Abstract
Purpose
Gefitinib targeting of the epidermal growth factor receptor (EGFR) has shown limited activity in clinical trials of head and neck squamous cell carcinoma (HNSCC). To investigate the underlying molecular mechanism, the proteomic signatures and responses of EGFR and downstream signals have been studied in a panel of HNSCC cell lines and tumor specimens pre- and post-gefitinib treatment.
Experimental Design
The IC50 of gefitinib for HNSCC cell lines were determined using 3-(4,5-dmethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide proliferation assay. The effects of gefitinib on activation of EGFR and downstream signaling molecules were determined by Western blot, ELISA, and reverse-phase protein microarray (RPMA). The biomarkers involved in the signaling pathways were examined in HNSCC tumor specimens from patients in a phase I gefitinib trial.
Results
In vitro, gefitinib inhibited cell proliferation with differing IC50, and suppressed activation of EGFR and downstream signaling molecules protein kinase B (AKT), extracellular signal-regulated kinase 1/2, signal transducer and activator of transcription 3 (STAT3), and nuclear factor κB. The drug sensitivity was statistically correlated with activation of phosphorylated AKT (p-AKT) and phosphorylated STAT3 (p-STAT3) detected by ELISA, and consistent with results measured by RPMA. In patient samples, a broad suppression of activation of EGFR and downstream signaling molecules was observed in a molecular responder patient, in contrast to a lack of inhibition or increased activation of biomarkers in different pathways in nonresponder patients.
Conclusions
Gefitinib sensitivity is correlated with p-AKTand p-STAT3 activation in HNSCC cell lines and tumor specimens. p-AKTand p-STAT3 could serve as potentially useful biomarkers and drug targets for further development of novel therapeutic agents for HNSCC.
Increased understanding of molecular alterations in head and neck squamous cell carcinoma (HNSCC) and other cancers has ushered in efforts to develop molecularly targeted therapies. Among these alterations, overexpression of the epidermal growth factor receptor (EGFR) has been identified in many cancers, including 80% to 100% of HNSCC (1–3), where it has also been implicated in a more aggressive phenotype, increased resistance to treatment, and poorer clinical outcome (4, 5). These observations have led to interest in the development of molecularly targeted small-molecule inhibitors of EGFR for the treatment of HNSCC. Gefitinib (Iressa), an EGFR tyrosine kinase inhibitor, has been tested in clinical trials in solid tumors, including HNSCC, as a single agent, or in the combination with other chemotherapies or radiation, but has shown limited clinical efficacy with response rates of 10% to 15% (6–9). Molecular biomarkers that could identify patients responsive to gefitinib or other EGFR inhibitors would be useful in the selection of patients for therapy.
The EGFR is a membrane-bound glycoprotein with an extracellular cysteine-rich ligand-binding domain linked by a short single transmembrane sequence to an intracellular tyrosine kinase and carboxyl-terminal scaffolding domains (1). EGFR (ErbB1) is a member of the ErbB receptor family of tyrosine kinases, which also includes ErbB2 (HER-2/neu), ErbB3, and ErbB4 (1, 4). The activation of EGFR family members may be either ligand-dependent or independent as a result of mutation or overexpression-induced homodimerization, or heterodimerization with other ErbB receptor family members (1). The binding by ligands such as epidermal growth factor (EGF), transforming growth factor α, amphiregulin, and heparin binding-EGF results in autophosphorylation of multiple tyrosine residues at the receptor carboxyl terminus of EGFR, which acts as scaffolding for SRC and other adaptor proteins that interact with the receptor and transduce mitogenic signals mediated by downstream molecular cascades (1, 4, 5). Ligand-independent activation of EGFR in HNSCC has been shown to result from a truncation mutation, EGFR variant III (EGFRvIII; ref. 10). Ligand-dependent and independent activation play a vital role in promoting cancer cell growth, cell migration, aberrant metabolism and differentiation, and enhanced cell survival (1, 4).
A number of pharmacologic agents that block EGFR signaling have been developed and evaluated in clinical trials. Gefitinib is the first of such molecules with oral bioavailability that inhibits EGFR activation by competing with ATP binding at the catalytic tyrosine kinase domain of the EGFR. Gefitinib was originally approved for the third-line treatment of non–small cell lung cancer, but has yielded limited results in several clinical trials in a variety of solid tumors (6–9). Gefitinib has been studied as monotherapy for patients with recurrent HNSCC (11), and in combination with paclitaxel (Taxol) and radiation in patients with local-regionally advanced HNSCC by our group.5 Consistent with the 10% to 15% response rates observed from larger monotherapy trials (11), only 1 of 7 patients whose tumors were studied showed molecular responses to single-agent gefitinib as identified by immunohistochemistry. A lack of understanding of downstream molecular mechanisms activated by EGFR and suitable biomarkers for selection of molecular responders has been a major hurdle for clinical trials and the rational selection of patients for treatment with EGFR inhibitors.
Molecular heterogeneity in subsets of tumors and/or patient populations has been shown to contribute to variable responses of non–small cell lung cancer to gefitinib and other EGFR inhibitors. It was previously shown that there is a beneficial clinical effect of gefitinib in Asian populations with non–small cell lung cancer, who had specific EGFR gene mutations and amplifications (12–16). However, molecular analysis of HNSCC tumor samples has not revealed the same spectrum of amplifications or mutations (17–19). Recently, an in-frame deletion in exon 19 of the EGFR gene was reported in 7.3% of HNSCC in the Korean population (20), and a deletion of exons 2 to 7 resulting in a truncated extracellular domain and constitutive tyrosine kinase activation, designated as EGFRvIII, was found in 42% of HNSCC (10). Because of the different genetic alterations of EGFR observed in HNSCC, the molecular mechanism(s) determining the therapeutic sensitivity to gefitinib and other tyrosine kinase inhibitors may not be the same as observed in lung cancer, and therefore need to be more thoroughly investigated.
In this study, we explored the effects of gefitinib on activation of the EGFR and downstream signal pathways, and determined if there was a relationship between these biomarkers and the anticancer effects in HNSCC cell lines and patients’ specimens. In addition, we cross-validated the biomarkers identified by several methods, including Western blot analysis, ELISA, and reverse-phase protein microarray (RPMA), a newly developed proteomic platform that is capable of measuring numerous specific phosphorylated proteins that are involved in important signaling pathways from a small tissue biopsy or a few thousand cells (21, 22). We found that gefitinib inhibited cell proliferation in vitro with different IC50, and suppressed activation of EGFR and downstream activation of protein kinase B (AKT), extracellular signal-regulated kinase 1/2 (ERK1/2), signal transducer and activator of transcription 3 (STAT3), and nuclear factor κB (NF-κB). Our observations indicate that these proteomic signatures to EGFR inhibitor could be useful in the development of more accurate molecular diagnosis, selection of therapy, and determinations of HNSCC prognosis. Cross-validation of results from in vitro and in vivo using RPMA by Western blot, quantitative ELISA, and immunohistochemistry provides an essential step for future application of these technologies in clinical trials.
Materials and Methods
Cell lines and reagents
HNSCC cell lines UMSCC-6, -9, -11A, and -11B were kind gifts from Dr. T.E. Carey (University of Michigan, Ann Arbor, MI), and their characteristics were previously described (23). The UMSCC cell lines were cultured in MEM (Invitrogen) with 10% fetal bovine serum and penicillin/streptomycin, and maintained in a humidified incubator at 37°C with 5% CO2. Gefitinib was provided for studies by AstraZeneca, under a Materials Cooperative Research and Development Agreement and Clinical Trials Agreement with the National Cancer Institute (NCI), and was dissolved in DMSO when reconstituted. Recombinant human EGF was purchased from R&D Systems.
3-(4,5-dmethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide cellular proliferation assay and calculation of IC50
The cell lines UMSCC-6, -9, -11A, and -11B cells were plated in triplicate at 5 × 103 cells per well onto 96-well plates. The cells were cultured overnight and then exposed to gefitinib at varying concentrations. Cell proliferation was measured using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltretrazolium bromide (MTT) Cell Proliferation Kit (Roche Diagnostics). The optical density was measured by a microplate reader at a wavelength of 570 nm. The IC50 of the drug was calculated using MTT data at day 5 comparing treatment samples versus control samples. The percentage changes of MTT values between controls and treated samples were calculated individually for each dose and cell line using the following equation: (control-treated sample/control) × 100. The percent change was plotted on a graph against drug dose. A best fit logarithmic curve was plotted and the IC50 was determined as the 50% inhibition point.
