Abstract
Background
Hepatocellular carcinoma (HCC) is a prevalent malignant tumor. Long non-coding RNAs (lncRNAs) have been demonstrated to be abnormally expressed in many tumors and act as crucial regulators in various biological processes. However, the expression and function of the recently identified macrophage-associated lncRNA ELMO1 antisense RNA 1 (ELMO1-AS1) in HCC are unclear.
Methods
The expression of ELMO1-AS1 was determined in HCC tissues and adjacent nontumorous tissues by quantitative real-time polymerase chain reaction (qRT-PCR). The Kaplan-Meier survival analysis and Cox regression analysis were performed to establish the correlation between the expression level and survival of HCC patients in a training set and a validation set, respectively. The overexpression experiments were also conducted to investigate the biological role of ELMO1-AS1 in HCC cells.
Results
We uncovered that ELMO1-AS1 was significantly downregulated in HCC tissues, and high expression of ELMO1-AS1 is correlated with optimistic treatment outcome suggesting its potential as an independent prognostic biomarker for HCC. It was also found that overexpression of ELMO1-AS1 in HCC cells suppressed cell proliferation, migration and invasion and engulfment and cell motility 1 (ELMO1) may be a target of ELMO1-AS1.
Conclusion
Our results suggested that macrophage-associated lncRNA ELMO1-AS1 could be a crucial regulator involved in HCC progression and considered as a potential prognostic biomarker and therapeutic target for HCC.
Keywords: long noncoding RNA, ELMO1-AS1, HCC, prognosis
Introduction
Hepatocellular carcinoma (HCC), as the most common type of primary liver cancer, is one of the most common malignant tumors, which is in the top three leading causes of tumor-related death around the world.1 HCC is particularly prevalent in Asia,2,3 and approximately half of the new HCC patients come from China.4 Although various risk factors of HCC occurrence and development have been revealed,5 and the concepts of precision medicine (PM)6 and multidisciplinary team (MDT)7 have been applied in HCC therapy, poor prognosis and high mortality rate still persist.8 New approaches to predict the outcome and further study to disclose the mechanism of HCC are urgently needed.
Long non-coding RNAs (lncRNAs), with more than 200 nucleotides in length and barely protein-coding ability, have been identified in the recent years. It is increasingly recognized that lncRNAs interact with DNA, RNA, protein molecules and/or their combinations, involve in serial steps of tumor development, and act as the crucial regulators in various biological processes.9,10 In addition, detection of lncRNAs as the biomarkers in tumors is instrumental to early diagnosis, progression monitoring, prognosis prediction and targeted therapy.11–13 However, the molecular mechanisms of lncRNAs for the occurrence and development of HCC remain unclear.
Our previous microarray profiling14 also found that lncRNA ELMO1 antisense RNA 1 (ELMO1-AS1) was significantly upregulated in classically activated macrophages (M1 macrophages) and downregulated in alternatively activated macrophages (M2 macrophages; >10 fold change, P=0.002). To date, the relevance of ELMO1-AS1 to HCC, a novel macrophage-associated lncRNA, has not been investigated. In the present study, we aimed to examine the expression of ELMO1-AS1 in HCC tissues and their adjacent nontumorous tissues and explore its potential for prognosis. The functional studies were also carried out to assess the molecular mechanisms of ELMO1-AS1 in the HCC cells. This study shall offer new insights into the roles of ELMO1-AS1 in the development and progression of HCC.
Materials and methods
Patients and tissue specimens
Two independent sets comprising a total of 222 pairs of surgical specimens, tumor tissues and adjacent nontumorous tissues from HCC patients, were enrolled in this study from the Department of Hepatobiliary Surgery of the Affiliated Tumor Hospital of Guangxi Medical University in China. The training set enrolled 110 cases from January 2014 to December 2014, and the validation set contained 112 patients from January 2015 to August 2016. All patients in the present study underwent radical hepatectomy and had pathologically diagnosed with hepatocellular carcinoma. None of them had received radiotherapy or chemotherapy before surgery. The collected tissue specimens were immediately stored at −80 °C before RNA isolation. In addition, the clinicopathological parameters were retrieved, including gender, age, tumor number, tumor diameter, Alpha-fetoprotein (AFP), portal vein tumor thrombus (PVTT), microsatellite, microvascular invasion (MVI) and TNM stage. The follow-up data were obtained through telephone calls or outpatient visits. This study was approved by the Ethics Committee of the Affiliated Tumor Hospital of Guangxi Medical University.
Cell culture
Two human liver cancer cell lines, SMMC-7721 and HepG2, were preserved in our central laboratory, and identified by short tandem repeat (STR) profile generated by GENEWIZ Inc. Cells were maintained in Dulbecco’s Modified Eagle’s Media (DMEM) medium, supplemented with 10% fetal bovine serum (FBS; Gibco, CA, USA) with 100 U/ml penicillin and 100 µg/ml streptomycin and cultured in a humidified incubator with 5% CO2 at 37 °C.
RNA isolation, reverse transcription and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA from the enrolled samples and cell lines was isolated by TRIzol reagent (Invitrogen, CA, USA) according to the protocol of manufacturer. After purity and integrity test, RNA that met the requirements were reverse transcribed into cDNA using the PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, Dalian, China). Real-time PCR was performed to detect the expression of RNAs using FastStart Universal SYBR Green Master (ROX) kit (Roche, Germany). Relative expression level was normalized to an endogenous control (β-actin) by relative quantification method (2−∆∆Ct). The primers for ELMO1-AS1 were 5′- TCGGGTGGAAGTCGTTGC-3′ and 5′- CCTCCGCTTGTCTCCCTTT-3′; the primers for ELMO1 were 5′- CCACGACAGATCCTTTGAGGAG-3′ and 5′- CTCATAACCTGCTCCTTCACCAC-3′; the primers for β-actin were 5′-TGCGTGACATTAAGGAGAAG-3′ and 5′- GTCAGGCAGCTCGTAGCTCT-3′.
