Abstract
Antimicrobial peptides are essential components of innate immune systems that protect hosts from infection. They are also useful candidates for developing a new generation of antibiotics to fight antibiotic-resistant pathogens. Human innate immune peptide LL-37 can inhibit biofilm formation, but suffers from high cost due to a long peptide length and rapid protease degradation. To improve the peptide, we previously identified the major active region and changed the peptide backbone structure. This study designed two families of new peptides by altering peptide side chains. Interestingly, these peptides displayed differential potency against various ESKAPE pathogens in vitro and substantially reduced hemolysis. Further potency test in vivo revealed that 17tF-W eliminated the burden of methicillin-resistant Staphylococcus aureus (MRSA) USA300 in both mouse-embedded catheters and their surrounding tissues. In addition, peptide treatment suppressed the level of chemokine TNFα, and boosted the levels of chemokines MCP-1, IL-17A and IL-10 in the surrounding tissues of the infected catheter embedded in mice. In conclusion, we have designed a set of new LL-37 peptides with varying antimicrobial activities, opening the door to potential topical treatment of infections involving different drug-resistant pathogens.
Keywords: Biofilms, cathelicidin, catheter, cytotoxicity, LL-37, peptide design
Graphical Abstract

1. INTRODUCTION
The rising crisis of antibiotic resistance is a serious health-related issue because untreatable infections lead to a high death rate [1–4]. The ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) can escape the killing of multiple drugs [5]. In addition, these pathogens can form biofilms covered with extracellular matrices. Conventional antibiotics are not effective in penetrating such biofilm matrices [6–7]. In contrast, cationic peptides designed based on naturally occurring antimicrobial peptides (AMPs) are effective [818]. According to the antimicrobial peptide database (APD; http://aps.unmc.edu/AP), such peptides are usually small (10–50 amino acids) and cationic (+1 to +7), ideal to target pathogens with anionic membranes [19,20]. Membrane damage is difficult to repair, thereby decreasing the likelihood of bacterial resistance [21–23].
Humans also produce innate immune peptides to fend off invading pathogens. Over 100 human AMPs have been documented [24]. The major members include lysozyme, defensins, histatins, cathelicidin, RNases, dermcidin, and antimicrobial chemokines [25–30]. Unlike defensins [25], there is only one cathelicidin gene in the human genome. The precusors of cathelicidins share a highly conserved N-terminal “cathelin” domain, but the C-terminal region encodes a highly variable antimicrobial region [31]. Interestingly, the same human cathelicidin precursor can be cleaved into different antimicrobial peptides, such as LL-37, ALL-38 and TLN-58, depending on the type of tissues and processing proteases [32–34]. LL-37 is one of the widely studied host defense peptides. It displays direct inhibitory activity against bacteria, viruses, fungi and parasites [18,21,27]. In addition, LL-37 is known to regulate immune response by interaction with various cell receptors [35–37].
The antimicrobial significance of LL-37 to human health is firmly established. It protects human urinary tract from infection [38]. Mice are more susceptible to infections when the cathelicidin gene is knocked out [39]. To meet the challenge of antimicrobial resistance, there is a great interest in developing human LL-37 into novel antibiotics. However, LL-37, with 37 amino acids, is costly to make and the peptide can be rapidly degraded by proteases. Multiple labs tried to identify the active regions of LL-37 using both peptide library and structure-based approaches [40–50]. For instance, Nell and colleagues used the library approach to identify P60.4, a variant of IG-24 (24-residue) corresponding to residues 13–36 of LL-37 [49]. In 2018, they reported SAAP-148, a modified version of this peptide with reduced plasma binding [13]. However, SAAP-148 consists of natural amino acids and can be cleaved by host and pathogen proteases. In 2006, we used a structural approach to identification of the major antimicrobial region of LL-37 corresponding to residues 17–32 [50]. GF-17 was obtained by appending a glycine in front of residues 17–32 (i.e., the FK-16 peptide). GF-17 showed a broad antimicrobial activity with superior antibiofilm activity to LL-37 [12]. To improve peptide stability, we subsequently engineered GF-17 into 17BIPHE2, which retains bacterial killing even in the presence of chymotrypsin [10]. 17BIPHE2 showed potent antibiofilm activity against S. aureus USA300. When combined with antibiotics, 17BIPHE2 demonstrated even better activity against the preformed biofilms of P. aeruginosa [11]. However, it is not clear how attachment of aliphatic or aromatic moieties to the aromatic ring of phenylalanines of the peptide template will impact peptide activity and toxicity. The GF-17d3 peptide template we used here retained two (F17 and F27) out of the four phenalalanines of LL-37 and contains three D-amino acids at positions 20, 24, and 28 (Table 1 as named in LL-37). This relationship between LL-37 and GF-17d3 is illustrated in the graphic for the Table of Contents. In this study, we designed two families of protease-resistant peptides to address this question by replacing F17 and/or F27 with various analogs (Table 1). Here we report differential effects of the newly designed LL-37 peptides on each of the drug-resistant ESKAPE pathogens in vitro. We also report in vivo efficacy for one of the peptides in a murine catheter-associated biofilm model against methicillin-resistant Staphylococcus aureus (MRSA). Our results indicate that the newly designed peptides not only kill bacteria but also regulate immune response in a catheter biofilm model.
Table 1.
Amino acid sequences of the LL-37 derived peptides
| Peptide | Sequence | Notes |
|---|---|---|
| LL-37 | LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES | FK-16 underlined; |
| GF-17 | GFKRIVQRIKDFLRNLV-amide | G+FK-16=GF-17 |
| GF-17d31 | GFKRlVQRlKDFlRKLV-amide | I20L and I24L; l is a D amino acid; |
| Group 1 | ||
| 17tF-W | GX1KRlVQRlKDWlRKLV-amide | l is a D amino acid; X1=4-t-butylphenylalanine |
| 17mF-W | GX1KRlVQRlKDWlRKLV-amide | l is a D amino acid; X1=4-methylphenylalanine |
| 17W2 | GX1KRlVQRlKDWlRKLV-amide | l is a D amino acid |
| Group 2 | ||
| 17tF2 | GX1KRlVQRlKDWlRKLV-amide | l is a D amino acid; X1 = X2 =4-t-butylphenylalanine |
| 17B-tF | GX1KRlVQRlKDWlRKLV-amide | l is a D amino acid; X1 = biphenylalanine; X2 = 4-tbutylphenylalanine |
| 17BIPHE2 | GX1KRlVQRlKD X2lRKLV-amide | l is a D amino acid; X1 = X2 = biphenylalanine |
First reported in ref [50].
2. RESULTS
2.1. Role of the fused aromatic and aliphatic moieties in bacterial killing
Our previous simultaneous 96-well plate activity and stability screening led to the identification of the GF-17D3 template (Table 1) [50], which is active against E. coli even in the presence of chymotrypsin but not active against S. aureus USA300 [10]. This template was used to design two families of peptides in Table 1. All the peptides are of high quality as indicated by a single HPLC peak (Supporting Fig. S1). Group 1 peptides include 17tF-W (also called HC10) [51], 17mF-W, and 17W2. In these peptides, F27 of GF-17D3 was replaced with W, while F17 was changed to 4-t-butyl-phenylalanine (tF), 4-methyl- phenylalanine (mF), and tryptophan (W), respectively. The second group includes 17tF2, 17B-tF, and 17BIPHE2. While 17tF2 possesses a 4-t-butyl-phenylalanine at both positions 17 and 27, the same two positions are occupied by biphenylalanines (B) in the case of 17BIPHE2 [10]. 17B-tF has a biphenylalanine at position 17 and a 4-t-butyl-phenylalanine at position 27 (Table 1). These two groups of peptides enabled us to compare the effects of an aliphatic vs. aromatic moieties attached to the aromatic ring of phenylalanines on peptide properties.
The antimicrobial activities of these peptides were evaluated by using the microdilution method (Table 2). GF-17 showed higher activity than LL-37 against these ESKAPE pathogens. Compared to GF-17, 17W2 showed increased minimal inhibitory concentrations (MIC) against most of the pathogens. Substituting F17 with mF17 did not gain activity, either. Nevertheless, 17tF-W, with a change of F17 to tF17, showed enhanced activity against E. cloacae (MIC 3.1 μM). Moreover, 17tF-W retained the MIC values of 17BIPHE2 against E. faecium, S. aureus, and P. aeruginosa, although it became 2–4 fold less active against K. pneumoniae and A. baumannii (Table 2).
Table 2.
