Significance
The network of genes and molecules that govern sleep and wakefulness remains largely unknown. Forward genetics is a powerful approach to elucidate important biological phenomena that cannot be predicted from the function of known genes. We established a large-scale screening system using EEG/EMG-based sleep/wake monitoring with reliable parameters. The search for novel genes has succeeded in identifying several sleep regulatory genes. By furthering the study, a more comprehensive picture of the regulatory pathways of sleep/wakefulness will become clear.
Keywords: ENU mutagenesis, dominant screening, linkage analysis, C57BL/6 substrains, Cacna1a
Abstract
The regulatory network of genes and molecules in sleep/wakefulness remains to be elucidated. Here we describe the methodology and workflow of the dominant screening of randomly mutagenized mice and discuss theoretical basis of forward genetics research for sleep in mice. Our high-throughput screening employs electroencephalogram (EEG) and electromyogram (EMG) to stage vigilance states into a wake, rapid eye movement sleep (REMS) and non-REM sleep (NREMS). Based on their near-identical sleep/wake behavior, C57BL/6J (B6J) and C57BL/6N (B6N) are chosen as mutagenized and counter strains, respectively. The total time spent in the wake and NREMS, as well as the REMS episode duration, shows sufficient reproducibility with small coefficients of variance, indicating that these parameters are most suitable for quantitative phenotype-driven screening. Coarse linkage analysis of the quantitative trait, combined with whole-exome sequencing, can identify the gene mutation associated with sleep abnormality. Our simulations calculate the achievable LOD score as a function of the phenotype strength and the numbers of mice examined. A pedigree showing a mild decrease in total wake time resulting from a heterozygous point mutation in the Cacna1a gene is described as an example.
Nearly one-third of human life is spent sleeping; sleep has a profound effect on mental and physical well-being. No animal species with a central nervous system has been demonstrated to sustain life without a sleep-like quiescent state. Mammalian sleep exhibits a cyclic transition between REMS and NREMS with characteristic brain activity and muscle tone, as measured by EEG and EMG, respectively. Although recent advances in the optogenetic and chemogenetic manipulations of neurons have led to the accumulation of information about executive neural circuitries regulating sleep and wake states (1), the molecular and cellular mechanisms that drive the switch between wakefulness, NREMS, and REMS are not elucidated to date.
Forward genetic screening based on a heritable phenotype is useful for searching the causal genetic change without requiring a specific working hypothesis (2–4). In behavioral neuroscience, forward genetic studies led to a groundbreaking series of discoveries using fruit flies (5, 6). The discovery of the period gene fueled application of forward genetics in both flies (7) and mice (8) to identify many fundamental genes that contribute to the current knowledge of the clock mechanism (9–11). Ever since the discovery that flies exhibit behavior states that qualify as sleep/wakefulness (12, 13), an intensive effort has been made to decipher sleep through the fly genetics (14). Currently in flies, however, sleep can only be assessed through monitoring the locomotive activities. It is unclear whether sleep in flies is truly analogous to mammalian sleep, in which the gold standard in defining and assessing sleep is through EEG/EMG analysis.
The advantages of sleep research using mice over fruit fly include reliable staging of three vigilance states, wakefulness, NREMS, and REMS, through EEG/EMG-based somnography analysis. Furthermore, the high similarity of the genome and brain structure between mouse and human predicts a conserved mechanism regulating sleep/wakefulness. We expect that findings in mice can be directly applied to human sleep physiology and pathology. For example, our research on orexin-deficient mice led to the understanding of the pathophysiology of the human sleep disorder narcolepsy (15, 16) and the development of a novel class of drug for insomnia (17). Thus, the use of mouse models has facilitated extension of research into human disease and drug discoveries, further emphasizing the utility of the murine model in translational studies.
Nonetheless, there are major obstacles when undertaking forward genetics research using adult mice in comparison with fruit flies. Mice have generation time of at least 3 mo, which is considerably longer than the fruit fly (10–12 d), and must be kept under a tightly controlled environment. This requires a large amount of labor and is costly. Also, the large-scale sleep analysis needs well-trained research staff for implantation surgery and EEG/EMG-based sleep staging. Despite these difficulties, advances in bioinformatics and genome editing over the last 10 y have improved the speed and feasibility of the forward genetics research in mice. Traditionally, counter strains genetically very distant from the mutagenized strain were preferred because polymorphic markers were more readily available. However, the availability of the complete mouse genome sequence with genome-wide list of genetic markers between B6J and B6N (18), together with the next-generation sequencing, has made use of such distant counter strains and fine-grained linkage analyses unnecessary. This allows us to use mutagenized and counter strains with genetic proximity and near-identical phenotype (19, 20). Whole-exome sequencing combined with coarse linkage analyses can immediately provide a list of candidate gene mutations. Importantly, the recently developed CRISPR/Cas9 system is extremely useful for the examination of genetic causality since the identified mutations are usually single nucleotide substitutions.
