Significance
Accurate chromosome segregation at mitosis and meiosis is crucial to prevent genome instability, birth defect, and cancer. Accordingly, separase, the protease that triggers chromosome distribution, is tightly regulated by a direct inhibitor, the securin. However, securin has not been identified, neither functionnally nor by sequence similarity, in other clades that fungi and animals. This raised doubts about the conservation of this mechanism in other branches of eukaryotes. Here, we identify and characterize the securin in plants. Despite extreme sequence divergence, the securin kept the same core function and is likely a universal regulator of cell division in eukaryotes.
Keywords: meiosis, mitosis, separase, securin
Abstract
Chromosome distribution at anaphase of mitosis and meiosis is triggered by separase, an evolutionarily conserved protease. Separase must be tightly regulated to prevent the untimely release of chromatid cohesion and disastrous chromosome distribution defects. Securin is the key inhibitor of separase in animals and fungi, but has not been identified in other eukaryotic lineages. Here, we identified PATRONUS1 and PATRONUS2 (PANS1 and PANS2) as the Arabidopsis homologs of securin. Disruption of PANS1 is known to lead to the premature separation of chromosomes at meiosis, and the simultaneous disruption of PANS1 and PANS2 is lethal. Here, we show that PANS1 targeting by the anaphase-promoting complex is required to trigger chromosome separation, mirroring the regulation of securin. We showed that PANS1 acts independently from Shugosins. In a genetic screen for pans1 suppressors, we identified SEPARASE mutants, showing that PANS1 and SEPARASE have antagonistic functions in vivo. Finally, we showed that the PANS1 and PANS2 proteins interact directly with SEPARASE. Altogether, our results show that PANS1 and PANS2 act as a plant securin. Remote sequence similarity was identified between the plant patronus family and animal securins, suggesting that they indeed derive from a common ancestor. Identification of patronus as the elusive plant securin illustrates the extreme sequence divergence of this central regulator of mitosis and meiosis.
Balanced segregation of chromosomes at both mitosis and meiosis requires that chromatid cohesion complexes be removed after proper chromosome alignment in the spindle. Inaccuracies in this process cause chromosome missegregation and aneuploidy, contributing to cancer and birth defects. Separase, which is conserved in fungi, animals, and plants, triggers cohesion release by cleaving the kleisin subunit of the cohesin complex, opening the cohesin ring and allowing chromosome segregation (1). Securin is the primary regulator of separase activity in animals and fungi, forming a complex with separase and blocking substrate access to its active site. Securin, which is defined by its functional and biochemical properties, is a largely unstructured protein whose sequence is conserved only between closely related species (2–6). At the onset of anaphase, the anaphase-promoting complex (also known as the cyclosome) (APC/C) triggers the degradation of securin, releasing separase activity and allowing chromosome separation. In vertebrates, the Cdk1–CyclinB1 complex is an additional regulator of separase activity (7). At meiosis I, the cleavage of cohesins is also spatially controlled: Pericentromeric cohesins are protected from separase cleavage by shugoshin-PP2A through dephosphorylation of the kleisin subunit (8, 9).
In Arabidopsis, separase is essential for cohesion release and chromosome segregation at both meiosis and mitosis (10, 11), APC/C regulates the progression of division (12, 13) and the shugoshins SGO1 and SGO2 and PP2A are required for cohesion protection at meiosis (14–16). However, securin is missing from the picture, raising the intriguing possibility that securin has been lost in the green lineage and that plant separase is regulated by a different mechanism.
We previously characterized the two paralogues PATRONUS1 and PATRONUS2 in Arabidopsis (PANS1, PANS2) (14). PANS1 is essential for the protection of sister chromatid cohesion between the two meiotic divisions, through an unknown mechanism. In the pans1 mutant, sister chromatid cohesion is lost before metaphase II, leading to chromosome segregation defects at meiosis II (14, 17, 18). In addition, the pans1 mutant also has a slight growth defect, which is exacerbated under stress conditions, associated with a certain level of mitotic defects and aneuploidy in somatic cells (14, 17, 19). The pans2 mutant is indistinguishable from the wild type but has synthetic lethal interaction with pans1, suggesting that PANS1 and PANS2 have an essential but redundant role at mitosis (14). PANS1 and PANS2 share 42% identity and encode proteins of unknown function. PANS1 has been shown to interact with APC/Cthrough its destruction-box (d-box) and KEN-box domains. Patronus proteins are well-conserved in dicots, one of the two major clades of flowering plants, with most species having one or two homologs (14). In monocots, the other major clade of flowering plants, PANS proteins share limited similarity with RSS1 (RICE SALT SENSITIVE 1, also known as Os02g39390), a protein that regulates cell cycle under stress conditions in rice (20). Although PANS1, PANS2, and RSS1 clearly play important role in cell division, the molecular function of PANS1, PANS2, and RSS1 has remained elusive.
