Abstract
Tandem microsatellite repeats are common throughout the human genome and intrinsically unstable, exhibiting expansions and contractions both somatically and across generations. Instability in a small subset of these repeats are currently linked to human disease, although recent findings suggest more disease-causing repeats await discovery. These nucleotide repeat expansion disorders (NREDs) primarily affect the nervous system and commonly lead to neurodegeneration through toxic protein gain-of-function, protein loss-of-function, and toxic RNA gain-of-function mechanisms. However, the lines between these categories have blurred with recent findings of unconventional Repeat Associated Non-AUG (RAN) translation from putatively non-coding regions of the genome. Here we review two emerging topics in NREDs: 1) The mechanisms by which RAN translation occurs and its role in disease pathogenesis and 2) How nucleotide repeats as RNA and translated proteins influence liquid-liquid phase separation, membraneless organelle dynamics, and nucleocytoplasmic transport. We examine these topics with a particular eye on two repeats: the CGG repeat expansion responsible for Fragile X syndrome and Fragile X-associated Tremor Ataxia Syndrome (FXTAS) and the intronic GGGGCC repeat expansion in C9orf72, the most common inherited cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Our thesis is that these emerging disease mechanisms can inform a broader understanding of the native roles of microsatellites in cellular function and that aberrations in these native processes provide clues to novel therapeutic strategies for these currently untreatable disorders.
Repetitive elements in the genome
Repetitive regions of DNA comprise over half of the human genome, and can span millions of bases (Treangen and Salzberg 2011; Lander et al. 2001). The exact composition and variation of repeat DNA within the genome is unknown, as current sequencing technology is suboptimal for resolving these sequences (Treangen and Salzberg 2011). Within this repetitive genome (the “repeatome”) exist two broad categories of repetitive elements: Tandem repeats and interspersed repeats (Hannan 2018b). Tandem repeats are regions of DNA with sequential blocks of patterns of nucleotides confined to a specific locus (Dumbovic, Forcales, and Perucho 2017). An estimated 17% of human genes contain tandem repeats within open reading frames (Gemayel et al. 2010) and they are common components of intragenic regions such as introns and 5’ and 3’ untranslated regions (UTRs). As such, tandem repeats are of particular interest as a potential source of genetic variation and function. The other, larger category of repetitive DNA, interspersed repeats, are noncontiguous structures dispersed throughout the genome, and include transposons and retropseudo genes (Smit 1999). Interspersed repeats will not be further discussed in this review.
Satellite DNA is a subcategory of tandem repeats. The term “satellite” was given based on the experimental finding that a distinct band of DNA separated from the principle DNA band after density gradient centrifugation of mouse genomic DNA (Kit 1962). It was hypothesized that the distinct “satellite” band arose due to differences in base composition, which is directly related to the buoyant density of native DNA (Kit 1962; Jones 1973). Through biochemical analysis, these satellite sequences were discovered to be largely repetitive (Britten and Kohne 1968; Waring and Britten 1966). It was soon concluded that repetitive regions were common but species-specific in prokaryotes and eukaryotes, including humans, and had discrete subcellular localization patterns at sites of heterochromatin (Britten and Kohne 1968; Yunis and Yasmineh 1971; Holmquist and Dancis 1979). Satellite DNA is further categorized into microsatellites, minisatellites, and macrosatellites by size of the repeat unit (Dumbovic, Forcales, and Perucho 2017). Macrosatellites primarily occupy telomeres and centromeres (Jones 1970; Pardue and Gall 1970), while minisatellites and microsatellites are present throughout the genome (Lander et al. 2001).
Human genome sequencing allowed for further characterization and quantification of these repetitive regions (Lander et al. 2001). Microsatellites (which we define here as 1–8 or more bases per repeat unit) reside in both coding and noncoding regions, and comprise around 3% of the genome (Lander et al. 2001; Richard, Kerrest, and Dujon 2008). These repetitive units are also known as short tandem repeats (STR) or simple sequence repeats (SSR). Microsatellites likely arose through polymerase template slippage during transcription, and they are considered unstable due to their tendency to change size both somatically and between generations (Lander et al. 2001; Kruglyak et al. 1998). Thus, distinct microsatellites are often polymorphic within populations and can be expanded or contracted within families. The degree of variance and instability across all microsatellites in humans is currently unknown. Microsatellite polymorphisms in coding regions of eukaryotic genes have been linked to a number of roles from modulating cell-cell interactions in S. cerevisiae, circadian rhythms in D. melanogaster, body morphology in canines, and more recently an association between the length of a CAG trinucleotide repeat in the huntingtin gene (HTT) and cognitive performance in humans (Verstrepen et al. 2005; Gemayel et al. 2010; Lee et al. 2018; Hannan 2018a; Fondon and Garner 2004). While microsatellites in promoter regions of genes can greatly influence gene expression (Gemayel et al. 2010; Vinces et al. 2009; Sawaya et al. 2013), the full range of mechanisms by which variations in repeat size influence gene and genome biology is an area of active study (Bagshaw 2017; Usdin 2008).
Microsatellites in neurodegenerative disease: shared and divergent molecular mechanisms that depend on repeat size, content and context
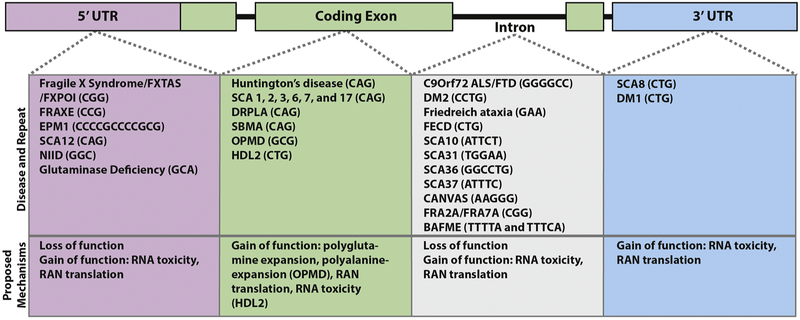
Instability and expansion of specific microsatellites is the genetic basis of over 40 nucleotide repeat expansion diseases (NREDs) (Figure 1). The vast majority of NREDs affect the nervous system, often leading to neurodegeneration (Paulson 2018a). Disease causing microsatellites are traditionally categorized into three groups: dominantly inherited repeat expansions within protein coding sequences, dominantly inherited repeat expansions outside of known protein coding sequences, and recessive or X-linked repeat expansions that impact the expression of the genes in which they reside. Most of the disease-causing repeats present within protein coding exons are CAG trinucleotides encoding for glutamine. These polyglutamine diseases include Huntington’s Disease (HD), spinal and bulbar muscular atrophy (SBMA), and several spinocerebellar ataxias (SCA). The resulting polyglutamine-containing proteins form insoluble aggregates, which place a proteotoxic burden on cells (La Spada et al. 1991; Trottier, Devys, et al. 1995; Trottier, Lutz, et al. 1995; Paulson 2018a; Bates 1996). Microsatellite expansion mutations can also result in RNA mediated toxicity, with myotonic dystrophy type 1 (DM1) as the standout example. Symptoms and severity in DM1 patients range from mild myotonia and cataracts to congenital forms which are often fatal due to respiratory deficits (Kamsteeg et al. 2012). In DM1, a CTG repeat resides in the 3’ UTR of the DMPK gene. This mutation does not meaningfully alter the abundance of DMPK, rather it causes RNA foci formation and sequestration of key proteins (Pettersson et al. 2015; Miller et al. 2000; Mankodi et al. 2001; Jiang et al. 2004). Microsatellite expansions can also elicit a loss of function in the genes in which they reside. Friedreich’s ataxia (FRDA) is the most common autosomal recessive ataxia, characterized by early adulthood onset of symptoms such as progressive ataxia with sensory loss (Paulson 2018a). In Friedreich’s ataxia, a GAA repeat expansion in an intron of the FXN gene leads to a change in the epigenetic state of the FXN locus that strongly suppresses the mRNA transcription (Campuzano et al. 1996). This leads to a partial, but not complete, loss of FXN mRNA and Frataxin protein, and impaired mitochondrial function and iron homeostasis (Akbar and Ashizawa 2015). Unlike the dominant disorders described above, Friedreich’s ataxia requires expansion or mutations in both FXN alleles, and the size of the repeat correlates inversely with the degree of mRNA expression and the severity of the clinical phenotype (Castaldo et al. 2008).
Figure 1: Nucleotide repeat expansion disorders.
Disorders are listed by their genomic location on an illustrated simplified gene. Each disorder is accompanied by the unstable repeat sequence that elicits disease. The prevailing disease mechanism is listed below each group to illustrate that genomic location of the expansion influences but does not completely determine the potential effects of any given repeat. Abbreviations: Fragile X-associated tremor/ataxia syndrome (FXTAS), Fragile X-primary ovarian insufficiency (FXPOI), progressive myoclonic epilepsy type 1/Unverricht-Lundborg disease (EPM1), spinocerebellar ataxia (SCA), neuronal intranuclear inclusion disease (NIID), dentatorubral-pallidoluysian atrophy (DRPLA), spinal-bulbar muscular atrophy (SBMA), oculopharyngeal muscular dystrophy (OPMD), Huntington disease-like 2 (HDL2), amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), myotonic dystrophy (DM), Fuchs endothelial corneal dystrophy (FECD), cerebellar ataxia, neuropathy and vestibular areflexia yndrome (CANVAS), benign adult familial myoclonic epilepsy (BAFME), and FRAXE, FRA2A and FRA7A refer to disease associated fragile sites in the genome that result from CGG repeat expansions.
While certain microsatellite expansions likely share underlying pathologic mechanisms, the convergence and divergence of phenotypes and genotypes across a range of microsatellites presents intriguing questions about these diverse disorders. The 28 dominantly inherited spinocerebellar ataxias are all progressive disorders characterized by cerebellar degeneration and clinically by loss of motor coordination (Soong and Paulson 2007). Six of the SCAs are caused by a CAG repeat expansion in coding regions of the genes in which they reside (SCA1, SCA2, SCA3, SCA7, DRPLA and SCA17) leading to polyglutamine expansions within six different protein coding genes, and an additional two SCAs are caused by CAG/CTG repeat expansions in putatively noncoding regions (SCA8 and SCA12) (Paulson et al. 2017). Early in the course of disease, specific clinical features can be used to separate the six polyglutamine SCAs phenotypically with varying degrees of certainty (Shakkottai and Fogel 2013). In addition, classic late-onset HD exhibits a very different clinical and pathological phenotype compared to the SCAs. As all of these genes are expressed ubiquitously within the nervous system, their divergent phenotypes are thought to emerge through specific aberrations in the native functions of the proteins in which they reside. Molecular evidence for this arises from differences in the repeat length thresholds for disease between different CAG repeat loci as well as the ability of specific mutations within these same proteins (most notably Ataxin 1) to recapitulate key features of disease absent expansion of the repeat element (Duvick et al. 2010). Perhaps the clearest example of native gene context influence on CAG repeat toxicity is seen in SBMA, where expansions within the androgen receptor gene only elicit toxicity in the setting of exposure to the receptor ligand, testosterone (Lieberman, Shakkottai, and Albin 2019; Katsuno et al. 2003; Katsuno et al. 2002). However, all polyglutamine NREDs share similar pathological features, including intranuclear ubiquitinated neuronal inclusions of the polyglutamine protein, and inclusions are often observed in brain regions which do not exhibit marked neurodegeneration or significant clinical phenotypes (Lieberman, Shakkottai, and Albin 2019). Moreover, at larger repeat sizes, a series of shared clinical features (dystonia, dementia, and spasticity) emerge in both HD and SCAs (Paulson 2018b). This convergence of clinical phenotypes may reflect a larger contribution of gene context-independent toxicity elicited by very large polyglutamine stretches. Consistent with this, expression of large polyglutamine expansions outside of a native gene context is sufficient to elicit inclusions and neurodegeneration across a series of model systems (Morley et al. 2002; Warrick et al. 1998; Ordway et al. 1997). Thus, for each polyglutamine NRED, it is likely the combinatorial effects of gene context dependent toxicity and polyglutamine expansion context independent toxicity that drive neurodegeneration, with the relative influence of each component dependent on repeat size and the strength of the effects of the native gene context.
Like DM1, DM2 (myotonic dystrophy type 2) is caused by a microsatellite repeat expansion in a noncoding region, however in this case it is a CCTG repeat in the first intron of the CNBP gene (Liquori et al. 2001). In their adult forms, there is significant overlap in the clinical features of DM2 and DM1, with some specific differences (such as the distribution of muscle weakness and dystrophy) usually allowing for clinical distinction of the two conditions. However, molecularly, the length of the DM1 CUG repeat expansion directly correlates with disease severity, which shows marked genetic anticipation and qualitatively different phenotypes in congenital cases with large expansions (Tsilfidis et al. 1992). This is not the case for DM2 which has a mean expansion length of 5,000 repeats but no clear congenital or juvenile form of the disease and a much less clear relationship between repeat size and age of onset (Kamsteeg et al. 2012; Paulson 2018a). Despite this clear clinical discrepancy, the mechanism of toxicity in DM2 is thought to be the same as that for DM1, with the repeat binding to and sequestering the RNA binding protein Muscleblind (Margolis et al. 2006). Similar splicing changes are observed in both conditions and in disease models expressing the two repeats (Nakamori et al. 2013). The underlying cause of the differences in these two disorders despite apparently shared disease mechanisms remains unclear, with potential roles of the neighboring genes in each repeat locus as well as non-muscleblind interacting partners for each repeat postulated as possible contributors (Meola and Cardani 2015).
A striking example of genetic convergence with phenotypic divergence is the G4C2 hexanucleotide expansion in C9Orf72 which is the most prevalent genetic cause of the two clinically separate diseases amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (Renton et al. 2011; DeJesus-Hernandez et al. 2011). ALS is a progressive neurodegenerative disease affecting neurons in the brain and spinal cord, whereas clinical phenotypes of FTD are most often forms of cognitive impairment such as disinhibition, apathy, changes in personality, and language dysfunction (Orr 2011; Paulson 2018a). These two diseases were known to be linked prior to this genetic finding through the study of families with ALS-FTD, and are also genetically linked through single nucleotide polymorphisms not associated with a microsatellite in the UBQLN2 gene (Deng et al. 2011). We will discuss major findings associated with the G4C2 hexanucleotide expansion in this review, but it is worth noting that the molecular underpinnings of this clinical divergence between ALS and FTD remains mysterious.
While much focus has been placed on the pathogenicity of microsatellites, the fact that their portion of the genome is about three-times greater than that of protein-coding regions suggests that microsatellites are likely to also influence the normal functions of the genes in which they reside (Consortium 2012). Our knowledge and understanding of microsatellites has grown recently with new discoveries related to the ways in which these repetitive regions can affect cellular function. This includes the topics of liquid-liquid phase separation to dictate membraneless organelle dynamics, nucleocytoplasmic transport and Repeat Associated Non-AUG (RAN) translation mediated production of cryptic proteins from repeat containing transcripts. In this review, we will outline the recent research on disease causing microsatellites, and discuss how this research has expanded our understanding of how microsatellites can function in the cell.