In this study, we describe the methods and workflow for the large-scale, high-throughput screening of randomly mutagenized mice for sleep/wake behaviors. We also provide a theoretical discussion on the relationship among the LOD score, statistical significance, and the extent of the sleep phenotype. In addition, a pedigree, Drowsy, is described which carries a mutation in the Cacna1a gene. Our screening scheme and statistical simulation of the phenotype are modifiable for any type of behavior and physiological screening other than sleep/wakefulness.
Results and Discussion
Screening Strategy.
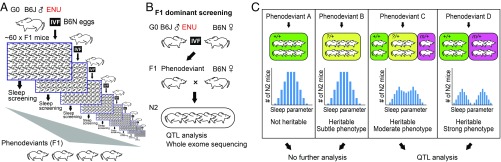
In our dominant screening, F1 offspring from a cross between the mutagenized G0 father (B6J) and wild-type counter strain mother (B6N) are screened for sleep/wake phenotype. The screening is set to examine up to 60 male F1 mice per week (Fig. 1A). The F1 phenodeviant is subsequently backcrossed with wild-type B6N female to obtain N2 progeny for assessing heritability of the sleep abnormality (Fig. 1B) (SI Appendix, Choice of Age and Sex of the Animals). In most cases (43 out of 57 pedigrees), the N2 generation did not inherit the presumptive phenotype seen in F1 and showed normal sleep/wake behavior (Fig. 1C, phenodeviant A). This may be due to a deviation by chance in F1 mice or a summed weak effect diluted by mating with wild-type mouse. Depending on the phenotype strength, we can partially or completely categorize the N2 males into affecteds and unaffecteds (Fig. 1C). We proceed to quantitative trait loci (QTL) analysis when at least 20% of the male N2 littermates are determined as phenodeviants.
Fig. 1.
Scheme of dominant screening in mice for sleep/wakefulness. (A) Sperms from mutagenized G0 males are harvested for IVF with eggs from B6N females. For each ENU-treated male mouse, up to 60 F1 male mice are examined for sleep abnormality. Less than 1% of the screened mice are selected as phenodeviants. (B) Each F1 phenodeviant is crossed with female wild-type B6N mice to obtain N2 mice. Male N2 mice are examined whether the sleep abnormality is heritable. If heritability is confirmed, N2 mice are used for QTL analysis and whole-exome sequencing to identify the gene mutation associated with the sleep phenotype. (C) Examples of heritability test results. In phenodeviant A, if the sleep parameter histogram in N2 mice is not deviated from the histogram for all mice screened (normal population), the sleep phenotype is considered not heritable (green). In phenodeviant B, if the sleep phenotype is heritable but subtle, the sleep parameter histogram cannot be distinguished from the normal population (yellow). In phenodeviant C, if the histogram in N2 mice is broader than and deviated from the normal population, the sleep phenotype is heritable and weak. Many N2 mice (?/+) cannot be determined as either phenodeviant (m/+) or normal (+/+) based on the sleep behavior. In phenodeviant D, if the histogram has two peaks that are not overlapped, the sleep phenotype of phenodeviant D is heritable and strong. Each N2 mouse can be assigned as either phenodeviant (m/+) or normal (+/+).
Given that sleep parameters follow normal distribution, the expected incidence of mice outside 3 SDs is 0.003. At the early stage of our screening, no more than 5 phenodeviants were identified for every 1,000 mice, indicating that many of these phenodeviants happened by chance. The chance of establishing heritable phenodeviant pedigree would increase with the higher number of mice screened. Thus far, we have screened 10,024 mice and identified 57 phenodeviants. Out of the 57 pedigrees, 14 are confirmed for the heritability of the sleep phenotype and mutations mapped to 10 loci, including 5 pedigrees carrying the same Sik3Slp allele (20). So far, at least 4 independent causative gene mutations have been validated.
Strategically, dominant screening presents advantages over recessive screening that include simpler workflow and substantially less requirement of resources for mouse propagation and maintenance. The inherent challenge in dominant screening is the low frequency of obtaining mice showing the heritable phenotypes compared with recessive screening. This may partly be due to the presence of the wild-type allele or the low penetrance of the gene mutation (21). Gain-of-function mutations may sometimes fail to reflect the endogenous function of the gene of interest, especially in disease manifestation by a neomorphic type of allele. On the other hand, screens for dominant mutants will, in many instances, reveal semidominance of the allele. Some of these mutations are antimorphic in nature and can be critical in understanding the gene’s function as in the case of the Clock ENU mutant; the function of the Clock gene would never have been considered based on the knockout phenotype alone (8, 22). In addition, homozygous mutations often lead to lethality or severe disease, preventing examination of sleep/wake phenotype, as in the case of the Cacna1a (Drowsy) mutation (see below) (23). Indeed, genetic mutations identified in the heritable sleep disorder, such as familial advance sleep phase, show autosomal-dominant inheritance (24). Several genome-wide association studies (GWAS) also show heterozygous carriers of the polymorphism having altered sleep architectures (25).