Results
Expression of Destruction-Box-Less PATRONUS1 Prevents Cohesion Release and Chromosome Segregation.
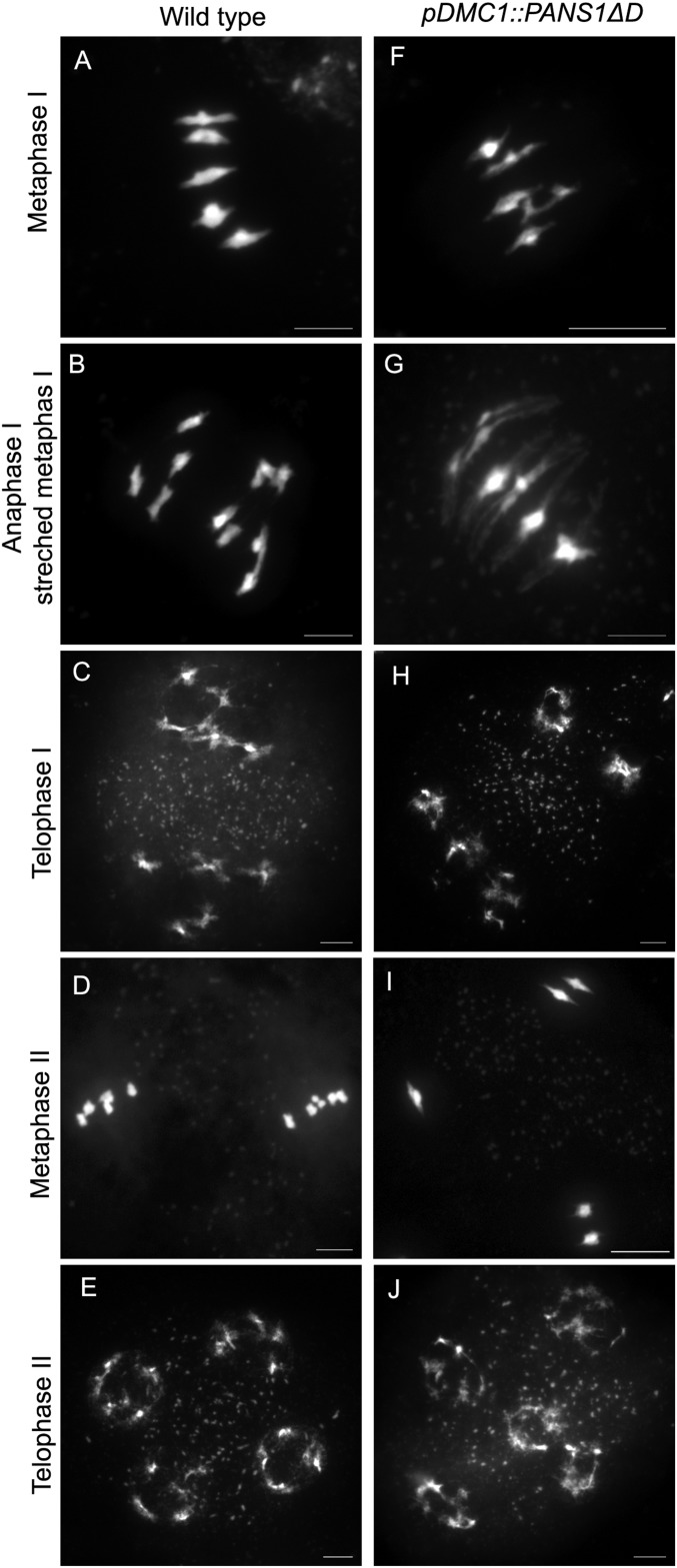
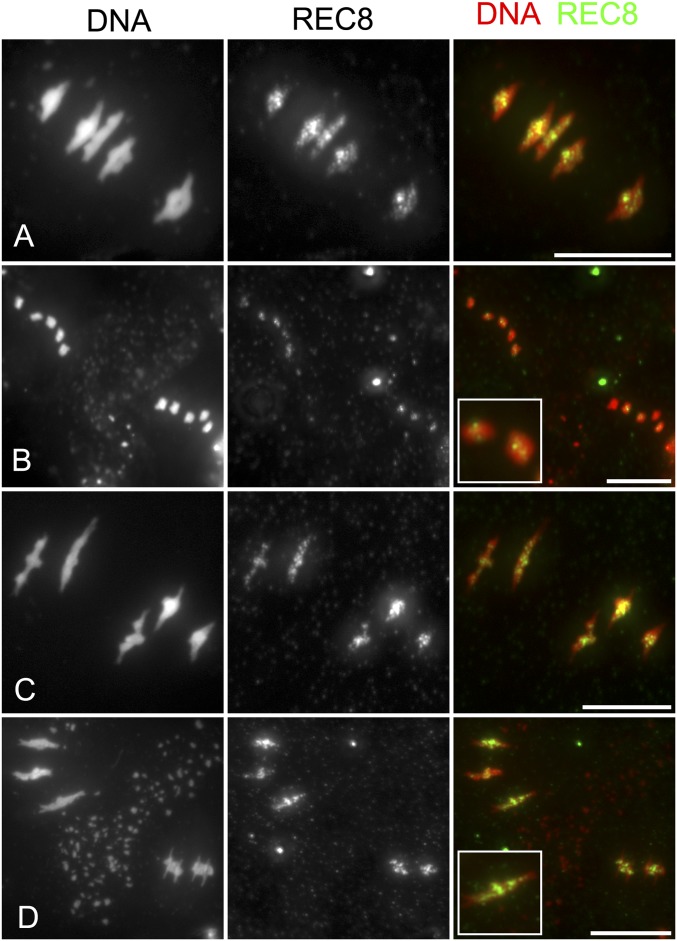
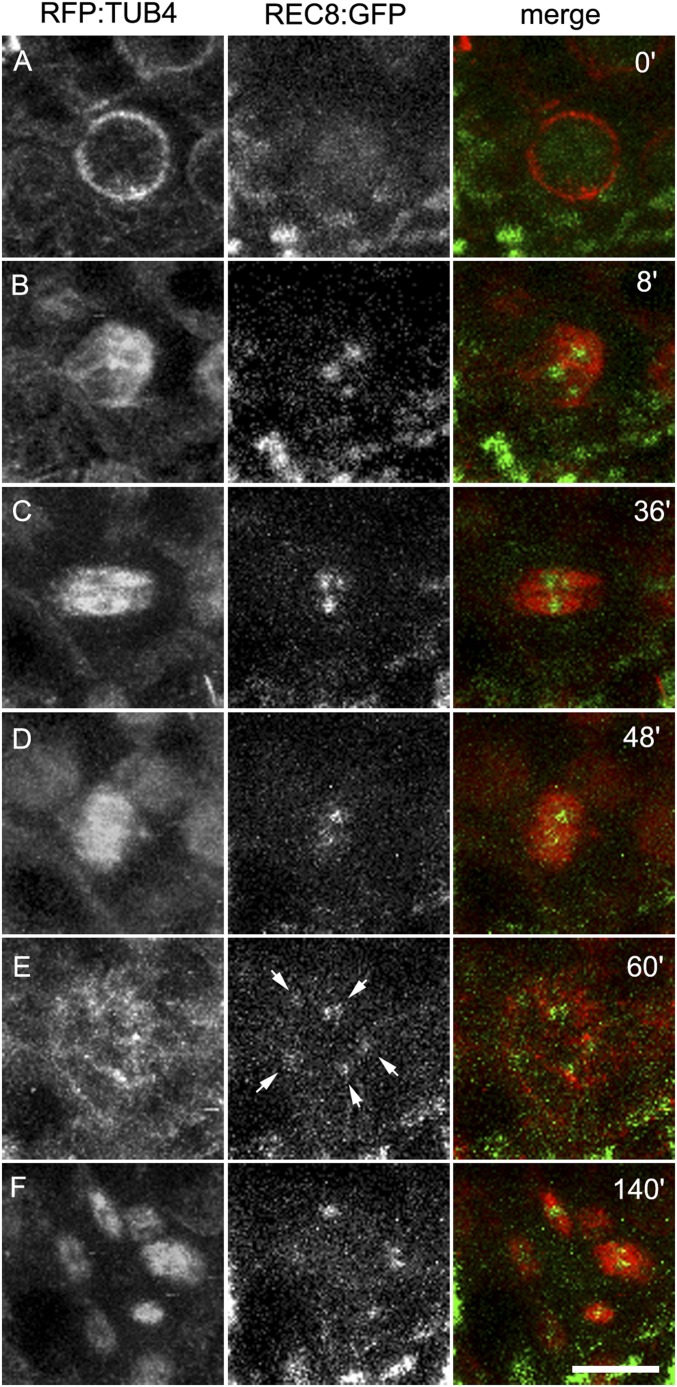
The sequences of PANS proteins contain a conserved d-box domain (RxxLxxxN), recognized by the anaphase-promoting complex (APC/C), which triggers the destruction of the targeted protein by the proteasome. A mutant version of PANS1 mutated in its d-box (PANS1ΔD; RxxL → LxxV) loses its capacity to interact with the APC/C activator subunit CDC20 in yeast two-hybrid (Y2H) assays (14). Expression of PANS1ΔD under its own promoter is lethal, suggesting that PANS1 accumulation prevents plant development (14). To assess the function of the PANS1 d-box at meiosis, we expressed PANS1ΔD under the meiosis-specific promoter DMC1. Wild-type PANS1 expressed under the DMC1 promoter was able to complement the meiotic defects of the pans1 mutant (n = 2/4) (SI Appendix, Fig. S1). In contrast pDMC1::PANS1ΔD caused full sterility when transformed in wild-type or pans1 plants (n = 15/15 and 2/2, respectively). In addition, pDMC1::PANS1ΔD plants showed a variable growth defect, from barely developing plants to wild-type-like plants (SI Appendix, Fig. S2), suggesting that the pDMC1 promoter can drive variable expression in somatic tissues, but all plants showed complete sterility. In pDMC1::PANS1ΔD plants, meiotic chromosome spreads did not reveal any defect in prophase or early metaphase I (Fig. 1). However, among the 119 postprophase cells observed, none showed the configuration typical of anaphase I, metaphase II, anaphase II, or telophase II, suggesting that the meiotic cells do not progress beyond metaphase I. We observed normal metaphase I with five bivalents aligned on the metaphase plate (compare Fig. 1F with Fig. 1A), but also metaphase I with five overstretched bivalents (Fig. 1G). We also observed configurations resembling metaphase II, with chromosomes distributed in at least two groups separated by a dense band of organelles. However, a total of five bivalents aligned on the two metaphase II plates in pDMC1::PANS1ΔD (e.g., three bivalents on one plate, and two on the other) (Fig. 1I), instead of five pairs of chromatids on each metaphase plates as observed in the wild type (Fig. 1D). At telophase II, we observed five nuclei, presumably each containing a decondensed bivalent. This observation suggests that pDMC1::PANS1ΔD abolishes the separation of homologous chromosomes at meiosis I. Immunostaining revealed that the bivalents in pDMC1::PANS1ΔD at a metaphase I- or metaphase II-like configuration were entirely decorated with the cohesin REC8, suggesting that the inhibition of chromosome separation is due to an incapacity to remove cohesins (Fig. 2). We then observed the effect on pDMC1::PANS1ΔD on male meiocytes with live imaging of cells expressing a red fluorescent protein (RFP)-tagged tubulin (RFP:TUB4) and a green fluorescent protein (GFP)-tagged REC8 (21) (Figs. 3–5 and Movies S1–S3). In the wild type (Movie S1), we observed the progression of male meiosis from prophase, with tubulin surrounding the nucleus (Fig. 3A), to metaphase I with the formation of the spindle and alignment of the chromosomes (Fig. 3C), and to anaphase I with the disappearance of the REC8 signal and reorganization of the spindle (Fig. 3 D and E). The first division, from the end of prophase (nuclear break down, Fig. 3B) to the onset of anaphase I, lasted 38 ± 5 min (mean ± SD, n = 29 cells). When analyzing pDMC1::PANS1ΔD with live imaging (Movies S2 and S3), we observed cells progressing normally from prophase to metaphase I (Fig. 4 A–C). However, the length of metaphase I was very variable, from 40 min (Movie S2 and Fig. 4) to more than 4 h (Movie S3). When anaphase occurred, as observed regarding microtubule reorganization (Fig. 4 D and E and Movie S2), the REC8 signal was still detected. Further, five REC8:GFP bodies, presumably representing the five bivalents, were still observed at late anaphase I (arrows in Fig. 4E), and following interphase, a spindle polymerized around each of the five REC8:GFP bodies (Fig. 4F and Movie S2). Thus, live imaging confirmed that the expression of a d-box-less PANS1 at meiosis prevents the release of REC8 and the separation of chromosomes.
