Abstract
The liver is the central metabolic organ in the human body, and also plays an essential role in innate and adaptive immunity. While mouse models offer significant insights into immune-inflammatory liver disease, human immunology differs in important respects. It is not easy to address those differences experimentally. Therefore, to improve the understanding of human liver immunobiology and pathology, we have established precision-cut human liver slices to study innate immunity in human tissue. Human liver slices collected from resected livers could be maintained in ex vivo culture over a two-week period. Although an acute inflammatory response accompanied by signs of tissue repair was observed in liver tissue following slicing, the expression of many immune genes stabilized after day 4 and remained stable until day 15. Remarkably, histological evidence of pre-existing liver diseases was preserved in the slices for up to 7 days. Following 7 days of culture, exposure of liver slices to the toll-like receptor (TLR) ligands, TLR3 ligand Poly-I:C and TLR4 ligand LPS, resulted in a robust activation of acute inflammation and cytokine genes. Moreover, Poly-I:C treatment induced a marked antiviral response including increases of interferons IFNB, IL-28B and a group of interferon-stimulated genes. Therefore, precision-cut liver slices emerge as a valuable tool to study human innate immunity.
Keywords: Liver slice culture, Innate immunity, Hepatocyte, Antiviral
1. Introduction
The liver is critical in innate and adaptive immunity in humans (Crispe, 2016; Heymann and Tacke, 2016). The human liver contains diverse populations of liver cells, the majority of which are the parenchymal cells, or hepatocytes. In addition, the resident non-parenchymal cells include liver sinusoidal endothelial cells (LSECs), hepatic stellate cells (HSCs), and hepatic macrophages (i.e. Kupffer cells), as well as vascular endothelial cells and bile duct epithelial cells. Furthermore, trafficking monocytes, dendritic cells (DCs), natural killer (NK) cells and NK T cells are also present in the human liver (Crispe, 2016). These cell populations together form a complex immunological network.
Despite the recent advances in liver research, liver diseases continue to be a major cause of morbidity and mortality worldwide (Blachier et al., 2013; Wang et al., 2014). In 2010 alone, 31 million people globally were afflicted with liver cirrhosis, and one million of those patients succumbed to the disease (Mokdad et al., 2014). Likewise, liver cancer is one of the leading causes of cancer deaths globally, accounting for > 600,000 deaths each year. According to statistics available from the American Cancer Society, liver cancer cases have been rising on average 2.7% each year over the last 10 years, and death rates have been rising on average 2.6% each year from 2005 to 2014. Therefore, improved understanding and treatment of liver diseases is urgently needed.
One approach to study liver biology that is gaining popularity is the precision-cut liver slice culture (PCLS) (de Graaf et al., 2010; Lagaye et al., 2012). The tissue preparation and culturing conditions for PCLS have steadily evolved (Olinga et al., 1997; Graaf et al., 2007). It is likely to be important that slices are established in culture within three hours of the in vivo excision, because the viability of hepatocytes in conventional cell culture is strongly associated with the time delay between hepatectomy and liver cell isolation (Bhogal et al., 2011). Compared with other liver research methods, including cell monocultures and patterned tissue cultures, the PCLS method has the advantage that hepatocytes and the major subsets of non-parenchymal cells are cultured simultaneously, enabling the analysis of liver cell function in the context of diverse liver cell types. Gene expression profile of PCLS was found to have a higher degree of similarity to intact liver compared to mono cell cultures of primary hepatocytes and cell lines after 24 h of ex vivo culturing (Boess et al., 2003). Furthermore, since each set of liver slice cultures originates from an individual donor, over time a diverse human population is sampled using the PCLS approach. This advantage will facilitate the development of precision medicine and personalized therapy for liver diseases in the era of personal genomics (Sitia, 2015; Li and Wang, 2016). To date, the PCLS method has been developed to study the metabolism (Janssen et al., 2015; Ijssennagger et al., 2016; Starokozhko et al., 2017), toxicology (Graaf et al., 2007; Hadi et al., 2013; Karim et al., 2013) and development of liver cells (Kasper et al., 2005; Westra et al., 2016). However, to our knowledge there has been little exploitation of PCLS to study human immunology.
In the present study, we used the PCLS approach to investigate innate immunity in human liver tissues. First, we developed a modified culture protocol that does not depend on elevated oxygen concentration in the incubator. Second, we documented the innate immune response in liver slices associated with tissue slicing. Third, we analyzed the antiviral response in liver slices induced by the Toll-Like Receptor 3 (TLR3) pathway.
2. Materials and methods
2.1. Liver samples, preparation and culturing
Fresh liver tissues were obtained from patients undergoing liver resection at the University of Washington Medical Center (Seattle, WA, USA). All patients in this study prospectively consented to donate liver tissue for research under the Institutional Review Board protocols #31281 and #51710. Clinical information regarding the patients is provided in Table S1. All patients in this study were free from active hepatitis and cirrhosis, which was confirmed by molecular or histologic examination of core needle biopsies of the liver. Liver cores of 6 mm diameter were excised from the resected liver tissue using a biopsy punch (Integra Miltex, York, PA, USA), stored in BELZER-UW solution (Bridge to Life Ltd., Columbia, SC, USA), and transferred to the research laboratories typically within 1 h of tissue excision.
Liver slices of 250 μm thickness were cut using a vibrating microtome, Leica VT1200 S (Nussloch, Germany), using Dulbecco’s Modified Eagles Medium (DMEM) as the cutting medium (de Graaf et al., 2010; Lagaye et al., 2012). Liver slices were cultured individually on 0.4 μm millicell organotypic inserts in 24 well plates (Millipore Corporation, Billerica, MA, USA). The culturing medium comprised 1 × advanced-DMEM medium, 5% Fetal Bovine Serum (FBS), 1 × GlutaMAX, 0.5 × Penicillin-Streptomycin, 1 × Insulin-Transferrin-Selenium supplement and 15 mM HEPES (pH 7.2–7.5) (all from Gibco, Grand Island, NY, USA). The liver cultures were maintained on a rocking platform at 17 rpm in a humidified incubator at 5% CO2 and atmospheric concentration of O2 at 37 °C. The medium was renewed every two to three days. An experimental schematic of the liver slice culture procedure is provided in Fig. S1.
2.2. MTS cell viability assay
Individual slices were placed in a 48-well plate with four hundred microliters of DMEM medium and eighty microliters of MTS assay reagent (Promega, Fitchburg, WI, USA). MTS is a tetrazolium compound [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt]. MTS is bioreduced by cells into a formazan product that is soluble in tissue culture medium. The absorbance of the formazan product at 490 nm can be measured directly from 96-well assay plates without additional processing. The conversion of MTS into the aqueous soluble formazan product is accomplished by dehydrogenase enzymes found in metabolically active cells. Reactions were incubated at 37 °C for 1 h with shaking. The OD490 nm of the supernatants was measured, using a Synergy H1 microplate reader (Biotek, Shoreline, WA, USA).
