Significance
Feeding the increasing world population continues to be a great challenge in agriculture. In this study, we demonstrate that OsRac1 Rho-like GTPase from plants (ROP GTPase), an important signaling protein, promotes rice grain yield at least in part through its promotion of grain size. OsRac1 increases grain size by regulating OsMAPK6 and enhancing cell division, another signaling molecule recently shown to promote cell division and grain size in rice. Hence, this study has established a ROP GTPase signaling pathway that regulates grain size in rice, and has potentially illuminated a strategy for enhancing agricultural productivity.
Keywords: ROP, OsRac1, grain size, grain yield, cell division
Abstract
Grain size is a key factor for determining grain yield in crops and is a target trait for both domestication and breeding, yet the mechanisms underlying the regulation of grain size are largely unclear. Here we show that the grain size and yield of rice (Oryza sativa) is positively regulated by ROP GTPase (Rho-like GTPase from plants), a versatile molecular switch modulating plant growth, development, and responses to the environment. Overexpression of rice OsRac1ROP not only increases cell numbers, resulting in a larger spikelet hull, but also accelerates grain filling rate, causing greater grain width and weight. As a result, OsRac1 overexpression improves grain yield in O. sativa by nearly 16%. In contrast, down-regulation or deletion of OsRac1 causes the opposite effects. RNA-seq and cell cycle analyses suggest that OsRac1 promotes cell division. Interestingly, OsRac1 interacts with and regulates the phosphorylation level of OsMAPK6, which is known to regulate cell division and grain size in rice. Thus, our findings suggest OsRac1 modulates rice grain size and yield by influencing cell division. This study provides insights into the molecular mechanisms underlying the control of rice grain size and suggests that OsRac1 could serve as a potential target gene for breeding high-yield crops.
To meet the food demands of a rapidly growing world population, it is critical to increase crop productivity through efficient breeding and biotechnology (1). Rice is the most important staple food crop, acting as the primary food source for ∼50% of the world’s population (2). Thus, investigation of the genetic basis and molecular mechanism for rice grain yield regulation is of great significance. An important yield trait is grain size, a complex agronomical trait. In recent years, many quantitative loci (QTLs) and genes modulating rice grain size and shape have been identified. For example, GRAIN SIZE 3 (GS3), GL3.1, Grain Length on Chromosome 7 (GL7), GLW7/OsSPL13, and DEP1 have been reported to regulate grain length via the regulation of cell division and expansion (3–7), whereas GRAIN WIDTH AND WEIGHT 2 (GW2), qSW5, GW5, GS5, GW7, and GW8 regulate grain width through activation of cell division (8–13). Other genes such as OsMKK4, OsMAPK6, and OsBG1 promote the increase in both grain length and width to improve rice yield (14–16). GS3, a major QTL for grain length, is a negative factor for grain length by regulating cell division (3). The homolog of GS3 in Arabidopsis is AGG3, which encodes an atypical heterotrimeric G protein subunit. AGG3 also can control seed and organ size by regulating cell division (17). GL7, which is the homolog of Arabidopsis LONGIFOLIA protein, is a positive factor for grain length by regulating cell expansion (5). GW2, which, first being found as a major QTL for grain width, encodes an E3 ubiquitin ligase including the new type RING domain. GW2 functions as a negative factor for grain width by mediating the degradation of substrate in cell division (8). GW8, a QTL that is synonymous with OsSPL16, positively regulates cell division for improving rice grain width and yield (13). However, none of these studies has clearly delineated a pathway that regulates grain size and shape.
ROP GTPases act as molecular switches in cellular signaling and play a critical role in the regulation of plant growth and development, mostly based on the studies on ROP signaling pathways in Arabidopsis, including cell polarity, cell morphogenesis, root hair development, root hair elongation, pollen tube elongation, phytohormone responses, abiotic stress responses, and so on (18–27). Little is known about the function of ROPs in rice growth and development, although OsROPs have been implicated in the regulation of disease resistance (28–30). Among the 7 OsROP members in rice, OsRac1 has been reported to act as a positive regulator of rice disease resistance by regulating OsMAPK6; however, OsRac4, OsRacD, and OsRacB are negative regulators in rice disease responses (28–30). Here we showed that OsRac1 regulates the protein level and activity of OsMAPK6, and then promotes cell division in rice panicle to regulate spikelet size. Thus, OsRac1-OsMAPK6 delineates a signaling pathway that promotes rice grain size and yield, which provides a strategy for improving grain crops.
