Significance
Nicotinamide adenine dinucleotide (NAD) is best known as an essential cofactor of biochemical reactions. In addition, it is involved in the regulation of virtually all major cellular events. These NAD-dependent regulatory functions are mediated by enzymes (e.g., sirtuins, poly–adenosine -diphosphate [ADP] ribose polymerases, ADP-ribosyl cyclases) that cleave the molecule to liberate nicotinamide (Nam). We show that diversification of NAD-dependent signaling in Deuterostomia was accompanied by an optimization of NAD biosynthesis to ensure efficient high-affinity recycling of Nam into NAD through Nam phosphoribosyltransferase (NamPT). In addition, a Nam N-methyltransferase (NNMT) emerged, which facilitates high NAD-dependent signaling turnover by preventing accumulation of inhibitory Nam. This unexpected kinetic interplay between NamPT and NNMT needs to be considered in therapeutic strategies targeting these enzymes.
Keywords: NAD-dependent signaling and biosynthesis, nicotinamide N-methyltransferase (NNMT), nicotinamide phosphoribosyltransferase (NamPT), NAD pathway dynamics and evolution, mathematical modeling
Abstract
Nicotinamide adenine dinucleotide (NAD) provides an important link between metabolism and signal transduction and has emerged as central hub between bioenergetics and all major cellular events. NAD-dependent signaling (e.g., by sirtuins and poly–adenosine diphosphate [ADP] ribose polymerases [PARPs]) consumes considerable amounts of NAD. To maintain physiological functions, NAD consumption and biosynthesis need to be carefully balanced. Using extensive phylogenetic analyses, mathematical modeling of NAD metabolism, and experimental verification, we show that the diversification of NAD-dependent signaling in vertebrates depended on 3 critical evolutionary events: 1) the transition of NAD biosynthesis to exclusive usage of nicotinamide phosphoribosyltransferase (NamPT); 2) the occurrence of nicotinamide N-methyltransferase (NNMT), which diverts nicotinamide (Nam) from recycling into NAD, preventing Nam accumulation and inhibition of NAD-dependent signaling reactions; and 3) structural adaptation of NamPT, providing an unusually high affinity toward Nam, necessary to maintain NAD levels. Our results reveal an unexpected coevolution and kinetic interplay between NNMT and NamPT that enables extensive NAD signaling. This has implications for therapeutic strategies of NAD supplementation and the use of NNMT or NamPT inhibitors in disease treatment.
Nicotinamide adenine dinucleotide (NAD) metabolism has received increasing attention, as a number of pathological states, including neurodegeneration (1), diabetes (2, 3), obesity (4–7), heart diseases (8, 9), muscle dystrophy (10), renal dysfunction (11), and different types of cancer (12–14), have been associated with changes in this complex network. It has been established that a gradual decline in NAD during aging is one of the major driving forces of these age-related pathologies (15–18). In addition, NAD metabolism has been identified to be a key regulator for axonal integrity (19–21). It is therefore not surprising that NAD metabolism has emerged as a promising pharmacological target for disease treatment (22–25). However, to fully exploit the therapeutic potential of NAD metabolism, the dynamic and functional interplay between the individual NAD pathway components needs to be established.
NAD represents one of the most critical links between cellular signal transduction and energy metabolism. Even though it is best known as cofactor for a multitude of redox reactions, NAD is involved in a number of signaling processes that consume NAD by cleaving NAD+ to nicotinamide (Nam) and adenosine-diphosphate (ADP) ribose (ADPR) (14). These NAD-dependent signaling reactions include poly- and mono-ADP ribosylation (26, 27), sirtuin-mediated protein deacylation (28), and the synthesis of calcium-mobilizing molecules such as cyclic ADP ribose (29), and they participate in the regulation of virtually all cellular activities. The enzymes involved in these processes are sensitive to the available NAD concentration. Therefore, NAD-dependent signaling can act as a transmitter of changes in cellular energy homeostasis, for example, to regulate gene expression or metabolic activity (30).
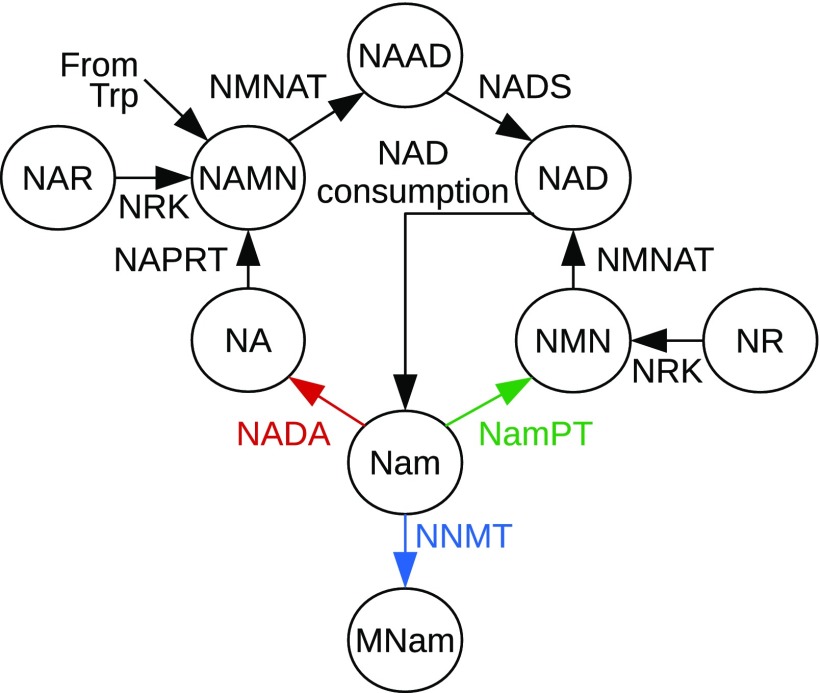
The significance of NAD-dependent signaling for NAD homeostasis has long been underestimated. It has now become clear that inhibition of NAD biosynthesis leads to a rapid decline of the cellular NAD concentration (13, 31). This observation documents that NAD-dependent signaling reactions consume substantial amounts of NAD. The resulting NAD turnover differs in a cell type-specific manner. Measurements of cellular NAD half-life have revealed that it can be as short as 15 min (32). To maintain the NAD concentration at physiological levels, NAD biosynthesis needs to act at an equally rapid rate. Imbalances in NAD homeostasis have been associated with a number of different diseases. In this context, it is conceivable that several recent studies have demonstrated impressive health benefits of dietary supplementation with intermediates of NAD biosynthesis, including, Nam (4), Nam mononucleotide (NMN) (16), and Nam riboside (NR) (2, 6, 17). Apparently, the exploitation of physiologically less active NAD biosynthetic routes, in addition to the use of Nam as a precursor (Fig. 1), results in increased NAD concentrations that stimulate beneficial NAD-dependent signaling processes, particularly protein deacetylation by sirtuins (3, 33).
Fig. 1.
Schematic overview of NAD biosynthesis pathways. NAD can be synthesized from tryptophan (Trp), Nam, NA, and the corresponding ribosides NR and NA riboside (NAR). Nam is the main precursor in humans and also the product of NAD-consuming signaling reactions by enzymes such as sirtuins (NAD-dependent deacylases) or PARPs. For the recycling of Nam, 2 different pathways exist. The pathway found in yeast, plants, and many bacteria starts with the deamidation of Nam by NADA. Further biosynthesis via the Preiss–Handler pathway, which also exists in vertebrates, requires 3 subsequent enzymatic steps catalyzed by NAPRT, NMNAT, and NADS. In vertebrates, Nam is directly converted to NMN by NamPT. The NNMT degrades MNam, which, in mammals, is excreted with the urine. The color code for the 3 different enzymes utilizing Nam is used in subsequent figures to denote the presence of these enzymes in different organisms.
Owing to the continuous release of Nam through NAD-consuming signaling reactions, NAD salvage using Nam as a precursor is the most important NAD synthesis pathway. There are 2 principal pathways that recycle Nam. Vertebrates use a direct 2-step pathway starting with the conversion of Nam into the mononucleotide NMN catalyzed by Nam phosphoribosyltransferase (NamPT) using phosphoribosyl pyrophosphate (PRPP) as a cosubstrate. At least in mammals, a nearly complete recycling of Nam by NamPT is achieved by an extraordinarily high substrate affinity to Nam, with the Michaelis–Menten constant (KM) being in the low nanomolar range (34). This appears to be mediated by an adenosine triphosphate (ATP)–dependent phosphorylation of a histidine residue in the catalytic core (35). Despite the importance of its salvage, Nam can also be marked for excretion by methylation. This reaction is catalyzed by Nam N-methyltransferase (NNMT). The presence of this enzyme in vertebrates (36) is among the most enigmatic and counterintuitive features of NAD metabolism. While NamPT is seemingly optimized to recycle even the faintest amounts of Nam back into NAD synthesis, NNMT seems to have no metabolic function other than to remove Nam from NAD metabolism. However, since NNMT uses the general methylation source S-adenosylmethionine (SAM), it has been suggested that Nam methylation may act as a metabolic methylation sink (37).
In most prokaryotes, as well as in plants and fungi, another pathway consisting of 4 reactions, starting with the deamidation of Nam to nicotinic acid (NA) by the Nam deamidase (NADA), is used (Fig. 1). The 3 enzymes that act downstream of NADA belong to the Preiss–Handler pathway that also exists in vertebrates. In this pathway, NA is converted into the corresponding NA-specific mononucleotide (NAMN) in a reaction performed by the NA-specific phosphoribosyltransferase (NAPRT). The conversion of both mononucleotides, NMN and NAMN, into their corresponding dinucleotides, NAD and NA adenine dinucleotide (NAAD), is catalyzed by the Nam/NA adenylyltransferases (NMNATs) that are essential in all organisms (38). The recycling pathway via NA finally requires reamidation of NAAD by NAD synthase (NADS). This final reaction includes an enzyme adenylation step that consumes ATP. Therefore, the Nam recycling by NADA appears to be energetically less efficient than the recycling pathway starting with NamPT.
