Significance
Despite decades of research, no approved drugs have been discovered for KRAS. Recently, a pocket occurring on the surface of the active and inactive form of KRAS was found, but, due to its comparatively shallow, polar nature, this pocket has been assumed to be “undruggable.” Starting from very weakly binding fragments and using structure-based drug design, we discovered BI-2852 (1), a nanomolar inhibitor to this pocket which is mechanistically distinct to covalent KRASG12C inhibitors; 1 modulates pERK and pAKT and has an antiproliferative effect in KRAS mutant cells. This work demonstrates the druggability of this so-called switch I/II pocket and provides the scientific community with a chemical probe that directly inhibits the active and inactive forms of KRAS.
Keywords: KRAS, NMR, oncology, structure-based drug design, fragment-based drug design
Abstract
The 3 human RAS genes, KRAS, NRAS, and HRAS, encode 4 different RAS proteins which belong to the protein family of small GTPases that function as binary molecular switches involved in cell signaling. Activating mutations in RAS are among the most common oncogenic drivers in human cancers, with KRAS being the most frequently mutated oncogene. Although KRAS is an excellent drug discovery target for many cancers, and despite decades of research, no therapeutic agent directly targeting RAS has been clinically approved. Using structure-based drug design, we have discovered BI-2852 (1), a KRAS inhibitor that binds with nanomolar affinity to a pocket, thus far perceived to be “undruggable,” between switch I and II on RAS; 1 is mechanistically distinct from covalent KRASG12C inhibitors because it binds to a different pocket present in both the active and inactive forms of KRAS. In doing so, it blocks all GEF, GAP, and effector interactions with KRAS, leading to inhibition of downstream signaling and an antiproliferative effect in the low micromolar range in KRAS mutant cells. These findings clearly demonstrate that this so-called switch I/II pocket is indeed druggable and provide the scientific community with a chemical probe that simultaneously targets the active and inactive forms of KRAS.
The 3 human RAS genes, KRAS, NRAS, and HRAS, encode 4 different RAS proteins (KRAS-4A, KRAS-4B, NRAS, and HRAS) which belong to the protein family of small GTPases that function as binary molecular switches involved in cell signaling (1). Activating mutations in RAS like the glycine 12 mutations are among the most common oncogenic drivers in human cancers. KRAS is the most frequently mutated oncogene, with mutation rates of 86 to 96% in pancreatic cancers (2), 40 to 54% in colorectal cancers (3), and 27 to 39% in lung adenocarcinomas (4). NRAS is predominantly mutated in melanoma and hematological malignancies (5, 6), while HRAS mutations are found in salivary gland and urinary tract cancers (7, 8).
The RAS family is known to cycle through 2 different conformational states that are defined by differential binding to nucleotides. In the “off” state, RAS proteins are bound to the nucleotide guanosine diphosphate (GDP), while in the “on” state they are bound to the nucleotide guanosine triphosphate (GTP). The γ-phosphate of GTP holds 2 regions, switch I and switch II (9), in a compact conformation that allows interaction with downstream effectors, such as CRAF, PI3Kα, and RALGDS, as well as with the allosteric site of SOS1 and SOS2. Hydrolysis of the γ-phosphate to produce GDP-RAS causes a conformational change in the switch regions, leading to the formation of an inactive state which is unable to bind effector molecules (10, 11). RAS itself has an intrinsic, but weak, GTPase activity that is enhanced by GTPase-activating proteins (GAPs) catalyzing RAS inactivation. The exchange of the bound nucleotide GDP into GTP is facilitated by guanine nucleotide exchange factors (GEFs) which, in the case of KRAS, is performed by SOS1 and SOS2 (12). GEFs catalyze the release of GDP from RAS in the cytoplasm and replace it with the more abundant intracellular GTP. Oncogenic mutations in RAS impair GTP hydrolysis, leading to stabilization of the activated GTP-RAS form and enhanced RAS signaling. The most common mutations occur as single-point mutations at codons 12, 13, and 61 (13).
Although KRAS could serve as an excellent drug target for many cancers, direct inhibition of oncogenic RAS has proven to be challenging. Despite decades of research, no therapeutic agent directly targeting RAS has been clinically approved. The main reason for this is the lack of druggable pockets on the surface of RAS. However, in recent years, there has been a resurgence of research around RAS, driven by the growing belief that RAS might be able to be drugged with low molecular weight organic molecules. This belief was sparked by the discovery of 2 pockets on the surface of RAS that could potentially be amenable to small-molecule drug discovery. The S.W.F. group at Vanderbilt (14), researchers at Genentech (15), and, more recently, the Rabbitts group (16, 17) discovered small molecules that bind to a shallow pocket between the switch I and II regions of KRAS. This pocket will be referred to as the switch I/II pocket (SI/II-pocket). In addition, the Shokat group discovered covalently linked small molecules which bind to a second pocket on RAS positioned above the switch II loop in GDP-KRASG12C, called the switch II pocket (SII-pocket) (11).
In this paper, we describe the discovery of nanomolar inhibitors that directly target the small, polar SI/II-pocket present on both the active and inactive form of KRAS. To discover small molecules that bind to KRAS, we conducted several fragment-based screens using uniformly 15N-labeled guanosine-5′-[(β,γ)-methyleno]triphosphate (GCP)-bound KRASG12D for validation. From these screens, we identified fragments that weakly bind to GCP-KRASG12D that were optimized using structure-based design. This was accomplished by developing a robust system for crystallizing small molecules bound to GTP-KRASG12D. The most potent KRAS inhibitor, BI-2852 (1), binds with nanomolar affinity to the active and inactive form of KRAS. Compound 1 blocks the interaction between GDP-KRAS and the catalytic site of SOS1, but, in contrast to covalent KRASG12C inhibitors, also inhibits the interactions between GTP-KRAS and the allosteric site of SOS1 as well as its effectors (CRAF and PI3Kα). In cells, 1 inhibits SOS1-catalyzed exchange of GDP to GTP as well as GAP-catalyzed exchange of GTP to GDP, which results in no net change in cellular GTP-RAS levels upon treatment. Compound 1 reduced pERK and pAKT levels in a dose-dependent manner, leading to an antiproliferative effect in NCI-H358 cells. The effects of 1 were confirmed to be KRAS-driven and not unspecific effects, through the consistent data generated for the 10-fold weaker distomer 44. Compound 1 demonstrates that the SI/II-pocket is indeed druggable and provides an ideal starting point for the design of more potent and selective RAS inhibitors. Compound 1 will also serve as a useful chemical probe for the scientific community in the study of RAS biology of simultaneous inhibition of active and inactive RAS in an in vitro setting.