Minimal inhibitory concentrations (μM) of the designed peptides against the ESKAPE pathogens
| Peptide | E 1 | S | K | A | P | E | HL50 |
|---|---|---|---|---|---|---|---|
| LL-37 2 | 6.25 | >50 | 6.25 | >50 | >50 | 50 | 170 |
| GF-17 | 3.1 | 3.1 | 3.1 | 3.1 | 12.5 | 3.1 | 180 |
| 17tF-W | 3.1 | 3.1 | 12.5 | 6.25 | 6.25 | 3.1 | >440 |
| 17mF-W | 12.5 | 12.5 | 25 | 6.25 | 25 | 6.25 | >440 |
| 17W2 | 6.25 | 6.25 | 6.25 | 12.5 | 25 | 3.1 | >440 |
| 17tF2 | 6.25 | 3.1 | 12.5 | 3.1 | 6.25 | 6.25 | >440 |
| 17B-tF | 6.25 | 3.1 | 12.5 | 6.25 | 6.25 | 6.25 | >440 |
| 17BIPHE2 | 3.1 | 3.1 | 3.1 | 3.1 | 6.25 | 6.25 | 180 |
E: E. faecium ATCC 51559; S, S. aureus USA300; K, K. pneumoniae ATCC 13883; A, A. baumannii B28–16, P, P. aeruginosa PAO1; E, E. cloacae B2366–12.
MIC were determined in 10% tryptic soy broth (TSB).
We also compared the antibacterial activity of 17tF2, 17B-tF, and 17BIPHE2. Interestingly, these peptides showed identical antimicrobial activity against MRSA, P. aeruginosa, and E. cloaca, indicating these appended aliphatic and aromatic portions are equivalent. In the case of 17B-tF, there was a two-fold increase in the MIC against E. faecium and A. baumannii compared to17BIPHE2. In addition, the activity of both 17tF and 17B-tF against K. pneumoniae was four fold less than 17BIPHE2, indicating that the appended aromatic rings are more important to kill this pathogen.
We then made a comparison between the two designed peptide groups (Table 1). When biphenylalanines were replaced with tryptophans, peptide 17W2 showed reduced activity against the ESKAPE pathogens except for E. cloaca (last E in Table 2). Such a difference may originate from the aromatic ring differences. Biphenyl, being entirely hydrophobic, might be able to better interdigitate bacterial membranes than the amphipathic indole ring of W. Also, aromatic side chains of both 17W2 and 17BIPHE2 appeared to be important to inhibit K. pneumoniae because those peptides with an aliphatic portion showed decreased activity (Table 2). In the case of S. aureus USA300, however, the appended aromatic portion did not show any advantage over the aliphatic portion as we observed the same MIC for the group 2 peptides (Table 2). Indeed, all these peptides are active against Staphylococcal clinical strains, including S. aureus Newman, Mu50, SA546, and UAMS (Table 3). While 17mF-W and 17W2 showed moderate activity (MIC up to 25 μM), the other four peptides were more potent (MIC 3.1–6.2 μM). To further understand the differences of the appended aromatic and aliphatic portions, we also compared the killing kinetics of the group 2 peptides against S. aureus USA300 by colony counting. While 17B-tF and 17BIPHE2 showed identical killing, 17tF2 was more potent at 120 min (Fig. S2), indicating preference of an aliphatic portion at position 17 for MRSA killing. The preference of an aliphatic moiety attached to F17 perhaps explains why 17tF-W has broad-spectrum activity against the ESKAPE pathogens similar to 17BIPHE2.
Table 3.
Antimicrobial activity of the designed peptides against clinical strains of S. aureus
| Peptide | Minimal inhibitory concentration (MIC, μM) | |||
|---|---|---|---|---|
| S. aureus Newman | S. aureus Mu50 | S. aureus SA546 | S. aureus UAMS-1 | |
| LL-371 | >50 | >50 | NA | >50 |
| GF-171 | 1.6 | 1.6 | NA | 3.1 |
| 17tF-W | 3.1 | 6.25 | 3.1 | 3.1–6.25 |
| 17mF-W | 25 | 25 | 12.5 | 12.5 |
| 17W2 | 12.5 | 12.5 | 6.25 | 6.25–12.5 |
| 17tF2 | 3.1 | 6.25 | 3.1 | 3.1 |
| 17B-tF | 6.25 | 3.1–6.25 | 3.1 | 3.1 |
| 17BIPHE2 | 3.1 | 3.1–6.25 | 3.1 | 3.1 |
Data from ref. [12] determined in 25% TSB. NA: not assayed.
2.2. Peptide hydrophobic side chain modification reduced hemolytic activity
We then compared the hemolytic ability of the two groups of designer peptides (Fig. 1). 17BIPHE2 showed a 50% hemolytic concentration (HL50) of ~180 μM similar to that of GF-17 or LL-37 (Table 2). Appending aliphatic sidechains to the phenylalanine ring, however, substantially reduced the hemolytic ability of both 17tF2 and 17B-tF (HL50 >440 μM) in Group 2. When the biphenylalanine at position 27 was changed to Trp, there was a substantial decrease in hemolysis for Group 1 peptides (less than 20% at 440 μM). To shed light on this, we also measured the HPLC retention times of these peptides on a reverse-phase C8 symmetry column. Notably, there is a logarithmic correlation between hemolysis at 220 μM and the peptide retention time (correlation coefficient 0.9 in Table S1): the longer the retention time (proportional to hydrophobicity), the more toxic the peptide. Taken together, our antibacterial and hemolytic assays identified an excellent peptide 17tF-W with a desired combination: an aliphatic moiety attached to F17 for antimicrobial potency and an aromatic Trp at position 27 for reduced hemolysis.
Figure 1.
Cytotoxicity of the peptides to human red blood cells (hRBCs). Changes of peptide side chains can drastically reduce their hemolysis.
We also compared the toxic effects of 17tF-W with 17BIPHE2 on other human cell lines, including skin, kidney and macrophages. Interestingly, these two peptides showed similar LC50 values (the peptide concentration that caused 50% cell death) (Table S2), indicating the basis for toxicity to hRBCs differs from that to other human cells. These results underscore the importance of the use of different cell lines for a more complete understanding of peptide toxicity.
2.3. Effects of proteases on peptide stability
The stability of peptides to proteases is regarded as a critical factor to achieve their antimicrobial task in vivo. Although GF-17 is active against various pathogens (Table 1), it can be rapidly degraded by proteases [10]. We compared the band intensity of newly designed peptides on SDS-PAGE after 24 h incubation with various proteases at 37°C. They are all similar to 17BIPHE2 in size at ~ 2 kD (Fig. S3A). It is clear that both 17tF-W and 17mF-W were stable in the presence of chymotrypsin, S. aureus V8 protease, or fungal proteinase K (Fig. 2A). Likewise, 17tF2, 17B-tF, and 17W2 were also stable similar to 17BIPHE2 (Fig. S3), although they were cleaved by trypsin. To substantiate the stability of 17tF-W, we also evaluated the antiMRSA activity of the peptide after incubation with chymotrypsin for one hour followed by overnight incubation with the S. aureus USA300 culture. As a control, we also tested GF-17, the natural peptide template for 17tF-W. If the peptide is stable, there is no bacterial growth above the MIC. Otherwise, the plate will be turbid due to bacterial growth [10]. In the case of 17tF-W, we only saw a small difference in the presence of chymotrypsin (Fig. 2B), implying only a minor degradation, leading to a two-fold increase in MIC against MRSA only at 3.1 μM. In the absence of chymotrypsin, S. aureus USA300 was unable to grow at 3.1 μM of GF-17 (grey curve in Fig. 2C). In the presence of the protease, the bacteria grew in all the wells, indicating rapid peptide degradation (black curve in Fig. 2C). These experiments established the important role of nonstandard amino acids in improving the stability of the new peptides designed based on a human LL-37 derived template.
Figure 2.
(A) LL-37 designer peptides to the action of proteases. The peptides 17tF-W and 17mF-W, after incubation with different proteases for 24 h at 37°C, were subjected to SDS-PAGE analysis: Lanes 1 & 6, the untreated peptides; 2 & 7, chymotrypsin (C) treated; 3 & 8, trypsin (T) treated; 4 & 9, S. aureus V8 (V8) protease treated; 5 & 10, fungal proteinase K (P) treated. (B,C) Effects of Chymotrypsin on peptide activity. GF17 (A), the major antimicrobial region of LL-37, and its engineered peptide 17tF-W (B) were exposed to chymotrypsin at a peptide: chymotrypsin molar ratio of 40:1 and incubated for 1 h at 37°C. Subsequently, the peptide solutions with (+C) and without (-C) chymotrypsin were mixed with the S. aureus USA300 culture exactly as in the microdilution antibacterial assays and incubated overnight. The curves represent the means of duplicate experiments ± standard deviation.