RAN translation of microsatellite repeats in non-coding and coding regions of mRNA
Microsatellites are capable of forming stable structures in RNA. While this can confer a toxic gain-of-function through direct binding with proteins (Renoux and Todd 2012; Todd and Paulson 2010), it also supports a newly recognized unconventional mode of translation initiation. This process is termed Repeat Associated Non-AUG (RAN) translation, whereby proteins are synthesized from microsatellite containing mRNA in the absence of an AUG start codon (Zu et al. 2011). This discovery was paradigm shifting as diseases caused by microsatellite expansions in noncoding regions were now looked at as having the potential to encode repetitive stretches of amino acids like those in polyglutamine disorders. RAN translation was first described by Zu etal. (2011) through work on spinocerebellar ataxia 8 (SCA8) where expansion of a CTG repeat in the 3’ UTR of ATXN8 is transcribed into both sense and antisense transcripts (Zu et al. 2011; Daughters et al. 2009; Moseley et al. 2006). The antisense transcript from ATXN8 (ATXN8OS) contains a short AUG-driven ORF encompassing the CAG repeat and encoding polyglutamine. Removal of the AUG or placement of stop codons just upstream of the repeat did not preclude synthesis of the polyglutamine product. Instead, translation persisted in all three repeat reading frames to produce polyalanine and polyserine products, in addition to a non-AUG initiated polyglutamine protein (Zu et al. 2011). The polyglutamine, polyalanine, and polyserine products were found to accumulate in cerebellar Purkinje cells of SCA8 patients (Zu et al. 2011; Ayhan et al. 2018). RAN translation of all three products increased with repeat size, and replacement of the CAG repeat with CAA prevented RAN translation in all three frames (Zu et al. 2011). Similarly, polyglutamine, polyalanine, and polyserine products were detected in the context of the antisense DM1 transcript (in the CAG orientation), where deletion of the only in-frame AUG (for polyserine) did not preclude translation. Polyglutamine aggregates were detected in myoblasts, skeletal muscle, and blood of DM1 patients (Zu et al. 2011). Since this initial finding, unconventional translation of repeats has been described in a variety of other nucleotide repeat expansion disorders, including C9orf72 in ALS/FTD, HTT in HD, TCF4 in Fuchs’ corneal dystrophy, along with several (but not all) of the spinocerebellar ataxia genes (Green, Linsalata, and Todd 2016; Ash et al. 2013; Mori et al. 2013; Banez-Coronel et al. 2015; Soragni et al. 2018; Ishiguro et al. 2017; Scoles et al. 2015). The exact mechanism by which this unconventional translation occurs is an area of active research and studies to date suggest that multiple mechanisms for initiation may occur at different repeats, or even in different reading frames of the same repeat (Gao, Richter, and Cleveland 2017). For the purposes of this review, we refer to all of these unconventional translation initiation events as RAN translation, with the understanding that future studies will be needed to truly delineate the mechanism(s) by which this process occurs.
RAN translation of CGG repeats in FXTAS
The gene FMR1 contains a polymorphic CGG repeat in its 5’ leader. This repeat is conserved among mammals, and increases in size in higher order primates (Eichler et al. 1995). In humans, this CGG element can expand to intermediate or “premutation” lengths of between 55 and 200 repeats. When this occurs, there is an enhancement in FMR1 transcription, but inefficient Fragile X protein (FMRP) production (Tassone, Hagerman, Taylor, et al. 2000). Clinically, premutation expansions result in the neurodegenerative disorder Fragile X-associated Tremor/Ataxia Syndrome (FXTAS) (Hagerman 2013). FXTAS occurs in ~1:5000 men over the age of 50 and in a lower percentage of female premutation carriers. In addition, premutation sized repeat expansions also cause premature ovarian failure, or Fragile X-associated Primary Ovarian Insufficiency (FXPOI), which is the most common genetic cause of early menopause. In both cases, the repeat is thought to elicit disease primarily through gain-of-function mechanisms, as expression of the repeat element in isolation is sufficient to elicit toxicity and enhancing or suppressing FMRP expression fails to modulate repeat elicited phenotypes in model systems (Lu et al. 2012; Arocena et al. 2005; Jin et al. 2007; Hashem et al. 2009). In contrast to FXTAS and FXPOI, Fragile X Syndrome (FXS) results from expansion of CGG repeats in FMR1 to greater than 200. This leads to hypermethylation of the promoter region and CGG repeats and heterochromatization of the FMR1 locus. This epigenetic change leads to a partial or complete transcriptional shut down of the FMR1 locus (Wang, Berry-Kravis, and Hagerman 2010). FXS is the most common known monogenic cause of autism and intellectual disability. FXS results from the loss of FMRP, as point mutations and deletions in FMR1 also elicit the clinical syndrome independent of repeat expansions. There is no evidence for neurodegeneration as a major feature in FXS cases, which suggests that the loss of FMRP is not a driver of the clinical or pathological features observed in FXTAS. Thus, while these diseases are allelic and result from expansion of the same repeat, they appear to have largely separate pathogenic mechanisms.
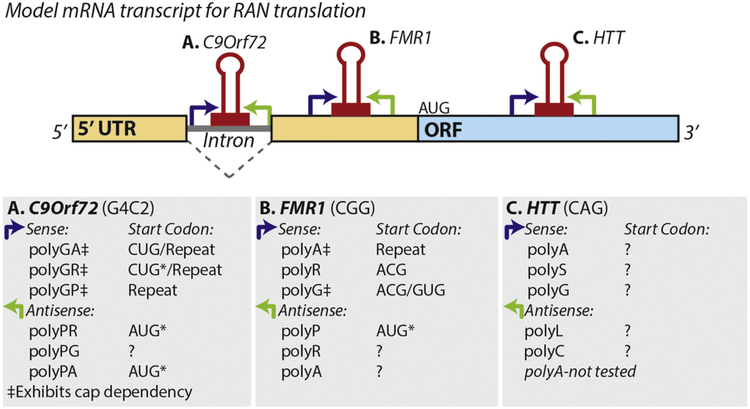
The FMR1 CGG repeat was found to support RAN translation in all three potential reading frames on the sense strand in the absence of an AUG (Krans, Kearse, and Todd 2016; Todd et al. 2013; Kearse et al. 2016), albeit with differing efficiencies. RAN translation also occurs at CCG repeats generated by antisense transcription through the beginning of the FMR1 gene (Krans, Kearse, and Todd 2016). Distinct from what was described in ATXN8, RAN translation in two of the three potential reading frames of the 5’ leader of FMR1 relies on initiation at near-cognate start codons just upstream of the repeat (Kearse et al. 2016; Todd et al. 2013). On the sense strand, the 0-frame RAN initiation event uses a ACG initiation site, and the +1-frame uses both an ACG and a GUG for initiation (Figure 2) (Kearse et al. 2016; Todd et al. 2013). Peaks upstream of the repeat were detected in RP datasets from mouse and man (Todd et al. 2013). Interestingly, an earlier study found that the Drosophila homologue to FMR1, dFMR1, encodes a functional N-terminal expansion that initiates at a CUG start site (Beerman and Jongens 2011). In CGG RAN translation, the +2-frame does not appear to rely on a start codon upstream of the repeat, making it more akin to what is observed at CAG repeat expansions with initiation occurring within the repeat (Kearse et al. 2016). Interestingly, both the +1-frame and +2-frame products increase as a function of increased repeat size, and 0-frame RAN translation is attenuated at normal and expanded repeat lengths (Todd et al. 2013; Kearse et al. 2016).
Figure 2: RAN translation in repeat expansion disorders.
Repeat associated non-AUG translation at three repeat expansions are laid out schematically. Experimentally determined initiation sites are listed for individual reading frames where determined, although in all cases initiation may also occur within the repeat itself. A) A G4C2 hexanucleotide repeat in the first intron of C9Orf72 is bidirectionally transcribed. Whether the repeat is translated as a spliced intron or as part of a retained intron or other aberrant transcript is not known. A CUG located just 5’ to the repeat is utilized for initiation of poly(GA). It may also be utilized for poly(GR) translation with subsequent frameshifting. AUG codons reside upstream of poly(PR) and poly(PG) in the most common antisense transcripts. B) A CGG repeat in the 5’ leader/ 5’ UTR of FMR1 is also bidirectionally transcribed. An AUG in the poly(P) reading frame of ASFMR1 is utilized for initiation in this reading frame but it can also undergo RAN translation in this reading frame. FMRpolyR expression is nearly undetectable from reporter constructs. C) The CAG repeat in Huntington’s disease is located in the coding region of the HTT transcript and is also bidirectionally transcribed. ?: initiation site unknown, ‡: initiation in this frame exhibits cap-dependency.
The scanning mechanism of translation initiation broadly refers to the process whereby the small ribosomal subunit with its associated factors interacts with the modified guanosine cap (m7G) on the 5’ end of mRNA transcripts, then progresses in the 3’ direction to a start codon where it joins with the large subunit to initiate translation (Hinnebusch 2011; Hinnebusch, Ivanov, and Sonenberg 2016). This process begins when a free 40S ribosomal subunit binds the ternary complex which is composed of the methionine initiator tRNA bound to eIF2, forming the 43S preinitiation complex (PIC). The PIC is recruited to the 5’ end of the transcript through interaction with the eIF4F complex, which binds the m7G cap. The PIC complex then scans along the mRNA, proceeding in a 5’ to 3’ direction, while sampling the sequence for pairing with the initiator methionine tRNA at an AUG initiation codon. Prior to experimentation, it was unknown whether RAN translation used a cap-dependent mechanism or an internal ribosome initiation site (IRES)-like mechanism for initiation (Kearse and Todd 2014). IRES initiation occurs on highly structured transcripts, and directly recruits the 43S PIC to the transcript independent of the cap (Pelletier and Sonenberg 1988; Jang et al. 1988; Pestova, Shatsky, and Hellen 1996; Pestova, Hellen, and Shatsky 1996). While originally described for viral elements, this same process also occurs from a subset of native mRNA transcripts in human cells (Ray, Grover, and Das 2006; Vagner et al. 1995; Coldwell et al. 2000; Gan and Rhoads 1996). Kearse et al. (2016) showed that translation of CGG repeats in RAN reporter transcripts required a functional 5’ cap and interaction with the cap-binding protein eIF4E, a component of the eIF4F complex that binds to the cap, and that blocking eIF4A, the helicase critical for PIC scanning, significantly impaired RAN translation in all three reading frames (Kearse et al. 2016). These data point to a canonical scanning mechanism of translation initiation at the CGG repeat, with a primary failure in start codon fidelity leading to RAN initiation (Green, Linsalata, and Todd 2016).
There is significant evidence supporting a role for RAN translation in the pathogenesis of FXTAS. RAN translation of the +1-frame CGG RAN protein, FMRpolyG, as a GFP fusion construct elicits formation of large intranuclear ubiquitinated inclusions in multiple model systems, including Drosophila, rodent neurons, mice and human cells (Sellier et al. 2017; Todd et al. 2013). These inclusions are highly reminiscent of the pathological hallmark of the disease observed in patients (Greco et al. 2006). Three different groups have now generated antibodies against different epitopes on FMRpolyG, which contains an expanded polyglycine stretch near its N-terminus. These antibodies recognize recombinant FMRpolyG protein and stain inclusions in multiple mouse and cell based models of the disease and, more importantly, in pathological samples from FXTAS cases (Todd et al. 2013; Sellier et al. 2017; Buijsen et al. 2016). FMRpolyG aggregates are also identified in ovarian stromal cells from FXPOI patients (Buijsen et al. 2016). FMRpolyG staining co-localizes with p62 and ubiquitin positive inclusions in patient brains, whereas pre-immune sera or peptide blocked sera do not recognize such inclusions (Todd et al. 2013; Buijsen et al. 2016; Sellier et al. 2017)(Amy Krans, personal communication).
In mice, ubiquitous transgenic expression or neuronal specific expression of the 5’UTR of FMR1 leads to neuronal inclusion formation, motoric dysfunction, Purkinje cell loss, and a decrease in survival (Sellier et al. 2017). In contrast, expression of this same CGG repeat sequence lacking the near-AUG initiation sites that support FMRpolyG production has no phenotype, no ubiquitinated inclusions and normal survival(Sellier et al. 2017). Similar results are observed in Drosophila models of FXTAS and in cultured neurons (Todd et al. 2013). Driving expression of FMRpolyG causes impaired proteasome function in Drosophila and human cells (Oh et al. 2015) and nuclear lamina integrity is affected by FMRpolyG expression, which directly interacts with the inner nuclear membrane protein Lap2ß (Sellier et al. 2017). In CGG repeat expansion “knock-in” mouse models, only those with 5’UTR sequences that support FMRpolyG production form neuronal ubiquitinated inclusions (Todd et al. 2013). Taken together, these data suggest that FMRpolyG production is necessary for CGG repeats to elicit neuronal toxicity and pathology across a variety of model systems. However, as CGG knock-in mice lack significant neurodegeneration (Krans and Todd, unpublished results), a direct link between RAN translation and inclusion formation with toxicity in endogenous systems lacking overexpression has not yet been established.
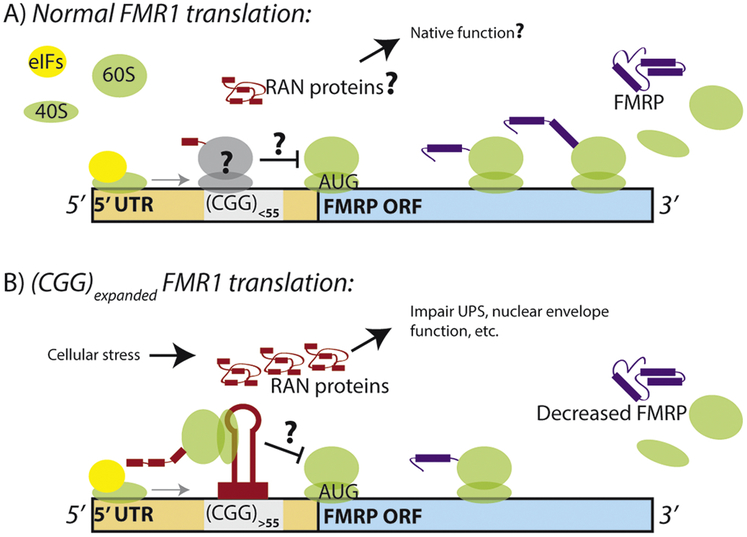
FMR1 RAN translation is also interesting because it appears to occur at normal repeat lengths in the unaffected human population. This is supported by both ribosome profiling data in cell lines with a normal repeat length and reporter studies (Todd et al. 2013; Kearse et al. 2016). This is important for two reasons: 1) it suggests that RAN proteins from this transcript can be synthesized under normal conditions and thus could have normal functions, and 2) translation in the 5’ leader of an mRNA transcript could have a significant negative impact on the translation of the main protein-coding ORF, suggesting that RAN translation on FMR1 could act as an upstream open reading frame (uORF) (Figure 3). uORFs can implement a regulatory role on its resident transcript through several overarching mechanisms: through sequence-dependent interactions with the translation machinery, through cis- and trans-interactions with uORF-derived peptides, and through influencing RNA stability (Hinnebusch, Ivanov, and Sonenberg 2016). Considering the translational efficiency of FMR1 and the levels of FMRP are both known to be decreased in FXTAS where RAN translation occurs most, RAN translation may act as a uORF to repress downstream FMRP synthesis (Tassone, Hagerman, Loesch, et al. 2000; Feng et al. 1995). uORF translation is intricately tied to environmental stressors that act upon translation machinery. Moreover, uORFs have been implicated in neuronal response to receptor activity as the transcripts that show a higher level of translation after pharmacological activation of metabotropic glutamate receptors are enriched in uORFs (Di Prisco et al. 2014). FMR1 is an important neuronal transcript whose regulation is likely central to normal neuronal function and plasticity. Thus elucidating how its 5’ leader might regulate its own translation and produce novel synaptic proteins could reveal a lot about gene expression in the nervous system.
Figure 3: RAN translation as a regulatory element.
A) Translation from the FMR1 transcript produces both RAN translation products and FMRP. Reporter constructs and ribosome profiling data suggests that this process occurs at the normal repeat size in humans. As both cistrons utilize a scanning- and cap-dependent initiation mechanism, preinitiation complexes must bypass RAN initiation sites and the CGG repeat to make FMRP. As such, RAN translation of FMRpolyG might serve a regulatory upstream open reading frame (uORF) to control FMRP synthesis. B) At expanded repeats, RAN translation is more efficient, which is correlates with less FMRP translation per transcript.
RAN translation in HD and ALS/FTD
Two other nucleotide repeat expansion disorders where RAN translation may contribute to overall pathology are HD and ALS. HD is a polyglutamine disease caused by CAG repeat expansions in the coding region of HTT, the Huntingtin gene. Patients generally exhibit involuntary movements and cognitive decline (Group 1993; Dickey and La Spada 2018). Individuals with larger expanded repeats are prone to earlier onset HD (Paulson 2018a; Andrew et al. 1993). While the polyglutamine containing protein defines the pathology of this disease, Banez-Coronel and colleagues (2015) showed that it is not the only homopolymeric expansion protein derived from this locus. RAN translation was shown to occur in both the sense and antisense direction through the CAG/CTG repeat, despite its residence within a protein coding open reading frame. In addition to the polyglutamine product, both poly-serine and polyalanine products (sense) as well as poly-leucine and poly-cysteine products (antisense) were detected (Banez-Coronel et al. 2015). The RAN peptides aggregate in patient brains, and are increased in abundance in early onset HD (Banez-Coronel et al. 2015). Intriguingly, HD RAN peptides are linked to nucleocytoplasmic export disruption in neurons (Grima et al. 2017).
