Abstract
Histone modifications represent an innate cellular mechanism to link nutritional status to gene expression. Metabolites such as acetyl-CoA and S-adenosyl methionine influence gene expression by serving as substrates for modification of histones. Yet, we lack a predictive model for determining histone modification levels based on cellular metabolic state. The numerous metabolic pathways that intersect with histone marks makes it highly challenging to understand their interdependencies. Here, we highlight new systems biology tools to unravel the impact of nutritional cues and metabolic fluxes on histone modifications.
Keywords: Metabolism, acetylation, methylation, histone modifications, metabolic modeling, systems biology, HDACs, HDAC inhibitors
Commentary on: Shen F, Boccuto L, Pauly R, Srikanth S, Chandrasekaran S. Genome-scale network model of metabolism and histone acetylation reveals metabolic dependencies of histone deacetylase inhibitors. Genome Biol. 2019;20(1):49. doi:10.1186/s13059-019-1661-z. PubMed PMID: 30823893. PubMed Central PMCID: PMC6397465. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6397465/.
Introduction
Cells rely on chemical modification of histones and DNA to change chromatin accessibility and regulate their transcriptional program. The levels of these modifications in a cell involve the interplay between “writers” such as histone methyltransferases and acetyltransferases and “erasers” such as histone demethylases and deacetylases. The writer and eraser enzymes differ in substrate and target specificity, making them sensitive to different metabolic stimuli1,2 (Figure 1). For example, DNA and histone methyltransferases use the metabolite S-adenosyl methionine (SAM) as a methyl donor and are highly responsive to substrate levels.3 Thus, alterations to metabolism of the cell can affect substrate levels in the nucleus resulting in altered histone modifications.2,4 However, the interconnectedness of the metabolic network makes it highly challenging to predict the impact of nutrient changes on histone modifications. For example, acetylation is sensitive to acetyl-CoA and methylation is sensitive to SAM1,2; these metabolites are involved in hundreds of metabolic reactions that can compete with histone modification enzymes for these substrates. A systems biology approach is necessary to comprehend the impact of cellular metabolic state on the epigenome.
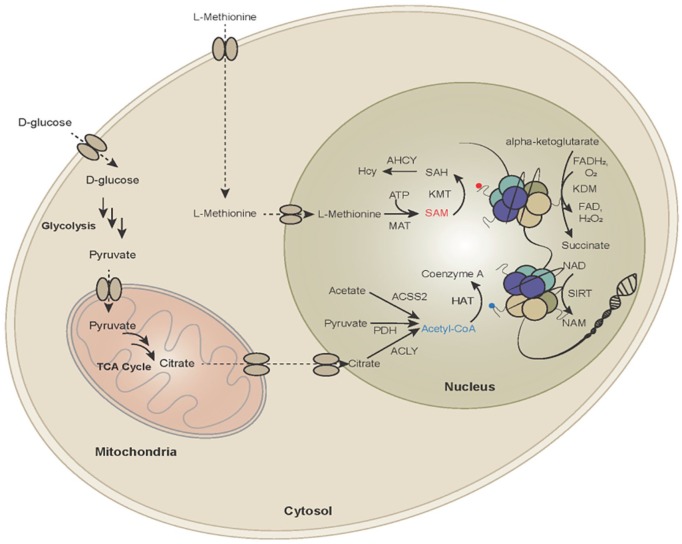
Figure 1.
Metabolic dependencies of histone acetylation and methylation. Enzymes that catalyze post-translational modifications of histone tails (writers—histone methyltransferases [KMT] and acetyltransferases (HAT or KAT), erasers—histone demethylases (KDM) and deacetylases (HDAC, SIRT)) use key metabolic intermediates (SAM, Acetyl-CoA) as substrates). The availability and flux of these metabolic substrates in the nucleus impact levels of histone modifications, which subsequently influences gene expression, thus linking gene regulation with the metabolic status of the cell.
Genome scale network modeling of metabolic-epigenetic interactions
To comprehensively account for the complexities of cellular metabolism and its impact on acetylation, we recently developed a network model of metabolism and histone acetylation.3 Our model provides a mechanistic picture of how nutrient shifts or mutations in metabolic enzymes affect this epigenetic mark. Our analysis makes use of genome-scale metabolic network models, which represent the mechanistic relationships between genes, proteins, and metabolites within a biological system.5 The genome-scale network models are manually curated from the literature and represent a map of all known metabolic reactions that happen in an organism. For example, the human network model by Duarte et al6 contains 3744 reactions, 1496 genes, 2004 proteins, and 2766 metabolites. Genome scale metabolic modeling has been used successfully to predict the metabolic state of various mammalian cell types, including cancer cells and stem cells, using transcriptomics or metabolomics data.7,8 However, these models cannot predict the impact of metabolism on other cellular processes, especially epigenetic modifications.
