Significance
Exosomes have been implicated in intercellular communication in cancer and neurodegenerative disorders. We explored their function in brain development. Proteomic analysis demonstrated that exosomes from isogenic control cultures contain neurodevelopmental signaling proteins, which are lacking in exosomes from MECP2 loss-of-function (MECP2LOF) cultures. Treating MECP2LOF neural cultures with control exosomes rescues neurodevelopmental deficits, increasing neurogenesis, synaptogenesis, and network activity. Exosomes function similarly in vivo: injecting purified exosomes into the lateral ventricles of P4 mouse brains increased hippocampal neurogenesis. These findings significantly advance the field by demonstrating that neural exosomes contain diverse protein cargo predicted to affect multiple outcome measures of neural development and that exosomes signal between cells in developing neural circuits to promote neural circuit development and function.
Keywords: exosomes, Rett syndrome, neuronal development, synaptogenesis, extracellular vesicle
Abstract
Exosomes are thought to be released by all cells in the body and to be involved in intercellular communication. We tested whether neural exosomes can regulate the development of neural circuits. We show that exosome treatment increases proliferation in developing neural cultures and in vivo in dentate gyrus of P4 mouse brain. We compared the protein cargo and signaling bioactivity of exosomes released by hiPSC-derived neural cultures lacking MECP2, a model of the neurodevelopmental disorder Rett syndrome, with exosomes released by isogenic rescue control neural cultures. Quantitative proteomic analysis indicates that control exosomes contain multiple functional signaling networks known to be important for neuronal circuit development. Treating MECP2-knockdown human primary neural cultures with control exosomes rescues deficits in neuronal proliferation, differentiation, synaptogenesis, and synchronized firing, whereas exosomes from MECP2-deficient hiPSC neural cultures lack this capability. These data indicate that exosomes carry signaling information required to regulate neural circuit development.
Exosomes are small vesicles secreted by all cells in the brain, including neurons, and have been hypothesized to play a critical role in cell–cell communication (1, 2). Exosomes can signal over short range within brain tissue (3), and can signal widely throughout the brain through the cerebrospinal fluid (4, 5). Strong evidence indicates that exosomes impart biological activity to neurons (3, 6). For instance, in the Drosophila larval neuromuscular junction, exosome-mediated protein transport is required for coordinated development of pre- and postsynaptic components of the neuromuscular junction (7, 8). Exosomes secreted by oligodendrocytes affect firing rate, signaling pathways, and gene expression in cultured primary neurons (9, 10). Although there is evidence of biological roles of exosomes secreted by neurons and other cell types in the brain, their function in the development of human neural circuits is largely unexplored.
To investigate the role of exosomes in neural circuit development, we developed a reductionist experimental paradigm in which we added purified exosomes isolated from human induced pluripotent stem cell (hiPSC)-derived neurons onto human primary neural cultures to assay their capacity to influence neuronal and circuit development. We found that treatment with exosomes increased neurogenesis by promoting cell proliferation and neuronal differentiation, suggesting that exosomes carry signaling components that influence cell fate in developing neural circuits. We injected purified rodent exosomes into the lateral ventricle of P4 mice to test their role in hippocampal neurogenesis in an in vivo model. Consistent with in vitro observations, exosome treatment increases in proliferation in the granule cell layer (GCL) of dentate gyrus. Based on these observations that exosomes affect neural circuit development in vitro and in vivo, we hypothesized that conditions that disrupt neural circuit development may arise from defective exosome signaling. The loss of function of the X-linked Methyl-CpG-binding protein 2 (MECP2) gene results in aberrant neural circuit development, leading to Rett syndrome (11–14). As disruption of a single protein, MECP2, leads to widespread pleiotropic deficits in neural circuit development, we further hypothesized that altered exosome-mediated intercellular communication may contribute to the impaired neural circuit development seen with MECP2 loss of function (MECP2 LOF). We had previously shown that neurons derived from MECP2 LOF hiPSC-derived neurons have fewer synapses, reduced spine density, smaller soma size, altered calcium signaling, and electrophysiological defects compared with control neurons (15), whereas CRISPR/Cas9 isogenic rescue of the MECP2 mutation (referred to as isogenic control) results in normal neural cultures (16, 17). Here, we compared exosomes released from MECP2 LOF donor neural cultures with exosomes from isogenic control donor neural cultures by using quantitative proteomic analysis. Isogenic control exosomes differed significantly from MECP2 LOF exosomes in their protein signaling content. Bioinformatic analyses revealed signaling complexes in control exosomes capable of eliciting neurodevelopmental outcomes, such as neural cell proliferation, neurogenesis, and synaptic development, that were lacking in MECP2 LOF exosomes. Treating recipient MECP2 LOF cultures with exosomes from isogenic control donor cultures increased cell proliferation, neurogenesis, synaptogenesis, and synchronized firing, whereas treating control recipient neural cultures with exosomes from MECP2 LOF donor cultures did not have adverse effects on neural phenotypes. These data show that exosomes play a significant role in neuronal circuit development and can be used to reverse deficits in neurological disease models.
Results
Exosomes Increase Cell Number in Developing Neural Cultures.
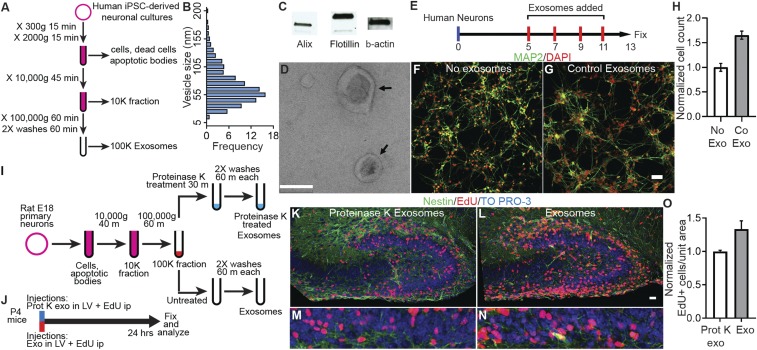
To test the hypothesis that exosomes play a physiological role in neuronal development, we established neural cultures derived from hiPSCs as a source for exosomes because of the demonstrated value of iPSCs in modeling human diseases (15–17) and the capacity to harvest large volumes of exosomes. We cultured hiPSC-derived neural cultures in serum-free growth and differentiation media to avoid potential contamination from exosomes from serum. The hiPSC-derived neural cultures used in this study have been well characterized and contain approximately 90% neurons and 10% glia (15). Exosomes were purified by using a differential centrifugation protocol (18), slightly modified for our cultures (Fig. 1A). Western blot analysis showed that the purified exosomes contain the typical exosome markers, Alix and Flotillin (Fig. 1B). Electron microscopic analysis demonstrated that the purified exosomes had typical disk-shaped morphology (Fig. 1C and SI Appendix, Fig. S1) and ranged in size from 30 to 200 nm; ∼60% of exosomes were between 40 and 80 nm and >80% were <120 nm (Fig. 1D).
Fig. 1.
Exosomes increase proliferation in vitro and in vivo. (A) Protocol to purify exosomes by sequential ultracentrifugation. (B) Western blot analysis showing exosomal markers Alix and Flotillin in exosomes purified from human iPSC-derived neural cultures. (C) Electron micrograph of purified exosomes. Arrows point to exosomes with typical disk-shaped morphology. (D) Size distribution of exosomes purified from human iPSC-derived neural cultures. Vesicle diameter was calculated from electron micrographs. (E) Protocol for treatment of human primary neural cultures with purified control exosomes (Co Exo) or an equal volume of media (No Exo) added on DIV 5, 7, 9, and 11. Cultures were fixed on DIV 13, labeled, and analyzed. (F and G) Images of human neural cultures treated with media (No Exo, F) or control exosomes (Co Exo, G) labeled with MAP2 antibodies (green) or DAPI (red). (H) Graph of the total cell number in cultures treated with control exosomes compared with media alone (No Exo). Exosome treatment increased total cell number 1.65× ± 0.08 (P = 0.001, n = 4 wells per group, 2-way ANOVA). (I and J) Protocols for preparation of exosomes purified from E18 rat primary neural cultures (I) and treatment of P4 mice (J). Exosomes were purified from serum-free media collected from DIV9 rat neural cultures, and the 100,000 × g exosome pellet was equally divided into 2 tubes. One tube was treated with 100 µg/mL Proteinase K at 37 °C for 30 min to cleave surface proteins, and the other tube was left untreated. P4 mice received exosome injections into the lateral ventricles (LV), followed by intraperitoneal (ip) injections of 25 mg/kg of EdU. The mouse brains were fixed and processed to detect EdU+ cells. (K–N) Images of hippocampal dentate gyrus of P4 mice injected with protein-depleted exosomes (K and L) and untreated exosomes (M and N) showing EdU+ cells (red), Nestin (green) immunolabeling, and TO-PRO-3 nuclear stain (blue). Low-magnification images were used to match identical brain regions (SI Appendix, Fig. S3). EdU+ cells were counted in the boxed regions in dentate gyrus region (K–M), shown at 3.3× zoom in L–N. Proliferation in the granule cell layer (GCL) of the dentate gyrus was quantified by counting EdU+ cells normalized to the area counted. (O) Exosome treatment increased Edu+ cells per unit area 1.34× ± 0.12 (n = 3 mice each) compared with proteinase K-treated exosomes. (Scale bars: C, 0.1 µm; F, G, K, and M, 20 µm.)