Sequencing and reverse transcription-PCR analysis of mutations in EGFR coding regions
RNA was isolated from UMSCC cells, and cDNA was reverse-transcribed and sequenced for coding regions of EGFR exon 18 to 21 bidirectionally by SeqWright Inc., according to the hot spots identified in the literature (24). The EGFRvIII deletion mutation was analyzed by reverse transcription-PCR, using the primers described by Dr. Grandis’ group (10).
Whole cell extraction and Western blot analysis
Cultured UMSCC cells at 75% confluence were treated with 1 or 10 μmol/L gefitinib for 4 h and then 10 ng/mL recombinant EGF (R&D systems) were added into half of the dishes for each treatment condition for 1 h. Cultured cells were washed with ice-cold PBS, lysed, and harvested with ice-cold cell extraction buffer (Invitrogen). The whole cell lysates were incubated on ice on a rocking platform for 30 min and vortexed every 10 min and then spun at 13,000 g at 4°C for 10 min. The supernatants were collected and the protein concentrations were quantified by BCA protein assay kit (Pierce).
For Western blot analysis, whole cell lysate was mixed with an SDS protein gel loading solution (Quality Biological, Inc.) and heated at 85°C for 2 min. The samples were loaded onto Tris-glycine precast gels (Invitrogen), and electrophoresis was completed at 125V for 105 min. The Invitrogen iBlot apparatus was used to complete the transfer (Invitrogen). The membranes were preblocked in a preblocking solution composed of 1% Tween-20 in TBS mixed with either 7.5% bovine serum albumin or 5% nonfat dry milk. Primary antibodies were diluted in either 5% nonfat powdered milk or 5% bovine serum albumin prepared from Tween-20 in TBS for: phospho-EGFR (Tyr845) and phospho-EGFR (Tyr1068), 1:1000 (Cell Signaling Technology); and phospho-EGFR (Tyr1173), phospho-EGFR (Tyr1148), and total EGFR, 1:1000 (Invitrogen). Each blot was incubated with Pierce Super Signal West Pico Substrate (Pierce) and exposed to Kodak X-OMAT film.
ELISA assays
ELISA assays that were used to quantify total EGFR, phospho-EGFR (Tyr 1173), total STAT3, phospho-STAT3 (Tyr 705), total ERK1/2, phospho-ERK1/2 (Tyr185/187), total AKT, and phosphor-AKT (Ser 473), were done according to the manufacturer’s protocol (Invitrogen). The ELISA assays used to detect phospho-EGFR Tyr 1068, phospho-EGFR Tyr 845, and NF-κB phospho-p65 (Ser 536) were done according to the manufacturer’s protocol (Cell Signaling Technologies). The whole cell lysates were isolated as described for the Western blot analysis for all ELISA assays, except that nuclear extracts were used for testing NF-κB p-p65 (Ser 536; Active Motif). Samples were measured for each condition against a respective standard provided by the kits. The optical density was measured by a microplate reader at a wavelength of 450 nm, and the data were calculated and presented as mean plus SD from triplicate assays.
Real time reverse transcription-PCR
RNA isolation and cDNA synthesis were done as previously described (25). Real Time reverse transcription-PCR for EGFR and 18S rRNA was achieved with Assays-on-Demand Gene Expression Assay from Applied Biosystems. Amplification conditions were as follows: activation of enzymes for 2 min at 50°C and 10 min at 95°C, followed by 40 cycles at 15 sec at 95°C and 1 min at 60°C. Thermal cycling and fluorescence detection was done using an ABI Prism 7700 Sequence Detection System (Applied Biosystems). EGFR values were normalized to 18S rRNA. Each condition was assayed in triplicate and data were presented as mean plus SD.
Patient treatment and tumor specimens
Patients with histologically proven HNSCC provided informed consent before being enrolled into a phase I study of gefitinib, paclitaxel, and radiation for patients with locoregionally advanced HNSCC, in a protocol that was approved by the Institutional Review Board of the NCI (NCI trial 04-C-0141). Gefitinib was produced by AstraZeneca Pharmaceuticals and supplied through the NCI’s Cancer Treatment Evaluation Program. Gefitinib (250 mg orally) was administered daily for 1 wk prior to paclitaxel and radiation, so that tumor specimens pretreatment and after 1 wk of treatment with gefitinib alone could be obtained from tumors accessible by transoral biopsy.
The patients received a total of 8 wk of gefitinib, including 7 wk in combination with weekly doses of paclitaxel and 35 fractions of radiation. Biopsies of patients’ normal mucosa and tumor pretreatment and 1 wk post-gefitinib treatment were obtained from 7 of 10 patients. Specimens were immediately embedded in optimum cutting temperature media and frozen at −80°C. Frozen tissues were sectioned at a thickness of 10 um at the largest area and placed on silanated glass slides (Histoserv) for immunostaining. The complete clinical results and analysis of correlative immunohistochemical studies for EGFR, selected phospho-proteins, proliferation (Ki-67), and apoptosis [terminal deoxynucleotidyl transferase biotin-dUTP nick end labeling (TUNEL)] markers have been described in a separate report.5
RPMA
The cell line lysates in SDS-based buffer were quantified with BCA protein assay kit (Pierce), and adjusted to 1μg/μL. The lysates were printed onto Whatman FAST nitrocellulose coated slides (Schleicher & Schuell) using a solid-pin 2470 Arrayer (Aushon Biosystems). Lysates were printed in a serial dilution curve, in duplicate.
Immunostaining and analysis of RPMAs
The antibodies used in RPMA were first tested to ensure that a single band was generated in Western blot on control HeLa cell line lysates that were treated or untreated with EGF. The rabbit polyclonal antibodies from Cell Signaling were affinity-purified against EGFR, phospho-EGFR (Tyr1068), phospho-EGFR(Tyr1148), AKT, phospho-AKT (Ser473), ERK1/2, phospho-ERK1/2 (Thr202/Tyr204), phospho-ERK1/2 (Tyr185/187), NF-κB p65, phospho-p65 (Ser536), STAT3 and phospho-STAT3 (Tyr705), mitogen-activated protein kinase kinase (MEK), and phospho-MEK (Ser 217/221).
Each array was incubated with a specific primary antibody, which was detected by using the catalyzed signal amplification system (DAKO). Briefly, each slide was blocked with I-block (Tropix) for 1 h. The slides were incubated with primary and biotin-labeled secondary antibodies, and then sequentially incubated with streptavidin-biotin complex for 15 min, biotinyl tyramide (for amplification) for 15 min, streptavidin-peroxidase for 15 min, and 3,3′-diaminobenzidine tetrahydrochloride chromogen for 2 min. Between steps, the slide was washed with catalyzed signal amplification buffer. The signal was scanned with a Perfection 1200S scanner (Epson America) with 256-shade gray scale at 1,200 dots per inch. For detection of total protein, arrays were stained with colloidal gold solution (Bio-Rad) and scanned similarly. Spot images were converted to raw pixel values using ImageQuant software (v. 5.2, Molecular Dynamics). Raw pixel values for the 1:2 dilutions of all samples were used for subsequent analysis.
For RPMA study in human patient samples, frozen sections of HNSCC or normal tissues were scraped, lysed, arrayed, and stained the same ways as described above. Total protein of each spotted lysate was determined using Sypro Ruby Protein Blot Stain (Molecular Probes) and a fluorescence imaging system (Kodak). Stained arrays were scanned on a flatbed scanner and spot images were converted to raw pixel values by ProteinScan/P-SCAN software [Peak quantification with Statistical Comparative Analysis, (http://abs.cit.nih.gov/pscan)], and were normalized to total protein.