Number of F1 Mice Derived from a Single ENU-Treated Male.
We considered the maximum number of F1 mice derived from each G0 male based on the diversity of the mutations in a single G0 mouse sperm. Since the number of stem cell-like type A-single spermatogonia (AS) is 35,000 per testis (26) and a single injection of 100 mg/kg ENU kills more than 90% AS spermatogonia (27), 3 serial treatments result in the survival of only 70 AS spermatogonia. Since all sperm derived from a single spermatogonium have the same set of induced mutations, the number of surviving spermatogonia determines the number of different sets of mutations. We calculated the number of nonoverlapping mutation sets with varying number of 10–200 F1 mice derived from an ENU-treated male (SI Appendix, Fig. S1). When the number of F1 mice is small, the nonoverlapping mutation sets increase proportionally to the number of F1 mice. As the number of F1 mice increases, the rate of increase is strongly suppressed by overlapping mutations. For this reason, we selected a moderate number of F1 mice (n = 60). Since the effect of ENU treatment on G0 mice may not be constant, the number of overlapped mutations may show an upward trend. In our experience, the identical mutation in the Sik3 genes was confirmed in 5 pedigrees derived from 5 founder mice among 60 offspring of a single G0 male (20). This was an exceptional case; no such duplication has been observed in other pedigrees. ENU might have particularly strong effect on this G0 mouse leaving very few spermatogonia surviving, which severely limited the diversity of mutations in sperms.
Choice of Inbred Strain.
The proper choice of inbred strains for mutagenized fathers and counter strain mothers is one of the most crucial factors for a successful screening. Each inbred strain shows a characteristic sleep/wake behavior (28–30), which is attributed to the presence of many QTLs. A wide variance may mask the effect of ENU-induced mutations if 2 strains are not chosen carefully. We selected B6J as mutagenized and B6N as counter strain because they are very close to each other (separated only ∼70 y ago) (SI Appendix, Fig. S2), yet the whole-genome sequences and an initial list of single nucleotide polymorphism (SNP) markers were available (18). However, we noticed inconsistent SNPs in each substrain derived from different breeders. Multiple substrains were sequenced to generate an evenly distributed list of 96 SNPs (SI Appendix, Table S1 and Material and Methods) (31) (Jackson Laboratory, https://www.jax.org/).
To evaluate the similarity in the sleep/wakefulness between B6J and B6N, we examined 2 strains against B10J. Consistent with the genetic proximity, B6J and B6N showed similar time spent in wakefulness, NREMS, and REMS, whereas B10J exhibited shorter daily wake time and longer daily NREMS time (Fig. 2 A–C). Although B6J and B6N showed similar episode duration over 24 h in wakefulness, NREMS, and REMS, the REMS episode durations in B6N mice were longer during the light phase and shorter during the dark phase (Fig. 2 D–F). EEG power spectrum analysis showed a similar trend in B6J, B6N, and B10J during NREMS and REMS (Fig. 2 G and H). There was no significant difference in the delta power (1–4 Hz) density during NREMS or the theta power (6–9 Hz) density during REMS (Fig. 2 I and J). We also examined the response to sleep deprivation in each substrain. During the 18-h recovery sleep following the 6-h sleep deprivation from ZT0-6, the total time spent in wake during dark phase was slightly but significantly longer in B6N with decrease in both NREMS and REMS (SI Appendix, Fig. S3 A–C). The NREMS EEG delta power during the first 6 h after sleep deprivation was slightly but significantly smaller in B6N (SI Appendix, Fig. S3D). Although this suggests that B6N may have marginally blunted homeostatic response to sleep deprivation, it would be difficult to map the causal loci because of the relatively poor reproducibility of these parameters (see below). The increase in the REMS time during dark phase was similar in B6J and B6N but significantly lower in B10J (SI Appendix, Fig. S3E).
Fig. 2.
Sleep parameters in B6J, B6N, and B10J. Total time spent in (A) wake, (B) NREMS, and (C) REMS in 24-h day, 12-h light phase, and 12-h dark phase. Episode duration of (D) wake, (E) NREMS, and (F) REMS. EEG power spectra during (G) NREMS and (H) REMS. (I) The delta density (1–4 Hz) during NREMS and (J) the theta density (6–9 Hz) during REMS. Male B6J (n = 20), B6N (n = 20), and B10J (n = 20). All data are shown as the mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (A–H) Two-way ANOVA followed by Tukey’s test. (I and J) One-way ANOVA followed by Tukey’s test.
Since the difference in sleep/wakefulness between B6J and B6N strains is very small, the use of the 2 strains as mutagenized and counter strains is optimal for forward genetics of sleep and presumably suitable for other behavioral and metabolic traits (18). Indeed, this combination enabled us to identify obesogenic mutations through dominant screening (19).
Sleep/Wake Parameters Used for Screening.