To investigate the effect of exosomes on neural development we first characterized proliferation and differentiation in the human primary neural cultures. Proliferation, assessed by EdU incorporation, was maintained over the first 9 d in vitro (DIV), after which proliferation declined (SI Appendix, Fig. S2). We isolated exosomes from 2-d conditioned media harvested from donor neural cultures derived from differentiated hiPSCs as in Fig. 1A. Purified exosomes were washed and resuspended in fresh growth media and applied to recipient neuronal cultures on 5, 7, and 11 DIV (Fig. 1E). Purified exosomes, labeled with fluorescent membrane label, ATTO 488 mCLING (19), are taken up by neurons and distributed to the cell body and different cellular compartments, including synapses (SI Appendix, Fig. S3). Recipient primary human neural cultures, labeled with DAPI and MAP2 (Fig. 1 F and G), showed a statistically significant increase in cell number in exosome-treated cultures compared with control (1.65× ± 0.08; P = 0.001, Fig. 1H).
Exosomes Increase Proliferation In Vivo in Mouse Dentate Gyrus.
To test whether exosomes display bioactivity in vivo and can influence cell proliferation in developing neural circuits analogous to neural cultures, we measured neural proliferation in the GCL of dentate gyrus in postnatal P4 mice, where granule cell proliferation continues during this early postnatal period (20, 21). We harvested exosomes from DIV9 rodent primary neural cultures and injected them into the lateral ventricles of P4 mice (Fig. 1 I and J). As a control, we injected exosomes treated with Proteinase K, which are depleted of protein (SI Appendix, Fig. S5). Exosome injections in each cerebral hemisphere were followed by i.p. administration of 5-ethynyl-2′-deoxyuridine (EdU; 25 mg/kg of body weight). The brains of treated mice were fixed after 24 h, and sagittal vibratome sections were labeled with DAPI, Nestin, and EdU. Anatomically matching sections were identified in low-magnification images (SI Appendix, Fig. S4), and EdU+ cells were counted in the GCL (Fig. 1 K–N). Mice injected with DIV19 exosomes had significantly greater EdU+ cell density in the GCL compared with pups with control protein-depleted exosomes (1.34× ± 0.12; P < 0.05; Fig. 1O). These data indicate that exosomes display bioactivity in vivo, increasing neural cell proliferation, analogous to their effect in neural cultures.
Proteomic Analysis Indicates That Exosomes Contain Signaling Proteins That Are Lacking in the Absence of MECP2.
To explore the idea that proteins may be involved in exosome intercellular signaling capacity in neural circuit development, we analyzed the proteomes of exosomes in a model of the neurodevelopmental disorder Rett syndrome, in which functional disruption of a single protein, MECP2, has devastating effects on brain development. We conducted an unbiased quantitative mass spectrometry (MS) proteomic study of exosomes using TMT isobaric tags. We compared purified exosomes from donor hiPSC-derived neural cultures from a male individual, in which MECP2 protein was completely absent (MECP2LOF), with control isogenic hiPSC-derived neural cultures that were corrected for the MECP2 mutation using CRISPR/Cas9 technology (isoCONTROL exosomes) (16). We detected a total of 2,572 proteins from MECP2LOF and control exosomes in 2 independent MS experiments (SI Appendix, Fig. S6A and Datasets S1–S3), of which 739 were annotated for function in neurons. Analysis of “cellular pathways” and “downstream effects on biological functions in the nervous system” using the manually curated Ingenuity database (22) indicated that exosomes released by hiPSC-derived neural cultures are enriched in proteins that impact neuronal development and function (SI Appendix, Fig. S6 B and E and Datasets S6 and S8). Specifically, they contain signaling components for protein translation, axonal guidance, integrin signaling, ephrin signaling, and cytoskeletal regulation (Rho family, Actin cytoskeleton), among the most significant categories (SI Appendix, Fig. S6B and Dataset S6), and have the capacity to influence downstream functions such as neuritogenesis, development, morphogenesis and proliferation of neurons, and synaptic development and function (SI Appendix, Fig. S6C and Dataset S8). We do not detect synaptic vesicular neurotransmitter transporters, early endosomal markers, and nuclear markers in the exosome proteome (SI Appendix, Fig. S7).
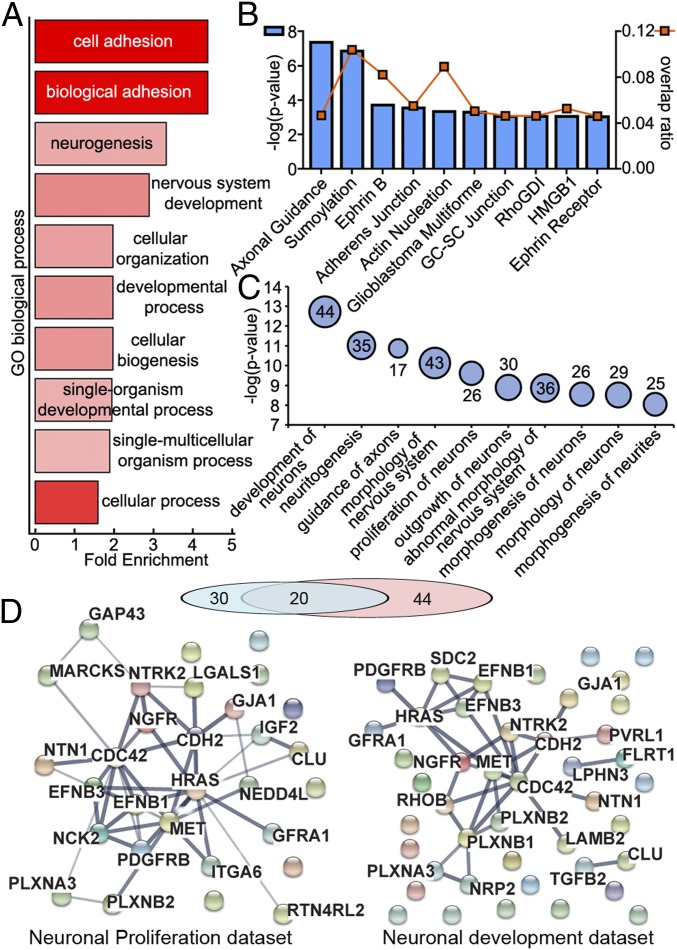
Of the 2,572 proteins present in MECP2LOF and isoCONTROL exosomes, 237 had an average fold difference of 1.5 or more between MECP2LOF and isoCONTROL samples (Fig. 2 and Dataset S4). We analyzed this dataset of 237 proteins by using the Ingenuity database and the PANTHER Overrepresentation Test (23) and GO enrichment using FunCoup (24). The PANTHER Overrepresentation Test using the GO “biological processes” complete annotation dataset indicated that “nervous system development” (+2.88×) and “neurogenesis” (+3.26×) were highly overrepresented compared with the normal human database (Fig. 2A and Dataset S5). Ingenuity analysis of the representation of components with a role in different cellular functions indicated that “axon guidance,” “ephrin signaling,” and “actin dynamics” are among the most significant (Fig. 2B and Dataset S7). The Ingenuity Pathway Analysis (IPA)-curated “downstream effects on biological functions in the nervous system” predicted roles in “neuronal development,” “neuritogenesis,” “axonal guidance,” “proliferation of neurons,” and “synaptic development” among the most significant categories (Fig. 2C and Dataset S9). We further analyzed the dataset of 30 proteins predicted to be involved in neuronal proliferation and dataset of 44 proteins predicted to be involved in neuronal development. There was significant overlap between the 2 datasets consistent with proteins annotated to perform function in neuronal proliferation as well as development (Fig. 2D). Both datasets showed robust functional protein association networks using STRING database (Fig. 2D). These quantitative proteomic and bioinformatic analyses indicate that exosomes contain proteins that interact in complex functional signaling networks that are significantly altered by MECP2 mutation. To test whether alterations of specific proteins in exosomes released from MECP2LOF donor cultures reflects similar alterations in cellular levels of the proteins, we compared protein expression in MECP2LOF donor cells and exosomes with isoControls using Western blots (SI Appendix, Fig. S2B). Differences in cargo between MECP2LOF and isoControl exosomes were largely independent of their cellular expression. Whereas HRAS and Alix showed similar levels in isoCONTROL and MECP2LOF cell lysates, Alix was reduced and HRAS was unchanged in MECP2LOF compared with isoCONTROL in exosomes. Flotillin, Cadherin 2, Calmodulin 1, GAP43, and L1CAM showed higher protein levels in cells with MECPLOF compared with isoCONTROL. In contrast, Flotillin and GAP43 were reduced in MECP2LOF exosomes, whereas Cadherin 2 and L1CAM were unchanged. These results indicate that MECP2 loss-of-function leads to specific protein alterations in exosomes and that exosome protein cargo is not merely a replica of cellular alterations. Mechanisms underlying the differential packaging of exosome cargo have yet to be elucidated.