Results
UMSCC cells exhibited differential sensitivity to gefitinib
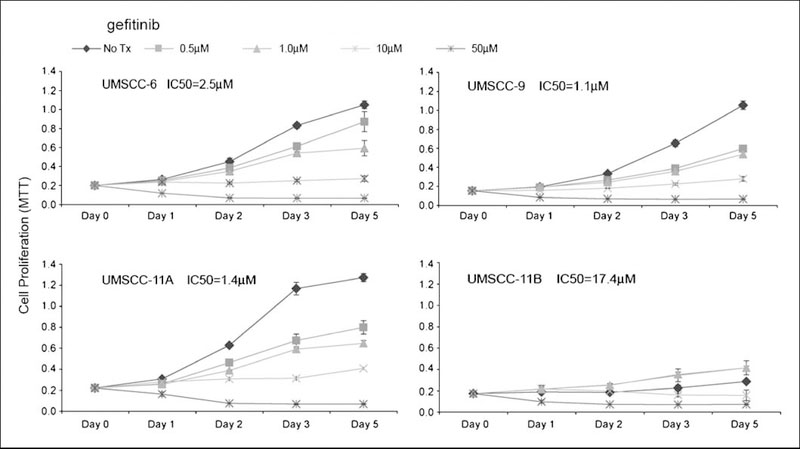
A phase I clinical trial combining gefitinib with paclitaxel and radiation in HNSCC patients was carried out at the NCI, and heterogeneous clinical responses were observed in 10 patients enrolled.5 To investigate the possible mechanism(s) for heterogeneous clinical responses observed in this and other clinical studies of HNSCC, we selected a panel of HNSCC cell lines, UMSCC-6, -9, -11A, and -11B. The selection criteria for these cell lines were based on our previous studies, in which we found that UMSCC-11A and -11B exhibited relatively higher EGFR expression and activation, and UMSCC-6 and -9 exhibited intermediate and lower levels (26, 27). In addition, multiple signal transduction pathways downstream from EGFR involving AKT, ERK, STAT3, and NF-κB have been extensively studied and characterized in these cell lines by our laboratory (23, 25–31). The effects of gefitinib on the activation of EGFR and downstream molecules were investigated in vitro. First, we tested gefitinib drug sensitivity of these cell lines using MTT assay (Fig. 1). UMSCC-6, -9, and -11A showed similar in vitro sensitivity to gefitinib with the IC50 range of ~1 to 2.5 μmol/L (Fig. 1). However, the UMSCC-11B cell line was less sensitive, with an IC50 of ~17 μmol/L, or about 10-fold more than that observed in the other cell lines (Fig. 1). In addition, a lower dose of gefitinib (0.5–1.0 μmol/L) showed a paradoxical stimulation of UMSCC-11B cell growth (Fig. 1).
Fig.1.
A dose-dependent antiproliferative effect of gefitinib in HNSCC cell lines. Cultured UMSCC cells were plated in 96-well microplates in quadruplicates, and the next day the cells were treated with different doses of gefitinib. The cell density was determined on days1, 2, 3, and 5 by MTT assay. The optical density (570 nm) was measured and presented as the mean plus SD from quadruplicates. The IC50 values were calculated based on the day 5 MTT data. The vehicle control was included as the NoTx condition.
EGFR mutations have been found to be rare among HNSCC from the United States or Europe (17–19), but to rule out the possibility that the gefitinib sensitivity is due to EGFR mutation, we sequenced EGFR exon 18 to 21, which code for the hot spots of EGFR mutations of ATP domains identified in lung cancer (24). We found a polymorphism in EGFR exon 18, which substitutes A for T at nucleotide 2133. This is the third codon for a threonine (aa# 629), where the T to A transversion is a silent change (Supplemental Fig. S1). The transversion was found to be homozygous in control HEKA and UMSCC-9 cells, T/A heterozygosity was found in UMSCC-6 and -11A cells, whereas the wild-type T residue presented in GENBANK was found only in UMSCC-11B cells. There was no sequence evidence for the previously identified in-frame deletion in exon 19 of the EGFR gene, which was reported in HNSCC of the Korean population (20). In addition, we did not find the deletion of EGFR exon 2 to 7 (EGFR vIII, PCR data not shown), which results in a truncated extracellular domain and constitutive tyrosine kinase activation (10).
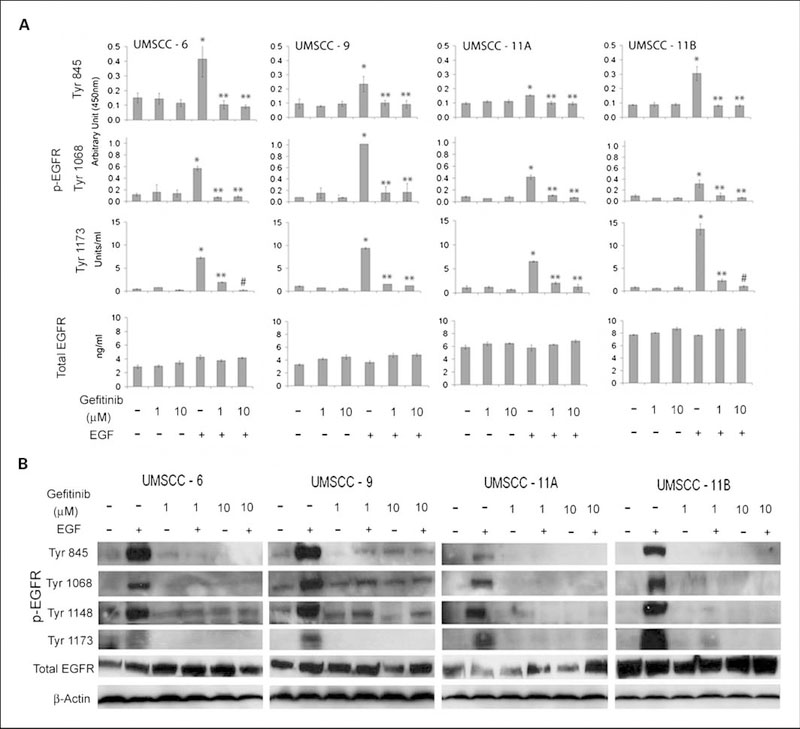
Gefitinib significantly inhibited EGF-induced EGFR phosphorylation
Gefitinib was designed to inhibit tyrosine kinase activity of EGFR. To test the specific phosphorylation sites of EGFR that could be affected by gefitinib in the HNSCC cell lines, we did a series of experiments to identify the status of known EGFR phosphorylation sites and elucidate the drug effects by ELISA (Fig. 2A) and Western blot analysis (Fig. 2B). The cultured cells exhibited relatively low basal levels of activated p-EGFR sites tested by both ELISA and Western blot, and gefitinib had little if any significant effect on the constitutive EGFR activation (Fig. 2). Upon EGF stimulation, the cell lines showed different responses in terms of phosphorylation of EGFR at different sites. UMSCC-6 and -11B exhibited relatively high levels of phosphorylation of p-EGFR at Tyr854; UMSCC-9 exhibited the highest level of phosphorylation of p-EGFR at Tyr1068; and UMSCC-11B exhibited the highest levels of p-EGFR at Tyr1173 (Fig. 2). Gefitinib at 1 to 10 times the pharmacologic serum concentration achieved with standard daily dosages of 250 to 500 mg orally (~1 and 10 μmol/L; ref. 32) showed completely inhibitory effects on each of the different EGFR phosphorylation sites in all cell lines, regardless of the drug sensitivity detected by MTT assay (Fig. 1). For total EGFR protein expression, both UMSCC-11A and -11B cells showed a relatively high level of expression when compared with UMSCC-6 and -9 cells (Fig. 2A). Neither EGF nor gefitinib significantly affected the total EGFR protein expression levels in any of the cell lines (Fig. 2).
Fig. 2.
Gefitinib effects on phosphorylated and total EGFR expressed by HNSCC cell lines. UMSCC cell lines were cultured in the log growth phase and treated with different doses of gefitinib for 4 h. Then the cells were treated with 10 ng/mL of recombinant EGF for 1 h followed by harvesting of the cell lysates. Phosphorylated and total EGFR were quantified by ELISA (A) or tested by Western blot analysis (B).The data for ELISA were calculated from triplicates from one of repeated experiments, and presented as the mean plus SD. The statistical differences are shown for comparisons with NoTx control (*), with EGF treatment (**), and with 1 μmol/L gefitinib and EGF (#, P < 0.05, Student’s t-test).
