Significance
The augmentation of memory through spaced learning has been shown across the animal kingdom. The animals’ brain would be equipped with the neural circuit that perceives spaced repetition and launches the molecular mechanism of memory consolidation, although those neural circuit-based mechanisms are unknown. In the Drosophila aversive spaced training paradigm, we proposed the neurons which could be differentially activated during spaced learning. Manipulation of the neural activity during spaced learning suggested that the identified neural circuit is significant in gene expression for long-term memory (LTM) formation. Thus, our study suggested the neural computation model which detects spaced learning and launches gene expression for memory augmentation.
Keywords: spaced learning, mushroom body, gene expression, long-term memory, Drosophila
Abstract
Memory consolidation is augmented by repeated learning following rest intervals, which is known as the spacing effect. Although the spacing effect has been associated with cumulative cellular responses in the neurons engaged in memory, here, we report the neural circuit-based mechanism for generating the spacing effect in the memory-related mushroom body (MB) parallel circuits in Drosophila. To investigate the neurons activated during the training, we monitored expression of phosphorylation of mitogen-activated protein kinase (MAPK), ERK [phosphorylation of extracellular signal-related kinase (pERK)]. In an olfactory spaced training paradigm, pERK expression in one of the parallel circuits, consisting of γm neurons, was progressively inhibited via dopamine. This inhibition resulted in reduced pERK expression in a postsynaptic GABAergic neuron that, in turn, led to an increase in pERK expression in a dopaminergic neuron specifically in the later session during spaced training, suggesting that disinhibition of the dopaminergic neuron occurs during spaced training. The dopaminergic neuron was significant for gene expression in the different MB parallel circuits consisting of α/βs neurons for memory consolidation. Our results suggest that the spacing effect-generating neurons and the neurons engaged in memory reside in the distinct MB parallel circuits and that the spacing effect can be a consequence of evolved neural circuit architecture.
Spaced learning, which consists of repeated learning with appropriate rest intervals, facilitates memory consolidation to a greater extent than repeated learning without rest. This augmentation of memory, known as the spacing effect, has been demonstrated in the animal kingdom (1–3). The central issue of this type of memory consolidation is how the neural circuit recognizes the temporally distributed same learning experience as spaced learning without recognizing each learning session as a novel experience and induce memory consolidation. Numerous studies have aimed to elucidate the mechanism by which the neurons recognize spaced learning through the cumulative cellular responses, such as the oscillatory activation of PKA (4) and mitogen-activated protein kinase (MAPK) (5–7, 8). However, animals encounter various sensory stimuli in the natural environment, and it remains unclear how repeated experiences among intermingled stimuli are specifically subjected to memory consolidation. A recent study has identified the neural correlates of novelty and familiarity in the olfactory system of Drosophila (9), raising another possibility that the spacing effect may be produced by distinguishing the initial novel training experience from subsequent training experiences at the neural circuit level.
The spacing effect in Drosophila has been demonstrated using an aversive training paradigm (3) in which an odor [the conditioned stimulus (CS)] is associated with electric shocks (the unconditioned stimulus). When flies are repeatedly subjected to aversive training with rest intervals, LTM formation occurs, depending on de novo gene expression (10). In contrast, single aversive training or repeated aversive training without rest intervals (massed training) does not induce LTM formation (3). Olfactory memory in flies is mediated by parallel circuits in the MB (11, 12), each of which circuit consists of different types of neurons, including ∼500 α/β surface (α/βs) neurons, 600 γmain (γm) neurons, and others (13). Given that retrieval of aversive LTM requires α/βs neurons (14), the spacing effect may target α/βs neurons for LTM formation. Importantly, MB axons are compartmentalized, and each compartment projects to a different single MB output neuron (MBON) (13). Each MBON exhibits projections to different brain areas, some of which are known to innervate dopamine neurons (DANs) and form feedback loops with MB neurons (13). This layered structure linking the MB parallel circuits may be important for producing the spacing effect.
In the present study, we explored the neural mechanisms underlying the spacing effect by focusing on the MB parallel circuits. Our findings suggested that the reduced activity of the MB parallel circuit consisting of γm neurons is important for LTM formation, which affects the activity of the downstream MBON-DAN network. Our results suggest that the spacing effect does not only solely depend on the cumulative cellular responses, but also relies on the neural circuit-based computation via the MB parallel circuits.
Results
Expression of pERK in γm Neurons Is Decreased during Spaced Training.
