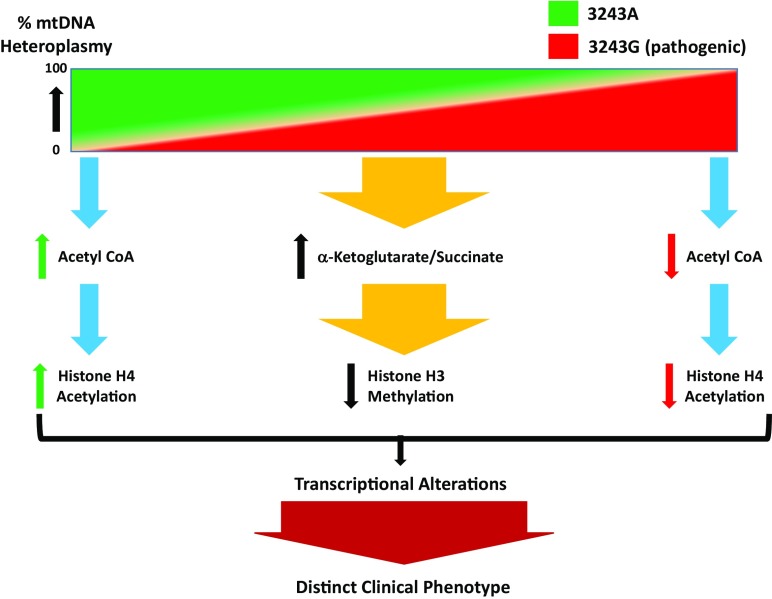
Mitochondria contain multiple copies of mitochondrial DNA (mtDNA), which encode genes essential for cellular bioenergetics. When more than one type of mtDNA genome exists within the mitochondrion, or between mitochondria, a condition termed heteroplasmy occurs. In this respect, it has been long observed that differences in mtDNA heteroplasmy involving pathogenic mtDNA mutations generate a broad range of clinical phenotypes. For instance, it is known that increasing levels of the transfer RNA leucine [tRNALeu(UUR)] 3243A > G mutant “result successively in diabetes, neuromuscular degenerative disease, and perinatal lethality” (1); however, the specific molecular mechanisms driving these diverse clinical phenotypes have been not clearly understood. In PNAS, Kopinski et al. (1) advance our understanding of this mystery by manipulating the levels of mtDNA containing either a normal (3243A) or pathogenic (3243G) mtDNA tRNALeu(UUR) mutation in a human bone osteosarcoma cybrid model. For these studies, they generated cell lines that had either 100% normal (3243A) or 100% pathogenic (3243G) mtDNA homoplasmies, in addition to a series of cybrids harboring different percentages of normal and pathogenic 3243A > G mtDNA heteroplasmies. Using these cell lines, they performed various measures of metabolism, including metabolic tracing and NAD+/NADH ratios, and, importantly, examined transcriptional and epigenetic changes in the nuclear genome. Another key feature of this work is that all cell lines (homoplasmic and heteroplasmic) shared the same nuclear genome, meaning that any observed metabolic and epigenetic changes were solely attributed to differences in mitochondrial genetics. This report provides direct evidence that changing levels of tRNALeu(UUR) 3243A > G heteroplasmy on the same nuclear background changes aspects of mitochondrial function (as anticipated), but, perhaps more importantly, also causes changes in nuclear gene expression via histone modifications modulated by levels of mitochondrially generated acetyl-CoA and α-ketoglutarate (αKG) levels. For example, Kopinski et al. find that, under conditions of high heteroplasmy (A3243G), acetyl-CoA levels decrease, which associates with decreased histone H4 acetylation (Fig. 1). Overall, they find that mitochondrial-derived metabolites correlate with histone posttranslational modifications, which differ across the different levels of heteroplasmy. Cybrids with 70 to 100% A3243G heteroplasmy have lower levels of acetyl-CoA that is associated with lower histone H4 acetylation, and additionally generate less acetyl-CoA from glucose, instead producing higher amounts of lactate, a phenotype recapitulated by inhibiting mitochondrial protein synthesis with chloramphenicol or complex I inhibition with rotenone. Additionally, cybrids with 30 to 70% A3243G have higher levels of αKG/succinate, which is associated with lower levels of histone 3 methylation (Fig. 1), likely due to αKG-dependent Jumonji C histone demethylases. Interestingly, the NAD+/NADH ratio is elevated in cybrids with 60 to 70% A3243G, which correlates with an up-regulation of NAD+ synthesis and mitochondrial oxidative phosphorylation genes—suggestive of a potential compensatory mechanism in response to declining mitochondrial function. Hence, the percent 3243A > G heteroplasmy impacts the metabolites generated through mitochondrial metabolic pathways, and these changes alter epigenetic pathways that account for the differential nuclear genome expression associated with heteroplasmy.