In 2011, expansion of the hexanucleotide repeat G4C2 in the gene C9Orf72 was discovered to be the most common known mutation underlying both ALS and FTD (Renton et al. 2011; DeJesus-Hernandez et al. 2011). Unaffected individuals normally have up to around 23 repeats, whereas individuals with ALS can have hundreds or thousands of this hexanucleotide repeat. G4C2 bearing transcripts are transcribed in the sense and antisense direction, and the repeat is present in an intron or promoter region of sense strand RNA transcripts. RAN translation through this repeat produces five potential dipeptide repeat (DPR) tract proteins—polyglycine/alanine (GA), -glycine/arginine (GR), -proline/alanine (PA), -proline/arginine (PR), and -glycine/proline (GP) (Green, Linsalata, and Todd 2016; Ash et al. 2013; Gendron et al. 2013; Zu et al. 2013; Mori et al. 2013). C9orf72-associated RAN proteins accumulate in the brains of patients in aggregates distinct from the TDP-43 positive aggregates classically seen in ALS/FTD. Several studies have concluded that GA-DPRs are the most abundant of the G4C2 RAN proteins pathologically (Gendron et al. 2015; Mackenzie et al. 2015; Mann et al. 2013). In vitro and cell based assays similarly demonstrate that translation in the GA reading frame of the sense strand repeat is the most efficient, with lesser expression in the GR and GP reading frames (Green et al. 2017; Sonobe et al. 2018; Westergard et al. 2019; Tabet et al. 2018). In addition, each frame exhibits different length-dependency and differential impedances based on initiation and elongation (Green, Linsalata, and Todd 2016; Tabet et al. 2018; Cheng et al. 2018). G4C2 RAN translation from linear mRNA transcripts appears to mainly depend on cap-binding of eIF4E and a canonical scanning mechanism involving eIF4A, similar to what has been reported for CGG repeats (Green et al. 2017; Tabet et al. 2018). However, G4C2 RAN can occur to at least some degree from bicistronic constructs and transcripts, suggesting a mechanism more aligned with internal ribosome entry (IRES) (Cheng et al. 2018; Sonobe et al. 2018). Translation in the GA reading frame primarily utilizes a near-cognate CUG codon 5’ to the repeat for initiation, as well as an AGG closer to the repeat (Green et al. 2017; Tabet et al. 2018). The dependency on a single start site for multiple reading frames suggests that frame-shifting may occur in certain contexts (Tabet et al. 2018). Frameshifting was previously reported at CAG repeats (Toulouse et al. 2005; Wills and Atkins 2006; Stochmanski et al. 2012; Girstmair et al. 2013; Wojciechowska et al. 2014; Saffert et al. 2016) but did not appear to be a major contributor to RAN translation product generation from CAG repeats on ATXN8OS (Zu et al, 2011). Thus, while many nucleotide repeat expansions appear to support RAN, the underlying mechanisms appear to have significant differences—not only between repeats but even in different frames of the same repeat.
The Integrated Stress Response (ISR) is activated by specific stimuli, namely ER stress, viral infection, oxidative stress and amino acid deprivation (Pakos-Zebrucka et al. 2016; Young and Wek 2016). These stimuli each act on specific kinases that phosphorylate eIF2α. eIF2 is comprised of three subunits. The γ subunit binds GTP. When GTP is hydrolyzed to GDP, eIF2 cannot bind methionine initiator tRNA. Phosphorylation of the α subunit of eIF2 inhibits the guanine exchange factor (GEF) eIF2B, which is responsible for adding GTP to eIF2 (Hinnebusch 2011). Thus, phosphorylation of the α subunit leads to a limiting number of ternary complexes and a global decrease in protein synthesis (Harding et al. 2003; Pakos-Zebrucka et al. 2016; Harding et al. 2000). Despite a global translational arrest, a subset of transcripts show a paradoxical increase in translation, and many of the gene products are associated with cellular response to mitigate stress and maintain homeostasis (Harding et al. 2000; Vattem and Wek 2004). Intriguingly, under ER stress and phosphorylation of eIF2α, CGG RAN translation of the +1− and +2-frames increases despite the global decrease in translation (Green et al. 2017). This increase was dependent on the presence of a near-cognate start site, and was also observed at the ALS/FTD G4C2 repeat (Green et al. 2017; Cheng et al. 2018; Sonobe et al. 2018; Westergard et al. 2019). Importantly, expression of either expanded CGG or G4C2 repeats induces stress granule formation in cells, which is dependent on eIF2α phosphorylation (Green et al. 2017; Cheng et al. 2018; Rossi et al. 2015). In addition, expression of GA DPRs triggers ISR activation independent of RNA repeats through a PERK-dependent mechanism(Zhang et al. 2014). This ISR activation, in concert likely with direct effects of DPRs on nucleolar function and ribosomes, leads to a suppression of global protein translation in cells expressing repeats or arginine-rich DPRs(Kwon et al. 2014; Zhang et al. 2014; Rossi et al. 2015; Tao et al. 2015; Yamakawa et al. 2015; Kanekura et al. 2016; Lee et al. 2016; Green et al. 2017; Mizielinska et al. 2017; Hartmann et al. 2018; Zhang, Gendron, et al. 2018; Moens et al. 2019; Haeusler et al. 2014). Translation of the GA reading frame of the G4C2 repeat was also found to be partly dependent on eIF2A, an initiation factor with roles in unconventional translation initiation (Sonobe et al. 2018; Starck et al. 2012). In iPS spinal motor neurons derived from patients with the G4C2expansion both cellular stress and neuron-specific excitotoxic stress, through application of homocysteine or excess glutamate, increased endogenous poly(GA), (GP), and (GR) at variable levels (Westergard et al. 2019). This was also observed in rat cortical neurons expressing reporters for each protein (Westergard et al. 2019). Taken together, these data support the model of toxicity where RAN translation is upregulated in the presence of cellular stress. In turn, both repeats as RNA and translated RAN proteins independently induce stress, creating a feed-forward loop that leads to further RAN translation and a global suppression of protein translation that contributes to neurodegeneration (Green et al. 2017).
Microsatellites impact on liquid-liquid phase separation of membraneless organelles
Membraneless organelles are organizational centers formed within the cell to perform a variety of functions in close proximity and shielded from the rest of the cellular milieu but in the absence of a structural membrane. For example, Cajal bodies and the nucleolus are nuclear organelles that perform specialized tasks which require a very specific set of molecular components within a restricted area (Boisvert et al. 2007; Gall 2003). Membraneless organelles are forged through liquid-liquid phase separation (LLPS), the physical process by which two liquids, when mixed together, spontaneously separate. The simplest way to explain LLPS is the example of oil and vinegar (Hyman, Weber, and Julicher 2014). When oil and vinegar are added together, discrete droplets of each liquid form through de-mixing to accomplish a lower free energy state (Gomes and Shorter 2018). In the cell, local molecular interactions can drive phase separation between the cytosol and its various components, which are classified as liquids (Hyman and Brangwynne 2011). These liquids can take many forms such as gels and colloidal solutions (Hyman, Weber, and Julicher 2014). LLPS ultimately both drives the formation of and maintains specialized compartments that can perform specific cellular tasks.
Proteins susceptible to phase separation commonly have intrinsically disordered regions (IDR). IDRs typically have low sequence complexity, and are enriched in amino acids that promote low hydrophobicity and a high net charge including Ala, Arg, Gly, Gln, Ser, Glu, Lys, and Pro (Uversky and Dunker 2010). An important outcome of such attributes is a failure to form rigid 3-D structures, and as a result they maintain a high level of flexibility (Uversky and Dunker 2010). IDRs permit a wide range of molecular interactions, and thus allow for both homomeric and heteromeric protein complex formations (Dyson and Wright 2005). In the crowded cytoplasm of a cell, binding of an IDR-containing protein to its molecular interactors drives up the local concentration of these proteins, which in turn favors phase separation a mechanism of reaching higher thermodynamic stability through multivalent interactions (Boeynaems et al. 2018). Importantly, some proteins with IDRs can self-assemble (Malinovska, Kroschwald, and Alberti 2013). However, when the local concentration of IDR-containing proteins is too high, or their relative affinities and interactions are stronger, they form more rigid conformations that are less dynamic and harder to disassemble (Dougan et al. 2009; Crick et al. 2006; Vitalis, Wang, and Pappu 2007; Walters and Murphy 2009). As discussed above, a hallmark of diseases in which microsatellite repeats are encoded into proteins is the presence of aggregates. It is thought that pathologically expanded repeat proteins prevent proper liquid-liquid phase transitions by forming solid, insoluble aggregates (Malinovska, Kroschwald, and Alberti 2013; Dobra et al. 2018; Boeynaems et al. 2018). Intramolecular interactions may also play a role. For example, as the size of a polyglutamine stretch within a protein expands, the monomer is more likely to adopt a structured conformation, increasing the propensity to form aggregates both with itself and with other proteins (Walters and Murphy 2009; Haran 2012; Vitalis and Pappu 2011; Williams and Paulson 2008).
Ribonucleoprotein (RNP) granules make up one category of membraneless organelles. These are dense bodies of RNA and RNA binding proteins (RBP) (Spector 2006). Messenger RNP (mRNP) granules form in the cytoplasm and are composed of translationally-silenced mRNA (Anderson and Kedersha 2009). In neurons, mRNPs are important for regulation of translation and trafficking of mRNAs out to synapses. The fragile X protein FMRP is an RNA binding protein and a key component of neuronal mRNP granules within dendrites. FMRP binds to ~4% of the brain transcriptome, with enrichment for transcripts related to neuronal function and whose dysfunction are associated with autism and intellectual disability (Siomi et al. 1994; Siomi et al. 1993; Brown et al. 2001; Darnell et al. 2011; Ascano et al. 2012). Loss of FMRP in models of FXS leads to altered global protein translation and mRNA transport within neurons, and as a consequence, dysregulated activity-dependent translation at synapses (Thomson et al. 2017; Darnell et al. 2001; Laggerbauer et al. 2001; Li et al. 2001; Todd, Mack, and Malter 2003; Brown et al. 2001; Zalfa et al. 2003; Huber et al. 2002; Liu et al. 2018; Udagawa et al. 2013; Banerjee et al. 2018; Ifrim, Williams, and Bassell 2015; Gross and Bassell 2012; Liu-Yesucevitz et al. 2011; Gross et al. 2011). This uncoupling of synaptic activity and local protein synthesis is believed to underlie learning and memory deficits in FXS. Ribosome profiling, a next generation sequencing assay to survey the location of all translating ribosomes, was implemented in adult neural stem cells of FMR1 knockout mice and compared to wild-type cells (Liu et al. 2018). This work corroborated aspects of FMRP’s proposed role as a translational repressor of mRNA transcripts to which it binds by describing increased translational efficiency of FMRP CLIP (cross-linking immunoprecipitation) targets in FMR1 knock-out cells. However, global translational efficiency changes were largely buffered at the mRNA level suggesting that compensatory changes may bypass some of FMRP’s roles as a translational regulator (Liu et al. 2018). What remains to be explored is how the absence of FMRP and this compensated state influences activity-dependent translation shifts in specific cellular compartments such as dendrites and axons.
Stress granules are a specialized type of mRNP with roles in a diverse set of neurodegenerative diseases (Wolozin 2012; Kedersha et al. 1999). They contain mRNAs stalled at translation initiation, along with small ribosomal subunits, select initiation factors, and RBPs such as G3BP, DDX3X, FMRP and TIA1 (Protter and Parker 2016; Buchan and Parker 2009; Teixeira et al. 2005; Hilliker et al. 2011). Stress granules form as a consequence of cellular stress that induces eIF2α phosphorylation and translational inhibition. This blockade of initiation without a concomitant blockade of elongation creates “naked” stretches of mRNA on protein-coding transcripts that would normally be coated with translating ribosomes. This unassociated RNA facilitates specific protein-protein interactions presumably by creating a nidus upon which abundant RBPs with low sequence binding specificity can assemble (Molliex et al. 2015; Lin et al. 2015). RNA-RNA interactions, RNA-protein interactions as well as protein-protein mediated phase separation of factors such as G3BP through IDR domains all contribute to this process (Molliex et al. 2015; Lin et al. 2015).
Stress granules are generally thought of as a way for cells to redirect their energy expenditure elsewhere in the setting of a transient insult to regain their healthy set point. While stress granules are generally adaptive, alterations in their assembly and disassembly dynamics may contribute meaningfully to multiple neurodegenerative disorders (Li et al. 2013; Wolozin 2012; Becker and Gitler 2018). Overexpression of C9 FTD/ALS DPRs modulates SG granule formation and dynamics (Zhang et al. 2014; Zhang, Gendron, et al. 2018; Lee et al. 2016; Kanekura et al. 2016; Tao et al. 2015; Rossi et al. 2015), with poly(GA) contributing to SG activation and poly(GR) and poly(PR) impacting SG stability and disassembly.
The polyglutamine tract in the ataxin-2 protein regulates membraneless organelles in the cytoplasm
Ataxin-2 is a common eukaryotic mRNP granule component that can bind directly to RNA and other RBPs (Kozlov et al. 2010; Swisher and Parker 2010; Nonhoff et al. 2007; Lastres-Becker et al. 2016). It helps promote, but is not required for, stress granule assembly in yeast and human cells, and is necessary for proper formation of RNP granules positive for the stress granule marker Me3B1 in Drosophila projection neurons (Buchan, Muhlrad, and Parker 2008; Becker et al. 2017; Bakthavachalu et al. 2018). Ataxin-2 has a polyglutamine tract in its IDR. Deletion of the IDR in the ataxin-2 protein, including the polyglutamine tract, disrupts Me3B1-positive RNP granule formation (Bakthavachalu et al. 2018). The ataxin-2 polyglutamine tract, encoded by a CAG microsatellite, is polymorphic in humans and can become expanded as a result of somatic and intergenerational instability (Pulst, Nechiporuk, and Starkman 1993; Sanpei et al. 1996; Pulst et al. 1996; Imbert et al. 1996). The mean repeat size in the population is 25 CAGs. Expansions beyond 34 CAGs causes spinocerebellar ataxia type 2 (SCA2), a late onset neurodegenerative disorder characterized by cerebellar dysfunction, spasticity, and some motor neuron disease-like features (Pulst 1993). In contrast, expansion of the repeat in the intermediate range (~27-33 CAGs) increases the risk for ALS without any association with SCA2 (Elden et al. 2010). Ataxin-2 interacts with FUS and TDP-43, RBPs that are commonly mutated and mislocalized in ALS. Intermediate expansions in ataxin-2 can enhance FUS mislocalization from the nucleus to the cytoplasm and ER-Golgi disruption in the setting of an ALS-causing FUS mutation (Farg et al. 2013). Deletion of the disordered region of ataxin-2, including the polyglutamine tract, partially rescues the eye degeneration phenotype in a Drosophila model expressing human FUS (Bakthavachalu et al. 2018). Additionally, intermediate length repeat expansions increase caspase 3 activation in cells expressing mutant TDP-43, and patient motor neurons with an intermediate length repeat show increased activated caspase 3 linking this moderate microsatellite repeat expansion directly to motor neuron death (Hart and Gitler 2012). Decreasing wild-type Ataxin-2 levels rescues mutant TDP-43 toxicity in yeast, fly, and mouse models of ALS (Elden et al. 2010; Becker et al. 2017) while decreasing expression of expanded Ataxin-2 alleles suppresses SCA2-relevant phenotypes in mice (Scoles et al. 2017). While the exact mechanism by which intermediate length expansion increases the risk for motor neuron disease is yet to be deciphered, insight from research on ataxin-2 in various RNP granules makes disrupted RNP granule dynamics a likely contributor (Zhang, Daigle, et al. 2018).
