manor U, Petralia RS, Brose RD, Wu RT, Ott C, et al. (2017). Sonic hedgehog pathway activation increases mitochondrial abundance and activity in hippocampal neurons. Molecular Biology of the Cell, 28, 387–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao PJ, Petralia RS, Ott C, Wang YX, Lippincott-Schwartz J, & Mattson MP (2015). Dendrosomatic sonic hedgehog signaling in hippocampal neurons regulates axon elongation. Journal of Neuroscience, 35, 16126–16141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.