Results
OsRac1 Modulates Rice Grain Size and Promotes Rice Yield.
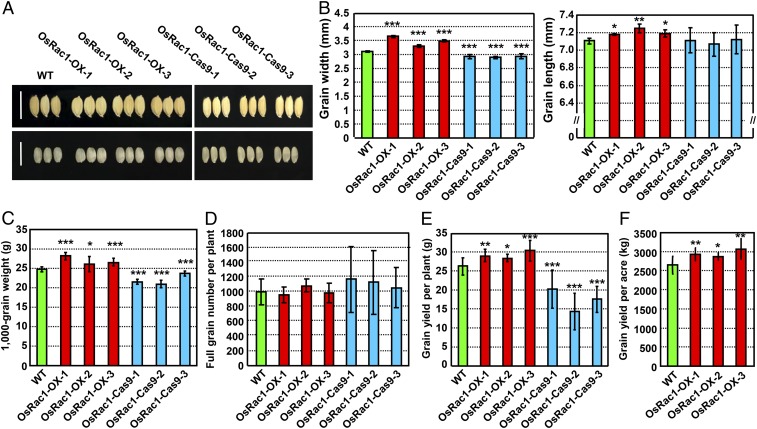
Given a role for Arabidopsis ROP2 in promoting the activation of TOR, a central regulator of growth (24), we speculated that the overexpression of ROPs may improve rice plant growth and grain yield. We generated overexpression and CRISPR-Cas9 knockout lines for various OsROPs in the Nipponbare background (Oryza sativa L. ssp. japonica). In this study, we focused on the OsRac1 overexpression and CRISPR-Cas9 knockout lines (SI Appendix, Fig. S1). Quantitative RT-PCR and promoter::GUS analyses showed that OsRac1 is expressed in spikelets and during the panicle development (SI Appendix, Fig. S2), hinting at its potential role in rice grain development. Phenotypic analyses of OsRac1 transgenic lines suggest that OsRac1 regulates rice grain size, a very important trait for determining rice yield (Fig. 1A). OsRac1-overexpressing lines exhibit larger grains. The width of grains from 3 independent overexpression lines, OsRac1-OX-1, OsRac1-OX-2, and OsRac1-OX-3, was increased by ∼17.3%, 6.2%, and 12.2%, respectively, and the grain length increased by ∼0.9%, 2%, and 1.2%, respectively (Fig. 1B and SI Appendix, Fig. S1A). In contrast, the width of grains from 3 independent CRISPR-Cas9 lines, OsRac1-Cas9-1, OsRac1-Cas9-2, and OsRac1-Cas9-3, was reduced by ∼6%, 6.8%, and 5.4%, respectively, but their grain length was essentially unchanged (Fig. 1B and SI Appendix, Fig. S1B). As a result, compared with WT, the 1,000-grain weight from OsRac1-OX-1, OsRac1-OX-2, and OsRac1-OX-3 increased by 13.6%, 5.3%, and 7.4%, respectively, and those of OsRac1-Cas9-1, OsRac1-Cas9-2, and OsRac1-Cas9-3 deceased by 13.2%, 15.5%, and 4.5%, respectively (Fig. 1C and SI Appendix, Fig. S1). Because the OsRac1-overxpressing lines produced a similar full grain number per plant to WT (Fig. 1D and SI Appendix, Fig. S1A), the grain size changes in these lines are the primary factor for the increased yield. Furthermore, we found the grain yield per plant in the OsRac1-overexpressing lines, OsRac1-OX-1, OsRac1-OX-2, and OsRac1-OX-3, increased by 10.3%, 8.1%, and 15.8%, respectively (Fig. 1E and SI Appendix, Fig. S1A), and those of OsRac1 CRISPR-Cas9 lines, OsRac1-Cas9-1, OsRac1-Cas9-2, and OsRac1-Cas9-3, decreased by 22.8%, 44.9%, and 33.1%, respectively (Fig. 1E and SI Appendix, Fig. S1B). Field trials showed that grain yield per acre in the OsRac1-overexpressing lines increased ∼10.4%, 8.2%, and 15.8%, respectively (Fig. 1F and SI Appendix, Fig. S1A). Therefore, OsRac1 is a novel positive regulator that controls grain size to improve rice yield.
Fig. 1.