The metabolic network interfaces with the epigenetic machinery through key intermediate metabolites like acetyl-CoA. To integrate epigenetic modifications with the metabolic model, we added biochemical reactions corresponding to acetylation, as a proof of principle. Similar to using Google Maps, with genome-scale modeling, we can identify an optimal path through the metabolic network from nutrients to epigenetic substrates based on reaction stoichiometry. Changes in enzyme activity due to gene knockouts, differential expression, or drug inhibition will result in the use of alternate metabolic routes through the network leading to differential flux through epigenetic reactions. Although this network modeling framework was used to study acetylation in our study, this can be easily extended to other histone marks. This network model provided new fundamental insights on metabolic-epigenetic interactions, as detailed below.
Excess carbon flux supports acetylation
Given a unit of nutrient carbon, where would the cell channel it? To biomass components or to acetylation? By analyzing the impact of various nutrient sources on the levels of histone acetylation in mammalian cells, our model revealed that histone acetylation levels reflect the level of excess carbon flux that is available beyond the requirement for biomass synthesis. The metabolic model recapitulated known effect of addition or depletion of nutrients, including glucose, amino acids and fatty acids, on acetylation. Surprisingly, certain starvation conditions resulted in increased acetylation. For example, lack of amino acids in the culture media did not reduce acetylation. However, lack of glucose and pyruvate greatly impaired acetylation levels. These results suggest that excess carbon availability relative to nitrogen is predictive of acetylation levels. Cells primarily use nutrient carbon and nitrogen sources for biomass synthesis first, and excess carbon, if available, is used for supporting acetylation.
Metabolic cost of acetylation
There are a billion potential acetylation sites in the mammalian nucleus.9 Although this is a very big number, an equal number of acetyl-coA units are synthesized and consumed by the cell in an hour in the cytoplasm.10 As only a small number of histones are acetylated,11 redirecting a small fraction of cytosolic carbon flux used for acetyl-coA metabolism to the nucleus is sufficient to support acetylation. Thus, the burden of histone acetylation on cellular metabolism is relatively small. In contrast to the flux of acetyl-coA, the steady-state concentration of acetyl-coA is relatively low.10 Hence, increased acetylation will likely be accompanied by rewiring of metabolic fluxes.
Metabolic state influences drug sensitivity
As acetylation depends on the metabolic state of the cell, it is foreseeable that the efficacy of drugs that disrupt histone acetylation will also be influenced by cellular metabolic state. Histone deacetylase inhibitors are widely used for treating many cancers, neurodegenerative diseases, and immune disorders.12 Their primary mode of action is by blocking histone deacetylation, resulting in hyper acetylation, and subsequently cell death.
We found that the growth inhibition of HeLa cells by Vorinostat—a deacetylase inhibitor—changed significantly when cells were grown in different nutrient conditions. Strikingly, the extent of growth inhibition was directly proportional to the extent of acetylation flux predicted by our metabolic model in each growth condition. This effect was also observed in a panel of diverse cell lines from different tissue lineages with inherently distinct metabolic activities. Cell lines predicted by the model to have high acetylation were more sensitive to various deacetylase inhibitors. This observation can be potentially used to selectively target specific cancer cells based on their metabolism.
Impact of metabolic fluxes on histone methylation
Similar to acetylases, the kinetic properties of histone methyltransferases make them sensitive to levels of their substrate, SAM.4 Hence, histone methylation is also sensitive to cellular metabolism. A similar genome-scale modeling framework was used to predict the impact of metabolic state on methylation in a related study by Chandrasekaran et al.7 As histone methylation plays a central role in early embryonic development, genome scale metabolic modeling was applied to characterize the metabolism of naïve and primed murine pluripotent stem cells. The naïve and primed stem cells represent two fundamental steps during embryonic development with distinct pluripotency potential (i.e. ability to differentiate into various cell types). By combining metabolomics data from each stem cell type with metabolic network models, this study revealed the activation of the one-carbon metabolic pathway in embryonic stem cells transitioning from naïve to primed pluripotent state. The metabolic model predicted that the activation of the one-carbon metabolic pathway would result in increased flux through the synthesis of SAM. The model revealed that this flux supports extensive histone methylation. The model prediction was validated using tracing of 13C labeled glucose and serine. Furthermore, perturbing SAM flux using inhibitors of SAM synthesis lead to altered histone methylation levels in naïve and primed cells consistent with the model predictions. Thus, genome scale modeling enables the simulation of the influence of various metabolic alterations on SAM synthesis and ultimately histone methylation.