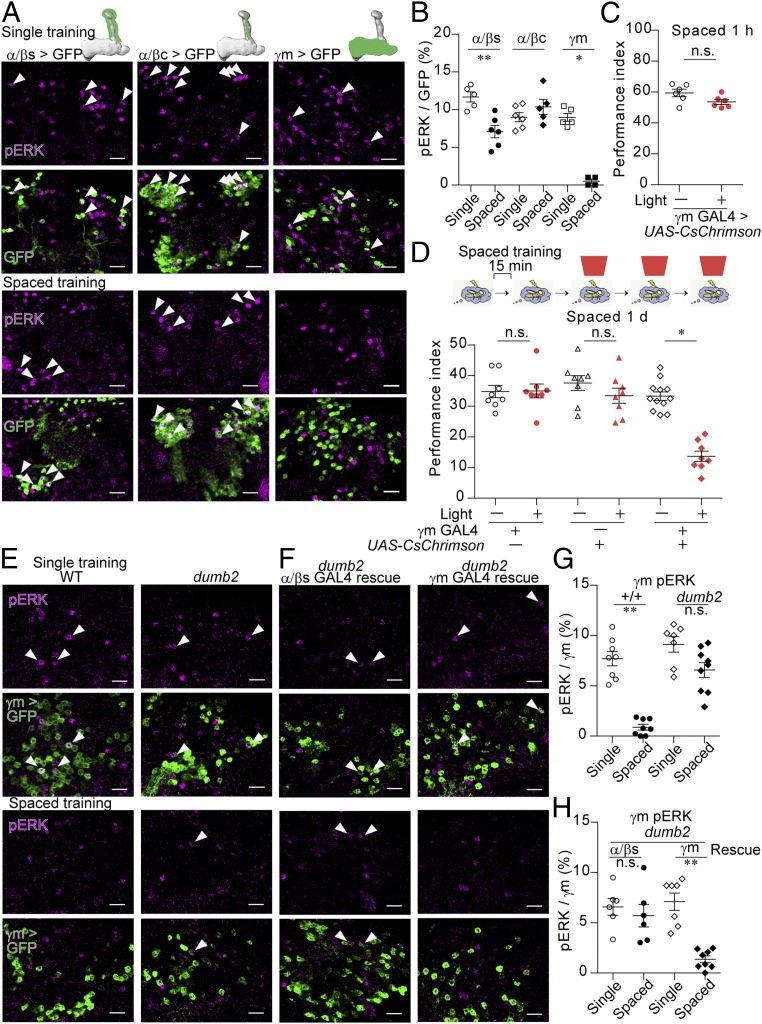
To understand the neural mechanism by which spaced learning induces memory consolidation, we investigated the MB neurons in the flies subjected to olfactory aversive single training or spaced training (3). Previous research has indicated that olfactory training induces the pERK in the nucleus (15), which could map the neurons activated by the training session. Following single training, MB neurons exhibited sparse nuclear expression of pERK (SI Appendix, Fig. S1A), which was absent in the flies with knockdown of Drosophila ERK, rl (SI Appendix, Fig. S1B). The frequency of pERK (SI Appendix, Fig. S1A) was consistent with the findings of studies utilizing whole-cell recording and calcium imaging (5–10%) (16, 17). We then labeled the individual types of MB neurons by expressing GFP using split-Gal4 drivers (13) (SI Appendix, Fig. S2) to determine whether expression of pERK is altered in specific MB subtypes after spaced training. Expression of pERK following single training was observed in α/βs, α/βc, and γm neurons (Fig. 1 A and B) but not in other types of MB neurons (SI Appendix, Fig. S1 C and D). Although α′/β′ neurons exhibit the highest baseline firing rates among MB neurons and the most vigorous responses to odors (17), expression of pERK was not detected in α′/β′ neurons (SI Appendix, Fig. S1 C and D), suggesting that the threshold of pERK expression depends on the cell type, probably due to the high intrinsic activity of α′/β′ neurons (17, 18) that could elevate the threshold of pERK expression. Expression of pERK in α/βs neurons was induced following spaced training, albeit to a lesser extent than following single training, while pERK expression remained unchanged in α/βc neurons following spaced training (Fig. 1 A and B). However, a remarkable decrease in pERK expression was observed in γm neurons following spaced training (Fig. 1 A and B). This decrease was not observed following massed training (SI Appendix, Fig. S1E), suggesting that rest intervals between the training sessions are important to decrease pERK expression in γm neurons. The same type of repetition is important for reducing pERK expression in γm neurons since the pERK expression was recovered by an additional training with a different odor from spaced training (SI Appendix, Fig. S1F). Expression of pERK in γm neurons was also observed in the flies exposed to an odor alone without electric shocks, which was increased by exposure to the second odor (SI Appendix, Fig. S1G), suggesting that expression of pERK correlates with the olfactory experience. Consistent with the previous finding in which γm neurons respond to electric shocks (19), electric shocks alone induced pERK expression in γm neurons (SI Appendix, Fig. S1H). The odor-induced and shock-induced pERK expression was decreased when flies were repeatedly exposed to the odor (SI Appendix, Fig. S1I) or electric shocks (SI Appendix, Fig. S1H) with rest intervals. When backward training was applied in which electric shocks precede odor exposure and, therefore, associative learning did not occur, pERK expression was induced in γm neurons, although it was reduced by repeating backward training with rest intervals (SI Appendix, Fig. S1J). We noted that, in contrast to γm neurons, α/βs and α/βc neurons did not show pERK expression by electric shocks (SI Appendix, Fig. S1 K and L). Thus, pERK expression in γm neurons would depend on the sensory stimulus including electric shocks, which is suppressed by its repetition with rest intervals, suggesting its possible role in spaced training-dependent LTM formation.
Fig. 1.
Expression of pERK is decreased during spaced training. (A and B) Nuclear pERK in subsets of MB neurons following single (A, Upper) or spaced training (A, Lower). GFP fused to the nuclear localization signal (nlsGFP) was expressed using the split-GAL4 drivers (SI Appendix, Fig. S2): MB185B for α/βs (P = 0.0087; n = 5 to 6), MB594B for α/βc (P = 0.3602; n = 5–6), and MB131B for γm (P = 0.0184; n = 4 to 5) (Scale bar, 10 μm). (C and D) Activation of γm neurons by pulsed red light (5 Hz, 1 min) during the shock periods in the last three sessions of spaced training impaired 1-d memory (Kruskal–Wallis test, P = 0.0005; n = 8–12) (D) without affecting 1-h memory after spaced training (P = 0.1320; n = 6) (C). Light was illuminated during paring of CS+ odor with electric shock. CsChrimson was expressed in γm neurons using MB131B. (E–H) Dopamine signaling was required for the decrease in pERK expression in γm neurons. Dumb2 mutant flies carry an upstream activating sequence (UAS) insertion in the first intron, which disrupts the expression of DopR1 but allows expression of DopR1 by crossing with the GAL4 driver. GFP was expressed by γm-LexA (R16A06-LexA) (SI Appendix, Fig. S2F). (E and F, Upper) Single training. (E and F, Lower) Spaced training. (G) (Kruskal–Wallis test, P = 0.0001; n = 6). (F and H) DopR1 was rescued in α/βs neurons using MB477B (P = 0.3939; n = 6) and in γm neurons using MB131B (P = 0.0014; n = 6) (Scale bar, 10 μm). The arrowheads indicate neurons expressing pERK. Data are represented as a mean ± SEM. n.s., not significant, P > 0.05; *P < 0.05; **P < 0.01.