Plasmid construction, lentivirus generation and cell infection
Full-length ELMO1-AS1 cDNA was compounded and inserted into the lentiviral expression plasmid pCDH commercially (Sagene, China). HEK293T cells were co-transfected with the lentiviral plasmid packaging system and pCDH-ELMO1-AS1 or pCDH-empty control using Lipofectamine 2000 (Invitrogen, USA). After transfection for 48 hrs, lentiviral particles in the medium were collected and centrifuged at 3000 g for 15 min and then filtered through a 0.22 µm filter. HCC cells were transfected with LV-pCDH-ELMO1-AS1 or LV-pCDH-empty control, respectively. After 48 h, the stable lines were selected by puromycin and used for the functional assay.
Cell proliferation assay
The cell counting kit-8 (CCK-8) assay was employed to detect the proliferation of HCC cells. Approximately 3,000 cells/well HCC cells, ELMO1-AS1 and control, were planked in 96-well plates. The cells were incubated with 100 μl fresh culture medium mixed with 10 μl CCK-8 (7sea, China) for 2 h at 0 h, 24 h, 48 h, 72 h and 96 h after seeded. The viability of HCC cells was detected by the optical density (OD) at a wavelength of 450 nm by using an enzyme-labeled analyzer.
Colony formation assay
500 HCC cells were planked per well in 6-well plates. The culture medium was replaced every 3 days. When macroscopic colonies appeared, the colonies were fixed with methyl alcohol for 30 min and dyed with 0.1% crystal violet solution for 30 min. Colonies in every well were counted independently.
Wound healing assay
HCC cells were seeded into 6-well plates and cultured in the humidified incubator overnight. Artificial wounds were performed by scratching with the 200 µl pipette tips and then incubated with serum-free medium to proceed. The artificial wounds were imaged at 0 h and 24 h by an inverted microscope.
Migration and invasion assays
According to assay protocol, 24-well plate with 8-µm pore size transwell chamber (Costar, USA) with or without Matrigel-coated membrane (BD, USA) was used to measure HCC cell migration or invasion. 5×104 and 1×105 cells were suspended into the upper chamber per well, respectively. In both assays, HCC cells were suspended in 200 µl of DMEM medium without FBS. 700 µl of DMEM containing 10% FBS was added in the lower chamber. After 18–24 hrs of culture in the incubator, the cells migrated to the bottom surface of the membrane, were fixed with methyl alcohol, stained with 0.1% crystal violet solution for 30 min and photographed and counted under an inverted microscope.
Statistical analysis
All statistical tests for this study were performed using SPSS 22.0 software (Chicago, IL, USA). Graphs were constructed using GraphPad Prism 5 software (La Jolla, CA, USA).
The data about continuous variables from experiments are presented as the mean ± SEM. Student’s t-test was carried out to evaluate ELMO1-AS1 RNA level between human HCC tissues and the matched non-tumorous samples. Qualitative variables were compared using χ2 tests. The Kaplan-Meier survival analysis and Cox regression analysis test were performed to compare the correlation between the expression level and survival. Two-sided P-value less than 0.05 was considered as the statistical significance.
Results
Downregulation of ELMO1-AS1 expression in HCC patients
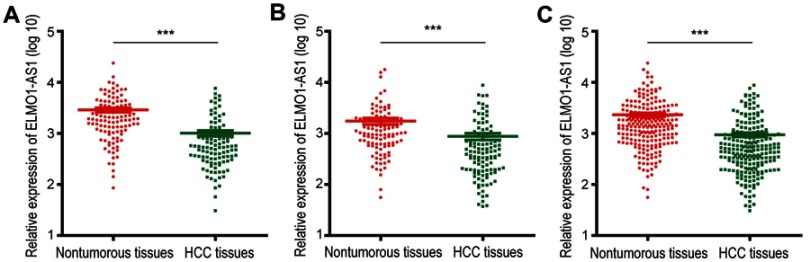
We measured ELMO1-AS1 in 222 HCC tissue samples and the adjacent nontumorous tissues by qRT-PCR. In order to verify the reliability of the results, a combined set uniting the training set and the validation set was also analyzed. To establish the correlation between ELMO1-AS1 and clinicopathological features, the samples in each set were categorized into two groups (high or low) depending on the median values of ELMO1-AS1 expression in HCC tissues. For the training set, the expression level of ELMO1-AS1 was evidently down-regulated in HCC tissues compared with adjacent nontumorous tissues (P<0.001, Figure 1A). For the validation set, ELMO1-AS1 expression was also dramatically reduced in HCC tissues (P<0.001, Figure 1B). These results are consistent with the observation from the combined set (P<0.001, Figure 1C).
Figure 1.
Relative expression of ELMO1-AS1 was frequently downregulated in HCC tissues. ELMO1-AS1 expression, as measured by qRT-PCR, was downregulated in HCC tissues when compared with adjacent nontumorous tissues in the training set (A), validation set (B) and combined set (C). ***P<0.001.
Abbreviations: HCC, hepatocellular carcinoma; qRT-PCR, quantitative real-time polymerase chain reaction.
Correlations between aberrant expression of ELMO1-AS1 and clinical pathological features of HCC
Baseline characteristics of HCC patients are shown in Table 1. 110 patients and 112 patients were included in the training set and validation set, respectively. In the training set, the ELMO1-AS1 expression level was strikingly lower in HCC tissues with portal vein tumor thrombus (PVTT, P=0.039). In the validation set, the expression level of ELMO1-AS1 was obviously correlated with microvascular invasion (MVI, P=0.038).
Table 1.