2.3. Liver perfusion and isolation of liver cells
Human liver cell isolation procedures were adapted from several sources (Bhogal et al., 2011; Mohar et al., 2015). Cell isolation was performed on wedges of resected tissue rather than cores. Perfusion buffer contained 1 × Hank’s Balanced Salt Solution (HBSS, without Ca ++, Mg++, or phenol red, from Gibco), 10 mM HEPES (pH 7.2–7.5) and 0.5 mM EDTA (pH 8.0). Collagenase buffer contained 1 × HBSS (Gibco), 5 mM MgCl2, 5 mM CaCl2, 5 mM HEPES (pH 7.2–7.5), 0.5% w/v Collagenase IV (Sigma-Aldrich, St. Louis, MO, USA), 0.25% w/v Protease (Sigma), 0.125% w/v Hyaluronidase (Sigma), 0.05% w/v DNase I (Sigma). Fresh aliquots of enzymes were added to the buffer on the day of the perfusion experiment. Forty mL of Perfusion buffer and 20 mL of Collagenase buffer were used for each 10 g of liver tissue. Buffers were pre-warmed to 37 °C prior to the perfusion step.
Before the perfusion step, liver tissue samples were rinsed with PBS (pH 7.4) until the fluid ran clear. Liver tissue wedges were sequentially perfused with Washing buffer (1 × HBSS and 10 mM HEPES, pH 7.2–7.5), Perfusion buffer, Washing buffer, and the recirculating Collagenase buffer at a flow rate of 12 mL/min (Gilson’s MINIPULS 3, Middleton, WI, USA). The perfused liver tissue samples were gently mashed with a sterile syringe plunger through a sterile mesh strainer in ice-cold DMEM medium. Cell extracts were filtered through a 100 μm sterile strainer and centrifuged at 50 ×g at 4 °C for 3 min to enrich for hepatocytes in the pellets and non-parenchymal cells in the supernatant. The supernatants were transferred to a new tube and kept on ice. The pellets were washed three times with ice-cold DMEM medium, and were pelleted each time at 50 ×g at 4 °C for 3 min. To further purify the live hepatocytes, cell pellets were resuspended with 5 mL of ice-cold PBS, overlaid with 10 mL of 25% Percoll gradient solution. The mixture was centrifuged at 1400 ×g (no brake) at 4 °C for 20 min. The pellet contained the purified live hepatocytes. The viability of the isolated hepatocytes was determined with the trypan blue exclusion assay (Thermo Fisher Scientific). If the viability was > 50%, the isolated hepatocytes were stored for RNA extraction.
For the non-parenchymal cells, the reserved centrifuged supernatants were further centrifuged at 500 ×g at 4 °C for 7 min. The pellets were resuspended in 5 mL of ice-cold PBS, and overlaid on top with 50% and 25% Percoll gradients (10 mL layers each), and were centrifuged at 1400 ×g (no brake) at 4 °C for 20 min. Cell layers were collected into 20 mL PBS each, and centrifuged again at 500 ×g at 4 °C for 7 min. The pellets were resuspended with the ice-cold Flow buffer containing 1 × PBS, 2% FBS, and 1 mM EDTA.
2.4. Flow cytometry and cell sorting
The isolated non-parenchymal cells were stained with the antibody mixture that included eBioscience (San Diego, CA, USA): anti-CD45 (Cat. No. 15–0459-42); BioLegend (San Diego, CA, USA): anti-CD3 (Cat. No. 317330), anti-CD11b (Cat. No. 553310), anti-CD14 (Cat. No. 301834), anti-CD31 (Cat. No. 303120), anti-CD32 (Cat. No. 303206), anti-CD68 (Cat. No. 333814), and anti-CD271 (Cat. No. 345110). In addition, cells were also stained with LIVE/DEAD Fixable Far Red Dead Cell Stain Kit (Cat. No. L10120, Life Technologies, Carlsbad, CA, USA). The incubation mixture was kept at 4 °C for 30 min in the dark on a rocking platform. The mixture was centrifuged with 500 ×g at 4 °C for 7 min. Cells were washed once with Flow buffer.
The antibody-labeled cells were sorted with a BD Aria III (BD Biosciences, San Jose, CA, USA). Analysis of cell populations was performed using FlowJo software, version 9.8.5 (FlowJo, LLC, Ashland, OR, USA). Kupffer cells were selected as the CD45+, CD3−, CD14+, CD68+, CD32+ populations (Alabraba et al., 2007; Ikarashi et al., 2013). LSECs were selected as CD45−, CD31+, CD11b− (Elvevold et al., 2008). HSCs were selected as CD45−, CD271+, auto-fluorescence positive with the emission wavelength at 460 nm (Buhring et al., 2007; D’Ambrosio et al., 2011) (Fig. S2).
2.5. RNA isolation and multiplex qRT-PCR analysis
The RNA of liver slices or the purified liver cells was isolated with TRIzol and the Direct-zol RNA MiniPrep Kit (Zymo Research, Irvine, CA, USA). The cDNA was synthesized with the QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany).
A pre-amplification step was included before the multiplex qRT-PCR assay (Brempelis et al., 2017), which included PCR reactions of cDNA as templates, and a primer mixture of TaqMan assays of interest (Thermo Fisher Scientific, Waltham, MA, USA) and the BIO-X-ACT Short Mix reagents (Bioline USA Inc., Taunton, MA, USA). The pre-amplified samples were diluted five-fold with RNase- and DNase-free H2O. Samples were analyzed with a 48 × 48 dynamic array and a BioMark HD microfluidics system (Fluidigm, San Francisco, CA, USA). The Fluidigm Real-Time PCR Analysis software was used to calculate Ct thresholds, using the settings of quality threshold 0.65, baseline correction linear, Ct threshold method auto detection. ACTB, HPRT, and GAPDH were chosen as the reference genes.
2.6. Microarray analysis
Microarray analysis was performed in the Janssen R&D microarray core facility using Affymetrix human genome U219 array strip kit. Data were analyzed with Transcriptome Analysis Console (TAC) Software (version 4.0, Thermo Fisher Scientific), using Robust Multi-array Average (RMA) summarization method (Irizarry et al., 2003). Heat map and principal component analysis were performed with Cluster 3.0 (de Hoon et al., 2004).
2.7. Histology of liver slices
Liver slices were fixed with the 10% neutral-buffered formalin (Sigma) at room temperature for 24 h. The fixed liver slices were embedded in paraffin and were sliced into 4 μm-thick sections for hematoxylin and eosin (H&E) stain, following the standard protocol in the Histology and Imaging Core (HIC) at Department of Comparative Medicine at UW. The H&E stained liver slides were scanned with Hamamatsu Nanozoomer Whole Slide Scanner (Hamamatsu City, Shizuoka Pref., Japan), and analyzed with NDP.view2 software (Hamamatsu).