2.4. 17tF-W is effective in membrane permeation
To provide evidence for the mechanism of action of the peptide, we conducted membrane permeation experiments. This experiment uses a membrane non-permeable dye such as propidium iodide (PI) as an indicator for membrane damage. Without peptide treatment, there was no fluorescence increase. Likewise, tobramycin, which binds bacterial 30S ribosome, was unable to damage bacterial membranes. However, there was a clear increase in fluorescence when the S. aureus USA300 culture was treated with peptide 17tF-W, 17W2, or 17BIPHE2. 17tF-W rapidly permeabilized the membranes of S. aureus in a time-dependent manner (Fig. 3A green). We also compared the ability of 17tF-W and 17BIPHE2 in permeating the membranes of a clinical P. aeruginosa strain. Both are potent although 17BIPHE2 appeared to be slightly more efficient (Fig. 3B). In the case of A. baumannii, we found similar membrane permeating abilities for 17B-tF and 17tF2 at 3.1 μM in 10 mM PBS, implying their potential use to treat this pathogen (Fig. S4). It appears that tF at position 27 is preferred for killing A. baumannii. At the same peptide concentration, however, 17BIPHE2 and 17B-tF were most effective to permeate the membranes of K. pneumoniae followed by 17tF2 (Fig. S5). These results suggest the importance of a biphenylalanine at position 17 to kill this pathogen.
Figure 3.
Measurement of membrane integrity using a non-permeable dye: (A) S. aureus USA300 treated with various peptides at 3.12 μM and (B) P. aeruginosa treated at 1.56 μM of peptides; Simultaneous probing permeation of the (C) outer (OM) and (D) inner membranes (IM) of E. coli ML35p by 17tF-W and other peptides (1 cycle = 3 min). The peptides treated at 6.25 μM and the absorbance are recorded for 40 min. There is an increase in absorbance in both cases relative to the untreated bacteria (red). Experimental data processed, and plotted using Graphpad prism 7 software.
To provide additional evidence for membrane damage, we also used E. coli ML-35p, which is constitutive for β-galactosidase expressed in the cytoplasm and produces a β-lactamase in the periplasm. This E. coli strain also lacks lactose permease [52]. The measurement of β-lactamase and β-galactosidase activities thus enables the detection of the outer and inner membrane permeabilization simultaneously. The peptide 17W2 is superior to 17tF-W and 17BIPHE2 in permeating the outer membrane of E. coli (Fig. 3C). In contrast, vancomycin, a S. aureus cell wall targeting antibiotic, was unable to. Likewise, 17W2 and 17tF-W showed a slightly better permeation of the inner membrane of E. coli than 17BIPHE2 (Fig. 3D). It appears that the amphipathic structure of the aromatic Trp is better suited for permeating the membranes. Collectively, Trp-containing peptides are preferred in membrane permeation of E. coli (at least in this experiment) but less toxic to human cells than biphenyl-containing peptides.
We also measured membrane potential changes due to membrane depolarization [53] using S. aureus USA300. It seems that 17BIPHE2 was more effective than 17tF-W, which is similar to the Triton X-100 control (Fig. 4). In contrast, daptomycin was very weak since the curve was only slightly deviated from the untreated control close to 60 min. It appears that peptide hydrophobicity is important for membrane polarization of S. aureus.
Figure 4. S. aureus USA300 membrane depolarization by 17tF-W, 17BIPHE2, and daptomycin treated at 12.5 μM.
Triton X-100 (0.1%) was used as positive control. RFU = Relative Fluorescence Unit.
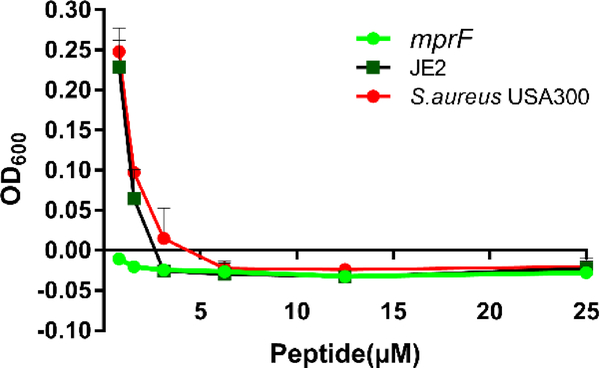
To provide additional evidence for membrane targeting, we also compared the MIC values of the peptides using both the wild type and an mprF transposon mutant of S. aureus USA300 [54]. This enzyme can modify the membrane surface of S. aureus by transferring a cationic lysine to anionic phosphatidylglycerol (PG), making the surface less negatively charged. Because most of the antimicrobial peptides are cationic (+2 to +3 in the APD database) [19], this bacterial surface modification as a mechanism of resistance could reduce peptide potency. We evidenced that the peptide 17tF-W inhibited the growth of both wild type and mprF mutant strains at 6.1 μM. Below this concentration, S. aureus USA300 and JE2 started to grow, whereas the mprF mutant was unable to (Fig. 5). Thus, electrostatic interactions between cationic peptides and anionic membranes play an important role in bacterial killing. Likewise, a decrease in the basic charge of the peptide can achieve a similar outcome. GF-17 becomes much less active against E. coli when R23 is altered to an alanine [55].
Figure 5: Comparison of antimicrobial activity of 17tF-W on membrane mutant and wild type strains.
Antibacterial assay using the wild type JE2 and mprF transposon mutant treated with various concentration of 17tF-W. The plot represents the mean ± SD of samples using Prism7, GraphPad software.
2.5. Assessment of antibiofilm activity of 17tF-W
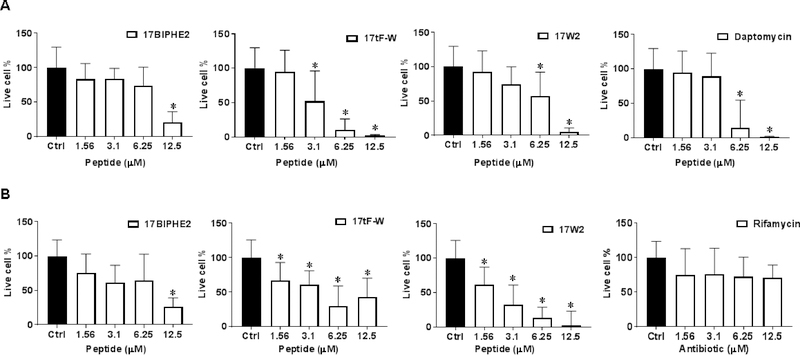
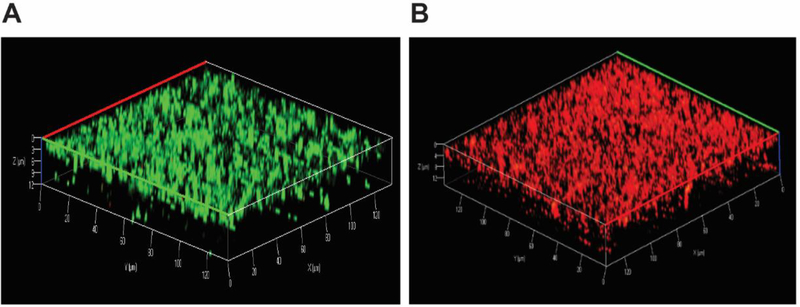
Most bacteria live in the biofilm form, which is more difficult to kill by conventional antibiotics. To evaluate antibiofilm activity, we compared the ability of the designed peptides in disrupting the 24 h preformed biofilms of S. aureus USA300 (Fig. 6A) and E. coli ATCC 25922 (Fig. 6B). In the case of S. aureus USA300 (Fig. 6A), 17tF-W was most potent in bacterial killing with a dose-dependent antibiofilm ability starting at a low concentration of 3.1 μM. This was followed by a known bacterial antimicrobial peptide daptomycin, which essentially eliminated biofilms at 6.25 μM. Both 17BIPHE2 and 17W2 became active at 12.5 μM. In the case of E. coli, all the three peptides showed a dose-dependent antibiofilm effect although 17W2 was more effective. In contrast, the antibiotic rifamycin was ineffective (Fig. 6B). Fig. 7 shows the confocal laser scanning microscopy images of MRSA biofilms treated with 17tF-W at 12.5 μM. The biofilms were stained with a live (green) and dead kit (red). Indeed, bacteria in the biofilms were killed (red cells in panel B) as a consequence of peptide treatment.
Figure 6.
Disruption of 24 h established biofilm of (A) S. aureus USA300 and (B) E. coli ATCC 25922 by newly designed LL-37 peptides. Determination of the anti-biofilm ability of peptides when compared with daptomycin and rifamycin. The live cell counts in biofilms were determined by XTT assay. A two-tailed unpaired student t test was conducted with a p value < 0.05 being considered statistically significant.
Figure 7.
Antibiofilm activity of the LL-37 engineered peptide 17tF-W against the 24 h established biofilms of S. aureus USA300 assessed by confocal laser scanning microscopy using live and dead staining: (A) control biofilms without peptide treatment and (B) biofilms treated with 17tF-W at 12.5 μM. Live cells are green (A), while dead cells are red (B).