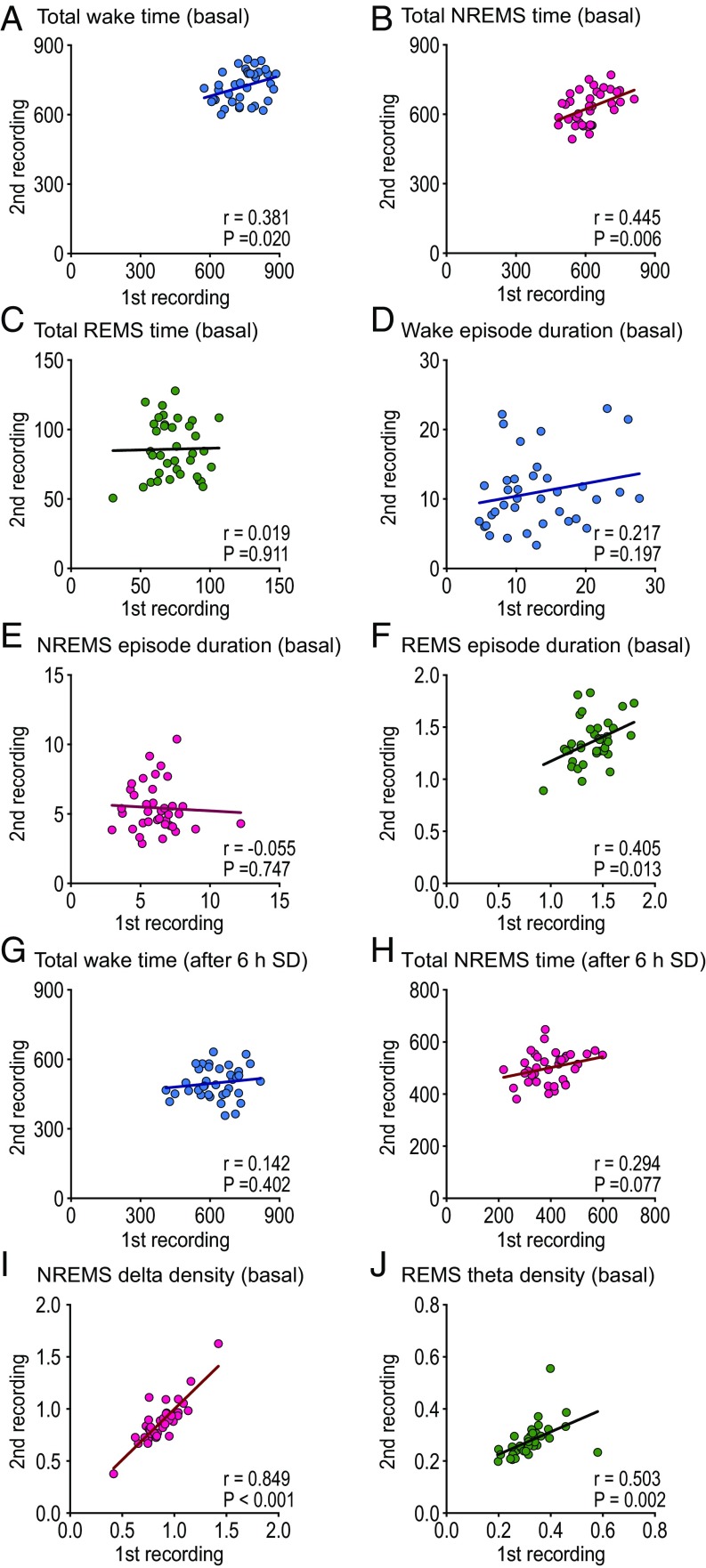
Sleep/wake parameters that are suitable for sleep phenotyping must be highly reproducible in each mouse. Based on our experience, qualitative findings such as loss of atonia during REMS or disappearance of the theta wave during REMS tend to be stable in the same mouse. For quantitative sleep parameters, such as the total time and episode duration, we tested reproducibility in B6J mice by recording twice at more than 4-wk intervals. The total time spent in wakefulness and NREMS of each mouse in the first recording significantly correlated with that in the second recording (Fig. 3 A and B) but not in the total REMS (Fig. 3C). Episode duration of wakefulness and NREMS did not reproduce between the first and second recordings (Fig. 3 D and E). Interestingly, the REMS episode duration in the first recording significantly correlated with that in the second recording (Fig. 3F). The total time spent in wakefulness and NREMS after 6-h sleep deprivation by shaking the cages was not correlated between the first and second recordings (Fig. 3 G and H). The good reproducibility of the total wake time, total NREMS time, and REMS episode duration was consistent with our experience that Sik3Sleepy mutant and NalcnDreamless mutant pedigrees were isolated based on total wake/NREM sleep time and REM sleep episode duration, respectively (20).
Fig. 3.