OsRac1 promotes grain yield by controlling grain size. (A) Grain morphology of WT and OsRac1 transgenic lines. (Scale bars: 1 cm.) (B) Statistical analysis of the grain width and grain length of WT and OsRac1 transgenic lines. (C–E) Statistical analysis of the 1,000-grain weight (C), full grain number per plant (D), and grain yield per plant (E) of WT and OsRac1 transgenic lines. (F) Statistical analysis of the grain yield per acre of WT and OsRac1 overexpression transgenic lines. The OsRac1 overexpression, OsRac1 CRISPR-Cas9 transgenic lines, and WT control were grown in the same location with at least three replicates, and the mature seeds were harvested at the same time. The two images shown in A were taken from one representative replicate with the identical magnification and exposure. Means ± SD are shown in B (n > 3 × 200), C (n > 10), D (n > 15), E (n > 20), and F (n = 4 × 400). *P < 0.1; **P < 0.01; ***P < 0.001 (t test).
OsRac1 also regulates various agronomic traits of rice, including the productive tillers, seed setting rate, panicle length, primary branches, secondary branches per panicle, and plant height (SI Appendix, Fig. S3). OsRac1 CRISPR-Cas9 lines had more productive tillers per plant (+47.5%, 47.3%, 12.2%), lower seed setting rate (−22.3%, 22.7%, 16.1%), shorter panicle length (−9.9%, 8%, 5.2%), fewer secondary branches per panicle (−24.1%, 15.7%, 18.8%), and shorter plant height (−12.8%, 7.1%, 6.3%) (SI Appendix, Fig. S3). OsRac1-overexpressing lines had less productive tillers per plant (−12.7%, 1.5%, 15.4%), shorter panicle length (−3.7%, 3.4%, 5.9%), more primary branches per panicle (+11.9%, 11.2%, 11.7%), and shorter plant height (−2.2%, 0, 2.7%). However, changes in these traits, together with similar full grain number per plant, made no contribution for OsRac1-overexpressing lines to increase grain yield. Thus, grain size is the main factor for OsRac1 to improve rice yield (Fig. 1D and SI Appendix, Fig. S3).
OsRac1 Modulates Grain Size by Regulating Cell Number in Grain Spikelet.
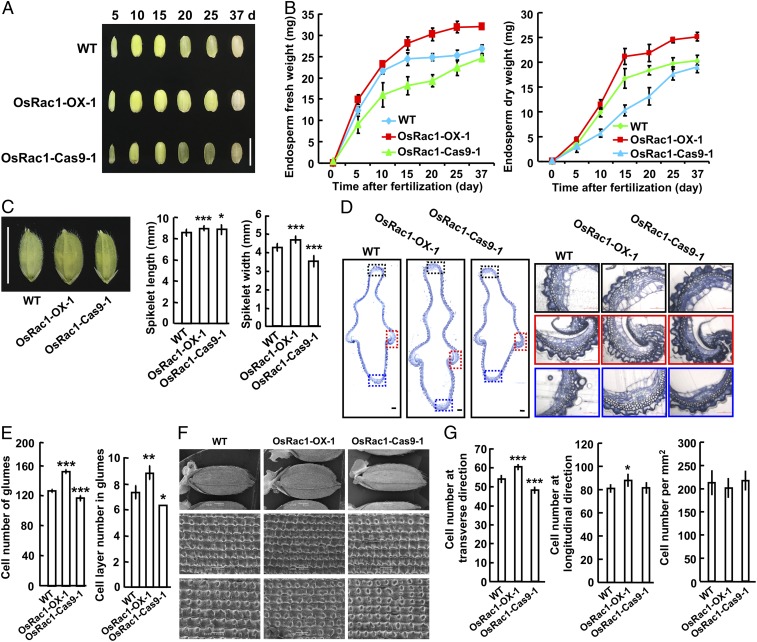
Grain milk filling rate indicates the increase in fresh weight or dry weight per day during the grain filling stage, and is a key factor that controls rice grain size (8, 11, 13). We performed a time course measurement of both fresh and dry weights of filling grains (Fig. 2A) and found an increase in the filling rate in the OsRac1-overexpressing lines, especially during the late filling stage. In contrast, the grain filling rate in the OsRac1-Cas9 lines was lower compared with WT (Fig. 2B). Therefore, OsRac1 promotes rice grain milk filling rate to increase seed plumpness.
Fig. 2.