2.6. Peptide efficacy in a catheter associated biofilm mouse model
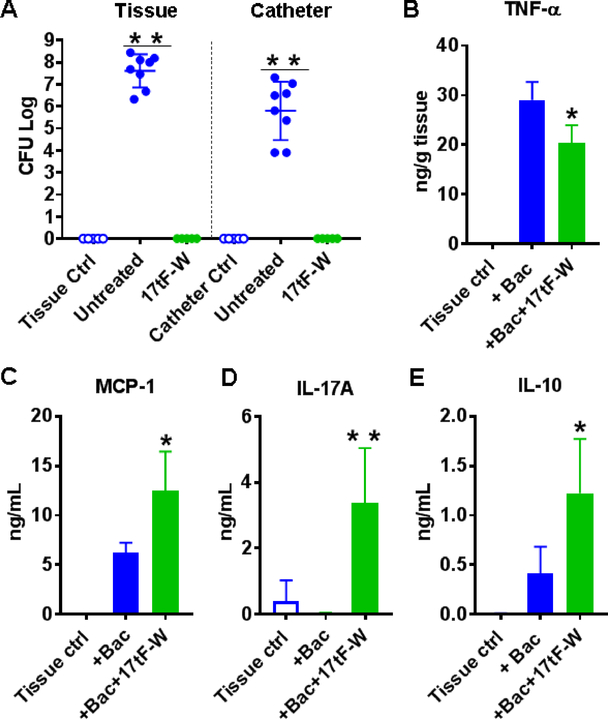
Because 17tF-W is rather effective in disrupting the S. aureus biofilms in vitro (Fig. 6A), we also tested its antibiofilm capability in a murine catheter-associated biofilm model. The S. aureus inoculum [1×103 colony forming units (CFU)/mouse] was injected into the lumen of the catheter embedded subcutaneously [56]. Without treatment, S. aureus USA300 formed biofilms with CFU reached 6–9 logs in tissues and 4–7 logs in catheters after three days. As a negative control, S. aureus was not detected in catheter or tissues without bacterial infection (Fig. 8A, open dots). Remarkably, daily subcutaneous injections of peptide 17tF-W for three days eliminated S. aureus USA300 in both catheters and surrounding tissues at day 3 (Fig. 8A, green), indicative of peptide potency in vivo.
Figure 8. Antibiofilm efficacy of 17tF-W in a murine catheter-associated S. aureus USA300 infection model.
(A) animals (n=5) without infection and without peptide treatment, animals with infection but without peptide treatment (n = 8 for the control group merged from two experiments), and animals infected and treated with 17tF-W. Cytokine levels with and without peptide treatment for (B) TNF-α (C) MCP-1, and (D) IL-17A, and (E) IL-10 in catheter-associated tissue homogenates. The plots, made using Prism software Graphpad 7, represent the averages of the experimental groups. P <0.05 is considered to be statistically significant (determined by Mann-Whitney test).
We also measured the cytokine levels in the tissues surrounding the catheters. At day 3, 17tF-W suppressed TNF-α (Fig. 8B), induced MCP-1/CCL2 (Fig. 8C), IL-17A (Fig. 8D), and IL-10 (Fig. 8E). While MCP-1 is known to recruit monocytes [10], IL-17A is known to attract neutrophils and other immune cells to the site of wound infection [57]. Accumulation of monocytes, neutrophils and lymphocytes at the infective site produced inflammatory responses, which were mediated by cytokine expression. The basis for the anti-inflammatory effect of 17tF-W may be due to contributions from several related mechanisms, including that of IL-10 [49]. Overall, these results indicate that the 17tF-W is able to kill S. aureus and modulates immune responses to clear the bacterial load around the catheter.
3. DISCUSSION
Unlike horse, sheep and cattle, humans possess only one cathelicidin gene. The significance of LL-37 in protecting humans from infection is well documented [38,39]. In addition, LL-37 is known to have biofilm inhibitory effect [8–18]. The antibacterial and antibiofilm properties are determined by the 3D structure of LL-37 [58]. Membrane targeting is achieved via a long amphipathic helix covering residues 2–31 [59]. NMR studies also reveals that all the four phenylalanines (F5, F6, F17 and F27) are important for targeting anionic lipid PG [59]. The segregation of this long helix into two hydrophobic domains due to the deployment of a hydrophilic serine at position 9 on the hydrophobic surface of LL-37 [59] provides one possible explanation for the synergistic binding of LL-37 to lipopolysaccharides (LPS) [60]. Such an LL-37 design appears to be important for host defense since an attempt to convert S9 to V9 to make the hydrophobic surface continual led to reduced antibacterial activity [61]. This LL-37 structure is responsible for eventual membrane disruption by a carpet model or pore formation [62,63].
It is known that antibacterial activity of LL-37 does not require the entire molecule. In fact, removal of the N-terminal fragment of LL-37 can enhance peptide activity [48]. While the N-terminal fragment LL-23 has poor antibacterial activity [61], the central helix between residues 10 and 31 contains the major antimicrobial region [58]. Peptides from this region has been widely studied [40–50]. The central region of LL-37 also contains the smallest antibacterial peptide KR-12 [59], which has been utilized by several labs to engineer and fabricate new peptide antimicrobials [16,64–67]. A recent study designed peptides beyond the core region to the N-terminus of the central region [17], while SAAP-148 was designed beyond the central region by including the C-terminally disordered tail of LL-37 [13]. We demonstrated that, GF-17, a peptide derived from the central antimicrobial region of LL-37, is superior to LL-37 in inhibiting the ESKAPE pathogens (Table 2), bacterial adhesion, and biofilm formation. It is also more potent in disrupting biofilms [12]. In addition, GF-17 is more potent than other LL-37 fragments in inhibiting MRSA, making it an excellent template for peptide engineering [10].
17BIPHE2, engineered based on GF-17 to gain stability to proteases [10], has antibiofilm capability similar to GF-17 against S. aureus USA300 [12]. This study has further improved our understanding by designing new peptides with varying combinations of aliphatic and aromatic side chains attached to phenylalanines of GF-17d3. The changes of biphenylalanines (17BIPHE2 in Table S1) to tryptophans (17W2) slightly reduced peptide activity against the ESKAPE pathogens in most of the cases (Table 1), probably due to a decrease in peptide hydrophobicity (Table S1). It is notable that appending an aliphatic moiety to phenylalanine clearly reduced the activity of these two peptides against K. pneumoniae. These results underscore the significance of 17BIPHE2 in fighting such nightmare pathogens, which our society has run short of useful antibiotics. However, 17W2 showed better antibiofilm activity against E. coli ATCC 25922 than other peptides. We also anticipate that 17tF2 is superior to 17BIPHE2 in controlling A. baumannii due to the same MIC value (Table 1) but reduced hemolysis of 17tF2 (Fig. 1). To combat P. aeruginosa, we previously found a better antibiofilm outcome by combining 17BIPHE2 with antibiotics [11]. We propose that both 17tF-W and 17W2 are better candidates to control E. cloacae than 17BIPHE2 in terms of potency as well as reduced hemolysis. Significantly, 17tF-W gained activity against the MRSA biofilms with reduced hemolysis compared to 17BIPHE2. Because 17tF-W and 17BIPHE2 are equally active against E. faecium, we propose that 17tF-W is a better candidate to combat both MRSA and E. faecium than 17BIPHE2 (Table 1). Thus, through our studies, a useful picture starts to emerge that the major antimicrobial region of human LL-37 could be tuned to better treat a specific ESKAPE pathogen (Table 2, with 3.1 μM bolded). These candidates are highly desirable because they retained the activity of 17BIPHE2 against a select pathogen with reduced hemolysis.
In addition, basic amino acids also play a role in modulating peptide activity. Biophysical and molecular simulation studies revealed preferential interaction of cationic LL-37 with anionic lipid vesicle models [68–70]. Among the three arginines, only R23 in the central antimicrobial region of LL-37 directly interacts with dioctanoyl phosphatidylglycerol [41]. Consistent with this observation, our alanine scan and antimicrobial studies reveal that the five cationic amino acids in the major antimicrobial peptide GF-17 are not equal and R23 is critical in initial recognition, membrane permeation, and bacterial killing [55]. Interestingly, R23 in 17BIPHE2 is also important in inhibiting bacteria as well as Zika virus, although GF-17 and 17BIPHE2 have different backbone structures [71,72]. This is largely determined by the type of the basic amino acid and its interfacial location in the amphipathic structure because even a swap of the positions between R23 and K25 can reduce peptide membrane permeation ability [73]. In a tryptophanrich peptide, the arginine analog is more effective in bacterial killing, but the lysine analog is more active against fungi such as Candida albicans [74]. Our study here also validated that deficiency in bacterial anionic lipid modification with lysine made MRSA more susceptible to killing (Fig. 6). Taken together, both hydrophobic and basic amino acids of LL-37 peptides are key components in determining peptide antimicrobial potency as well as toxicity to mammalian cells. Modulation of hydrophobicity is a general approach to reducing peptide toxicity [50]. This study presents new examples for tuning hydrophobicity.