Compare between ELMO1-AS1 expression and clinicopathological characteristics in HCC patients
| Training set | Validation set | Combined set | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinicopathologic Parameters | Total (n=110) | Low (n=55) | High (n=55) | P-value | Total (n=112) | Low (n=56) | High (n=56) | P-value | Total (n=222) | Low (n=111) | High (n=111) | P-value |
| Gender | 0.801 | 0.341 | 0.239 | |||||||||
| Female | 19 | 9 | 10 | 11 | 4 | 7 | 30 | 12 | 18 | |||
| Male | 91 | 46 | 45 | 101 | 52 | 49 | 192 | 99 | 93 | |||
| Age (years) | 0.057 | 0.280 | 1.000 | |||||||||
| <55 | 88 | 40 | 48 | 96 | 50 | 46 | 184 | 92 | 92 | |||
| ≥55 | 22 | 15 | 7 | 16 | 6 | 10 | 38 | 19 | 19 | |||
| Tumor number | 0.606 | 0.645 | 0.304 | |||||||||
| <3 | 92 | 47 | 45 | 88 | 43 | 45 | 180 | 93 | 87 | |||
| ≥3 | 18 | 8 | 10 | 24 | 13 | 11 | 42 | 18 | 21 | |||
| Tumor diameter | 0.189 | 0.357 | 0.113 | |||||||||
| <5 cm | 28 | 11 | 17 | 24 | 10 | 14 | 52 | 21 | 31 | |||
| ≥5 cm | 82 | 44 | 38 | 88 | 46 | 42 | 170 | 90 | 80 | |||
| AFP | 1.000 | 0.057 | 0.178 | |||||||||
| <400 ng/ml | 58 | 29 | 29 | 50 | 20 | 30 | 108 | 49 | 59 | |||
| ≥400 ng/ml | 52 | 26 | 26 | 62 | 36 | 26 | 114 | 62 | 52 | |||
| PVTT | 0.039 | 1.000 | 0.178 | |||||||||
| No | 97 | 45 | 52 | 94 | 47 | 47 | 191 | 92 | 99 | |||
| Yes | 13 | 10 | 3 | 18 | 9 | 9 | 31 | 19 | 12 | |||
| Microsatellite | 0.185 | 0.324 | 0.239 | |||||||||
| No | 100 | 48 | 52 | 92 | 44 | 48 | 192 | 93 | 99 | |||
| Yes | 10 | 7 | 3 | 20 | 12 | 8 | 30 | 18 | 12 | |||
| MVI | 0.340 | 0.038 | 0.060 | |||||||||
| No | 55 | 25 | 30 | 55 | 22 | 33 | 110 | 48 | 62 | |||
| Yes | 55 | 30 | 25 | 57 | 34 | 23 | 112 | 63 | 49 | |||
| TNM stage | 0.152 | 0.287 | 0.139 | |||||||||
| I+II | 75 | 34 | 41 | 72 | 34 | 38 | 147 | 69 | 78 | |||
| III+IV | 35 | 21 | 14 | 38 | 22 | 16 | 73 | 42 | 31 | |||
Abbreviations: AFP, alpha-fetoprotein; PVTT, portal vein tumor thrombus; MVI, microvascular invasion.
Association between ELMO1-AS1 expression and outcomes of patients with HCC
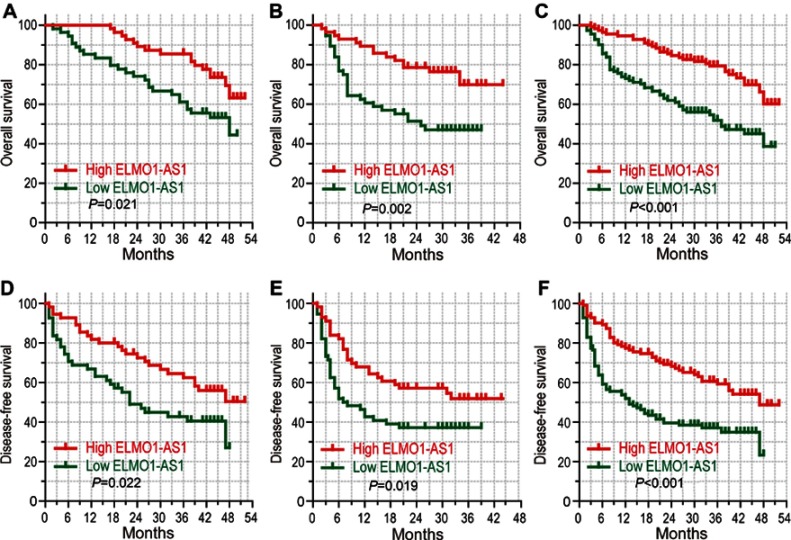
We compared the overall survival (OS) and disease-free survival (DFS) between different groups based on ELMO1-AS1 level. Kaplan-Meier survival analysis suggested that patients with low ELMO1-AS1 expression had a poor OS compared to those with high ELMO1-AS1 expression in the training set, validation set and combined set (46.4% vs 63.2%, P=0.021; 47% vs 69.8%, P=0.002 and 38.6% vs 60.1%, P<0.001, respectively; Figure 2A–C). Consistently, patients with low ELMO1-AS1 showed a poor DSF in all three sets (27.0% vs 50.4%, P=0.022; 37.1% vs 51.8%, P=0.019 and 23.2% vs 48.7%, P<0.001, respectively; Figure 2D–F). Taken together, low ELMO1-AS1 expression is correlated with pessimistic outcomes of HCC.
Figure 2.
High level of ELMO1-AS1 expression indicated better prognosis. Kaplan-Meier survival analysis comparing time to overall survival and disease-free survival of HCC patients with high or low ELMO1-AS1 expression in training cohort (n=110; A, D), validation cohort (n=112; B, E) and combined set (n=222; C, F).
Abbreviation: HCC, hepatocellular carcinoma.
Univariate and multivariate regression analyses of overall survival and disease-free survival
Univariate and multivariate analysis was implemented for OS (Table 2) and DFS (Table 3) to further investigate the clinical relevance of ELMO1-AS1. In the training set, high ELMO1-AS1 expression was disclosed to be an independent protective factor for OS and DFS (HR: 0.518; 95% CI: 0.277–0.968; P=0.039 for OS; HR: 0.557; 95% CI: 0.323–0.960; P=0.035 for DFS). In the validation set, high ELMO1-AS1 expression was an independent protective factor for OS (HR: 0.430; 95% CI: 0.225–0.824; P=0.011), but not DFS (HR: 0.616; 95% CI: 0.363–1.045; P=0.072). Moreover, when combined the two sets, ELMO1-AS1 expression was an independent prognostic factor for both OS and DFS (HR: 0.407; 95% CI: 0.257–0.643; P<0.001 for OS; HR: 0.476; 95% CI: 0.324–0.699; P<0.001 for DFS). HCC patients with high ELMO1-AS1 expression is likely to have a better prognosis.
Table 2.