Fig. 1.
Meiotic spreads in the wild type (A–E) and pDMC1::PANS1ΔD (F–J). Male meiocytes were spread and stained with DAPI. (Scale bar, 10 µm.)
Fig. 2.
REC8 persists on chromosomes in pDMC1::PANS1ΔD. Immunolocalization of the cohesion REC8 was performed on meiotic chromosome spreads stained with DAPI (white). (A) Wild-type metaphase I. The bivalents under tension aligned on metaphase I are decorated with REC8. (B) Wild-type metaphase II. Five pairs of chromatids are aligned on both metaphase II plates. A faint REC8 signal is detected in the middle of each chromosome. (C) Metaphase I in pDMC1::PANS1ΔD. The bivalents are decorated with REC8, as in the wild type. (D) Aberrant metaphase II in pDMC1::PANS1ΔD. Two groups of bivalents are separated by an organelle band. The bivalents are under tension and are still decorated with REC8. (Scale bar, 10 µm.)
Fig. 3.
Time course of male meiosis I in wild type. Anthers at meiotic stages were observed in plant expressing the TUBULIN BETA CHAIN 4 fused with the RED FLUORESCENT PROTEIN (RFP:TUB4) and the cohesin REC8 fused with the GREEN FLUORESCENT protein (REC8:GFP). Images were acquired every 3 min (Movie S1). The time is indicated on the snapshots in minutes, starting with the beginning of the movie. (A) Prophase. (B) Tubulin surrounds the nucleus at prometaphase. (C) The nucleus disappears and the spindle is forming. Metaphase I chromosomes are aligned in the spindle. (D) Early anaphase I, the REC8:GPF signal suddenly decreases. (E) Late anaphase I. The spindle has reorganized. (Scale bar, 10 µm.)
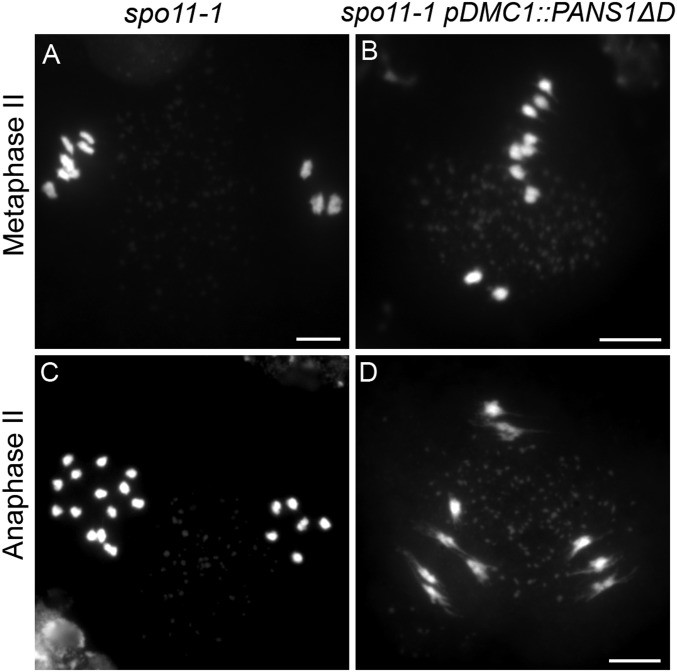
Fig. 5.
pDMC1::PANS1ΔD triggers arrest at metaphase II in spo11-1. Male meiocytes were spread and stained with DAPI. (A) Metaphase II in spo11-1. In this cell, three chromosomes are aligned on one plate, and seven on the other. (B) Metaphase II in spo11-1 pDMC1::PANS1ΔD. Similarly, uneven numbers of chromosomes are aligned on the two metaphase plates. (C) Anaphase II in spo11-1. Anaphase II with separation of sister chromatids (3, 3, 7, 7). (D) Pseudoanaphase II in spo11-1 pDMC1::PANS1ΔD with stretched chromosomes and no separation of sister chromatids. (Scale bars, A and B, 10 µm; D, 5 µm.)
Fig. 4.
Time course of male meiosis in pDMC1::PANS1ΔD. Anthers at meiotic stages were observed in plants expressing RFP:TUB4, REC8:GFP, and pDMC1::PANS1ΔD. Images were acquired every 4 min (Movie S2). The time is indicated on the snapshots in minutes, starting with the beginning of the movie (Movie S2). (A) Prophase. (B) Prometaphase. (C) Metaphase I. (D) Early anaphase I. (E) Abnormal late anaphase I with five REC8:GFP, indicated by arrows. (F) Aberrant metaphase II with five spindles. This time course corresponds to Movie S2. Movie S3 is an independent acquisition in the same background. (Scale bar, 10 µm.)
In Arabidopsis spo11-1 mutants, the formation of crossovers is eliminated and the resulting unconnected chromosomes (univalents) segregate randomly at anaphase I and progress through metaphase II and anaphase II with segregation of sister chromatids (22) (Fig. 5). Thus, the spo11-1 mutation should allow segregation of chromosomes at meiosis I in pDMC1::PANS1ΔD and entry into meiosis II, making it possible to assess the effect of PANS1ΔD on chromatid pairs at anaphase II. Our results indicated that spo11-1 pDMC1::PANS1ΔD meiocytes progressed to metaphase II (Fig. 5B), but appeared to be arrested at that stage with stretched chromosomes (Fig. 5D), showing that PANS1ΔD can prevent chromosome segregation of both homologs at meiosis I and sister chromatids at meiosis II. The lethality of PANS1ΔD expressed under its own promoter suggests that it can also prevent chromosome segregation at mitosis. Altogether, these results suggest that targeting of PANS by APC/C is required for the release of cohesion and chromosome distribution at anaphase of mitosis and meiosis, mimicking the regulation and function of securin (7, 23, 24).
PATRONUS1 Acts Independently of SHUGOSHINs.