Reproducibility of sleep parameters. Reproducibility of basal sleep parameters between first recording (first) and second recording more than 4 wk later (second). Total time (min/24 h) in (A) wake, (B) NREMS, and (C) REMS. Episode duration (min) of (D) wake, (E) NREMS, and (F) REMS. Total time during 18-h period after 6-h sleep deprivation (min) in (G) wake and (H) NREMS. (I) Relative delta density (1–4 Hz) during basal NREMS and (J) relative theta density (6–9 Hz) during REMS. B6J (n = 37). The regression lines are drawn on each graph. Pearson’s correlation coefficient (r) and probability (P) are indicated.
Moreover, sleep parameters suitable for forward genetics should exhibit minimal individual variability, indicated as the coefficient of variation (CV). For quantitative parameters, we selected mice with phenotypes that deviated from the average by ∼3 SD with good reproducibility in the second recording. A parameter with a high CV is not suitable for behavioral screening because the distribution of mutants overlaps with normal mice. For example, the circadian period length was very tightly regulated with CV = 0.0072, which is 50-fold lower than that of fear conditioning and the psychostimulant response (2), leading to successful application of forward genetics. The CVs of total wake time, total NREMS time, and REMS time were 0.11, 0.13, and 0.21, respectively. For episode duration of the wake, NREMS, and REMS, CVs were 0.48, 0.27, and 0.13, respectively. Thus, the smaller variation in total wake time, total NREMS time, and REMS episode duration may further corroborate the successful establishment of Sik3Sleepy mutant and NalcnDreamless mutant pedigrees (20).
The EEG delta density during NREM sleep and the theta density during REM sleep also correlated well between the first and second recordings (Fig. 3 I and J). However, the CVs of the NREMS delta density and REMS theta density were 0.23 and 0.24, respectively, thus making the two sleep parameters less suitable for forward genetic screening.
Identification of Responsible Gene Mutation and Proof of Causality.
Almost all phenotype-inducing gene mutations identified in ENU mutagenesis are amino acid changing mutations identified in the coding regions (3, 32). Thus, whole-exome sequencing should detect a majority of responsible mutations in mutant pedigrees. Still, even though all candidate mutations in the heritable pedigrees so far mapped to coding regions in our screening, we must consider the possibility that there may be causal mutations outside coding regions.
In some cases, whole-exome sequencing may reveal more than 1 candidate mutation within the mapped region. Although it is usually easy to select 1 gene from the candidate genes linked to the phenotype, proving that the other genes do not affect the phenotype could be a challenge. For example, we identified mutations in the Sim1 gene (Chr10: 50908536) and Sec63 gene (Chr10: 42816394) in an obesity pedigree (19). Since SIM1 is known to cause obesity and mutant SIM1 protein did not function as a transcription factor, the Sim1 mutation alone could explain the obesity. However, we could not positively deny the possibility that mutant SEC63 protein, which is involved in protein translocation at the ER membrane, may also affect body weight. In such cases, one way to establish causality would be to generate animals that are recombinants across this region; only 1 mutation will consistently segregate with the phenotype.
Simulation of the LOD Score and Probability of Reaching Significance.
High LOD score indicates a high probability of a responsible gene mutation located in a particular chromosomal region (33). To examine the relationship among the strength of sleep phenotype, number of N2 mice examined, and LOD score, we conducted a numerical simulation of the LOD score with varying number of N2 mice (50% nonmutants [+/+], 50% mutants [m/+]) and varying mean total wake time of mutant mice. The mean wake time of the control (+/+) mice was set to 740 min according to our data. The SD of the wake time was set to 0.1 of the mean wake time based on the sleep data of our F1 and N2 males. The simulation showed the LOD score as a function of the number of mice examined and of how far the mutant sleep parameter deviated from the mean sleep parameter of the control mice (SI Appendix, Fig. S4 A and B). Importantly, the simulation also showed moderate variability in the LOD score even under the same condition, which is due to chance. One of the Sleepy pedigrees, B021, showed an LOD score of 32 for 118 N2 mice (20). The simulation predicted that a total wake time of a mutant pedigree showing an LOD score of 32 was ∼500 min, which is indeed consistent with the sleep phenotype of Sleepy pedigrees (SI Appendix, Fig. S4A).
To confirm the causality of the gene mutation, we compare the sleep/wakefulness of mutant mice with control mice. A real-world setting was applied in the simulation of the probability of reaching statistical significance (P < 0.05) using 10 control mice and 10 mutant mice. We set the mean wake time of the control group to 740 min and the SD to mean wake time multiplied by CV = 0.10 according to our data. The mutant group showing a mean daily wake time of ∼650 and 675 min showed 80 and 50% probability of reaching statistical significance (P < 0.05), respectively (SI Appendix, Fig. S4C). However, when we set the CV to 0.15, which can be found in the literature (34, 35), the mutant mean wake time changed to 600 min (80% chance) and 640 min (50% chance), indicating the importance of highly consistent sleep measurements.
Weak Hypersomnia Phenotype in the B016 Pedigree Carrying a Cacna1a Mutation.