Univariate and multivariate Cox regression analyses of overall survival
| Characteristic | Training set | Validation set | Combined set | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Univariate analyses | ||||||
| Gender | 1.071 (0.476–2.413) | 0.868 | 5.316 (0.732–38.629) | 0.099 | 1.747 (0.844–3.619) | 0.133 |
| Age | 0.761 (0.338–1.714) | 0.761 | 0.582 (0.208–1.626) | 0.302 | 0.650 (0.345–1.224) | 0.182 |
| Tumor number | 1.379 (0.658–2.892) | 0.395 | 2.291 (1.213–4.329) | 0.011 | 1.835 (1.137–2.963) | 0.013 |
| Tumor diameter | 1.943 (0.863–4.374) | 0.109 | 3.529 (1.259–9.889) | 0.016 | 2.582 (1.371–4.865) | 0.003 |
| AFP | 1.197 (0.652–2.199) | 0.562 | 2.176 (1.138–4.163) | 0.019 | 1.650 (1.069–2.545) | 0.024 |
| PVTT | 1.694 (0.749–3.832) | 0.205 | 1.776 (0.876–3.601) | 0.111 | 1.840 (1.081–3.134) | 0.025 |
| Microsatellite | 4.082 (1.911–8.720) | <0.001 | 2.357 (1.209–4.594) | 0.012 | 3.201 (1.938–5.287) | <0.001 |
| MVI | 1.349 (0.734–2.479) | 0.334 | 2.519 (1.335–4.754) | 0.004 | 1.831 (1.185–2.829) | 0.006 |
| TNM stage | 2.011 (1.092–3.702) | 0.025 | 2.340 (1.295–4.230) | 0.005 | 2.197 (1.438–3.359) | <0.001 |
| ELMO1-AS1 expression | 0.490 (0.262–0.914) | 0.025 | 0.386 (0.206–0.724) | 0.003 | 0.403 (0.258–0.629) | <0.001 |
| Multivariate analyses | ||||||
| Gender | ||||||
| Age | ||||||
| Tumor number | 1.514 (0.687–3.336) | 0.304 | 1.438 (0.822–2.514) | 0.203 | ||
| Tumor diameter | 2.381 (0.808–7.022) | 0.116 | 1.811 (0.932–3.520) | 0.080 | ||
| AFP | 1.293 (0.626–2.669) | 0.487 | 1.367 (0.868–2.152) | 0.178 | ||
| PVTT | 1.075 (0.604–1.914) | 0.805 | ||||
| Microsatellite | 2.794 (1.311–5.954) | 0.008 | 1.337 (0.663–2.912) | 0.465 | 2.013 (1.159–3.496) | 0.013 |
| MVI | 1.382 (0.663–2.880) | 0.387 | 1.271 (0.782–2.064) | 0.333 | ||
| TNM stage | 1.825 (0.987–3.374) | 0.055 | 1.486 (0.764–2.891) | 0.243 | 2.359 (0.828–2.233) | 0.225 |
| ELMO1-AS1 expression | 0.518 (0.277–0.968) | 0.039 | 0.430 (0.225–0.824) | 0.011 | 0.407 (0.257–0.643) | <0.001 |
Abbreviations: AFP, alpha-fetoprotein; PVTT, portal vein tumor thrombus; MVI, microvascular invasion.
Table 3.
Univariate and multivariate Cox regression analyses of disease-free survival
| Characteristic | Training set | Validation set | Combined set | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Univariate analyses | ||||||
| Gender | 1.110 (0.543–2.267) | 0.775 | 8.868 (1.228–64.043) | 0.030 | 1.982 (1.036–3.792) | 0.039 |
| Age | 1.031 (0.544–1.953) | 0.925 | 0.830 (0.394–1.746) | 0.624 | 0.921 (0.569–1.490) | 0.736 |
| Tumor number | 1.501 (0.774–2.910) | 0.229 | 1.836 (1.058–3.185) | 0.031 | 1.713 (1.124–2.609) | 0.012 |
| Tumor diameter | 1.745 (0.901–3.378) | 0.099 | 1.637 (0.830–3.230) | 0.155 | 1.732 (1.079–2.782) | 0.023 |
| AFP | 1.127 (0.667–1.905) | 0.655 | 1.798 (1.064–3.037) | 0.028 | 1.461 (1.013–2.106) | 0.042 |
| PVTT | 2.162 (1.052–4.443) | 0.036 | 1.656 (0.894–3.065) | 0.109 | 1.905 (1.194–3.039) | 0.007 |
| Microsatellite | 1.351 (0.578–3.162) | 0.488 | 2.030 (1.143–3.604) | 0.016 | 1.886 (1.182–3.009) | 0.008 |
| MVI | 1.709 (1.005–2.906) | 0.048 | 1.872 (1.119–3.133) | 0.017 | 1.773 (1.226–2.565) | 0.002 |
| TNM stage | 1.296 (0.744–2.259) | 0.360 | 1.933 (1.160–3.220) | 0.011 | 1.1623 (1.117–2.356) | 0.011 |
| ELMO1-AS1 expression | 0.546 (0.320–0.930) | 0.026 | 0.556 (0.334–0.926) | 0.024 | 0.469 (0.323–0.681) | <0.001 |
| Multivariate analyses | ||||||
| Gender | 7.808 (1.066–57.205) | 0.043 | 1.762 (0.916–3.389) | 0.090 | ||
| Age | ||||||
| Tumor number | 1.195 (0.609–2.346) | 0.605 | 1.605 (0.968–2.661) | 0.067 | ||
| Tumor diameter | 1.462 (0.884–2.416) | 0.139 | ||||
| AFP | 1.448 (0.798–2.627) | 0.224 | 1.237 (0.835–1.832) | 0.290 | ||
| PVTT | 1.574 (0.741–3.345) | 0.238 | 1.246 (0.745–2.084) | 0.403 | ||
| Microsatellite | 1.369 (0.703–2.665) | 0.356 | 1.067 (0.623–1.827) | 0.812 | ||
| MVI | 1.642 (0.947–2.849) | 0.077 | 1.072 (0.574–2.002) | 0.826 | 1.466 (0.973–2.210) | 0.067 |
| TNM stage | 1.535 (0.863–2.730) | 0.145 | 0.999 (0.642–1.556) | 0.998 | ||
| ELMO1-AS1 expression | 0.557 (0.323–0.960) | 0.035 | 0.616 (0.363–1.045) | 0.072 | 0.476 (0.324–0.699) | <0.001 |
Abbreviations: AFP, alpha-fetoprotein; PVTT, portal vein tumor thrombus; MVI, microvascular invasion.