2.8. Treatment of liver slices with Poly-I:C and LPS
Liver slices were cultured ex vivo for 7 days. Final concentrations of 15 μg/mL polyinosinic–polycytidylic acid (Poly-I:C) (Sigma, Cat. No. P1530), 1.5 μg/mL lipopolysaccharide endotoxin (LPS) (Sigma, Cat. No. L2630) or 1 × PBS (control, matched by volume) were added to the growth medium. During the stimulation, liver slices were maintained on transwell inserts placed on a rocking platform at 37 °C. Three liver slices were harvested at each time point, including 0 h (time zero control), 4 h, 8 h, 12 h or 24 h. The concentrations of Poly-I:C and LPS treatments were based on our previous studies with isolated liver leukocytes (Tu et al., 2008).
3. Results
3.1. Viability and evolution of human liver slice ex vivo
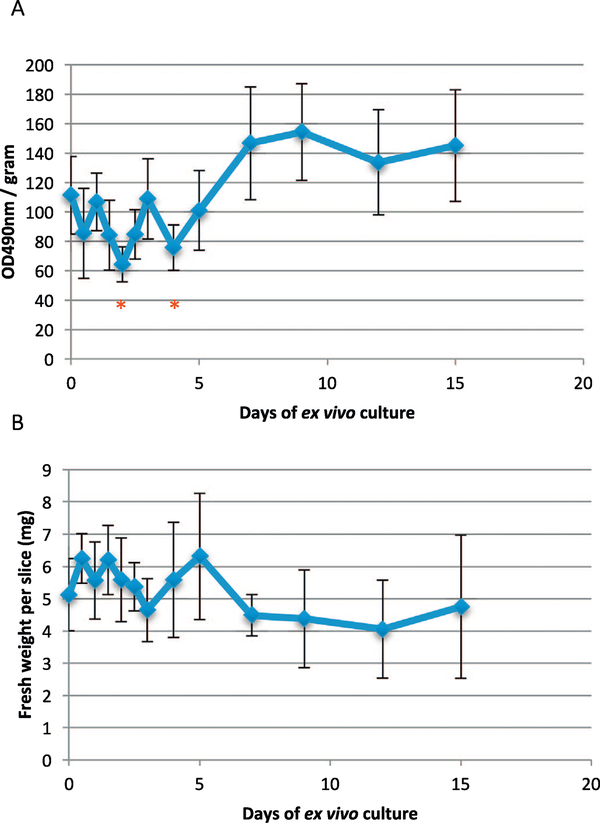
Previous analyses focus on liver slices cultured for a short time period (< 5 days) (de Graaf et al., 2010; Lagaye et al., 2012; Karim et al., 2013). To determine the viability of the liver slices cultured ex vivo over an extended period, we measured the metabolic activity of human liver slices with the MTS assay over a time course of 15 days (Fig. 1AB). Similar to previous reports, we observed a decrease in the MTS activity of liver slices in the first 2 days (48 h) (Karim et al., 2013). However, the MTS activity of liver slices rebounded after day 2, and stabilized after day 4. After day 7 and until day 15, MTS activity was sustained. These results indicate that human liver slices could be cultured ex vivo for more than two weeks.
Fig. 1.
Metabolic activity of human liver slices after ex vivo culturing for a 15-day period. (A) MTS activity of liver slices normalized by the fresh cell weight of the liver slices. (B) Measurements of fresh weight of liver slices. Error bars indicate the 95% confidence interval of standard deviation of seven replicates at each time point. Asterisks indicate the statistically significant differences compared with time zero using the two-way ANOVA test (*, P < 0.05).
To determine the changes of liver specific cell types in liver slices in the ex vivo culture, we monitored the cell type-specific gene expression over the period of culture. First, to identify the cell type-specific genes, we isolated the major cell subsets from human liver, using liver perfusion, differential centrifugation and FACS-sorting (Bhogal et al., 2011; Mohar et al., 2015). Individual populations of hepatocytes, Kupffer cells, LSECs, and HSCs were purified (Fig. S2).
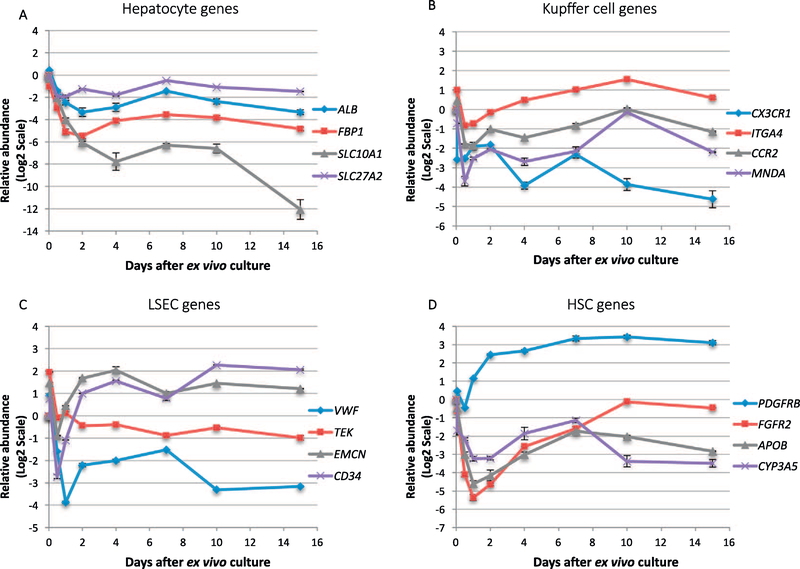
We used microarray analysis to identify a set of cell type-specific genes for each type of liver cell populations. These genes were consistent across cells purified from three separate donors (Fig. S3A). A principal component analysis across all genes also confirmed that gene expression of the same cell type was more similar to each other, compared with the other purified cell types (Fig. S3B). Based on the magnitude of differential expression, the functional relevance of the gene to the particular liver cell type, and previous reports of liver cell type-specific gene expression analysis (Azimifar et al., 2014; Ding et al., 2016), a subset of genes were selected for inclusion in a time course analysis (Tables S2–S4). The Kupffer cell markers were chemokine (C- × 3-C motif) receptor 1 (CX3CR1), chemokine (C-C motif) receptor 2 (CCR2), integrin alpha-4 (ITGA4), and myeloid cell nuclear differentiation antigen (MNDA). The LSEC markers were von Willebrand factor (VWF), angiopoietin-1 receptor (TEK), endomucin (EMCN), and hematopoietic progenitor cell antigen CD34. The HSC markers were platelet-derived growth factor receptor β (PDGFRB), fibroblast growth factor receptor 2 (FGFR2), apolipoprotein B (APOB), and cytochrome P450 3A5 (CYP3A5). In addition, albumin (ALB), fructose-1,6-bisphosphatase 1 (FBP1), sodium/bile acid cotransporter (SLC10A1, also known as HBV receptor gene NTCP), and very long-chain acyl-CoA synthetase (SLC27A2) were selected as hepatocyte-specific genes, according to previous hepatocyte-specific gene expression analyses (Mohar et al., 2015; Wisniewski et al., 2016; Brempelis et al., 2017).