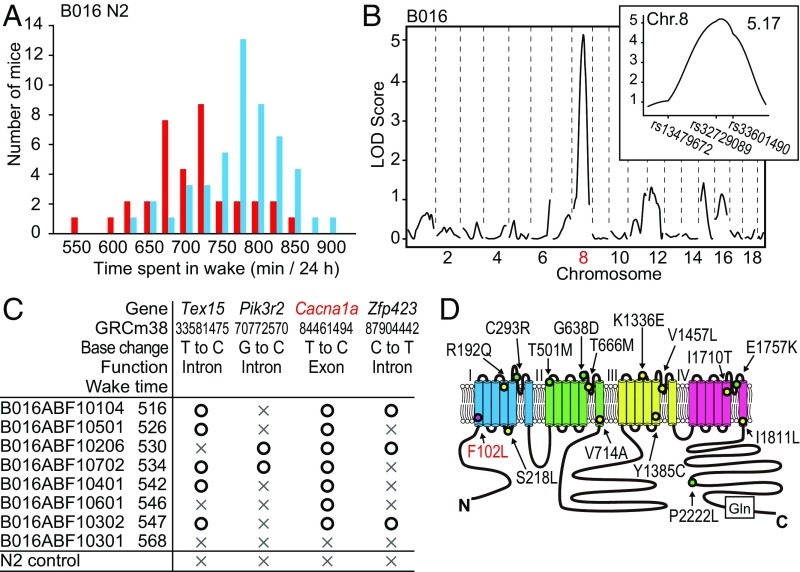
The founder male of the B016 (Drowsy) pedigree had a total wake time of 502 min/24 h. Distribution of total wake time in affected Drowsy N2 males was shifted to the left compared with unaffecteds (Fig. 4A). QTL analysis showed a single LOD score peak of 5.17 for total wake time located around rs32729089 (chr8:74953357) and rs33601490 (chr8:91507617) (n = 81; Fig. 4B). According to our simulation (SI Appendix, Fig. S4B), the total wake time of the pedigree showing an LOD score of 5.2 (n = 81) was expected to be 645–680 min, which was 60–95 min shorter than the wild-type group mean (740 min). Consistently, the difference in the total time between the Drowsy mutant group and the wild-type group was ∼70 min.
Fig. 4.
Identification of the Cacna1a gene mutation in the Drowsy pedigree showing a heritable but weak sleep phenotype. (A) Wake time distribution of retrospectively genotyped Drowsy N2 littermates. Cacna1a+/+ (blue) and Cacna1am/+ (red) mice. (B) QTL analysis of the Drowsy pedigree for total wake time (n = 81). (Inset) LOD score peak between rs13479672 and rs33601490 on chromosome 8. (C) Mutational analysis of affected mice (bottom 20% in total wake time) and unaffected mice (top 20% in total wake time) within the Drowsy pedigree. (D) Structure of the CACNA1A protein indicating the F102L mutation (this study; pink), together with examples of human missense mutations reported in familial hemiplegic migraine (yellow) and episodic ataxia type2 (green), as well as the polyglutamine repeat found in spinocerebellar ataxia type 6 (Gln).
Haplotype distribution of affected Drowsy N2 mice (total wake time in bottom 20%) was different from that of unaffected group (total wake time in top 20%) (SI Appendix, Fig. S5A). Whole-exome sequencing of the Drowsy pedigree identified a heterozygous single nucleotide substitution (chr8: 84461494) of the Cacna1a gene in 7 out of 8 Drowsy F1 mice with short total wake time (Fig. 4C), and direct sequencing confirmed the nucleotide change (SI Appendix, Fig. S5B). CACNA1A is a voltage-dependent calcium channel that is highly expressed in cerebellar Purkinje cells and many other types of neurons (36). The mutation results in an amino acid substitution F102L in the first transmembrane region of domain I (Fig. 4D), well conserved among vertebrates and invertebrates (SI Appendix, Fig. S5C). Many missense mutations in Cacna1a are associated with two autosomal dominant diseases, episodic ataxia type2 and familial hemiplegic migraine (36, 37). Besides, a CAG repeat expansion in Cacna1a causes spinocerebellar ataxia 6 (Fig. 4D) (36). As predicted, the homozygous Drowsy mutant suffered from severe abnormalities in locomotion and posture which was reported in Cacna1a-deficient mice (Movie S1) (38, 39). Notably, a recent computational simulation-based study showed the potential role of voltage-dependent calcium channel in sleep/wake maintenance (40).
Conclusions
We described the method for EEG/EMG-based sleep screening of randomly mutagenized mice and discussed the selection of inbred mouse strains. Statistically examined reproducibility of the selected screening parameters proved the precision of the sleep/wake measurement. Furthermore, we showed the workflow for identifying the causative gene, as an example, the identification of a Cacna1a mutation in the Drowsy pedigree showing a mild hypersomnia phenotype.