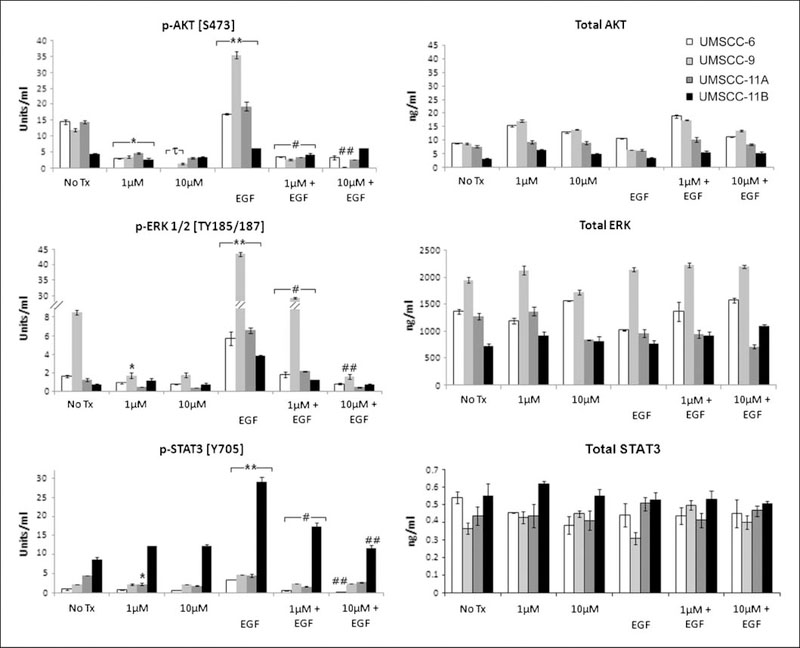
Gefitinib significantly inhibited EGFR-regulated downstream pathways
Next, we tested the effects of gefitinib on downstream pathways potentially regulated by EGFR, such as AKT, ERK, and STAT3, by assessing the level of the total and phosphorylated forms of these proteins (Fig. 3). These pathways were constitutively activated, with varying levels of basal phosphorylated and total proteins observed in the different cell lines. Gefitinib sensitivity of cell lines UMSCC-6, -9, and -11A corresponded with elevated basal levels of phospho- and total AKT relative to gefitinib-resistant UMSCC-11B cells. By contrast, UMSCC-11B cells showed the highest basal levels of phosphorylated STAT3. UMSCC-9 cells showed the highest basal phosphorylated and total ERK proteins (Fig. 3). Under EGF stimulation, UMSCC-9 cells showed the strongest induction in p-AKT (S473) and ERK1/2 (TYR185/187), and UMSCC-11B cells exhibited the largest induction in phosphor-STAT3 (Y705; Fig. 3). Gefitinib significantly inhibited the basal p-AKT, as well as EGF inducible phosphorylation of AKT, ERK1/2, and STAT3 in the cell lines, in a dose-dependent fashion (Fig. 3). The levels of total proteins were not affected by either EGF stimulation or by gefitinib (Fig. 3).
Fig. 3.
Gefitinib effects on phosphorylated and total proteins of signal pathways downstream of EGFR. UMSCC cells were treated and the whole cell lysates were harvested the same way as indicated in Fig. 2.The phosphorylated and total proteins were quantified using ELISA and the data were calculated from triplicates from one of repeated experiments with the mean plus SD. The statistical differences are shown for comparisons with NoTx controls (*), with 1 μmol/L gefitinib (τ), EGF treated with NoTx conditions (**), EGF treated with 1 μmol/L gefitinib and EGF-treated conditions (#), and between different doses of gefitinib when both treated with EGF (##), P < 0.05 (Student’s t-test).
Next we tested if there were correlations between IC50 values and EGFR downstream molecules tested. The IC50 were statistically and negatively correlated with levels of basal p-AKT (R2 = −0.93; P = 0.022) and EGF-induced p-AKT (R2 = −0.89; P = 0.037), and positively correlated with EGF-induced p-STAT3 (R2 = 0.98; P = 0.008). The other correlations, such as basal and EGF-induced p-ERK, as well as basal p-STAT3, did not reach statistical significance, possibly due to the smaller sample sizes.
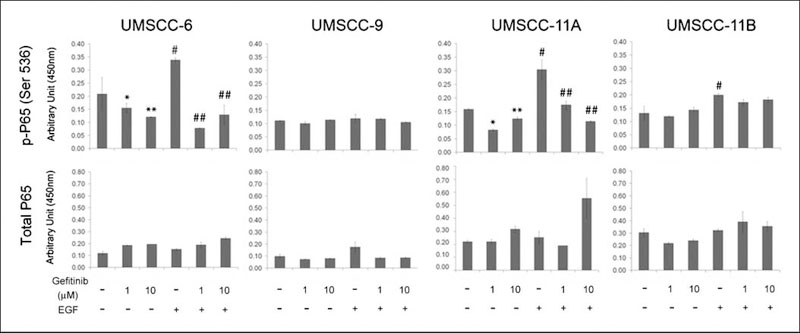
Gefitinib significantly inhibits the EGFR-regulated NF-kB pathway
In addition, we also tested the effects of the drug on the NF-κB pathway, because our laboratory has shown that NF-κB promotes HNSCC pathogenesis, and EGFR can contribute to aberrant activation of NF-κB in HNSCC cell lines (26), which was inhibited in the tumor specimen of the molecular responder to gefitinib.5 We tested the effects of EGF and gefitinib on the NF-κB subunit p65 (S536), implicated as the most important site for p65 transactivation by IκB kinase β of the canonical IκB kinase α/β/γ complex (33). EGF induced significant activation of NF-κB p65 (S536) in 3 of 4 cell lines (Fig. 4). Gefitinib partially inhibited constitutive and significantly inhibited EGF-induced activation of NF-κB p65 (S536) in 2 of 3 of the gefitinib-sensitive lines, UMSCC-6 and -11A (Fig. 4). In general, EGF and gefitinib did not show inhibitory effects on total p65 protein expression levels.
Fig. 4.
EGF and gefitinib effects on NF-κB activation in HNSCC. UMSCC cells were treated the same way as indicated in Fig. 3, and nuclear extracts were harvested. The phosphorylated and total NF-κB p65 proteins were quantified in triplicates, and the data were calculated as mean plus SD. The statistical significances are shown for comparisons between gefitinib-treated samples with untreated controls (*), EGF-treated samples with controls (IκB), and EGF and gefitinib-treated samples with EGF-treated samples (##), P < 0.05 (Student’s t-test).
Cross-comparison of the results from RPMA with those detected by ELISA and Western blot analyses
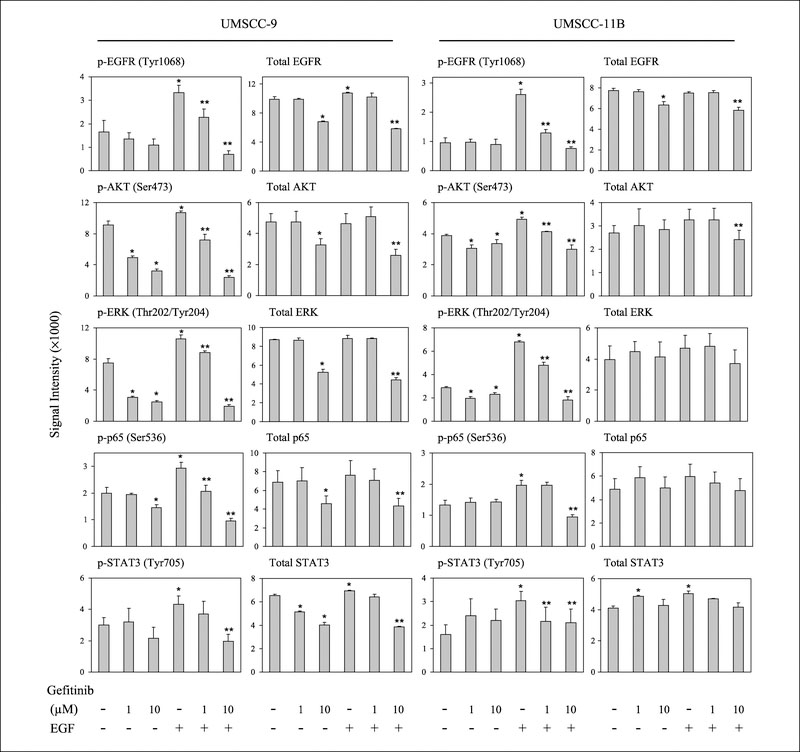
RPMA is a recently developed proteomic microarray platform that is capable of simultaneously quantifying concentrations of numerous proteins and posttranslational modifications such as phosphorylation from small amounts of tissue or cells using validated antibodies (21, 22). To compare the results obtained by RPMA with results previously obtained using the classic Western blot and ELISA methods, RPMA was done with the same whole cell and nuclear lysates as used in Figs. 2–4. We observed similar trends of EGF induction and gefitinib inhibition of the activation of EGFR and downstream molecules in UMSCC-9 and -11B cells (Fig. 5), although there was some variability in results among RPMA, Western blot, and ELISA. More significant alteration in phosphorylation of signaling proteins than in abundance of total signaling proteins was observed, consistent with the findings observed by the classic methods.