Results
GTP-KRAS Fragment Screening.
We adopted multiple approaches to identify inhibitors of KRAS. Initial attempts to find starting points for GTP-KRAS using high-throughput screening of 1.7 million compounds with a luminescent oxygen channeling immunoassay (18), as well as a mammalian protein−protein interaction (PPI) trap cellular assay (19, 20), failed to deliver any hits which could be validated to bind to KRAS in a dose-dependent manner.
Next, we attempted to identify compounds that bind to GTP-KRAS via fragment-based screening (21–23). A library of 1,800 fragments was screened using both saturation transfer difference NMR (24, 25) and microscale thermophoresis (26) using KRASG12V-phosphomethylphosphonic acid guanylate ester (GCP-KRASG12V), from which 16 fragments were found and subsequently validated (hit rate 0.9%) by the observation of cross-peak shifts in the 2D 1H/15N heteronuclear single-quantum correlation (HSQC) NMR spectra of GCP-KRASG12D. We also screened a library of 13,800 compounds using uniformly 15N-labeled GNP-KRAS by HSQC NMR experiments. Representatives of some of the fragments which bound the active form of KRAS from the 2 screens are depicted in Fig. 1A. Despite the large number of hits obtained from the screens (55 in total), the binding affinity of the fragments identified all displayed dissociation constants (KD) of greater than 1 mM as measured via HSQC NMR (SI Appendix, Fig. S1).
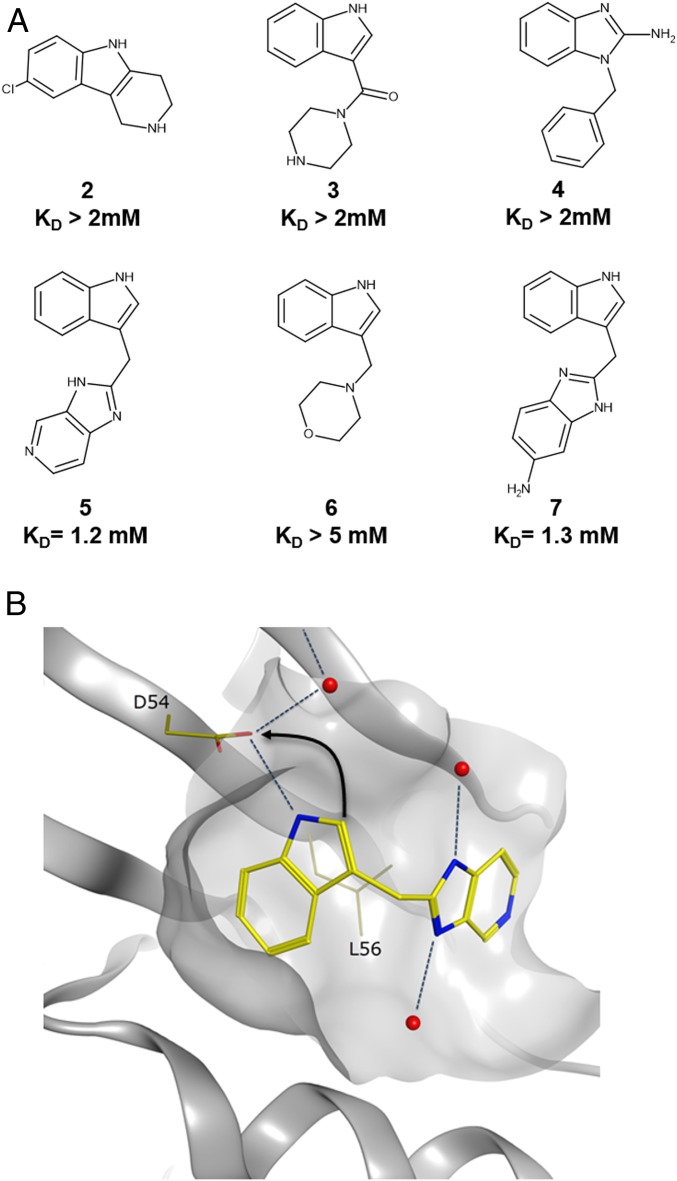
Fig. 1.
Fragments identified from 2 separate fragment screens. (A) Representative indole and benzimidazole fragments identified from the fragment screens. (B) The binding mode of indole 5 in GDP-KRAS (Protein Data Bank [PDB] ID code 4EPV) showing the H bond between the indole NH and the side chain of D54. Indole 5 shown in yellow, water molecules shown in red. Arrow indicates the strategy of forming an additional charge−charge interaction with the side chain of D54 from the indole 2 position.
Following the fragment screen, we conducted a follow-up screen of commercially and internally available compounds with high structural similarity to the initial fragment hits, often called “structure activity relationship (SAR) by catalogue” (27, 28). We biased the follow-up SAR by a catalog screen with available indoles bearing a pendent group containing a basic amine at the 2 position, as we hypothesized that forming a charged interaction with D54 in addition to the existing hydrogen bond (H bond) formed by the indole NH (Fig. 1B) would lead to a significant improvement in binding affinity. Indeed, indoles containing a methylamino functionality at the 2 position showed a high propensity to bind to GCP-KRAS with measureable KDs in the range of 1 mM (SI Appendix, Table S1). However, our GCP-KRAS cocrystallization efforts were unsuccessful using any of these more potent fragments from the SAR by catalog screen.
Further Optimization Leading to Cocrystal Structures.
Although protein NMR is the only biophysical method capable of robustly measuring KDs in the millimolar range, the approach is protein-intensive. To facilitate this, a reliable and high-yielding procedure (∼10 mg/L purified yield) for obtaining large amounts of uniformly 15N-labeled GCP-KRASG12D was developed. This was achieved by first expressing the GDP-KRAS from Escherichia coli, hydrolyzing GDP to guanidine, and then performing the exchange with the stable GTP analog phosphomethylphosphonic acid guanylate ester (GCP). With this modified approach, we could achieve better yields of uniformly nucleotide-exchanged KRAS compared with the previously described methods using the less stable GTP gamma S or GMPPNP (29). In addition, we used an artificial KRAS T35S mutant for the first optimization steps, as this point mutant was described to display better in HSQC NMR spectra (30).
The SI/II-pocket on KRAS (Fig. 2B) is around 7 times smaller in volume compared with the druggable acetyl lysine-binding pocket on the bromodomain containing protein BRD4 (Fig. 2A). As such, optimizing ligands for this small, shallow, and polar pocket represents a daunting challenge for medicinal chemistry. Our strategy to overcome this was to use structure-based drug design to precisely target polar interactions within the binding site while minimizing the ligand desolvation penalty (31). The design of molecules capable of forming an H bond to the side-chain hydroxyl of T74 was prioritized to gain further binding affinity, and the dihydroisoindolinone moiety linked with the S configuration to the 3 position of the indol-2-ylmethylamino fragments was chosen. Importantly, this compound series led to an affinity improvement of 5- to 10-fold (SI Appendix, Table S2), with the pyrrolidine analog 15 displaying an NMR KD of 440 µM.