We and others have shown earlier that the 2 NAD biosynthesis pathways starting from Nam (Fig. 1) coexist in some eukaryotes (36, 39), as well as in some bacterial species (40). Why these pathways coexist in some organisms and over a very long evolutionary time frame, and why NADA nevertheless disappeared in vertebrates, is not known. Whether the occurrence of NNMT may have contributed to these evolutionary processes has also remained unexplored.
In the present study, we performed a comprehensive phylogenetic analysis of the NAD pathways using 793 eukaryotic and 7,892 prokaryotic genomes. This large-scale analysis revealed that there has been an evolutionary transition resulting in the coexistence of NamPT and NNMT in Deuterostomia, while the deamidation pathway, which is dominant in bacteria, became superfluous. Importantly, this selection for NamPT and NNMT was accompanied by a marked increase in the number of NAD-consuming signaling enzymes. Mathematical modeling of the pathway revealed an unexpected positive kinetic role of NNMT for NAD-consuming signaling fluxes, through prevention of accumulation of Nam, the product of NAD-dependent signaling reactions. In addition, our model predicts that NNMT likely exerted an evolutionary pressure on NamPT to develop a high affinity toward its substrate Nam. Indeed, we identified a short sequence insertion in NamPT, which first occurs in Deuterostomia and appears to modulate the affinity of NamPT. Simulating resource competition, we furthermore show that the presence of high-affinity NamPT together with NNMT makes the NADA-dependent pathway obsolete, providing a rationale for the evolutionary transition of the pathway in Metazoa.
Taken together, our analyses suggest that the coexistence of NamPT and NNMT has been a prerequisite to enable the evolutionary development of versatile NAD-dependent signaling mechanisms present in vertebrates.
Results
Paradoxical Evolutionary Correlation between NAD-Dependent Signaling and Precursor Metabolism.
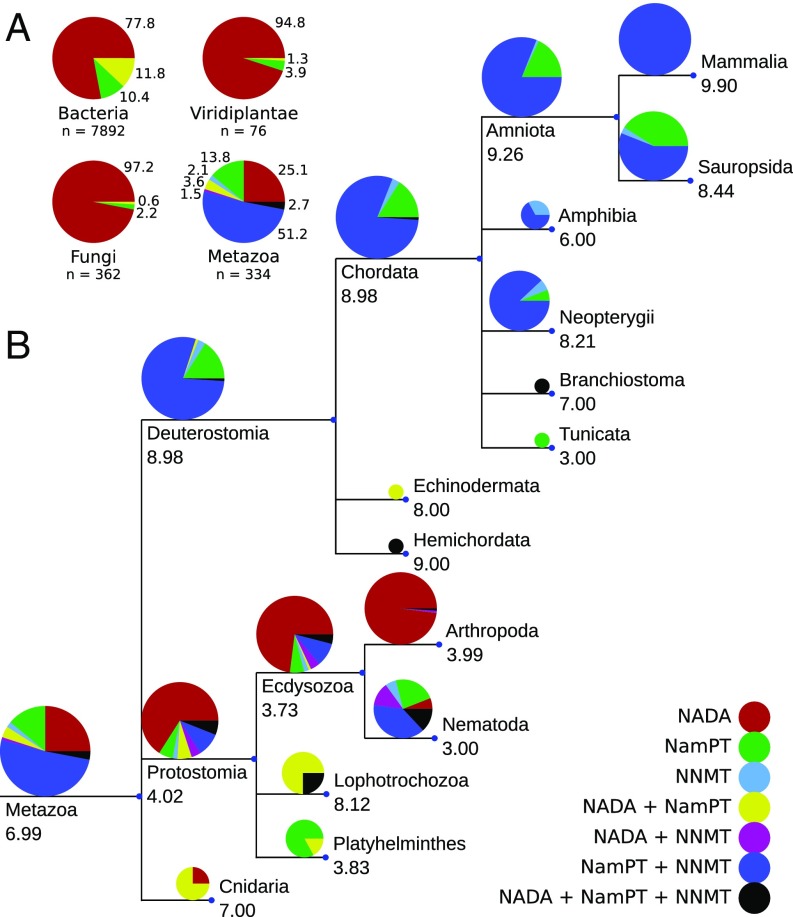
To understand the functional roles and potential interplay between the 3 known enzymes that use Nam as a substrate (NamPT, NADA, and NNMT; Fig. 1), we conducted a comprehensive analysis of the phylogenetic distribution of these 3 enzymes. As shown in Fig. 2A, bacteria, fungi, and plants predominantly possess NADA, and only a very limited number of species harbor NamPT. In contrast, most Metazoa lost NADA, and rather possess NamPT together with NNMT. NADA and NamPT, the 2 enzymes that initiate the 2 different NAD salvage pathways, show a scattered distribution in bacteria. Co-occurrence of these enzymes is rather rare, and has occasionally been found in bacteria (40) and some marine invertebrates (36).
Fig. 2.
Phylogenetic distribution of NADA, NNMT, and NamPT and their relation to the number of NAD consumers. (A) Distribution of NADA, NNMT, and NamPT in selected clades. NADA is dominant in bacteria, fungi, and plants (Viridiplantae), whereas NamPT, together with NNMT, is dominant in Metazoa. Numbers outside the pie charts show the percentage of species per clade, which possess the respective enzyme combinations indicated by the color code explained in the lower right of the figure (n = number of species per clade included in the analysis). (B) Common tree of selected clades within Metazoa, including 334 species. The pie charts indicate the distribution of species within the respective clade that encode the enzyme combination indicated by the different colors. The size of the pie charts is proportional to the logarithm of the number of species analyzed in the particular clade. The numbers below the clade names indicate the average number of NAD-consuming enzyme families found in all species of that clade. The branch length is arbitrary. A detailed analysis of birds is provided in SI Appendix, Fig. S1, and the template sequences used for the analysis are listed in SI Appendix, Table S1.
NNMT seems to have arisen de novo or diverged rapidly in the most recent common ancestor of Ecdysozoa and Lophotrochozoa (Fig. 2B). We were unable to find any indication for the presence of NNMT in fungi or plants (protein Blast [Blastp] e-value cutoff of 0.1). Interestingly, NA can be methylated to trigonelline in plants and bacteria (41), but the corresponding enzyme has no homology to NNMT or any other enzyme in Metazoa. Nematodes are the only organisms in which we observed a concomitant presence of NADA and NNMT. In Deuterostomia, the only large clade that possesses only NamPT and seems to have lost NNMT is the Sauropsida clade, and among sauropsids, especially birds. The reason why about half of the sequenced bird genomes do not seem to harbor NNMT remains unclear. The distribution of NNMT in birds is quite scattered (SI Appendix, Fig. S1). It is possible that detection of NNMT in some bird genomes failed because of their high guanine-cystosine content (42) or because of difficulties in assembling very small chromosomes commonly found in birds. The absence of NNMT might, alternatively, be related to the differences in the excretion systems. In mammals, the product of NNMT, methyl-Nam, is excreted with the urine. There are few metazoan species for which we could not find NamPT or NADA, while NNMT was detected. We assume that this is due to incomplete genomes in the database, as these species are scarce and their distribution appears to be randomly scattered.
In addition to the phylogenetic distribution of the 2 Nam salvage enzymes NADA and NamPT, we analyzed the phylogenetic diversity of enzymes catalyzing NAD-dependent signaling reactions. To do so, we used the previously established classification into 10 different families of NAD-consuming signaling enzymes (36), including poly–ADP ribose polymerase 1 (PARP1) to PARP3, PARP4, PARP6/8, PARP7/9 to PARP15, PARP16, sirtuins, tankyrases, ADPR-cyclases, mono-ADP ribosyltransferases, and t-RNA–phosphotransferases. The detailed list of templates used for the phylogenetic analyses can be found in SI Appendix, Table S1. The numbers shown in Fig. 2B denote the average number of NAD-dependent signaling enzyme families found in each clade (a detailed distribution is provided in SI Appendix, Table S2). With the exception of Cnidaria and Lophotrochozoa, we noted an average of 3 to 4 families in protostomes, whereas most deuterostome species have, on average, more than 8 families, with an increasing diversification of enzymes within some of these families, especially PARPs (43).
Taken together, we found that NADA is lost in vertebrates but strongly preserved in most other organisms, despite the higher energetic requirement of this pathway. Moreover, the transition to having both NamPT and NNMT coincides with a considerable diversification of NAD-dependent signaling. This observation seems counterintuitive, as one would expect that increased NAD-dependent signaling should be compensated for by an increased efficiency of substrate (Nam) utilization for NAD biosynthesis. Since NNMT removes Nam from recycling into NAD, it is not obvious how this enzyme could contribute to higher NAD turnover.
Functional Properties of NamPT and NNMT Have Evolved to Maximize NAD-Dependent Signaling.
To resolve this apparent contradiction, we turned to modeling approaches permitting us to simulate the behavior of the complex NAD metabolic network under different conditions. We built a dynamic model of NAD metabolism based on ordinary differential equations using previously reported kinetic data (details are provided in Methods and Materials and SI Appendix, Table S3).