We used these candidate genes to analyze the evolution of liver slices during the ex vivo culture. For the hepatocyte-specific gene candidates, we observed that ALB, FBP1, SLC10A1 and SLC27A2 decreased dramatically in the first 48 h. Nonetheless, the decrease stabilized after day 4, and only SLC10A1 still showed a substantial decreased expression from day 10 to day 15 (Fig. 2A). These changes may result from a decreased expression of hepatocyte-specific genes in liver slices after ex vivo culturing, or that hepatocytes survived poorly in the ex vivo culture relative to other cell types. Previous studies with murine primary hepatocytes also reported a reduction of hepatocyte-specific gene expression associated with the ex vivo culture (Clayton and Darnell, 1983).
Fig. 2.
Expression of liver cell-type-specific genes in the ex vivo culture. Genes are classified as (A) Hepatocyte, (B) Kupffer cell, (C) LSEC, (D) HSC. Data are expression based on qRT-PCR and relative to time zero. Error bars indicates the standard deviation of three liver slices cultured from the same patients at each time point. Similar trend of expression changes was also observed in the other two analyzed patients.
For the Kupffer cell-specific genes (CX3CR1, CCR2, ITGA4, MNDA), decreased expression was also observed for the initial 24 h. Their expression changes stabilized after day 2 (Fig. 2B). The decreased expression of CX3CR1 is also associated with the decreased abundance of its ligand CX3CL1 (Fractalkine) (Fig. 3A), which may suggest the differentiation of monocytes or macrophages in favor of liver fibrosis (Karlmark et al., 2010). For LSEC genes (VWF, TEK, EMCN, CD34), a rapid decrease was also observed for the initial 24–48 h culturing period. Two of the genes (EMCN, CD34) quickly increased in expression between day 2 to day 4. After day 4, the expression level of all four genes was stabilized, with EMCN and CD34 at the level greater than day 0, while VWF and TEK maintained at the decreased level compared to day 0 (Fig. 2C). For HSC gene candidates (PDGFRB, FGFR2, APOB, CYP3A5), the initial decrease was also apparent in the first 24–48 h post ex vivo culture (Fig. 2D). PDGFRB, which is also an activation marker for HSC activation during liver fibrosis, quickly increased in abundance between day 2 to day 4. The expression levels of all four genes became stabilized after day 4 until day 15.
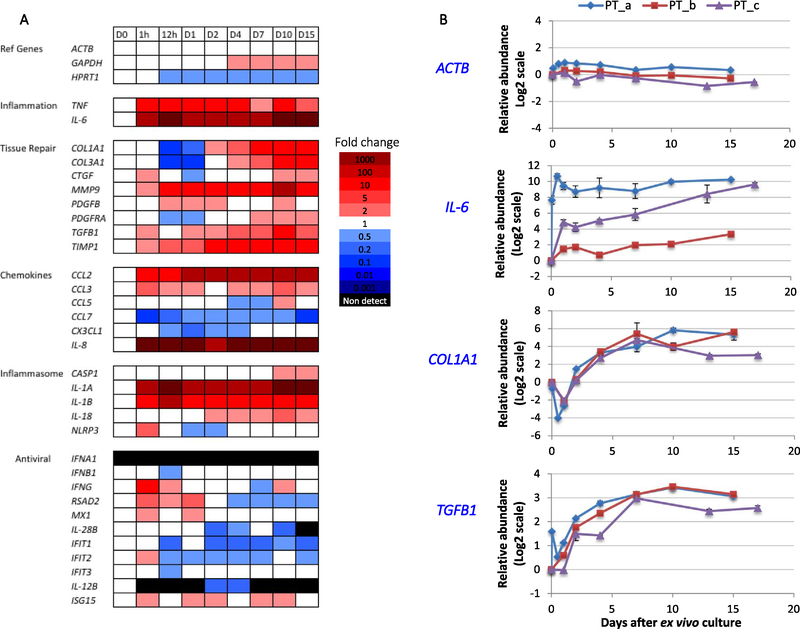
Fig. 3.
Innate immune response following liver slicing. (A) Heat map showing gene expression changes of a panel of innate immune genes in liver slices after vibratome slicing and ex vivo culturing. The time points include day 0, 1 h and 12 h, days 1, 2, 4, 7, 10 and 15. Red color indicates increased abundance, and blue color indicates decreased abundance. Black bar means the gene was not detected. (B) Detailed time course changes of ACTB, IL-6, COL1A1, and TGFB1. Data of three individual donors are identified as PT_a, PT_b and PT_c. Error bars are the standard deviation of three liver slices cultured from the same patient at each time point. Also see Fig. S4. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Hence, the expression profiles of these cell type-specific candidate genes suggest that there is likely a readjustment of liver cell populations in liver slices associated with ex vivo culturing. However, after 4 days of ex vivo culturing the liver cell condition appeared to be stabilized in liver slices.
3.2. Innate immune response to slicing
To further investigate the adaptation of liver slices in the ex vivo culture, we analyzed the expression of innate immune genes in liver slices. First, we observed a rapid activation of acute inflammatory genes (Gabay and Kushner, 1999) encoding interleukin-1β (IL-1B), interleukin-6 (IL-6), interleukin-1α (IL-1A), and Tumor Necrosis Factor-α (TNF) in liver slices immediately after the slicing and culturing (< 1 h) (Fig. 3AB). Furthermore, monocyte-recruiting chemokine gene CCL2 and neutrophil-recruiting chemokine gene CXCL8 (IL-8) also markedly increased expression in liver slices in < 1 h. These increases stabilized by day 4, and the elevated level was sustained until day 15 (Figs. 3AB, S4). In contrast, the reference genes ACTB and HPRT1 did not show significant increases.
Second, a group of tissue repair genes was also significantly increased after tissue slicing. Collagen synthesis is one of the hallmarks of liver fibrosis. Collagen genes COL1A1 and COL3A1 decreased in expression for the initial 24 h, but began to increase at day 2, and the increased level (compared to day 0) was sustained to day 15 (Figs. 3AB, S4). Delayed activation of COL1A1 was also observed in other studies with the liver slice culture (van de Bovenkamp et al., 2008; Westra et al., 2016). Furthermore, expression of platelet-derived growth factor receptor PDGFRA and PDGFRB is an activation marker for HSC during liver fibrosis (Wong et al., 1994; Hayes et al., 2014). Both PDGFRA and PDGFRB decreased in expression for the initial 24–48 h, but increased later (Figs. 2D, S4), resembling the pattern of changes of COL1A1 and COL3A1. In addition, other activation markers of liver fibrosis including genes encoding the profibrogenic cytokine transforming growth factor TGF-β, matrix metalloproteinase MMP-9 and metalloproteinase inhibitor TIMP1 (Xu et al., 2012) also increased rapidly in the first 4 days, but reached a plateau after day 4 (Figs. 3AB, S4).
The above results indicate that liver slices that are cultured ex vivo have increased level of cytokine genes and tissue repair genes from the beginning of the ex vivo culturing process. Nevertheless, because the initial inflammation and activation of repair genes stabilize after day 4 until day 15, we are able to investigate the immune response of liver slices to exogenous stimuli, using day 7 as the baseline. Unlike many pro-inflammatory genes, the inflammasome genes NLRP3 and CASP1, interferon genes IFNA1, IFNB1, IFNG and IL-28B, and interferon-stimulated genes were not increased in expression in response to tissue slicing (Fig. 3A).