Overexpression of ELMO1-AS1 inhibited HCC cell proliferation
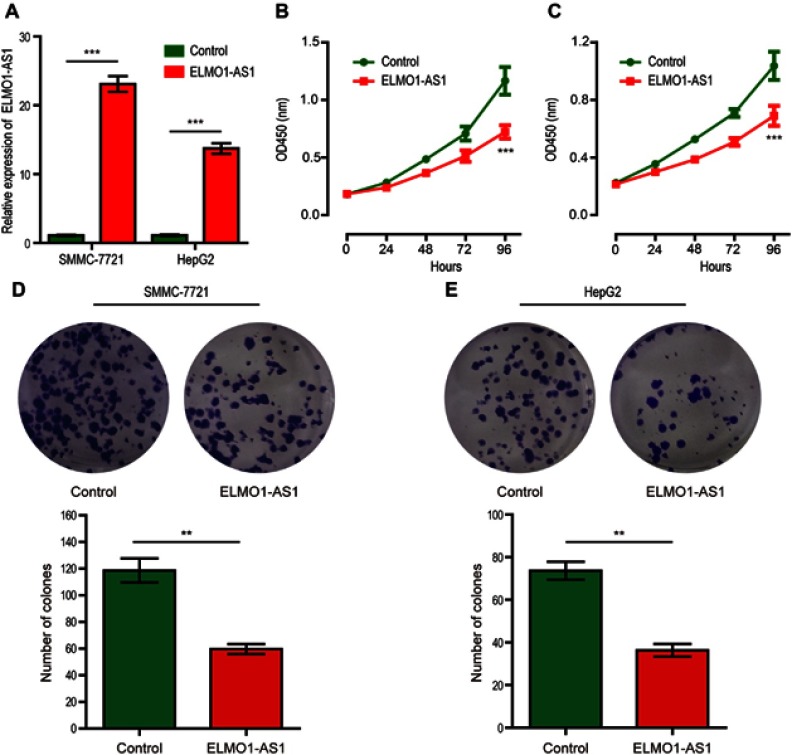
SMMC-7721 and HepG2 cells were infected with lentivirus containing the plasmid for stable expression of ELMO1-AS1. After being selected by puromycin, the transfection efficiency of the cells was assessed by qRT-PCR and results showed that the relative expression level of ELMO1-AS1 transfected with LV-pCDH-ELMO1-AS1 is significantly higher compared with LV-pCDH-empty control (Figure 3A). Then CCK-8 assay revealed that upregulated ELMO1-AS1 expression markedly inhibited the growth of both SMMC-7721 and HepG2 cells (Figure 3B and C). Similarly, colony formation assay disclosed that the number of HCC cell colonies were distinctly decreased by overexpression of ELMO1-AS in SMMC-7721 and HepG2 cells (Figure 3D and E).
Figure 3.
Overexpression of ELMO1-AS1 inhibits HCC cells proliferation. Expression of ELMO1-AS1 in SMMC-7721 and HepG2 cells was significantly upregulated after ELMO1-AS1 lentiviral transfection compared to control (A). The CCK-8 assay was performed to determine the proliferation of ELMO1-AS1 or control-transfected SMMC-7721 (B) and HepG2 (C) cells. Colony formation assay was performed to determine the proliferation of ELMO1-AS1 or control-transfected SMMC-7721 (D) and HepG2 (E) cells. ** and *** represent P<0.01 and P<0.001, respectively.
Abbreviations: CCK-8, cell counting Kit-8; OD, optical density; HCC, hepatocellular carcinoma.
Overexpression of ELMO1-AS1 inhibited HCC cell migration and invasion
To investigate whether ELMO1-AS1 is involved in HCC cells migration and invasion, wound healing assay and transwell assay were performed. Cell wound healing assay showed that overexpression of ELMO1-AS1 in HCC cells led to slow wound healing rate (Figure 4A and B). Transwell assays without or with Matrigel both found that HCC cells overexpressed with ELMO1-AS1 had less active migration and invasion than control cells (Figure 4C–F). These results indicated that ELMO1-AS1 had an important role in inhibiting HCC cells migration and invasion in vitro.
Figure 4.
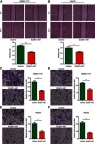
Overexpression of ELMO1-AS1 inhibited HCC cell migration and invasion in vitro. Wound healing assay showed that the migratory potential of SMMC-7721 (A) and HepG2 (B) cells were significantly reduced in the ELMO1-AS1-transfected group than in the control-transfected group. Transwell migration assay showed that the number of migrated SMMC-7721 (C) and HepG2 (E) cells was significantly lower in the ELMO1-AS1-transfected group than in the control-transfected group. Transwell invasion assay showed that the number of invaded SMMC-7721 (E) and HepG2 (F) cells was significantly lower in the ELMO1-AS1-transfected group than in the control-transfected group. ** and *** represent P<0.01 and P<0.001, respectively.
Abbreviation: HCC, hepatocellular carcinoma.