Given the rather limited information about species-specific expression levels of enzymes, we first assumed equal expression of all enzymes, thereby enabling an initial comparison of metabolic features in rather different organisms. Moreover, due to the lack of specific kinetic data from most organisms, we mainly relied on kinetic constants found for human or yeast enzymes. Wherever possible, we included substrate affinities and known product inhibitions as well as inhibition by downstream metabolites, such as the inhibition of NamPT by NAD (34). Finally, the models assumed that cell growth and consecutive cell divisions are a major driving force for NAD biosynthesis, besides NAD-consuming reactions.
First, we addressed the observed phylogenetic correlation between the transition to the co-occurrence of NamPT and NNMT and the increase in the number of NAD-consuming enzymes. We calculated steady-state NAD concentrations and NAD consumption fluxes by simulating NAD biosynthesis via NamPT in the presence or absence of NNMT (Fig. 3 A and B). Due to the very low turnover number of NamPT (∼0.01 per second), we used 40-fold higher NamPT levels compared with the other enzymes to achieve free NAD concentrations in the range reported in the literature (44). NAD concentrations can be further increased with higher NamPT levels (SI Appendix, Fig. S3 A and B). Since NamPT limits the flux, changing NMNAT levels had no effect under the conditions tested (SI Appendix, Fig. S3 A and B).
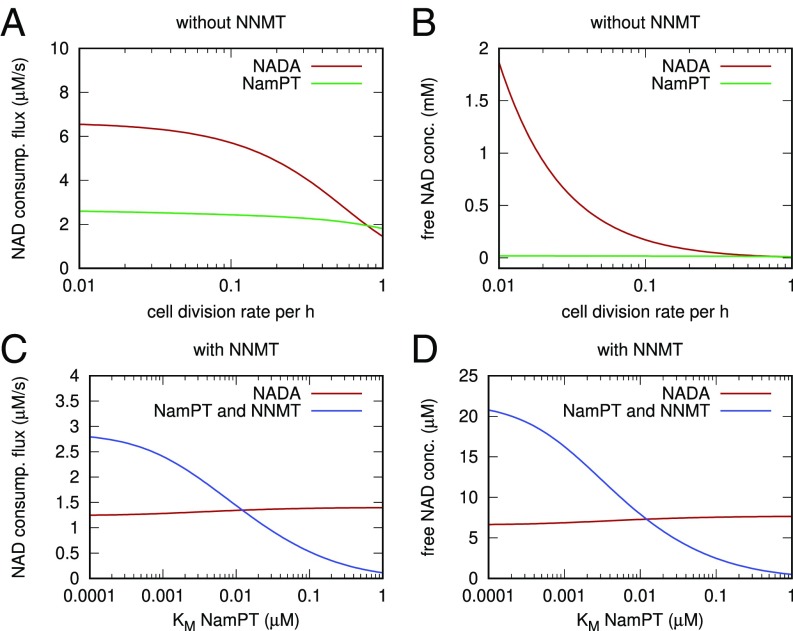
Fig. 3.
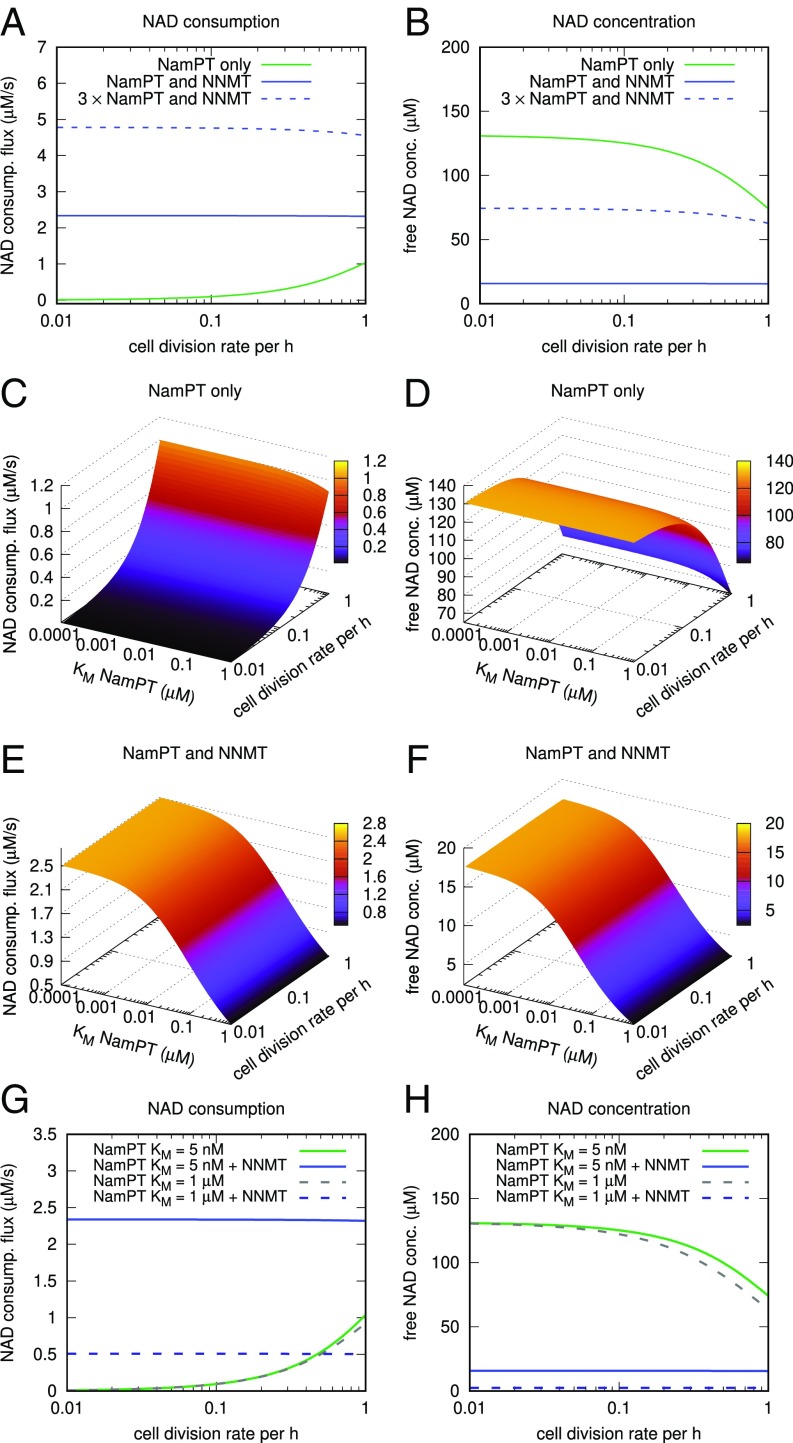
NNMT enables high NAD consumption flux and is a potential driver of NamPT affinity increase. A dynamic model of NAD biosynthesis and consumption (details are provided in Methods and Materials and SI Appendix, Table S3) was used to simulate steady-state NAD consumption (consump.) flux (A) and NAD concentration (conc.) (B). Except for the results shown as dashed lines in A and B, the enzyme amounts were kept constant for all simulations shown. In the presence of NNMT (blue curves), steady-state NAD consumption rates are higher despite reduced NAD concentration. Increasing the amount of NamPT in the simulation 3-fold (dotted blue curves) partially compensates for the decreased NAD concentration caused by Nam degradation through NNMT. The potential effect of different affinities of NamPT for Nam (inversely proportional to the KM) on the steady-state NAD consumption flux and NAD concentration was simulated at different cell division rates. In the absence of NNMT, the affinity of NamPT has little influence on NAD consumption (C) and NAD concentration (D), but both are strongly influenced by cell division rates. (E and F) In the presence of NNMT, increasing affinity of NamPT enables increasing NAD consumption flux and NAD concentration. The presence of NNMT makes both NAD consumption flux and concentration almost independent of cell division rates. (G and H) Calculated NAD consumption fluxes and free NAD concentrations, respectively, are shown for the assumption of high affinity of NamPT (KM = 5 nM, as found in the human enzyme) and low affinity (1 μM, dashed lines). Comparing the situation with and without NNMT reveals that at a low substrate affinity of NamPT and high cell division rates, NNMT no longer enables higher NAD consumption rates compared with NamPT alone (green curves and dashed gray curves).
Surprisingly, our simulations predict that the presence of NNMT enables higher rather than lower NAD consumption fluxes (Fig. 3A), although diminishing the steady-state concentration of NAD (Fig. 3B). The decline in NAD concentration can be compensated for by a higher expression of NamPT, which also further increases NAD consumption flux (dashed lines in Fig. 3 A and B). These results indicate a stimulatory role for NNMT solely on the basis of the enzyme kinetics, without having to invoke any regulatory mechanism (e.g., signaling events). It turns out that these results can be explained on the basis of the kinetic parameters of NamPT and NAD-consuming enzymes such as Sirtuin 1 (Sirt1). Most NAD-consuming enzymes are inhibited by their product, Nam. Thus, the presence of NNMT enables higher NAD consumption fluxes by removing excess Nam from the system (SI Appendix, Fig. S3 D and E). At the same time, a high substrate affinity of NamPT ensures the maintenance of a sufficiently high NAD concentration, although the concentration is, as expected, lower than in the system without NNMT. To verify that the release of Nam inhibition is indeed responsible for the increase in NAD consumption flux, we simulated the network with varying inhibition constant (Ki) values for the NAD consuming reaction. As can be inferred from SI Appendix, Fig. S3 E and F, increasing the Ki for Nam in a system without NNMT mimics the situation when NNMT is included. In contrast, changing the Ki of NamPT for NAD has no effect; that is, decreased NamPT inhibition by lowered NAD concentrations does not cause the flux increase observed in the presence of NNMT. However, if the NAD concentration is strongly reduced due to high expression of NNMT, the NAD consumption declines again (SI Appendix, Fig. S3 C and D).