At the scale of ∼10,000 mice, we identified 10 heritable loci to date. While low occurrence of sleep phenodeviant was expected, possibly due to redundant regulatory mechanisms in sleep/wakefulness and low penetrance of the phenotype, our screening output seems to be comparable to other trials where a single biological trait is screened. The Clock ENU mutant is a lucky example that found the causal mutation from screening 304 G1 progeny (41). Daxinger et al. reported identification of a single candidate gene from a population of 2,045 G1 mice (42). Another study showed 20 mutant progeny that carry 10 unique gene mutations from screening of ∼5,000 G1 offspring, of which 9 were previously known genes in epigenetic regulation (43).
The identified genes in our study usually encode proteins with large molecular weights, probably because the cumulative frequency of ENU-induced mutation is proportional to the gene size (44); some of known sleep regulatory genes, such as the very compact orexin and orexin receptor genes, have not been detected in this screening. Also, for unknown reasons, the majority of our sleep phenodeviants has been long-sleep mutants.
Phenotype-driven screening at such large scale, indeed, is labor-intensive and costly. It is not recommended for an attempt to furthering the knowledge of largely defined mechanisms since there is a high chance of identifying already known genes, as it was in the case of our obesity screening (19). Also, in a dominant screening we recommend a highly focused screen rather than trying to examine multiple biological traits in a workflow; after all, the most expensive step in the screening is phenotyping, not the production of mutagenized mice. It is difficult to draw estimation on further identification of candidate genes and whether/when saturation can be reached in a dominant screen (21). Nevertheless, we should find more genes in sleep/wake regulation by enlarging the scale and continuing the search. The screening strategy is easily modifiable for studying other physiological phenomena in mice with careful selection of the screening criteria and the biology to be studied.
Material and Methods
See SI Appendix for detailed materials and methods, including data analyses, numerical simulation, and instrumental specifications.
Animals.
All procedures were conducted in accordance with the Guidelines for Animal Experiments of University of Tsukuba and approved by the Institutional Animal Care and Use Committee of University of Tsukuba (approved protocol ID no. 140200). For animal source and husbandry condition, refer to SI Appendix, Material and Methods.
Supplementary Material
Acknowledgments
We thank all M. Yanagisawa and H.F. laboratory members and International Institute for Integrative Sleep Medicine members for discussion and comments on this manuscript. We appreciate the support of The Institute of Physical and Chemical Research (Japan) BioResource Center Mouse Clinic team. This work was supported by the World Premier International Research Center Initiative from Ministry of Education, Culture, Sports, Science and Technology (MEXT) to M. Yanagisawa; Japan Society for the Promotion of Science (JSPS) KAKENHI (Grant Number 17H06095 to M. Yanagisawa, H.F.; 16K15187, 17H04023, and 17H05583 to H.F.; and 26507003 to C.M. and H.F.), MEXT KAKENHI (Grant Number 15H05935 to H.F.), CREST (A3A28043 to M. Yanagisawa); Funding Program for World-Leading Innovative R&D on Science and Technology from JSPS to M. Yanagisawa; Uehara Memorial Foundation research grant to M. Yanagisawa; and Takeda Science Foundation research grant (to M. Yanagisawa).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906774116/-/DCSupplemental.
References
- 1.Weber F., Dan Y., Circuit-based interrogation of sleep control. Nature 538, 51–59 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Takahashi J. S., Shimomura K., Kumar V., Searching for genes underlying behavior: Lessons from circadian rhythms. Science 322, 909–912 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beutler B., Du X., Xia Y., Precis on forward genetics in mice. Nat. Immunol. 8, 659–664 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Brown S. D. M., Nolan P. M., Mouse mutagenesis-systematic studies of mammalian gene function. Hum. Mol. Genet. 7, 1627–1633 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Konopka R. J., Benzer S., Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 68, 2112–2116 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddy P., et al. , Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell 38, 701–710 (1984). [DOI] [PubMed] [Google Scholar]
- 7.Sehgal A., Price J. L., Man B., Young M. W., Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science 263, 1603–1606 (1994). [DOI] [PubMed] [Google Scholar]
- 8.King D. P., et al. , Positional cloning of the mouse circadian clock gene. Cell 89, 641–653 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubowy C., Sehgal A., Circadian rhythms and sleep in Drosophila melanogaster. Genetics 205, 1373–1397 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi J. S., Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ripperger J. A., Jud C., Albrecht U., The daily rhythm of mice. FEBS Lett. 585, 1384–1392 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Hendricks J. C., et al. , Rest in Drosophila is a sleep-like state. Neuron 25, 129–138 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Shaw P. J., Cirelli C., Greenspan R. J., Tononi G., Correlates of sleep and waking in Drosophila melanogaster. Science 287, 1834–1837 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Tomita J., Ban G., Kume K., Genes and neural circuits for sleep of the fruit fly. Neurosci. Res. 118, 82–91 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Chemelli R. M., et al. , Narcolepsy in orexin knockout mice: Molecular genetics of sleep regulation. Cell 98, 437–451 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Hara J., et al. , Genetic ablation of orexin neurons in mice results in narcolepsy, hypophagia, and obesity. Neuron 30, 345–354 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Coleman P. J., Gotter A. L., Herring W. J., Winrow C. J., Renger J. J., The discovery of suvorexant, the first orexin receptor drug for insomnia. Annu. Rev. Pharmacol. Toxicol. 