Transcribed and translated C9ORF72 hexanucleotide repeat impairs nuclear phase separation
In the setting of the C9ORF72 expanded G4C2 repeat, the arginine-containing RAN proteins, poly(GR) and poly(PR), contribute to impaired membraneless organelle dynamics. Poly(GR) and poly(PR) peptides are extremely toxic to cells when put directly into culture medium or expressed from a plasmid (Kwon et al. 2014; Wen et al. 2014; Tao et al. 2015; Lee et al. 2016; Flores et al. 2016), and cause neurodegeneration in Drosophila models (Mizielinska et al. 2014; Lee et al. 2016; Wen et al. 2014). Poly(PR) and poly(GR) cellular interactors were identified by mass spectrometry; their interactomes were enriched in low-complexity domains, a common attribute of IDRs that can contribute to liquid-liquid phase separation, and many poly(PR) interactors fall into the category of membraneless cellular puncta (Lee et al. 2016; Lin et al. 2016; Zhang, Gendron, et al. 2018; Hartmann et al. 2018). One such protein was hnRNPA1, an RBP component of stress granules with a prion-like domain in which mutations can lead to impaired phase transition and aberrant fibrillization (Guo et al. 2018; Lin et al. 2016; Kim et al. 2013). Through mutational analysis, the low-complexity domain of hnRNPA1 was determined to be necessary and sufficient for this poly(PR) interaction providing evidence that poly(PR) can directly interact with these domains (Lin et al. 2016). Poly(PR) and poly(GR) binding also promotes hnRNPA1 phase separation in vitro (Lee et al. 2016). Both RAN peptides promote the assembly of aberrant stress granules, and lower the critical concentration of hnRNPA1 and TIA-1, another stress granule protein, required for phase separation in vitro (Lee et al. 2016; Boeynaems et al. 2017). Poly(GA) peptides also independently trigger SG formation (Tao et al. 2015), and their co-expression with arginine rich repeat influences the toxicity and behavior of these charged DPRs (Yang et al. 2015; Darling et al. 2019; Lee et al. 2017).
Overexpression of poly(GR) and poly(PR) localize directly to the nucleolus, and poly(GR) also localizes to the cytoplasm (Lee et al. 2016; Wen et al. 2014; Tao et al. 2015; Kwon et al. 2014). Poly(GR) and poly(PR) impair the behavior of proteins that help organize the nucleolus through liquid-liquid phase transition. Ribosome biogenesis is also impaired in their presence, which takes place in the granular component of the nucleolus where both RAN peptides localize (Lee et al. 2016; Tao et al. 2015; Suzuki et al. 2018). Additionally, poly(GR) and poly(PR) alter the assembly of two other nuclear membraneless organelles: Cajal bodies and nuclear speckles (Lee et al. 2016). Taken together, these data demonstrate that arginine-containing RAN peptides can drive toxicity in cells and neurons by disrupting the phase separation properties of membraneless organelles. However, it is important to acknowledge that almost all studies to date have been done with constructs where poly(PR) and poly(GR) are generated from AUG-driven translation in the absence of the repeat element and usually fused to a fluorescent protein tag such as GFP. Given that both of these proteins are present at very low levels in C9 ALD/FTD neurons, these overexpression systems may not recapitulate what happens endogenously. In postmortem patient brain tissue, aggregates of poly(PR) and poly(GR) appear mostly perinuclear (Zu et al. 2013; Mori et al. 2013). Moreover, there is evidence that the other DPRs generated by RAN translation may counteract or influence the behavior of these arginine rich proteins in terms of their distribution and relative toxicity (Darling et al. 2019). Thus, future studies are needed which selectively decrease expression of these arginine-rich DPRs from endogenous loci to better delineate impact they have in the disease state.
G4C2 repeat RNAs form G-quadraplex structures in vitro and there is some evidence to suggest that these same structures form in vivo (Conlon et al. 2016; Fay, Anderson, and Ivanov 2017; Reddy et al. 2013; Haeusler et al. 2014). G4C2 repeat-containing RNAs can also undergo phase separation (Moens et al. 2018). Purified (G4C2)n RNA form gels in vitro in a repeat-dependent manner, and G4C2 RNAs form foci when transfected into cells similar to what is observed for (CAG)n and (CUG)n RNA (Jain and Vale 2017; Fay, Anderson, and Ivanov 2017). Foci are also present in patient cells, including brain tissue and motor neurons (Lee et al. 2013; Lagier-Tourenne et al. 2013; Sareen et al. 2013; Donnelly et al. 2013). Of note, another study used the MS2 system to tag both (G4C2)n and (CAG)n RNA, both showed repeat-dependent neurite localization with the (CAG)n RNA increasing in neurite localization at larger repeats, with the opposite pattern of localization occurring with expansion of (G4C2)n RNA (Burguete et al. 2015). Neuritic expanded G4C2 and CAG repeat-containing RNA was shown to be actively transported in mRNP granules, and was correlated with neurite branching defects. Intriguingly, neurite-localized G4C2RNA puncta were detected in iPSC-derived neurons from C9orf72 hexanucleotide expansion carriers (Burguete et al. 2015).
Sequestration of RBPs is a common pathologic mechanism underlying microsatellite expansions categorized as RNA gain-of-function mutations (Todd and Paulson 2010). G4C2 nuclear RNA foci sequester a number of RBPs including ADARB2, SRSF2, hnRNP H1/F, Aly/REF, hnRNP A1, Pur α, hnRNP H, among others (Donnelly et al. 2013; Cooper-Knock et al. 2014; Conlon et al. 2016; Xu et al. 2013; Sareen et al. 2013; Lee et al. 2013). Sequestration of hnRNP H, specifically, can lead to mis-splicing of its targets (Lee et al. 2013; Conlon et al. 2016). However, the exact mechanism by which such repetitive elements might bind to and sequester RNA binding proteins and thus drive cellular dysfunction is less clear (Renoux and Todd 2012). The working model for RNA dominant toxicity has been that sequestered proteins would need to have a very high affinity for the repeat element sequence, otherwise their interactions would be dynamic and thus functional sequestration would be unlikely. One would anticipate that highly abundant proteins would in this context be very difficult to functionally sequester. Yet, in most contexts, the interactions between repeat RNAs and their target RBPs are reversible in vitro and with binding affinities that would make functional sequestration difficult to achieve through classically understood RNA-protein binding interaction relationships. Proteins such as Muscle blind and hnRNP H are abundant in cells. In this context, recent studies demonstrating that repeat RNAs themselves can phase separate are quite intruiging (Jain and Vale 2017; Fay, Anderson, and Ivanov 2017). Moreover, RNA-mediated phase separation can intersect with protein-mediated phase separation as the addition of polyuridine RNA to purified poly(PR) protein in vitro promotes de-mixing in a dose-dependent manner, and this relationship influences stress granule formation and dynamics (Boeynaems et al. 2017; Van Treeck et al. 2018). If microsatellite expansion mutations drive phase separation of RNAs, then perhaps functional sequestration is achieved through a synergistic combination of classic repetitive RNA motif-protein interactions and phase separation that could be aided by both the intrinsically disordered domains of the protein and the phase separation propensities of the RNA. In such a context, even moderate affinity interactions with the repetitive motif sequence could potentially push RNA-protein complexes into a phase separated or even gelated state that would be very stable, making the protein component inaccessible for normal cellular functions.
Aberrant phase transitions cause nucleocytoplasmic transport defects
The central transport system between the cytoplasm and the nucleus is the nuclear pore complex (NPC). It functions as a highly regulated channel to pass macromolecules between the two segregated compartments (Hoelz, Debler, and Blobel 2011). Its function is central to proper RNA trafficking, and thus proper gene expression. The NPC is a large macromolecular complex with a mass of around 66MDa in eukaryotes. It is composed of multiple molecules of approximately 30 different nucleoporins or nups that are classified by their position within the larger NPC (Reichelt et al. 1990). The greater structure of the NPC is formed by both symmetrical nucleoplasmic and cytoplasmic rings, and an inner ring complex. In the center is a channel which can dilate and contract to permit cargo through (Hoelz, Debler, and Blobel 2011). The barrier to this channel is formed by phenylalanine-glycine repeat containing nups (FG-nups) with low sequence complexity that must remain flexible and responsive to cellular conditions (Kim and Taylor 2017). The FG motifs interact with each other to create a meshwork barrier (Schmidt and Gorlich 2016). The “selective phase model” describes the FG-nup barrier as a hydrogel that forms reversible crosslinks between FG domains to act as a sieve (Frey and Gorlich 2007; Mohr et al. 2009; Kim and Taylor 2017; Ribbeck and Gorlich 2001; Schmidt and Gorlich 2015, 2016). The FG-nups undergo phase transitions through multivalent interactions of their IDRs in order to maintain compartmental fidelity.
The FG motifs interact with nuclear transport receptors that shuttle through the pore to facilitate the translocation of larger molecules. Importins and exportins, both different types of nuclear transport receptors, regulate the transport of molecules in and out of the nucleus with the help of the RanGTP-RanGDP system. Ran protein, which stands for Ras-related nuclear protein, regulates the assembly and disassembly of transport complexes (Hoelz, Debler, and Blobel 2011). Ran guanine exchange factor (RanGEF) protein is located in the nucleus, while Ran GTPase activating protein (RanGAP) is located exclusively in the cytoplasm (Kim and Taylor 2017). Segregation of RanGEF and RanGAP sets up a gradient of RanGTP that dictates the directionality of transport through the NPC and the integrity of the FG barrier is crucial for maintaining this barrier (Debler, Blobel, and Hoelz 2009).
A role for nucleocytoplasmic transport machinery in C9Orf72 ALS emerged from several coincident reports across model systems. First, a genetic screen was performed in a Drosophila model of C9Orf72 ALS that expressed a (G4C2)58 repeat within the 5’UTR of GFP, leading to an eye degeneration phenotype (Freibaum et al. 2015). Two key NPC proteins modified the eye phenotype. Deletion of Nup50 enhanced degeneration, while deletion of Ref1 mitigated G4C2 toxicity (Freibaum et al. 2015). Human NUP50 promotes nuclear import, whereas Aly/REF (the human orthologue of Ref1) assists in mRNP export (Makise et al. 2012; Kim and Taylor 2017; Viphakone et al. 2012). Fly salivary gland cells expressing the (G4C2)58 repeat exhibited a “wrinkled” nuclear envelope and an accumulation of nuclear RNA, which was also observed in patient iPSC-derived neurons (Freibaum et al. 2015). Work done by a separate group using a similar fly model with 30 repeats showed that a dominant, gain-of-function mutation in RanGAP, that works similar to RanGAP overexpression, suppressed repeat-induced toxicity (Zhang et al. 2015). RanGAP1 binds directly to G4C2-containing RNA, and RanGAP1 is mislocalized in repeat expressing cells and in C9Orf72 patient iPSCs and brain tissue (Zhang et al. 2015). This mislocalization of RanGAP disrupts the nucleocytoplasmic Ran gradient and impairs nucleocytoplasmic transport (Zhang et al. 2015).
Studies on the effect of peptides synthesized through RAN (Repeat Associated Non-AUG) translation of the G4C2 repeat in C9Orf72 revealed an interaction between several dipeptide repeat proteins and the nucleocytoplasmic transport machinery. A large-scale screen in Saccharomyces cerevisiae for modifiers of poly(PR) toxicity revealed importins as suppressors of toxicity when overexpressed (Jovicic et al. 2015). Further studies in a Drosophila model of RAN peptide toxicity whereby the transgene was codon-optimized to remove the G4C2 repeat in the RNA but still encode the same amino acids similarly identified multiple nucleocytoplasmic transport factors as modifiers of poly(PR) toxicity, corroborating several of the previously mentioned genetic interactors such as Nup50 and RanGAP1 (Boeynaems et al. 2016). Poly(PR) binds to the nup FG domain and disrupts the nuclear pore complex in Xenopus (Shi et al. 2017). In a mouse model overexpressing poly(GA) by viral injection, RanGAP1 as well as the nuclear pore protein POM121 showed aberrant accumulation (Zhang et al. 2016). Both often formed aggregates colocalizing with poly(GA) (Zhang et al. 2016). These reports point to a RAN peptide-mediated disruption of the nucleocytoplasmic machinery in the setting of the expanded G4C2 hexanucleotide repeat.
Conclusion
Over almost thirty years, research on a relatively rare series of NREDs has played a tremendous role in adding to our understanding of many aspects of neurobiology and neurodegeneration. The discovery of the G4C2 repeat in C9Orf72 in particular has been a recent catalyst for such research, some of which is highlighted here. However, our understanding of the native functions of tandem microsatellites themselves has seen only limited progress, despite a great potential for contributing to normal function and human disease. Current deep-sequencing technology do not adequately resolve most microsatellites, and thus we do not have an accurate map of the variance in repeating elements in the human genome to even begin predicting which repeats might be important. Given recent discoveries—even in the past year— of new repeats that cause human disease, it seems likely that we are overlooking many causing disease-causing microsatellites (LaCroix et al. 2019; Ishiura et al. 2018; Cortese et al. 2019; Sone et al. 2019).
Improved technologies for long read and single cell deep-sequencing may provide new insights into both somatic mosaicism and population level variance of repeat sizes in brain tissue (McConnell et al. 2017). In this context, many of the repeat-dependent effects described here could affect different cells in different ways, including actions mediated by more moderate “risk-factor” effects akin to what occurs with Ataxin-2. A greater understanding of the basics of repeat biology in normal cellular functions will be required to understand their aberrancy in disease pathogenesis. This becomes more apparent as new disease associated NREs are uncovered. For example, a dominantly inherited intronic AAGGG repeat expansion in the poly(A) tail of the RFC1 gene was recently identified a major cause of cerebellar ataxia, neuropathy, and vestibular areflexia syndrome (CANVAS) (Cortese et al. 2019). Histological examination of patient tissue reveals Purkinje cell loss, however neither RFC1 transcript nor protein abundance appears affected and no inclusions or RNA foci were identified (Cortese et al. 2019). Thus, at first pass, this new repeat does not seem to fit within our current conceptualization of how dominantly inherited repeat expansions cause disease. This mystery may open us to new insights into repeat biology, opening up exploration of how repeats may trigger distant epigenetic effects or alter the functions of non-coding RNAs. As with our evolving understanding of how repeats elicit toxic gain-of-function as RNA or through RAN translation, it will be important to keep an open mind on the possibilities for how such a repeat might cause disease.
Along these lines, the finding that microsatellites in noncoding regions of RNA can in fact be translated has the potential to dramatically alter how we view a series of normal cellular processes. Not only does this change the way expansion diseases are viewed, it changes our understanding of mRNA translation generally. Structured regions are quite common in RNA, and their ability to promote translation has long been overlooked (Kozak 1984). The finding that RAN translation is supported at shorter repeat lengths in reporter constructs suggests potential biological functions of these processes in healthy cells (Todd et al. 2013). Not only has this been detected with the CGG repeat, but certain reading frames of the C9Orf72 repeat as well (Green et al. 2017). As many of these transcripts are highly expressed in neurons, their roles in neuronal function may be particularly interesting. Such non-canonical ORFs might greatly expand the known human proteome and perform distinct functions. Given how pathological expansions of repetitive elements influence the assembly and disassembly of membraneless organelles, it will be particularly important to evaluate how transcribed and translated microsatellite repeats impact mRNP trafficking and temporal and spatial regulation of translational dynamics in neurons, which are highly compartmentalized and rely on precise regulation of such events to allow for circuit formation and synaptic plasticity.
Similarly, the ability of repetitive elements as RNA and protein to trigger phase separation and influence the formation of membraneless organelles has revealed an important component of cellular organization that becomes dysfunctional in NREDs. Yet, at normal repeat sizes these same elements likely enhance the ability of the proteins and transcripts in which they reside to become components of these complexes and thus may aid in their function rather than acting as a hindrance. For example, the CGG Repeat in FMR1 as RNA may influence its entry into mRNPs that allow its transport out into distal dendrites where it is translated in response to activity (Muslimov et al. 2011). As proteins, this is especially apparent in the nuclear pore, where phenylalanine-glycine repeat elements in specific nup proteins play important roles in the pore function that is actively impaired by charged poly(PR) proteins generated by RAN translation (Kim and Taylor 2017). Understanding these native functions and interactions elicited by repeats will allow a clearer delineation of the mechanisms by which expanded repeats impair the behavior of these macromolecular complexes and thus what cellular compensatory mechanisms might be co-opted as therapeutic targets in disease.