OsRac1 controls grain filling rate and spikelet size. (A) The filling grains of WT and OsRac1 transgenic lines at 5, 10, 15, 20, 25, and 37 d. (Scale bar: 1 cm.) (B) Time course of WT and OsRac1 transgenic lines endosperm fresh weight and endosperm dry weight. (C) The morphology and statistical analysis of spikelet hulls length and width of WT and OsRac1 transgenic lines. (Scale bar: 1 cm.) (D) Cross-sections of the spikelet hulls of WT and OsRac1 transgenic lines. (Scale bar: 50 µm.) (E) Statistical analysis of the cell number and layer number at the outer parenchyma layer of the spikelet hulls of WT and OsRac1 transgenic lines. (F) Microscope scanning of the glume outer surfaces of WT and OsRac1 transgenic mature seeds. (Upper) Glumes. (Scale bar: 1 mm.) (Middle) Palea. (Scale bar: 100 µm.) (Bottom) Lemma. (Scale bar: 100 µm.) (G) Statistical analysis of the cell number in the transverse, longitudinal direction and per millimeter squared of the outer surfaces of WT and OsRac1 transgenic grains. Means ± SD are shown in B (n > 3), C (n = 18), E (n > 3), and G (n =10). *P < 0.1; **P < 0.01; ***P < 0.001 (t test).
We next investigated the cellular mechanism by which OsRac1 modulates grain size. Spikelet size is another key factor tightly associated with rice grain size. First, we observed the size and cell number of grain spikelets just before fertilization and grain filling (Fig. 2C and SI Appendix, Fig. S4). The spikelet width and length in the OsRac1-overexpressing lines greatly increased (e.g., the spikelet width and length in OsRac1-OX-1 increased by 10.7% and 4.8%, respectively). At the same time, the spikelet width in the OsRac1-Cas9 lines greatly decreased (e.g., by 17.1% in OsRac1-Cas9-1), but the spikelet length in OsRac1-Cas9-1 increased by 3.3% (Fig. 2C). Thus, OsRac1 levels affect the spikelet size before grain filling. We then examined the cross-sections of the central parts of grain spikelet hulls before heading stage and found that OsRac1 overexpression greatly increased the number and layer of parenchyma cells in spikelets, whereas CRISPR-mediated OsRac1 knockout had an opposite effect (Fig. 2 D and E). Scanning electron microscopy analysis showed that the epidermal cell number in both transverse and longitudinal directions of seeds after grain filling was increased by 12% and 8.6% in OsRac1-OX-1, respectively, whereas the epidermal cell number in the transverse direction of seed epidermal in OsRac1-Cas9-1 decreased by 10.8% (Fig. 2 F and G). However, the cell number per millimeter squared in the seed epidermis did not show any difference between WT and OsRac1 transgenic lines (Fig. 2G), implying a role for OsRac1 in the promotion of cell division, but not cell expansion, in influencing the spikelet size.
OsRac1 Promotes Cell Division in the Young Panicle.
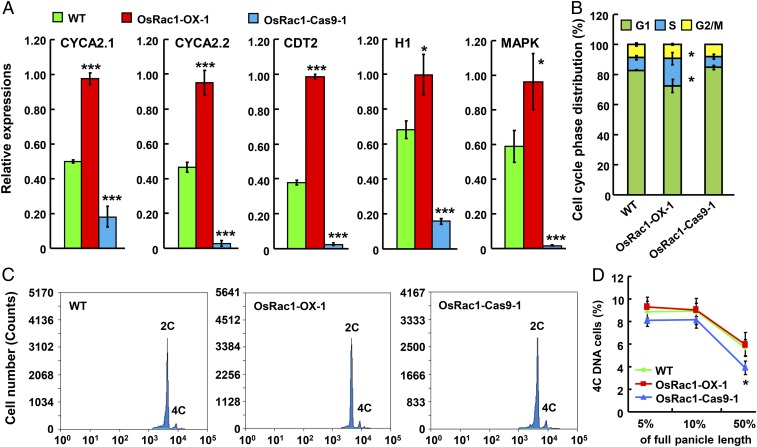
We next investigated whether OsRac1 promotes cell division. Analysis of transcriptome profiles during rice young panicle growth from 0.5 to 6 cm in length showed that different from WT (SI Appendix, Fig. S5A), genes related to DNA metabolic processes, cellular component organization, biosynthetic processes, and signal transduction were obviously up-regulated in 6-cm panicles in the OsRac1-overexpressing lines (SI Appendix, Fig. S5B), supporting a role for OsRac1 in promoting cell division. Furthermore, qRT-PCR analysis confirmed the changes in the expression of cell cycle-related genes including CYCA2.1, CYCA2.2, CDT2, H1, and MAPK in the OsRac1 transgenic lines, which were significantly up-regulated or suppressed under OsRac1 overexpression or knockout, respectively (Fig. 3A).