Fig. 5.
Reverse phase protein array detection of phosphrylated and total proteins in UMSCC cells lines. Reverse phase protein array was done using the same whole cell and nuclear extracts as used in the previous experiments. The images of immunohistochemical staining were scanned, and data were extracted by ImageQuant5.2 software from replicated slides. * indicates the statistical significance when compared with NoTx conditions; ** indicates the difference when compared with EGF-treated conditions (P < 0.05, Student’s t-test).
Identification of potential biomarkers in HNSCC patients’ samples from a phase I clinical trial using RPMA
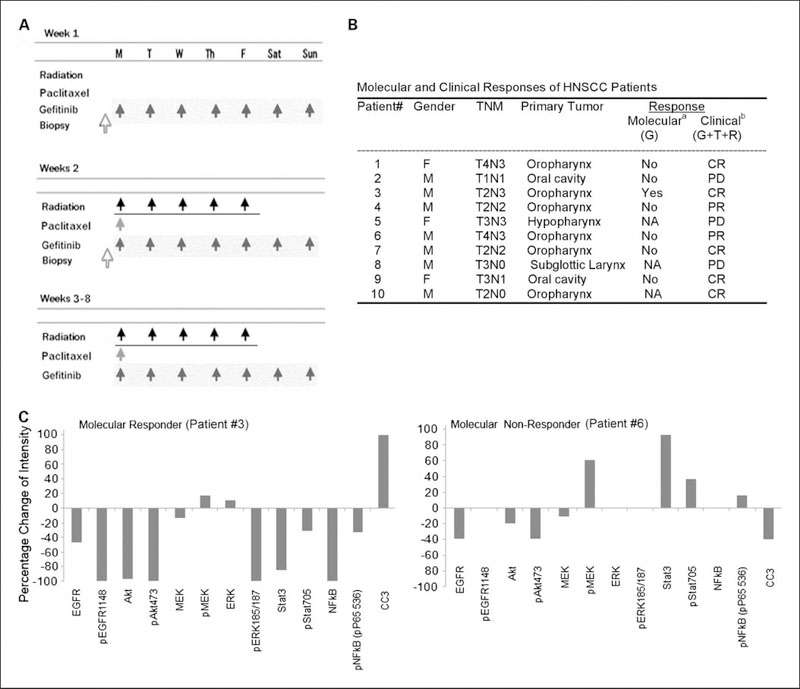
A phase I clinical trial combining gefitinib with paclitaxel and radiation in HNSCC patients was concluded at the NCI, and heterogeneous clinical responses were observed in 10 patients enrolled (Fig. 6A and B). The details of clinical responses, and the selection and results of the immunohistochemistry biomarker studies will be presented elsewhere.5 In biopsies obtained from patients on treatment day 8 of gefitinib alone, significant inhibition of EGFR, multiple-signal phospho-proteins, and proliferation marker Ki-67 was observed, which was accompanied with increased apoptosis (TUNEL) staining in 1 of 7 subjects. This patient was one of the five patients who had a complete response to the combination (patient 3; Fig. 6A and B) and was identified as a molecular responder. Of the remaining six patients defined as molecular nonresponders to gefitinib based on the biomarkers detected by immunohistochemistry, most of the patients responded to standard modalities included in the combined therapy of gefitinib, paclitaxel, and radiation. As expected for a chemotherapy and radiation therapy combination of known activity for previously untreated patients, the 6 of 10 (60%) complete clinical response observed is close to that seen with paclitaxel and radiation without gefitinib in our previous trial (34). It is expected that the response to paclitaxel and radiation exceeded the molecular response to gefitinib, because clinical responders received 8 weeks of gefitinib, including 7 weeks in combination with weekly doses of paclitaxel and 35 fractions of radiation, whereas the patient samples collected for the biomarker study to define the molecular responders were obtained after receiving gefitinib alone for a run-in period of only 7 days, as indicated in Fig. 6A and B.
Fig. 6.
Clinical information and reverse phase protein array detection of phosphorylated and total proteins in HNSCC patient tumor samples. Ten patients with HNSCC were enrolled in the phase I clinical trial combining gefitinib, radiation, and paclitaxel. A, biopsy was done prior to treatment and 1 wk after 7 doses of gefitinib treatment. Gefitinib was given weeks 2–8 concurrent with weekly paclitaxel on Monday and radiation Monday through Friday, as indicated. B, molecular response and clinical responses of HNSCC patients. TNM, tumor-node-metastasis staging. Molecular response (with gefitinib treatment alone, a), indicated by inhibition of phospho-signaling and increased apoptosis after treatment (yes); no consistent inhibition of signaling or apoptosis (no); not available (NA). Clinical response, determined 3 mo after treatment (gefitinib + paclitaxel + radiation, b). Complete response (CR); Partial response (PR); progressive disease (PD). C, reverse phase protein array was done using the lysates from tumor tissues of this clinical trial. Left panel, data from patient 3, who was identified as the molecular responder and a complete responder clinically. Right panel, data from patient 6, who was identified as the molecular nonresponder. However, clinically, after the combination treatment, the patient showed partial response. The data presented are the percentage changes in intensity, which were calculated from the intensity ratio between the stained samples before versus after treatment.
We did RPMA from tumor lysates obtained pre-gefitinib and after 7 days of gefitinib alone and compared the results with the immunohistochemistry biomarker analysis done on the same biopsy specimen.5 Protein microarrays were generated from tissue lysates of patient biopsy samples, and 13 biomarkers related to EGFR and its downstream pathways were tested. In patient 3 (Fig. 6B), who was determined to be a molecular responder based on effects on EGFR signaling, apoptosis, and proliferation biomarkers by immunohistochemistry, we also observed consistent molecular responses by RPMA (Fig. 6C, left panel). Consistent with immunohistochemistry data, a broad decrease in RPMA staining of EGFR and downstream signal molecules (10 of 13 biomarkers) was observed in this patient after gefitinib treatment, including the molecules involved in the AKT, ERK, STAT3, and NF-κB pathways (Fig. 6C, left panel). In addition, the only molecular responder identified in this study did show significantly higher levels of total AKT, p-AKT (Ser473), and total STAT3 in the pretreated samples, but did not show higher levels of total and p-EGFR, p-AKT (Ser308), and total and p-ERKs and MEKs (data not shown). An increased staining in cleaved caspase 3 was only observed in specimen from this patient, consistent with the increase in apoptosis detected by TUNEL assay in the same tumor specimen.5 In contrast, in a representative sample of a molecular nonresponder (Fig. 6C, right panel), there was only a ~40% decrease of total EGFR and p-AKT, a 10% to 20% decrease of total AKT and MEK, but ~60% increase of p-MEK, ~100% increase of STAT3, ~40% increase of p-STAT3, and a slight increase of p-NF-κB. No increase in cleaved caspase 3 was observed in this or other RPMA nonresponder samples. In addition, an increase of either total or phospho-proteins involved in each of the ERK/MEK, STAT3, or NF-κB pathways was observed in 4 of the 5 patients as the molecular nonresponders with evaluable RPMA data. For each of the molecular nonresponders, increased staining of >2 molecules involved in different pathways was observed. Thus, the data are consistent between RPMA and immunohistochemistry, showing that lack of response to gefitinib may have been due to activation of the ERK, STAT3, and NF-κB pathways, either by or in compensatory response to gefitinib, or other mechanisms independent of gefitinib or EGFR pathway.