Artificial Activation of γm Neurons Impairs LTM Formation.
Expression of pERK may indicate the neural plasticity or the neural activity. If decreases in pERK expression in γm neurons are related to the reduced neural activity during spaced training, their activation should impair LTM formation. To examine this hypothesis, we optogenetically activated γm neurons in the later training sessions via expression of the red-shifted channelrhodopsin CsChrimson (20) (Fig. 1 C and D). Optogenetic stimulation of γm neurons with pulses of red light-induced robust nuclear expression of pERK, comparing to the genetic control flies (SI Appendix, Fig. S1 M and N). Stimulating γm neurons as the flies received electric shocks during the last three sessions did not affect 1-h memory (Fig. 1C) but significantly impaired 1-d memory after spaced training (Fig. 1D). Therefore, activation of γm neurons in the later training sessions specifically impairs LTM formation without affecting short-term memory (STM).
Dopamine Signaling Is Required for the Decrease in pERK Expression in γm Neurons.
Given that dopamine is essential for associative aversive learning in flies (21, 22), we investigated whether dopamine is also required for the reduction of pERK expression in γm neurons. We used DopR1 mutant flies (dumb2) carrying a UAS insertion in the intronic region, which impairs DopR1 expression and aversive memory formation (22). In contrast to pERK expression in wild-type (WT) flies, pERK expression in γm neurons of dumb2 mutant flies was not significantly reduced after spaced training (Fig. 1 E and G). DopR1 expression can be induced by the UAS sequence inserted in dumb2 mutant flies via crossing with GAL4 driver lines (22). Decreased pERK expression in γm neurons following spaced training was observed in dumb2 mutant flies in which the expression of DopR1 was reintroduced in γm neurons but not in α/βs neurons (Fig. 1 F and H). These findings indicate that the decreased pERK expression in γm neurons following spaced training is associated with dopamine signaling in γm neurons.
We further sought to identify the dopamine neurons responsible for the decrease in pERK expression in γm neurons. The dopamine neurons, PPL1-γ2α′1 [also known as MB-MV1 (23)], and protocerebral anterior medial (PAM)-γ3 neurons [also known as MB-M2 (23)] innervate γm neurons and are activated by electric shocks (24). Another dopamine neuron, the PPL1-γ1pedc neuron [also known as MB-MP1 (23)], is required for STM (25, 26). These dopamine neurons may be responsible for LTM formation via suppression of γm neurons. Inactivation of either the PPL1-γ2α′1 or the PPL1-γ1pedc neuron via expression of a human inwardly rectifying K+ channel (Kir2.1) (27) impaired LTM formation (SI Appendix, Fig. S3 A–C), whereas inactivation of the PAM-γ3 neuron did not (SI Appendix, Fig. S3D). The inactivation of the PPL1-γ1pedc neuron also impaired STM (SI Appendix, Fig. S3E), whereas the inactivation of the PPL1-γ2α′1 neuron did not affect STM and 1-d after massed training (SI Appendix, Fig. S3 F and G), which induces anesthesia-resistant memory independent of gene expression. Thus, the PPL1-γ2α′1 neuron is specifically required for LTM formation. Importantly, the decreased pERK expression in γm neurons following spaced training was not observed when the PPL1-γ2α′1 neuron was inactivated (SI Appendix, Fig. S3H). The inactivation of the PPL1-γ2α′1 neuron via expression of Kir2.1 was supported by the finding that pERK expression was suppressed in the PPL1-γ2α′1 neuron following the training (SI Appendix, Fig. S3 I and J). These results suggest that dopamine release from the PPL1-γ2α′1 neuron is required for a decrease in pERK expression in γm neurons.
Artificial Activation of γm Neurons Impairs Gene Expression Associated with LTM Formation.