Kinetic parameters of NamPT were previously reported for the human enzyme (34) as well as for some bacterial enzymes (45), with the latter having a much lower substrate affinity for Nam. We thus simulated the potential effect of NamPT affinity (KM) on NAD steady-state concentration and NAD consumption flux. In the absence of NNMT, a variation of the substrate affinity of NamPT for Nam is predicted to have very little effect on steady-state NAD concentration and NAD consumption flux (Fig. 3 C and D). In the presence of NNMT, however, NAD consumption flux and NAD concentration would rise with increasing affinity of NamPT (Fig. 3 E and F).
Remarkably, NAD concentration and consumption flux are both considerably affected by cell division rates in a system without NNMT. Our simulations predict a tradeoff between sustainable NAD concentration and consumption flux in the absence of NNMT. In the presence of NNMT, however, NAD consumption rates and concentrations are almost independent of cell division rates. These observations point to a role of NNMT for NAD homeostasis at varying cell division and consumption rates.
Given that a lower affinity of NamPT has been described for the bacterial enzyme (45) where NNMT is not present, we were wondering if the advantage provided by NNMT is dependent on a high affinity of NamPT. In Fig. 3 G and H, we show the direct comparison of simulations assuming different affinities of NamPT in the presence or absence of NNMT. Interestingly, a low affinity that is in the range of the affinity of NADA for Nam, and far above those measured for bacterial NamPTs, leads to higher NAD consumption flux in the presence of NNMT, but only when cell division rates are low (Fig. 3G). However, if the affinity of NamPT is high enough (KM << 1 μM), consumption rates are always higher with NNMT than without. The NAD concentration is, as would be assumed, always lower with NNMT (Fig. 3H).
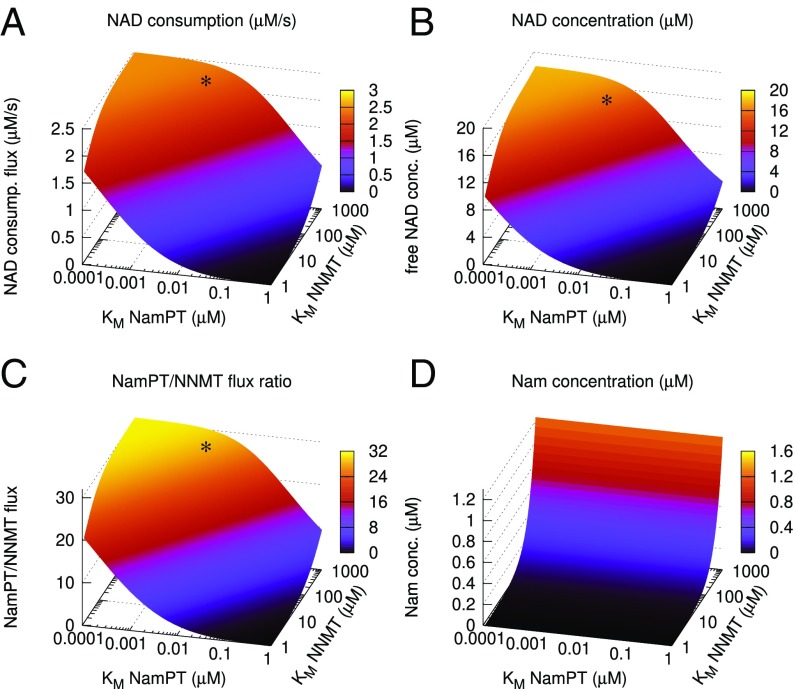
To understand the interplay and competition for Nam between NamPT and NNMT, we conducted simulations in which we scanned a wide range of possible substrate affinities for both enzymes. As shown in Fig. 4, these simulations indicate that both NAD consumption flux and NAD concentration would be minimal in case of a low substrate affinity of NamPT and high affinity of NNMT. Conversely, increasing the affinity of NamPT increases NAD consumption, NAD concentration, and the flux ratio between NamPT and NNMT, reaching a plateau when the substrate affinity of NNMT is sufficiently low. Remarkably, as indicated by the asterisks in Fig. 4 A–C, the reported substrate affinities for human NamPT and NNMT (KM of 5 nM and KM of 400 μM, respectively) are within the predicted optimal range, where further adjustment would lead to little or no increase of NAD consumption flux, NAD concentration, or NamPT-to-NNMT flux ratio.
Fig. 4.
Evolutionary optimality of the substrate affinities of human NNMT and NamPT. We simulated the impact of changes in the substrate affinities of both NamPT and NNMT on NAD consumption (consump.) rates (A), free NAD concentration (conc.) (B), NamPT/NNMT flux ratio (C), and Nam concentration (D). With increasing affinity of NamPT (decreasing KM), but decreasing affinity of NNMT (increasing KM), NAD consumption rates and free NAD concentrations, as well as the ratio between NamPT and NNMT flux, are augmented. The affinities reported for human enzymes (indicated by a black asterisk) appear to be in the optimal range according to our simulations. The steady-state concentration of Nam is largely independent of the substrate affinity of NamPT, but strongly dependent on the affinity of NNMT.
Sequence Variance Acquired in Metazoan NamPT Enhances Substrate Affinity.
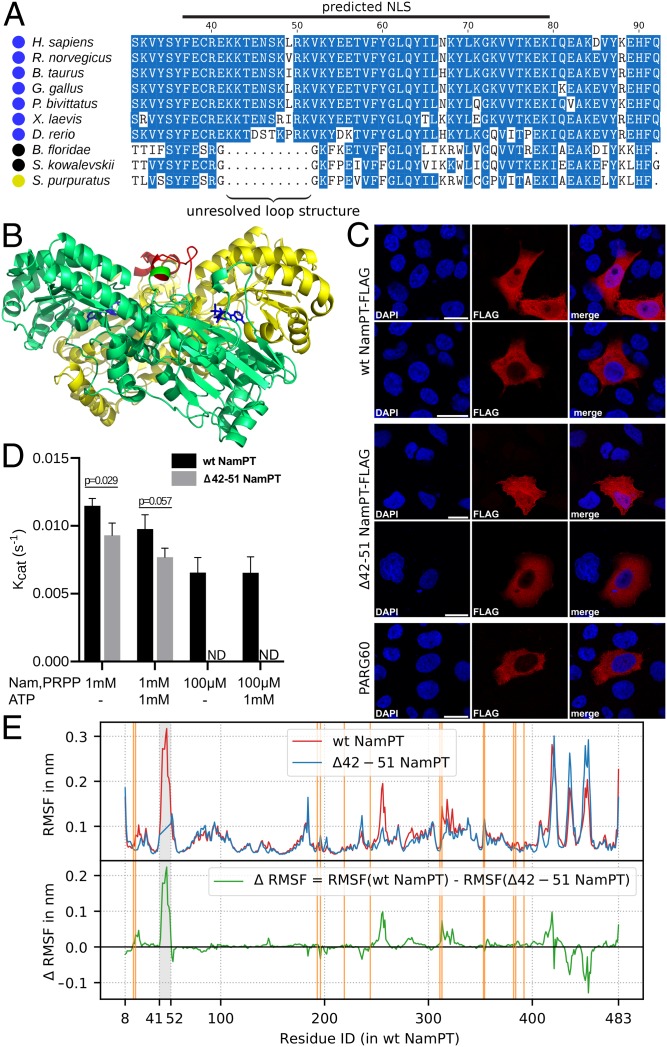
Given the kinetic interdependence of NNMT and NamPT revealed above, it seemed possible that NNMT exerted an evolutionary pressure on the development of NamPT. In this case, one would expect to observe adaptations that are reflected in the NamPT protein sequence arising in conjunction with the occurrence of NNMT. To explore this, we created a multiple sequence alignment of NamPT protein sequences from Metazoa. An alignment of selected sequences is shown in Fig. 5A, and a more comprehensive multiple sequence alignment containing a larger number of species can be found in SI Appendix, Fig. S2. We found an insert of 10 amino acids in most Deuterostomia that possess only NamPT and NNMT (indicated by blue circles in Fig. 5A). This insert corresponds to positions 42 to 51 in the human enzyme and overlaps with a predicted weak nuclear localization signal (NLS). The NLS prediction is lost when the insert is removed. The 10-amino acid insert has so far not been resolved in any of the available crystal structures obtained for NamPT. When modeling this stretch into the known homodimeric structure, the predicted loop (depicted in red in Fig. 5B), is connected to 1 of the β-sheets involved in substrate binding (35). Intriguingly, the loops of the 2 subunits are in close proximity.
Fig. 5.