Our results suggests that 17tF-W is a superior anti-MRSA peptide since it showed antibiofilm activity better than daptomycin. In vitro, it also showed a higher membrane permeation and biofilm disruption capability than 17BIPHE2. This peptide was less hemolytic than 17BIPHE2 and retained the stability to a panel of proteases. 17tF-W used less non-standard amino acids, presumably making it less costly to synthesize. Importantly, this peptide also demonstrated in vivo efficacy against MRSA in a catheter model, indicative of its potential topical applications. In addition, 17tF-W was able to regulate immune response presumably to help clear the infection. In conclusion, our newly engineered peptide 17tF-W gained merits in fighting MRSA-involved biofilm infection compared to 17BIPHE2.
4. CONCLUSION
This article demonstrated successful design of new LL-37 peptides against the ESKAPE pathogens, which are responsible for 90% hospital-associated infections. Our study revealed one feasible mechanism to modulate the antimicrobial potency of a chymotrypsin-resistant peptide template by altering the hydrophobic moieties to achieve optimal inhibition of various pathogens. While aromatic moieties are preferred for K. pneumoniae, aliphatic moieties at position 17 are effective against S. aureus. In contrast, aliphatic moieties at position 27 are preferred for A. baumannii. Also in this template, the Trp indole ring confers cell selectivity to peptides and is more effective against E. coli. Such knowledge may be harnessed to engineer peptides with desired properties. Collectively, our engineered peptides have multiple desired features: broad antimicrobial activity, antibiofilm, poor toxicity, and stability to proteases. Therefore, they constitute new candidates for topical use to combat resistant infections caused by the ESKAPE pathogens.
5. Materials and Methods
5.1. Peptides and chemicals
All peptides were chemically synthesized and purified to >95% purity (Genemed Synthesis, TX). Mass Spectroscopy and HPLC chromatograms indicate the quality of each peptide. For each experiment, fresh peptide stock solutions were prepared using autoclaved distilled water, and their concentrations were determined by UV spectroscopy [75]. Other chemicals were purchased from Sigma (MO, USA) unless specified.
5.2. HPLC retention time measurements
The retention time of each peptide was measured on a Waters HPLC system equipped with an analytical reverse-phase WATERS C8 symmetry column (3.9 × 150 mm). The peptide detected at 220 nm was eluted with a gradient of acetonitrile (containing 1% TFA) from 5% to 95% at a flow rate of 1 mL/min [76].
5.3. Bacterial strains and growth media
The bacterial strains used in this study included the Gram-positive methicillin-resistant Staphylococcus aureus USA300, Enterococcus faecium ATCC 51559, Gram-negative isolates Klebsiella pneumonia ATCC13883, Enterobacter cloacae B2366–12, Acinetobacter baumannii B28–16, and Pseudomonas aeruginosa PAO1. Also used are clinical strains P. aeruginosa #2, S. aureus Newman, S. aureus Mu50, S. aureus SA546, and S. aureus UAMS-1. Bacteria were cultivated in tryptic soy broth (TSB) from BD Bioscience MD, USA.
5.4. Antimicrobial assays
The antibacterial activity of peptides was evaluated using a standard broth micro-dilution protocol with minor modifications [10]. In brief, logarithmic phase bacterial cultures (i.e., optical density at 600 nm ≈ 0.5) were diluted to OD600 0.001 using TSB media and aliquoted into a 96-well polystyrene microplate at 90 μL per well. After treatment with 10 μL of peptide solutions two fold diluted to various concentrations, microplates were incubated at 37°C overnight and read on a ChroMate 4300 Microplate Reader at 600 nm (GMI, Ramsey, MN). The minimal inhibitory concentration (MIC) is the lowest peptide concentration that fully inhibited bacterial growth.
5.5. Peptide stability to the action of proteases
Peptide stability was evaluated based on our published protocol [10]. Aliquots (10 μL) of the reaction solutions (peptide/ protease molar ratio, 40:1) in 10 mM phosphate buffer saline (PBS, pH ~7.4) at 37°C were taken post 24 h incubation and immediately mixed with 10 μL of 2× SDS loading buffer followed by boiling for 5 min to stop the reactions. Samples were analyzed using 5% stacking/18% resolving tricine gel.
5.6. Effects of chymotrypsin on the 17tF-W peptide activity
Peptide antimicrobial activity was evaluated by incubating the peptide with chymotrypsin at 40:1 molar ratio for 1 h at 37 °C. Post incubation, the peptides in the presence or absence of chymotrypsin was mixed with bacterial culture and incubated overnight in the same manner as described in the antibacterial assays.
5.7. Hemolytic assays
Hemolytic assays were performed as described [76]. Briefly, human red blood cells (hRBCs) obtained from UNMC Blood Bank. Blood was washed three times using PBS and diluted to a 2% solution (v/v). After peptide treatment, incubation at 37°C for one hour, and centrifugation at 13,000 rpm, aliquots of the supernatant were carefully transferred to a fresh 96-well microplate. The amount of released hemoglobin was measured at 545 nm. The percent lysis was calculated by assuming 100% release when human blood cells were treated with 1% Triton X-100, and 0% release when incubated with PBS buffer. The peptide concentration that caused 50% lysis of hRBCs is defined as HL50.
5.8. Cytotoxicity to other mammalian cells
Peptides were assayed for potential in vitro toxicity using other human cells. Briefly, cells were seeded at a density of 3×104 per well in a 96-well plate in DMEM supplemented with 10% fetal bovine serum (FBS), and incubated at 37°C in a 5% CO2 atmosphere for 24 h. Wells were replaced with serum free media and treated with peptides at different concentrations for 1 h. After incubation, the cells were washed and incubated with 100 μL of DMEM media containing 20 μL of MTS for 2 h at 37°C. Absorbance was measured using the above ChroMate microplate reader.
5.9. Membrane permeation assay
The experiment was performed as described previously with minor modifications [71]. Serially diluted 10× peptides (10 μL each well) were made in 96-well microtiter plates. Propidium iodide (2 μL) at a fixed concentration of 20 μM was added to each well followed by the addition of 88 μL of the S. aureus USA300 or P. aeruginosa #2 culture (a final OD600 ~0.1 in 10 mM PBS). The plates were incubated at 37°C with continuous shaking at 100 rpm in a FLUOstar Omega microplate reader (BMG LABTECH Inc, NC, USA). The fluorescence from the plate was read every 5 min for a total duration of 2 h with an excitation and emission wavelengths of 584 nm and 620 nm, respectively. Plots were made using average values of duplicated experiments with the MARS software (BMG LABTECH Inc, NC, USA).
5.10. Ability of peptides to cross the outer and inner membranes of bacterial cells
Membrane permeation experiments were performed using a mutant E. coli strain developed by Lehrer and colleagues [52]. Bacteria were grown from a single colony overnight in TSB at 37°C. After washing in PBS three times, the culture was diluted to ~106 CFU/mL in PBS containing 0.3 mg/mL TSB and was added to all wells in a non-culture-treated polystyrene microplate, together with increasing concentrations of peptide solutions (0 to 12.5 μM) in phosphate buffer. Each well also contained 30 μM nitrocefin or 2.5 mM ONPG in phosphate buffer. Absorbance was followed simultaneously at 490 nm and at 420 nm (these were closest to the maximum value that the fixed-wavelength microplate allowed us to use) for 40 min at 37°C by using FLUOstar Omega microplate reader (BMG LABTECH Inc, NC, USA).
5.11. Membrane depolarization of bacteria
Membrane depolarization was measured as described [53]. In brief, S. aureus USA300 was sub-cultured in TSB media to the exponential phase. Cells were then spun, pelleted, washed with PBS twice, and re-suspended in twice the volume of PBS containing 25 mM glucose for 15 min at 37°C to energize the bacterial cells. For membrane depolarization measurements, 250 nM (final concentration) of the dye DiBAC4(3) [bis-(1,3-dibutylbarbituric acid) trimethineoxonol] (ANASPEC, CA, USA) was added, and vortexed gently. The energized 90 μL of bacterial cells was aliquoted in a 96 well microtiter plate (Corning COSTAR). The plate was immediately placed in a FLUOstar Omega microplate reader (BMG LABTECH Inc., NC, USA) to record readings for 20 min to get dye normalization. After 20 min, 10 μL of the peptide solution or positive control Triton X-100 (0.1%) was added. Fluorescence was read for 40 min at excitation and emission wavelengths of 485 nm and 520 nm, respectively.