Future directions
A key limitation of the model of metabolic-epigenetic interactions in Shen et al3 and Chandrasekaran et al7 is that it is predictive of only bulk histone marks and does not account for modifications in specific histone sites such as K9 or K27. Different histone sites can have differing sensitivities to metabolic substrates. Furthermore, although this model focuses on “writers,” we lack a similar model for deacetylation or demethylation enzymes that are sensitive to the redox molecules NADH and FADH (Figure 1). Future models need to account for the dynamic interplay between writers and erasers in each metabolic condition. Finally, another limitation is that the current model does not account for other chromatin marks such as succinylation, malonylation, crotonylation, and sugar modifications.13 These modifications also use metabolic substrates and are potentially sensitive to metabolic state. Expanding this modeling framework to other modifications and incorporating the competition between different modifications can provide a comprehensive picture of how histone modifications sense and respond to metabolic alterations. Furthermore, understanding the transcriptomic and chromatin state of the cell along with its metabolism can help unravel the feedback regulation of metabolism by histone modifications. Two recent studies have simulated the impact of transcriptome changes due to altered chromatin modifications on metabolic pathways using genome-scale network modeling.14,15 Hence, in combination with the model used in Shen et al, these tools can help simulate the reciprocal regulation of metabolic and epigenetic machinery.
Acknowledgments
I thank Scott Campit for making the figure.
Footnotes
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of Conflicting Interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: SC wrote the manuscript.
ORCID iD: Sriram Chandrasekaran  https://orcid.org/0000-0002-8405-5708
https://orcid.org/0000-0002-8405-5708
References
- 1. Kaelin WG, Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Su X, Wellen KE, Rabinowitz JD. Metabolic control of methylation and acetylation. Curr Opin Chem Biol. 2016;30:52-60. doi: 10.1016/j.cbpa.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shen F, Boccuto L, Pauly R, Srikanth S, Chandrasekaran S. Genome-scale network model of metabolism and histone acetylation reveals metabolic dependencies of histone deacetylase inhibitors. Genome Biol. 2019;20:49. doi: 10.1186/s13059-019-1661-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reid MA, Dai Z, Locasale JW. The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat Cell Biol. 2017;19:1298-1306. doi: 10.1038/ncb3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O’Brien EJ, Monk JM, Palsson BO. Using genome-scale models to predict biological capabilities. Cell. 2015;161:971-987. doi: 10.1016/j.cell.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Duarte NC, Becker SA, Jamshidi N, et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci U S A. 2007;104:1777-1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chandrasekaran S, Zhang J, Sun Z, et al. Comprehensive mapping of pluripotent stem cell metabolism using dynamic genome-scale network modeling. Cell Rep. 2017;21:2965-2977. doi: 10.1016/j.celrep.2017.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shlomi T, Cabili MN, Herrgard MJ, Palsson BO, Ruppin E. Network-based prediction of human tissue-specific metabolism. Nat Biotechnol. 2008;26:1003-1010. doi: 10.1038/nbt.1487. [DOI] [PubMed] [Google Scholar]
- 9. Martinez-Pastor B, Cosentino C, Mostoslavsky R. A tale of metabolites: the cross-talk between chromatin and energy metabolism. Cancer Discov. 2013;3:497-501. doi: 10.1158/2159-8290.CD-13-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fan J, Kamphorst JJ, Mathew R, et al. Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Mol Syst Biol. 2013;9:712. doi: 10.1038/msb.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leroy G, Dimaggio PA, Chan EY, et al. A quantitative atlas of histone modification signatures from human cancer cells. Epigenetics Chromatin. 2013;6:20. doi: 10.1186/1756-8935-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eckschlager T, Plch J, Stiborova M, Hrabeta J. Histone deacetylase inhibitors as anticancer drugs. Int J Mol Sci. 2017;18:E1414. doi: 10.3390/ijms18071414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wellen KE, Thompson CB. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol. 2012;13:270-276. doi: 10.1038/nrm3305. [DOI] [PubMed] [Google Scholar]
- 14. Pacheco MP, John E, Kaoma T, et al. Integrated metabolic modelling reveals cell-type specific epigenetic control points of the macrophage metabolic network. BMC Genomics. 2015;16:809. doi: 10.1186/s12864-015-1984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Salehzadeh-Yazdi A, Asgari Y, Saboury AA, Masoudi-Nejad A. Computational analysis of reciprocal association of metabolism and epigenetics in the budding yeast: a genome-scale metabolic model (GSMM) approach. PLoS ONE. 2014;9:e111686. doi: 10.1371/journal.pone.0111686. [DOI] [PMC free article] [PubMed] [Google Scholar]