Fig. 2.
Exosomes contain signaling proteins that are altered with MECP2LOF. (A) Quantitative mass spectrometry comparing exosomes from MECP2LOF or isogenic CRISPR control hiPSC-derived neural cultures identified 2,572 proteins. The PANTHER overrepresentation test was performed on the dataset of 237 proteins with >1.5× change between MECP2LOF or control exosomes using the GO “biological processes” complete annotation dataset. The 237 candidates were classified into annotated GO biological process categories and compared with the normal human database to determine whether they are overrepresented or underrepresented for a given GO biological process. Fold enrichment (x-axis) is the ratio of proteins classified in each GO category from the experimental dataset relative to the number of proteins predicted to be in the GO category from the reference normal human dataset. Positive values indicate overrepresentation of the GO category in the experimental dataset. The y-axis shows the top 10 GO biological processes by P values plotted as a heat map, where color intensity depicts −log(P value) from 15.98 (bright red) to 10.38 (light red). “Neurogenesis” and “nervous system development” are among the most significantly enriched GO categories. (B) Top 10 canonical pathways from Ingenuity analysis using the dataset of 237 proteins. Blue bars, associated with the left y axis, show significance values plotted as −log(P value), and square orange markers, associated with the right y-axis, show the overlap ratios of the number of proteins from our dataset relative to the total number of proteins annotated to each canonical pathway. (C) Bubble chart showing Ingenuity analysis of the relative strength of downstream effects for biological functions and diseases in the nervous system using the dataset of 237 proteins showing >1.5× difference. The top 10 predicted biological functions are plotted vs. significance values plotted as −log(P values), where the bubble size represents the number of proteins from the dataset annotated for given function. The data values for bubble sizes are shown on the bubbles. (D) Functional protein association networks of 2 predicted biological functions from C, neuronal development and neuronal proliferation, are plotted by using the STRING database. The exosome proteins involved in 2 functions form robust networks and show significant overlap. The line thickness represents the confidence of association from highest (0.9) to medium (0.4).
Control Exosomes Increase Proliferation and Neuronal Differentiation Whereas MECP2LOF Exosomes Have No Effect.
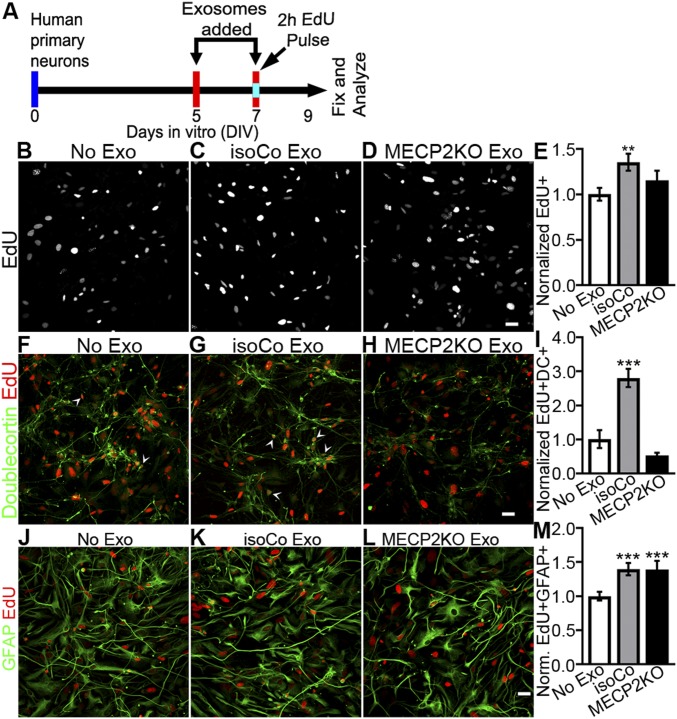
The proteomic analysis just described predicts that MECP2LOF and control exosomes may have different effects on cell proliferation and neuronal differentiation. To test these hypotheses, we treated recipient human primary neural cultures with exosomes from MECP2LOF or control donor cultures on DIV 5 and 7 (Fig. 3A). On DIV 7, cultures received a 2-h pulse of 10 µM EdU before addition of exosomes. The cultures were fixed on DIV 9, and EdU-labeled NPC progeny were analyzed to assess cell proliferation. The isoCONTROL exosome treatment increased EdU labeled progeny by a factor of 1.35× ± 0.09 (P = 0.006) compared with no exosome treatment, whereas treatment with MECP2LOF exosomes was comparable to addition of media alone (Fig. 3 B–E). These data indicate that isogenic control exosomes promote NPC proliferation and survival of EdU-labeled progeny, whereas these proliferation and survival signals are lacking in MECP2LOF exosomes, consistent with our proteomic data analysis.
Fig. 3.
Isogenic control exosomes increase proliferation and neuronal fate in developing neural cultures, whereas MECP2LOF exosomes have no effect. (A) Protocol for treatment of human primary neural cultures with exosomes to assay proliferation, survival, and cell fate. Human primary neural cultures were treated with media alone or exosomes on DIV 5 and 7. On DIV 7, cultures were exposed to 10 µM EdU for 2 h just before the second exosome or media treatment. Cultures were fixed and immunolabeled for analysis on DIV 9. (B–E) Isogenic control exosomes increase cell proliferation. (B–D) Confocal images show EdU-labeled human neural cultures treated with media (B, No exo), isogenic control exosomes (C, IsoCo exosomes), and MECP2LOF exosomes (D). (E) Treatment with isogenic control exosomes (gray bars) increased EdU-labeled progeny by 1.35× ± 0.09 (P = 0.006) compared with no exosomes (white bar), whereas MECP2LOF exosome treatment (black bars) had no effect (E). Edu+ cell numbers normalized to no-media condition. (F–I) Isogenic control exosomes increase neuronal differentiation. Confocal images of EdU-labeled (red) and doublecortin-labeled (green) cultures treated with media (F), isogenic control exosomes (G), and MECP2LOF exosomes (H). Control exosomes (gray bars) increased neuronal progeny 2.8× ± 0.27 (P = 0.005), whereas MECP2LOF exosomes (black bars) had no effect (I). (J–M) Isogenic control exosomes and MECP2LOF exosomes increase astrocyte differentiation. Confocal images of EdU-labeled (red) and GFAP-labeled (green) cultures treated with media (J), isogenic control exosomes (K), and MECP2LOF exosomes (L). Control as well as MECP2LOF exosome treatments increase Edu+GFAP+ astroglial progeny by 1.4× ± 0.09 (P = 0.006) and 1.4× ± 0.13 (P = 0.009), respectively (M). n = 4 wells per group. Statistics computed with 2-way ANOVA with Bonferroni correction. (Scale bar, 20 µm.)
To investigate whether cells generated in response to exosome treatment differentiate into neurons, we birth-dated cells with EdU, then immunolabeled the cultures with doublecortin (DC), a marker of immature neurons, and GFAP, to identify astrocytes. The isoCONTROL exosome treatment increased EdU+DC+ double-labeled neurons 2.8× ± 0.27 (P = 0.005) compared with control conditions (media alone, no exosomes), whereas MECP2LOF exosome treatment did not affect numbers of EdU+DC+ double-labeled neurons (Fig. 3 F–I). The isoCONTROL and MECP2LOF exosome treatments both increased EdU+GFAP+ double-labeled astroglia compared with addition of media alone (Fig. 3 J–M), indicating that increased astroglia generation was proportional to the increase in progeny. These results show that isoCONTROL exosomes enhance neuronal fate and expand the neuronal pool. Whereas MECP2LOF and control exosomes had an equivalent capacity to enhance astroglial formation, MECP2LOF exosomes lacked signals that influence neuronal fate. These results are consistent with our proteomic data analysis predicting neurogenic effects of exosome cargo (Fig. 2D). The selective effect of MECP2LOF exosomes on increased astrocyte fate supports previous studies demonstrating a beneficial role for noncell autonomous signaling from astrocytes to neurons in a mouse model of Rett syndrome (25).
isoCONTROL Exosomes Restore Number of Neurons in MECP2-Knockdown Neural Cultures.