Prediction of ELMO1-AS1
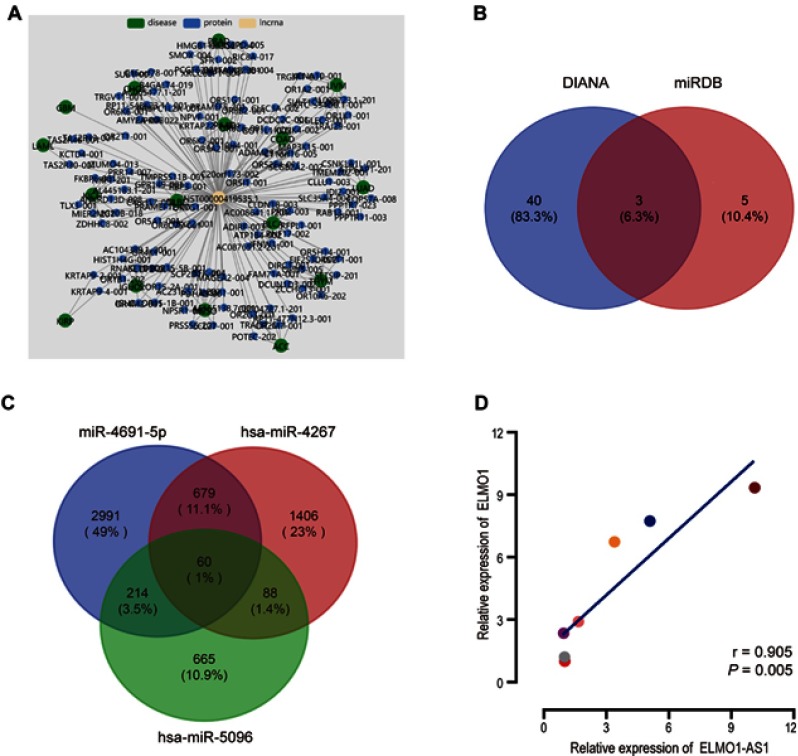
A co-expression network using Lnc2Catlas15 indicated that ENST00000419535.1, the transcript of ELMO1-AS1 is implicated with various diseases and physiology processes (Figure 5A). Further research by lncLocator16 discovered that cytoplasm is the dominant subcellular location of ELMO1-AS1, suggesting that ELMO1-AS1 may target miRNAs to modulate genes expression and biological functions as a competitive endogenous RNA (ceRNA). The potential targets of ELMO1-AS1 were identified based on the intersection of DIANA tools-LncBase v.217 and miRDB databases.18 Three miRNAs, hsa-miR-4267, hsa-miR-5096 and hsa-miR-4691-5p, appeared in both sets (Figure 5B). We searched these three miRNAs in TargetScan19 to find the feasible mRNA targets, based on total context++ score and a cutoff value less than −0.10. Intriguingly, 60 identical mRNAs were found in the intersection of miRNAs targets, including engulfment and cell motility 1 (ELMO1), the neighboring intrachromosomal coding gene of ELMO1-AS1 (Figure 5C). The relationship between ELMO1-AS1 and ELMO1 was assessed in HCC tissues by qRT-PCR and showed a positive correlation (r =0.905, P=0.005, Figure 5D).
Figure 5.
ELMO1 could be one of the potential targets of ELMO1-AS1. The co-expression network showed ELMO1-AS1 associated with multiple diseases and proteins (A). Three differently expressed target miRNAs of ELMO1-AS1 were integrated from DIANA and miRDB databases (B). ELMO1 was one of the intersections of ELMO1-AS1 associated miRNAs targets (C). Pearson analysis of the relationship between ELMO1-AS1 and ELMO1 expression (D).
Discussion
For the past few years, surgical resection, transplantation, radiotherapy, transarterial chemoembolization and molecular targeted therapy have become the main means of treatment for HCC patients, but the outcome remains pessimistic.20 It is an enormous clinical challenge to investigate the internal mechanisms of HCC development and excavate novel prognostic indicators.21 Mounting evidence indicates a vital role for long non-coding RNAs in the development and progression of human tumors. Localizing to chromatin, interacting with protein, modulating RNA metabolism and driving cancer phenotypes are some functions of lncRNAs but not all.22 Moreover, It also has been used for prediction of tumor prognosis among multiple tumors. Sadek et al23 suggested in their research that lncRNAs, including ecCEBPA and UCA1, may be helpful as prognostic of HCC. The results of Fang et al24 showed overexpression of lncRNA DGCR5 inhibits bladder cancer and correlates with better prognosis via transcriptionally facilitating P21 expression. As a matter of fact, the value of lncRNAs on estimating the outcome of HCC patients could provide an accurate direction in the postoperative adjuvant therapy,25,26 and there are few reports on the relationship between ELMO1-AS1 and HCC.
We investigated the expression of ELMO1-AS1 in two independent sets collected at different times with 222 pairs of tumor tissues and adjacent nontumorous tissues from HCC patients. Evidently, the expression of ELMO1-AS1 is significantly lower in tumor tissues compared to adjacent nontumorous tissues. Statistical analyses suggest that high ELMO1-AS1 expression is closely correlated with a longer overall survival and a longer disease-free survival of HCC patients. Univariate and multivariate analyses of clinicopathological features also show that ELMO1-AS1 expression is an independent protective factor for OS and DFS in patients with HCC, implying that it could be employed as a prognostic biomarker for HCC.
However, the biological role of ELMO1-AS1 in HCC cells remained poorly clear. In this work, we found that stable upregulated ELMO1-AS1 expression in HCC cells influenced their biological behavior. Cell proliferation assay and colony formation assay showed that overexpression of ELMO1-AS1 inhibited HCC cells growth. On the other hand, the results of wound healing assay and transwell assays indicated that ectopic expression of ELMO1-AS1 suppressed the migration and invasion of HCC cells in vitro. These results suggested that ELMO1-AS1 played an important regulatory role of in HCC.
Moreover, bioinformatics analysis and molecular biology experiment were taken to explore the function and probable molecular mechanism. The co-expression network showed a huge three-dimensional space network among ELMO1-AS1, diseases and proteins, which indirectly imply the importance of ELMO1-AS1 in biology. Accumulating studies have demonstrated that lncRNAs regulate cell functions through interacting with 3′-UTR of miRNAs in the competitive endogenous RNAs (ceRNA) regulatory networks.27 For example, Huang et al28 found that lncRNA IGF2-AS affects gastric adenocarcinoma development by SHOX2 via sponging miR-503. Sun et al29 suggested that high expression of lncRNA HOTTIP is associated with small cell lung cancer progression through the ceRNA network HOTTIP/miR-574-5p/EZH1. Therefore, we hypothesized that ELMO1-AS1 may also serve as a ceRNA. DIANA tools-LncBase v.2 and miRDB databases were used to predict the potential binding sites between ELMO1-AS1 and miRNAs, indicated that ELMO1-AS1 might target one or more of 43 and 8 miRNAs, respectively. We took the intersection of these two miRNA groups and three miRNAs were identified. TargetScan database was performed to research the connections with ELMO1-AS1. ELMO1, the neighboring intrachromosomal coding gene of ELMO1-AS1, which could function in apoptosis and cell migration,30 was appeared to be a possible target of miRNAs as described, reminding ELMO1 might be one of the potential target genes of ELMO1-AS1. Moreover, the strong positive correlation between ELMO1-AS1 and ELMO1 shall support this suspect using qRT-PCR array. Multiple studies have shown that neighboring intrachromosomal coding genes can be regulated by lncRNA transcript.31,32 For instance, Li et al disclosed that lncRNA ZEB1-AS1 is bound up with ZEB1 and positively mediates the ZEB1 expression.33 Taken together, ELMO1-AS1 shall act as a tumor suppressor in HCC by targeting its neighboring intrachromosomal coding gene ELMO1 thus affect HCC biological behavior. However, the exact mechanism remains to be further studied.