Fig. 3.
OsRac1 regulates cell division in rice panicle. (A) Quantitative real-time PCR analysis of cell cycle-related gene expression in the 4-cm-long young panicles of WT and OsRac1 transgenic lines. (B) Percentage comparison of the distribution of cells in different phases of the cell cycle in young panicle cells (the panicle length reached 5% of full panicle). The G1, S, and G2/M phases are shown in colored boxes. (C) The cell number (counts) that contain 2C DNA and 4C DNA in young panicles (the panicle length reached 5% of full panicle) of WT and OsRac1 transgenic lines by flow cytometry analysis. (D) Flow cytometry analysis of the young panicles at 3 developmental stages (the panicle length reached 5%, 10%, and 50% of full panicle) of WT and OsRac1 transgenic lines. The percentage of cells with 4C DNA content was determined as a proportion of the total number of isoploid and tetraploid nuclei. Means ± SD are shown in A (n = 3), B (n = 3), and D (n = 3). *P < 0.1; **P < 0.01; ***P < 0.001 (t test).
To confirm that increased cell division is responsible for the larger spikelet hulls of OsRac1-overexpressing lines, we analyzed the cell division rate of young panicles at different developmental stages by flow cytometry (4, 31). Measurement of DNA contents in the cells at the stages when the panicle length reached 5%, 10%, and 50% of full panicle showed that at the early stage (5% length), the percentage of S and G2/M phase cells with higher DNA content was elevated in OsRac1-OX-1, whereas the percentage of G1 phase cells with 2C DNA content decreased after new cell cycles were initiated (Fig. 3 B and C). Consistently, the percentage of S and G2/M phase cells with higher DNA content was decreased in OsRac1-Cas9-1, and the percentage of G1 phase cells with 2C DNA content was increased (Fig. 3 B and D). This change was greater at the late stage (50% length; Fig. 3D). Thus, we conclude that cell division rate was increased in the OsRac1-overexpressing lines and decreased in the OsRac1 CRISPR mutant lines during spikelet development compared with the WT. These results confirmed that OsRac1 promotes cell division at the early stage of spikelet development and demonstrated that OsRac1 positively regulates cell division, but not cell elongation, in rice panicles to increase spikelet size.
OsRac1 Regulates OsMAPK6 to Control Grain Size.
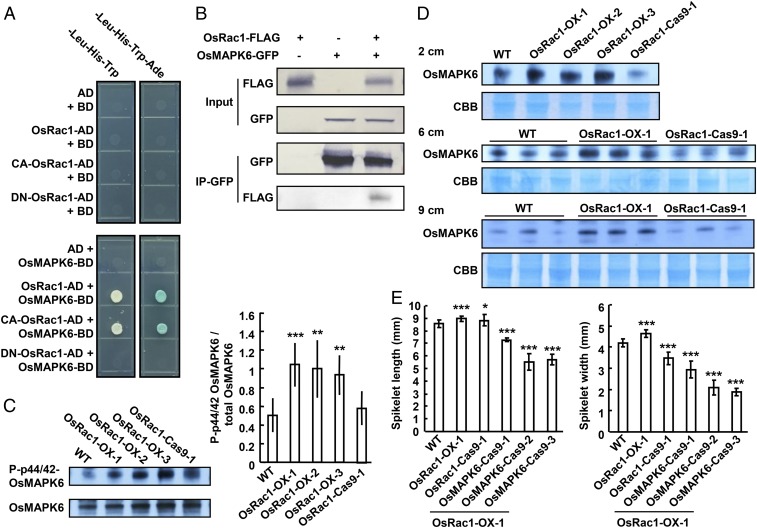
To investigate the molecular mechanism by which OsRac1 regulates cell division, we sought to identify OsRac1-interacting proteins using yeast 2-hybrid screening, and obtained 10 candidates. Among those candidates, OsMAPK6 has been reported to be an important factor in regulating rice grain size (15). OsMAPK6 interacted with OsRac1, especially a constitutively active form of OsRac1 (CA-OsRac1) in yeast cells (Fig. 4A). Coimmunoprecipitation assay using rice protoplasts transiently expressing the fusion proteins confirmed that OsMAPK6 interacts with OsRac1 in vivo (Fig. 4B). In addition, the OsMAPK6 transcript is ubiquitously expressed in all tissues examined with a relatively higher expression level in panicles (SI Appendix, Fig. S6).
Fig. 4.