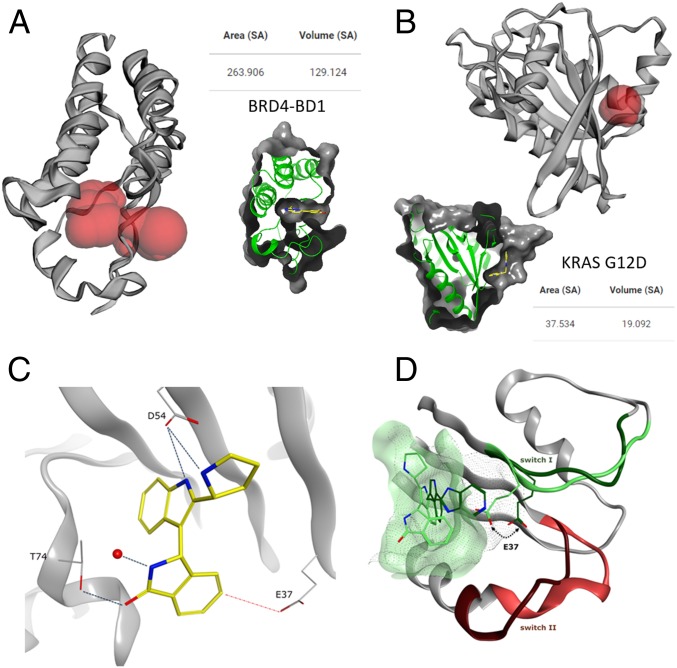
Fig. 2.
Analysis of protein pockets and X-ray structure of 15 in GCP-KRASG12D. A Computer Atlas of Surface Topography of proteins (48) analysis of pockets in proteins for calculating the solvent-accessible surface area (Area SA) and volume (Volume SA) with a radius probe of 1.4 Å showing differences in pocket size for (A) BRD4-BD1 (PDB ID code 5M39) as an example of a highly druggable pocket and (B) KRAS G12D (PDB ID code 6GJ5) with a very small volume. Calculated pockets are shown in red. Insets show ligand binding in the respective pocket, for comparison reasons. (C) Polar interactions formed by 15 to T74, D54, and a conserved water. The ideal position of E37 for introducing a further polar interaction is highlighted with the red dotted line (PDB ID code 6GJ5). (D) Comparison of SI/II-pocket in GDP (PDB ID code 4EPV) and GTP-KRAS showing the significantly reduced pocket size in GTP-KRAS. The switch I and switch II regions are colored dark green and dark red in GDP-KRAS and light green and light red for GTP-KRAS, respectively.
Given the significant improvement in binding potency of these compounds, we reverted to the use of the naturally occurring T35 construct, with NMR KDs being easily measured despite the reduction in visible cross-peaks for the G12D, C118S double mutant construct (SI Appendix, Table S2). C118S was introduced for stability reasons in NMR as described before (14). More importantly, we were successful in obtaining an X-ray structure of 15 in the new GCP-KRASG12D,C118S construct. As designed, the carbonyl oxygen of the dihydroisoindolinone forms a H bond to T74 at a distance of 2.4 Å, and H bonds involving the pyrrolidine and indole nitrogens of 15 are formed with D54 (Fig. 2C). In addition, a conserved water molecule is coordinated to the indolinone nitrogen. Interestingly, E37 blocks part of the SI/II-pocket in GCP-KRAS, compared with the GDP-KRAS structure 4EPV, even further reducing the size of the SI/II-pocket in the GTP form of KRAS (Fig. 2D). Important for further optimization was the observation that the side-chain position of E37 was ideally placed to form a fourth polar interaction with isoindolinones substituted at the 5 position.
Discovery of the Micromolar RAS Inhibitor 18.
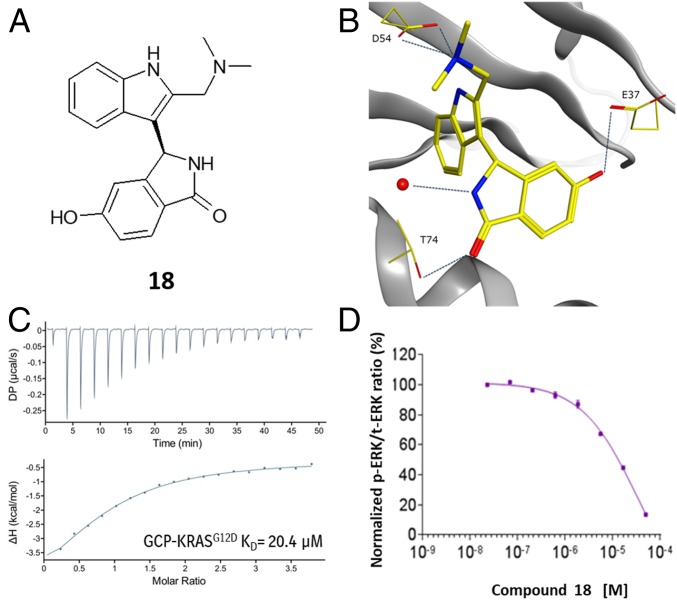
Based on the X-ray structure of 15 bound to GCP-KRAS, we chose to introduce a phenolic oxygen as a H-bond donor at the 5 position of the isoindolinone to interact with E37. The 5-hydroxy-isoindolinone 18 (Fig. 3A) was 6-fold more potent than the nonphenolic matched molecular pair 17 (SI Appendix, Table S2), with an isothermal titration calorimetry (ITC) (32) KD of 20 µM (Fig. 3C). X-ray crystallography confirmed the desired phenolic H bond to E37 at a distance of 2.7 Å and, all 3 previously discovered polar interactions to D54 and T74 were maintained (Fig. 3B).
Fig. 3.
X-ray, biophysical and cellular data for 18. (A) Chemical structure of 18. (B) X-ray structure of 18 in GCP-KRASG12D, highlighting the polar interactions formed with D54, T74, and E37 (PDB ID code 6GJ6). (C) ITC dose titration curve for 18 and GCP-KRASG12D. (D) Meso Scale Discovery analysis of pERK levels in NCI-H358 cells after 2-h treatment of 18.