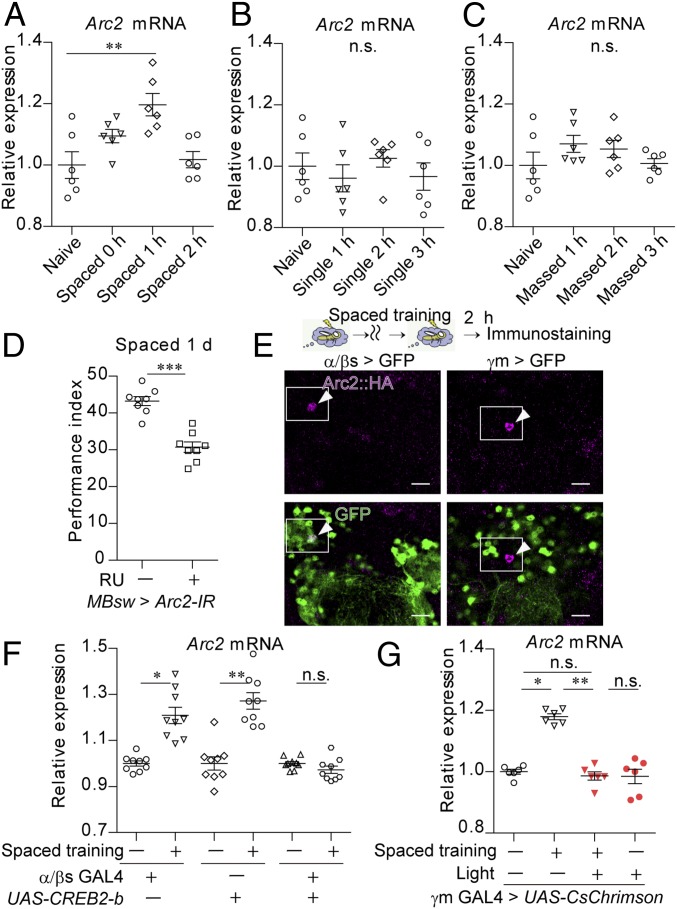
We then examined whether the optogenetic activation of γm neurons inhibits the expression of genes associated with LTM formation (Fig. 2). In mammals, levels of c-fos, homer, and Arc expression are well known to increase following neuronal activation (28). In flies, their orthologs kayak (kay) (29), homer, and Arc2, respectively, also exhibited significant increases in expression following spaced training (Fig. 2A and SI Appendix, Fig. S4 B and C). However, single training also increased expression of kay and homer (SI Appendix, Fig. S4 D and E). In contrast, Arc2 expression was not induced after single or massed training (Fig. 2 B and C). Given that Arc2 in the MBs—but not another Arc ortholog, Arc1 (SI Appendix, Fig. S4F)—is specifically required for LTM formation (Fig. 2D and SI Appendix, Fig. S4 G–I), Arc2 may represent a genetic marker of LTM as its expression is specific to spaced training. Using the flies carrying an HA-tag insertion at the C terminus of endogenous Arc2, we observed that the Arc2 protein is expressed in α/βs neurons of flies subjected to spaced training (Fig. 2E; 1.32 ± 0.61%, mean ± SEM, n = 9) but not in naive flies (SI Appendix, Fig. S4J; 0 ± 0%, mean ± SEM, n = 10). Accordingly, blocking the activity of the LTM-related transcription factor cAMP response element binding protein (CREB) via the expression of its repressor isoform CREB2-b (10) in α/βs neurons impaired induction of Arc2 mRNA in the head following spaced training (Fig. 2F) and expression of Arc2 protein in α/βs neurons (SI Appendix, Fig. S4K), indicating that spaced training induces Arc2 mRNA expression predominantly in α/βs neurons. Despite this fact, Arc2 protein was also observed in neurons other than α/βs neurons (SI Appendix, Fig. S4J), suggesting that the amounts of protein and mRNA of Arc2 may not well correlated, or Arc2 mRNA expression in α/βs neurons is robust, comparing to the basal Arc2 mRNA expression in nonα/βs neurons. There is a possibility that other neurons than α/βs neurons also induce Arc2 mRNA to a lesser extent, which is undetectable by RT-PCR using whole heads. Importantly, optogenetic activation of γm neurons during the last three sessions inhibited increases in Arc2 expression following spaced training (Fig. 2G and SI Appendix, Fig. S4L). In the flies where the decreased pERK expression in γm neurons was not observed via inactivation of the PPL1-γ2α′1 neuron, Arc2 expression was not induced following spaced training (SI Appendix, Fig. S4M). These data suggest that suppression of γm neurons in spaced training is involved in Arc2 expression for LTM formation.
Fig. 2.
Artificial activation of γm neurons impairs Arc2 expression in LTM formation. (A–C) Arc2 mRNA was specifically induced by spaced training. The flies were subjected to spaced training (Kruskal–Wallis test, P = 0.0048; n = 6) (A), single training (Kruskal–Wallis test, P = 0.6700; n = 6) (B), or massed training (Kruskal–Wallis test, P = 0.3313; n = 6) (C) (SI Appendix, Fig. S4A for the experimental schedules). RNA extracted from the fly heads was analyzed via RT-qPCR. (D) Knockdown of Arc2 impaired 1-d memory after spaced training. RNAi-based knockdown of Arc2 (Arc2-IR) in the whole MBs was performed using MBsw (38) by feeding the flies RU486 for 3 d (P = 0.0003; n = 8). (E) Arc2 protein was expressed in α/βs neurons at 2 h after spaced training. HA tags were inserted at the C terminus of Arc2 (Arc2::HA). NlsGFP was expressed in α/βs neurons using MB477B and in γm neurons using MB131B (Scale bar, 10 μm). The arrowheads indicate neurons expressing Arc2. (F and G) Arc2 mRNA expression at 1 h after spaced training was inhibited by expressing CREB2-b in α/βs neurons using MB477B (Kruskal–Wallis test, P < 0.0001; n = 9) (F) and by activation of γm neurons during the last three sessions of spaced training (Kruskal–Wallis test, P = 0.0039; n = 6) (G). Pulsed red light (5 Hz, 1 min) was delivered to flies expressing CsChrimson using MB131B (γm GAL4) as they received electric shocks during the last three sessions (G). Data are represented as a mean ± SEM. n.s., not significant, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.
A GABAergic Neuron Postsynaptic to γm Neurons (MBON-γ1pedc) Mediates Gene Expression Required for LTM Formation.