5.12. Effects on established bacterial biofilms
The anti-biofilm activity of the peptide against 24 h established biofilms was evaluated [9,11, 12]. In short, S. aureus USA300 (105 CFU/ mL) was made from the exponential phase culture in TSB media and 200 μL was distributed to each well of the microtiter plate (Corning Costar Cat No. 3595). The plates were incubated at 37°C for 24 h to form biofilms. Media were then pipetted out and the attached biofilms were washed with 1× PBS to remove the planktonic bacteria. Each well of the plate was then filled with 20 μL of 10× peptide solution and 180 μL of fresh TSB media and the plates were further incubated at 37°C for 24 h. Media were then pipetted out and the wells were washed with 1× PBS. Biofilms treated with water served as a positive control while media without bacterial inoculation served as the negative control. Live cells in the biofilms were then quantitated using XTT [2,3-bis (2-methyloxy-4-nitro-5sulfophenyl)-2H-tertazolium-5-carboxanilide] assay by following the manufacturer’s instructions with modifications. 180 μL of fresh TSB and 20 μL of XTT solution was added to each well and the plates were again incubated at 37°C for 2 h. Absorbance at 450 nm (only media with XTT containing wells served as the blank) was obtained using a ChromateTM microtiter plate reader. Percentages of biofilm growth with and without peptide treatment were plotted by assuming 100% biofilm growth achieved on the bacterial wells without peptide treatment.
5.13. Assessment of peptide antibiofilm activity using confocal imaging
S. aureus USA300 (105 CFU/ mL) was made from an exponential phase bacterial culture. To the glass slide chambers (Borosilicate cover glass systems, Nunc Cat. No: 155380) 2.0 mL of the culture was added and incubated at 37°C for 24 h to allow biofilm formation. Media were aspirated and chambers were washed with 1× PBS to remove non-adhered cells. To test the peptide effect on the preformed biofilms, 200 μL of peptide 10× stocks was added to 1.8 mL of TSB. Control cuvettes were treated with water instead of peptide. The cuvettes were further incubated for another 24 h at 37°C. The next day the supernatant was pipetted out and the chambers were washed with 1× PBS. The biofilms were stained with 10 μL of the LIVE/DEAD kit (Invitrogen Molecular Probes, USA) according to the manufacturer’s instructions. The samples were examined with a confocal laser-scanning microscope (Zeiss 710) and the data were processed using the Zen 2010 software.
5.14. Catheter-associated biofilm mouse model
The animal studies were performed by following protocols and guidelines approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Nebraska Medical Center (UNMC). On the study termination day, animals were euthanized humanely using CO2 followed by harvesting the catheter and surrounding tissues for laboratory analysis. All efforts were made to minimize animal pain and suffering.
Female C57BL/6 mice (8 weeks old) were purchased from Charles River. Animals (5 per cage) were kept in entilated cages (IVCs) at temperatures of 20–24°C, humidity of 50–60%, 60 air exchanges per hour and a 12/12-hour light/dark cycle. Mice were fed with standardized food (Teklad Laboratory diet for rodents) and water (Hydropac® Alternative Watering System) from an animal’s ad libitum (free feeding). All materials, including IVCs, lids, feeders, bottles, bedding, and water, were autoclaved before use. Experimental and control mice were kept in IVCs under a negative pressure under the conditions stated above. All animal manipulations were conducted in a class II laminar flow biological safety cabinet.
Compound potency was evaluated as described [10, 77]. On the day of infection, mice were anesthetized using ketamine/xylazine (100/10 mg/kg). A small incision was made on the left flank region of the mouse using a small blunt spatula, a pouch was made for catheter insertion. Sterile catheters with a length of 1 cm were inserted into the pouch aseptically. The cut was sealed using wound closure VetBond glue. S. aureus USA300 LAC (20 μL with 103 CFU/catheter) was injected into the lumen of the catheter. Two, 24, and 48 hours post infection, peptide (250 μg) was injected (250 μL in and around the catheter lumen, 50 μl per site). Mice were allowed for full recovery from anesthesia in an oxygen-enriched chamber. Three days after infection, untreated and treated animal groups were CO2 euthanized and the tissues around the catheter were harvested along with the catheter. The catheter was sonicated (15 minutes, 37 kHz) to release the bacteria in biofilms and the tissue was homogenized. Appropriate dilutions were made for plating onto the blood agar plates, which were incubated at 37°C overnight. The CFU of each mouse was plotted as an individual dot and error bars represent the deviation within the experimental group. Statistical significances were determined by the Mann-Whitney test (*, p<0.05 and **, p <0.01).
5.15. ELISA quantification of cytokines in animal tissues
Enzyme-linked immunosorbent assay (ELISA) development kits for detection of cytokines were purchased from Biolegend (San Diego, CA, USA). The animal skin tissues were homogenized and centrifuged at 13,000 rpm for 10 minutes. The supernatants were removed and stored at −80°C. Cytokines [TNF-α, interleukin (IL)-17, IL-10 and MCP-1] in the supernatants were examined using ELISA as described by the manufacturer. Cytokine levels were derived from the standard curve and expressed as mean ± SD. Plots were generated using Prism 7.0, Graphpad software and a p value <0.05 was considered significant.
5.16. Statistical analysis
Data analysis was performed using the Prism Version 7.0 software (GraphPad). Data were represented as mean ± SD from two independent experiments. The significance between two groups was determined by two-tailed unpaired student t tests in vitro and Mann-Whitney test in vivo. The p value < 0.05 was considered statistically significant.
Supplementary Material
Highlights.
A peptide of LL-37 is more potent against resistant pathogens, including biofilms.
The peptide template is engineered into protease-resistant peptides.
Aromatic and aliphatic moieties are useful factors to modulate peptide activity.
One designed peptide shows in vivo efficacy in a catheter mouse model.
Acknowledgements
This study was supported by the United States National Institutes of Health (AI105147 to GW). We are very grateful to Dr. Tammy Kielian’s lab, especially Dr. Cortney E. Heim, for teaching us the catheter model used in this study. We also thank Drs. Kenneth Bayles and Paul Fey for providing us bacterial strains, and the UNMC confocal core for assistance with the images.
Footnotes
Conflicts of Interest
The authors declare no conflict of interest. The funding agency played no role in the design, execution, interpretation, and writing of this study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Allison KR, Brynildsen MP & Collins JJ, Metabolite-enabled eradication of bacterial persisters by aminoglycosides, Nature 473 (2011) 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conlon BP, Nakayasu ES, Fleck LE, LaFleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN & Lewis K, Activated ClpP kills persisters and eradicates a chronic biofilm infection, Nature 503 (2013) 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies J & Davies D, Origins and evolution of antibiotic resistance, Microbiol. Mol. Biol. Rev 74 (2010) 417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lohner K & Staudegger E, In: Lohner K (Ed.), Development of Novel Antimicrobial Agents: Emerging Strategies,1st ed.; Horizon Scientific Press, Norfolk, United States of America, 2001, 284. [Google Scholar]
- 5.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B & Bartlett J, Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America, Clin. Infect. Dis 48 (2009) 1–12. [DOI] [PubMed] [Google Scholar]
- 6.Flemming HC & Wingender J, The biofilm matrix, Nat. Rev. Microbiol 8 (2010) 623–633. [DOI] [PubMed] [Google Scholar]
- 7.Gerdes K & Semsey S, Microbiology: Pumping persisters, Nature 534 (2016) 41–42. [DOI] [PubMed] [Google Scholar]