3.3. Morphology of liver slices in the ex vivo culture
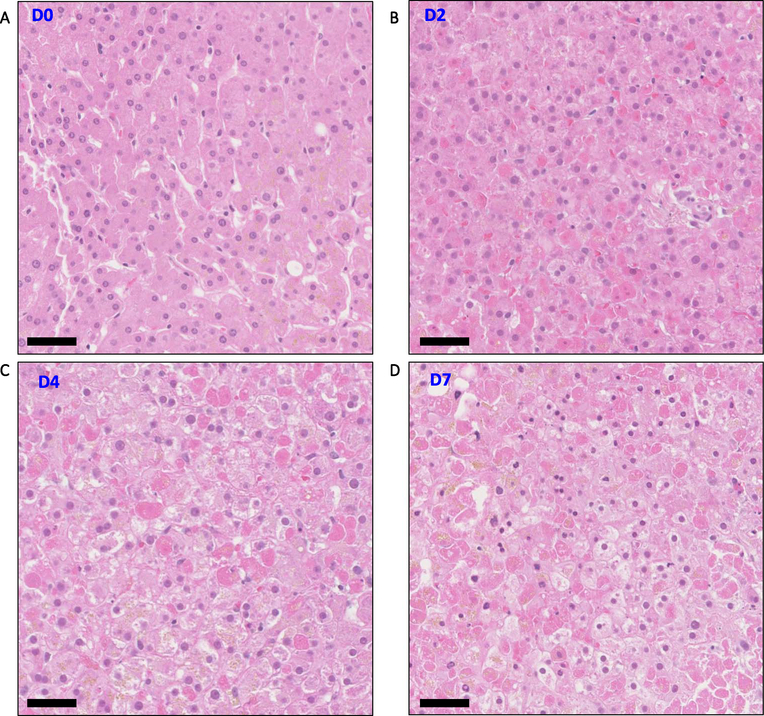
We visualized the cellular morphology of liver slices with H&E staining. Liver slices at day 0 (time zero) maintain typical anatomical structural features of a cross-section of liver lobe. The portal tracts, including the portal vein, hepatic artery and bile duct, were clearly defined. Hepatocytes were arranged in cords (Figs. 4A, S5A). Liver slices from some donors also contained patches of lymphocytes, suggesting prior inflammation in the liver or at the time of surgery (Fig. S5B).
Fig. 4.
Hematoxylin and eosin (H&E) stain of liver slices. (A–D) Morphology of liver slices at days 0, 2, 4 and 7. Scale bar represents 50 μm length. Injured hepatocytes appeared by day 2 but viable hepatocytes were also present at all time points. Also see Figs. S5–7.
Liver slices that were cultured ex vivo for 2 days, 4 days, and 7 days contained a substantial number of viable hepatocytes (Fig. 4BCD), consistent with the detected MTS activity in the slices. Remarkably, hepatocytes with pre-existing pathology such as steatosis or cholestasis (Mills et al., 2012) survived the tissue slicing, and these features were preserved in liver slice culture for up to seven days (Fig. S6AB). On the other hand, hepatocyte death was also observed in liver slices. Hepatocyte apoptosis and necrosis (Elmore et al., 2016) appeared as early as 12 h (day 0.5) post ex vivo culturing and were also present in day 2, day 4 and day 7 slices (Fig. S7AB).
3.4. Innate immune response to Poly-I:C and LPS stimuli
Human liver slices were viable at day 7 and the expression of pro-inflammatory genes was broadly stabilized. To test the innate immune properties of liver slices, we exposed them to the TLR3 stimulant Poly-I:C and the TLR4 stimulant LPS on day 7. Although overlap exists in TLR3 and TLR4 immune activation and intracellular signaling (Lester and Li, 2014), TLR3 mediates a more potent antiviral response than TLR4 in macrophages (Doyle et al., 2003). Concentrations of 15 μg/mL for Poly-I:C and 1.5 μg/mL for LPS were chosen, because the two concentrations induce high level of cytokines (IL-10 and IL-18) in human Kupffer cells (Tu et al., 2008).
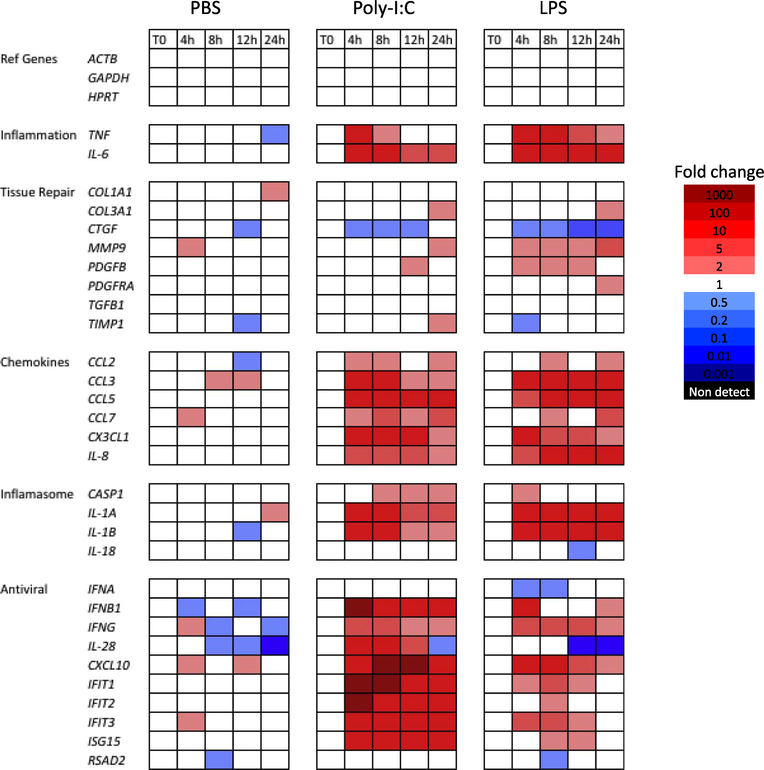
We confirmed that both TLR3 and TLR4 genes were expressed in human liver slices in the ex vivo culture. Both Poly-I:C and LPS treatments resulted in a marked response of inflammation genes (TNF, IL-6), chemokine genes (CCL2, CCL3, CCL5, CCL7, CX3CL1, IL-8), and inflammasome genes (IL-1A, IL-1B) (Fig. 5). In comparison, PBS as a control did not induce the systematic increases in these genes. Interestingly, the tissue repair genes only showed a weak increase or no increase in response to Poly-I:C or LPS, which is different from the innate immune response to the slicing (Fig. 3).
Fig. 5.