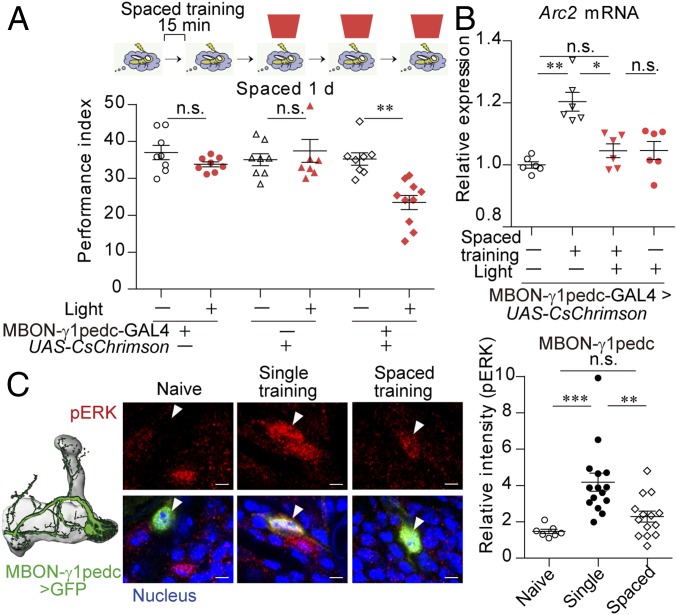
We hypothesized that the postsynaptic neuron to γm neurons is involved in the spacing effect. If this is the case, activation of those neurons during spaced training would disrupt LTM formation, similar to the activation of γm neurons. Although optogenetic stimulation of most of the neurons postsynaptic to γm neurons during the last three sessions of spaced training did not affect LTM formation (SI Appendix, Fig. S5A), the stimulation of a single GABAergic neuron postsynaptic to γm neurons, MBON-γ1pedc>α/β (MBON-γ1pedc) (13), also known as MB-MVP2, impaired LTM formation (Fig. 3A) without affecting STM (SI Appendix, Fig. S5B). The optogenetic activation of the MBON-γ1pedc neuron also impaired the increase in Arc2 expression after spaced training (Fig. 3B and SI Appendix, Fig. S5C). Consistent with this finding, nuclear pERK expression was induced in the MBON-γ1pedc neuron following single training (Fig. 3C) and massed training (SI Appendix, Fig. S5D) but not following spaced training (Fig. 3C), although ERK expression itself was not changed by spaced training (SI Appendix, Fig. S5 E and F). These results suggest that a decrease in the MBON-γ1pedc activity in the latter sessions of spaced training is required for gene expression in LTM formation. As observed for γm neurons, an additional training session using a different odor significantly induced pERK expression in the MBON-γ1pedc neuron (SI Appendix, Fig. S5G), and pERK expression in the MBON-γ1pedc neuron was also observed following the exposure to an odor, electric shocks, and backward training, which was reduced by their repetition with rest intervals (SI Appendix, Fig. S5 H–J).
Fig. 3.
A GABAergic neuron (MBON-γ1pedc) postsynaptic to γm neurons mediates gene expression required for LTM formation. (A and B) Activation of the MBON-γ1pedc neuron by pulsed red light (40 Hz, 1 min) during the shock periods of the last three sessions of spaced training impaired 1-d memory (Kruskal–Wallis test, P = 0.0001; n = 8–10) (A), and Arc2 mRNA expression at 1 h after spaced training (Kruskal–Wallis test, P = 0.0025; n = 6) (B). CsChrimson was expressed in the MBON-γ1pedc neuron using MB112C (SI Appendix, Fig. S6C). (C) Nuclear pERK expression was decreased in the MBON-γ1pedc neuron after spaced training. The MBON-γ1pedc neuron was labeled with nlsGFP using MB112C (Scale bar, 2 μm). n = 7–14 for all data. Data are represented as a mean ± SEM. n.s., not significant, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.
The Postsynaptic Neuron to the GABAergic MBON-γ1pedc Neuron (PPL1-α′2α2 Neuron) Is Required for Gene Expression in LTM Formation.
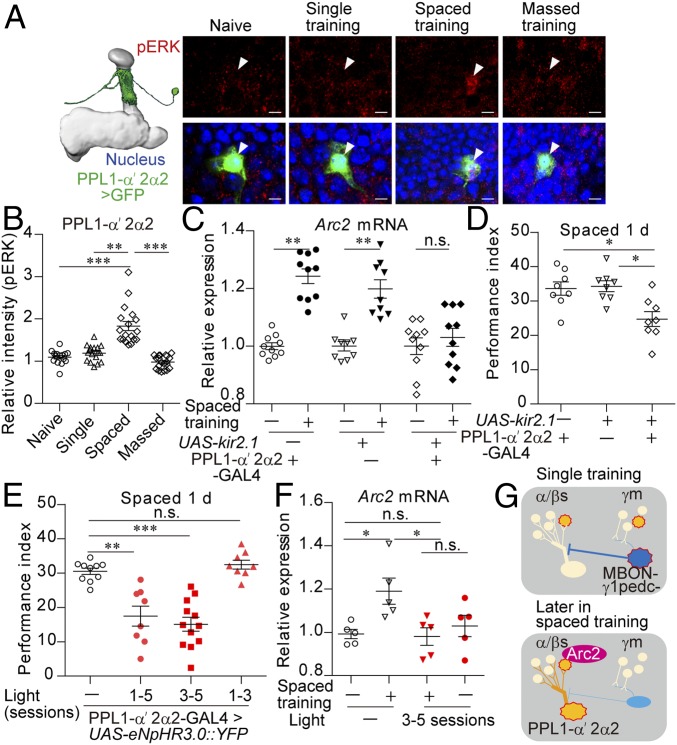
A decrease in GABAergic release from the MBON-γ1pedc neuron would disinhibit the postsynaptic neurons, which may be involved in gene expression in α/βs neurons. Among the previously identified neurons postsynaptic to the MBON-γ1pedc neuron (30) and innervating α/βs lobes, a dopaminergic neuron PPL1-α′2α2 (13) [also known as MB-V1 (23)] exhibited nuclear expression of pERK following spaced training only (Fig. 4 A and B) without alteration in ERK expression itself (SI Appendix, Fig. S6A), whereas another candidate, the PPL1-α3 neuron (13), did not (SI Appendix, Fig. S6B). Optogenetic activation of the MBON-γ1pedc neuron in the latter sessions of spaced training suppressed pERK expression in the PPL1-α′2α2 neuron (SI Appendix, Fig. S6G), suggesting that, although there are few synapses between MBON-γ1pedc and PPL1-α′2α2 neurons (30), the MBON-γ1pedc neuron can directly or indirectly regulate the activity of the PPL1-α′2α2 neuron, similar to other DAN neurons that are previously shown to be regulated by the MBON-γ1pedc neuron (31).