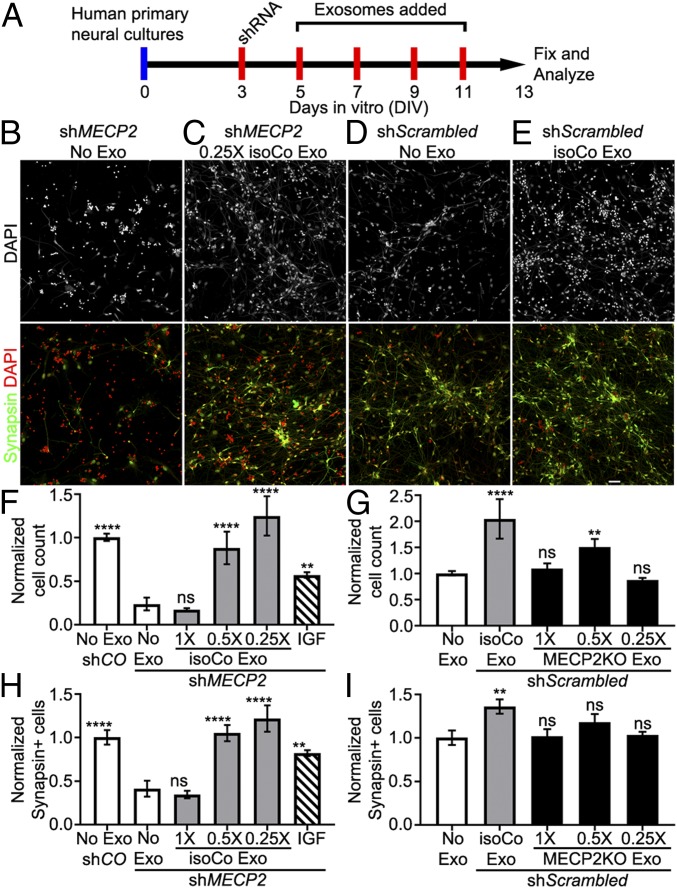
To test the effect of control exosome treatment on MECP2LOF neural cultures, we knocked down MECP2 in human neural cultures by using MECP2 shRNA lentivirus, as in our previous study (15). MECP2 shRNA or scrambled control lentivirus (shMECP2 or shScrambled) was added to human primary neural cultures on DIV 3, followed by treatment with isoCONTROL exosomes on DIV 5, 7, 9, and 11 (Fig. 4A). We tested the effect of 3 doses of isoCONTROL exosomes on MECP2-knockdown cultures by serially diluting the purified exosomes (1× dose) to 0.5× and 0.25× relative concentrations to investigate the relationship of dose with bioactivity. The cultures were fixed on DIV 13 and immunolabeled with the neuronal marker Synapsin and DAPI (Fig. 4 B–E and SI Appendix, Fig. S7). We used insulin like growth factor-1 (IGF1) as a positive control (Fig. 4E), as it has been shown to partially restore some of the MECP2 loss-of-function phenotypes by increasing expression of synaptic proteins, enhancing excitatory synaptic transmission, and restoring dendritic spine densities (15). Without exosome treatment, MECP2KD decreased total cell numbers and neurons compared with shScrambled control cultures (Fig. 4 F–H and SI Appendix, Fig. S7A). Treatment with isoCONTROL exosomes rescued the decreased total cell number (Fig. 4 B, C, and F and SI Appendix, Fig. S7 A–D) and the decrease in neurons in the MECP2KD cultures (Fig. 4 B, C, and G and SI Appendix, Fig. S7 A–D) and increased total cell number and neurons in shScrambled control cultures (Fig. 4 G and I). Whereas the 1× dose of control exosomes was ineffective, the 0.5× and 0.25× doses produced complete rescue, in contrast with partial rescue with IGF treatment.
Fig. 4.
Control exosomes rescue the reduced number of neurons resulting from MECP2 knockdown, but MECP2LOF exosomes have no adverse affect. (A) Protocol for treatment of human primary neural cultures with exosomes to assay changes in neurons. Human primary neural cultures were infected with lentivirus expressing shRNA to knock down MECP2 (green bars in F–H) or control scrambled shRNA (blue bars in F–I). Cultures were treated with media alone (No exosomes, white bars in F–I), 1×, 0.5×, or 0.25× dose of control exosomes (gray bars in F–I) or MECP2LOF exosomes (black bars in F–I), or 10 ng/mL IGF (striped bars in F–H) on DIV 5, 7, 9, and 11. On DIV 13, cultures were fixed and immunolabeled with Synapsin1 and counterstained with DAPI to quantify the number of neurons and total cells, respectively. (B–E) DAPI labeling (grayscale; Top) and results from independent experiments (Bottom) with separate DAPI (red) and Synapsin1 (green) double-labeled wide-field fluorescent images from cultures with MECP2 knockdown (MECP2KD) treated with media alone (no Exo; B) and 0.25× control exosomes (C). Images from cultures with control shRNA and no exosomes (D) or isogenic control exosomes (E). (F and G) Plots of cell counts normalized to control cultures with no exosomes (no Exo shCO). MECP2KD (shMECP2, no exosomes) reduces the number of cells to 0.24× ± 0.07 (P = 0.0001) of control conditions (scrambled shRNA control, No exosomes). Treatment with 0.5× and 0.25× doses of control exosomes rescued the decreased cell number seen with MECP2KD without exosomes to 0.88× ± 0.19 (P = 0.0001 vs. MECP2KD, no exosomes) and 1.25× ± 0.22 (P = 0.0001 vs. MECP2KD, no exosomes) of control, respectively. IGF treatment showed partial rescue to 0.57× ± 0.03 (P = 0.007 vs. MECP2KD, no exosomes) of control. (G) MECP2KD (No exosome) decreases the number of neurons to 0.42× ± 0.09 control (CO-No exosome; P = 0.0001). Treatment with 0.5× control exosomes and 0.25× control exosomes rescued the number of neurons to 1.05× ± 0.09 and 1.22× ± 0.15 of control, respectively (P = 0.0001 each vs. MECP2KD, no exosomes). IGF treatment showed partial rescue to 0.8× ± 0.03 of control (P = 0.003 vs. MECP2KD, no exosomes). (H and I) In human primary neurons infected with lentivirus expressing control scrambled shRNA, treatment with isogenic control exosomes increased the number of cells 2.05× ± 0.69 (P = 4.4 × 10−4) compared with controls (no exosome) (H). The treatment with 0.5× MECP2LOF exosomes increased the number of cells by 1.48× ± 0.38 (P = 0.0001), whereas treatments with 1× MECP2LOF and 0.25× exosome doses did not show statistical differences from control. (I) Control exosome treatment increased the number of neurons 1.36× ± 0.08 (P = 0.004) compared with control (no exosomes), but none of the MECP2LOF exosome treatments altered neuron numbers compared with control (n = 3 for each group). Statistics computed with 2-way ANOVA with Bonferroni correction. (Scale bar, 20 µm.) ns, not significant.
To test whether MECP2LOF exosomes could contain negative signals that contribute to MECP2LOF phenotypes (15), we treated neural cultures with the control scScrambled virus and then added MECP2LOF exosomes harvested from the hiPSC-derived neural donor cultures on DIV 5, 7, 9, and 11 (Fig. 4A). We tested 3 doses of MECP2LOF exosomes by serially diluting the 1× stock to 0.5× and 0.25×. The cultures were fixed on DIV 13 and immunolabeled with Synapsin and DAPI to determine numbers of neurons and total cells respectively. Whereas treatment with isoCONTROL exosomes increased total cell number and the number of neurons compared with addition of media alone (Fig. 4 D, E, G, and I and SI Appendix, Fig. S7 E–H), MECP2LOF exosomes did not show an adverse effect on total cell number or numbers of neurons (Fig. 4 G and I). On the contrary, the 0.5× dose of MECP2LOF exosomes increased the total cell number compared with control cultures without exosome addition (Fig. 4G). These results suggest that MECP2LOF exosomes are capable of signaling. The data further suggest that MECP2LOF exosomes do not have a dominant acute deleterious effect on cell proliferation or neuronal differentiation. Together with our proteomic analysis, these results suggest that isoControl exosomes contain signaling codes that are likely to have different outcomes depending on dose and the state of recipient cells. They further suggest that MECP2LOF exosomes lack signals that increase neuronal differentiation.