Innate immune response of liver slices to Poly-I:C and LPS stimuli. Fold changes are referenced to the day 7 time zero slices. Red color indicates increased abundance, and blue color indicates decreased abundance. The PBS treatment is included as the control. Solid blocks of red indicate increased expression of chemokine genes and anti-viral genes, starting at 4 h after treatment. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
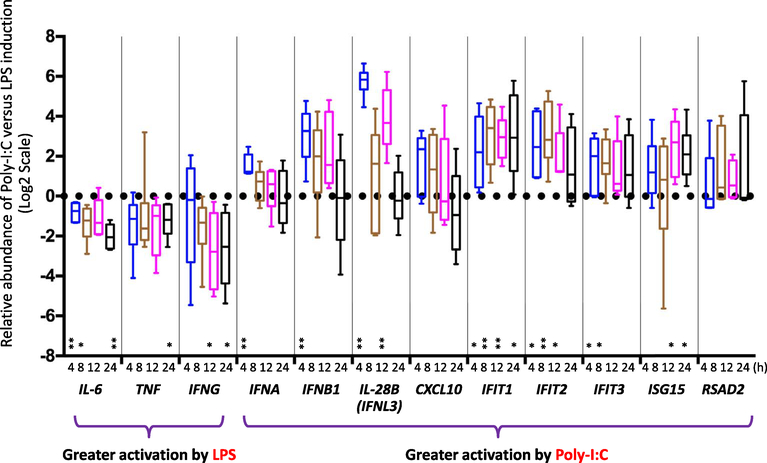
We observed a distinctive antiviral response induced by Poly-I:C compared with LPS (Fig. 6). In contrast to the stronger activation of IL-6, TNF and IFNG by LPS treatment, Poly-I:C induced a greater response in antiviral genes in general including IFNA, IFNB, IL-28B, IFIT½/3, and ISG15. For the interferons IFNA, IFNB, and IL-28B, the enhanced activation by Poly-I:C was most notable at 4 h. The differences diminished for IFNA, IFNB after 4 h, but IL-28B still showed greater induction at 12 h. For the interferon stimulated genes (ISGs), changes appear to be more gradual and sustained. IFIT1 showed more significant activation by Poly-I:C at all four time points (4 h, 8 h, 12 h, 24 h). IFIT2 exhibited differences at 4 h, 8 h and 12 h. IFIT3, also showed increased induction by Poly-I:C at two time points (4 h and 8 h), and ISG15 showed differences at a later time points 12 h and 24 h. Previous study with the Huh7.5 hepatocyte-derived cell line also found a sustained-24 h induction of ISGs contributed by IFN-λ induction (Kohli et al., 2012).
Fig. 6.
Antiviral response preferentially induced by Poly-I:C compared with LPS. Box-and-whisker plot shows the inter-quartile range, the median value, and the 95% confidence interval of the standard deviation of data representing six patient samples. Responses were examined at four time points, including 4 h, 8 h, 12 h, 24 h. Fold changes are referenced to the time zero for each patient. A positive value indicates a more marked increase induced by Poly-I:C compared with LPS; conversely a negative value indicates a more marked increase induced by LPS compared with Poly-I:C. Statistical significance is indicated (*, < 0.05; **, < 0.01; two-way ANOVA test).
4. Discussion
Here we describe an improved method for the sustained culture of human liver slices, and the use of such slices in the investigation of human liver immunology. One key issue for liver slice cultures has been the provision of sufficient oxygen. A slice submerged in tissue culture media may benefit from either hyperbaric conditions, or an elevated oxygen concentration in the incubator. We approached the problem of gas exchange a different way, by culturing the liver slices at a media-gas phase interface supported on a trans-well tissue culture insert, and also by placing the culture vessel on a rocking platform. The combination of exposure to the 5% CO2 in air gas mixture, and the media movement caused by rocking, evidently favored gas and nutrient diffusion since these measures helped us obtain prolonged survival of the slices without the need for an elevated partial pressure of O2.
We demonstrated that liver slices produce a strong response in acute inflammatory and tissue repair response upon tissue slicing. However, such induction stabilizes after day 4 until day 15. Exposure of liver slices at day 7 of culture to Poly-I:C and LPS resulted in a robust activation of acute inflammation and cytokine genes. In particular, Poly-I:C treatment led to a marked antiviral response including increases of interferons IFNB, IL-28B and a group of interferon-stimulated genes.
Our histological analysis of slices from different tissue donors also showed us that the patterns of liver disease that are present in fresh tissue are maintained over time in the slice cultures. For example, steatotic liver slices retained fat droplets, and cholestatic liver slices were manifested with yellow-green pigment in hepatocytes. This opens the possibility to investigate innate immunity in the context of these abnormalities over time.
Scientific areas in which these culture will be useful also include the study of the innate immune response to injury, which is readily approached in mice (Brempelis et al., 2017) but relatively inaccessible in humans. The slices also manifest features of the acute response to sterile liver injury, and the mechanisms involved could be studied directly. Notably, Lagaye and colleagues reported the successful ex vivo infection of liver slices with hepatitis C virus (HCV) strains JFH-1, H77/C3 and Con1/C3, and with HCV-positive patient serum (Lagaye et al., 2012). Thus, liver slice culture holds a great promise to study liver immunity against HCV and other viral infections.
Furthermore, liver slices have been applied to study the microenvironment of liver cancer cells (Zimmermann et al., 2009; Koch et al., 2014). One major advantage of the liver slice culture approach is that it can be reliably applied to liver tissue from essentially anyone undergoing liver resection surgery, whether this is in the context of a primary liver cancer, or a metastasis, or a non-cancerous lesion. Over time, data can be accumulated from a large pool of individuals reflecting both human genetic diversity and patterns of disease. In the context of cancer, new immunotherapeutic approaches have often been remarkably effective, but only in a subset of patients (Pardee and Butterfield, 2012; Boyiadzis et al., 2016), and choosing the correct immunological intervention for each individual may be key for the success of future therapy. With the recent advancement of immunotherapy in hepatocellular carcinoma, we may be able to use liver slices to characterize patient-specific liver immunity to help design a patient-oriented therapy. A similar approach may apply to other solid tumors (Jiang et al., 2017).
As with any model system, there are also limitations inherent to the PCLS method. First, some cytokine genes and tissue repair genes are activated in liver slices during the preparations of human liver slices. Therefore, identification of further small fold changes for those cytokine and tissue repair genes in intended treatments could be difficult, unless the stimulus overcomes the elevated baseline. However, we were able to ameliorate this problem by allowing the cultures to achieve equilibrium over 7 days in vitro, before adding innate immune stimuli. Second, there was evidence of hepatocyte death and changes in the expression of cell-type-specific genes, which may suggest changes in the balance of cell populations in slices during the culture. In consequence, certain findings with the PCLS method may not reflect the full range of the in vivo function of liver cells. Third, the PCLS method requires immediate access to human clinical tissues, and we do not know if tissue transport from remote sites is compatible with our approach. Despite these limitations, PCLS offers a human liver immunology platform with distinct advantages, as we show here.
5. Conclusion
We have developed a modified culture protocol for human liver slices that is suitable to study human liver innate immunity. Future work could utilize PCLS method to study intra-hepatic immunology during disease progression and the effects of novel immune modulators.