Function of the structurally unresolved loop of NamPT. (A) Multiple sequence alignment of NamPT revealed a sequence insertion in the N-terminal region of this enzyme in most Deuterostomia that possess NamPT and NNMT. The relevant sequence section is shown for selected organisms. A more comprehensive alignment can be found in SI Appendix, Fig. S2. Colored circles indicate the enzymes present in the respective species: NamPT and NNMT (blue); NamPT, NADA, and NNMT (black); NamPT and NADA (yellow). (B) Structure visualization of human NamPT is based on a structure prediction by SWISS-MODEL (78, 79) using the PDB model 2H3D of the human NamPT as a template (69). The region corresponding to amino acids 42 to 51 (shown in red) in the human enzyme is not resolved in any of the currently available crystal structures of NamPT, and thus appears to be a flexible loop at the surface of the NamPT dimer. (C) Confocal laser scanning micrographs of HeLaS3 cells expressing C-terminally FLAG-tagged WT NamPT and mutant Δ42 to 51 NamPT lacking the unresolved loop. Both proteins showed a heterogeneous nuclear-cytosolic localization. Nuclei were stained with DAPI. The C-terminally FLAG-tagged human poly-ADP ribose glycohydrolase isoform PARG60 was used as a control for exclusive cytosolic localization (Scale bars, 20 μm). (D) Measurement of NamPT (WT and Δ42 to 51 mutant) enzymatic activity in the presence of 1 mM or 100 μM substrate (Nam and PRPP, respectively) with or without 1 mM ATP. ND, no detection of the reaction product (NMN). The P value was calculated using a nonparametric 1-tailed Mann–Whitney U test. The His-tagged proteins were expressed in E. coli and purified as described in Methods and Materials (also SI Appendix, Fig. S4). (E) Molecular dynamics simulations were performed for WT NamPT (red) and Δ42 to 51 NamPT (blue). (Top) Root mean square fluctuations (RMSFs) for every residue of chain A are shown. (Bottom) Difference RMSF for every residue is shown (green). For better comparison, the residue IDs for Δ42 to 51 NamPT are aligned to accord with the WT structure and the average RMSFs of residues 42 and 51 are displayed in the blue curve between these residues. For the RMSF calculation, the first 100 ns of the simulation is omitted to allow equilibration. In addition, root mean square deviation values between different published structures of human NamPT structures were calculated and are presented in SI Appendix, Fig. S5 and Table S4. The orange lines indicate the position of residues involved in substrate binding, according to Burgos et al. (35).
From these observations, we derived 2 possible hypotheses regarding the role of the loop in NamPT function. The first hypothesis is that the presence of the loop could affect the subcellular localization of NamPT, as it is overlapping with a predicted NLS. To test this hypothesis, we created a mutant NamPT lacking the loop and recombinantly expressed FLAG-tagged wild-type (WT) and mutant NamPT in HeLa S3 cells. Immunofluorescence imaging showed a mixed cytosolic/nuclear localization for both the WT and mutant NamPT (Fig. 5C). Thus, deletion of the loop did not compromise nuclear localization.
The second hypothesis is based on our model simulations that predict that the presence of NNMT might have exerted evolutionary pressure on NamPT kinetics and that the sequence insertion might therefore have an effect on substrate binding of NamPT. To analyze this possibility, we expressed and purified the WT and mutant enzymes, which lack the stretch of amino acids 42 to 51, in Escherichia coli, N-terminally fused to a 6xHis-tag. The size exclusion chromatography (SEC) profile showed that both WT and mutant proteins were expressed as dimers (SI Appendix, Fig. S4C), indicating that the mutant protein was likely folded correctly. The enzymatic activity was assessed by measuring NMN formation. Upon incubation with the NamPT inhibitor FK866 (31) for 30 min, neither WT nor mutant NamPT synthesized NMN, suggesting that binding of FK866 is not affected by the mutation (SI Appendix, Fig. S4D). Using Nam and PRPP at 100 μM as substrates, the WT showed a turnover rate of 0.0065 ± 0.0010 s−1 and 0.0077 ± 0.0006 s−1 without and with ATP, respectively, while the mutant did not have any detectable activity (Fig. 5D). With both substrates at 1 mM, the turnover rate of the WT enzyme increased to 0.0115 ± 0.0005 s−1 and 0.0098 ± 0.0010 s−1 without and with ATP, respectively. Under these conditions, the turnover rate of the mutant enzyme was 0.0093 ± 0.0008 s−1 and 0.0077 ± 0.0006 s−1 without and with ATP, respectively. When keeping both PRPP and ATP constant at 1 mM, the WT enzyme reaches its maximal rate in the low micromolar range of Nam concentrations (SI Appendix, Fig. S4E). Under these conditions, activity of the mutant is detectable only at submillimolar Nam concentrations and still rises between 0.5 and 1 mM (SI Appendix, Fig. S4E), indicating a far lower affinity toward Nam compared with the WT. The decrease in activity with ATP at high concentrations of substrates has been observed earlier (34) and has been attributed to the competitive binding of ATP and PRPP (35). Overall, our observations suggest that human NamPT lacking the stretch from amino acid 42 to 51 is catalytically active and retains its dimeric state and sensitivity to FK866. However, it has a lower activity and affinity to Nam. These observations lend support to the conclusion that the acquisition of this loop in the NamPT of higher vertebrates has led to an increased affinity to Nam, as predicted by our metabolic modeling approach.
To see whether we could find a molecular explanation for the reduced affinity of the mutant enzyme, we analyzed different available protein structures of NamPT and tested whether the loop insertion could potentially lead to dynamic structural rearrangements. To this end, we applied homology modeling (Fig. 5B) and molecular dynamics simulations for structures with and without the loop insertion (Fig. 5E). We did not observe substantial structural rearrangements, and the molecular dynamics simulations showed only limited structural changes upon loop deletion. Rather, we observed a mostly structurally stable catalytic core. This might be based on the fact that all available protein structures of NamPT differ very little even at the catalytic site (between 0.33 and 0.95 Å; SI Appendix, Table S4). Some residues close to the catalytic site showed slightly elevated mobilities in the WT structure. However, these elevated mobilities were dominated by rare events during the simulation time of 1 μs. Therefore, they do not appear as a robust change of structural dynamics upon loop insertion.
NNMT Made NADA Obsolete in Vertebrates.
Finally, we wished to understand whether NADA may have been lost in vertebrates due to kinetic constraints. As shifts in evolutionary selection pressure may result from competition for resources, we built a 2-compartment model based on the pathway model described above. One compartment contains NADA, while the other contains either NamPT alone or together with NNMT. Both compartments share a limited Nam source (model details are provided in SI Appendix, Table S3). Without NNMT, the compartment containing NADA shows a higher NAD consumption rate (Fig. 6A), and is able to maintain much higher NAD concentrations, especially at low cell division rates (Fig. 6B). At high cell division rates, steady-state concentrations and NAD consumption rates in both compartments are similar. As bacteria often have relatively high growth rates and a low number of NAD-consuming enzymes, this might explain why, at least in some bacteria, both systems may coexist.
Fig. 6.
NNMT provides a competitive advantage and makes NADA obsolete in vertebrates. To simulate competition for common resources, a 2-compartment model was created (Methods and Materials and SI Appendix, Table S3). In this model, 1 compartment contained NADA, but no NamPT, and the other compartment contained NamPT either with or without NNMT, but no NADA. NADA and NamPT were simulated to be present at equal amounts. In the absence of NNMT, the compartment containing NADA has a higher NAD consumption (consump.) rate (A) and a considerably higher steady-state NAD concentration (conc.) (B). In the presence of NNMT, however, both NAD consumption (C) and NAD concentration (D) are lower in the NADA compartment. This effect is dependent on a high affinity of NamPT for Nam.
In the presence of NNMT, the NamPT compartment has both higher NAD consumption rates and higher steady-state NAD concentrations than the compartment containing NADA (Fig. 6 C and D). This is, however, dependent on the affinity of NamPT for Nam. If the substrate affinity of NamPT is too low, the NADA compartment is able to maintain higher NAD concentrations and consumption flux. As shown in SI Appendix, Fig. S6, expression of NADA in addition to NNMT and NamPT provides a slight advantage over NADA alone as long as NamPT has a low affinity. Taken together, the results suggest that the NADA pathway might have helped in the transition to high-affinity NamPT, potentially explaining its co-occurrence with NamPT and NNMT in some invertebrates. Eventually, NADA became obsolete upon emergence of a high-affinity NamPT. This, in turn, might have been induced by the appearance of NNMT.
Discussion
The present study has revealed fundamental insights into the evolution and dynamic interplay of the enzymes in NAD metabolism. Our results show that the occurrence of NNMT enabled the enormous diversification of NAD-consuming signaling enzymes in Deuterostomia. NNMT promotes the removal of excessive Nam produced in the signaling reactions. This is necessary to overcome Nam inhibition of the corresponding enzymes. To enable both high NAD turnover and continuous salvage of Nam into NAD, the kinetic parameters of both human NamPT and NNMT have attained values that are in the optimal range predicted through our simulations (Fig. 4). Our analyses have identified a stretch of 10 amino acids in the structure of NamPT which contributes to the unusually high substrate affinity of this enzyme in higher vertebrates. While it remains unclear why lower organisms and plants primarily use the Nam salvage pathway through NADA, our analyses demonstrate that the combination of NamPT and NNMT outcompetes this alternative when high turnover of NAD is required for signaling processes. Consequently, NADA has been lost in vertebrates.
The positive effect of NNMT on NAD consumption flux, especially on sirtuins, is in line with a lifespan extension observed in worms overexpressing NNMT (46). The effect of NNMT overexpression or silencing has been controversially discussed and is presumably tissue- and context-specific (37). In this regard, it should be noted that although we predict an overall positive effect of NNMT on NAD consumption rates, excessive expression of NNMT can lead to adverse effects (SI Appendix, Fig. S3C). Moreover, our analyses predict that the presence of NNMT generally lowers the Nam concentration and reduces cellular NAD concentrations. This effect can, however, be counterbalanced by increased NamPT expression (47, 48). The moderate effects of NNMT expression changes on NAD concentrations (SI Appendix, Fig. S3D) might explain why some experimental results reported in the literature seem inconsistent. For example, a considerable decrease of liver NAD levels has been reported upon NNMT overexpression (49), while no changes in NAD concentrations could be detected in other studies (5, 50).
It has been shown that methyl-Nam (MNam) excretion is mostly proportional to Nam uptake (51), supporting our findings that NNMT contributes to NAD pathway homeostasis. As shown in several recent studies, this homeostatic control by NNMT can be circumvented by supplying NR (2, 52–54), which is not a substrate of NNMT. At the cellular level, NNMT is presumably mainly advantageous when high NAD consumption rates are required for tissue function or, even more likely, might be important to prevent spatiotemporal accumulation of Nam within cells due to temporally increased NAD consumption (e.g., PARP activation through DNA damage).