Given the improved potency of 18 and the high conservation of the SI/II-pocket across RAS isoforms, selectivity was evaluated using ITC, and 18 was found to bind with similar affinity to both the GCP and GDP forms of KRAS, HRAS, and NRAS, including both mutant and wild-type KRAS (SI Appendix, Table S3 and Fig. S3). Compound 18 also showed biochemical inhibition of GTP-KRAS with SOS1 (EC50 = 33 µM; see SI Appendix, Table S4) using a time-resolved fluorescence energy transfer assay.
The cellular activity of 18 was evaluated in the lung cancer cell line NCI-H358, which bears a heterozygous KRASG12C mutation. Active RAS signals through RAF and MEK to induce phosphorylation of ERK (phospho-ERK). Phospho-ERK levels were quantified using a plate-based electrochemiluminescent assay (MESO SCALE DISCOVERY). A dose-dependent decrease in phospho-ERK levels was observed 2 h after treatment of NCI-H358 cells with 18, leading to almost complete inhibition at 50 µM (Fig. 3D).
Discovery of the Nanomolar RAS Inhibitor BI-2852 (1).
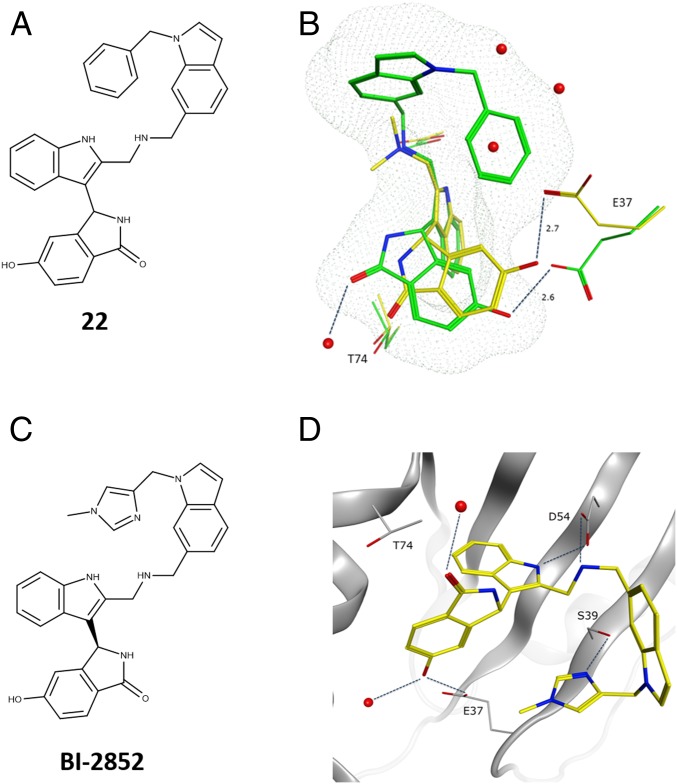
To further improve the binding affinity of the compounds, we explored a variety of substituents at the 2-methylamino position (SI Appendix, Table S4). While a benzyl substituent 19 showed no improvement in potency, indole substituted compounds 20 and 21 showed a 5-fold improvement in biochemical activity. Thus, we explored larger substituents at the pendant indole nitrogen and discovered that the racemic N-benzylindole derivative 22 (Fig. 4A, SI Appendix, Table S4) exhibited submicromolar activity in the GTP-KRAS::SOS1 fluorescence resonance energy transfer assay with an IC50 of 870 nM.
Fig. 4.
GCP-KRASG12D X-ray structures of compound 22 and BI-2852 (1). (A) Chemical structure of 22. (B) Overlay of the binding modes of 18 and 22. The relative orientation of 18 and E37 in the X-ray with KRASG12D is depicted in yellow. The binding mode of 22, E37, and T74 are depicted in green. Dotted mesh depicts the van der Waals radii around 22, showing overlap with 3 waters from the X-ray structure of 18 (PDB ID code 6GJ8). (C) Chemical structure of 1. (D) X-ray structure of 1 in GCP-KRASG12D, highlighting the polar interactions formed with D54, T74, S39, and E37 (PDB ID code 6GJ7). The racemate 23 was used for soaking, and eutomer 1 was crystallized.
The X-ray structure of 22 in complex with GCP-KRAS showed that the N-benzylindole moiety folds back on itself to displace 3 water molecules present in this area of the pocket when unoccupied (Fig. 4B). Despite the side-chain rotation of E37 to avoid a clash with the benzyl group, the H bond with E37 was maintained. However, the direct interaction with T74 was lost, and the carbonyl forms a H bond to a water molecule instead. Due to the high lipophilicity (ClogP of 4.8), 22 displayed poor solubility (<1 µg/mL at pH 6.8) and, as such, did not constitute a molecule of sufficient quality for reliably investigating KRAS biology.
To overcome the solubility limitations of 22, the chiral N-methyl imidazole derivative 1 (Fig. 4C) was prepared. This compound exhibited a significantly reduced ClogP of 2.6 and had a solubility of 18 µg/mL at pH 6.8 while maintaining nanomolar binding affinity to GTP-KRASG12D (KD = 750 nM) as measured by ITC and with an IC50 of 450 nM in the Alpha Screen (SI Appendix, Tables S5 and S6). All interactions observed for 22 were maintained, with an additional H bond formed between the imidazole nitrogen and the side-chain oxygen of S39 at a distance of 2.7 Å (Fig. 4D). As observed for compound 18, 1 also binds with similar affinity to KRAS, NRAS, and HRAS (SI Appendix, Fig. S4 and Table S5), with the exception of a small selectivity window (5- to 10-fold) to active KRASwt and inactive NRASwt.
Characteristics of the RAS Inhibitor BI-2852 (1).