The ELMO1-AS1, located at chromosome 7p14.2, is identified as Homo sapiens engulfment and cell motility 1 antisense RNA 1. The previous microarray profiling experiments, which detecting the differential expression of lncRNAs between diverse phenotypes of macrophage, showed that ELMO1-AS1 was significantly upregulated in M1 macrophages compared to M2 macrophages. Many studies have shown that tumor-associated macrophages (TAMs), mainly M2 macrophages, are recruited to the tumor sites whereby to promote tumor angiogenesis and tumor cell proliferation.34–36 They also had a close relationship with poor prognosis of patients with different tumors.37,38 Consistently, our results showed that macrophage-associated lncRNA ELMO1-AS1 was upregulated in adjacent nontumorous tissues compared to tumor tissues and high expression of ELMO1-AS1 could inhibit tumor cell progression and predict favorable prognosis in HCC patients. ELMO1, a possible target of ELMO1-AS1 as demonstrated by us before, have been shown to mediate bacterial internalization and intestinal inflammation in macrophages by Sarkar et al39 Shi et al40 disclosed that CC chemokine ligand 18 (CCL18) predominantly secreted by TAMs, could promote migration and invasion of non-small cell lung cancer cells through activating ELMO1-integrin β1 signaling. Those results further suggest that ELMO1-AS1 could affect the occur and development progress of HCC through ELMO1. Moreover, the oncolytic adenovirus expressing tumor suppressor gene, such as SNORD44, GAS5 or TSLC1, could enhance the anti‐tumor effect through targeting some crucial signaling pathways.41,42 Genome editing using clustered regularly interspaced short palindromic repeats-associated endonuclease 9 (CRISPR-Cas9) is an efficient approach to not only delete or block lncRNAs expression by the targeted interruption, but also upregulate lncRNA expression by targeting transcriptional activator complexes or inserting a promoter.32 These indicated to us that overexpression of ELMO1-AS1 by oncolytic adenovirus or CRISPR-Cas9 provides a hopeful method for HCC targeted therapy. It was found for the first time that macrophage-associated lncRNA ELMO1-AS1 could regulate behaviors of HCC cells and predict the prognosis of HCC patients. However, further studies are required. First, in the validation set, high ELMO1-AS1 expression was an independent protective factor for DFS only under univariate analysis model, but failed to reach significance under multivariate analysis model. That might be explained by patient heterogeneity, because significance was found in the training set and combined set. Second, the exact mechanism of how ELMO1-AS1 regulate its neighboring intrachromosomal coding gene ELMO1 by targeting miRNAs need to be further studied. Third, in vivo experiments are needed to confirm some conclusions drawn from in vitro experiments.
Conclusion
Our results showed that macrophage-associated lncRNA ELMO1-AS1 was significantly downregulated in HCC tissues and high expression of ELMO1-AS1 associated with optimistic outcome in HCC patients. ELMO1-AS1 could inhibit HCC cells proliferation, migration and invasion and ELMO1 may be a target of ELMO1-AS1. ELMO1-AS1 could be a potential prognostic biomarker and therapeutic target for HCC.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81860512), the Guangxi Natural Science Foundation (2016GXNSFBA380194 and 2018GXNSFAA138006), the Guangxi Science and Technology Project (AB18126032), and the Innovation Project of Guangxi Graduate Education (YCSW2017109).
Ethical approval and informed consent
All patients provided written informed consent, and that this was conducted in accordance with the Declaration of Helsinki. This study was approved by the Ethics Committee of the Affiliated Tumor Hospital of Guangxi Medical University.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2.Yoon SK, Chun HG. Status of hepatocellular carcinoma in South Korea. Chin Clin Oncol. 2013;2(4):39. [DOI] [PubMed] [Google Scholar]
- 3.Chow PK, Choo SP, Ng DC, et al. National Cancer Centre Singapore consensus guidelines for hepatocellular carcinoma. Liver Cancer. 2016;5(2):97–106. doi: 10.1159/000367759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Q, Qin SK. Features and treatment options of Chinese hepatocellular carcinoma. Chin Clin Oncol. 2013;2(4):38. [DOI] [PubMed] [Google Scholar]
- 5.Severi T, van Malenstein H, Verslype C, van Pelt JF. Tumor initiation and progression in hepatocellular carcinoma: risk factors, classification, and therapeutic targets. Acta Pharmacol Sin. 2010;31(11):1409–1420. doi: 10.1038/aps.2010.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15(10):599–616. doi: 10.1038/s41571-018-0073-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charriere B, Muscari F, Maulat C, et al. Outcomes of patients with hepatocellular carcinoma are determined in multidisciplinary team meetings. J Surg Oncol. 2017;115(3):330–336. doi: 10.1002/jso.24500 [DOI] [PubMed] [Google Scholar]
- 8.Ozakyol A. Global epidemiology of hepatocellular carcinoma (HCC epidemiology). J Gastrointest Cancer. 2017;48:238–240. doi: 10.1007/s12029-017-9959-0 [DOI] [PubMed] [Google Scholar]
- 9.Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097–1109. doi: 10.1016/j.bbagrm.2014.08.012 [DOI] [PubMed] [Google Scholar]
- 10.Bassett AR, Akhtar A, Barlow DP, et al. Considerations when investigating lncRNA function in vivo. Elife. 2014;3:e03058. doi: 10.7554/eLife.03058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang HM, Lu JH, Chen WY, Gu AQ. Upregulated lncRNA-UCA1 contributes to progression of lung cancer and is closely related to clinical diagnosis as a predictive biomarker in plasma. Int J Clin Exp Med. 2015;8(7):11824–11830. [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Yang B, Zhang M, et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell. 2018;33(4):706–720 e709. doi: 10.1016/j.ccell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol Med. 2018;24(3):257–277. doi: 10.1016/j.molmed.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo HL, Chen J, Luo T, et al. Downregulation of macrophage-derived T-UCR uc.306 Associates with poor prognosis in hepatocellular carcinoma. Cell Physiol Biochem. 2017;42(4):1526–1539. doi: 10.1159/000479269 [DOI] [PubMed] [Google Scholar]