OsRac1 regulates rice panicles through OsMAPK6. (A) Analysis of protein interactions by yeast 2-hybrid system. Proteins OsRac1, CA-OsRac1, DN-OsRac1, and OsMAPK6 are analyzed. AD, pGADT7; BD, pGBKT7; CA, constitutively active; DN, dominant negative; -Leu-His-Trp SD medium, 3AT = 1 mg/L; -Leu-His-Trp-Ade SD medium, 3AT = 1 mg/L. (B) Coimmunoprecipitation between OsRac1 and OsMAPK6 in rice protoplasts. (C) The phosphorylation level of OsMAPK6 in WT and OsRac1 transgenic lines. The loading control was OsMAPK6 protein. (D) OsMAPK6 protein levels in 2-, 6-, and 9-cm young panicles of WT and OsRac1 transgenic lines. The loading control was total protein. CBB, Coomassie brilliant blue. (E) Statistical date of spikelet hulls length and width of WT, OsRac1 transgenic lines, and OsMAPK6-Cas9 transgenic lines (under the OsRac1-OX-1 background). Means ± SD are shown in C (n > 3) and E (n > 3). *P < 0.1; **P < 0.01; ***P < 0.001 (t test).
The preferential interaction of OsMAPK6 with the active form of OsRac1 suggests OsMAPK6 may act as a downstream effector of OsRac1, because GTPase effectors generally interact with the active form of GTPase (32). Interestingly, the previous study suggests that OsMAPK6 also positively controls rice grain size by regulating cell division (15). Thus, we tested the hypothesis that OsMAPK6 acts downstream of OsRac1. We compared the phosphorylation level of OsMAPK6 in young panicles between WT and the OsRac1 transgenic lines by using the phospho-p44/42 MAPK antibody (31), which can detect the phosphorylation level of OsMAPK6 on Thr202/Tyr204 or Thr185/Tyr187 sites in vivo. As shown in Fig. 4C and SI Appendix, Fig. S7A, the phosphorylation level of OsMAPK6 was elevated in the panicles of the OsRac1-overexpressing lines compared with WT. OsRac1 overexpression also increased OsMAPK6 protein level (Fig. 4D), but not its transcript level (SI Appendix, Fig. S7B). To further test our hypothesis that OsMAPK6 acts downstream of OsRac1, we knocked out OsMAPK6 in an OsRac1-overexpressing line by the CRISPR-CAS9 system (SI Appendix, Fig. S7C). In those osmapk6 mutant lines, the spikelet size was smaller than that of the WT and OsRac1-overexpressing line (Fig. 4E), confirming that OsRac1 modulates rice grain size through OsMAPK6 by regulating cell division. In conclusion, the OsRac1-OsMAPK6 signaling pathway plays a vital role both in promoting rice growth and grain yields.
Discussion
Recent studies have identified a series of genes that affect rice grain yields through regulating the cell division of glumes in the spikelet (3, 4, 8–13), suggesting a link between the cell division and seed yield. However, little is known about the mechanisms behind the connection between cell division and seed yields, as well as the regulatory mechanisms underlying cell division that affect seed yield. As an important source organ, spikelet hull plays crucial roles in grain development, filling, and hence yield. Studies have shown that size and shape of rice grains are strictly controlled by spikelet hull (33). In this study, we found that OsRac1, a member of the highly conserved ROP/Rac small GTPase family in plants, promotes rice grain size and yield (Fig. 1). Importantly OsRac1 promotes cell division in the rice young spikelet to increase the spikelet size and then the grain filling rate (Fig. 2), resulting in the increased grain size and yield. Hence, our findings not only identify OsRac1 as a key regulator of grain size by acting on cell division control but also suggest a strategy for grain size modification in a wide range of cereal crops for yield improvement.
Our biochemical and genetic studies show that OsRac1 acts directly upstream of OsMAPK6 to promote cell division and rice grain size (Fig. 4). These findings establish a signaling pathway that promotes grain size and yield in rice via the regulation of cell division in the rice young spikelet. This pathway will provide an important stepping stone for identifying signals and downstream mechanisms that control cell division, and consequently seed yields. ROP/Rac GTPases are activated by the plant-specific RopGEFs (Rop guanine nucleotide exchange factor), which are directly regulated by receptor-like kinases (RLKs) such as TMKs, PRKs, and CrRLK1Ls, the most important plant cell membrane receptors (19, 34–37). It would be interesting to know whether RLKs regulate OsRac1 to affect cell division and yield in rice. MAPKs play a key role in regulating cell cycle/cell division (both meiosis and mitosis, refs. 38 and 39). MAPK6 is localized to mitotic microtubules, secretory TGN (trans-Golgi network), and the plasma membrane, and is involved in cell division plane control in Arabidopsis (40). Interestingly, recent reports showed that the function of OsMAPK6 in regulating grain size is dependent on its phosphorylation activity. Rice lines with deficiency in MAPK phosphorylation activity display decreased grain size and reduced expression levels of some cell division effectors (15, 31). Thus, future studies should be directed to understand exactly how OsMAPK6 and its phosphorylation activity regulate cell division in rice panicles, and whether the Rac1-MAPK6 pathway provides a conserved mechanism for promoting cell division and grain yields in different plant species.