Fig. 4.
The postsynaptic neuron to the GABAergic MBON-γ1pedc neuron (PPL1-α′2α2) is required for gene expression in LTM formation. (A and B) Nuclear pERK expression was increased in the PPL1-α′2α2 neuron following spaced training. The PPL1-α′2α2 neuron was labeled with nlsGFP using MB058B (SI Appendix, Fig. S6E) (Kruskal–Wallis test, P < 0.0001; n = 15–20). (C and D) Inactivation of the PPL1-α′2α2 neuron impaired Arc2 mRNA expression at 1 h after spaced training (Kruskal–Wallis test, P < 0.0001; n = 9 to 10) (C) and 1-d memory after spaced training (Kruskal–Wallis test, P = 0.0109; n = 8) (D). Kir2.1 was expressed in the PPL1-α′2α2 neuron using MB058B. (E and F) Optogenetic inactivation of the PPL1-α′2α2 neuron impaired 1-d memory after spaced training (Kruskal–Wallis test, P < 0.0001; n = 8) (E), and Arc2 mRNA expression at 1 h after spaced training (Kruskal–Wallis test, P = 0.0128; n = 6) (F). MB058B was used to express eNpHR3.0 in the PPL1-α′2α2 neuron. Flies were illuminated by red light at 40 Hz during the shock periods of the indicated sessions of spaced training. (G) Model: Arc2 expression is induced in α/βs neurons via simultaneous activation of α/βs and PPL1-α′2α2 neurons. Spaced training allows activation of the PPL1-α′2α2 neuron due to reduced activity in γm and GABAergic MBON-γ1pedc neurons. Data are represented as a mean ± SEM. n.s., not significant, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.
We then examined whether the PPL1-α′2α2 neuron is involved in gene expression for LTM formation. Inactivation of the PPL1-α′2α2 neuron via expression of Kir2.1 impaired Arc2 expression (Fig. 4C) and LTM formation after spaced training (Fig. 4D) without affecting STM (SI Appendix, Fig. S6H) and 1-d memory after massed training (SI Appendix, Fig. S6I). To inactivate the PPL1-α′2α2 neuron specifically during the conditioning, eNpHR3.0 (32) was expressed in the PPL1-α′2α2 neuron, which successfully suppressed pERK expression following spaced training with pulses of red light (SI Appendix, Fig. S6J). Optogenetic inactivation of the PPL1-α′2α2 neuron in the latter three sessions of spaced training but not the former three sessions of spaced training impaired LTM formation (Fig. 4E) without affecting STM (SI Appendix, Fig. S6K), although the light illumination itself did not affect LTM formation in the genetic control flies (SI Appendix, Fig. S6L). Optogenetic suppression of the PPL1-α′2α2 neuron in the latter three sessions of spaced training also impaired Arc2 expression (Fig. 4F and SI Appendix, Fig. S6 M and N). These data suggest that the activation of the PPL1-α′2α2 neuron in the latter sessions of spaced training is required for gene expression in LTM formation.
Artificial Activation of the PPL1-α′2α2 Neuron and α/βs Neurons Induces Gene Expression Related to LTM Formation.
We next addressed whether activation of the PPL1-α′2α2 neuron leads to gene expression. To examine this hypothesis, we expressed the thermosensitive cation channel dTRPA1 (33) in either or both the PPL1-α′2α2 and the α/βs neurons, which enables artificial activation of the expressed neuron when flies are subjected to high temperatures (SI Appendix, Fig. S7 A and B). Although activation of either the PPL1-α′2α2 neuron or the α/βs neurons did not, their simultaneous activation induced expression of Arc2 mRNA (SI Appendix, Fig. S7C) and Arc2 protein in α/βs neurons (SI Appendix, Fig. S7D, 1.41 ± 0.42%, mean ± SEM, n = 5). Activation of PPL1-α3, which is one of the dopamine neurons innervating the tips of the MB α/β lobes, did not induce Arc2 mRNA when combined with activation of α/βs neurons (SI Appendix, Fig. S7E), suggesting that the PPL1-α′2α2 neuron has a specific role for Arc2 expression.
In agreement with the decreased pERK expression in the MBON-γ1pedc neuron by repetition of exposure to an odor, electric shocks, and backward training (SI Appendix, Fig. S5 H–J), pERK expression in the PPL1-α′2α2 neuron increased in these conditions (SI Appendix, Fig. S7 F–H). However, backward spaced training, which contains the context of repeated exposure to the odors and electric shocks but no association between them, did not induce Arc2 mRNA expression (SI Appendix, Fig. S7I), suggesting that association of an odor with electric shocks is necessary for Arc2 mRNA expression in addition to the PPL1-α′2α2 activation.
Given that massed training induces gene expression-independent memory, artificial activation of the PPL1-α′2α2 neuron, or artificial inactivation of the MBON-γ1pedc or γm neurons during massed training may enhance memory. However, optogenetic activation of the PPL1-α′2α2 neuron and optogenetic inactivation of the MBON-γ1pedc neuron or γm neurons in the latter three training sessions in massed training did not affect 1-d memory (SI Appendix, Fig. S7 J and K). Given that STM was reduced by optogenetic inactivation of the MBON-γ1pedc neuron (SI Appendix, Fig. S7L), which is consistent to the previous finding (26), optogenetic inactivation via expression of eNpHR3.0 was effective in this experimental condition. Thus, activation of the PPL1-α′2α2 neuron would not be sufficient to induce LTM, suggesting that spaced training also employs other factors, such as ones related to the synaptic plasticity for LTM formation.