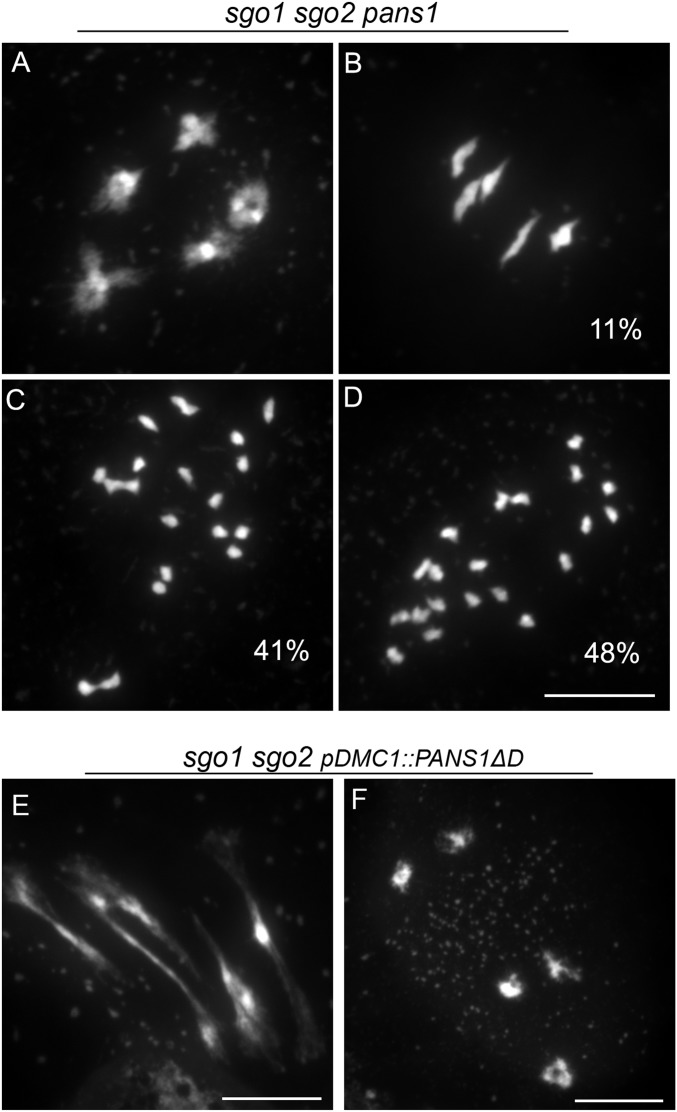
Shugoshin (SGO) is an evolutionarily conserved protein that protects sister chromatid cohesion against separase (8, 25). In Arabidopsis, both shugoshin genes (SGO1 and SGO2) are involved in the protection of pericentromeric cohesion during meiosis, with the corresponding double-mutant leading to the complete loss of sister chromatid cohesion at anaphase I (14), without apparent somatic function. The pans1 mutant loses pericentromeric cohesion during interkinesis. In both pans1 and sgo1 sgo2, metaphase I appears normal with five aligned bivalents (14). To test if PANS1 acts through or independently of SGO, we first combined sgo1 sgo2 and pans1 mutations (Fig. 6 A–D). In the pans1 sgo1 sgo2 triple mutant, only a small proportion of metaphase I showed five bivalents (11%, n = 321 cells, Fig. 6B), whereas the majority showed an almost complete (41%, Fig. 6C) or complete (48%, Fig. 6D) loss of cohesion, with 20 free chromatids. At diakinesis, the stage of prophase that immediately precedes metaphase I, five bivalents were systematically observed showing that crossovers occurred and sister chromatid cohesion was established (Fig. 6A). Thus, in pans1 sgo1 sgo2, sister chromatid cohesion was lost at prometaphase I or early metaphase I, dismantling the bivalent into free chromatids. This result shows that PANS1 and SGOs act in parallel to protect sister chromatid cohesion at metaphase I. This also reveals that SGO1 and SGO2 protect cohesion not only at pericentromeres, but also along the chromosome arms, redundantly with PANS1.
Fig. 6.
PANS1 acts independently of SGOs. (A–D) Chromosome spreads in sgo1 sgo2 pans1 male meiosis. (A) Diakinesis. No defect is detected; five bivalents are observed. (B–D) Metaphases I, with five bivalents (B), almost complete loss of cohesion (C), or complete loss of cohesion leading to 20 free chromatids (D). (E and F) Chromosome spreads in sgo1 sgo2 pDMC1::PANS1ΔD male meiosis. (E) Metaphase I with stretched bivalents. (F) Aberrant metaphase II with bivalents. (Scale bars, 10 µm.)
We then expressed pDMC1::PANS1ΔD in the sgo1 sgo2 mutant (n = 4 plants). In this context, meiotic chromosome spreads showed metaphase I with stretched bivalents (Fig. 6E) and aberrant metaphase II with five bivalents (Fig. 6F), similarly to what was observed when pDMC1::PANS1ΔD was expressed in the wild type (Fig. 1). This shows that the expression of PANS1ΔD at meiosis prevents chromosome segregation even in the absence of SGOs, confirming that the PANS1 function is independent of SGOs.
Mutations in SEPARASE Can Restore Sister Chromatid Cohesion in patronus1.
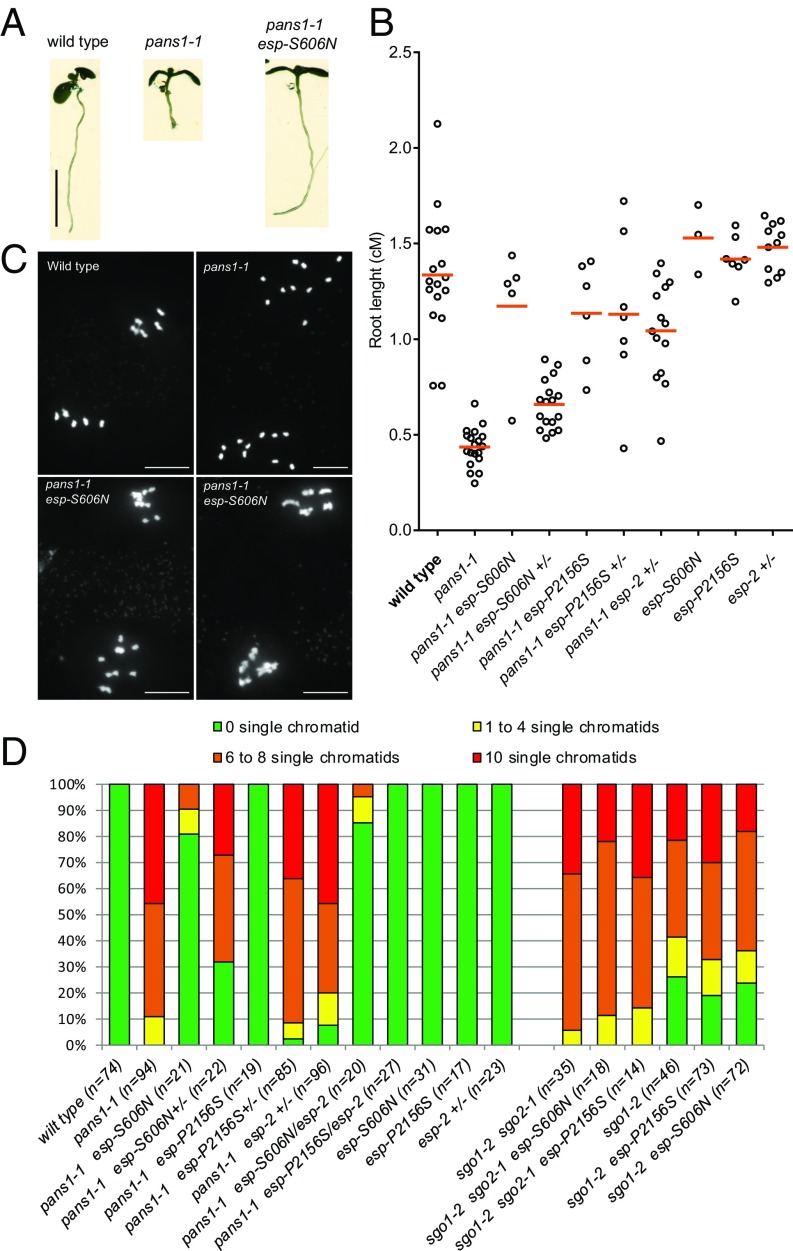
With the aim of identifying antagonists of PATRONUS and to shed light on its function, we set up a genetic suppressor screen. We took advantage of the root-growth defect of pans1 when cultivated on medium supplemented with NaCl (14, 19). pans1-1 seeds were mutated with ethyl methanesulfonate and the M2 families obtained by self-fertilization of individual mutagenized plants were screened for (i) longer roots than pans1-1 on NaCl medium and, subsequently, (ii) longer fruits than pans1-1 after transfer to the greenhouse. Plants satisfying both criteria were identified in 8 of the 200 independent families screened. Whole-genome sequencing of these plants revealed that four of the eight suppressors had a missense mutation in the Arabidopsis SEPARASE gene (AtESP; At4g22970, SI Appendix, Table S1), that we hereafter call esp-S606N, esp-P1946L, esp-A2047T, and esp-P2156S (Figs. 7 and 8 and SI Appendix, Fig. S3). Bulk genome sequencing of a segregating population identified esp-S606N as the mutation most strongly linked to the growth phenotype among the mutations segregating in that line, further supporting the conclusion that the mutations in ESP are causal. The esp-P1946L, esp-A2047T, and esp-P2156S mutations affected well-conserved residues of the protease domain of ESP (SI Appendix, Fig. S3). The S606 residue is in a less conserved helical domain and belongs to a stretch of serine, which may suggest regulation of ESP by phosphorylation. The previously described esp-2 mutation is null and lethal (10) but can restore pans1 growth in a dominant manner (Fig. 7C), confirming that decreasing ESP activity can suppress the pans1 somatic defect. Single mutants esp-S606N, esp-P1946L, esp-A2047T, and esp-P2156S are viable when homozygous, without apparent defects in growth or development, suggesting that they are hypomorphs. Quantification of root growth in pans1 mutants segregating for the esp-S606N or esp-P2156S mutations showed that these mutations restore root growth in a semidominant and dominant manner, respectively (Fig. 7D).