Our findings that OsMAPK6 acts as a direct effector of OsRac1 are significant with regard to signaling mechanisms. MAPK is well known to be the last component of the MAPK cascade (41). The direct regulation of OsMAPK6 by OsRac1 represents a mechanism for the regulation of the MAPKs, which have been shown to control a large number of developmental and physiological responses in plants (15, 38–44). An interesting question is whether the potential RLK-ROP/Rac-MAPK pathway provides a widespread signaling mechanism in plants.
It is worthy to note that the OsRac1-mediated RbohB-ROS signaling pathway and the OsMAPK6-PAL1-OsWRKY19 signaling pathway are known to promote rice disease resistance (29, 45). How can OsRac1 positively affect both yield and disease traits, as genes conferring disease resistance are generally considered to have a grain yield penalty. For general signaling proteins such as ROPs and MAPKs, pathways compartmentation is a typical mechanism for their functional specialization (i.e., OsRac1 and OsMAPK6 may be partitioned into different physically separated complexes that, respectively, control cell division and disease resistance). It is possible that they could be shifted toward the disease resistance pathway when rice plants are under pathogen attack, causing yield penalty. Thus, it would be important to determine whether the OsRac1-overexpressing lines are able to produce higher yield in the presence of pathogen infection than that of WT control in the absence of a pathogen. Nonetheless, understanding how the OsRac1-OsMAPK6 pathway is involved in coordinating and balancing disease responses and yield improvement of rice should be an exciting future direction.
Materials and Methods
Details of experimental procedures, including transformation method, trait measurements, genes expression detection, RNA-seq and GO term analysis, GUS staining, nucleus isolation and assessment of ploidy, yeast 2-hybrid screening and assay, coimmunoprecipitation, and analysis of OsMAPK6 phosphorylation level are available in SI Appendix, Materials and Methods. All primers and antibodies used in the study are listed in SI Appendix, Tables S1–S5.
Supplementary Material
Acknowledgments
We thank professor Hong-Xuan Lin at Shanghai Institute of Plant Physiology and Ecology (SIPPE) for helpful discussions and suggestions. We also thank Yanlin Liu at Fujian Agriculture and Forestry University and Zhongtian Xu at Shanghai Center for Plant Stress Biology, CAS, for their help on OsMAPK6 activity detection, and RNA-Seq data analysis, respectively. We thank Ji-Qin Li and Wen-Fang Zhao at SIPPE for assistance with spikelet paraffin section and flow cytometry, respectively. The study is supported by the National Transformation Science and Technology Program (2016ZX08001006-009) and the National Key Research and Development Program of China (2016YFD0100501, 2016YFD0100902).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.J.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902321116/-/DCSupplemental.