Discussion
We adopted an olfactory spaced training paradigm in Drosophila to investigate the neural circuit underlying the spacing effect. We took the advantage of immunohistochemistry by monitoring phosphorylation of MAPK (ERK), which allowed us to map the neurons activated in the normal training paradigm. Although an increase or decrease in pERK expression may result from either the change in the neural activation or of the ERK-signaling pathway, the optogenetic manipulation in this study suggested that the neural activity change in the MB-MBON-DAN network is significant in LTM formation. While previous studies have demonstrated that γm neurons are actively involved in memory formation (19, 34, 35), the present study suggests that a decrease in γm activation is also required for LTM formation. As a result, a single GABAergic neuron (MBON-γ1pedc) postsynaptic to γm neurons became inactivated, which, in turn, led to activation of a dopamine neuron (PPL1-α′2α2). Our findings further revealed that the PPL1-α′2α2 neuron innervates another MB parallel circuit consisting of α/βs neurons to induce gene expression required for LTM (Fig. 4G). Our study suggests the model in which the multistep linear circuit in the MB would be significant to index spaced learning of the environment. This neural circuit may act in concert with the cumulative cellular responses, such as the previously proposed oscillatory kinase activity during spaced learning (7, 29). Dopamine-dependent synaptic suppression between MB neurons and MBON as previously demonstrated (9, 24, 36) may also affect the MBON-DAN network.
PPL1-α′2α2 activation in the latter sessions of spaced training was required for gene expression in LTM formation. PPL1-α′2α2 activation was observed via calcium imaging during single training (37). However, increases in PPL1-α′2α2 activation during spaced training via MBON-γ1pedc inactivation may be necessary to provide sufficient signaling for inducing gene expression. Backward spaced training significantly increased pERK expression in the PPL1-α′2α2 neuron (SI Appendix, Fig. S7H), although Arc2 mRNA was not induced (SI Appendix, Fig. S7I), suggesting that association of an odor and electric shocks is also required for Arc2 expression. Consistently, although dTRPA1-dependent activation induced pERK in all α/βs neurons (SI Appendix, Fig. S7A), artificial activation of the PPL1-α′2α2 neuron, and α/βs neurons induced Arc2 protein expression in only a few α/βs neurons (SI Appendix, Fig. S7D), which would be the result of bypassing the requirement of the association due to the artificial activation. Thus, the multiple mechanisms for gene expression should be converged during spaced training, which include activation of the PPL1-α′2α2 neuron (spacing effect information), α/βs neurons (odor information), and other dopamine neurons (electric shock information). A previous study demonstrated that the cfos-expressing neurons show pERK expression upon memory retrieval (29). In contrast, we never found pERK expression in the Arc2-expressing neurons upon retraining, memory retrieval, or reverse training (SI Appendix, Fig. S8). Accordingly, we found that the pERK-expressing α/βs neurons were slightly reduced following spaced training, compared to single training (Fig. 1B). There are 2 possibilities. First, the neural activity of the Arc2-expressing neurons could be suppressed by spaced training. Given that synaptic depression between MBs and MBONs has been proposed as the neural correlates of memory (24, 36), the decreased activity of the Arc2-expressing neurons may play an important role in LTM. Second, the Arc2-expressing neurons could undergo down-regulation in the ERK signaling, although the neurons are activated during memory retrieval. These should be examined in the future study to understand the physiological role of gene expression involved in LTM.
Previous studies have suggested that olfactory information relies on sparse coding in the parallel circuits of the MB (16, 17), although the plasticity of these sparse codings has yet to be explored. In the present study, we demonstrated that spaced learning preferentially targets sparse coding in the MB parallel circuit consisting of γm neurons via dopamine signaling, leading to memory consolidation in another MB parallel circuit consisting of α/βs neurons. Thus, the neurons responsible for generating the spacing effect and the neurons engaged in memory reside in the different MB parallel circuits. This neural circuit-based computation is accomplished by the MBON-DAN network linking these parallel circuits. This may be generalized to other types of sensory input in Drosophila and may provide insight into the neural representations within parallel neural circuits in other animals.
Materials and Methods
Culture Conditions.
Flies were raised under a 12-h light:dark cycle at a temperature of 24 °C and humidity of 60%. For other methods, see SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the members of SK Laboratories for helpful discussions and T. Kitamura, Y. Aso, C. Yokoyama, J. Horiuchi, M. Saitoe, T. Tabata, Y. Hayashi, H. Okuno, and D. Yamazaki for a critical reading of the paper. We thank G. M. Rubin, M. Saitoe, and R. Davis for materials and T. Miyashita for the technical advice. The present study was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grants JP17H04984 and JP16H01274, the Ichiro Kanehara Foundation, the Senri Life Science Foundation, and Shionogi & Co., Ltd. (to Y.H.), and by JSPS KAKENHI Grant JP17K17823 (to H.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901292116/-/DCSupplemental.