- 8.Overhage J, Campisano A, Bains M, Torfs EC, Rehm BH & Hancock RE, Human host defense peptide LL-37 prevents bacterial biofilm formation, Infect. Immun 76 (2008) 4176–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean SN, Bishop BM & van Hoek ML, Natural and synthetic cathelicidin peptides with anti-microbial and anti-biofilm activity against Staphylococcus aureus, BMC Microbiol. 11 (2011) 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang G, Hanke ML, Mishra B, Lushnikova T, Heim CE, Chittezham Thomas V, Bayles KW & Kielian T, Transformation of human cathelicidin LL-37 into selective, stable, and potent antimicrobial compounds, ACS Chem. Biol 9 (2014) 1997–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishra B & Wang G, Individual and Combined Effects of Engineered Peptides and Antibiotics on Pseudomonas aeruginosa Biofilms, Pharmaceuticals (Basel) 10 (2017) 10.3390/ph10030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishra B, Golla RM, Lau K, Lushnikova T & Wang G, Anti-Staphylococcal Biofilm Effects of Human Cathelicidin Peptides, ACS Med. Chem. Lett. 7 (2015) 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Breij A, Riool M, Cordfunke RA, Malanovic N, de Boer L, Koning RI, Ravensbergen E, Franken M, van der Heijde T, Boekema BK, Kwakman PHS, Kamp N, El Ghalbzouri A, Lohner K, Zaat SAJ, Drijfhout JW & Nibbering PH, The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms, Sci. Transl. Med 10 (2018) 10.1126/scitranslmed.aan4044. [DOI] [PubMed] [Google Scholar]
- 14.Amer LS, Bishop BM, & van Hoek ML Antimicrobial and antibiofilm activity of cathelicidins and short, synthetic peptides against Francisella. Biochem Biophys Res Commun. 396 (2010) 246–51. [DOI] [PubMed] [Google Scholar]
- 15.Feng X, Sambanthamoorthy K, Palys T & Paranavitana C The human antimicrobial peptide LL-37 and its fragments possess both antimicrobial and antibiofilm activities against multidrug-resistant Acinetobacter baumannii. Peptides. 49 (2013) 131–7. [DOI] [PubMed] [Google Scholar]
- 16.Luo Y, McLean DT, Linden GJ, McAuley DF, McMullan R & Lundy FT The Naturally Occurring Host Defense Peptide, LL-37, and Its Truncated Mimetics KE-18 and KR-12 Have Selected Biocidal and Antibiofilm Activities Against Candida albicans, Staphylococcus aureus, and Escherichia coli In vitro. Front Microbiol. 8 (2017) 544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z, Yang G, Lu S, Chen D, Fan S, Xu J, Wu B & He J Design and antimicrobial activities of LL-37 derivatives inhibiting the formation of Streptococcus mutans biofilm. Chem Biol Drug Des. 2019. January 11. doi: 10.1111/cbdd.13419. [DOI] [PubMed] [Google Scholar]
- 18.Wang G, Narayana JL, Mishra B, Zhang Y, Wang F, Wang C, Zarena D, Lushnikova T & Wang X, Design of Antimicrobial Peptides: Progress Made with Human Cathelicidin LL-37. Adv Exp Med Biol. 1117 (2019) 215–240. [DOI] [PubMed] [Google Scholar]
- 19.Wang G, Li X & Wang Z, APD3: the antimicrobial peptide database as a tool for research and education, Nucleic Acids Res. 44 (2016) D1087–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z & Wang G, APD: the antimicrobial peptide database, Nucleic Acids Res. 32 (2004) D590–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oren Z, Lerman JC, Gudmundsson GH, Agerberth B & Shai Y, Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes: relevance to the molecular basis for its non-cell-selective activity, Biochem. J 341 ( Pt 3) (1999) 501–513. [PMC free article] [PubMed] [Google Scholar]
- 22.Zasloff M, Antimicrobial peptides of multicellular organisms, Nature 415 (2002) 389–395. [DOI] [PubMed] [Google Scholar]
- 23.Hancock RE & Lehrer R, Cationic peptides: a new source of antibiotics, Trends Biotechnol. 16 (1998) 82–88. [DOI] [PubMed] [Google Scholar]
- 24.Wang G, Human antimicrobial peptides and proteins, Pharmaceuticals (Basel) 7 (2014) 545–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehrer RI & Lu W α-Defensins in human innate immunity. Immunol Rev. 245 (2012) 84–112. [DOI] [PubMed] [Google Scholar]
- 26.Kavanagh K & Dowd S Histatins: antimicrobial peptides with therapeutic potential. J Pharm Pharmacol. 56 (2004) 285–9. [DOI] [PubMed] [Google Scholar]
- 27.Vandamme D, Landuyt B, Luyten W & Schoofs L A comprehensive summary of LL-37, the factotum human cathelicidin peptide. Cell Immunol. 280 (2012) 22–35. [DOI] [PubMed] [Google Scholar]
- 28.Becknell B & Spencer JD A Review of Ribonuclease 7’s Structure, Regulation, and Contributions to Host Defense. Int J Mol Sci. 17 (2016) 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burian M & Schittek B The secrets of dermcidin action. Int J Med Microbiol. 305 (2015) 283–6. [DOI] [PubMed] [Google Scholar]
- 30.Yang D, Chen Q, Hoover DM, Staley P, Tucker KD, Lubkowski J & Oppenheim JJ Many chemokines including CCL20/MIP-3alpha display antimicrobial activity. J Leukoc Biol. 74 (2003) 448–55. [DOI] [PubMed] [Google Scholar]
- 31.Zanetti M, Gennaro R, Skerlavaj B, Tomasinsig L & Circo R, Cathelicidin peptides as candidates for a novel class of antimicrobials, Curr. Pharm. Des 8 (2002) 779–793. [DOI] [PubMed] [Google Scholar]
- 32.Gudmundsson GH, Agerberth B, Odeberg J, Bergman T, Olsson B & Salcedo R, The human gene FALL39 and processing of the cathelin precursor to the antibacterial peptide LL-37 in granulocytes, Eur. J. Biochem 238 (1996) 325–332. [DOI] [PubMed] [Google Scholar]
- 33.Sorensen OE, Gram L, Johnsen AH, Andersson E, Bangsboll S, Tjabringa GS, Hiemstra PS, Malm J, Egesten A & Borregaard N, Processing of seminal plasma hCAP-18 to ALL-38 by gastricsin: a novel mechanism of generating antimicrobial peptides in vagina, J. Biol. Chem 278 (2003) 28540–28546. [DOI] [PubMed] [Google Scholar]
- 34.Murakami M, Kameda K, Tsumoto H, Tsuda T, Masuda K, Utsunomiya R, Mori H, Miura Y & Sayama K, TLN-58, an Additional hCAP18 Processing Form, Found in the Lesion Vesicle of Palmoplantar Pustulosis in the Skin, J. Invest. Dermatol 137 (2017) 322–331. [DOI] [PubMed] [Google Scholar]
- 35.Nijnik A & Hancock RE, The roles of cathelicidin LL-37 in immune defences and novel clinical applications, Curr. Opin. Hematol 16 (2009) 41–47. [DOI] [PubMed] [Google Scholar]
- 36.Morizane S, Yamasaki K, Muhleisen B, Kotol PF, Murakami M, Aoyama Y, Iwatsuki K, Hata T & Gallo RL, Cathelicidin antimicrobial peptide LL-37 in psoriasis enables keratinocyte reactivity against TLR9 ligands, J. Invest. Dermatol 132 (2012) 135143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh D, Vaughan R & Kao CC, LL-37 peptide enhancement of signal transduction by Toll-like receptor 3 is regulated by pH: identification of a peptide antagonist of LL-37, J. Biol. Chem 289 (2014) 27614–27624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chromek M, Slamova Z, Bergman P, Kovacs L, Podracka L, Ehren I, Hokfelt T, Gudmundsson GH, Gallo RL, Agerberth B & Brauner A, The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection, Nat. Med 12 (2006) 636–641. [DOI] [PubMed] [Google Scholar]
- 39.Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, Pestonjamasp V, Piraino J, Huttner K & Gallo RL, Innate antimicrobial peptide protects the skin from invasive bacterial infection, Nature 414 (2001) 454–457. [DOI] [PubMed] [Google Scholar]
- 40.Turner J, Cho Y, Dinh NN, Waring AJ, Lehrer RI, Activities of LL-37, a cathelin-associated antimicrobial peptide of human neutrophils, Antimicrob Agents Chemother 42 (1998) 2206–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang G, Determination of solution structure and lipid micelle location of an engineered membrane peptide by using one NMR experiment and one sample, Biochim. Biophys. Acta 1768 (2007) 3271–81. [DOI] [PubMed] [Google Scholar]
- 42.Ciornei CD, Sigurdardottir T, Schmidtchen A, Bodelsson M, Antimicrobial and chemoattractant activity, lipopolysaccharide neutralization, cytotoxicity, and inhibition by serum of analogs of human cathelicidin LL-37, Antimicrob Agents Chemother. 49 (2005) 2845–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanthawong S, Bolscher JG, Veerman EC, van Marle J, Nazmi K, Wongratanacheewin S, Taweechaisupapong S, Antimicrobial activities of LL-37 and its truncated variants against Burkholderia thailandensis, Int J Antimicrob Agents 39 (2012) 39–44. [DOI] [PubMed] [Google Scholar]
