Photostable dye improves superresolution imaging of mitochondria
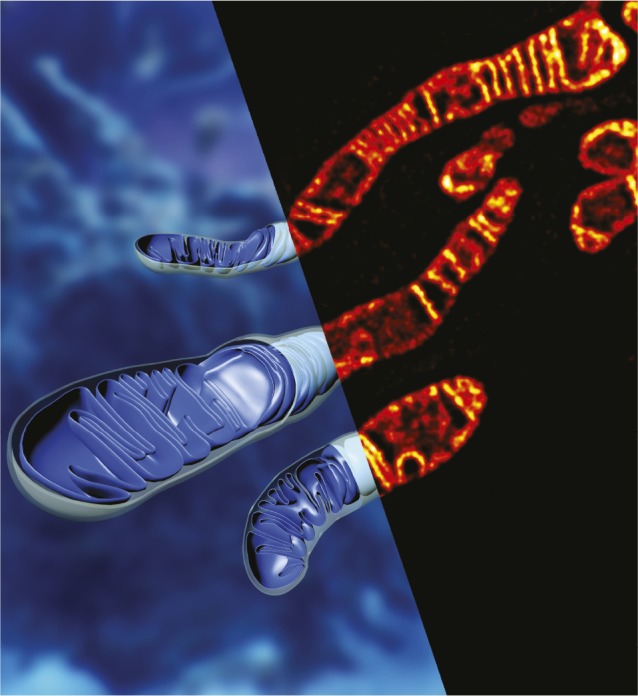
STED image and schematic of mitochondria labeled with MitoPB Yellow.
Stimulated emission depletion (STED) microscopy is a superresolution imaging method that can enable live cell imaging at high spatial and temporal resolution. However, the strong lasers used in STED microscopy rapidly bleach fluorescent dyes used to visualize mitochondria, which are the powerhouses of eukaryotic cells. Chenguang Wang et al. (pp. 15817–15822) fashioned a fluorescent probe named MitoPB Yellow, whose high photostability, long fluorescence lifetime, and specificity for the mitochondrial inner membrane render it a superior tool for live STED imaging of the organelle, compared with conventional dyes such as MitoTracker Green FM, MitoTracker Deep Red FM, and Rhodamine 123. MitoPB Yellow enabled visualization of ultrastructural features of mitochondrial cristae—folds of the inner membrane that serve to maximize surface area for energy generation—at 60-nm resolution while using a 660-nm depletion laser. The dye revealed fine details of the mitochondrial inner membrane in live mammalian cells previously visible only through electron microscopy and allowed the authors to derive accurate counts of cristae from the fluorescence intensity. Additionally, MitoPB Yellow helped observe the dynamic remodeling of cristae in cells exposed to starvation and apoptotic stress as well as the merging of cristae within individual mitochondria and fusion between mitochondria. According to the authors, the findings demonstrate the advantages of MitoPB Yellow for STED imaging of mitochondrial membrane structure and dynamics in living cells. — P.N.
Conceptual knowledge and neural response to facial emotion
Mounting behavioral evidence suggests that the perception of emotions may be shaped by conceptual knowledge, including memories and expectations, leading to variability in perception across individuals. Jeffrey Brooks et al. (pp. 15861–15870) combined functional MRI and behavioral experiments to examine the responses of 40 adult participants to 6 emotion categories: anger, disgust, fear, happiness, sadness, and surprise. The participants rated each category based on its conceptual relationship to 40 emotion features—including thoughts, sensations, and actions—such as crying, heart racing, and clenching fists. For each participant, the authors measured the conceptual similarity between pairs of the 6 different emotion categories by comparing the overlap in patterns of responses to the 40 items. In addition, neural similarity ratings between pairs of the different emotion categories were generated for each participant by comparing patterns of neural responses to images of emotional faces. At the individual level, conceptual similarity predicted neural similarity, specifically in the right fusiform gyrus, a brain region that plays a role in face perception. According to the authors, the findings suggest that conceptual knowledge can flexibly influence the brain’s representations of face categories. — J.W.
Native, type I CRISPR-Cas system for genome editing

L. crispatus. Image courtesy of North Carolina State University/Claudio Hidalgo-Cantabrana and Valerie Lapham.
Current CRISPR-Cas genome editing tools predominantly target eukaryotes by repurposing class 2 single effector nuclease enzymes such as Cas9 and Cas12. Far less attention has been paid to in situ editing in bacteria using native class 1 systems such as CRISPR-Cas3, abundant in bacteria and archaea. Claudio Hidalgo-Cantabrana et al. (pp. 15774–15783) describe a type I-E CRISPR-Cas system in Lactobacillus crispatus, a commensal species associated with human vaginal health and poultry intestinal health. By reprogramming the endogenous type I-E machinery and devising custom repair templates, the authors show that the native CRISPR-Cas3 system in L. crispatus can be used to induce genome edits ranging from insertions and deletions to single nucleotide substitutions. Numerous studies have documented the ability of L. crispatus to outcompete pathogens, produce antimicrobial compounds, and interact beneficially with the host immune system. However, the molecular basis of the bacterial probiotic effects are poorly understood, due to the bacterium’s genetic recalcitrance. The findings offer both a proof of concept for in situ repurposing of native CRISPR-Cas systems in bacteria and a potential pathway for engineered probiotics, according to the authors. — T.J.
Genetic history of Nunavik Inuit
The Nunavik Inuit of Canada are a small and isolated population adapted to the extreme Arctic environment. The genetic characteristics of the population, including factors associated with an increased risk for cardiovascular diseases, are poorly understood. Sirui Zhou et al. (pp. 16012–16017) analyzed genome-wide DNA variants called single nucleotide polymorphisms of 165 Nunavik Inuit, in addition to whole-exome sequences, which are protein-coding regions of the genome, of 114 of such individuals. The results suggest that the genetic background of the Nunavik Inuit is homogenous and distinct from that of any known present-day population. The Nunavik Inuit underwent a potential bottleneck approximately 10,000 years ago and likely split from their closest relatives, the Greenlandic Inuit and the Siberian Eskimos, approximately 10,500 and 11,000 years ago, respectively. Moreover, the genetic profile of the Nunavik Inuit shows evidence of adaptations in pathways involving fatty acid metabolism and cellular adhesion, as well as a variant in the OR4C3 gene associated with an increased risk of intracranial aneurysms, a complex cerebrovascular disorder characterized by weakness of the intracranial artery walls. According to the authors, the unique genetic background of the Nunavik Inuit may underlie adaptations to their extreme environment as well as increased susceptibility to cardiovascular diseases. — J.W.