References
- 1.Ebbinghaus H. E., Memory: A Contribution to Experimental Psychology (Dover, Newyork, 1885). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carew T. J., Pinsker H. M., Kandel E. R., Long-term habituation of a defensive withdrawal reflex in aplysia. Science 175, 451–454 (1972). [DOI] [PubMed] [Google Scholar]
- 3.Tully T., Preat T., Boynton S. C., Del Vecchio M., Genetic dissection of consolidated memory in Drosophila. Cell 79, 35–47 (1994). [DOI] [PubMed] [Google Scholar]
- 4.Parsons R. G., Davis M., A metaplasticity-like mechanism supports the selection of fear memories: Role of protein kinase A in the amygdala. J. Neurosci. 32, 7843–7851 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye X., Shobe J. L., Sharma S. K., Marina A., Carew T. J., Small G proteins exhibit pattern sensitivity in MAPK activation during the induction of memory and synaptic facilitation in Aplysia. Proc. Natl. Acad. Sci. U.S.A. 105, 20511–20516 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y., et al. , Computational design of enhanced learning protocols. Nat. Neurosci. 15, 294–297 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pagani M. R., Oishi K., Gelb B. D., Zhong Y., The phosphatase SHP2 regulates the spacing effect for long-term memory induction. Cell 139, 186–198 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naqib F., Farah C. A., Pack C. C., Sossin W. S., The rates of protein synthesis and degradation account for the differential response of neurons to spaced and massed training protocols. PLoS Comput. Biol. 7, e1002324 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hattori D., et al. , Representations of novelty and familiarity in a mushroom body compartment. Cell 169, 956–969 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin J. C., et al. , Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell 79, 49–58 (1994). [DOI] [PubMed] [Google Scholar]
- 11.Guven-Ozkan T., Davis R. L., Functional neuroanatomy of Drosophila olfactory memory formation. Learn. Mem. 21, 519–526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cognigni P., Felsenberg J., Waddell S., Do the right thing: Neural network mechanisms of memory formation, expression and update in Drosophila. Curr. Opin. Neurobiol. 49, 51–58 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aso Y., et al. , The neuronal architecture of the mushroom body provides a logic for associative learning. eLife 3, e04577 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang C., Wang P., Xie Z., Wang L., Zhong Y., The differential requirement of mushroom body α/β subdivisions in long-term memory retrieval in Drosophila. Protein Cell 4, 512–519 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Q., et al. , Importin-7 mediates memory consolidation through regulation of nuclear translocation of training-activated MAPK in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 113, 3072–3077 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honegger K. S., Campbell R. A., Turner G. C., Cellular-resolution population imaging reveals robust sparse coding in the Drosophila mushroom body. J. Neurosci. 31, 11772–11785 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner G. C., Bazhenov M., Laurent G., Olfactory representations by Drosophila mushroom body neurons. J. Neurophysiol. 99, 734–746 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Inada K., Tsuchimoto Y., Kazama H., Origins of cell-type-specific olfactory processing in the Drosophila mushroom body circuit. Neuron 95, 357–367(2017). [DOI] [PubMed] [Google Scholar]
- 19.Akalal D. B., Yu D., Davis R. L., A late-phase, long-term memory trace forms in the γ neurons of Drosophila mushroom bodies after olfactory classical conditioning. J. Neurosci. 30, 16699–16708 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klapoetke N. C., et al. , Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwaerzel M., et al. , Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J. Neurosci. 23, 10495–10502 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y. C., Lee H. G., Han K. A., D1 dopamine receptor dDA1 is required in the mushroom body neurons for aversive and appetitive learning in Drosophila. J. Neurosci. 27, 7640–7647 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka N. K., Tanimoto H., Ito K., Neuronal assemblies of the Drosophila mushroom body. J. Comp. Neurol. 508, 711–755 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Cohn R., Morantte I., Ruta V., Coordinated and compartmentalized neuromodulation shapes sensory processing in Drosophila. Cell 163, 1742–1755 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aso Y., et al. , Three dopamine pathways induce aversive odor memories with different stability. PLoS Genet. 8, e1002768 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aso Y., et al. , Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. eLife 3, e04580 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baines R. A., Uhler J. P., Thompson A., Sweeney S. T., Bate M., Altered electrical properties in Drosophila neurons developing without synaptic transmission. J. Neurosci. 21, 1523–1531 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minatohara K., Akiyoshi M., Okuno H., Role of immediate-early genes in synaptic plasticity and neuronal ensembles underlying the memory trace. Front. Mol. Neurosci. 8, 78 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyashita T., Kikuchi E., Horiuchi J., Saitoe M., Long-term memory engram cells are established by c-Fos/CREB transcriptional cycling. Cell Rep. 25, 2716–2728 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Takemura S. Y., et al. , A connectome of a learning and memory center in the adult Drosophila brain. eLife 6, e26975 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ueoka Y., Hiroi M., Abe T., Tabata T., Suppression of a single pair of mushroom body output neurons in Drosophila triggers aversive associations. FEBS Open Bio 7, 562–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Q., et al. , Gap junction networks in mushroom bodies participate in visual learning and memory in Drosophila. eLife 5, e13238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamada F. N., et al. , An internal thermal sensor controlling temperature preference in Drosophila. Nature 454, 217–220 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin H., et al. , Gamma neurons mediate dopaminergic input during aversive olfactory memory formation in Drosophila. Curr. Biol. 22, 608–614 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamazaki D., et al. , Two parallel pathways assign opposing odor valences during Drosophila memory formation. Cell Rep. 22, 2346–2358 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Hige T., Aso Y., Modi M. N., Rubin G. M., Turner G. C., Heterosynaptic plasticity underlies aversive olfactory learning in Drosophila. Neuron 88, 985–998 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cervantes-Sandoval I., Phan A., Chakraborty M., Davis R. L., Reciprocal synapses between mushroom body and dopamine neurons form a positive feedback loop required for learning. eLife 6, e23789 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao Z., Roman G., Zong L., Davis R. L., Pharmacogenetic rescue in time and space of the rutabaga memory impairment by using Gene-Switch. Proc. Natl. Acad. Sci. U.S.A. 101, 198–203 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.