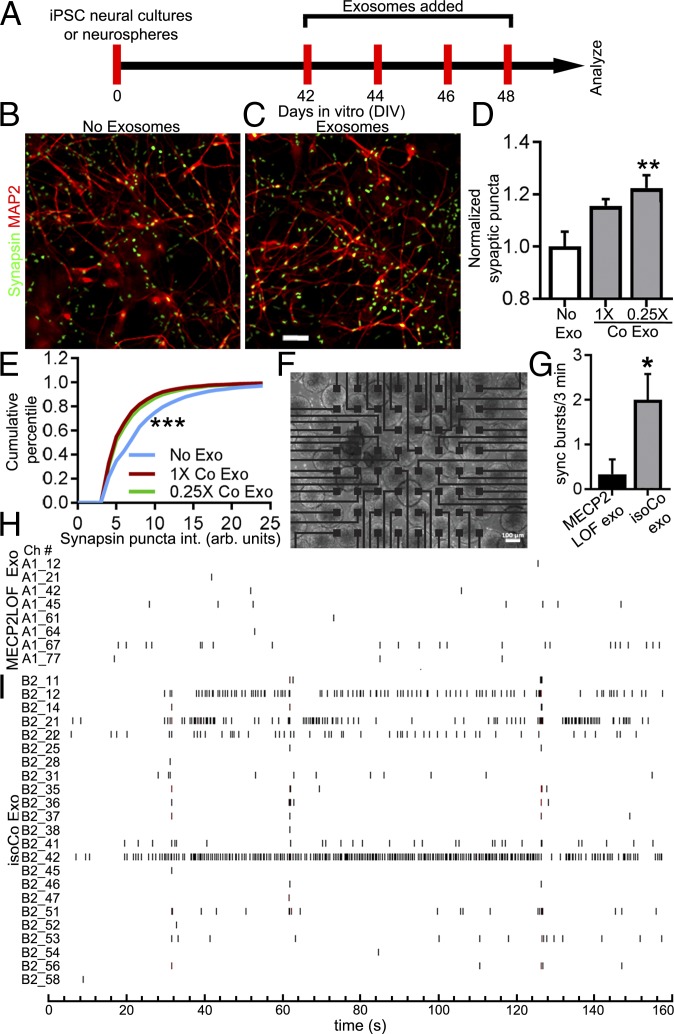
Control Exosome Treatment Increases Synapse Density and Neuronal Firing in MECP2LOF hiPSC-Derived Neurospheres.
MECP2 knockdown in human neuronal cultures decreases synapses and activity-driven calcium transients compared with control neurons (15). We tested whether isogenic control exosomes have the capacity to rescue these synaptic and circuit deficits. To assay the effect of exosome treatment on synaptogenesis, we differentiated MECP2LOF hiPSC-derived NPCs into mature 6-wk-old neural cultures and treated the cultures 4 times with 1× or 0.25× doses of control exosomes over 8 d, as described in Fig. 5A. After exosome treatments, the cultures were fixed and labeled with antibodies against MAP2 to label neurons and Synapsin1 to label presynaptic puncta. Images were analyzed for quantitative changes in synaptic puncta number and intensity. Neural cultures treated with the low dose of control exosomes had significantly higher puncta density compared with cultures without exosome treatment (Fig. 5 B–D). Furthermore, treatments with control exosomes shift the distribution of puncta intensities toward lower intensities (Fig. 5E), suggesting that the increase in synaptic density seen in Fig. 5D may be caused by an increase in lower-intensity puncta. Together, these data suggest that control exosomes increase synaptogenesis in MECP2LOF hiPSC-derived neural cultures.
Fig. 5.
Control exosome treatment increases synapse density and neuronal firing in MECP2LOF hiPSC-derived neurospheres. (A) Protocol for treatment of MECP2LOF hiPSC-derived neural cultures or neurospheres with control exosomes or media alone to assay synapses and synchronized firing. (B–D) Control exosomes increased synapse density in MECP2LOF iPSC-derived neural cultures. Cultures were fixed on DIV 50 and labeled with the neuronal antibody MAP2 and Synapsin1 to identify synaptic puncta. Synaptic puncta density and intensity were quantified. Images of MAP2 (red) and Synapsin1 (green) labeling in MECP2LOF neural cultures without exosome treatment (B) or with control exosome treatment (C). (D) Exosome treatments increased synapse density. Treatment with control exosomes (0.25× dose) increased presynaptic puncta density to 1.22× ± 0.05 of control (no exosome) values (P = 0.006, n = 4 wells each, 2-way ANOVA with Bonferroni correction). (E) Kolmogorov–Smirnoff plot of cumulative frequency of synaptic puncta intensity shows that control exosome treatments (1× control [red line, D-stat = 0.24, P < 0.0001] and 0.25× control [green line, D-stat = 0.21, P < 0.0001]) increase the fraction of low-intensity puncta compared with no exosome treatment (blue line). (F–I) Treatment of MECP2LOF neurospheres with control exosomes increased neural circuit activity. MECP2LOF hiPSC-derived neurospheres were plated on 64-channel multielectrode array (MEA; F) and treated with MECP2LOF exosomes or control exosomes. (G) Graph showing that synchronized bursts of activity occur with a greater frequency in MECP2LOF neurospheres treated with isogenic control exosomes compared with neurospheres treated with MECP2LOF exosomes (MECP2LOF exo, 0.33 ± 0.33 bursts per 3 min; control exo, 2.0 ± 0.68 bursts per 3 min; P = 0.03, n = 3 arrays each, 2-tailed t test). (H and I) Aligned raster plots of spiking activity over 3 min of recordings from active channels. MECP2LOF neurospheres treated with isogenic control exosomes (I) have more overall activity and more synchronized activity across different electrodes compared with neurospheres treated with MECP2LOF exosomes (H). Note also the fewer active channels in neurospheres treated with MECP2LOF exosomes. (Scale bars: B and C, 50 µm; F, 100 µm.)
Neurospheres generated from hiPSC-derived neural cells are a convenient 3-dimensional experimental system for measuring neuronal activity and connectivity in a network using multielectrode arrays (MEAs). Neurospheres mature into a network that displays synchronized neuronal firing (26). To test whether exosome treatments affect circuit development, we analyzed neuronal firing in neurospheres treated with control exosomes compared with MECP2LOF exosomes, which do not appear to have deleterious effects on neuronal differentiation (Fig. 4). We generated neurospheres from hiPSC MECP2LOF-derived NPCs and plated them onto MEAs (Fig. 5F). Neurospheres were treated with exosomes 4 times over 8 d (Fig. 5A), and activity across the MEA was evaluated. Raster plots of spike recordings from individual electrodes show that neuronal firing in MECP2LOF neurospheres treated with MECP2LOF exosomes is sparse and activity across electrodes is not synchronized (Fig. 5H). By contrast, treatment with control exosomes increased neuronal activity in MECP2LOF neurospheres compared with treatment with MECP2LOF exosomes. In particular, neurospheres treated with control exosomes had more activity per channel and more active channels than neurospheres treated with MECP2LOF exosomes (Fig. 5 H and I). Furthermore, the aligned raster plots showed more synchronized activity across multiple channels in neurospheres treated with control exosomes compared with neurospheres treated with MECP2LOF exosomes (Fig. 5 H and I), indicating that control exosome treatment increases neuronal and network activity in MECP2LOF neurospheres. Together, these results indicate that control exosome treatments are capable of increasing presynaptic puncta density and increasing activity in neural circuits.
Discussion
The potential function of exosomes in neural circuit development is unclear. Here we show that exosomes play a profound role in neural circuit development leading to enhanced neural progenitor proliferation, neuronal differentiation, and circuit connectivity. Furthermore, we show that exosomes from MECP2LOF hiPSC-derived neural cultures fail to promote neural circuit development, whereas exosomes from isogenic control hiPSC-derived neural cultures ameliorate developmental deficits, including neurogenesis, synaptogenesis, and circuit connectivity, in human neural cultures or neurospheres with MECP2LOF. Quantitative proteomic analysis showed that exosomes from human neural cultures are enriched in proteins known to play diverse roles in neuronal circuit development. Exosomes from MECP2LOF and control neural cultures differed significantly in signaling proteins involved in neuronal circuit development. Together our data demonstrate that exosomes facilitate the development of human neural circuits, that treatments with control exosomes rescue neurodevelopmental defects in a model of Rett syndrome, and that mutation of a single gene, MECP2, results in large-scale changes in exosome protein cargo and exosome signaling bioactivity.