Supplementary Material
Acknowledgements
We thank the helpful discussion of Ms. Radika Soysa at University of Washington. We thank the Flow Cytometry Core and the Histology Core Facilities of the Department of Pathology at UW, and the Histology and Imaging Core (HIC) of the Department of Comparative Medicine at UW. This work was supported by Janssen Research and Development, the Seattle Foundation, and by the Department of Pathology, University of Washington.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jim.2018.01.012.
Reference
- Alabraba EB, Curbishley SM, Lai WK, Wigmore SJ, Adams DH, Afford SC, 2007. A new approach to isolation and culture of human Kupffer cells. J. Immunol. Methods 326, 139–144. [DOI] [PubMed] [Google Scholar]
- Azimifar SB, Nagaraj N, Cox J, Mann M, 2014. Cell-type-resolved quantitative proteomics of murine liver. Cell Metab. 20, 1076–1087. [DOI] [PubMed] [Google Scholar]
- Bhogal RH, Hodson J, Bartlett DC, Weston CJ, Curbishley SM, Haughton E, Williams KT, Reynolds GM, Newsome PN, Adams DH, Afford SC, 2011. Isolation of primary human hepatocytes from normal and diseased liver tissue: a one hundred liver experience. PLoS One 6, e18222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachier M, Leleu H, Peck-Radosavljevic M, Valla DC, Roudot-Thoraval F, 2013. The burden of liver disease in Europe: a review of available epidemiological data. J. Hepatol 58, 593–608. [DOI] [PubMed] [Google Scholar]
- Boess F, Kamber M, Romer S, Gasser R, Muller D, Albertini S, Suter L, 2003. Gene expression in two hepatic cell lines, cultured primary hepatocytes, and liver slices compared to the in vivo liver gene expression in rats: possible implications for toxicogenomics use of in vitro systems. Toxicological Sciences 73, 386–402. [DOI] [PubMed] [Google Scholar]
- van de Bovenkamp M, Groothuis GM, Meijer DK, Olinga P, 2008. Liver slices as a model to study fibrogenesis and test the effects of anti-fibrotic drugs on fibrogenic cells in human liver. Toxicology In Vitro 22, 771–778. [DOI] [PubMed] [Google Scholar]
- Boyiadzis M, Bishop MR, Abonour R, Anderson KC, Ansell SM, Avigan D, Barbarotta L, Barrett AJ, Van Besien K, Bergsagel PL, Borrello I, Brody J, Brufsky J, Cairo M, Chari A, Cohen A, Cortes J, Forman SJ, Friedberg JW, Fuchs EJ, Gore SD, Jagannath S, Kahl BS, Kline J, Kochenderfer JN, Kwak LW, Levy R, de Lima M, Litzow MR, Mahindra A, Miller J, Munshi NC, Orlowski RZ, Pagel JM, Porter DL, Russell SJ, Schwartz K, Shipp MA, Siegel D, Stone RM, Tallman MS, Timmerman JM, Van Rhee F, Waller EK, Welsh A, Werner M, Wiernik PH, Dhodapkar MV, 2016. The Society for Immunotherapy of Cancer consensus statement on immunotherapy for the treatment of hematologic malignancies: multiple myeloma, lymphoma, and acute leukemia. J. Immunother. Cancer 4, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brempelis KJ, Yuen SY, Schwarz N, Mohar I, Crispe IN, 2017. Central role of the TIR-domain-containing adaptor-inducing interferon-beta (TRIF) adaptor protein in murine sterile liver injury. Hepatology 65, 1336–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhring HJ, Battula VL, Treml S, Schewe B, Kanz L, Vogel W, 2007. Novel markers for the prospective isolation of human MSC. Ann. N. Y. Acad. Sci 1106, 262–271. [DOI] [PubMed] [Google Scholar]
- Clayton DF, Darnell JE Jr., 1983. Changes in liver-specific compared to common gene transcription during primary culture of mouse hepatocytes. Mol. Cell. Biol 3, 1552–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispe IN, 2016. Hepatocytes as immunological agents. J. Immunol 196, 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrosio DN, Walewski JL, Clugston RD, Berk PD, Rippe RA, Blaner WS, 2011. Distinct populations of hepatic stellate cells in the mouse liver have different capacities for retinoid and lipid storage. PLoS One 6, e24993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C, Li Y, Guo F, Jiang Y, Ying W, Li D, Yang D, Xia X, Liu W, Zhao Y, He Y, Li X, Sun W, Liu Q, Song L, Zhen B, Zhang P, Qian X, Qin J, He F, 2016. A cell-type-resolved liver proteome. Molecular & Cellular Proteomics: MCP 15, 3190–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SE, O’Connell R, Vaidya SA, Chow EK, Yee K, Cheng G, 2003. Toll-like receptor 3 mediates a more potent antiviral response than toll-like receptor 4. J. Immunol 170, 3565–3571. [DOI] [PubMed] [Google Scholar]
- Elmore SA, Dixon D, Hailey JR, Harada T, Herbert RA, Maronpot RR, Nolte T, Rehg JE, Rittinghausen S, Rosol TJ, Satoh H, Vidal JD, Willard-Mack CL, Creasy DM, 2016. Recommendations from the INHAND apoptosis/necrosis working group. Toxicol. Pathol 44, 173–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvevold K, Smedsrod B, Martinez I, 2008. The liver sinusoidal endothelial cell: a cell type of controversial and confusing identity. Am. J. Physiol. Gastrointest. Liver Physiol 294, G391–400. [DOI] [PubMed] [Google Scholar]
- Gabay C, Kushner I, 1999. Acute-phase proteins and other systemic responses to inflammation. N. Engl. J. Med 340, 448–454. [DOI] [PubMed] [Google Scholar]
- Graaf IA, Groothuis GM, Olinga P, 2007. Precision-cut tissue slices as a tool to predict metabolism of novel drugs. Expert Opin. Drug Metab. Toxicol 3, 879–898. [DOI] [PubMed] [Google Scholar]
- de Graaf IAM, Olinga P, de Jager MH, Merema MT, de Kanter R, van de Kerkhof EG, Groothuis GMM, 2010. Preparation and incubation of precision-cut liver and intestinal slices for application in drug metabolism and toxicity studies. Nat. Protoc 5, 1540–1551. [DOI] [PubMed] [Google Scholar]
- Hadi M, Westra IM, Starokozhko V, Dragovic S, Merema MT, Groothuis GM, 2013. Human precision-cut liver slices as an ex vivo model to study idiosyncratic drug-induced liver injury. Chem. Res. Toxicol 26, 710–720. [DOI] [PubMed] [Google Scholar]
- Hayes BJ, Riehle KJ, Shimizu-Albergine M, Bauer RL, Hudkins KL, Johansson F, Yeh MM, Mahoney WM Jr., Yeung RS, Campbell JS, 2014. Activation of platelet-derived growth factor receptor alpha contributes to liver fibrosis. PLoS One 9, e92925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann F, Tacke F, 2016. Immunology in the liver—from homeostasis to disease. Nat. Rev. Gastroenterol. Hepatol 13, 88–110. [DOI] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S, 2004. Open source clustering software. Bioinformatics 20, 1453–1454. [DOI] [PubMed] [Google Scholar]