- 44.Sieprawska-Lupa M, Mydel P, Krawczyk K, Wojcik K, Puklo M, Lupa B, Suder P, Silberring J, Reed M, Pohl J, Shafer W, McAleese F, Foster T, Travis J, Potempa J, Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases, Antimicrob Agents Chemother. 48 (2004) 4673–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sigurdardottir T, Andersson P, Davoudi M, Malmsten M, Schmidtchen A, Bodelsson M, In silico identification and biological evaluation of antimicrobial peptides based on human cathelicidin LL-37, Antimicrob Agents Chemother. 50 (2006) 2983–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molhoek EM, den Hertog AL, de Vries AM, Nazmi K, Veerman EC, Hartgers FC, Yazdanbakhsh M, Bikker FJ, van der Kleij D, Structure-function relationship of the human antimicrobial peptide LL-37 and LL-37 fragments in the modulation of TLR responses, Biol Chem 390 (2009) 295–303. [DOI] [PubMed] [Google Scholar]
- 47.Nagant C, Pitts B, Nazmi K, Vandenbranden M, Bolscher JG, Stewart PS, Dehaye JP, Identification of peptides derived from the human antimicrobial peptide LL-37 active against biofilms formed by Pseudomonas aeruginosa using a library of truncated fragments, Antimicrob Agents Chemother. 56 (2012) 5698–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Braff MH, Hawkins MA, Di Nardo A, Lopez-Garcia B, Howell MD, Wong C, Lin K, Streib JE, Dorschner R, Leung DY & Gallo RL, Structure-function relationships among human cathelicidin peptides: dissociation of antimicrobial properties from host immunostimulatory activities, J. Immunol 174 (2005) 4271–4278. [DOI] [PubMed] [Google Scholar]
- 49.Nell MJ, Tjabringa GS, Wafelman AR, Verrijk R, Hiemstra PS, Drijfhout JW & Grote JJ, Development of novel LL-37 derived antimicrobial peptides with LPS and LTA neutralizing and antimicrobial activities for therapeutic application, Peptides 27 (2006) 649–660. [DOI] [PubMed] [Google Scholar]
- 50.Li X, Li Y, Han H, Miller DW & Wang G, Solution structures of human LL-37 fragments and NMR-based identification of a minimal membrane-targeting antimicrobial and anticancer region, J. Am. Chem. Soc 128 (2006) 5776–5785. [DOI] [PubMed] [Google Scholar]
- 51.Mishra B, Lakshmaiah Narayana J, Lushnikova T, Wang X & Wang G, Low cationicity is important for systemic in vivo efficacy of database-derived peptides against drug-resistant Gram-positive pathogens, Proc Natl Acad Sci U S A. 2019. June 17 pii: 201821410. doi: 10.1073/pnas.1821410116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lehrer RI, Barton A & Ganz T, Concurrent assessment of inner and outer membrane permeabilization and bacteriolysis in E. coli by multiple-wavelength spectrophotometry, J. Immunol. Methods 108 (1988) 153–158. [DOI] [PubMed] [Google Scholar]
- 53.Marks LR, Clementi EA & Hakansson AP, Sensitization of Staphylococcus aureus to methicillin and other antibiotics in vitro and in vivo in the presence of HAMLET, PLoS One 8 (2013) e63158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL & Bayles KW, A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes, Mbio 4 (2013) e00537–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang G, Epand RF, Mishra B, Lushnikova T, Thomas VC, Bayles KW & Epand RM, Decoding the functional roles of cationic side chains of the major antimicrobial region of human cathelicidin LL-37, Antimicrob. Agents Chemother 56 (2012) 845–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heim CE, Hanke ML & Kielian T, A mouse model of Staphylococcus catheter-associated biofilm infection, Methods Mol. Biol 1106 (2014) 183–191. [DOI] [PubMed] [Google Scholar]
- 57.Bagheri N, Azadegan-Dehkordi F, Shirzad H, Rafieian-Kopaei M, Rahimian G & Razavi A, The biological functions of IL-17 in different clinical expressions of Helicobacter pylori-infection, Microb. Pathog 81 (2015) 33–38. [DOI] [PubMed] [Google Scholar]
- 58.Wang G, Mishra B, Epand RF & Epand RM, High-quality 3D structures shine light on antibacterial, anti-biofilm and antiviral activities of human cathelicidin LL-37 and its fragments, Biochim. Biophys. Acta 1838 (2014) 2160–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang G, Structures of human host defense cathelicidin LL-37 and its smallest antimicrobial peptide KR-12 in lipid micelles, J. Biol. Chem 283 (2008) 32637–32643. [DOI] [PubMed] [Google Scholar]
- 60.Turner J, Cho Y, Dinh NN, Waring AJ & Lehrer RI, Activities of LL-37, a cathelin-associated antimicrobial peptide of human neutrophils, Antimicrob. Agents Chemother 42 (1998) 2206–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang G, Elliott M, Cogen AL, Ezell EL, Gallo RL & Hancock RE, Structure, dynamics, and antimicrobial and immune modulatory activities of human LL-23 and its single-residue variants mutated on the basis of homologous primate cathelicidins, Biochemistry 51 (2012) 653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shai Y, Mode of action of membrane active antimicrobial peptides, Biopolymers 66 (2002) 236–48. [DOI] [PubMed] [Google Scholar]
- 63.Henzler Wildman KA, Lee DK & Ramamoorthy A, Mechanism of lipid bilayer disruption by the human antimicrobial peptide, LL-37, Biochemistry 42 (2003) 6545–58. [DOI] [PubMed] [Google Scholar]
- 64.Jacob B, Park IS, Bang JK & Shin SY, Short KR-12 analogs designed from human cathelicidin LL-37 possessing both antimicrobial and antiendotoxic activities without mammalian cell toxicity, J Pept Sci. 19 (2013) 700–7. [DOI] [PubMed] [Google Scholar]
- 65.da Silva BR, Conrado AJS, Pereira AL, Evaristo FFV, Arruda FVS, Vasconcelos MA, Lorenzón EN, Cilli EM & Teixeira EH, Antibacterial activity of a novel antimicrobial peptide [W7]KR12-KAEK derived from KR-12 against Streptococcus mutans planktonic cells and biofilms, Biofouling 33 (2017) 835–846. [DOI] [PubMed] [Google Scholar]
- 66.Caiaffa KS, Massunari L, Danelon M, Abuna GF, Bedran TBL, Santos-Filho NA, Spolidorio DMP, Vizoto NL, Cilli EM & Duque C, KR-12-a5 is a non-cytotoxic agent with potent antimicrobial effects against oral pathogens, Biofouling. 33 (2017) 807–818. [DOI] [PubMed] [Google Scholar]
- 67.Liu M, Liu T, Zhang X, Jian Z, Xia H, Yang J, Hu X, Xing M, Luo G & Wu J, Fabrication of KR-12 peptide-containing hyaluronic acid immobilized fibrous eggshell membrane effectively kills multi-drug-resistant bacteria, promotes angiogenesis and accelerates re-epithelialization, Int J Nanomedicine. 14 (2019) 3345–3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao L, Cao Z, Bian Y, Hu G, Wang J & Zhou Y, Molecular Dynamics Simulations of Human Antimicrobial Peptide LL-37 in Model POPC and POPG Lipid Bilayers. Int J Mol Sci.19 (2018). pii: E1186. doi: 10.3390/ijms19041186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Neville F, Cahuzac M, Konovalov O, Ishitsuka Y, Lee KY, Kuzmenko I, Kale GM & Gidalevitz D, Lipid headgroup discrimination by antimicrobial peptide LL-37: insight into mechanism of action. Biophys J. 90 (2006) 1275–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sood R, Domanov Y, Pietiäinen M, Kontinen VP & Kinnunen PK, Binding of LL-37 to model biomembranes: insight into target vs host cell recognition, Biochim Biophys Acta. 1778 (2008) 983–96. [DOI] [PubMed] [Google Scholar]
- 71.Wang X, Mishra B, Lushnikova T, Narayana JL & Wang G, Amino Acid Composition Determines Peptide Activity Spectrum and Hot-Spot-Based Design of Merecidin, Adv. Biosyst 2 (2018) pii:1700259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He M, Zhang H, Li Y, Wang G, Tang B, Zhao J, Huang Y & Zheng J, Cathelicidin-Derived Antimicrobial Peptides Inhibit Zika Virus Through Direct Inactivation and Interferon Pathway, Front. Immunol 9 (2018) 722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang X, Junior JCB, Mishra B, Lushnikova T, Epand RM & Wang G, Arginine-lysine positional swap of the LL-37 peptides reveals evolutional advantages of the native sequence and leads to bacterial probes, Biochim. Biophys. Acta Biomembr 1859 (2017) 1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mishra B, Lushnikova T, Golla RM, Wang X & Wang G, Design and surface immobilization of short anti-biofilm peptides, Acta Biomater. 49 (2017) 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Waddell WJ, A simple ultraviolet spectrophotometric method for the determination of protein, J. Lab. Clin. Med 48 (1956) 311–314. [PubMed] [Google Scholar]
- 76.Mishra B & Wang G, Ab initio design of potent anti-MRSA peptides based on database filtering technology, J. Am. Chem. Soc 134 (2012) 12426–12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Menousek J, Mishra B, Hanke ML, Heim CE, Kielian T & Wang G, Database screening and in vivo efficacy of antimicrobial peptides against methicillin-resistant Staphylococcus aureus USA300, Int. J. Antimicrob. Agents 39 (2012) 402–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.