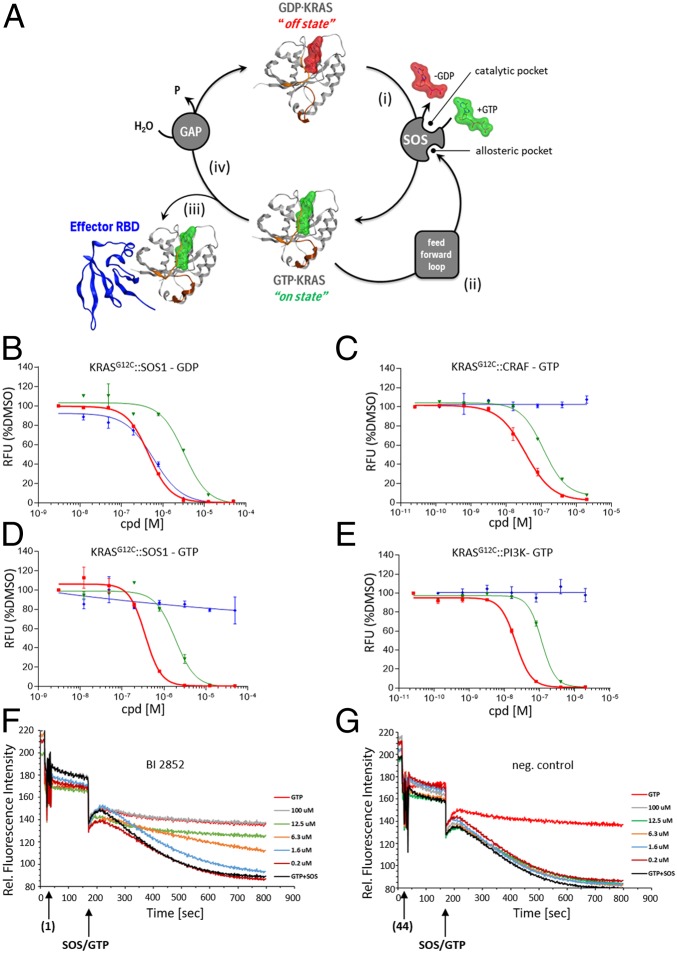
Using biochemical assays, we investigated whether 1 inhibited 3 of the 4 PPIs important for KRAS cycling (Fig. 5 A, i–iii). Namely, 1) GDP-KRAS interaction with the catalytic site of SOS (33), 2) GTP-KRAS interaction with the allosteric site of SOS (34), and 3) GTP-KRAS interaction with downstream effectors (CRAF and PI3Kα). We were unable to establish a biochemical assay for the fourth intervention point, namely, 4) GTP-KRAS interaction with its GAPs (Fig. 5 A, iv). As a reference compound for these assays, we used the KRASG12C-specific covalent inhibitor ARS-1620 (35). Compound 1 inhibited all 3 RAS cycle intervention points (1 to 3) in a dose-dependent manner (Fig. 5 B–E) in the range of 100 to 770 nM (SI Appendix, Table S6). ARS-1620 displayed activity on the GDP-dependent KRASG12C::SOS1 interaction, but was inactive in all GTP-dependent KRAS assays, in line with the lack of accessibility of the pocket when GTP is bound to KRAS. Moreover, when we exchanged the G12C mutation in KRAS against G12D, ARS-1620 lost the ability to inhibit the GDP-dependent interaction, while 1 maintained the inhibitory function (SI Appendix, Table S6).
Fig. 5.
Biochemical assay dose–response curves for BI-2852 (1), distomer 44, and ARS-1620. (A) KRAS cycle is depicted with KRAS in Channing Der‘s “beating heart of cancer” orientation switching between its “off state” with the nucleotide GDP bound (red surface) and its “on state” with GCP bound (green surface). The 4 PPI intervention points in the KRAS cycle are shown. (i) The interaction between GDP-KRAS and the catalytic site of its GEF; here SOS is depicted. (ii) GTP-KRAS binding to the allosteric site of SOS which constitutes the feed forward loop. (iii) GTP-KRAS binding to downstream effectors; CRAF (in blue) is shown as an example. (iv) GTP-KRAS binding to GAPs. KRAS is depicted in gray, with the switch I region colored in orange and the switch II region in brown. (B−E) Biochemical assay dose–response curves for 1 (red), distomer 44 (green), and ARS-1620 (blue) for (B) GDP-KRASG12C::SOS1 alpha screen assay, (C) GTP-KRASG12C::CRAF alpha screen assay, (D) GTP-KRASG12C::SOS1 alpha assay, and (E) GTP-KRASG12C::PI3Kα alpha screen assay. All values shown are normalized to DMSO (= 100%) for better comparability. Error bars indicated show the SD of duplicates measured. Shown are representative examples of multiple repetitions with identical results. (F and G) The ability of test compounds to affect SOScat-catalyzed nucleotide exchange on RAS was assessed at several concentrations. The addition of SOScat and excess GTP (at 120 s) initiates the exchange of labeled boron-dipyrromethene-GDP (BODIPY-GDP) already loaded into RAS. The BODIPY-GDP to GTP exchange mediated by SOScat (black curve) is observed as a decrease in relative fluorescence units (RFUs) over time. While the negative control distomer 44 (G) shows no effect, increasing concentrations of BI-2852 (1) (F) show a slower decrease in RFU over time, representing a slower exchange rate. The highest concentrations show full inhibition of SOScat-mediated exchange, matching the rate in the absence of SOScat (red curve).
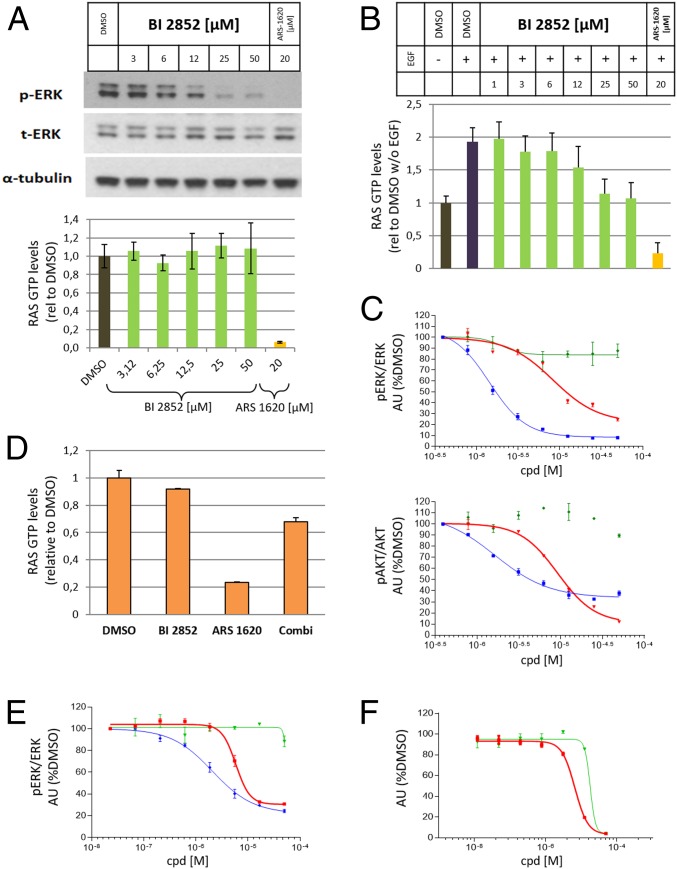
Compound 1 also inhibits the rate of nucleotide exchange in a dose-dependent manner as measured by an SOS1 nucleotide exchange assay (36) (Fig. 5F). Despite this fact and a dose-dependent reduction of pERK, 1 does not change steady-state RAS GTP levels in NCI-H358 cells under high growth factor conditions, while the covalent GDP-KRASG12C inhibitor ARS-1620 strongly reduced RAS-GTP levels (Fig. 6A). To demonstrate in cells that 1 inhibits SOS-catalyzed nucleotide exchange, we starved NCI-H358 cells for 24 h in low serum, causing a decrease of GTP-loaded KRAS. When we added 1 for 2 h and then added epidermal growth factor (EGF) to artificially increase RAS GTP levels, we observed a dose-dependent inhibition of the formation of GTP-loaded KRAS, which, at high concentrations, stayed at the levels of the dimethyl sulfoxide (DMSO)-treated sample (Fig. 6B). This result is in line with a blockade of the SOS-catalyzed conversion of KRAS from GDP to GTP. ARS-1620 reduced the RAS GTP levels even below the DMSO control. Analysis of downstream signaling events revealed that, under the described conditions, pERK and pAKT levels were dose-dependently reduced (Fig. 6C). Next, we explored whether 1 could also interfere with the GAP-catalyzed conversion of GTP-loaded KRAS to inactive GDP-bound KRAS in cells, as we were unable to investigate this biochemically. To our knowledge, there is no physiological condition available to rapidly convert GTP-KRAS to the inactive form; hence we made use of ARS-1620, which, when applied in sufficient concentration, causes a near-complete depletion of active GTP KRASG12C. When we incubated NCI-H358 cells with 50 µM 1 for 1 h, as previously observed, no change in GTP-KRAS was observed. In contrast, 20 µM ARS-1620 caused an 80% reduction of the GTP-bound RAS pool. When the 2 compounds were combined, only a 30% reduction of GTP-RAS was observed, which is in line with inhibiting the binding of GAPs with KRAS (Fig. 6D).