- Ijssennagger N, Janssen AW, Milona A, Ramos Pittol JM, Hollman DA, Mokry M, Betzel B, Berends FJ, Janssen IM, van Mil SW, Kersten S, 2016. Gene expression profiling in human precision cut liver slices in response to the FXR agonist obeticholic acid. J. Hepatol 64, 1158–1166. [DOI] [PubMed] [Google Scholar]
- Ikarashi M, Nakashima H, Kinoshita M, Sato A, Nakashima M, Miyazaki H, Nishiyama K, Yamamoto J, Seki S, 2013. Distinct development and functions of resident and recruited liver Kupffer cells/macrophages. J. Leukoc. Biol 94, 1325–1336. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP, 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264. [DOI] [PubMed] [Google Scholar]
- Janssen AW, Betzel B, Stoopen G, Berends FJ, Janssen IM, Peijnenburg AA, Kersten S, 2015. The impact of PPARalpha activation on whole genome gene expression in human precision cut liver slices. BMC Genomics 16, 760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Seo YD, Chang JH, Coveler A, Nigjeh EN, Pan S, Jalikis F, Yeung RS, Crispe IN, Pillarisetty VG, 2017. Long-lived pancreatic ductal adenocarcinoma slice cultures enable precise study of the immune microenvironment. Oncoimmunology 6, e1333210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim S, Liaskou E, Hadley S, Youster J, Faint J, Adams DH, Lalor PF, 2013. An in vitro model of human acute ethanol exposure that incorporates CXCR3- and CXCR4-dependent recruitment of immune cells. Toxicological Sciences 132, 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlmark KR, Zimmermann HW, Roderburg C, Gassler N, Wasmuth HE, Luedde T, Trautwein C, Tacke F, 2010. The fractalkine receptor CX(3)CR1 protects against liver fibrosis by controlling differentiation and survival of infiltrating hepatic monocytes. Hepatology 52, 1769–1782. [DOI] [PubMed] [Google Scholar]
- Kasper HU, Dries V, Drebber U, Kern MA, Dienes HP, Schirmacher P, 2005. Precision cut tissue slices of the liver as morphological tool for investigation of apoptosis. In Vivo 19, 423–431. [PubMed] [Google Scholar]
- Koch A, Saran S, Tran DD, Klebba-Farber S, Thiesler H, Sewald K, Schindler S, Braun A, Klopfleisch R, Tamura T, 2014. Murine precision-cut liver slices (PCLS): a new tool for studying tumor microenvironments and cell signaling ex vivo. Cell Communication and Signaling: CCS 12, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli A, Zhang X, Yang J, Russell RS, Donnelly RP, Sheikh F, Sherman A, Young H, Imamichi T, Lempicki RA, Masur H, Kottilil S, 2012. Distinct and overlapping genomic profiles and antiviral effects of interferon-lambda and -alpha on HCV-infected and noninfected hepatoma cells. J. Viral Hepat 19, 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagaye S, Shen H, Saunier B, Nascimbeni M, Gaston J, Bourdoncle P, Hannoun L, Massault P-P, Vallet-Pichard A, Mallet V, Pol S, 2012. Efficient replication of primary or culture hepatitis C virus isolates in human liver slices: a relevant ex vivo model of liver infection. Hepatology 56, 861–872. [DOI] [PubMed] [Google Scholar]
- Lester SN, Li K, 2014. Toll-like receptors in antiviral innate immunity. J. Mol. Biol 426, 1246–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang H, 2016. Heterogeneity of liver cancer and personalized therapy. Cancer Lett. 379, 191–197. [DOI] [PubMed] [Google Scholar]
- Mills SE, Carter D, Greenson JK, Reuter VE, Stoler MH, 2012. Sternberg’s Diagnostic Surgical Pathology. Wolters Kluwer Health. [Google Scholar]
- Mohar I, Brempelis KJ, Murray SA, Ebrahimkhani MR, Crispe IN, 2015. Isolation of non-parenchymal cells from the mouse liver. Methods Mol. Biol 1325, 3–17. [DOI] [PubMed] [Google Scholar]
- Mokdad AA, Lopez AD, Shahraz S, Lozano R, Mokdad AH, Stanaway J, Murray CJ, Naghavi M, 2014. Liver cirrhosis mortality in 187 countries between 1980 and 2010: a systematic analysis. BMC Med. 12, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinga P, Meijer DK, Slooff MJ, Groothuis GM, 1997. Liver slices in in vitro pharmacotoxicology with special reference to the use of human liver tissue. Toxicology In Vitro 12, 77–100. [DOI] [PubMed] [Google Scholar]
- Pardee AD, Butterfield LH, 2012. Immunotherapy of hepatocellular carcinoma: unique challenges and clinical opportunities. Oncoimmunology 1, 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitia G, 2015. Towards personalized medicine in chronic HBV patients? Liver International 35, 1783–1785. [DOI] [PubMed] [Google Scholar]
- Starokozhko V, Vatakuti S, Schievink B, Merema MT, Asplund A, Synnergren J, Aspegren A, Groothuis GMM, 2017. Maintenance of drug metabolism and transport functions in human precision-cut liver slices during prolonged incubation for 5 days. Arch. Toxicol 91, 2079–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z, Bozorgzadeh A, Pierce RH, Kurtis J, Crispe IN, Orloff MS, 2008. TLR-dependent cross talk between human Kupffer cells and NK cells. J. Exp. Med 205, 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FS, Fan JG, Zhang Z, Gao B, Wang HY, 2014. The global burden of liver disease: the major impact of China. Hepatology 60, 2099–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra IM, Mutsaers HA, Luangmonkong T, Hadi M, Oosterhuis D, de Jong KP, Groothuis GM, Olinga P, 2016. Human precision-cut liver slices as a model to test antifibrotic drugs in the early onset of liver fibrosis. Toxicology In Vitro 35, 77–85. [DOI] [PubMed] [Google Scholar]
- Wisniewski JR, Vildhede A, Noren A, Artursson P, 2016. In-depth quantitative analysis and comparison of the human hepatocyte and hepatoma cell line HepG2 proteomes. J. Proteome 136, 234–247. [DOI] [PubMed] [Google Scholar]
- Wong L, Yamasaki G, Johnson RJ, Friedman SL, 1994. Induction of beta-platelet-derived growth factor receptor in rat hepatic lipocytes during cellular activation in vivo and in culture. J. Clin. Invest 94, 1563–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Zhang Z, Wang FS, 2012. Liver fibrosis: mechanisms of immune-mediated liver injury. Cell. Mol. Immunol 9, 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, Armeanu S, Smirnow I, Kupka S, Wagner S, Wehrmann M, Rots MG, Groothuis GM, Weiss TS, Konigsrainer A, Gregor M, Bitzer M, Lauer UM, 2009. Human precision-cut liver tumor slices as a tumor patient-individual predictive test system for oncolytic measles vaccine viruses. Int. J. Oncol 34, 1247–1256. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.