Fig. 7.
Mutations in SEPARASE suppress pans1. (A) Nine-day-old plantlets of wild type, pans1-1 and pans1-1 esp-S606N grown on NaCl medium, as quantified in B. (B) Quantification of roots length of 9-d-old plantlets. (C) Metaphase II plates of wild-type, pans1-1, and pans1-1 esp-S606N plants as quantified in D. (D) Quantification of sister chromatid cohesion. Metaphase II plates were sorted into classes according to the number of single chromatids detected, from 0 to 10. Zero means full cohesion as observed in wild type; 10 indicates a complete loss of cohesion. The number of metaphase plates analyzed is indicated in parentheses. (Scale bars, A, 0.5 cm; C, 5 µm.)
Fig. 8.
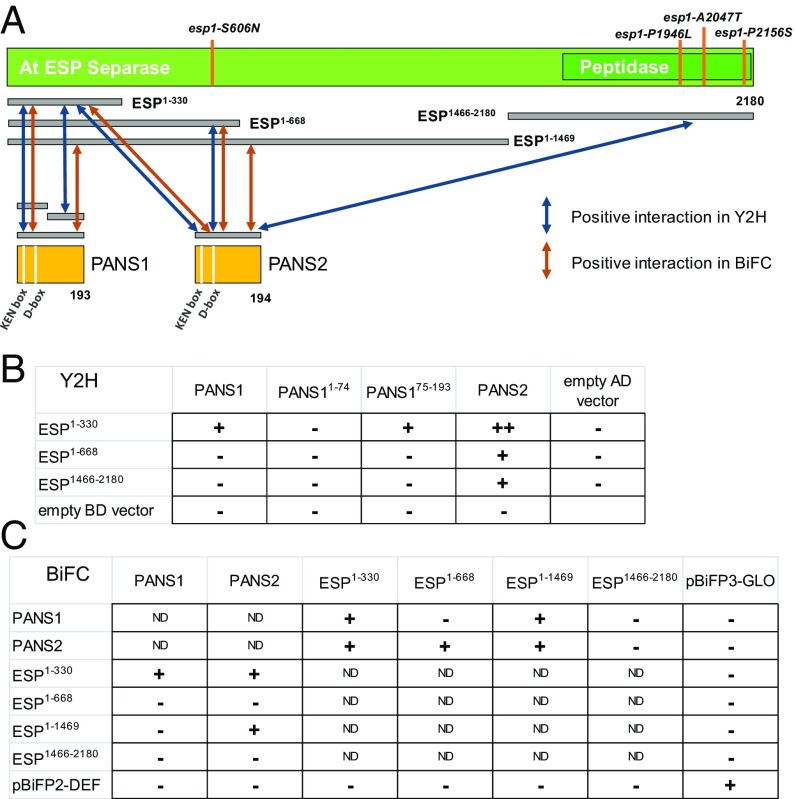
ESP and PANS1/2 interact in Y2H and BiFC assays. (A) Schematic representation of the ESP protein and its interactions with PANS1 and PANS2. The positions of the four mutations identified in this study and of the peptidase domain are indicated (SI Appendix, Fig. S3). Positive interactions detected in Y2H and BiFC are indicated by arrows. (B) Results of Y2H essays. PANS1 and PANS2 proteins were fused with the activating domain (AD), and ESP was fused with the binding domain (BD). Growth on LWH is indicated by +, and growth on LWHA is indicated by ++. Raw data are shown in SI Appendix, Fig. S4. (C) Results of BiFC essays. The C terminus fusions with the C-YFP (pBiFP3) are listed in rows and the N terminus fusion with N-YFP (pBiFP2) are listed in columns. +, the detection of a YFP signal (SI Appendix, Fig. S5); −, the absence of signal; ND, that the interaction was not tested.
To test if the mutations in ESP suppress the pans 1 meiotic defects, we quantified sister chromatid cohesion at metaphase II (Fig. 7 C and D). Although cohesion is almost completely lost in pans1 (Fig. 7D, 10 free chromatids indicating complete absence of cohesion), it is partially restored in a semidominant manner by the esp-S606N mutation and fully restored by the esp-P2156S mutation in a recessive manner. The esp-2 heterozygous mutation did not restore the pans1 sister chromatid defect, but pans1 esp-2/esp-P2156S plants had restored cohesion, further confirming that esp mutations cause the suppression of the pans1 phenotype. Thus, mutations in ESP can suppress the meiotic sister chromatid defect of pans1. However, the esp mutations were not able to restore the gametophytic lethality of the pans1 pans2 double mutant, suggesting that mutating ESP cannot compensate for the total absence of PANS (SI Appendix, Table S2). In addition, esp-S606N and esp-P2156S mutations were not able to suppress the meiotic sister chromatid defect of sgo1 or of sgo1 sgo2 (Fig. 7D). This result suggests that PANS1 and ESP have a specific antagonistic function in regulating the release of sister chromatid cohesion at meiosis.
PATRONUS Interacts Directly with SEPARASE and APC/C.
To better understand the role of PANS1, we searched for interacting partners using pull-down protein purification coupled with mass spectrometry using overexpressed GSrhino-tagged PANS1 as bait in Arabidopsis cell culture (26). After filtering copurified proteins for false positives (see Methods and ref. 27), we recovered peptides from PANS1 itself and a series of additional proteins in three replicate experiments (Table 1). We recovered 10 subunits of the APC/C complex, confirming previous findings (14). Most importantly, the PANS1 pulldown identified numerous peptides of SEPARASE (ESP1) in all three purification experiments, showing that PANS1 interacts with SEPARASE in vivo. Y2H assays and bimolecular fluorescent complementation (BiFC) experiments both confirmed that PANS1 and PANS2 interact with SEPARASE (Fig. 8 and SI Appendix, Fig. S4). Y2H experiments with truncated PANS1 showed that the C-terminal half of PANS1 (which does not contain the conserved KEN and D boxes) is sufficient to mediate an interaction with SEPARASE. In yeast and animals, the securin C-terminal region also mediates the interaction with separase (4, 5) (Fig. 9). The N-terminal domain of Arabidopsis SEPARASE showed the strongest interaction with PANS1 and PANS2. In yeast and human, securin interacts along the entire length of separase (4, 5, 7, 28).