Fig. 6.
Cellular data for BI-2852 (1), distomer 44, and ARS-1620. (A) Western blot of pERK levels versus total ERK and alpha-tubulin under high serum conditions in NCI-H358 cells upon increasing doses of 1 and a fixed concentration of ARS-1620 (Upper) and G-LISA assay measuring GTP-RAS levels under high serum conditions in NCI-H358 cells upon increasing doses of 1 and a fixed concentration of ARS-1620 (Lower). (B) GTP-RAS levels measured with a G-LISA assay in NCI-H358 cells starved for 24 h in low serum conditions (−EGF), followed by 2-h treatment with increasing concentrations of 1 or 20 µM treatment of ARS-1620 and then EGF addition. For quantification, RAS GTP levels without EGF and in the presence of DMSO were set to 1 (A and B). (C) pERK and pAKT levels in NCI-H358 cells starved for 24 h in low serum conditions (−EGF), followed by 2-h treatment with increasing concentrations of 1 (red), 44 (green), and ARS-1620 (blue); then EGF addition were quantified. DMSO-treated samples after EGF stimulation were set to 100%. (D) GTP-RAS levels measured with a G-LISA assay in NCI-H358 cells with treatment of 50 µM compound 1, 20 µM ARS-1620 and simultaneous treatment of 1 and ARS-1620. For quantification, RAS GTP levels in the presence of DMSO were set to 1. (E) pERK dose–response curves for 1 (red), 44 (green), and ARS-1620 (blue) in NCI-H358 cells under high serum conditions. (F) Antiproliferative dose–response curves for NCI-H358 cells in soft agar and low serum conditions for 1 (red) and 44 (blue). DMSO-treated samples were set to 100% (E and F). Error bars indicate SDs. Indicated experiments were performed 2 or more times with similar results.
The cellular signaling activity of 1 was tested in NCI-H358 cells. NCI-H358 cells cultured under high growth factor conditions were treated with increasing concentrations of 1 and ARS-1620 for 2 h. A dose-dependent inhibition of pERK relative to total ERK (EC50 = 5.8 µM) was observed after treatment with 1, with a similar, albeit more extended, inhibition of pERK being observed for ARS-1620 (Fig. 6E). A rebound in pERK inhibition was observed (SI Appendix, Fig. S5) similar to what was observed for BRAF inhibitors in BRAFV600E mutated cell lines (37). In contrast to low serum conditions, no reduction of pAKT levels was observed under high serum conditions in NCI-H358 cells. We then assessed whether the observed pERK reduction after 2 h would lead to an antiproliferative effect on cells. We plated NCI-H358 cells in soft agar and low serum conditions and indeed observed a dose-dependent antiproliferative effect of 1 at an EC50 of 6.7 µM (Fig. 6F). No antiproliferative effect was observed under standard 2D culture conditions, in line with recent observations that KRAS proliferation inhibition is predominantly measurable under 3D, nonadherent conditions (35).
To convince ourselves that the effects of 1 were due to direct KRAS inhibition and not unspecific effects, the properties of the distomer 44 were investigated. The distomer 44 inhibited the PPIs between the GTP and GDP forms of KRAS ∼10-fold more weakly than the eutomer 1 (Fig. 5 B–E and SI Appendix, Table S6). The distomer 44 also did not inhibit SOS1-catalyzed nucleotide exchange (Fig. 5G). This difference in activity is consistently maintained in the cellular assays with no pERK reduction observed at concentrations up to 50 µM (Fig. 6E) and a significant antiproliferative effect observed only at 50 µM (Fig. 6F). Further, we have tested 1 and 44 in 4 BRAF(v600E) cell lines which signal in a RAS-independent manner (38). No antiproliferative effect was observed for 1 and 44 in any of the 4 cell lines under low serum, soft agar, or high serum, adherent conditions, and no inhibition of pERK was observed (SI Appendix, Fig. S6). This clearly demonstrates that BI-2852 does not exhibit off-target antiproliferative effects. Taken together, these data support the interpretation that the biochemical and cellular effects observed for 1 are the result of direct inhibition of KRAS.
Discussion
Here, we describe the discovery of 1, an inhibitor of both the active and inactive form of KRAS, with nanomolar binding affinity to the SI/II-pocket, a small, shallow, and polar pocket deemed by many to be “undruggable.” Compound 1 is the first RAS inhibitor for the SI/II pocket with KRAS-driven cellular activity, displaying low micromolar pERK modulation and antiproliferative effects on a KRAS mutant cell line. Recent compounds (16, 17) claiming cellular activity do not provide negative control data and display antiproliferative effects under 2D cell culture conditions which should not be interpreted as KRAS-driven, given that KRAS antiproliferative effects are predominantly only observed under 3D, nonadherent conditions (35).