The main healthy tissues expressing NNMT are the liver and adipose tissues, while no or only little expression of NNMT is observed in most other organs (55). Increased NNMT expression has been found in several types of cancer (56, 57), and might serve to remove Nam produced by increased NAD-dependent signaling. To maintain high NAD concentrations in the tumors, a concomitant increase of NamPT expression is required, which has indeed been described for some cancers (47, 48). It is worth noticing that NNMT is only advantageous as long as NamPT affinity and activity are sufficiently high. This suggests that certain types of cancers, which express NNMT at a high level, could potentially be more susceptible to inhibitors of NamPT. Several such inhibitors are currently being tested in clinical studies (23, 58). Based on our analyses, it might be reasonable to test patients for NNMT expression in the tumor tissue. Non–NNMT-expressing tumors might respond less to competitive NamPT inhibitors, because deficient Nam degradation in those cancer cells would potentially lead to an accumulation of Nam that could outcompete the inhibitor.
Neither the scattered distribution of NamPT and NADA that is especially pronounced in bacteria (40) nor the loss of NADA in the ancestor of vertebrates has been understood earlier. Our combined phylogenetic modeling analysis provides a potential explanation for both observations. Using simulated competition between 2 compartments that share the same limited source of Nam, we show that the compartment that contains NamPT and NNMT can maintain a higher steady-state NAD concentration and NAD consumption rate than the compartment containing NADA (Fig. 6). This is, however, only the case, if NamPT’s substrate affinity is sufficiently high. The dominant enzyme combination found in vertebrates, a high-affinity NamPT along with NNMT, thus seems to provide a competitive advantage when high NAD turnover rates are needed. This is not necessarily the case in organisms that use Nam recycling through NADA.
In our analyses, we did not consider the potential effects of most cosubstrates of the investigated pathway. These cosubstrates include targets of the NAD-consuming enzymes, such as acylated proteins for sirtuins or PRPP and ATP required for NMN synthesis by NamPT. However, we did perform an analysis of the effect of concentration changes in the methyl donor SAM. Its precursor methionine has been shown to potentially limit the effect of NNMT (57). As shown in SI Appendix, Fig. S3 G and H, SAM can have both positive and negative effects on the NAD consumption flux, depending on the SAM concentration range. The effects observed in the physiological SAM concentration range (59) are, however, much smaller than those observed for expression changes in NamPT or NNMT (SI Appendix, Fig. S3 A and C, respectively). Nevertheless, under some conditions, changes in methionine metabolism might influence cellular NAD concentration and NAD consumption rates. As NNMT, in turn, consumes SAM, NNMT might not only provide a kinetic advantage for NAD metabolism but is likely to have a role in regulating other cellular processes through its impact on SAM availability (37). For example, it has been shown that the product MNam can induce the expression of sirtuins (46, 50, 60). However, the underlying mechanisms are still unknown.
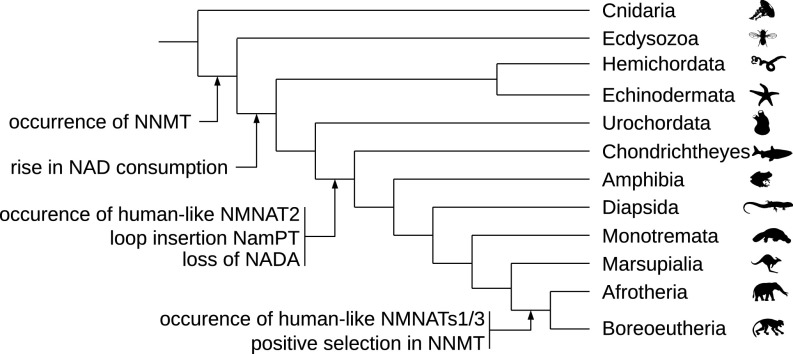
In conclusion, we have comprehensively analyzed the functional coevolution of several enzymes of the NAD pathway. The appearance of NNMT apparently initiated and drove complex alterations of the pathway such as an increase in and diversification of NAD-dependent signaling, paralleled by an increase in NamPT substrate affinity. Remarkably, when analyzing the possibility of coevolutionary developments within the NAD pathway, we also noted that the loss of NADA and the loop insertion in NamPT co-occurred with the appearance of a human-like NMNAT2 (a schematic overview is provided in Fig. 7, details are provided in SI Appendix, Figs. S7–S9). Moreover, the occurrence of human-like NMNAT1 and NMNAT3, and thus the further compartmentalization of NAD metabolism (61), coincided with a site-specific positive selection event in NNMT (SI Appendix, Figs. S7 and S8). This might point to a role of NNMT in NAD pathway compartmentalization in addition to spatiotemporal regulation of the pathway in general.
Fig. 7.
Schematic representation of evolutionary events in the NAD biosynthesis pathway. The scheme illustrates major evolutionary events in Metazoa detected in our phylogenetic analyses of NAD metabolism. The time of occurrence of human-like NMNAT1 and NMNAT3 has been reported previously (61), and it has been identified that human-like NMNAT2 most likely originated in the last common ancestor (LCA) of vertebrates, while human-like NMNAT1/3 can be traced back to the LCA of Placentalia (SI Appendix, Fig. S9). To test whether the rise of human-like NMNAT1/3 was associated with an event of rapid sequence diversification in NNMT, we conducted a test of positive selection specific to the branch leading to the LCA of Placentalia using a coding DNA substitution rate ratio model. Indeed, we obtain a strong signature of positive selection for NNMT in the tested branch and can pinpoint residue 171 as being significantly associated with the signature of positive selection (SI Appendix, Figs. S7 and S8). Specific events in the evolution of NMNATs coincide with those of NamPT or NNMT, indicating a coevolution of functions beyond those identified in the present study. The tree is a schematic representation of selected taxa and is based on information provided by the Tree of Life Web Project (80).
Methods and Materials
Phylogenetic Analysis.
Functionally verified sequences of NNMT, NADA, NamPT, and NAD-consuming enzymes were used as sequence templates for a Blastp analysis against the National Center for Biotechnology Information (NCBI) nonredundant protein sequence database. A list of template sequences is provided in SI Appendix, Table S1. Blastp parameters were set to yield a maximum of 20,000 target sequences, using the BLOSUM62 matrix with a word size of 6 and gap opening and extension costs of 11 and 1, respectively. Low-complexity filtering was disabled. To prevent cross-hits, a matrix was created in which the lowest e-values were given at which Blast yielded the same result for each query protein pair. With help of the matrix, the e-value cutoff was set to 1e-30 for all enzymes. To further prevent false-positive results, a minimal length limit was set based on a histogram of the hit lengths found for each query protein, excluding peaks much lower than the total protein length. Length limits are given in SI Appendix, Table S1. In addition, obvious sequence contaminations were removed by manual inspection of the results. The taxonomy IDs of the species for each enzyme were derived from the accession2taxonomy database provided by the NCBI. Scripts for creating, analyzing, and visualizing the phylogenetic tree were written in Python 3.5, using the ETE3 toolkit (62) and are accessible through the following GitHub repository https://github.com/MolecularBioinformatics/Phylogenetic-analysis (63).
Dynamic Modeling.
Kinetic parameters (substrate affinity [KM] and turnover rates, substrate and product inhibitions) were retrieved from the enzyme database BRENDA and additionally evaluated by checking the original literature, especially with respect to measurement conditions. Parameter values from mammalian species were used if available. For enzymes not present in mammals, values from yeast were integrated. The full list of kinetic parameters, including references to original literature, can be found in SI Appendix, Table S3. For NMNAT, the previously developed rate law for substrate competition was used (64). Otherwise, Henri–Michaelis–Menten kinetics were applied for all reactions except the import and efflux of Nam, which were simulated using constant flux and mass action kinetics, respectively. Steady-state calculation and parameter scan tasks provided by COPASI 4.25 (65) were used for all simulations. The model files are accessible through the Biomodels database (accession nos. MODEL1905220001 and MODEL1905220002). Related figures were generated using Gnuplot 5.0 (66, 67).
Generation of Expression Vectors Encoding WT and Mutant Human NamPT.
For eukaryotic expression with a C-terminal FLAG-epitope, the open reading frame (ORF) encoding human NamPT was inserted into pFLAG-CMV-5a (Sigma–Aldrich Merck) via EcoRI/BamHI sites. Using a PCR approach, this vector provided the basis for the generation of a plasmid encoding a NamPT deletion mutant lacking amino acid residues 42 to 51 (Δ42 to 51 NamPT). For prokaryotic expression with an N-terminal 6xHis-tag, the WT and mutant ORFs were inserted into pQE-30 (Qiagen) via BamHI and PstI sites. All cloned sequences were verified by DNA sequence analysis.
Transient Transfection, Immunocytochemistry, and Confocal Laser Scanning Microscopy.
HeLa S3 cells cultivated in Ham’s F-12 medium supplemented with 10% (vol/vol) fetal calf serum, 2 mM l-glutamine, and penicillin/streptomycin were seeded on coverslips in a 24-well plate. After 1 d, cells were transfected using Effectene transfection reagent (Qiagen) according to the manufacturer’s recommendations. Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) 24 h posttransfection, permeabilized (0.5% [vol/vol] Triton X-100 in PBS), and blocked for 1 h with complete culture medium. After overnight incubation with primary FLAG-antibody (mouse M2; Sigma–Aldrich) diluted 1:2,500 in complete medium, cells were washed and incubated for 1 h with secondary Alexa Fluor 594-conjugated goat anti-mouse antibody (Invitrogen, Thermo Fisher Scientific) diluted 1:1,000 in complete culture medium. Nuclei were stained with DAPI, and the cells were washed. The coverslips were mounted onto microscope slides using ProLong Gold (Invitrogen, Thermo Fisher Scientific). Confocal laser scanning microscopy of cells was performed at the Molecular Imaging Center at the Department of Biomedicine, University of Bergen, using a Leica TCS SP8 STED 3× microscope equipped with a 100× oil immersion objective (numerical aperture of 1.4).