Rapid progress in our understanding of microsatellite expansions has led to a series of recent clinical trials in patients, some of which appear quite promising. Knockdown of repeat expanded mRNA transcripts using antisense oligonucleotides has been quite successful in animal models and is currently in trials in multiple repeat expansion disorders where gain-of-function mechanisms are thought to drive disease pathogenesis (Kordasiewicz et al. 2012). However, in diseases like FXTAS, where loss of the patent transcript itself would likely be problematic, alternative approaches will need to be developed. Such cases highlight the importance of further research on these topics in order to develop therapeutics that act downstream on affected pathways and not solely on the mutated gene. For example, reduction of wild-type Ataxin-2 in a mouse model of ALS provides one example of how targeted therapies aimed a more general component of the neurodegenerative process could prove helpful (Becker et al. 2017). With continued research, more discoveries like this will bring a greater understanding of microsatellite expansion diseases and novel strategies by which we might cure these diseases.
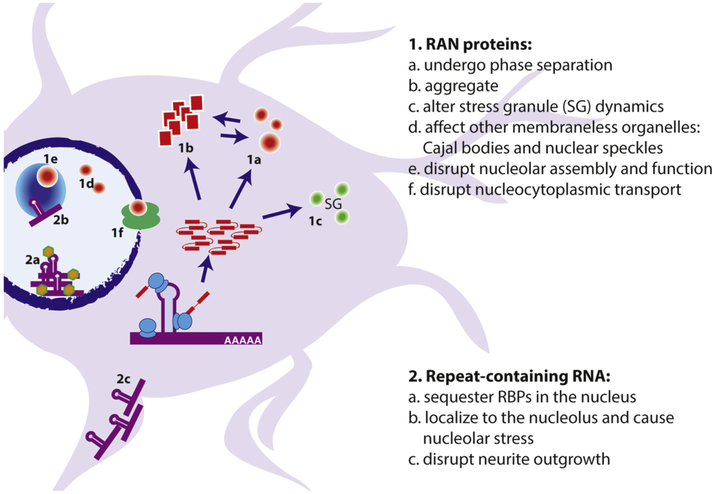
Figure 4: Mechanisms by which transcribed repeats elicit neurodegeneration.
Expanded repeats can be toxic as RNA or as protein. 1) poly(GR) and poly(PR) containing RAN DPR proteins undergo phase separation (1a) into liquid-like droplets (LLPS) in vitro and in vivo. Poly(GA), poly(GR), and poly(PR) RAN proteins can also alter stress granule assembly and dynamics (1b) and form insoluble aggregates (1c). In the nucleus, RAN proteins can impair the dynamics of Cajal bodies and membraneless organelles (1d), and disrupt nucleolar function and ribosomal biogenesis (1e). They can also disrupt the nuclear envelope and nuclear pore complex (1f). Repeat containing RNAs can sequester important RNA-binding proteins in the nucleus and can themselves undergo LLPS, perhaps in concert with RBPs (2a). Repeat RNAs that exit the nucleus can be exported to neuronal processes where they can impair neurite outgrowth and potentially alter mRNP dynamics (2b).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akbar U, and Ashizawa T. 2015. 'Ataxia', Neurol Clin, 33: 225–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, and Kedersha N. 2009. 'RNA granules: post-transcriptional and epigenetic modulators of gene expression', Nat Rev Mol Cell Biol, 10: 430–6. [DOI] [PubMed] [Google Scholar]
- Andrew SE, Goldberg YP, Kremer B, Telenius H, Theilmann J, Adam S, Starr E, Squitieri F, Lin B, Kalchman MA, and et al. 1993. 'The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington's disease', Nat Genet, 4: 398–403. [DOI] [PubMed] [Google Scholar]
- Arocena DG, Iwahashi CK, Won N, Beilina A, Ludwig AL, Tassone F, Schwartz PH, and Hagerman PJ. 2005. 'Induction of inclusion formation and disruption of lamin A/C structure by premutation CGG-repeat RNA in human cultured neural cells', Hum Mol Genet, 14: 3661–71. [DOI] [PubMed] [Google Scholar]
- Ascano M Jr., Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, Williams Z, Ohler U, and Tuschl T. 2012. 'FMRP targets distinct mRNA sequence elements to regulate protein expression', Nature, 492: 382–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW 3rd, Rademakers R, Boylan KB, Dickson DW, and Petrucelli L. 2013. 'Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS', Neuron, 77: 639–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayhan F, Perez BA, Shorrock HK, Zu T, Banez-Coronel M, Reid T, Furuya H, Clark HB, Troncoso JC, Ross CA, Subramony SH, Ashizawa T, Wang ET, Yachnis AT, and Ranum LP. 2018. 'SCA8 RAN polySer protein preferentially accumulates in white matter regions and is regulated by eIF3F', Embo j, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagshaw Andrew T.M. 2017. 'Functional Mechanisms of Microsatellite DNA in Eukaryotic Genomes', Genome Biol Evol, 9: 2428–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakthavachalu B, Huelsmeier J, Sudhakaran IP, Hillebrand J, Singh A, Petrauskas A, Thiagarajan D, Sankaranarayanan M, Mizoue L, Anderson EN, Pandey UB, Ross E, VijayRaghavan K, Parker R, and Ramaswami M. 2018. 'RNP-Granule Assembly via Ataxin-2 Disordered Domains Is Required for Long-Term Memory and Neurodegeneration', Neuron, 98: 754–66 e4. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Ifrim MF, Valdez AN, Raj N, and Bassell GJ. 2018. 'Aberrant RNA translation in fragile X syndrome: From FMRP mechanisms to emerging therapeutic strategies', Brain Res, 1693: 24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banez-Coronel M, Ayhan F, Tarabochia AD, Zu T, Perez BA, Tusi SK, Pletnikova O, Borchelt DR, Ross CA, Margolis RL, Yachnis AT, Troncoso JC, and Ranum LP. 2015. 'RAN Translation in Huntington Disease', Neuron, 88: 667–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates G 1996. 'Expanded glutamines and neurodegeneration--a gain of insight', Bioessays, 18: 175–8. [DOI] [PubMed] [Google Scholar]
- Becker LA, and Gitler AD. 2018. 'A neurodegenerative-disease protein forms beneficial aggregates in healthy muscle', Nature, 563: 477–78. [DOI] [PubMed] [Google Scholar]
- Becker LA, Huang B, Bieri G, Ma R, Knowles DA, Jafar-Nejad P, Messing J, Kim HJ, Soriano A, Auburger G, Pulst SM, Taylor JP, Rigo F, and Gitler AD. 2017. 'Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice’, Nature, 544: 367–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman RW, and Jongens TA. 2011. 'A non-canonical start codon in the Drosophila fragile X gene yields two functional isoforms', Neuroscience, 181: 48–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L, Tompa P, and Fuxreiter M. 2018. 'Protein Phase Separation: A New Phase in Cell Biology', Trends Cell Biol, 28: 420–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, Volkov A, Guharoy M, De Decker M, Jaspers T, Ryan VH, Janke AM, Baatsen P, Vercruysse T, Kolaitis RM, Daelemans D, Taylor JP, Kedersha N, Anderson P, Impens F, Sobott F, Schymkowitz J, Rousseau F, Fawzi NL, Robberecht W, Van Damme P, Tompa P, and Van Den Bosch L. 2017. 'Phase Separation of C9orf72 Dipeptide Repeats Perturbs Stress Granule Dynamics', Mol Cell, 65: 1044–55 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Bogaert E, Michiels E, Gijselinck I, Sieben A, Jovicic A, De Baets G, Scheveneels W, Steyaert J, Cuijt I, Verstrepen KJ, Callaerts P, Rousseau F, Schymkowitz J, Cruts M, Van Broeckhoven C, Van Damme P, Gitler AD, Robberecht W, and Van Den Bosch L. 2016. 'Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD', Sci Rep, 6: 20877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, and Lamond AI. 2007. 'The multifunctional nucleolus', Nat Rev Mol Cell Biol, 8: 574–85. [DOI] [PubMed] [Google Scholar]
- Britten RJ, and Kohne DE. 1968. 'Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms', Science, 161: 529–40. [DOI] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, and Warren ST. 2001. 'Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome', Cell, 107: 477–87. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, and Parker R. 2008. 'P bodies promote stress granule assembly in Saccharomyces cerevisiae', J Cell Biol, 183: 441–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, and Parker R. 2009. 'Eukaryotic stress granules: the ins and outs of translation', Mol Cell, 36: 932–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buijsen RA, Visser JA, Kramer P, Severijnen EA, Gearing M, Charlet-Berguerand N, Sherman SL, Berman RF, Willemsen R, and Hukema RK. 2016. 'Presence of inclusions positive for polyglycine containing protein, FMRpolyG, indicates that repeat-associated non-AUG translation plays a role in fragile X-associated primary ovarian insufficiency', Hum Reprod, 31: 158–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burguete AS, Almeida S, Gao FB, Kalb R, Akins MR, and Bonini NM. 2015. 'GGGGCC microsatellite RNA is neuritically localized, induces branching defects, and perturbs transport granule function', Elife, 4: e08881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, Zara F, Canizares J, Koutnikova H, Bidichandani SI, Gellera C, Brice A, Trouillas P, De Michele G, Filla A, De Frutos R, Palau F, Patel PI, Di Donato S, Mandel JL, Cocozza S, Koenig M, and Pandolfo M. 1996. 'Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion', Science, 271: 1423–7. [DOI] [PubMed] [Google Scholar]
- Castaldo I, Pinelli M, Monticelli A, Acquaviva F, Giacchetti M, Filla A, Sacchetti S, Keller S, Avvedimento VE, Chiariotti L, and Cocozza S. 2008. 'DNA methylation in intron 1 of the frataxin gene is related to GAA repeat length and age of onset in Friedreich ataxia patients', J Med Genet, 45: 808–12. [DOI] [PubMed] [Google Scholar]
- Cheng W, Wang S, Mestre AA, Fu C, Makarem A, Xian F, Hayes LR, Lopez-Gonzalez R, Drenner K, Jiang J, Cleveland DW, and Sun S. 2018. 'C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2alpha phosphorylation', Nat Commun, 9: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coldwell MJ, Mitchell SA, Stoneley M, MacFarlane M, and Willis AE. 2000. 'Initiation of Apaf-1 translation by internal ribosome entry', Oncogene, 19: 899–905. [DOI] [PubMed] [Google Scholar]
- Conlon EG, Lu L, Sharma A, Yamazaki T, Tang T, Shneider NA, and Manley JL. 2016. 'The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains', Elife, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, ENCODE project. 2012. 'An integrated encyclopedia of DNA elements in the human genome', Nature, 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper-Knock J, Walsh MJ, Higginbottom A, Robin Highley J, Dickman MJ, Edbauer D, Ince PG, Wharton SB, Wilson SA, Kirby J, Hautbergue GM, and Shaw PJ. 2014. 'Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions', Brain, 137: 2040–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese A, Simone R, Sullivan R, Vandrovcova J, Tariq H, Yan YW, Humphrey J, Jaunmuktane Z, Sivakumar P, Polke J, Ilyas M, Tribollet E, Tomaselli PJ, Devigili G, Callegari I, Versino M, Salpietro V, Efthymiou S, Kaski D, Wood NW, Andrade NS, Buglo E, Rebelo A, Rossor AM, Bronstein A, Fratta P, Marques WJ, Zuchner S, Reilly MM, and Houlden H. 2019. 'Biallelic expansion of an intronic repeat in RFC1 is a common cause of late-onset ataxia', Nat Genet, 51: 649–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick SL, Jayaraman M, Frieden C, Wetzel R, and Pappu RV. 2006. 'Fluorescence correlation spectroscopy shows that monomeric polyglutamine molecules form collapsed structures in aqueous solutions', Proc Natl Acad Sci U S A, 103: 16764–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AL, Breydo L, Rivas EG, Gebru NT, Zheng D, Baker JD, Blair LJ, Dickey CA, Koren J 3rd, and Uversky VN. 2019. 'Repeated repeat problems: Combinatorial effect of C9orf72-derived dipeptide repeat proteins', Int J Biol Macromol, 127: 136–45. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, and Darnell RB. 2001. 'Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function', Cell, 107: 489–99. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, and Darnell RB. 2011. 'FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism', Cell, 146: 247–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, Swanson MS, and Ranum LP. 2009. 'RNA gain-of-function in spinocerebellar ataxia type 8', PLoS Genet, 5: e1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debler EW, Blobel G, and Hoelz A. 2009. 'Nuclear transport comes full circle', Nat Struct Mol Biol, 16: 457–9. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, and Rademakers R. 2011. 'Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS', Neuron, 72: 245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, Jiang H, Hirano M, Rampersaud E, Jansen GH, Donkervoort S, Bigio EH, Brooks BR, Ajroud K, Sufit RL, Haines JL, Mugnaini E, Pericak-Vance MA, and Siddique T. 2011. 'Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia', Nature, 477: 211–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Prisco GV, Huang W, Buffington SA, Hsu CC, Bonnen PE, Placzek AN, Sidrauski C, Krnjevic K, Kaufman RJ, Walter P, and Costa-Mattioli M. 2014. 'Translational control of mGluR-dependent long-term depression and object-place learning by eIF2alpha', Nat Neurosci, 17: 1073–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey AS, and La Spada AR. 2018. 'Therapy development in Huntington disease: From current strategies to emerging opportunities', Am J Med Genet A, 176: 842–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobra I, Pankivskyi S, Samsonova A, Pastre D, and Hamon L. 2018. 'Relation Between Stress Granules and Cytoplasmic Protein Aggregates Linked to Neurodegenerative Diseases', Curr Neurol Neurosci Rep, 18: 107. [DOI] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, Maragakis N, Tienari PJ, Petrucelli L, Traynor BJ, Wang J, Rigo F, Bennett CF, Blackshaw S, Sattler R, and Rothstein JD. 2013. 'RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention', Neuron, 80: 415–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan L, Li J, Badilla CL, Berne BJ, and Fernandez JM. 2009. 'Single homopolypeptide chains collapse into mechanically rigid conformations', Proc Natl Acad Sci U S A, 106: 12605–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumbovic G, Forcales SV, and Perucho M. 2017. 'Emerging roles of macrosatellite repeats in genome organization and disease development', Epigenetics, 12: 515–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick L, Barnes J, Ebner B, Agrawal S, Andresen M, Lim J, Giesler GJ, Zoghbi HY, and Orr HT. 2010. 'SCA1-like disease in mice expressing wild-type ataxin-1 with a serine to aspartic acid replacement at residue 776', Neuron, 67: 929–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ, and Wright PE. 2005. 'Intrinsically unstructured proteins and their functions', Nat Rev Mol Cell Biol, 6: 197–208. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Kunst CB, Lugenbeel KA, Ryder OA, Davison D, Warren ST, and Nelson DL. 1995. 'Evolution of the cryptic FMR1 CGG repeat', Nat Genet, 11: 301–8. [DOI] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, Padmanabhan A, Clay-Falcone D, McCluskey L, Elman L, Juhr D, Gruber PJ, Rub U, Auburger G, Trojanowski JQ, Lee VM, Van Deerlin VM, Bonini NM, and Gitler AD. 2010. 'Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS', Nature, 466: 1069–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farg MA, Soo KY, Warraich ST, Sundaramoorthy V, Blair IP, and Atkin JD. 2013. 'Ataxin-2 interacts with FUS and intermediate-length polyglutamine expansions enhance FUS-related pathology in amyotrophic lateral sclerosis', Hum Mol Genet, 22: 717–28. [DOI] [PubMed] [Google Scholar]
- Fay MM, Anderson PJ, and Ivanov P. 2017. 'ALS/FTD-Associated C9ORF72 Repeat RNA Promotes Phase Transitions In Vitro and in Cells', Cell Rep, 21: 3573–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Zhang F, Lokey LK, Chastain JL, Lakkis L, Eberhart D, and Warren ST. 1995. 'Translational suppression by trinucleotide repeat expansion at FMR1', Science, 268: 731–4. [DOI] [PubMed] [Google Scholar]
- Flores BN, Dulchavsky ME, Krans A, Sawaya MR, Paulson HL, Todd PK, Barmada SJ, and Ivanova MI. 2016. 'Distinct C9orf72-Associated Dipeptide Repeat Structures Correlate with Neuronal Toxicity', PLoS One, 11: e0165084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondon JW 3rd, and Garner HR. 2004. 'Molecular origins of rapid and continuous morphological evolution', Proc Natl Acad Sci U S A, 101: 18058–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC, Petrucelli L, Kim HJ, Gao FB, and Taylor JP. 2015. 'GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport', Nature, 525: 129–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, and Gorlich D. 2007. 'A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes', Cell, 130: 512–23. [DOI] [PubMed] [Google Scholar]