Discussion
We investigated the effect of gefitinib on EGFR and related molecular signaling pathways in HNSCC cell lines and patient tumor samples. We showed that gefitinib suppressed EGF-induced EGFR phosphorylation to basal levels at three phosphorylation sites studied, and it inhibited the activation-specific phosphorylation of the downstream signal pathway components AKT, ERK, STAT3, and NF-κB to various degrees in different HNSCC cell lines and tumors. In vitro, a dose-dependent inhibition of tumor cell proliferation was observed that corresponded to higher basal AKT phosphorylation, and was associated with EGF-dependent phosphorylation of AKT and STAT3. In a clinical trial using gefitinib in HNSCC patients, consistent results were observed from tumor specimens of seven patients where the phosphorylation of coactivated downstream pathways and a marker of apoptosis were compared by immunohistochemistry and RPMA. Thus, our study identified alterations in a panel of specific EGFR and downstream signal pathway components of known biological and clinical significance in HNSCC, which could be used as early biomarkers for molecular responsiveness in clinical trials of EGFR-targeted therapeutic agents alone, or used in combination with other agents targeting these multiple pathways.
Using a panel of four UMSCC cell lines, we examined EGFR and downstream molecular pathways activated in HNSCC that are potentially related to EGFR sensitivity. The four cell lines exhibited various levels of total EGFR and EGF-induced receptor phosphorylation (Fig. 2), which are consistent with the reported overexpression of EGFR in HNSCC (1–5, 35). The UMSCC-11A and -11B cell lines exhibited the higher total EGFR than UMSCC-6 and -9 by ELISA (Fig. 2A), which is consistent with our previous reports (26, 27). The constitutive activation of EGFR was low among the sites tested, and variability in inducible EGFR phosphorylation was observed (Fig. 2). However, drug sensitivity of the cell lines (IC50) could not be simply explained by the level of total EGFR, basal or EGF-induced EGFR phosphorylation, in that gefitinib induced strong suppression of all EGFR phosphorylation sites in all of the cell lines tested (Figs. 1 and 2).
The gefitinib sensitivity in HNSCC lines (IC50) seemed to correlate most closely with the extent of phosphorylation of the individual EGFR sites and their mediated AKT, ERK, and STAT3 signal molecules (Figs. 2 and 3). Phosphorylation at EGFR Tyr845 of the kinase domain is involved in c-SRC activation and regulated by both EGF- and integrin-mediated EGFR activation (4, 5, 36). Tyr1068 serves as docking site for PIP3K and growth factor binding protein 2 (Grb2), and these molecules mediate activation of the AKT and ERK pathways (8, 37–39). The pair of phosphorylated residues, Tyr1148 and Tyr1173, provides a docking site for the Shc scaffold protein, and both sites are involved in mitogen-activated protein (MAP) kinase signaling activation (38, 39). In addition, Tyr1173 is the favored autophosphorylation site in ligand-activated wild-type EGFR and seems to be the major site of autophosphorylation in the mutant EGFRvIII (10, 40). Among these sites, UMSCC-9 cells exhibited the greatest level of activation of Tyr1068 after EGF stimulation, and also showed the highest basal ERK, as well as strong EGF-induced AKT and ERK phosphorylation (Figs. 2 and 3), consistent with the role of this EGFR site in phosphoinositide 3-kinase, AKT, and ERK pathway activation. UMSCC-9 was also the most sensitive cell line to gefitinib, with an IC50 of ~ 1.09μmol/L. In addition, the statistical correlations were observed between gefitinib sensitivity with basal and EGF-induced phosphorylation of AKT across the cell lines. The data from cell lines are consistent with the data from the molecular responder in the clinical study, who retained the highest total AKT and p-AKT Ser 473 prior to treatment (data not shown). Although we did not find known mutations previously detected in lung cancer, our data are consistent with previous preclinical studies in lung cancer cells that harbor EGFR mutations (12–16, 41), in which the drug sensitivity to gefitinib is closely correlated with EGFR-dependent AKT activation. However, the overexpression and activation of ERK observed did not reach statistical significance in the cell line study, and was not associated with the molecular responder in this small clinical trial. The significance of ERK activation as the biomarker for gefitinib sensitivity in HNSCC needs to be validated in a larger clinical correlative biomarker trial. Our results suggest that phosphorylation of EGFR and AKT may serve as useful early markers of EGFR activation and drug response, which may be due to different etiology.
In addition, a truncation mutation of EGFR, EGFRvIII, has been reported, which harbors an in-frame deletion that results in a truncated 150 kDa protein with constitutive phosphorylation (10, 42–44). These HNSCC with the EGFRvIII deletion showed increased proliferation and resistance to cisplatin and cetuximab treatment, but the response to gefitinib in the cells with such a mutation has not been well studied (10). In this study, however, the EGFR protein from the four UMSCC cell lines comigrate with EGFR of normal keratinocytes and recombinant EGFR protein on Western blot analysis,6 and sequencing and PCR analyses did not show such deletion mutation. Our data suggest that these truncated forms are unlikely to account for EGFR activation, as well as gefitinib sensitivity and resistance in these HNSCC cells. We sequenced the EGFR genes of these lines for ATP domain mutations identified in gefitinib-responsive lung cancers from Asian populations (17–19), but we did not find the mutations in the exon 18 to 21 of EGFR coding regions. We did find a polymorphism at the EGFR exon 18, but the T to A transversion is a silent sequence change which did not alter the coded amino acid (Supplemental Fig. S1). In addition, the constitutive EGFR tyrosine phosphorylations in UMSCC-9 cells were at the comparable or lower levels compared with those found in other UMSCC cells, suggesting that the gefitinib sensitivity observed in UMSCC-9 and other responsive HNSCC lines could be due to different mechanisms.
Gefitinib significantly suppressed basal AKT and ERK phosphorylation but not the basal EGFR phosphorylation, which suggests that the basal AKT and ERK activation could be due to other sites of EGFR phosphorylations or other receptor tyrosine kinases, such as hepatocyte growth factor receptor/c-Met (25). The observation is consistent with the fact that gefitinib may not be strictly specific for EGFR, but also be a competitor for the ATP site of other tyrosine kinases as well (45). We previously reported that the hepatocyte growth factor/c-Met pathway is constitutively activated in UMSCC cells, which modulates downstream ERK and PI3K pathways (28). In addition, the relatively high IC50 in UMSCC-11B correlated with the higher level of constitutive and EGF-induced activation of STAT3, in which gefitinib did not affect basal and only partially suppressed EGF-induced STAT3 activation (Figs. 3 and 5). As previously reported, STAT3 activation could be mediated by EGFR and interleukin-6R signaling in HNSCC (27, 46, 47), in which gefitinib does not affect interleukin-6R–activated STAT3 signal pathway (27). Consistent with this observation, the rest of the cell lines in the panel exhibited only a modest basal and increase in p-STAT3 when treated with EGF, indicating that EGF is not the major signal to induce STAT3 activation in these cells (Figs. 3 and 5). The data provided by the UMSCC cell lines are in good agreement with the data from RPMA from the HNSCC patient specimens, in that the molecular responder showed highest basal total STAT3, as well as suppressed total and phosphorylated STAT3 proteins after gefitinib treatment; however, in the molecular nonresponders, 4 of 5 patients showed increased total and phosphorylated STAT3 proteins after treatment (Fig. 1 and data not shown). These observations suggest that supplementing inhibition of EGFR with agents blocking interleukin-6 or STAT3 may have combinatorial or broader activity in patients with HNSCC.
Furthermore, we found evidence of inhibition and stimulation by EGFR of the NF-κB pathway. Previously we showed that EGFR signaling partially contributes to NF-κB activation (28), which is also inducible and important for cell survival in response to tumor necrosis factor-α, radiation, and chemotherapy (48), consistent with its known functions modulating the critical cellular processes involved in proliferation and apoptosis (48). In this study, EGF induced and gefitinib suppressed NF-κB p65 Ser536 phosphorylation in 2 of 3 of the gefitinib-sensitive UMSCC cell lines (Fig. 4). Our data are consistent with our previous report (26), showing that EGFR signals partially contribute to NF-κB activation, and are in good agreement with the studies from other laboratories that targeting EGF can sensitize renal cell carcinoma cell lines to NF-κB inhibition by bortezomib (49).