NamPT Expression.
E. coli BL21-CodonPlus (DE3)-RIL (Agilent) was transformed with pQE-30 WT NamPT or pQE-30 Δ42 to 51 NamPT, along with pREP4 (Qiagen) encoding the lac repressor. Bacterial cells were grown at 37 °C in 1 L of Luria–Bertani broth supplemented with 100 μg/mL ampicillin, 50 μg/mL kanamycin, and 32 μg/mL chloramphenicol. Protein expression was induced with 0.2 mM isopropyl-β-d-thiogalactoside at 0.4 to 0.6 optical density at 600 nm, and cells were subsequently grown overnight at 18 °C.
Purification of NamPT.
The cells were harvested by centrifugation and resuspended in lysis buffer (20 mM Tris⋅HCl [pH 8.0], 500 mM NaCl, 4 mM dithiothreitol (DTT), 1 mg/mL lysozyme, 1× cOmplete EDTA-free Protease Inhibitor mixture [Roche]). After sonication, the lysate was centrifuged at 13,000 × g for 30 min. The supernatant was incubated with 2 mL of nickel-nitrilotriacetic acid resin (Qiagen). Nonspecific protein binding was removed with washing buffer (20 mM Tris⋅HCl [pH 8.0], 500 mM NaCl, 1 mM DTT, 20 mM imidazole). The protein was eluted with 2.5 mL of elution buffer (20 mM Tris⋅HCl [pH 8.0], 500 mM NaCl, 1 mM DTT, 300 mM imidazole).
The eluted protein was immediately subjected to SEC using an ÄKTA Pure system (GE Healthcare) equipped with a HiLoad 16/60 Superdex 200-pg column (GE Healthcare). The chromatography was performed at a flow rate of 1 mL⋅min−1 with SEC buffer (20 mM Tris⋅HCl [pH 8.0], 500 mM NaCl). Fractions containing the recombinant protein were pooled and used for enzymatic assays. The purity and size of the protein were assessed by sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Enzymatic Assay.
In a final volume of 1.2 mL of reaction buffer (20 mM Tris⋅HCl [pH 8.0], 500 mM NaCl, 6 mM MgCl2, 0.03% [wt/vol] bovine serum albumin), 2 μM enzyme was incubated with PRPP and Nam (both at 100 μM or 1 mM). The reaction was incubated at 30 °C for 10 min and stopped by adding 100 μM FK866. Subsequently, the samples were frozen in liquid nitrogen. The amount of NMN produced was analyzed using NMR spectroscopy. To do so, the samples were dried with an Eppendorf Vacufuge Concentrator and then resuspended in 200 μL of NMR solvent containing 5% (vol/vol) deuterated H2O and 1 mM 4,4-dimethyl-4-silapentane-1-sulfonate (DSS). One-dimensional 1H NMR spectra were acquired on a 850-MHz Ascend Bruker spectrometer equipped with a 5-mm TCI triple-resonance CryoProbe and a pulse field gradient along the z axis. The spectra were acquired with the zgesgppe pulse sequence, allowing water suppression using excitation sculpting with gradients and perfect echo. The temperature was kept constant at 300 K. Data were acquired with 2,000 scans, a 1-s relaxation delay, and a 1.6-s acquisition time, and contained 65,000 data points with a spectral width of 14 parts per million (ppm). The spectra phase and baseline were automatically and manually corrected using TopSpin 3.5 software (Bruker Biospin). Quantification of NMN was done by the integration of the peak at 9.52 ppm, and DSS was used as an internal standard. The raw measurement data are accessible using the FAIRDOMHub (DOI: 10.15490/fairdomhub.1.datafile.2942.1) (68).
All experiments were conducted at the Norwegian NMR Platform (Grant 226244/F50).
Molecular Dynamics Simulations.
All-atom molecular dynamics simulations were performed with explicit solvent for WT NamPT and mutant Δ42 to 51 NamPT [Protein Data Bank (PDB) ID code 2H3D (69)]. The AMBER99SB-ILDN force field (70) was used with the standard water model TIP3P (transferable intermolecular potential with 3 points) (71) in GROMACS 5.1.2 (72). The structures were simulated each in a box of water with the distance between the solute and the box set to 0.2 nm at a temperature of 300 K for a total time of 1 μs. A time step of 2 fs and the stochastic dynamics integrator were used. For the evaluation of the root mean square fluctuation, the first 100 ns of the simulations was omitted.
Identification of Human-Like NMNATs and Test of Positive Selection in NNMNTs.
For the identification of human-like NMNATs, we clustered the retrieved sequences using BAli-Phy (73) (SI Appendix, Fig. S9). Human-like NMNAT1, NMNAT2, and NMNAT3 were identified based on the isoform-specific targeting and interaction domains described by Lau et al. (61).
We conducted a test of positive selection for orthologs of human NNMT from 60 vertebrate species. To calculate the nonsynonymous and synonymous substitution rates, dN and dS, respectively, we obtained coding sequences for all species and aligned the respective protein-translated sequences using MUSCLE (74), and prepared codon-based alignments for further processing with PAL2NAL (75). We used codeml from the PAML package (76) to conduct a branch-site model A test of positive selection. The species names and the underlying tree topology for the codeml runs are depicted in SI Appendix, Fig. S8B. As a null model, we assumed neutrality (e.g., diversifying site class with dN/dS = ω = 1), which then was compared with a model with positive selection (dN/dS = ω > 1). Significance between the 2 models is assessed using a likelihood ratio test assuming that twice the likelihood difference is χ2-distributed. The critical value is 3.84 at the 5% level. Additionally, we identified codons with a site-specific signal of positive selection using a Bayes empirical Bayes analysis with a probability value >0.9 (77).
Supplementary Material
Acknowledgments
I.H. and M.Z. are supported by grants from the Norwegian Research Council (Grant 250395/F20) and the European Union’s Horizon 2020 research and innovation programme under Grant 754688. We thank Heiko Stark for advice with respect to taxonomic classifications. We also thank the National Infrastructure for High Performance Computing and Data Storage in Norway for the computation time provided through UNINETT Sigma2. M.Z. was supported by a grant from the Norwegian Cancer Society (Grant 673238). I.R. and A.S. received support from the Helmholtz Association Initiative and Networking Fund under Project ZT-I-0003. T.I.G. is supported by a Leverhulme Early Career Fellowship (Grant ECF-2015-453) and Natural Environment Research Council Grant NE/N013832/1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Scripts for this work have been deposited in the GitHub repository (https://github.com/MolecularBioinformatics/Phylogenetic-analysis). Simulation model files have been deposited at the Biomodels database (accession nos. MODEL1905220001 and MODEL1905220002). The raw measurement data have been deposited at the FAIRDOMHub (DOI: 10.15490/fairdomhub.1.datafile.2942.1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902346116/-/DCSupplemental.