Fragment screens delivered hits in the millimolar binding affinity range which were optimized using structure-based design. Although highly resource-intensive, the application of NMR to measure dissociation constants for newly designed compounds in the millimolar range served as an important method for establishing structure activity relationships. Guided by the X-ray structures of cocomplexes, we were able to optimize binding, as evidenced by the discovery of the isoindolinone 18, which bound to GCP-KRASG12D at 20 µM by ITC and also displayed inhibition of the key PPI with KRAS in a similar range. This allowed us to switch from NMR KD measurements to biochemical PPI assays to further optimize the potency of the compounds.
Obtaining 3D crystallographic information on RAS proteins in the active form has been historically limited. The development of a reliable procedure to produce >10 mg/L of purified GCP-KRAS was instrumental in enabling crystallography, which, in turn, revealed critical information on the binding of the ligands to the SI/II-pocket of GCP-KRAS. Compound 1 maintained the polar interactions to D54 and E37 also addressed by 18 and, in addition, formed a H bond to S39 and displaced 3 water molecules, which are presumably responsible for the >100-fold improvement in potency. It should be noted that T74, which improves potency by 5- to 10-fold (SI Appendix, Tables S1 and S2), is not yet addressed by 1 and that 1 still contains 7 rotatable bonds. This highlights the potential for significant improvement beyond the current potency of 1 (e.g., IC50 of 180 nM for the PPI between active KRASG12D and CRAF) and indicates that the SI/II-pocket is indeed druggable.
Triple RAS knockout mice are not embryonically viable but can be rescued by reintroduction of an HRAS transgene, indicating functional redundancy among the RAS family (39) and suggesting that sparring at least one wild-type RAS isoform will be needed for a RAS drug. As the SI/II-pocket is conserved on both the inactive and active forms of all RAS isoforms, obtaining sufficient selectivity presents an additional significant challenge to drugging this pocket. Interestingly, 1 demonstrates a 10-fold selectivity of binding to GCP-KRASwt (SI Appendix, Table S5) which translates to a 4-fold selectivity of inhibition biochemically (inhibition of KRASwt versus KRASG12C binding to CRAF) (SI Appendix, Table S6). The relative lack of selectivity versus the KRASG12D::CRAF is expected due to the 5-fold weaker affinity of KRASG12D for CRAF, while KRASG12C and KRASwt maintain the same affinity for the RAS binding domain of CRAF (40). Also, a weaker affinity to GDP-NRAS was observed for 1. Together, this suggests that, despite the high conservation of the SI/II-pocket, it might be possible to design molecules with sufficient RAS isoform selectivity.
The SI/II-pocket is involved in interactions with GEFs (41), GAPs (42), and downstream effectors (43, 44), and we provide evidence that compound 1 inhibits all of these PPIs. Functionally, 1 inhibits SOS1-catalyzed exchange of GDP-KRAS to GTP-KRAS as well as GAP-catalyzed exchange of GTP-KRAS to GDP-KRAS, which results in no net change in cellular GTP-RAS levels upon treatment. E37 on switch II, to which the phenolic oxygen of 1 H-bonds, is also an important residue for RAS binding to downstream effectors (45), GEFs (41, 46), and GAPs (47), explaining mechanistically how 1 inhibits the binding of multiple key RAS interactions partners.
Compound 1 reduced pERK and pAKT levels in a dose-dependent manner in NCI-H358 cells, leading to an antiproliferative effect in NCI-H358 cells under nonadherent, low serum conditions. The effects of 1 were confirmed to be KRAS-driven and not off-target through the consistent data generated for the 10-fold less active distomer 44 and through the absence of any effects on BRAF(V600E) cell lines. We expect BI-2852 to serve as a useful chemical probe for the study of RAS biology in an in vitro setting, and it is available to the scientific community (https://opnme.com/molecules/kras-bi-2852). BI-2852 is also an ideal starting point for the design of more-potent and selective RAS inhibitors.
Supplementary Material
Acknowledgments
We thank Andreas Bergner, Helmut Berger, Matthias Klemencic, Norbert Kraut, Erik Patzelt, Jens Quant, Michaela Streicher, Diane Thompson, Ingrid Vorwahlner, Anika Weiss, and Piro Lito. Funding to support this work came from US National Institutes of Health (NIH) Grants P50A095103 (National Cancer Institute Specialized Programs of Research Excellence in Gastrointestinal Cancer) and RC2CA148375 (NIH American Recovery and Reinvestment Act Stimulus Grant), and through grants from the Lustgarten Foundation for Pancreatic Cancer Research to S.W.F. Additional Austrian governmental funding was provided by the Austrian Forschungsförderungsgesellschaft through Grants 854341 and 861507 (Basisprogramme). Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy Office of Science by the Argonne National Laboratory, was supported by US Department of Energy Contract DE-AC02-06CH11357. In addition, the X06SA beamline at the Swiss Light Source, Paul Scherrer Institut, Villigen, Switzerland, was used for crystallographic measurements, with special thanks to Dirk Reinert and Expose GmbH for data measurement. We thank the Vanderbilt High-Throughput Screening (HTS) Core, in which some of these experiments were performed. The HTS Core receives support from the Vanderbilt Institute of Chemical Biology and the Vanderbilt-Ingram Center (NIH Grant P30CA68485).
Footnotes
Conflict of interest statement: D.K., M.G., A.M., L.J.M., A.Z., M.M., A.G., D.C., S.F., T. Gerstberger, T. Gmashitz, P.G., D.H., W.H., J.H., J.K.-O., P.K., S.K., M.K., R.K., L.L., F.M., S.M.-M., C.P., J.R., C.S., Y.S., K.S., R.S., A.S., B.S., G.S., B.W., M.Z., M.P., and D.B.M. were employees of Boehringer Ingelheim at the time of this work.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6GJ5, 6GJ6, 6GJ7, and 6GJ8).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904529116/-/DCSupplemental.