57, 509–533 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Kumar V., et al. , C57BL/6N mutation in cytoplasmic FMRP interacting protein 2 regulates cocaine response. Science 342, 1508–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hossain M. S., et al. , Identification of mutations through dominant screening for obesity using C57BL/6 substrains. Sci. Rep. 6, 32453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Funato H., et al. , Forward-genetics analysis of sleep in randomly mutagenized mice. Nature 539, 378–383 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nolan P. M., Kapfhamer D., Bućan M., Random mutagenesis screen for dominant behavioral mutations in mice. Methods 13, 379–395 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Debruyne J. P., et al. , A clock shock: Mouse CLOCK is not required for circadian oscillator function. Neuron 50, 465–477 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Hayasaka N., et al. , Salt-inducible kinase 3 regulates the mammalian circadian clock by destabilizing PER2 protein. Elife 6, 1–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi G., Wu D., Ptáček L. J., Fu Y.-H. H., Human genetics and sleep behavior. Curr. Opin. Neurobiol. 44, 43–49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goel N., Genetic markers of sleep and sleepiness. Sleep Med. Clin. 12, 289–299 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Tegelenbosch R. A., de Rooij D. G., A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat. Res. 290, 193–200 (1993). [DOI] [PubMed] [Google Scholar]
- 27.Oakberg E. F., Crosthwait C. D., The effect of ethyl-, methyl- and hydroxyethyl-nitrosourea on the mouse testis. Mutat. Res. 108, 337–344 (1983). [DOI] [PubMed] [Google Scholar]
- 28.Franken P., Chollet D., Tafti M., The homeostatic regulation of sleep need is under genetic control. J. Neurosci. 21, 2610–2621 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tafti M., Chollet D., Valatx J. L., Franken P., Quantitative trait loci approach to the genetics of sleep in recombinant inbred mice. J. Sleep Res. 8 (suppl. 1), 37–43 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Franken P., Malafosse A., Tafti M., Genetic variation in EEG activity during sleep in inbred mice. Am. J. Physiol. Regul. Integr. Comp. Physiol., 275, R1127–R1137 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Mekada K., Hirose M., Murakami A., Yoshiki A., Development of SNP markers for C57BL/6N-derived mouse inbred strains. Exp. Anim. 64, 91–100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon M. M., et al. , Current strategies for mutation detection in phenotype-driven screens utilising next generation sequencing. Mamm. Genome 26, 486–500 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiltshire S., Cardon L. R., McCarthy M. I., Evaluating the results of genomewide linkage scans of complex traits by locus counting. Am. J. Hum. Genet. 71, 1175–1182 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graves L. A., et al. , Genetic evidence for a role of CREB in sustained cortical arousal. J. Neurophysiol. 90, 1152–1159 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Lee J., Kim D., Shin H.-S., Lack of delta waves and sleep disturbances during non-rapid eye movement sleep in mice lacking alpha1G-subunit of T-type calcium channels. Proc. Natl. Acad. Sci. U.S.A. 101, 18195–18199 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajakulendran S., Kaski D., Hanna M. G., Neuronal P/Q-type calcium channel dysfunction in inherited disorders of the CNS. Nat. Rev. Neurol. 8, 86–96 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Sintas C., et al. , Mutation spectrum in the CACNA1A gene in 49 patients with episodic ataxia. Sci. Rep. 7, 2514 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jun K., et al. , Ablation of P/Q-type Ca(2+) channel currents, altered synaptic transmission, and progressive ataxia in mice lacking the alpha(1A)-subunit. Proc. Natl. Acad. Sci. U.S.A. 96, 15245–15250 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fletcher C. F., et al. , Dystonia and cerebellar atrophy in Cacna1a null mice lacking P/Q calcium channel activity. FASEB J. 15, 1288–1290 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Tatsuki F., et al. , Involvement of Ca(2+)-dependent hyperpolarization in sleep duration in mammals. Neuron 90, 70–85 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Vitaterna M. H., et al. , Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264, 719–725 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daxinger L., et al. , An ENU mutagenesis screen identifies novel and known genes involved in epigenetic processes in the mouse. Genome Biol. 14, R96 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furuse T., et al. , Phenotypic characterization of a new Grin1 mutant mouse generated by ENU mutagenesis. Eur. J. Neurosci. 31, 1281–1291 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Bauer D. C., McMorran B. J., Foote S. J., Burgio G., Genome-wide analysis of chemically induced mutations in mouse in phenotype-driven screens. BMC Genomics 16, 866 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.