Exosomes have been hypothesized to play important roles in the nervous system and have been shown to be involved in neurodegenerative disorders (4, 27–29), axonal pathfinding (30), and synaptic pruning (31); however, the potential role of exosomes in normal neural development or neurodevelopmental disorders is unclear. Here, we provide quantitative analysis of differences in proteomic content of exosomes from MECP2LOF iPSC-derived neural cultures compared with CRISPR/Cas9-corrected isogenic control exosomes. The use of isogenic control neural cultures generated by correcting the MECP2 mutation minimizes effects of differences in genetic background to parse out differences specifically resulting from MECP2 mutation. Our proteomic analysis identified ∼240 differentially expressed neural proteins between MECP2LOF and control exosomes, which are predicted to affect proliferation, neuronal development, and synaptic maturation of neurons, leading to abnormal connectivity of neural circuits. Our data indicate that exosomes play diverse roles in neuronal circuit development. Given the spatial and temporal complexity of circuit development, we used an experimental design in which we examined the effects of exosome treatments at different developmental time points of circuit development. This allowed us to demonstrate effects of exosomes on cell proliferation, assayed at relatively early time points; neuronal differentiation and apoptosis, assayed at intermediate time points; and synaptogenesis and circuit connectivity, assayed at later time points. It is interesting to note that, for any given neurodevelopmental function, exosome cargo includes multiple components within candidate signaling pathways, and also includes both positive and negative regulators. This complexity of exosome cargo suggests that it is unlikely that any individual cargo would account for particular neurodevelopmental outcomes we have studied. In addition, many classes of signaling molecules, like Ephrins and Trk receptors, play distinct roles in neuronal development and function that change over time and place, consistent with our observations that different doses of exosomes affect different outcome measures, such as proliferation, differentiation, and synaptogenesis. The diverse outcomes of exosome treatments we observe are consistent with proteomic cargo identified in control exosomes. Taken together, these data suggest that the effect of exosome signaling is likely to be a combinatorial sum of overlapping and sometimes opposing signaling networks, consistent with our observation that different doses of exosomes generate different responses in neurodevelopmental outcome measures.
Our data indicate that treatment with exosomes released from control neural cultures rescue several phenotypes of deficient neural circuit development seen with MECP2KD in vitro (15), including deficits in neurogenesis, synaptogenesis, and neural circuit connectivity. These data are consistent with the proteomic data indicating that exosome cargo are predicted to affect multiple processes that occur over the course of neural development. An important feature of exosomes is that they can signal over a variety of length scales and act non–cell-autonomously. This may be particularly relevant to studies that have shown that restoration of MECP2 function in glia can rescue neuronal and behavioral deficits in mouse models, consistent with long-range non–cell-autonomous mechanisms underlying rescue (25).
Exosomes have been hypothesized to play a pathological role in several neurological disorders, particularly proteinopathies, by spreading pathological molecules to healthy tissue. Alternatively, exosomes have been postulated to play a protective role by dumping pathological molecules out of cells (32, 33). In the context of healthy human neural cultures, we find that exosome treatment promotes neural development. In the context of the neurodevelopmental deficits resulting from MECP2 loss of function, our data indicate that MECP2LOF exosomes do not play a pathogenic role in circuit development. Furthermore, MECP2LOF exosomes neither rescue nor exacerbate neuronal circuit defects resulting from MECP2LOF.
In conclusion, our data demonstrate that neural exosomes have intercellular signaling bioactivity that has functional impact in a neurodevelopmental disease model. Furthermore, we show that exosomes are able to reverse some of the pathological phenotypes observed in MECP2 mutant neurons, indicating that exosomes could have therapeutic applications in brain disorders.
Experimental Methods
All procedures and protocols have been approved by the Institutional Animal Care and Use Committees at The Scripps Research Institute and University of California San Diego.
Human iPSC-Derived Neural Cultures.
To generate neural progenitor cells (NPCs), cells were differentiated and maintained as previously described (15, 34). A male patient cell line with MECP2 Q83X (noted as MECP2 LOF) mutation, its male parental control, and an isogenic rescue, corrected using CRISPR/Cas9 genome editing, were used in this study (16). Two different clones of control (isogenic rescue or father of Q83X patient, WT83) iPSCs and MECP2LOF iPSCs were used in our studies to mitigate line-to-line variation. iPSCs lines maintained in mTESR media were switched to N2 (DMEM/F12 media supplemented with 1× N2 NeuroPlex Serum-Free Supplement [Gemini] with the dual SMAD inhibitors, 1 μM of dorsomorphin [Tocris], and 10 μM of SB431542 [Stemgent]) daily for 48 h. After 2 d, colonies were scraped off and cultured under agitation (95 rpm) as embryoid bodies (EB) for 7 d using N2 media with dorsomorphin and SB431542. Media was changed every other day. EBs were then plated on Matrigel-coated dishes and maintained in DMEM/F12 supplemented with 0.5× of N2 supplement, 0.5× Gem21 NeuroPlex Serum-Free Supplement (Gemini), 20 ng/mL basic fibroblast growth factor (bFGF; LifeTechnologies), and 1% penicillin/streptomycin (P/S). After 7 d in culture, rosettes from the plated EBs were manually selected, gently dissociated with StemPro Accutase (Life Technologies), and plated onto 10 μg/mL poly-l-ornithine (Sigma)/5 μg/mL Laminin (Life Technologies)-coated plates. NPCs were maintained in DMEM/F12 with 0.5× N2, 0.5× Gem21, 20 ng/mL bFGF, and P/S. NPCs were expanded as soon as they had reached confluence by using StemPro Accutase for 5 min at 37 °C, centrifuged, and replated with NGF with a 1:3 ratio in poly-l-ornithine/Laminin-coated plates. To induce cortical neuron differentiation, FGF was retrieved as previously described (15). The cortical neurons were allowed to differentiate for 4 wk before exosome collection. The media was collected every 2 d for exosome isolation. All media formulations used were serum-free.
All cell lines used have been authenticated. All cell lines have tested negative for mycoplasma. Cytogenetic analysis was performed in all clones to evaluate correct cell karyotype (Children’s Hospital Los Angeles). In addition, all cell lines have been genotyped to ensure the presence of the mutation. The absence of the MECP2 protein in the patient cell line has been verified by immunohistochemistry.
Human Primary Neural Cultures.
Human primary neurons were purchased from ScienCell (cat. no. 1520), without identification of sex. Cells were thawed and plated according to the manufacturer’s instructions. After thawing, cells were counted, and 20,000 cells per 96-well plate were plated in serum-free ScienCell Neuronal Medium (cat. no. 1521) onto 10 μg/mL poly-l-ornithine (Sigma)/5 μg/mL Laminin (Life Technologies)-coated plates. Cells were infected with lentivirus expressing shRNA targeted to MECP2 (shMECP2) or scrambled control shRNA (shControl) as described previously (15). As a positive control in the rescue experiments, 10 ng/mL of IGF1 was added to the cultures (cat. no. 100–11R3; PeproTech).
Exosome Purification from hiPSC-Derived Neural Cultures.
Exosomes were enriched from serum-free media from hiPSC-derived neural cultures as described in Fig. 1A (18). Fresh media was replaced in cultures 2 d before collecting exosomes. After 2 d, culture media was harvested for exosome isolation and replaced with fresh growth media. For multiple treatments, exosomes were harvested from the media collected from the same cultures every 2 d. The harvested media was centrifuged at 300 × g for 15 min at 4 °C, and the supernatant was collected and centrifuged at 2,000 × g for 15 min at 4 °C in a Beckman tabletop centrifuge. The second supernatant was further centrifuged using a Beckman ultracentrifuge at 10,000 × g for 45 min at 4 °C. The supernatant was collected and centrifuged at 100,000 × g for 1 h at 4 °C to pellet exosomes. The exosome pellet was washed twice with d-PBS solution containing Ca2+ and Mg2+ (GIBCO) at 100,000 × g for 1 h at 4 °C, followed by resuspension in normal serum-free growth media described earlier. Exosomes were used for assays immediately after purification, as we observed decreased bioactivity when exosomes were stored overnight at 4 °C. Exosome collection was consistently started at week 4 from identically and simultaneously plated isoCONTROL and MECP2LOF neural cultures. Purified exosomes were added to recipient cultures every 2 d as described in the figures. The protein content of 1× control exosomes was 0.190 ± 0.031 mg/mL, and that of 1× MECP2LOF was 0.184 ± 0.033 mg/mL. This is 500× enriched from conditioned media. Comparable amounts of exosomes were added at each time point. There was no statistical difference in exosome yield from healthy differentiated neural cultures from week 4 to week 6. The exosome preparation for proteomics was prepared in a similar way from 2-d media from 4- to 6-wk-old neural cultures.
Proteomic Analysis.
Twelve exosome preparations, half from MECP2LOF iPSC-derived neural cultures and half from isogenic control iPSC-derived neural cultures, were labeled with TMT isobaric tags for quantitative mass spectrometry (MS) analysis. Two independent MS experiments were performed, each with 3 different MECP2LOF preparations and 3 different control preparations obtained from at least 3 independent batches of neuronal differentiations of NPCs for each line; therefore, 6 biological replicates were analyzed together in each MS experiment. The TMT labeling was performed according to the manufacturer’s instructions (Thermo Fisher Scientific) with modifications as previously described (35). Before loading on a MudPIT trapping column, the labeled peptide mixture was diluted 5 times with buffer A (5% acetonitrile/0.1% formic acid) and then centrifuged at 12,000 × g for 30 min to remove particulates.