References
- 1.Ljungberg M. C., et al. , CREB-activity and nmnat2 transcription are down-regulated prior to neurodegeneration, while NMNAT2 over-expression is neuroprotective, in a mouse model of human tauopathy. Hum. Mol. Genet. 21, 251–267 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trammell S. A., et al. , Nicotinamide riboside opposes type 2 diabetes and neuropathy in mice. Sci. Rep. 6, 26933 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshino J., Mills K. F., Yoon M. J., Imai S., Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 14, 528–536 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell S. J., et al. , Nicotinamide improves aspects of healthspan, but not lifespan, in mice. Cell Metab. 27, 667–676.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraus D., et al. , Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature 508, 258–262 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantó C., et al. , The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 15, 838–847 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kannt A., et al. , A small molecule inhibitor of Nicotinamide N-methyltransferase for the treatment of metabolic disorders. Sci. Rep. 8, 3660 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu C. P., Oka S., Shao D., Hariharan N., Sadoshima J., Nicotinamide phosphoribosyltransferase regulates cell survival through NAD+ synthesis in cardiac myocytes. Circ. Res. 105, 481–491 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diguet N., et al. , Nicotinamide riboside preserves cardiac function in a mouse model of dilated cardiomyopathy. Circulation 137, 2256–2273 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryu D., et al. , NAD+ repletion improves muscle function in muscular dystrophy and counters global PARylation. Sci. Transl. Med. 8, 361ra139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poyan Mehr A., et al. , De novo NAD+ biosynthetic impairment in acute kidney injury in humans. Nat. Med. 24, 1351–1359 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiarugi A., Dölle C., Felici R., Ziegler M., The NAD metabolome–A key determinant of cancer cell biology. Nat. Rev. Cancer 12, 741–752 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Buonvicino D., et al. , Identification of the nicotinamide salvage pathway as a new toxification route for antimetabolites. Cell Chem. Biol. 25, 471–482.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Verdin E., NAD+ in aging, metabolism, and neurodegeneration. Science 350, 1208–1213 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Chini C. C. S., Tarragó M. G., Chini E. N., NAD and the aging process: Role in life, death and everything in between. Mol. Cell. Endocrinol. 455, 62–74 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mills K. F., et al. , Long-term administration of nicotinamide mononucleotide mitigates age-associated physiological decline in mice. Cell Metab. 24, 795–806 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H., et al. , NAD+ repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 352, 1436–1443 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Imai S. I., Guarente L., It takes two to tango: NAD+ and sirtuins in aging/longevity control. NPJ Aging Mech. Dis. 2, 16017 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Araki T., Sasaki Y., Milbrandt J., Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science 305, 1010–1013 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Beirowski B., et al. , Non-nuclear Wld(S) determines its neuroprotective efficacy for axons and synapses in vivo. J. Neurosci. 29, 653–668 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Stefano M., et al. , NMN deamidase delays wallerian degeneration and rescues axonal defects caused by NMNAT2 deficiency in vivo. Curr. Biol. 27, 784–794 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Yoshino J., Baur J. A., Imai S. I., NAD+ intermediates: The biology and therapeutic potential of NMN and NR. Cell Metab. 27, 513–528 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Espindola-Netto J. M., et al. , Preclinical efficacy of the novel competitive NAMPT inhibitor STF-118804 in pancreatic cancer. Oncotarget 8, 85054–85067 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajman L., Chwalek K., Sinclair D. A., Therapeutic potential of NAD-boosting molecules: The in vivo evidence. Cell Metab. 27, 529–547 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinclair D. A., Guarente L., Small-molecule allosteric activators of sirtuins. Annu. Rev. Pharmacol. Toxicol. 54, 363–380 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupte R., Liu Z., Kraus W. L., PARPs and ADP-ribosylation: Recent advances linking molecular functions to biological outcomes. Genes Dev. 31, 101–126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bütepage M., Eckei L., Verheugd P., Lüscher B., Intracellular mono-ADP-ribosylation in signaling and disease. Cells 4, 569–595 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osborne B., Bentley N. L., Montgomery M. K., Turner N., The role of mitochondrial sirtuins in health and disease. Free Radic. Biol. Med. 100, 164–174 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Lee H. C., Cyclic ADP-ribose and nicotinic acid adenine dinucleotide phosphate (NAADP) as messengers for calcium mobilization. J. Biol. Chem. 287, 31633–31640 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koch-Nolte F., Haag F., Guse A. H., Lund F., Ziegler M., Emerging roles of NAD+ and its metabolites in cell signaling. Sci. Signal. 2, mr1 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Hasmann M., Schemainda I., FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 63, 7436–7442 (2003). [PubMed] [Google Scholar]
- 32.Liu L., et al. , Quantitative analysis of NAD synthesis-breakdown fluxes. Cell Metab. 27, 1067–1080.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.North B. J., Verdin E., Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 5, 224 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burgos E. S., Schramm V. L., Weak coupling of ATP hydrolysis to the chemical equilibrium of human nicotinamide phosphoribosyltransferase. Biochemistry 47, 11086–11096 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burgos E. S., Ho M.-C., Almo S. C., Schramm V. L., A phosphoenzyme mimic, overlapping catalytic sites and reaction coordinate motion for human NAMPT. Proc. Natl. Acad. Sci. U.S.A. 106, 13748–13753 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gossmann T. I., et al. , NAD(+) biosynthesis and salvage–A phylogenetic perspective. FEBS J. 279, 3355–3363 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Pissios P., Nicotinamide N-methyltransferase: More than a vitamin B3 clearance enzyme. Trends Endocrinol. Metab. 28, 340–353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Figueiredo L. F., Gossmann T. I., Ziegler M., Schuster S., Pathway analysis of NAD+ metabolism. Biochem. J. 439, 341–348 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Carneiro J., et al. , The evolutionary portrait of metazoan NAD salvage. PLoS One 8, e64674 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gazzaniga F., Stebbins R., Chang S. Z., McPeek M. A., Brenner C., Microbial NAD metabolism: Lessons from comparative genomics. Microbiol. Mol. Biol. Rev. 73, 529–541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perchat N., et al. , Elucidation of the trigonelline degradation pathway reveals previously undescribed enzymes and metabolites. Proc. Natl. Acad. Sci. U.S.A. 115, E4358–E4367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hron T., Pajer P., Pačes J., Bartůněk P., Elleder D., Hidden genes in birds. Genome Biol. 16, 164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gossmann T. I., Ziegler M., Sequence divergence and diversity suggests ongoing functional diversification of vertebrate NAD metabolism. DNA Repair (Amst.) 23, 39–48 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cambronne X. A., et al. , Biosensor reveals multiple sources for mitochondrial NAD+. Science 352, 1474–1477 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sorci L., et al. , Genomics-driven reconstruction of acinetobacter NAD metabolism: Insights for antibacterial target selection. J. Biol. Chem. 285, 39490–39499 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmeisser K., et al. , Role of sirtuins in lifespan regulation is linked to methylation of nicotinamide. Nat. Chem. Biol. 9, 693–700 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bi T. Q., et al. , Overexpression of Nampt in gastric cancer and chemopotentiating effects of the Nampt inhibitor FK866 in combination with fluorouracil. Oncol. Rep. 26, 1251–1257 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Wang B., et al. , NAMPT overexpression in prostate cancer and its contribution to tumor cell survival and stress response. Oncogene 30, 907–921 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Komatsu M., et al. , NNMT activation can contribute to the development of fatty liver disease by modulating the NAD+ metabolism. Sci. Rep. 8, 8637 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hong S., et al. , Nicotinamide N-methyltransferase regulates hepatic nutrient metabolism through Sirt1 protein stabilization. Nat. Med. 21, 887–894 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang-Lee Y. A., et al. , Metabolic effects of nicotinamide administration in rats. J. Nutr. 113, 215–221 (1983). [DOI] [PubMed] [Google Scholar]
- 52.Gong B., et al. , Nicotinamide riboside restores cognition through an upregulation of proliferator-activated receptor-γ coactivator 1α regulated β-secretase 1 degradation and mitochondrial gene expression in Alzheimer’s mouse models. Neurobiol. Aging 34, 1581–1588 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu H. W., et al. , Pharmacological bypass of NAD+ salvage pathway protects neurons from chemotherapy-induced degeneration. Proc. Natl. Acad. Sci. U.S.A. 115, 10654–10659 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martens C. R., et al. , Chronic nicotinamide riboside supplementation is well-tolerated and elevates NAD+ in healthy middle-aged and older adults. Nat. Commun. 9, 1286 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aksoy S., Szumlanski C. L., Weinshilboum R. M., Human liver nicotinamide N-methyltransferase. cDNA cloning, expression, and biochemical characterization. J. Biol. Chem. 269, 14835–14840 (1994). [PubMed] [Google Scholar]
- 56.Okamura A., et al. , Increased hepatic nicotinamide N-methyltransferase activity as a marker of cancer cachexia in mice bearing colon 26 adenocarcinoma. Jpn. J. Cancer Res. 89, 649–656 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ulanovskaya O. A., Zuhl A. M., Cravatt B. F., NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat. Chem. Biol. 9, 300–306 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu T.-Y., et al. , Discovery and characterization of novel small-molecule inhibitors targeting nicotinamide phosphoribosyltransferase. Sci. Rep. 5, 10043 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reed M. C., Nijhout H. F., Sparks R., Ulrich C. M., A mathematical model of the methionine cycle. J. Theor. Biol. 226, 33–43 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Liu K. Y., et al. , Nicotinamide N-methyltransferase increases complex I activity in SH-SY5Y cells via sirtuin 3. Biochem. Biophys. Res. Commun. 467, 491–496 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Lau C., et al. , Isoform-specific targeting and interaction domains in human nicotinamide mononucleotide adenylyltransferases. J. Biol. Chem. 285, 18868–18876 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huerta-Cepas J., Serra F., Bork P., ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bockwoldt M., Heiland I., Scripts used for phylogenetic analysis of NAD metabolism. GitHub. https://github.com/MolecularBioinformatics/Phylogenetic-analysis. Deposited 3 February 2019.
- 64.Schäuble S., Stavrum A.-K., Puntervoll P., Schuster S., Heiland I., Effect of substrate competition in kinetic models of metabolic networks. FEBS Lett. 587, 2818–2824 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Hoops S., et al. , COPASI–A COmplex PAthway SImulator. Bioinformatics 22, 3067–3074 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Heiland I., NAD pathway model analysing the impact of NNMT on pathway dynamics and evolution. EBI BioModels. https://www.ebi.ac.uk/biomodels/MODEL1905220001. Deposited 22 May 2019.
- 67.Heiland I., Two compartment model of NAD biosynthesis and consumption. https://www.ebi.ac.uk/biomodels/MODEL1905220002. Deposited 22 May 2019.
- 68.Houry D., Ziegler M., Analysis of NAMPT WT and Δ42-51 by NMR. FairdomHub. 10.15490/fairdomhub.1.datafile.2942.1. Deposited 29 May 2019. [DOI]
- 69.Wang T., et al. , Structure of Nampt/PBEF/visfatin, a mammalian NAD+ biosynthetic enzyme. Nat. Struct. Mol. Biol. 13, 661–662 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Lindorff-Larsen K., et al. , Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78, 1950–1958 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 72.Abraham M. J., et al. , GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015). [Google Scholar]
- 73.Suchard M. A., Redelings B. D., BAli-Phy: Simultaneous Bayesian inference of alignment and phylogeny. Bioinformatics 22, 2047–2048 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Edgar R. C., MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suyama M., Torrents D., Bork P., PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Yang Z., Wong W. S., Nielsen R., Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118 (2005). [DOI] [PubMed] [Google Scholar]
- 78.Arnold K., Bordoli L., Kopp J., Schwede T., The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Biasini M., et al. , SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maddison D. R., Schulz K.-S., Maddison W., Tree of Life Web Project. Zootaxa, 1668, 19–40 (2007). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.