- 15.Ren C, An G, Zhao C, Ouyang Z, Bo X, Shu W. Lnc2Catlas: an atlas of long noncoding RNAs associated with risk of cancers. Sci Rep. 2018;8(1):1909. doi: 10.1038/s41598-018-20232-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao Z, Pan X, Yang Y, Huang Y, Shen HB. The lncLocator: a subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier. Bioinformatics. 2018;34(13):2185–2194. doi: 10.1093/bioinformatics/bty085 [DOI] [PubMed] [Google Scholar]
- 17.Paraskevopoulou MD, Vlachos IS, Karagkouni D, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44(D1):D231–D238. doi: 10.1093/nar/gkv1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu W, Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019;20(1):18. doi: 10.1186/s13059-019-1629-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stotz M, Gerger A, Haybaeck J, Kiesslich T, Bullock MD, Pichler M. Molecular targeted therapies in hepatocellular carcinoma: past, present and future. Anticancer Res. 2015;35(11):5737–5744. [PubMed] [Google Scholar]
- 21.El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142(6):1264–1273 e1261. doi: 10.1053/j.gastro.2011.12.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadek KM, Lebda MA, Nasr NE, Nasr SM, El-Sayed Y. Role of lncRNAs as prognostic markers of hepatic cancer and potential therapeutic targeting by S-adenosylmethionine via inhibiting PI3K/Akt signaling pathways. Environ Sci Pollut Res Int. 2018;25(20):20057–20070. doi: 10.1007/s11356-018-2179-8 [DOI] [PubMed] [Google Scholar]
- 24.Fang C, He W, Xu T, Dai J, Xu L, Sun F. Upregulation of lncRNA DGCR5 correlates with better prognosis and inhibits bladder cancer progression via transcriptionally facilitating P21 expression. J Cell Physiol. 2019;234(5):6254–6262. doi: 10.1002/jcp.27356 [DOI] [PubMed] [Google Scholar]
- 25.Hua L, Wang CY, Yao KH, Chen JT, Zhang JJ, Ma WL. High expression of long non-coding RNA ANRIL is associated with poor prognosis in hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8(3):3076–3082. [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X, Xie X, Xiao YF, et al. The emergence of long non-coding RNAs in the tumorigenesis of hepatocellular carcinoma. Cancer Lett. 2015;360(2):119–124. doi: 10.1016/j.canlet.2015.02.035 [DOI] [PubMed] [Google Scholar]
- 27.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang J, Chen YX, Zhang B. IGF2-AS affects the prognosis and metastasis of gastric adenocarcinoma via acting as a ceRNA of miR-503 to regulate SHOX2. Gastric Cancer. 2019. doi: 10.1007/s10120-019-00976-2 [DOI] [PubMed] [Google Scholar]
- 29.Sun Y, Zhou Y, Bai Y, et al. A long non-coding RNA HOTTIP expression is associated with disease progression and predicts outcome in small cell lung cancer patients. Mol Cancer. 2017;16(1):162. doi: 10.1186/s12943-017-0729-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han HB, Gu J, Ji DB, et al. PBX3 promotes migration and invasion of colorectal cancer cells via activation of MAPK/ERK signaling pathway. World J Gastroenterol. 2014;20(48):18260–18270. doi: 10.3748/wjg.v20.i48.18260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sahu A, Singhal U, Chinnaiyan AM. Long noncoding RNAs in cancer: from function to translation. Trends Cancer. 2015;1(2):93–109. doi: 10.1016/j.trecan.2015.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi: 10.1038/nm.3981 [DOI] [PubMed] [Google Scholar]
- 33.Li T, Xie J, Shen C, et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35(12):1575–1584. doi: 10.1038/onc.2015.223 [DOI] [PubMed] [Google Scholar]
- 34.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21(3):309–322. doi: 10.1016/j.ccr.2012.02.022 [DOI] [PubMed] [Google Scholar]
- 35.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141(1):39–51. doi: 10.1016/j.cell.2010.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun L, Chen B, Jiang R, Li J, Wang B. Resveratrol inhibits lung cancer growth by suppressing M2-like polarization of tumor associated macrophages. Cell Immunol. 2017;311:86–93. doi: 10.1016/j.cellimm.2016.11.002 [DOI] [PubMed] [Google Scholar]
- 37.Yuan A, Hsiao YJ, Chen HY, et al. Opposite effects of M1 and M2 macrophage subtypes on lung cancer progression. Sci Rep. 2015;5:14273. doi: 10.1038/srep14273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeuchi H, Tanaka M, Tanaka A, Tsunemi A, Yamamoto H. Predominance of M2-polarized macrophages in bladder cancer affects angiogenesis, tumor grade and invasiveness. Oncol Lett. 2016;11(5):3403–3408. doi: 10.3892/ol.2016.4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarkar A, Tindle C, Pranadinata RF, et al. ELMO1 regulates autophagy induction and bacterial clearance during enteric infection. J Infect Dis. 2017;216(12):1655–1666. doi: 10.1093/infdis/jix528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi L, Zhang B, Sun X, et al. CC chemokine ligand 18(CCL18) promotes migration and invasion of lung cancer cells by binding to Nir1 through Nir1-ELMO1/DOC180 signaling pathway. Mol Carcinog. 2016;55(12):2051–2062. doi: 10.1002/mc.22450 [DOI] [PubMed] [Google Scholar]
- 41.Yuan S, Wu Y, Wang Y, Chen J, Chu L. An oncolytic adenovirus expressing SNORD44 and GAS5 exhibits antitumor effect in colorectal cancer cells. Hum Gene Ther. 2017;28(8):690–700. doi: 10.1089/hum.2017.041 [DOI] [PubMed] [Google Scholar]
- 42.Zhang J, Lai W, Li Q, et al. A novel oncolytic adenovirus targeting Wnt signaling effectively inhibits cancer-stem like cell growth via metastasis, apoptosis and autophagy in HCC models. Biochem Biophys Res Commun. 2017;491(2):469–477. doi: 10.1016/j.bbrc.2017.07.041 [DOI] [PubMed] [Google Scholar]