- Gall JG 2003. 'The centennial of the Cajal body', Nat Rev Mol Cell Biol, 4: 975–80. [DOI] [PubMed] [Google Scholar]
- Gan W, and Rhoads RE. 1996. 'Internal initiation of translation directed by the 5'-untranslated region of the mRNA for eIF4G, a factor involved in the picornavirus-induced switch from cap-dependent to internal initiation', J Biol Chem, 271: 623–6. [DOI] [PubMed] [Google Scholar]
- Gao FB, Richter JD, and Cleveland DW. 2017. 'Rethinking Unconventional Translation in Neurodegeneration', Cell, 171: 994–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemayel R, Vinces MD, Legendre M, and Verstrepen KJ. 2010. 'Variable tandem repeats accelerate evolution of coding and regulatory sequences', Annu Rev Genet, 44: 445–77. [DOI] [PubMed] [Google Scholar]
- Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, Cosio DM, van Blitterswijk M, Lee WC, Rademakers R, Boylan KB, Dickson DW, and Petrucelli L. 2013. 'Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS', Acta Neuropathol, 126: 829–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, van Blitterswijk M, Bieniek KF, Daughrity LM, Jiang J, Rush BK, Pedraza O, Lucas JA, Murray ME, Desaro P, Robertson A, Overstreet K, Thomas CS, Crook JE, Castanedes-Casey M, Rousseau L, Josephs KA, Parisi JE, Knopman DS, Petersen RC, Boeve BF, Graff-Radford NR, Rademakers R, Lagier-Tourenne C, Edbauer D, Cleveland DW, Dickson DW, Petrucelli L, and Boylan KB. 2015. 'Cerebellar c9RAN proteins associate with clinical and neuropathological characteristics of C9ORF72 repeat expansion carriers', Acta Neuropathol, 130: 559–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girstmair H, Saffert P, Rode S, Czech A, Holland G, Bannert N, and Ignatova Z. 2013. 'Depletion of cognate charged transfer RNA causes translational frameshifting within the expanded CAG stretch in huntingtin', Cell Rep, 3: 148–59. [DOI] [PubMed] [Google Scholar]
- Gomes E, and Shorter J. 2018. 'The molecular language of membraneless organelles', J Biol Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco CM, Berman RF, Martin RM, Tassone F, Schwartz PH, Chang A, Trapp BD, Iwahashi C, Brunberg J, Grigsby J, Hessl D, Becker EJ, Papazian J, Leehey MA, Hagerman RJ, and Hagerman PJ. 2006. 'Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS)', Brain, 129: 243–55. [DOI] [PubMed] [Google Scholar]
- Green KM, Glineburg MR, Kearse MG, Flores BN, Linsalata AE, Fedak SJ, Goldstrohm AC, Barmada SJ, and Todd PK. 2017. 'RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response', Nat Commun, 8: 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KM, Linsalata AE, and Todd PK. 2016. 'RAN translation-What makes it run?', Brain Res, 1647: 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima JC, Daigle JG, Arbez N, Cunningham KC, Zhang K, Ochaba J, Geater C, Morozko E, Stocksdale J, Glatzer JC, Pham JT, Ahmed I, Peng Q, Wadhwa H, Pletnikova O, Troncoso JC, Duan W, Snyder SH, Ranum LPW, Thompson LM, Lloyd TE, Ross CA, and Rothstein JD. 2017. 'Mutant Huntingtin Disrupts the Nuclear Pore Complex', Neuron, 94: 93–107.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C, and Bassell GJ. 2012. 'Excess protein synthesis in FXS patient lymphoblastoid cells can be rescued with a p110beta-selective inhibitor', Mol Med, 18: 336–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C, Yao X, Pong DL, Jeromin A, and Bassell GJ. 2011. 'Fragile X mental retardation protein regulates protein expression and mRNA translation of the potassium channel Kv4.2', J Neurosci, 31: 5693–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Group, The Huntington's Disease Collaborative Research. 1993. 'A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes.', Cell, 72: 971–83. [DOI] [PubMed] [Google Scholar]
- Guo L, Kim HJ, Wang H, Monaghan J, Freyermuth F, Sung JC, O'Donovan K, Fare CM, Diaz Z, Singh N, Zhang ZC, Coughlin M, Sweeny EA, DeSantis ME, Jackrel ME, Rodell CB, Burdick JA, King OD, Gitler AD, Lagier-Tourenne C, Pandey UB, Chook YM, Taylor JP, and Shorter J. 2018. 'Nuclear-Import Receptors Reverse Aberrant Phase Transitions of RNA-Binding Proteins with Prion-like Domains', Cell, 173: 677–92.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, Rothstein JD, and Wang J. 2014. 'C9orf72 nucleotide repeat structures initiate molecular cascades of disease', Nature, 507:195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman P 2013. 'Fragile X-associated tremor/ataxia syndrome (FXTAS): pathology and mechanisms', Acta Neuropathol, 126: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan AJ 2018a. 'Tandem Repeats and Repeatomes: Delving Deeper into the 'Dark Matter' of Genomes', EBioMedicine, 31: 3–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan AJ 2018b. 'Tandem repeats mediating genetic plasticity in health and disease', Nat Rev Genet, 19: 286–98. [DOI] [PubMed] [Google Scholar]
- Haran G 2012. 'How, when and why proteins collapse: the relation to folding', Curr Opin Struct Biol, 22: 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, and Ron D. 2000. 'Regulated translation initiation controls stress-induced gene expression in mammalian cells', Mol Cell, 6: 1099–108. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, and Ron D. 2003. 'An integrated stress response regulates amino acid metabolism and resistance to oxidative stress', Mol Cell, 11: 619–33. [DOI] [PubMed] [Google Scholar]
- Hart MP, and Gitler AD. 2012. 'ALS-associated ataxin 2 polyQ expansions enhance stress-induced caspase 3 activation and increase TDP-43 pathological modifications', J Neurosci, 32: 9133–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann H, Hornburg D, Czuppa M, Bader J, Michaelsen M, Farny D, Arzberger T, Mann M, Meissner F, and Edbauer D. 2018. 'Proteomics and C9orf72 neuropathology identify ribosomes as poly-GR/PR interactors driving toxicity', Life Sci Alliance, 1: e201800070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem V, Galloway JN, Mori M, Willemsen R, Oostra BA, Paylor R, and Nelson DL. 2009. 'Ectopic expression of CGG containing mRNA is neurotoxic in mammals', Hum Mol Genet, 18: 2443–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker A, Gao Z, Jankowsky E, and Parker R. 2011. 'The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex', Mol Cell, 43: 962–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG 2011. 'Molecular mechanism of scanning and start codon selection in eukaryotes', Microbiol Mol Biol Rev, 75: 434–67, first page of table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Ivanov IP, and Sonenberg N. 2016. 'Translational control by 5'-untranslated regions of eukaryotic mRNAs', Science, 352: 1413–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelz A, Debler EW, and Blobel G. 2011. 'The structure of the nuclear pore complex', Annu Rev Biochem, 80: 613–43. [DOI] [PubMed] [Google Scholar]
- Holmquist GP, and Dancis B. 1979. 'Telomere replication, kinetochore organizers, and satellite DNA evolution', Proc Natl Acad Sci U S A, 76: 4566–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, and Bear MF. 2002. 'Altered synaptic plasticity in a mouse model of fragile X mental retardation', Proc Natl Acad Sci U S A, 99: 7746–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, and Brangwynne CP. 2011. 'Beyond stereospecificity: liquids and mesoscale organization of cytoplasm', Dev Cell, 21: 14–6. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, and Julicher F. 2014. 'Liquid-liquid phase separation in biology', Annu Rev Cell Dev Biol, 30: 39–58. [DOI] [PubMed] [Google Scholar]
- Ifrim MF, Williams KR, and Bassell GJ. 2015. 'Single-Molecule Imaging of PSD-95 mRNA Translation in Dendrites and Its Dysregulation in a Mouse Model of Fragile X Syndrome', J Neurosci, 35: 7116–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert G, Saudou F, Yvert G, Devys D, Trottier Y, Garnier JM, Weber C, Mandel JL, Cancel G, Abbas N, Durr A, Didierjean O, Stevanin G, Agid Y, and Brice A. 1996. 'Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats', Nat Genet, 14: 285–91. [DOI] [PubMed] [Google Scholar]
- Ishiguro T, Sato N, Ueyama M, Fujikake N, Sellier C, Kanegami A, Tokuda E, Zamiri B, Gall-Duncan T, Mirceta M, Furukawa Y, Yokota T, Wada K, Taylor JP, Pearson CE, Charlet-Berguerand N, Mizusawa H, Nagai Y, and Ishikawa K. 2017. 'Regulatory Role of RNA Chaperone TDP-43 for RNA Misfolding and Repeat-Associated Translation in SCA31', Neuron, 94: 108–24 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiura H, Doi K, Mitsui J, Yoshimura J, Matsukawa MK, Fujiyama A, Toyoshima Y, Kakita A, Takahashi H, Suzuki Y, Sugano S, Qu W, Ichikawa K, Yurino H, Higasa K, Shibata S, Mitsue A, Tanaka M, Ichikawa Y, Takahashi Y, Date H, Matsukawa T, Kanda J, Nakamoto FK, Higashihara M, Abe K, Koike R, Sasagawa M, Kuroha Y, Hasegawa N, Kanesawa N, Kondo T, Hitomi T, Tada M, Takano H, Saito Y, Sanpei K, Onodera O, Nishizawa M, Nakamura M, Yasuda T, Sakiyama Y, Otsuka M, Ueki A, Kaida KI, Shimizu J, Hanajima R, Hayashi T, Terao Y, Inomata-Terada S, Hamada M, Shirota Y, Kubota A, Ugawa Y, Koh K, Takiyama Y, Ohsawa-Yoshida N, Ishiura S, Yamasaki R, Tamaoka A, Akiyama H, Otsuki T, Sano A, Ikeda A, Goto J, Morishita S, and Tsuji S. 2018. 'Expansions of intronic TTTCA and TTTTA repeats in benign adult familial myoclonic epilepsy', Nat Genet, 50: 581–90. [DOI] [PubMed] [Google Scholar]
- Jain A, and Vale RD. 2017. 'RNA phase transitions in repeat expansion disorders', Nature, 546: 243–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, and Wimmer E. 1988. 'A segment of the 5' nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation', J Virol, 62: 2636–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Mankodi A, Swanson MS, Moxley RT, and Thornton CA. 2004. 'Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons', Hum Mol Genet, 13: 3079–88. [DOI] [PubMed] [Google Scholar]
- Jin P, Duan R, Qurashi A, Qin Y, Tian D, Rosser TC, Liu H, Feng Y, and Warren ST. 2007. 'Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome', Neuron, 55: 556–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KW 1970. 'Chromosomal and nuclear location of mouse satellite DNA in individual cells', Nature, 225: 912–5. [DOI] [PubMed] [Google Scholar]
- Jones KW 1973. 'Satellite DNA', J Med Genet, 10: 273–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW 3rd, Sun S, Herdy JR, Bieri G, Kramer NJ, Gage FH, Van Den Bosch L, Robberecht W, and Gitler AD. 2015. 'Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS', Nat Neurosci, 18:1226–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamsteeg EJ, Kress W, Catalli C, Hertz JM, Witsch-Baumgartner M, Buckley MF, van Engelen BG, Schwartz M, and Scheffer H. 2012. 'Best practice guidelines and recommendations on the molecular diagnosis of myotonic dystrophy types 1 and 2', Eur J Hum Genet, 20:1203–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekura K, Yagi T, Cammack AJ, Mahadevan J, Kuroda M, Harms MB, Miller TM, and Urano F. 2016. 'Poly-dipeptides encoded by the C9ORF72 repeats block global protein translation', Hum Mol Genet, 25: 1803–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuno M, Adachi H, Doyu M, Minamiyama M, Sang C, Kobayashi Y, Inukai A, and Sobue G. 2003. 'Leuprorelin rescues polyglutamine-dependent phenotypes in a transgenic mouse model of spinal and bulbar muscular atrophy', Nat Med, 9: 768–73. [DOI] [PubMed] [Google Scholar]
- Katsuno M, Adachi H, Kume A, Li M, Nakagomi Y, Niwa H, Sang C, Kobayashi Y, Doyu M, and Sobue G. 2002. 'Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy', Neuron, 35: 843–54. [DOI] [PubMed] [Google Scholar]
- Kearse MG, Green KM, Krans A, Rodriguez CM, Linsalata AE, Goldstrohm AC, and Todd PK. 2016. 'CGG Repeat-Associated Non-AUG Translation Utilizes a Cap-Dependent Scanning Mechanism of Initiation to Produce Toxic Proteins', Mol Cell, 62: 314–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, and Todd PK. 2014. 'Repeat-associated non-AUG translation and its impact in neurodegenerative disease', Neurotherapeutics, 11: 721–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, and Anderson P. 1999. 'RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules', J Cell Biol, 147: 1431–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, Kanagaraj AP, Carter R, Boylan KB, Wojtas AM, Rademakers R, Pinkus JL, Greenberg SA, Trojanowski JQ, Traynor BJ, Smith BN, Topp S, Gkazi AS, Miller J, Shaw CE, Kottlors M, Kirschner J, Pestronk A, Li YR, Ford AF, Gitler AD, Benatar M, King OD, Kimonis VE, Ross ED, Weihl CC, Shorter J, and Taylor JP. 2013. 'Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS', Nature, 495: 467–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, and Taylor JP. 2017. 'Lost in Transportation: Nucleocytoplasmic Transport Defects in ALS and Other Neurodegenerative Diseases', Neuron, 96: 285–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kit S 1962. 'Species differences in animal deoxyribonucleic acid as revealed by equilibrium sedimentation in density gradients', Nature, 193: 274–5. [DOI] [PubMed] [Google Scholar]
- Kordasiewicz HB, Stanek LM, Wancewicz EV, Mazur C, McAlonis MM, Pytel KA, Artates JW, Weiss A, Cheng SH, Shihabuddin LS, Hung G, Bennett CF, and Cleveland DW. 2012. 'Sustained therapeutic reversal of Huntington's disease by transient repression of huntingtin synthesis', Neuron, 74: 1031–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M 1984. 'Selection of initiation sites by eucaryotic ribosomes: effect of inserting AUG triplets upstream from the coding sequence for preproinsulin', Nucleic Acids Res, 12: 3873–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, Safaee N, Rosenauer A, and Gehring K. 2010. 'Structural basis of binding of P-body-associated proteins GW182 and ataxin-2 by the Mlle domain of poly(A)-binding protein', J Biol Chem, 285: 13599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krans A, Kearse MG, and Todd PK. 2016. 'Repeat-associated non-AUG translation from antisense CCG repeats in fragile X tremor/ataxia syndrome', Ann Neurol, 80: 871–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak S, Durrett RT, Schug MD, and Aquadro CF. 1998. 'Equilibrium distributions of microsatellite repeat length resulting from a balance between slippage events and point mutations', Proc Natl Acad Sci U S A, 95: 10774–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, and McKnight SL. 2014. 'Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells', Science, 345: 1139–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Spada AR, Wilson EM, Lubahn DB, Harding AE, and Fischbeck KH. 1991. 'Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy', Nature, 352: 77–9. [DOI] [PubMed] [Google Scholar]