References
- 1.Godfray H. C., et al. , Food security: The challenge of feeding 9 billion people. Science 327, 812–818 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Rosegrant M. W., Cline S. A., Global food security: Challenges and policies. Science 302, 1917–1919 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Mao H., et al. , Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. U.S.A. 107, 19579–19584 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qi P., et al. , The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 22, 1666–1680 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., et al. , Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47, 944–948 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Si L., et al. , OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Huang X., et al. , Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41, 494–497 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Song X. J., Huang W., Shi M., Zhu M. Z., Lin H. X., A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Shomura A., et al. , Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40, 1023–1028 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Weng J., et al. , Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 18, 1199–1209 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Li Y., et al. , Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Wang S., et al. , The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47, 949–954 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Wang S., et al. , Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Duan P., et al. , SMALL GRAIN 1, which encodes a mitogen-activated protein kinase kinase 4, influences grain size in rice. Plant J. 77, 547–557 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Liu S., et al. , OsMAPK6, a mitogen-activated protein kinase, influences rice grain size and biomass production. Plant J. 84, 672–681 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Liu L., et al. , Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc. Natl. Acad. Sci. U.S.A. 112, 11102–11107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S., et al. , Roles of the Arabidopsis G protein γ subunit AGG3 and its rice homologs GS3 and DEP1 in seed and organ size control. Plant Signal. Behav. 7, 1357–1359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones M. A., et al. , The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14, 763–776 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan Q., Kita D., Li C., Cheung A. Y., Wu H. M., FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. U.S.A. 107, 17821–17826 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu Y., Vernoud V., Fu Y., Yang Z., ROP GTPase regulation of pollen tube growth through the dynamics of tip-localized F-actin. J. Exp. Bot. 54, 93–101 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Li H., Lin Y., Heath R. M., Zhu M. X., Yang Z., Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11, 1731–1742 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin Y., Yang Z., Rapid tip growth: Insights from pollen tubes. Semin. Cell Dev. Biol. 22, 816–824 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu T., et al. , Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143, 99–110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X., et al. , Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes. Proc. Natl. Acad. Sci. U.S.A. 114, 2765–2770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poraty-Gavra L., et al. , The Arabidopsis Rho of plants GTPase AtROP6 functions in developmental and pathogen response pathways. Plant Physiol. 161, 1172–1188 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Z. L., et al. , Plasma membrane-associated ROP10 small GTPase is a specific negative regulator of abscisic acid responses in Arabidopsis. Plant Cell 14, 2787–2797 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu F., et al. , FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proc. Natl. Acad. Sci. U.S.A. 109, 14693–14698 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ono E., et al. , Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. U.S.A. 98, 759–764 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim S. H., et al. , The bHLH Rac Immunity1 (RAI1) is activated by OsRac1 via OsMAPK3 and OsMAPK6 in rice immunity. Plant Cell Physiol. 53, 740–754 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Chen L., et al. , Analysis of the Rac/Rop small GTPase family in rice: Expression, subcellular localization and role in disease resistance. Plant Cell Physiol. 51, 585–595 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Guo T., et al. , GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 30, 871–888 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Z., Small GTPases: Versatile signaling switches in plants. Plant Cell 14 (suppl), S375–S388 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li N., Li Y., Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 33, 23–32 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Trotochaud A. E., Hao T., Wu G., Yang Z., Clark S. E., The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell 11, 393–406 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai N., Wang W., Patterson S. E., Bleecker A. B., The TMK subfamily of receptor-like kinases in Arabidopsis display an essential role in growth and a reduced sensitivity to auxin. PLoS One 8, e60990 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu Y., et al. , Arabidopsis PRK6 interacts specifically with AtRopGEF8/12 and induces depolarized growth of pollen tubes when overexpressed. Sci. China Life Sci. 61, 100–112 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Kessler S. A., Lindner H., Jones D. S., Grossniklaus U., Functional analysis of related CrRLK1L receptor-like kinases in pollen tube reception. EMBO Rep. 16, 107–115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jonak C., Páy A., Bögre L., Hirt H., Heberle-Bors E., The plant homologue of MAP kinase is expressed in a cell cycle-dependent and organ-specific manner. Plant J. 3, 611–617 (1993). [DOI] [PubMed] [Google Scholar]
- 39.Calderini O., et al. , A cell cycle regulated MAP kinase with a possible role in cytokinesis in tobacco cells. J. Cell Sci. 111, 3091–3100 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Müller J., et al. , Arabidopsis MPK6 is involved in cell division plane control during early root development, and localizes to the pre-prophase band, phragmoplast, trans-Golgi network and plasma membrane. Plant J. 61, 234–248 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Garrington T. P., Johnson G. L., Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr. Opin. Cell Biol. 11, 211–218 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Wang P., Du Y., Li Y., Ren D., Song C. P., Hydrogen peroxide-mediated activation of MAP kinase 6 modulates nitric oxide biosynthesis and signal transduction in Arabidopsis. Plant Cell 22, 2981–2998 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.López-Bucio J. S., et al. , Arabidopsis thaliana mitogen-activated protein kinase 6 is involved in seed formation and modulation of primary and lateral root development. J. Exp. Bot. 65, 169–183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu Q., Jackson D., Detection of MAPK3/6 phosphorylation during hypersensitive response (HR)-associated programmed cell death in plants. Methods Mol. Biol. 1743, 153–161 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Nagano M., et al. , Plasma membrane microdomains are essential for Rac1-RbohB/H-mediated immunity in rice. Plant Cell 28, 1966–1983 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.