Fig. 1.
Influence of mtDNA heteroplasmy on clinical phenotype via changes on metabolic programming that impact the epigenome and nuclear gene expression. As shown in Kopinski et al. (1), different levels of heteroplasmic tRNALeu(UUR) m.3243 A > G mutation change levels of acetyl-CoA available for histone acetylation (histone H4). Decreased and increased levels of m.3243G heteroplasmy are linked with greater and lower levels of acetyl-CoA, respectively, which change levels of histone acetylation. Interestingly, intermediate levels of m.3243G heteroplasmy were associated with increased levels of αKG/succinate ratios and decreased histone (histone H3) methylation. Collectively, these epigenetic changes influence nuclear gene expression (transcriptional alterations) that conveys the distinct clinical phenotypes associated with heteroplasmic tRNALeu(UUR) m.3243 A > G mtDNA mutations.
While the basic concept of mitochondrial−nuclear communication is not new, these results provide an expansive viewpoint regarding the role of the mitochondrion, its related genetics, and overall impact upon cell function and response. Classically, signaling from the mitochondrion to the nucleus has been termed retrograde signaling—this type of signaling has largely focused on the role of mitochondrial oxidant generation to convey mitochondrial dysfunction to the nucleus (2, 3). Mitochondrial oxidants are increasingly recognized as key activators of JNK and ERK1/2 pathways, which activate either apoptosis or oxidant detoxification pathways, depending on the strength of the stimulus. The reverse, or signaling from nucleus to mitochondrion, has been typically referred to as anterograde signaling, which has been well studied as it pertains to mitochondrial biogenesis (2, 3); ADP/ATP and NAD+/NADH ratios are also known to regulate AMPK and sirtuin pathways, respectively, and collectively converge to coordinate mitochondrial biogenesis (2). Interestingly, while these types of mitochondrial−nuclear communication have been known for some time, they have generally been thought of as a means for affecting cell response to acute stressors from a pathway perspective. The concept of mitochondrial−nuclear communication via changes in mtDNA genetics (e.g., heteroplasmy) as a basic means for controlling nuclear gene expression from a metabolic homeostatic systems perspective, however, is likely more appropriate, and the findings of Kopinski et al. (1) raise and confirm this possibility, revealing that mitochondrial−nuclear communication is genetically influenced and is a continuous process that is not governed exclusively by acute events.
Studies examining nuclear gene expression relative to mtDNA copy number or haplogroup have shown that depletion of mtDNA in breast cancer and osteosarcoma cells altered the methylation status of CpG islands, and restoration of mtDNA levels to baseline partially restored some of the genomic methylation patterns, suggesting that the mtDNA serves as a proxy of mitochondrial function in regulating epigenomic pathways (4). The mtDNA haplotype in murine embryonic stem cell lines is also associated with patterns of genomic methylation, suggesting that mitochondrial genetic variants may play a role in epigenomic regulation of gene expression (5). Further, in human cybrids with mitochondrial haplogroup J, higher levels of genomic methylation were found relative to cybrids with other European haplogroups (6). In Drosophila melanogaster, exposure to hypoxia induced gene expression changes that differed by mitochondrial haplotype, suggesting that environmental exposures impact mitochondrial signaling responses differently based upon the mitochondrial haplotype (7). Similarly, a series of studies utilizing mitochondrial−nuclear exchange models, wherein different mtDNA genetic backgrounds were paired with the same nuclear genetic background, have directly shown that disease susceptibility, bioenergetic function, oxidant production, and, importantly, gene expression are significantly changed in vivo (8–10). Importantly, the findings of Kopinski et al. (1) provide a likely explanation for these observed differences in that they link mtDNA genetics with nuclear epigenomic modulation via changes in metabolic intermediates—further providing evidence of systemic molecular mechanisms that are defined by mtDNA genetics, which modulate transcriptional control of the nuclear genome.