- LaCroix AJ, Stabley D, Sahraoui R, Adam MP, Mehaffey M, Kernan K, Myers CT, Fagerstrom C, Anadiotis G, Akkari YM, Robbins KM, Gripp KW, Baratela WAR, Bober MB, Duker AL, Doherty D, Dempsey JC, Miller DG, Kircher M, Bamshad MJ, Nickerson DA, Mefford HC, and Sol-Church K. 2019. 'GGC Repeat Expansion and Exon 1 Methylation of XYLT1 Is a Common Pathogenic Variant in Baratela-Scott Syndrome', Am J Hum Genet, 104: 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, and Fischer U. 2001. 'Evidence that fragile X mental retardation protein is a negative regulator of translation', Hum Mol Genet, 10: 329–38. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li HR, Jiang J, Watt AT, Chun S, Katz M, Qiu J, Sun Y, Ling SC, Zhu Q, Polymenidou M, Drenner K, Artates JW, McAlonis-Downes M, Markmiller S, Hutt KR, Pizzo DP, Cady J, Harms MB, Baloh RH, Vandenberg SR, Yeo GW, Fu XD, Bennett CF, Cleveland DW, and Ravits J. 2013. 'Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration', Proc Natl Acad Sci U S A, 110: E4530–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, and Szustakowki J. 2001. 'Initial sequencing and analysis of the human genome', Nature, 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Lastres-Becker I, Nonis D, Eich F, Klinkenberg M, Gorospe M, Kotter P, Klein FA, Kedersha N, and Auburger G. 2016. 'Mammalian ataxin-2 modulates translation control at the pre-initiation complex via PI3K/mTOR and is induced by starvation', Biochim Biophys Acta, 1862: 1558–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Conrad A, Epping E, Mathews K, Magnotta V, Dawson JD, and Nopoulos P. 2018. 'Effect of Trinucleotide Repeats in the Huntington’s Gene on Intelligence', EBioMedicine, 31: 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A, Maxwell BA, Kim NC, Temirov J, Moore J, Kolaitis RM, Shaw TI, Bai B, Peng J, Kriwacki RW, and Taylor JP. 2016. 'C9orf72 Dipeptide Repeats Impair the Assembly, Dynamics, and Function of Membrane-Less Organelles', Cell, 167: 774–88.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YB, Baskaran P, Gomez-Deza J, Chen HJ, Nishimura AL, Smith BN, Troakes C, Adachi Y, Stepto A, Petrucelli L, Gallo JM, Hirth F, Rogelj B, Guthrie S, and Shaw CE. 2017. 'C9orf72 poly GA RAN-translated protein plays a key role in amyotrophic lateral sclerosis via aggregation and toxicity', Hum Mol Genet, 26: 4765–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, Troakes C, Nishimura AL, Scotter EL, Vance C, Adachi Y, Sardone V, Miller JW, Smith BN, Gallo JM, Ule J, Hirth F, Rogelj B, Houart C, and Shaw CE. 2013. 'Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic', Cell Rep, 5: 1178–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, King OD, Shorter J, and Gitler AD. 2013. 'Stress granules as crucibles of ALS pathogenesis', J Cell Biol, 201: 361–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Ku L, Wilkinson KD, Warren ST, and Feng Y. 2001. 'The fragile X mental retardation protein inhibits translation via interacting with mRNA', Nucleic Acids Res, 29: 2276–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman AP, Shakkottai VG, and Albin RL. 2019. 'Polyglutamine Repeats in Neurodegenerative Diseases', Annu Rev Pathol, 14: 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Mori E, Kato M, Xiang S, Wu L, Kwon I, and McKnight SL. 2016. 'Toxic PR Poly-Dipeptides Encoded by the C9orf72 Repeat Expansion Target LC Domain Polymers', Cell, 167: 789–802 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, and Parker R. 2015. 'Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins', Mol Cell, 60: 208–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, and Ranum LP. 2001. 'Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9', Science, 293: 864–7. [DOI] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, Warren ST, and Wolozin B. 2011. 'Local RNA translation at the synapse and in disease', J Neurosci, 31: 16086–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Li Y, Stackpole EE, Novak A, Gao Y, Zhao Y, Zhao X, and Richter JD. 2018. 'Regulatory discrimination of mRNAs by FMRP controls mouse adult neural stem cell differentiation', Proc Natl Acad Sci U S A, 115: E11397–e405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Lin L, Tan H, Wu H, Sherman SL, Gao F, Jin P, and Chen D. 2012. 'Fragile X premutation RNA is sufficient to cause primary ovarian insufficiency in mice', Hum Mol Genet, 21: 5039–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Frick P, Grasser FA, Gendron TF, Petrucelli L, Cashman NR, Edbauer D, Kremmer E, Prudlo J, Troost D, and Neumann M. 2015. 'Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers', Acta Neuropathol, 130: 845–61. [DOI] [PubMed] [Google Scholar]
- Makise M, Mackay DR, Elgort S, Shankaran SS, Adam SA, and Ullman KS. 2012. 'The Nup153-Nup50 protein interface and its role in nuclear import', J Biol Chem, 287: 38515–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovska L, Kroschwald S, and Alberti S. 2013. 'Protein disorder, prion propensities, and self-organizing macromolecular collectives', Biochim Biophys Acta, 1834: 918–31. [DOI] [PubMed] [Google Scholar]
- Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, and Thornton CA. 2001. 'Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2', Hum Mol Genet, 10: 2165–70. [DOI] [PubMed] [Google Scholar]
- Mann DM, Rollinson S, Robinson A, Bennion Callister J, Thompson JC, Snowden JS, Gendron T, Petrucelli L, Masuda-Suzukake M, Hasegawa M, Davidson Y, and Pickering-Brown S. 2013. 'Dipeptide repeat proteins are present in the p62 positive inclusions in patients with frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72', Acta Neuropathol Commun, 1: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis JM, Schoser BG, Moseley ML, Day JW, and Ranum LP. 2006. 'DM2 intronic expansions: evidence for CCUG accumulation without flanking sequence or effects on ZNF9 mRNA processing or protein expression', Hum Mol Genet, 15: 1808–15. [DOI] [PubMed] [Google Scholar]
- McConnell MJ, Moran JV, Abyzov A, Akbarian S, Bae T, Cortes-Ciriano I, Erwin JA, Fasching L, Flasch DA, Freed D, Ganz J, Jaffe AE, Kwan KY, Kwon M, Lodato MA, Mills RE, Paquola ACM, Rodin RE, Rosenbluh C, Sestan N, Sherman MA, Shin JH, Song S, Straub RE, Thorpe J, Weinberger DR, Urban AE, Zhou B, Gage FH, Lehner T, Senthil G, Walsh CA, Chess A, Courchesne E, Gleeson JG, Kidd JM, Park PJ, Pevsner J, and Vaccarino FM. 2017. 'Intersection of diverse neuronal genomes and neuropsychiatric disease: The Brain Somatic Mosaicism Network', Science, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meola G, and Cardani R. 2015. 'Myotonic Dystrophy Type 2: An Update on Clinical Aspects, Genetic and Pathomolecular Mechanism', J Neuromuscul Dis, 2: S59–S71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, and Swanson MS. 2000. 'Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy', Embo j, 19: 4439–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IOC, Pietrzyk J, Cleverley K, Nicoll AJ, Pickering-Brown S, Dols J, Cabecinha M, Hendrich O, Fratta P, Fisher EMC, Partridge L, and Isaacs AM. 2014. 'C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins', Science, 345: 1192–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S, Ridler CE, Balendra R, Thoeng A, Woodling NS, Grasser FA, Plagnol V, Lashley T, Partridge L, and Isaacs AM. 2017. 'Bidirectional nucleolar dysfunction in C9orf72 frontotemporal lobar degeneration', Acta Neuropathol Commun, 5: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens TG, Mizielinska S, Niccoli T, Mitchell JS, Thoeng A, Ridler CE, Gronke S, Esser J, Heslegrave A, Zetterberg H, Partridge L, and Isaacs AM. 2018. 'Sense and antisense RNA are not toxic in Drosophila models of C9orf72-associated ALS/FTD', Acta Neuropathol, 135: 445–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens TG, Niccoli T, Wilson KM, Atilano ML, Birsa N, Gittings LM, Holbling BV, Dyson MC, Thoeng A, Neeves J, Glaria I, Yu L, Bussmann J, Storkebaum E, Pardo M, Choudhary JS, Fratta P, Partridge L, and Isaacs AM. 2019. 'C9orf72 arginine-rich dipeptide proteins interact with ribosomal proteins in vivo to induce a toxic translational arrest that is rescued by eIF1A', Acta Neuropathol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr D, Frey S, Fischer T, Guttler T, and Gorlich D. 2009. 'Characterisation of the passive permeability barrier of nuclear pore complexes', Embo j, 28: 2541–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, and Taylor JP. 2015. 'Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization', Cell, 163: 123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, Haass C, and Edbauer D. 2013. 'The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS', Science, 339: 1335–8. [DOI] [PubMed] [Google Scholar]
- Morley JF, Brignull HR, Weyers JJ, and Morimoto RI. 2002. 'The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans', Proc Natl Acad Sci U S A, 99: 10417–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, Chen G, Weatherspoon MR, Clark HB, Ebner TJ, Day JW, and Ranum LP. 2006. 'Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8', Nat Genet, 38: 758–69. [DOI] [PubMed] [Google Scholar]
- Muslimov IA, Patel MV, Rose A, and Tiedge H. 2011. 'Spatial code recognition in neuronal RNA targeting: role of RNA-hnRNP A2 interactions', J Cell Biol, 194: 441–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamori Masayuki, Sobczak Krzysztof, Puwanant Araya, Welle Steve, Eichinger Katy, Pandya Shree, Dekdebrun Jeannne, Heatwole Chad R., McDermott Michael P., Chen Tian, Melissa Cline, Rabi Tawil, Osborne Robert J., Wheeler Thurman M., Swanson Maurice S., Moxley Richard T. 3rd, and Thornton Charles A.. 2013. 'Splicing biomarkers of disease severity in myotonic dystrophy', Ann Neurol, 74: 862–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonhoff U, Ralser M, Welzel F, Piccini I, Balzereit D, Yaspo ML, Lehrach H, and Krobitsch S. 2007. 'Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules', Mol Biol Cell, 18: 1385–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SY, He F, Krans A, Frazer M, Taylor JP, Paulson HL, and Todd PK. 2015. 'RAN translation at CGG repeats induces ubiquitin proteasome system impairment in models of fragile X-associated tremor ataxia syndrome', Hum Mol Genet, 24: 4317–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordway JM, Tallaksen-Greene S, Gutekunst CA, Bernstein EM, Cearley JA, Wiener HW, Dure LS, Lindsey R, Hersch SM, Jope RS, Albin RL, and Detloff PJ. 1997. 'Ectopically expressed CAG repeats cause intranuclear inclusions and a progressive late onset neurological phenotype in the mouse', Cell, 91: 753–63. [DOI] [PubMed] [Google Scholar]
- Orr HT 2011. 'FTD and ALS: genetic ties that bind', Neuron, 72: 189–90. [DOI] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, and Samali A. 2016. 'The integrated stress response', 17: 1374–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue ML, and Gall JG. 1970. 'Chromosomal localization of mouse satellite DNA', Science, 168: 1356–8. [DOI] [PubMed] [Google Scholar]
- Paulson H 2018a. 'Repeat expansion diseases', Handb Clin Neurol, 147: 105–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson HL, Shakkottai VG, Clark HB, and Orr HT. 2017. 'Polyglutamine spinocerebellar ataxias - from genes to potential treatments', Nat Rev Neurosci, 18: 613–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson Henry. 2018b. 'Chapter 9 - Repeat expansion diseases' in Geschwind Daniel H., Paulson Henry L. and Klein Christine (eds.), Handb Clin Neurol (Elsevier; ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, and Sonenberg N. 1988. 'Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA', Nature, 334: 320–5. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU, and Shatsky IN. 1996. 'Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry', Mol Cell Biol, 16: 6859–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, and Hellen CU. 1996. 'Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes', Mol Cell Biol, 16: 6870–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson OJ, Aagaard L, Jensen TG, and Damgaard CK. 2015. 'Molecular mechanisms in DM1 - a focus on foci', Nucleic Acids Res, 43: 2433–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DSW, and Parker R. 2016. 'Principles and Properties of Stress Granules', Trends Cell Biol, 26: 668–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulst SM 1993. 'Spinocerebellar Ataxia Type 2' in Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K and Amemiya A (eds.), GeneReviews((R)) (University of Washington, Seattle. GeneReviews is a registered trademark of the University of Washington, Seattle. All rights reserved.: Seattle (WA)) [Google Scholar]
- Pulst SM, Nechiporuk A, Nechiporuk T, Gispert S, Chen XN, Lopes-Cendes I, Pearlman S, Starkman S, Orozco-Diaz G, Lunkes A, DeJong P, Rouleau GA, Auburger G, Korenberg JR, Figueroa C, and Sahba S. 1996. 'Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2', Nat Genet, 14: 269–76. [DOI] [PubMed] [Google Scholar]
- Pulst SM, Nechiporuk A, and Starkman S. 1993. 'Anticipation in spinocerebellar ataxia type 2', Nat Genet, 5: 8–10. [DOI] [PubMed] [Google Scholar]
- Ray PS, Grover R, and Das S. 2006. 'Two internal ribosome entry sites mediate the translation of p53 isoforms', EMBO Rep, 7: 404–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K, Zamiri B, Stanley SY, Macgregor RB Jr., and Pearson CE. 2013. 'The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures', J Biol Chem, 288: 9860–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt R, Holzenburg A, Buhle EL Jr., Jarnik M, Engel A, and Aebi U. 1990. 'Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components', J Cell Biol, 110: 883–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renoux AJ, and Todd PK. 2012. 'Neurodegeneration the RNA way', Prog Neurobiol, 97: 173–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Holtta-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chio A, Restagno G, Borghero G, Sabatelli M, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, and Traynor BJ. 2011. 'A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD', Neuron, 72: 257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, and Gorlich D. 2001. 'Kinetic analysis of translocation through nuclear pore complexes', Embo j, 20:1320–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard Guy-Franck, Kerrest Alix, and Dujon Bernard. 2008. 'Comparative genomics and molecular dynamics of DNA repeats in eukaryotes', Microbiol Mol Biol Rev, 72: 686–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S, Serrano A, Gerbino V, Giorgi A, Di Francesco L, Nencini M, Bozzo F, Schinina ME, Bagni C, Cestra G, Carri MT, Achsel T, and Cozzolino M. 2015. 'Nuclear accumulation of mRNAs underlies G4C2-repeat-induced translational repression in a cellular model of C9orf72 ALS', J Cell Sci, 128:1787–99. [DOI] [PubMed] [Google Scholar]
- Saffert P, Adamla F, Schieweck R, Atkins JF, and Ignatova Z. 2016. 'An Expanded CAG Repeat in Huntingtin Causes +1 Frameshifting', J Biol Chem, 291: 18505–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanpei K, Takano H, Igarashi S, Sato T, Oyake M, Sasaki H, Wakisaka A, Tashiro K, Ishida Y, Ikeuchi T, Koide R, Saito M, Sato A, Tanaka T, Hanyu S, Takiyama Y, Nishizawa M, Shimizu N, Nomura Y, Segawa M, Iwabuchi K, Eguchi I, Tanaka H, Takahashi H, and Tsuji S. 1996. 'Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT', Nat Genet, 14: 277–84. [DOI] [PubMed] [Google Scholar]
- Sareen D, O'Rourke JG, Meera P, Muhammad AK, Grant S, Simpkinson M, Bell S, Carmona S, Ornelas L, Sahabian A, Gendron T, Petrucelli L, Baughn M, Ravits J, Harms MB, Rigo F, Bennett CF, Otis TS, Svendsen CN, and Baloh RH. 2013. 'Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion', Sci Transl Med, 5: 208ral49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya S, Bagshaw A, Buschiazzo E, Kumar P, Chowdhury S, Black MA, and Gemmell N. 2013. 'Microsatellite tandem repeats are abundant in human promoters and are associated with regulatory elements', PLoS One, 8: e54710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HB, and Gorlich D. 2015. 'Nup98 FG domains from diverse species spontaneously phase separate into particles with nuclear pore-like permselectivity', Elife, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HB 2016. 'Transport Selectivity of Nuclear Pores, Phase Separation, and Membraneless Organelles', Trends Biochem Sci, 41: 46–61. [DOI] [PubMed] [Google Scholar]