References
- 1.Colicelli J., Human RAS superfamily proteins and related GTPases. Sci. STKE 2004, RE13 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biankin A. V., et al. ; Australian Pancreatic Cancer Genome Initiative , Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neumann J., Zeindl-Eberhart E., Kirchner T., Jung A., Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol. Res. Pract. 205, 858–862 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network , Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colombino M., et al. , BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J. Clin. Oncol. 30, 2522–2529 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Bacher U., Haferlach T., Schoch C., Kern W., Schnittger S., Implications of NRAS mutations in AML: A study of 2502 patients. Blood 107, 3847–3853 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Yoo J., Robinson R. A., H-ras gene mutations in salivary gland mucoepidermoid carcinomas. Cancer 88, 518–523 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Jebar A. H., et al. , FGFR3 and Ras gene mutations are mutually exclusive genetic events in urothelial cell carcinoma. Oncogene 24, 5218–5225 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Milburn M. V., et al. , Molecular switch for signal transduction: Structural differences between active and inactive forms of protooncogenic ras proteins. Science 247, 939–945 (1990). [DOI] [PubMed] [Google Scholar]
- 10.Vetter I. R., Wittinghofer A., The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Ostrem J. M., Shokat K. M., Direct small-molecule inhibitors of KRAS: From structural insights to mechanism-based design. Nat. Rev. Drug Discov. 15, 771–785 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Liceras-Boillos P., et al. , Sos1 disruption impairs cellular proliferation and viability through an increase in mitochondrial oxidative stress in primary MEFs. Oncogene 35, 6389–6402 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Prior I. A., Lewis P. D., Mattos C., A comprehensive survey of Ras mutations in cancer. Cancer Res. 72, 2457–2467 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Q., et al. , Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew. Chem. Int. Ed. Engl. 51, 6140–6143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maurer T., et al. , Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl. Acad. Sci. U.S.A. 109, 5299–5304 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quevedo C. E., et al. , Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment. Nat. Commun. 9, 3169 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cruz-Migoni A., et al. , Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds. Proc. Natl. Acad. Sci. U.S.A. 116, 2545–2550 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ullman E. F., et al. , Luminescent oxygen channeling immunoassay: Measurement of particle binding kinetics by chemiluminescence. Proc. Natl. Acad. Sci. U.S.A. 91, 5426–5430 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He R., Li X., Mammalian two-hybrid assay for detecting protein-protein interactions in vivo. Methods Mol. Biol. 439, 327–337 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulrichts P., Lemmens I., Lavens D., Beyaert R., Tavernier J., MAPPIT (mammalian protein-protein interaction trap) analysis of early steps in toll-like receptor signalling. Methods Mol. Biol. 517, 133–144 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W., Discovering high-affinity ligands for proteins: SAR by NMR. Science 274, 1531–1534 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Hajduk P. J., Greer J., A decade of fragment-based drug design: Strategic advances and lessons learned. Nat. Rev. Drug Discov. 6, 211–219 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Jhoti H., Cleasby A., Verdonk M., Williams G., Fragment-based screening using X-ray crystallography and NMR spectroscopy. Curr. Opin. Chem. Biol. 11, 485–493 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Mayer M., Meyer B., Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J. Am. Chem. Soc. 123, 6108–6117 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Ludwig C., et al. , Evaluation of solvent accessibility epitopes for different dehydrogenase inhibitors. ChemMedChem 3, 1371–1376 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Jerabek-Willemsen M., et al. , MicroScale thermophoresis: Interaction analysis and beyond. J. Mol. Struct. 1077, 101–113 (2014). [Google Scholar]
- 27.Hubbard R. E., Fragment approaches in structure-based drug discovery. J. Synchrotron Radiat. 15, 227–230 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baurin N., et al. , Design and characterization of libraries of molecular fragments for use in NMR screening against protein targets. J. Chem. Inf. Comput. Sci. 44, 2157–2166 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Eberth A., Ahmadian M. R., In vitro GEF and GAP assays. Curr. Protoc. Cell. Biol. 43, 14.9.1−14.9.25 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Spoerner M., Herrmann C., Vetter I. R., Kalbitzer H. R., Wittinghofer A., Dynamic properties of the Ras switch I region and its importance for binding to effectors. Proc. Natl. Acad. Sci. U.S.A. 98, 4944–4949 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cramer J., Krimmer S. G., Heine A., Klebe G., Paying the price of desolvation in solvent-exposed protein pockets: Impact of distal solubilizing groups on affinity and binding thermodynamics in a series of thermolysin inhibitors. J. Med. Chem. 60, 5791–5799 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Jelesarov I., Bosshard H. R., Isothermal titration calorimetry and differential scanning calorimetry as complementary tools to investigate the energetics of biomolecular recognition. J. Mol. Recognit. 12, 3–18 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Hall B. E., Yang S. S., Boriack-Sjodin P. A., Kuriyan J., Bar-Sagi D., Structure-based mutagenesis reveals distinct functions for Ras switch 1 and switch 2 in Sos-catalyzed guanine nucleotide exchange. J. Biol. Chem. 276, 27629–27637 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Margarit S. M., et al. , Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell 112, 685–695 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Janes M. R., et al. , Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell 172, 578–589.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Burns M. C., et al. , High-throughput screening identifies small molecules that bind to the RAS:SOS:RAS complex and perturb RAS signaling. Anal. Biochem. 548, 44–52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lito P., et al. , Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell 22, 668–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao Z., et al. , BRAF mutants evade ERK-dependent feedback by different mechanisms that determine their sensitivity to pharmacologic inhibition. Cancer Cell 28, 370–383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura K., et al. , Partial functional overlap of the three ras genes in mouse embryonic development. Oncogene 27, 2961–2968 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Hunter J. C., et al. , Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol. Cancer Res. 13, 1325–1335 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Boriack-Sjodin P. A., Margarit S. M., Bar-Sagi D., Kuriyan J., The structural basis of the activation of Ras by Sos. Nature 394, 337–343 (1998). [DOI] [PubMed] [Google Scholar]
- 42.Scheffzek K., et al. , The ras-RasGAP complex: Structural basis for GTPase activation and its loss in oncogenic ras mutants. Science 277, 333–338 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Fetics S. K., et al. , Allosteric effects of the oncogenic RasQ61L mutant on Raf-RBD. Structure 23, 505–516 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker E. H., Perisic O., Ried C., Stephens L., Williams R. L., Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature 402, 313–320 (1999). [DOI] [PubMed] [Google Scholar]
- 45.Huang L., Weng X., Hofer F., Martin G. S., Kim S. H., Three-dimensional structure of the ras-interacting domain of RalGDS. Nat. Struct. Biol. 4, 609–615 (1997). [DOI] [PubMed] [Google Scholar]
- 46.Kano Y., Cook J. D., Lee J. E., Ohh M., “New structural and functional insight into the regulation of Ras” in Seminars in Cell & Developmental Biology, Nussinov R., Csermely P., Korcsmaros T., Eds. (Elsevier, 2016), pp. 70–78. [DOI] [PubMed] [Google Scholar]
- 47.Scheffzek K., et al. , Structural analysis of the GAP-related domain from neurofibromin and its implications. EMBO J. 17, 4313–4327 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian W., Chen C., Lei X., Zhao J., Liang J., 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 46, W363–W367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.