Table 1.
Pull-down protein purification using PANS1 as bait
| Gene ID | Function | No. of assigned peptide matches | |||
| IP1 | IP2 | IP3 | Total | ||
| AT3G14190 | PANS1 | 21 | 20 | 19 | 60 |
| AT4G22970 | SEPARASE | 27 | 31 | 36 | 94 |
| AT2G39090 | APC7 | 13 | 16 | 22 | 51 |
| AT5G05560 | APC1 | 13 | 16 | 20 | 49 |
| AT2G20000 | APC3b | 9 | 11 | 12 | 32 |
| AT3G48150 | APC8 | 9 | 11 | 10 | 30 |
| AT1G78770 | APC6 | 11 | 10 | 9 | 30 |
| AT4G21530 | APC4 | 3 | 6 | 12 | 21 |
| AT1G06590 | APC5 | 2 | 3 | 10 | 15 |
| AT2G04660 | APC2 | 3 | 2 | 3 | 8 |
| AT4G11920 | CCS52A2 | — | 3 | 2 | 5 |
| AT5G63135 | APC15 | — | — | 2 | 2 |
| AT1G48300 | Acyltransferase | 8 | 6 | 6 | 20 |
| AT3G13330 | PA200 | — | 2 | 3 | 5 |
| AT5G23900 | Ribosomal protein | 2 | 3 | — | 5 |
| AT3G07090 | Thiol peptidase | — | 2 | 2 | 4 |
| AT2G15270 | PRKRIP1 | 2 | — | — | 2 |
Fig. 9.
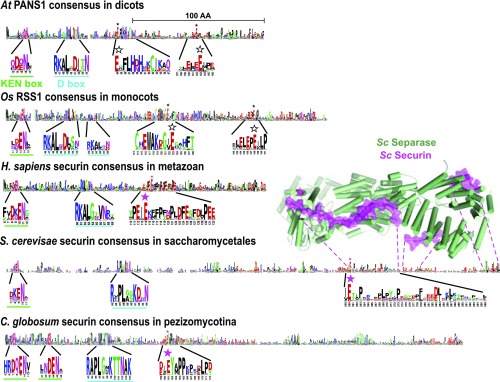
Conservation of securin organization in eukaryotes. Sequence consensus of securin homologs obtained from the multiple sequence alignments of A. thaliana PANS1 (PANS1_ARATH), Oryza sativa RSS1 (XP_015627424.1), H. sapiens securin (AAC69752.1), S. cerevisiae securin (NP_010398.3), Chaetomium globosum securin (XP_001225358.1) with their respective homologs represented using sequence logos (53). The homologs used to build the consensus span the dicot, monocot, metazoan, Saccharomycetales, and Pezizomycotina clades for AtPANS1, OsRSS1, Hs, Sc, and Cg securins, respectively. Sequence motifs corresponding to the KEN and D boxes are magnified and underlined in green and cyan, respectively. For all consensuses, the conserved stretches containing an invariant or almost-invariant glutamate residue (E) are magnified, with a star indicating the position of the conserved residue. Magenta stars report residues that were validated experimentally as acting as pseudosubstrates to inhibit separase activity (4, 5, 33). Unfilled black stars indicate the location of the potential pseudosubstrate glutamates in AtPANS1 and OsRSS1. The structure of Sc securin complexed to Sc separase is shown in magenta and green, respectively, with a magenta star indicating the position of the conserved glutamate and was obtained from the PDB ID code 5U1T.
We previously showed that PANS1 interacts with the APC/C subunit CDC20 through its D box (14) and showed here that PANS2 also interacts with CDC20 (SI Appendix, Fig. S4). Yeast and animal securins have also been shown to bind directly CDC20 in a D box-dependent manner (29, 30).
PATRONUS Has Remote Sequence Similarity with Securins.
The experimental data presented above suggest that PATRONUS may be the elusive plant securin. We thus investigated the degree to which PATRONUS is conserved and if any sequence similarity can be detected between PATRONUS and animal or yeast securin.
Position-specific iterated BLAST (PSI-BLAST) against the green lineage identified homologs of PATRONUS and RSS1 in flowering plants, gymnosperms, and basal vascular plants (e.g., mosses and ferns) as previously reported (14, 20). Further iterations then identified homologs in algae, including Ostreococcus lucimarinus (XP_001422400.1) (Methods). Reciprocal PSI-BLAST analyses starting from the O. lucimarinus protein recovered the entire plant protein family, including RSS1, PATRONUS1, and PATRONUS2, reinforcing the conclusion that the identified proteins are homologs (Methods and SI Appendix, Table S3). The slow convergence of the PSI-BLAST search likely arises from the high sequence divergence between the PATRONUS family members and from their intrinsically disordered character.
Next, we repeated the PSI-BLAST analyses using the O. lucimarinus sequence (XP_001422400.1) as bait and expanded the interrogation to the full eukaryotic tree. After eight iterations, a few proteins of unknown function from bivalves and gastropod species were identified, such as the bivalve mollusk Mizuhopecten yessoensis (XP_021355525.1). Remarkably, only a few PSI-BLAST iterations were sufficient to connect the latter sequence to homologs of securin in vertebrates, human, and mouse securins. Other homologs experimentally probed as securin in Caenorhabditis elegans, Drosophila melanogaster, or the fungi Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Chaetomium thermophilum were not retrieved in this search. Conversely, using these securins as bait of a PSI-BLAST search led to rapid convergence with homologs detected only in closely related species, suggesting that their sequences diverged too much to be recognized using this method. Hence, the fact that plant RSS1/PATRONUS can be connected to vertebrate securins in PSI-BLAST searches gives strong support to the suggestion that they share remote orthologous relationships, given that the lineage that led to plants has separated very early during eukaryotic evolution from the branch that led to animals and yeast (31).
Looking at the conservation patterns of the securin proteins within several clades, some striking features emerged (Fig. 9 and SI Appendix, Fig. S4). In each clade (monocots, dicots, Saccharomycetales, Pezizomycotina, Metazoa), the same profile appeared: conserved KEN and D boxes in the N-terminal and an invariable glutamic acid (E) within a relatively conserved patch. This glutamic acid has been shown to be pivotal for the inhibition of the separase cleavage site in the mammalian and the two fungus clades (pink stars in Fig. 9) and is also shared with separase substrates (4, 7, 32, 33). In plants, two glutamic acids (E) are well conserved (SI Appendix, Fig. S4, unfilled stars in Fig. 9) and are thus prime candidates important for separase inhibition. One is in the C-terminal third of the protein and is very well-conserved among the entire plant lineage (monocots, dicots, basal plants, and algae). However, it is not conserved in some groups of species (e.g., the Phalaenopsis orchids and several algae such as Micractinium conductix), making this candidate less likely. The other candidate glutamates lie in the middle of the proteins in domains conserved within dicots and within monocots in different regions (E-x-F-L-H-D/N-H and W–A–K/R–D/E/G–G–V/I–E, respectively; Fig. 9 and SI Appendix, Fig. S4) but of unknown function (14, 20). The monocot glutamate is extremely well-conserved in the Viridiplantae, ranging from monocots to very distant species such as algae (SI Appendix, Fig. S6), supporting its pivotal role. However, standard algorithms did not align the conserved E-containing patch from dicots with the conserved E-containing patch from monocots. We favor the scenario in which these E-containing domains represent the separase inhibition site, but it has diverged too much in dicots to be properly aligned using the current algorithms.