LC-MS/MS analysis was performed on an LTQ Orbitrap Velos instrument (Thermo Scientific) interfaced at the front end with a quaternary HP 1100 series HPLC pump (Agilent Technology) using an in-house–built electrospray stage. The SEQUEST search and DTASelect analysis were performed identically as previously described (35). To confirm the TMT labeling efficiency, the spectra were also searched without these modifications. Census, a software tool for quantitative proteomic analysis (36), was used to extract the relative intensities of reporter ions for each peptide from the MS2 spectra. Analysis of TMT quantification datasets with Census was performed as previously described (35).
Ingenuity Pathway Analysis software version 01–08 (QIAGEN Bioinformatics) was used for annotations and pathway analyses. PANTHER Overrepresentation Test (release 20170413) was performed by using GO Ontology database (released 4/24/2017) using Homo Sapiens database as a reference list and GO biological processes complete as an annotation dataset. Bonferroni correction for multiple testing was applied for statistics.
Exosome/EdU Treatments.
Purified control or MECP2LOF exosomes were resuspended in fresh serum-free growth media and added to lentivirus-infected human primary neural cultures or iPSC-derived neural cultures by completely replacing the media. For neurospheres, the exosomes were resuspended as 10× concentrated stock of doses and added onto neurosphere cultures without replacing media to achieve desired dose concentration. Exosome treatments were performed every second day 2 to 4 times as described in the schematics in the figures. For EdU labeling experiments, human primary neural cultures were treated with control or MECP2LOF exosomes on DIV 5 and 7. On day 7, a 2-h pulse of 10 µM EdU (Thermo Fisher Scientific) was provided before addition of exosomes. The 1× exosome was equivalent to 0.19 ± 0.03 mg/mL of protein content.
Generation of Neurospheres.
To generate functional neural networks, growing NPCs were dissociated with Accutase and plated on 6-well dishes (3–5 × 106 cell per well) under shaker agitation (95 rpm) at 37 °C. The media used was DMEM-F12 supplemented with 0.5× GEM21 (Gemini), 0.5× N2 (Gemini), and 20 ng/mL bFGF (Life Technologies). The next day (day 0), neural fate was induced by removing bFGFs and adding 10 μM Rock inhibitor Y-27632 (Tocris). Two days later (day 2), media was changed to fresh media without Rock inhibitor. The differentiation took place for 2 wk in suspension, with media changed every 4 d. On day 15 of differentiation, 3D neurospheres were plated on poly-ornithine/Laminin–precoated 12-well multielectrode array (MEA) plates (Axion) with DMEM-F12 media supplemented with 0.5× GEM21, 0.5× N2, and 1% FBS. Media was changed every 3 to 4 d. After 1 to 2 wk, media was replaced with Neurobasal media (Life Technologies) supplemented with 1× B-27 (Life Technologies) and 1:400 GlutaMAX (Life Technologies) and changed every 3 to 4 d. Experiments were performed after 4 to 5 media changes in serum-free medium.
Multielectrode Array Recordings.
Spontaneous action potential activity was recorded by using Axion Biosystems. Purified MECP2LOF or control exosomes were resuspended in serum-free neurosphere media and were added directly to MECP2LOF or control neurospheres that had been plated on the multielectrode arrays (MEAs). MEA activity in neurospheres without added exosomes is stable (26). Exosomes were added every second day to neurospheres for 1 to 2 wk. Neurosphere activity was recorded before and 24 h after each exosome addition. MEA channels with similar number and density of cells, and with more than 10 spikes in a 5-min interval were included in the analysis. Recordings were analyzed with the software Neuroexplorer (Nex Technologies) and MATLAB-based Neural Metric Tool Software Axis (Axion Biosystems). Raster plots were generated using Neuroexplorer.
Immunolabeling, Microscopy, and Image Analysis of Human Cultures.
After treatments, cultures were fixed in 4% paraformaldehyde at room temperature (RT) for 15 min and then permeabilized with 0.5% Triton X-100 in d-PBS solution with Ca2+ and Mg2+ at RT for 5 min. Cells were then blocked in 5% normal goat serum in d-PBS solution with Ca2+ and Mg2+ at RT for 1 h before incubation with primary antibody diluted in 5% normal goat serum overnight at 4 °C. After 3 washes with d-PBS solution containing Ca2+ and Mg2+, cells were incubated with Alexa 488, Alexa 568, or Alexa 633 goat anti-mouse IgG, goat anti-rabbit IgG, or goat anti-chicken IgY (depending on host for primary antibodies as listed in the SI Appendix) and (H+L) highly cross-adsorbed secondary antibodies (Invitrogen/Thermo Fisher), followed by 3 washes with d-PBS solution containing Ca2+ and Mg2+. During the last wash, the cultures were labeled with DAPI solution (3 ng/mL) for 10 min and mounted in Prolong Gold mounting media (Invitrogen/Thermo Fisher). Each experimental condition was blinded before imaging. Imaging was performed with Plan Apo 20× objective with 0.75 NA on a Zeiss LSM 710 or Nikon A1 confocal system with an Andor iXon EMCCD camera system. Images were collected from randomized areas in each dish and analyzed by using Metamorph software (Molecular Devices). Synaptic puncta analysis was performed by using a custom ImageJ plugin. A trimming procedure was applied to isolate and segregate individual puncta based on the inclusion of pixels with intensities greater than 0.3 of maximal intensity within a unit of connected pixels having intensity greater than the set threshold. This was achieved by an iterative procedure using a step size (0.01) for each iteration, which limits for the number of iterations of the trimming procedure. The resultant image consists of a background-subtracted image containing a set of well-defined puncta and provides quantification of net intensity per spot and total pixel area per spot.
Primary Rat Neuron Culture.
Cortices from E18 rat embryos were dissected in Hank’s buffered salt solution (HBSS) supplemented with Hepes (10 mM) and glucose (0.66 M; Sigma). Cortices were dissociated in papain (Worthington) supplemented with DNase I (100 μg/mL; Sigma) for 20 min at 37 °C, washed 3 times, and manually triturated in plating medium supplemented with DNase. Cells were then plated at 3.5 × 106 cells per 10 cm2 on dishes coated with poly-d-lysine (1 mg/mL; Sigma) in neurobasal medium supplemented with 2.5% FBS (Gemini), B27 (1×), l-glutamine (2 mM), and P/S (penicillin 2.5 U/mL/streptomycin 2.5 μg/mL). Unless otherwise indicated, all products were from Invitrogen. After 1 d in vitro, medium was replaced completely with serum-free neurobasal medium with B-27 (1×), l-glutamine (2 mM), P/S, and 1 µM FUDR (5-fluorodeoxyuridine), and one half of the medium was then changed every 5 d.
Exosome Purification from Rodent Cultures.
The media was harvested from DIV9 E18 rat primary neural cultures and centrifuged at 300 × g for 15 min at 4 °C. The supernatant was collected and centrifuged at 2,000 × g for 15 min at 4 °C in a Beckman tabletop centrifuge. The second supernatant was further centrifuged by using a Beckman ultracentrifuge at 10,000 × g for 45 min at 4 °C. The supernatant was collected and centrifuged at 100,000 × g for 1 h at 4 °C to pellet exosomes. The exosome pellet was resuspended in 1 mL PBS solution and divided equally into 2 tubes. The tubes were incubated at 37 °C for 30 min after 100 µg/mL of Proteinase K was added to one and an equal volume of PBS solution to the other. The exosomes were then washed twice with PBS solution (Gibco) at 120,000 × g for 1 h at 4 °C followed by resuspension in PBS solution to an effective enrichment of 2,400× compared with conditioned culture media. Proteinase K treatment depletes exosomes of proteins (SI Appendix, Fig. S5).
P4 Mice Injections.
A litter of P4 mice was divided into 2 groups, and 2 µL of 2,400×-enriched exosomes (∼3 ± 0.75 µg protein) from DIV9 E18 rat neural cultures were injected into the lateral ventricle (LV) of each cerebral hemisphere. One group was injected with proteinase K-treated exosomes, whereas the other group was injected with control exosomes. Injections were carried out manually with 1.0-mm glass needles (World Precision Instruments) pulled with P-97 flaming/brown micropipette puller (Sutter Instruments). A glass needle with graduated 1-µL volume marks was attached to the syringe with a tube. Intracerebroventricular injections were followed by peritoneal injections of EdU (5-ethynyl-2′-deoxyuridine; Lumiprobe) at a dose of 25 mg/kg by using a Hamilton syringe.
Sectioning and Immunolabeling.