We compared the results from this study using the traditional ELISA and Western blot (Figs. 2–4) with newly developed RPMA technology (Fig. 5). The development and improvement of RPMA technology provide a new platform for proteomic mapping of signaling pathways in tumor specimens that can help us to simultaneously identify and characterize multiple protein biomarkers that are critical for diagnosis, prognosis, and selection of targeted agents for personalized cancer treatment (21, 50). The advantages of this technology are the small sample size required, as well as the semiquantitative and sensitive protein measurements permitted with use of high-quality validated antibodies. A 1-cm long core or punch biopsy can yield up to 100 RPMA slides, which makes it possible to screen a panel of biomarkers (21, 22, 50), in contrast to the extensive usage of tissue specimen by classic immunohistochemistry staining or other testing methods. Secondly, this method is able to generate the quantitative measurement of the specific biomarkers using a fitting curve with serial dilution of the samples, whereas classic immunohistochemistry is primarily qualitative. The successful cross-comparison of biomarkers by RPMA technology identified in vitro and in small pilot clinical studies herein shows the feasibility of applying the newly developed technology in clinical trials in which a major obstacle includes the limited clinical samples from patients. In this study, we obtained data consistent with the results using classic immunohistochemistry5 and the studies from UMSCC cell lines (Figs. 2–4). We observed the best molecular response was associated with the highest level of pretreatment p-AKT Ser 473, total AKT, and total STAT3 (data not shown), as well as with the strong gefitinib-suppressed p-EGFR Tyr1148, p-AKT Ser473, p-ERK Ser185/187, and NF-κB p-65 Ser536 in the only molecular and clinical responder (Fig. 6). In addition, an increase of cleaved caspase 3 activation was also observed in the responder, which is consistent with the tumor apoptosis data.5 In contrast, gefitinib failed to inhibit but rather induced activation of the MEK, STAT3, and/or NF-κB pathways in the rest of nonresponder patients. Our study revealed the heterogeneity of cancer as a major hurdle that limits the effective treatment. Such heterogeneity usually remains undetected by standard histologic pathologic classification and clinical grading systems. To address this problem, we recently have identified such subgroup-specific gene expression signatures in HNSCC cell lines with increased frequency of promoter binding sites for transcription factors NF-κB, AP-1, and STAT3 or p53 (23, 30), that support conclusions drawn from this study. Our study suggests that by utilizing RPMA and other proteomic technologies, it may be feasible to identify clinically useful biomarkers associated with the higher risk for HNSCC recurrence and metastasis, and provide more accurate prognosis and selection of therapy based on the proteomic signatures.
Supplementary Material
Translational Relevance.
Gefitinib, the first small molecule drug targeting epidermal growth factor (EGF) receptor (EGFR), has not attained the efficacy anticipated in clinical trials. To address this problem, the molecular mechanism of gefitinib sensitivity was investigated in head and neck cancer lines and tumors. The differential sensitivity was associated with the basalor EGF-induced activation of downstream protein kinase B and signal transducer and activator of transcription 3, but not with the level of EGFR phosphorylation or known EGFR mutations. A similar panel of biomarkers was studied in tumor specimens from a phase I gefitinib trial, by immunohistochemistry and reverse-phase protein microarray (RPMA), and consistent results were observed. Our data suggest that the activation status of EGFR down stream signaling molecules, instead of EGFR itself, could serve as potentially useful biomarkers for predicting sensitivity of gefitinib, or other potential drugs targeting EGFR. The newly developed RPMA is a powerful and reliable method for detecting multiple biomarkers using clinical biopsy specimens.
Acknowledgments
We thank Justin L. Ricker, MD, PhD, medical director at Abbott, Abbott Park, IL, and John Sunwoo, MD, assistant professor in the Department of Otolaryngology, Washington University School of Medicine, for reading and providing useful comments on the manuscript.
Grant support: Clinical Research Training Program, a public-private partnership supported jointly by the NIH and a grant to the Foundation for the NIH from Pfizer Pharmaceuticals Group. Intramural Research Program of the NIH, NIDCD project Z01-DC-00016, and NCI, Center for Cancer Research.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors have received gefitinib for research and clinical trials from AstraZeneca Inc., under a cooperative research development and clinical trials agreement with the National Cancer Institute.
Van Waes C, et al., IntJRad Onc Biol Phys, in press.
Pernas F, unpublished observation.
Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
References
- 1.Thelemann A, Petti F, Griffin G, et al. Phosphotyrosine signaling networks in epidermal growth factor receptor overexpressing squamous carcinoma cells. Mol Cell Proteomics 2005;4:356–76. [DOI] [PubMed] [Google Scholar]
- 2.Santini J, Formento JL, Francoual M, et al. Characterization, quantification, and potential clinical value of the epidermal growth factor receptor in head and neck squamous cell carcinomas. Head Neck 1991;13: 132–9. [DOI] [PubMed] [Google Scholar]
- 3.Dassonville O, Formento JL, Francoual M, et al. Expression of epidermal growth factor receptor and survival in upper aerodigestive tract cancer. J Clin Oncol 1993;11:1873–8. [DOI] [PubMed] [Google Scholar]
- 4.Rogers SJ, Harrington KJ, Rhys-Evans P, O-Charoenrat P, Eccles SA. Biological significance of c-erbB family oncogenes in head and neck cancer. Cancer Metastasis Rev 2005;24:47–69. [DOI] [PubMed] [Google Scholar]
- 5.Kalyankrishna S, Grandis JR. Epidermal growth factor receptor biology in head and neck cancer. J Clin Oncol 2006;24:2666–72. [DOI] [PubMed] [Google Scholar]
- 6.Ranson M, Hammond LA, Ferry D, et al. ZD1839, a selective oral epidermal growth factor receptor tyrosine kinase inhibitor, is well tolerated and active in patients with solid, malignant tumors: results of a phase I trial. J Clin Oncol 2002;20:2240–50. [DOI] [PubMed] [Google Scholar]
- 7.Baselga J, Rischin D, Ranson M, et al. Phase I safety, pharmacokinetic, and pharmacodynamic trial of ZD1839, a selective oral epidermal growth factor receptor tyrosine kinase inhibitor, inpatients with five selected solid tumor types. J Clin Oncol 2002;20:4292–302. [DOI] [PubMed] [Google Scholar]
- 8.Cappuzzo F, Magrini E, Ceresoli GL, et al. Akt phosphorylation and gefitinib efficacy in patients with advanced non-small-cell lung cancer. J Natl Cancer Inst 2004;96:1133–41. [DOI] [PubMed] [Google Scholar]
- 9.Hirsch FR, Varella-Garcia M, Bunn PA Jr., et al. Molecular predictors of outcome with gefitinib in a phase III placebo-controlled study in advanced non-small cell lung cancer. J Clin Oncol 2006;24:5034–42. [DOI] [PubMed] [Google Scholar]
- 10.Sok JC, Coppelli FM, Thomas SM, et al. Mutant epidermal growth factor receptor (EGFRvIII) contributes to head and neck cancer growth and resistance to EGFR targeting. Clin Cancer Res 2006;12: 5064–73. [DOI] [PubMed] [Google Scholar]
- 11.Cohen EE, Rosen F, Stadler WM, et al. Phase II trial of ZD1839 in recurrent or metastatic squamous cell carcinoma of the head and neck. J Clin Oncol 2003; 21:1980–7. [DOI] [PubMed] [Google Scholar]
- 12.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129–39. [DOI] [PubMed] [Google Scholar]
- 13.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci US A 2004;101:13306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science New York NY 2004;304: 1497–500. [DOI] [PubMed] [Google Scholar]