In this respect, the findings of Kopinski et al. (1) have even a more global and lasting impact on our understanding of eukaryotic genetics. Despite the fact that 1) the ancestral mtDNA genome transferred the majority of its genes to the nucleus (thus making many of the genes in the nucleus of mitochondrial genetic origins) and 2) mitochondrial and nuclear genomes have coevolved for the past 1.5 billion years of eukaryotic genetic evolution, the idea of a continuous dialogue between the 2 genomes is only just emerging, and the concept that mitochondrial metabolites regulate nuclear gene expression has been limited. Moreover, that a genetic connection exists between nuclear gene expression and the mitochondrial genome via its programming of cellular metabolism has not been seriously considered—likely due to the challenging question of how a mtDNA mutant can have such broad effects upon what is heretofore thought of as “nonmitochondrial” functions. Kopinski et al.’s work demonstrates how this can occur, which directly couples mtDNA genetics → mitochondrial metabolism → nuclear gene expression. From this mito-Mendelian perspective, variants within mitochondrial genes in both genomes, nuclear and mitochondrial, likely modify the responses of the mitochondrion to stimuli such as endogenous or exogenous factors (e.g., cytokines or environmental factors, respectively), and therefore represent an area that, as of yet, remains largely unexplored. Further, these studies and others (8–10) strongly suggest the need for greater contemplation of how to interpret results from experiments utilizing genetic models having different mtDNA backgrounds. In fact, mitochondrial genetic diversity may be a key missing link in our understanding of how genetic–environmental interactions impact disease risk, in that it would now include a mito-Mendelian perspective. Our understanding of the intersections of bigenomic communication in response to environmental exposures and in the presence of pathology will advance as large datasets with multiple omics measures become more readily available. Additional studies such as those reported by Kopinski et al. are needed in order for us to gain a greater mechanistic understanding of the intersections of mitochondrial genetic determinants of epigenetic regulation in the setting of human diseases.
Footnotes
The authors declare no conflict of interest.
See companion article on page 16028.
References
- 1.Kopinski P. K., et al. , Regulation of nuclear epigenome by mitochondrial DNA heteroplasmy. Proc. Natl. Acad. Sci. U.S.A. 116, 16028–16035 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guha M., Avadhani N. G., Mitochondrial retrograde signaling at the crossroads of tumor bioenergetics, genetics and epigenetics. Mitochondrion 13, 577–591 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quirós P. M., Mottis A., Auwerx J., Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 17, 213–226 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Smiraglia D. J., Kulawiec M., Bistulfi G. L., Gupta S. G., Singh K. K., A novel role for mitochondria in regulating epigenetic modification in the nucleus. Cancer Biol. Ther. 7, 1182–1190 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee W. T., et al. , Mitochondrial DNA haplotypes induce differential patterns of DNA methylation that result in differential chromosomal gene expression patterns. Cell Death Discov. 3, 17062 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellizzi D., D’Aquila P., Giordano M., Montesanto A., Passarino G., Global DNA methylation levels are modulated by mitochondrial DNA variants. Epigenomics 4, 17–27 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Mossman J. A., et al. , Mitonuclear interactions mediate transcriptional responses to hypoxia in Drosophila. Mol. Biol. Evol. 34, 447–466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunham-Snary K. J., et al. , Mitochondrial - nuclear genetic interaction modulates whole body metabolism, adiposity and gene expression in vivo. EBioMedicine 36, 316–328 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feeley K. P., et al. , Mitochondrial genetics regulate breast cancer tumorigenicity and metastatic potential. Cancer Res. 75, 4429–4436 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fetterman J. L., et al. , Mitochondrial genetic background modulates bioenergetics and susceptibility to acute cardiac volume overload. Biochem. J. 455, 157–167 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]