Twenty-four hours after exosome and EdU injections, the brains were dissected and fixed in 4% paraformaldehyde for 24 h at 4 °C. Fixed brains were embedded in 4% low-melting agarose and sliced into 50-µm sections in sagittal plane by using a vibratome (Leica). EdU incorporated into proliferating cells was detected by conjugating with Sulfo-Cyanine3 azide dye (Lumiprobe) by using click chemistry. Briefly, sections were permeabilized with 0.5% Triton-X 100 for 30 min at room temperature, and click reaction with Sulfo-Cyanine3 azide was performed for 30 min with CuSO4 as catalyst and ascorbic acid as reductant for copper following the protocol provided by Lumiprobe. After click reaction, sections were washed 5 times with PBS solution for 5 min at room temperature and blocked in 5% normal goat serum (NGS) followed by immunohistochemistry (as described). Sections were labeled with anti-nestin antibody (MAB353; Millipore) and TO-PRO-3 (642/661; T3605; Thermo Fisher Scientific) and mounted in ProLong Gold Antifade (Thermo Fisher Scientific).
Image Processing.
Images were background-subtracted as described; 10× images were used to anatomically match sections through the GCL of the dentate gyrus of different animals. In 20× high-resolution images, regions of interest (ROI) were created around the GCL by using nestin and nuclear labeling as a guide. EdU+ cells were counted inside the GCL ROI by using “Manually Count Objects” dropin of Metamorph image-processing software. EdU-positive cells in the polymorphic layer that were adjacent to the GCL were also included.
Quantification and Statistical Analysis.
Statistical analysis was performed by using Microsoft Excel, Statistics Online Computational Resource (SOCR) of UCLA (socr.ucla.edu/), and GraphPad Prism 7.03. For statistics, 2-way ANOVA with Bonferroni correction applied for multiple comparisons or independent-sample Wilcoxon rank-sum test for pairwise comparisons was performed. P values are provided in figure legends. For cumulative frequency distribution, the Kolmogorov–Smirnoff test was performed. D-stat, the absolute maximum difference between 2 cumulative frequency distribution function, and P values are provided in figure legends.
Supplementary Material
Acknowledgments
We thank Kristin Baldwin and Ardem Patapoutian from comments on the manuscript and members of the laboratory of H.T.C. for helpful discussions. This work was supported by grants from the National Institutes of Health (R01MH108528, R01MH094753, R01MH109885, R01MH100175, and U19MH107367), SFARI Grant 345469, and a NARSAD Independent Investigator Grant to A.R.M.; an International Rett syndrome Foundation (IRSF) fellowship to P.M.; NIH grants (R01MH103134 and R01EY011261) and an endowment from the Hahn Family Foundation to H.T.C.; a fellowship from the California Institute of Regenerative Medicine (CIRM; TG2-01165) and a Fellowship from the Helen Dorris Foundation to P.S.; and NIH Grants 5R01MH067880 and 5R01MH100175 to J.R.Y.
Footnotes
Conflict of interest statement: The authors declare a conflict of interest. A.R.M. is a cofounder and has equity interest in TISMOO, a company dedicated to genetic analysis focusing on therapeutic applications customized for autism spectrum disorder and other neurological disorders with genetic origins. The terms of this arrangement have been reviewed and approved by the University of California San Diego, in accordance with its conflict of interest policies. The remaining authors declare that they have no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902513116/-/DCSupplemental.
References
- 1.Budnik V., Ruiz-Cañada C., Wendler F., Extracellular vesicles round off communication in the nervous system. Nat. Rev. Neurosci. 17, 160–172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma P., Schiapparelli L., Cline H. T., Exosomes function in cell-cell communication during brain circuit development. Curr. Opin. Neurobiol. 23, 997–1004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zappulli V., Friis K. P., Fitzpatrick Z., Maguire C. A., Breakefield X. O., Extracellular vesicles and intercellular communication within the nervous system. J. Clin. Invest. 126, 1198–1207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiasserini D., et al. , Proteomic analysis of cerebrospinal fluid extracellular vesicles: A comprehensive dataset. J. Proteomics 106, 191–204 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Coleman B. M., Hill A. F., Extracellular vesicles–Their role in the packaging and spread of misfolded proteins associated with neurodegenerative diseases. Semin. Cell Dev. Biol. 40, 89–96 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Rajendran L., et al. , Emerging roles of extracellular vesicles in the nervous system. J. Neurosci. 34, 15482–15489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korkut C., et al. , Trans-synaptic transmission of vesicular Wnt signals through Evi/Wntless. Cell 139, 393–404 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korkut C., et al. , Regulation of postsynaptic retrograde signaling by presynaptic exosome release. Neuron 77, 1039–1046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fröhlich D., et al. , Multifaceted effects of oligodendroglial exosomes on neurons: Impact on neuronal firing rate, signal transduction and gene regulation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130510 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frühbeis C., et al. , Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication. PLoS Biol. 11, e1001604 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldman D., Banerjee A., Sur M., Developmental dynamics of Rett syndrome. Neural Plast. 2016, 6154080 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leonard H., Cobb S., Downs J., Clinical and biological progress over 50 years in Rett syndrome. Nat. Rev. Neurol. 13, 37–51 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Zoghbi H. Y., Rett syndrome and the ongoing legacy of close clinical observation. Cell 167, 293–297 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Amir R. E., et al. , Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23, 185–188 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Marchetto M. C., et al. , A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell 143, 527–539 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z. N., et al. , Layered hydrogels accelerate iPSC-derived neuronal maturation and reveal migration defects caused by MeCP2 dysfunction. Proc. Natl. Acad. Sci. U.S.A. 113, 3185–3190 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Souza J. S., et al. , IGF1 neuronal response in the absence of MECP2 is dependent on TRalpha 3. Hum. Mol. Genet. 26, 270–281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thery C., Amigorena S., Raposo G., Clayton A., Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. Chapter 3, Unit 3.22 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Revelo N. H., et al. , A new probe for super-resolution imaging of membranes elucidates trafficking pathways. J. Cell Biol. 205, 591–606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlessinger A. R., Cowan W. M., Gottlieb D. I., An autoradiographic study of the time of origin and the pattern of granule cell migration in the dentate gyrus of the rat. J. Comp. Neurol. 159, 149–175 (1975). [DOI] [PubMed] [Google Scholar]
- 21.Altman J., Bayer S. A., Migration and distribution of two populations of hippocampal granule cell precursors during the perinatal and postnatal periods. J. Comp. Neurol. 301, 365–381 (1990). [DOI] [PubMed] [Google Scholar]
- 22.Calvano S. E., et al. ; Inflamm and Host Response to Injury Large Scale Collab. Res. Program , A network-based analysis of systemic inflammation in humans. Nature 437, 1032–1037 (2005). Correction in: Nature438, 696 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Mi H., Muruganujan A., Casagrande J. T., Thomas P. D., Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexeyenko A., Sonnhammer E. L., Global networks of functional coupling in eukaryotes from comprehensive data integration. Genome Res. 19, 1107–1116 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lioy D. T., et al. , A role for glia in the progression of Rett’s syndrome. Nature 475, 497–500 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nageshappa S., et al. , Altered neuronal network and rescue in a human MECP2 duplication model. Mol. Psychiatry 21, 178–188 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basso M., Bonetto V., Extracellular vesicles and a novel form of communication in the brain. Front. Neurosci. 10, 127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jayaram S., Gupta M. K., Polisetty R. V., Cho W. C., Sirdeshmukh R., Towards developing biomarkers for glioblastoma multiforme: A proteomics view. Expert Rev. Proteomics 11, 621–639 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Tomlinson P. R., et al. , Identification of distinct circulating exosomes in Parkinson’s disease. Ann. Clin. Transl. Neurol. 2, 353–361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong J., Körner R., Gaitanos L., Klein R., Exosomes mediate cell contact-independent ephrin-Eph signaling during axon guidance. J. Cell Biol. 214, 35–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bahrini I., Song J. H., Diez D., Hanayama R., Neuronal exosomes facilitate synaptic pruning by up-regulating complement factors in microglia. Sci. Rep. 5, 7989 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quek C., Hill A. F., The role of extracellular vesicles in neurodegenerative diseases. Biochem. Biophys. Res. Commun. 483, 1178–1186 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Howitt J., Hill A. F., Exosomes in the pathology of neurodegenerative diseases. J. Biol. Chem. 291, 26589–26597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chailangkarn T., et al. , A human neurodevelopmental model for Williams syndrome. Nature 536, 338–343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rauniyar N., Gao B., McClatchy D. B., Yates J. R. 3rd, Comparison of protein expression ratios observed by sixplex and duplex TMT labeling method. J. Proteome Res. 12, 1031–1039 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park S. K., Venable J. D., Xu T., Yates J. R. 3rd, A quantitative analysis software tool for mass spectrometry-based proteomics. Nat. Methods 5, 319–322 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.