Discussion
Securin is the central regulator of chromosome distribution at both mitosis and meiosis in animals and fungi. Securin inhibits the separase protease, thus preventing untimely release of sister chromatid cohesion. At the onset of anaphase, APC/C degrades securin, releasing separase and thereby allowing chromosome distribution. However, to date, securin counterparts in plants have been elusive, suggesting that an alternative mechanism may be at play. We provide five pieces of strong evidence that patronus are securin homologs in plants. First, at meiosis, depletion of PANS1 leads to the premature release of cohesion (14); conversely, expression of an APC/C-insensitive PANS1 abolishes cohesion release and chromatid separation, mimicking the depletion of separase (10). If expressed constitutively, the APC/C-insensitive PANS1 is lethal. The single pans1 mutation provokes some chromosome missegregation and aneuploidy (17, 19). No defects were detected in the pans2 mutant, but the pans1 pans2 double mutation is lethal (14). We propose that PANS1 and PANS2, whose duplication has been retained from the whole genome duplication at the origin of Brassicaceae, have redundant functions, but that expression and/or activity levels of PANS1 are higher than those of PANS2. Supporting that possibility, both genes are expressed in all dividing tissues with PANS2 being globally 6.3 times more expressed than PANS1 (34) (SI Appendix, Table S4). Our data support the conclusion that PATRONUS1 and/or PATRONUS 2 prevent cohesion release in both mitosis and meiosis and that degradation of PATRONUS by APC/C is required to lift this inhibition. Second, we showed that PANS1 controls chromosome segregation independently of SGOs, excluding the alternative hypothesis that patronus regulates shugoshin. Third, we demonstrated that the PATRONUS1 and PATRONUS2 proteins interact directly with SEPARASE and APC/C. Fourth, using a forward genetic screen, we identified mutations in SEPARASE that suppress the defects of patronus1 mutants, showing that PATRONUS1 and SEPARASE have antagonistic functions. Lastly, we identified remote sequence similarity between plant PATRONUS/RSS1 proteins and animal securins. Altogether, this strongly supports the conclusion that PATRONUS/RSS1 is the plant securin.
We propose that all plant PATRONUS homologs, including the rice RSS1 (35), represent plant securins. Consistent with this conclusion, RSS1 is expressed in dividing cells and is regulated by the APC/C in a D box-dependent manner (20). Further, expression of RSS1 deleted of its N-terminal domain that contains D and KEN boxes is lethal. RSS1 functions in the regulation of the cell cycle, but the rss1 mutant is viable and fertile (20). However, RSS1 has an uncharacterized paralogue in the rice genome (Os01g0898400) (20) (SI Appendix, Fig. S5), which may act redundantly with RSS1, as PANS1 and PANS2 do. RSS1 has been shown to interact with the PP1 phosphatase (20, 35, 36), suggesting that RSS1 is either regulated by PP1 or has an additional function than inhibiting securin.
It is intriguing that securin proteins have such a poorly conserved sequence although they play such a central role in cell division. The only conserved features are the presence of D and KEN boxes, which are involved in cell-cycle regulation, and a variable conserved patch containing glutamic acid (E), which is pivotal in separase inhibition. In the plant lineage, the phylogenetic analysis of PANS1/RSS1 suggests that, in addition to sequence divergence, the PANS family has a complex history of gene duplication/gene loss (14). Securin interacts with separase along all its intrinsically unstructured length (7) (SI Appendix, Fig. S9) and acts as a pseudosubstrate. It is likely that securin has no catalytic activity. One possibility is that the securin sequence simply drifts passively due to the absence of any selective pressure on its sequence, leading to relaxed purifying selection. Alternatively, securin being a key regulator of cell cycle, pivotal in development and the stress response, may evolve rapidly in response to selective pressures. In support of this idea, the rice rss1 and the Arabidopsis pans1 mutants are hypersensitive to a range of abiotic stresses (14, 19, 20) and intrinsically disordered protein regions are frequent targets of positive selection (37, 38). The rapid divergence of PATRONUS and its securin homologs may thus represent the accumulation of physiological and developmental responses to a constantly changing environment.
Methods
Plant Materials and Growth Conditions.
Arabidopsis thaliana plants were grown in greenhouses or growth chambers (16 h day/8 h night, 20 °C, 70% humidity) and Nicotiana benthamiana in greenhouses (13-h day 25 °C/11-h night 17 °C). For in vitro culture, Arabidopsis seeds were surface sterilized for 10 min in 70% ethanol + 0.05% sodium dodecyl sulfate and washed for 10 min in 70% ethanol and grown in Petri dishes with culture medium (Gamborg B5 medium-Duchefa supplied with 0.16% bromocresol purple and 0.1% sucrose). The culture medium was supplemented with 80 mM NaCl for the root-growth experiment and with 100 µg/mL kanamycin sulfate (Euromedex) or 25 µg/mL hygromycin B (Duchefa) for selection of transformants. Arabidopsis transformation was performed using the floral dip method: Agrobacterium tumefaciens (EHA105 strain) was grown at 28–30 °C to saturation, centrifuged, and resuspended in a 1% sucrose 0.05% Silwet l-77 solution. This solution is used for dipping A. thaliana plants. Plants were maintained under glass overnight to increase humidity (39). The A. tumefaciens (C58C1 strain) was used for N. benthamiana leaf infiltration (40).
The pans1-1 (Salk_070337), Atsgo1-2 (SK2556), Atsgo2-1 (line 34303), spo11-1–3 (Salk_146172), and esp-2 (Salk_037016) mutants were genotyped by PCR (30 cycles of 30 s at 94 °C, 30 s at 56 °C, and 1 min at 72 °C) and two primer pairs were used (SI Appendix, Table S5). The first pair is specific to the wild-type allele, and the second pair is specific to the left border of the inserted sequence. pans1-1: N570337U and N570337L, N570337L and LbSalk2. Atsgo1-2: SK2556U and SK2556L, SK2556U and pSKTail1. Atsgo2-1: SGO2U and SGO2L, SGO2U and GABI. spo11-1-3: N646172U and N646172L, N646172L and LbSalk2. esp-2: N537016U and N537016L, N537016U and LbSalk2.
The esp-S606N and esp-P2156S mutants were genotyped using PCR. The PCR products were digested by restriction enzymes. For esp-S606N, the 502-pb PCR product (esp606U and esp606L) was digested with the TasI (Thermo Fisher Scientific) restriction enzyme at 65 °C for 1 h. The wild-type allele yielded three DNA fragments (441, 49, and 12 pb) and the esp-S606N allele yielded four DNA fragments (300, 141, 49, and 12 pb). For esp-P2156S, the 1040-pb PCR product (esp2156U and esp2156L) was digested with the Hin6I (Thermo Fisher Scientific) restriction enzyme at 37 °C for 1 h. The wild-type allele yielded two DNA fragments (556 and 474 pb) and the esp-P2156S allele yielded one DNA fragment (1,040 pb).
The RFP-tagged tubulin (RFP:TUB4) line and the GFP-tagged REC8 (REC8:GFP) were provided by A.S. Homozygous lines for RFP:TUB4 and REC8:GFP were transformed using floral dip with the pDMC1::PANS1ΔD construct. Transformants were selected on Petri dishes containing in vitro culture medium supplied with kanamycin.
Suppressor Screen.
For the suppressor screen, homozygous pans1-1 seeds were incubated for 17 h at room temperature in 5 mL of 0.3% (vol/vol) ethyl methanesulfonate (EMS) (Sigma) with gentle agitation. EMS was neutralized by adding 5 mL of 1 M sodium thiosulfate for 5 min. Then, 3 mL of water was added to make the seeds sink. The supernatant was removed, and seeds were washed three times for 20 min with 15 mL of water. These M1 seeds were grown in the greenhouse and selfed to produce M2 seeds. M2 seeds were sterilized and sown in Petri dishes containing in vitro culture medium supplemented with 80 mM NaCl (∼15 M2 per M1). M2 plants with long roots were transferred in the greenhouse and visually scored for fruit length. Mutants or mutant populations were sequenced using Illumina technology (GET platform https://get.genotoul.fr/en). Mutations were identified using the MutDetect pipeline (41). The esp-S606N line identified in the suppressor screen was back-crossed with pans1-1, and 52 plants were selected in the F2 population for having long roots on an NaCl medium. Bulk genome sequencing of these plants identified the esp-S606N mutation as the most strongly linked (36/41 mutant reads) to the phenotype among the mutation segregating in that line, supporting that it is causal.