- Scoles DR, Ho MH, Dansithong W, Pflieger LT, Petersen LW, Thai KK, and Pulst SM. 2015. 'Repeat Associated Non-AUG Translation (RAN Translation) Dependent on Sequence Downstream of the ATXN2 CAG Repeat', PLoS One, 10: e0128769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoles DR, Meera P, Schneider MD, Paul S, Dansithong W, Figueroa KP, Hung G, Rigo F, Bennett CF, Otis TS, and Pulst SM. 2017. 'Antisense oligonucleotide therapy for spinocerebellar ataxia type 2', Nature, 544: 362–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellier C, Buijsen RA, He F, Natla S, Jung L, Tropel P, Gaucherot A, Jacobs H, Meziane H, Vincent A, Champy MF, Sorg T, Pavlovic G, Wattenhofer-Donze M, Birling MC, Oulad-Abdelghani M, Eberling P, Ruffenach F, Joint M, Anheim M, Martinez-Cerdeno V, Tassone F, Willemsen R, Hukema RK, Viville S, Martinat C, Todd PK, and Charlet-Berguerand N. 2017. 'Translation of Expanded CGG Repeats into FMRpolyG Is Pathogenic and May Contribute to Fragile X Tremor Ataxia Syndrome', Neuron, 93: 331–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakkottai VG, and Fogel BL. 2013. 'Clinical neurogenetics: autosomal dominant spinocerebellar ataxia', Neurol Clin, 31: 987–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi KY, Mori E, Nizami ZF, Lin Y, Kato M, Xiang S, Wu LC, Ding M, Yu Y, Gall JG, and McKnight SL. 2017. 'Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export', Proc Natl Acad Sci U S A, 114: E1111–E17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Choi M, Siomi MC, Nussbaum RL, and Dreyfuss G. 1994. 'Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome', Cell, 77: 33–9. [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi MC, Nussbaum RL, and Dreyfuss G. 1993. 'The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein', Cell, 74: 291–8. [DOI] [PubMed] [Google Scholar]
- Smit AF 1999. 'Interspersed repeats and other mementos of transposable elements in mammalian genomes', Curr Opin Genet Dev, 9: 657–63. [DOI] [PubMed] [Google Scholar]
- Sone Jun, Mitsuhashi Satomi, Fujita Atsushi, Mizuguchi Takeshi, Mori Keiko, Koike Haruki, Hashiguchi Akihiro, Takashima Hiroshi, Sugiyama Hiroshi, Kohno Yutaka, Takiyama Yoshihisa, Maeda Kengo, Doi Hiroshi, Koyano Shigeru, Takeuchi Hideyuki, Kawamoto Michi, Kohara Nobuo, Ando Tetsuo, Toshiaki leda Yasushi Kita, Kokubun Norito, Tsuboi Yoshio, Katsuno Masahisa, Iwasaki Yasushi, Yoshida Mari, Tanaka Fumiaki, Suzuki Ikuo K., Frith Martin C., Matsumoto Naomichi, and Sobue Gen. 2019. 'Long-read sequencing identifies GGC repeat expansion in human-specific NOTCH2NLC associated with neuronal intranuclear inclusion disease', bioRxiv: 515635. [DOI] [PubMed] [Google Scholar]
- Sonobe Y, Ghadge G, Masaki K, Sendoel A, Fuchs E, and Roos RP. 2018. 'Translation of dipeptide repeat proteins from the C9ORF72 expanded repeat is associated with cellular stress', Neurobiol Dis, 116: 155–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soong BW, and Paulson HL. 2007. 'Spinocerebellar ataxias: an update', Curr Opin Neurol, 20: 438–46. [DOI] [PubMed] [Google Scholar]
- Soragni E, Petrosyan L, Rinkoski TA, Wieben ED, Baratz KH, Fautsch MP, and Gottesfeld JM. 2018. 'Repeat-Associated Non-ATG (RAN) Translation in Fuchs' Endothelial Corneal Dystrophy', Invest Ophthalmol Vis Sci, 59: 1888–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL 2006. 'SnapShot: Cellular bodies', Cell, 127: 1071. [DOI] [PubMed] [Google Scholar]
- Starck SR, Jiang V, Pavon-Eternod M, Prasad S, McCarthy B, Pan T, and Shastri N. 2012. 'Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I', Science, 336: 1719–23. [DOI] [PubMed] [Google Scholar]
- Stochmanski SJ, Therrien M, Laganiere J, Rochefort D, Laurent S, Karemera L, Gaudet R, Vyboh K, Van Meyel DJ, Di Cristo G, Dion PA, Gaspar C, and Rouleau GA. 2012. 'Expanded ATXN3 frameshifting events are toxic in Drosophila and mammalian neuron models', Hum Mol Genet, 21: 2211–8. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Shibagaki Y, Hattori S, and Matsuoka M. 2018. 'The proline-arginine repeat protein linked to C9-ALS/FTD causes neuronal toxicity by inhibiting the DEAD-box RNA helicase-mediated ribosome biogenesis', Cell Death Dis, 9: 975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher KD, and Parker R. 2010. 'Localization to, and effects of Pbp1, Pbp4, Lsm12, Dhh1, and Pab1 on stress granules in Saccharomyces cerevisiae', PLoS One, 5: e10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabet R, Schaeffer L, Freyermuth F, Jambeau M, Workman M, Lee CZ, Lin CC, Jiang J, Jansen-West K, Abou-Hamdan H, Desaubry L, Gendron T, Petrucelli L, Martin F, and Lagier-Tourenne C. 2018. 'CUG initiation and frameshifting enable production of dipeptide repeat proteins from ALS/FTD C9ORF72 transcripts', Nat Commun, 9: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Wang H, Xia Q, Li K, Li K, Jiang X, Xu G, Wang G, and Ying Z. 2015. 'Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity', Hum Mol Genet, 24: 2426–41. [DOI] [PubMed] [Google Scholar]
- Tassone F, Hagerman RJ, Loesch DZ, Lachiewicz A, Taylor AK, and Hagerman PJ. 2000. 'Fragile X males with unmethylated, full mutation trinucleotide repeat expansions have elevated levels of FMR1 messenger RNA', Am J Med Genet, 94: 232–6. [DOI] [PubMed] [Google Scholar]
- Tassone F, Hagerman RJ, Taylor AK, Gane LW, Godfrey TE, and Hagerman PJ. 2000. 'Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome', Am J Hum Genet, 66: 6–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, and Parker R. 2005. 'Processing bodies require RNA for assembly and contain nontranslating mRNAs', Rna, 11: 371–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson SR, Seo SS, Barnes SA, Louros SR, Muscas M, Dando O, Kirby C, Wyllie DJA, Hardingham GE, Kind PC, and Osterweil EK. 2017. 'Cell-Type-Specific Translation Profiling Reveals a Novel Strategy for Treating Fragile X Syndrome', Neuron, 95: 550–63.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Mack KJ, and Malter JS. 2003. 'The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95', Proc Natl Acad Sci U S A, 100: 14374–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Oh SY, Krans A, He F, Sellier C, Frazer M, Renoux AJ, Chen KC, Scaglione KM, Basrur V, Elenitoba-Johnson K, Vonsattel JP, Louis ED, Sutton MA, Taylor JP, Mills RE, Charlet-Berguerand N, and Paulson HL. 2013. 'CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome', Neuron, 78: 440–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, and Paulson HL. 2010. 'RNA-mediated neurodegeneration in repeat expansion disorders', Ann Neurol, 67: 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulouse A, Au-Yeung F, Gaspar C, Roussel J, Dion P, and Rouleau GA. 2005. 'Ribosomal frameshifting on MJD-1 transcripts with long CAG tracts', Hum Mol Genet, 14: 2649–60. [DOI] [PubMed] [Google Scholar]
- Treangen TJ, and Salzberg SL. 2011. 'Repetitive DNA and next-generation sequencing: computational challenges and solutions', Nat Rev Genet, 13: 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottier Y, Devys D, Imbert G, Saudou F, An I, Lutz Y, Weber C, Agid Y, Hirsch EC, and Mandel JL. 1995. 'Cellular localization of the Huntington's disease protein and discrimination of the normal and mutated form', Nat Genet, 10: 104–10. [DOI] [PubMed] [Google Scholar]
- Trottier Y, Lutz Y, Stevanin G, Imbert G, Devys D, Cancel G, Saudou F, Weber C, David G, Tora L, and et al. 1995. 'Polyglutamine expansion as a pathological epitope in Huntington's disease and four dominant cerebellar ataxias', Nature, 378: 403–6. [DOI] [PubMed] [Google Scholar]
- Tsilfidis C, MacKenzie AE, Mettler G, Barcelo J, and Korneluk RG. 1992. 'Correlation between CTG trinucleotide repeat length and frequency of severe congenital myotonic dystrophy', Nat Genet, 1: 192–5. [DOI] [PubMed] [Google Scholar]
- Udagawa T, Farny NG, Jakovcevski M, Kaphzan H, Alarcon JM, Anilkumar S, Ivshina M, Hurt JA, Nagaoka K, Nalavadi VC, Lorenz LJ, Bassell GJ, Akbarian S, Chattarji S, Klann E, and Richter JD. 2013. 'Genetic and acute CPEB1 depletion ameliorate fragile X pathophysiology', Nat Med, 19: 1473–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usdin K 2008. 'The biological effects of simple tandem repeats: lessons from the repeat expansion diseases', Genome Res, 18: 1011–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, and Dunker AK. 2010. 'Understanding protein non-folding', Biochim Biophys Acta, 1804: 1231–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S, Gensac MC, Maret A, Bayard F, Amalric F, Prats H, and Prats AC. 1995. 'Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes', Mol Cell Biol, 15: 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, and Parker R. 2018. 'RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome', Proc Natl Acad Sci U S A, 115: 2734–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattem KM, and Wek RC. 2004. 'Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells', Proc Natl Acad Sci U S A, 101: 11269–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen KJ, Jansen A, Lewitter F, and Fink GR. 2005. 'Intragenic tandem repeats generate functional variability', Nat Genet, 37: 986–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinces MD, Legendre M, Caldara M, Hagihara M, and Verstrepen KJ. 2009. 'Unstable tandem repeats in promoters confer transcriptional evolvability', Science, 324: 1213–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viphakone N, Hautbergue GM, Walsh M, Chang CT, Holland A, Folco EG, Reed R, and Wilson SA. 2012. 'TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export', Nat Commun, 3: 1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitalis A, and Pappu RV. 2011. 'Assessing the contribution of heterogeneous distributions of oligomers to aggregation mechanisms of polyglutamine peptides', Biophys Chem, 159: 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitalis A, Wang X, and Pappu RV. 2007. 'Quantitative characterization of intrinsic disorder in polyglutamine: insights from analysis based on polymer theories', Biophys J, 93: 1923–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RH, and Murphy RM. 2009. 'Examining polyglutamine peptide length: a connection between collapsed conformations and increased aggregation', J Mol Biol, 393: 978–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LW, Berry-Kravis E, and Hagerman RJ. 2010. 'Fragile X: leading the way for targeted treatments in autism', Neurotherapeutics, 7: 264–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring M, and Britten RJ. 1966. 'Nucleotide sequence repetition: a rapidly reassociating fraction of mouse DNA', Science, 154: 791–4. [DOI] [PubMed] [Google Scholar]
- Warrick JM, Paulson HL, Gray-Board GL, Bui QT, Fischbeck KH, Pittman RN, and Bonini NM. 1998. 'Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila', Cell, 93: 939–49. [DOI] [PubMed] [Google Scholar]
- Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB, Pasinelli P, Ichida JK, and Trotti D. 2014. 'Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death', Neuron, 84: 1213–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergard Thomas, McAvoy Kevin, Russell Katelyn, Wen Xinmei, Pang Yu, Morris Brandie, Pasinelli Piera, Trotti Davide, and Haeusler Aaron. 2019. 'Repeat-associated non-AUG translation in C9orf72-ALS/FTD is driven by neuronal excitation and stress', EMBO Mol Med, 0: e9423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AJ, and Paulson HL. 2008. 'Polyglutamine neurodegeneration: protein misfolding revisited', Trends Neurosci, 31: 521–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills NM, and Atkins JF. 2006. 'The potential role of ribosomal frameshifting in generating aberrant proteins implicated in neurodegenerative diseases', RNA, 12: 1149–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowska M, Olejniczak M, Galka-Marciniak P, Jazurek M, and Krzyzosiak WJ. 2014. 'RAN translation and frameshifting as translational challenges at simple repeats of human neurodegenerative disorders', Nucleic Acids Res, 42: 11849–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin B 2012. 'Regulated protein aggregation: stress granules and neurodegeneration', Mol Neurodegener, 7: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Poidevin M, Li X, Li Y, Shu L, Nelson DL, Li H, Hales CM, Gearing M, Wingo TS, and Jin P. 2013. 'Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration', Proc Natl Acad Sci U S A, 110: 7778–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa M, Ito D, Honda T, Kubo K, Noda M, Nakajima K, and Suzuki N. 2015. 'Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS', Hum Mol Genet, 24: 1630–45. [DOI] [PubMed] [Google Scholar]
- Yang D, Abdallah A, Li Z, Lu Y, Almeida S, and Gao FB. 2015. 'FTD/ALS-associated poly(GR) protein impairs the Notch pathway and is recruited by poly(GA) into cytoplasmic inclusions', Acta Neuropathol, 130: 525–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SK, and Wek RC. 2016. 'Upstream Open Reading Frames Differentially Regulate Gene-specific Translation in the Integrated Stress Response', J Biol Chem, 291: 16927–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunis JJ, and Yasmineh WG. 1971. 'Heterochromatin, satellite DNA, and cell function. Structural DNA of eucaryotes may support and protect genes and aid in speciation', Science, 174: 1200–9. [DOI] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, and Bagni C. 2003. 'The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses', Cell, 112: 317–27. [DOI] [PubMed] [Google Scholar]
- Zhang K, Daigle JG, Cunningham KM, Coyne AN, Ruan K, Grima JC, Bowen KE, Wadhwa H, Yang P, Rigo F, Taylor JP, Gitler AD, Rothstein JD, and Lloyd TE. 2018. 'Stress Granule Assembly Disrupts Nucleocytoplasmic Transport', Cell, 173: 958–71.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, Gupta S, Thomas MA, Hong I, Chiu SL, Huganir RL, Ostrow LW, Matunis MJ, Wang J, Sattler R, Lloyd TE, and Rothstein JD. 2015. 'The C9orf72 repeat expansion disrupts nucleocytoplasmic transport', Nature, 525: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Gendron TF, Ebbert MTW, O'Raw AD, Yue M, Jansen-West K, Zhang X, Prudencio M, Chew J, Cook CN, Daughrity LM, Tong J, Song Y, Pickles SR, Castanedes-Casey M, Kurti A, Rademakers R, Oskarsson B, Dickson DW, Hu W, Gitler AD, Fryer JD, and Petrucelli L. 2018. 'Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis', Nat Med, 24: 1136–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu YF, Katzman RB, Gass J, Murray ME, Shinohara M, Lin WL, Garrett A, Stankowski JN, Daughrity L, Tong J, Perkerson EA, Yue M, Chew J, Castanedes-Casey M, Kurti A, Wang ZS, Liesinger AM, Baker JD, Jiang J, Lagier-Tourenne C, Edbauer D, Cleveland DW, Rademakers R, Boylan KB, Bu G, Link CD, Dickey CA, Rothstein JD, Dickson DW, Fryer JD, and Petrucelli L. 2016. 'C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins', Nat Neurosci, 19: 668–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Jansen-West K, Xu YF, Gendron TF, Bieniek KF, Lin WL, Sasaguri H, Caulfield T, Hubbard J, Daughrity L, Chew J, Belzil VV, Prudencio M, Stankowski JN, Castanedes-Casey M, Whitelaw E, Ash PE, DeTure M, Rademakers R, Boylan KB, Dickson DW, and Petrucelli L. 2014. 'Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress', Acta Neuropathol, 128: 505–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA, Nan Z, Forster C, Low WC, Schoser B, Somia NV, Clark HB, Schmechel S, Bitterman PB, Gourdon G, Swanson MS, Moseley M, and Ranum LP. 2011. 'Non-ATG-initiated translation directed by microsatellite expansions', Proc Natl Acad Sci U S A, 108: 260–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, Ostrow LW, Rothstein JD, Troncoso JC, and Ranum LP. 2013. 'RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia', Proc Natl Acad Sci U S A, 110: E4968–77. [DOI] [PMC free article] [PubMed] [Google Scholar]