- 15.Mitsudomi T, Kosaka T, Endoh H, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with post operative recurrence. J Clin Oncol 2005;23:2513–20. [DOI] [PubMed] [Google Scholar]
- 16.Mitsudomi T, Kosaka T, Yatabe Y. Biological and clinical implications of EGFR mutations in lung cancer. Int JClin Oncol Japan Soc Clin Oncol 2006;11:190–8. [DOI] [PubMed] [Google Scholar]
- 17.Temam S, Kawaguchi H, El-Naggar AK, et al. Epidermal growth factor receptor copy number alterations correlate with poor clinical outcome in patients with head and neck squamous cancer. J Clin Oncol 2007; 25:2164–70. [DOI] [PubMed] [Google Scholar]
- 18.Cohen EE, Lingen MW, Martin LE, et al. Response of some head and neck cancers to epidermal growth factor receptor tyrosine kinase inhibitors may be linked to mutation of ERBB2 rather than EGFR. Clin Cancer Res 2005;11:8105–8. [DOI] [PubMed] [Google Scholar]
- 19.Loeffler-Ragg J, Witsch-Baumgartner M, Tzankov A, et al. Low incidence of mutations in EGFR kinase domain in Caucasian patients with head and neck squamous cell carcinoma. Eur J Cancer 2006;42:109–11. [DOI] [PubMed] [Google Scholar]
- 20.Lee JW, Soung YH, Kim SY, et al. Somatic mutations of EGFR gene in squamous cell carcinoma of the head and neck. Clin Cancer Res 2005;11:2879–82. [DOI] [PubMed] [Google Scholar]
- 21.Calvo KR, Liotta LA, Petricoin EF. Clinical proteomics: from biomarker discovery and cell signaling profiles to individualized personal therapy. Biosci Rep 2005;25:107–25. [DOI] [PubMed] [Google Scholar]
- 22.Hingorani SR, Petricoin EF, Maitra A, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 2003;4: 437–50. [DOI] [PubMed] [Google Scholar]
- 23.Yan B, Yang X, Lee TL, et al. Genome-wide identification of novel expression signatures reveal distinct patterns and prevalence of binding motifs for p53,nuclear factor-κB and other signal transcription factors in head and neck squamous cell carcinoma. Genome Biol 2007;8:R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu D, Scaringe WA, Li K, et al. Database of somatic mutations in EGFR with analyses revealing indel hotspots but no smoking-associated signature. Hum Mutat 2007;28:760–70. [DOI] [PubMed] [Google Scholar]
- 25.Worden B, Yang XP, Lee TL, et al. Hepatocyte growth factor/scatter factor differentially regulates expression of proangiogenic factors through Egr-1in head and neck squamous cell carcinoma. Cancer Res 2005;65:7071–80. [DOI] [PubMed] [Google Scholar]
- 26.Bancroft CC, Chen Z, Yeh J, et al. Effects of pharmacologic antagonists of epidermal growth factor receptor, PI3K and MEK signal kinases on NF-κB and AP-1 activation and IL-8 and VEGF expression in human head and neck squamous cell carcinoma lines. Int J Cancer 2002;99:538–48. [DOI] [PubMed] [Google Scholar]
- 27.Lee TL, Yeh J, Van Waes C, Chen Z. Epigenetic modification of SOCS-1 differentially regulates STAT3 activation in response to interleukin-6 receptor and epidermal growth factor receptor signaling through JAK and/or MEK in head and neck squamous cell carcinomas. Mol CancerTher 2006;5:8–19. [DOI] [PubMed] [Google Scholar]
- 28.Dong G, Chen Z, Li ZY, Yeh NT, Bancroft CC, Van Waes C. Hepatocyte growth factor/scatter factor-induced activation of MEK and PI3K signal pathways contributes to expression of proangiogenic cytokines interleukin-8 and vascular endothelial growth factor in head and neck squamous cell carcinoma. Cancer Res 2001;61:5911–8. [PubMed] [Google Scholar]
- 29.Hong SH, Ondrey FG, Avis IM, et al. Cyclooxygenase regulates human oropharyngeal carcinomas via the proinflammatory cytokine IL-6: a general role for inflammation? FASEB J 2000;14:1499–507. [DOI] [PubMed] [Google Scholar]
- 30.Lee TL, Yang XP, Yan B, et al. A novel nuclear factor-κB gene signature is differentially expressed in head and neck squamous cell carcinomas in association with TP53 status. Clin Cancer Res 2007; 13:5680–91. [DOI] [PubMed] [Google Scholar]
- 31.Lee TL, Yeh J, Friedman J, et al. A signal network involving coactivated NF-κB and STAT3 and altered p53 modulates BAX/BCL-XL expression and promotes cell survival of head and neck squamous cell carcinomas. Int J Cancer 2008;122:1987–98. [DOI] [PubMed] [Google Scholar]
- 32.Mukohara T, Engelman JA, Hanna NH, et al. Differential effects of gefitinib and cetuximab on non-small-cell lung cancers bearing epidermal growth factor receptor mutations. J Natl Cancer Inst 2005;97: 1185–94. [DOI] [PubMed] [Google Scholar]
- 33.Hayden MS, Ghosh S. Signaling to NF-κB. Genes Dev 2004;18:2195–224. [DOI] [PubMed] [Google Scholar]
- 34.Sunwoo JB, Herscher LL, Kroog GS, et al. Concurrent paclitaxel and radiation in the treatment of locally advanced head and neck cancer. J Clin Oncol 2001; 19:800–11. [DOI] [PubMed] [Google Scholar]
- 35.Pomerantz RG, Grandis JR. The epidermal growth factor receptor signaling network in head and neck carcinogenesis and implications for targeted therapy. Semin Oncol 2004;31:734–43. [DOI] [PubMed] [Google Scholar]
- 36.Boerner JL, Biscardi JS, Silva CM, Parsons SJ. Transactivating agonists of the EGF receptor require Tyr 845 phosphorylation for induction of DNA synthesis. Mol Carcinog 2005;44:262–73. [DOI] [PubMed] [Google Scholar]
- 37.Franke TF, Hornik CP, Segev L, Shostak GA, Sugimoto C. PI3K/Akt and apoptosis: size matters. Oncogene 2003;22:8983–98. [DOI] [PubMed] [Google Scholar]
- 38.She QB, Solit DB, Ye Q, O’Reilly KE, Lobo J, Rosen N. The BAD protein integrates survival signaling by EGFR/MAPK and PI3K/Akt kinase pathways in PTEN deficient tumor cells. Cancer Cell 2005;8:287–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zebisch A, Czernilofsky AP, Keri G, Smigelskaite J, Sill H, Troppmair J. Signaling through RAS-RAF-MEKERK: from basics to bedside. Curr Med Chem 2007; 14:601–23. [DOI] [PubMed] [Google Scholar]
- 40.Okabe T, Okamoto I, Tamura K, et al. Differential constitutive activation of the epidermal growth factor receptor in non-small cell lung cancer cells bearing EGFR gene mutation and amplification. Cancer Res 2007;67:2046–53. [DOI] [PubMed] [Google Scholar]
- 41.Noro R, Gemma A, Kosaihira S, et al. Gefitinib (IRESSA) sensitive lung cancer cell lines show phosphorylation of Akt without ligand stimulation. BMC Cancer 2006;6:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Batra SK, Castelino-Prabhu S, Wikstrand CJ, et al. Epidermal growth factor ligand-independent, unregulated, cell-transforming potential of a naturally occurring human mutant EGFRvIII gene. Cell Growth Differ 1995;6:1251–9. [PubMed] [Google Scholar]
- 43.Huang HS, Nagane M, Klingbeil CK, et al. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem 1997;272:2927–35. [DOI] [PubMed] [Google Scholar]
- 44.Okamoto I, Kenyon LC, Emlet DR, et al. Expression of constitutively activated EGFRvIII in non-small cell lung cancer. Cancer Sci 2003;94:50–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ono M, Kuwano M. Molecular mechanisms of epidermal growth factor receptor (EGFR) activation and response to gefitinib and other EGFR-targeting drugs. Clin Cancer Res 2006;12:7242–51. [DOI] [PubMed] [Google Scholar]
- 46.Sriuranpong V, Park JI, Amornphimoltham P, Patel V, Nelkin BD, Gutkind JS. Epidermal growth factor receptor-independent constitutive activation of STAT3 in head and neck squamous cell carcinoma is mediated by the autocrine/paracrine stimulation of the interleukin 6/gp130 cytokine system. Cancer Res 2003;63:2948–56. [PubMed] [Google Scholar]
- 47.Quesnelle KM, Boehm AL, Grandis JR. STAT mediated EGFR signaling in cancer. J Cell Biochem 2007;102:311–9. [DOI] [PubMed] [Google Scholar]
- 48.VanWaes C Nuclear factor-Κb in development, prevention, and therapy of cancer. Clin Cancer Res 2007; 13:1076–82. [DOI] [PubMed] [Google Scholar]
- 49.An J, Rettig MB. Epidermal growth factor receptor inhibition sensitizes renal cell carcinoma cells to the cytotoxic effects of bortezomib. Mol Cancer Ther 2007;6:61–9. [DOI] [PubMed] [Google Scholar]
- 50.Nishizuka S, Charboneau L, Young L, et al. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc Natl Acad Sci US A 2003;100:14229–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.