Meiotic Chromosome Spreads and Immunolocalization.
Meiotic chromosome spreads and immunolocalization were performed following Ross et al (42) with modifications. Inflorescences were harvested in 3:1 fixative (3 vol EtOH:1 vol acetic acid). Fixative was replaced once. For slide preparation, inflorescences were washed twice in water and once in citrate buffer (10 mM trisodium-citrate, pH adjusted to 4.5 with HCl). They were digested for 3 h at 37 °C in a moist chamber with a digestion mix (0.3% [wt/vol] pectolyase Y-23 [MP Biomedicals], 0.3% [wt/vol] Driselase [Sigma] 0.3% [wt/vol] cellulase [Onozuka R10] [Duchefa] 0.1% sodium azide in 10 mM citrate buffer). Three ∼0.5-mm washed buds were transferred on a slide in a drop of water and dilacerated with thin needles to generate a cell suspension. After adding 10 µL of 60% acetic acid, the slide was incubated on a hot block at 45 °C for 1 min and the cell suspension was stirred with a hooked needle. Another 10 µL of 60% acetic acid was added and stirred during one more minute. The cell suspension drop was surrounded by fresh 3:1 fixative, and the slide was rinsed with fixative. Dry slides were ready for DAPI staining and immunolocalization.
For DAPI staining, a drop for DAPI solution (2 µg/mL) in Citifluor AF1 (Agar Scientific) was added. Slides were observed using a Zeiss Axio Imager Z2 microscope and Zen blue software. Images were acquired using a Plan-Apochromat 100×/1.40 Oil M27 objective, Optovar 1.25× Tubelens. DAPI was excited at λ 335–385 nm and detected at λ between 420 and 470 nm.
For immunolocalization, slides were microwaved in 10 mM citrate buffer pH 6 for 45 s at 850 W and immediately transferred to 0.1% Triton in PBS. Slides were incubated with primary antibodies diluted in 1% bovine serum albumin (BSA) in PBS for 48 h at 4 °C, then washed in 0.1% Triton in PBS three times for 15 min before adding the secondary antibodies in 1% BSA in PBS. After 1 h of incubation at 37 °C slides were washed in 0.1% Triton in PBS three times for 15 min and mounted in Vectashield antifade medium (Vector Laboratories) with 2 µg/mL DAPI. Slides were observed using a Zeiss Axio Imager Z2 microscope and Zen blue software. Images were acquired using a Plan-Apochromat 100×/1.40 Oil M27 objective, Optovar 1.25× Tubelens. DAPI was excited at λ 335–385 nm and detected at λ between 420 and 470 nm. Green fluorescence was excited at λ 484–504 nm and detected at λ between 517 and 537 nm. Red fluorescence was excited at λ 576–596 nm and detected at λ between 612 and 644 nm. The rat anti-AtREC8 polyclonal antibody has been described by ref. 14 and was used at a dilution of 1:250. The secondary antibody Alexa568 goat anti-rat (Thermo Fisher Scientific) was used at a dilution of 1:250.
Live Meiosis Image Acquisition and Analysis.
Live cell imaging was performed following Prusicki et al. (21) with modifications. Briefly, flowers buds of 0.4–0.6 mm were isolated on a slide. Buds were carefully dissected to isolate undamaged anthers. Anthers were transferred into a slide topped by a spacer (Invitrogen Molecular ProbesSecure-Seal Spacer, eight wells, 9 mm diameter, 0.12 mm deep) filled with 10 µL of water, and covered by a coverslip. Time lapses were acquired using a Leica SP8 confocal laser-scanning and LAS X 3.5.0.18371 software.
Images were acquired using harmonic compound system, plan apochromats, confocal scanning2 (HC PL APO CS2) 20×/0.75 IMM and HC PL APO CS2 63×/1.20 WATER objectives upon illumination of the sample with an argon laser and diode-pumped solid-state laser (561 nm) GFP was excited at λ 488 nm and detected at λ between 494 and 547 nm. RFP was excited at λ 561 nm and detected at λ between 570 and 629 nm. Detection was performed using Leica HyD detectors. Time lapses were acquired as a series of Z-stacks (between 15 and 20 μm distance). Interval time varied from 1.5 to 3 min depending on sample conditions. Deconvolution was performed using the lightning deconvolution option. Images processing were done with Fiji. Image drift was corrected by the Stack Reg plugin (Rigid Body option). Decrease in fluorescence was corrected using the bleach correction option (simple ratio- background intensity 0- or histogram matching).
Plasmid Construction.
To generate the pDMC1::PANS1 construct, the PANS1 genomic fragment was amplified using PCR with PANS1_XhoI and PANS1_SpeI primers. The amplification covered PANS1 from the ATG to 477 pb after the stop codon. The PCR product was cloned by restriction digest with XhoI and SpeI into the pPF408 vector (43) to fuse the PANS1 genomic fragment with the DMC1 promoter. The DMC1 promoter covered −2,940 pb before ATG and +205 pb after ATG and has been amplified from the Landsberg erecta accession. The pDMC1::PANS1 DNA fragment was amplified with DMC1_GTW and PANS1SpeI_GTW to introduce Gateway tail (Thermo Fisher Scientific) and cloned into pDONR207 to create pENTR- pDMC1::PANS1, on which directed mutagenesis was performed using the Stratagene QuikChange Site-Directed Mutagenesis Kit. The primer PANS1∆D was used to create the pDMC1::PANS1∆D. For plant transformation, the LR reaction was performed with the binary vector pGWB1 (44).
For Y2H experiments, cDNA was amplified using the corresponding primers (SI Appendix, Table S5). They were cloned using the Gateway cloning system (Thermo Fisher Scientific) into pDONR221 to generate pENTR clones and into pDEST22 (prey plasmid) and pDEST 32 (bait plasmid) Gateway, ProQuest (Thermo Fisher Scientific). To generate the pDONR221-ESP1466-2180 construct, a 4,474-pb fragment of ESP cDNA was obtained by DNA synthesis (GeneArt-Thermo Fisher Scientific) (sequence available in SI Appendix, Table S5). For BiFC experiments, the pENTR clones described above for Y2H were cloned by LR recombination reaction into the pBiFP2 and pBiFP3 vectors (40).
Yeast two-hybrid assay.
DNA product was cloned using Gateway (Invitrogen) into the pDONR221 vector (Invitrogen) to create pENTR. LR reactions were carried out on the pDEST32 (bait) and pDEST22 (prey) vectors (Invitrogen). Plasmids encoding the bait (pDEST32) and prey (pDEST22) were transformed into the yeast strain AH109 and Y187 (Clontech) by the LiAc method following the protocol in the MATCHMAKER GAL4 Two Hybrid System 3 manual (Clontech). The TDM protein self-interaction was used as positive controls (45). Transformed yeast cells were selected on synthetic dropout (SD) plates without Leu (SD-L) for bait or without Trp (SD-W) for prey. Interactions between proteins were assayed using the mating method. The resulting diploid cells were selected on synthetic dropout medium lacking a combination of amino acids, driven by the auxotrophy genes carried by the cloning vectors. Protein interactions were assayed by growing diploid cells on SD-LWH and SD-LWHA.
BIFC.
Protein interactions were tested in planta using BiFC assays (46) in leaf epidermal cells of N. benthamiana plants expressing a nuclear CFP fused to histone 2B (47). N-terminal fusions, using the pENTR clones described above for Y2H, with two yellow fluorescent protein (YFP) complementary regions (YFPN + YFPC) were coinfiltrated in N. benthamiana leaves and scored after 3 or 4 d for fluorescence as described in ref. 41. YFPN-DEFICIENS and YFPC-GLOBOSA, two interacting components of an Anthirrinum majus MADS box transcription factor (48), were used as positive control. Each experiment was replicated at least twice, each corresponding to the infiltration of two different plants.
Observations were made using a Leica SP8 confocal laser-scanning microscope. Optical sections were collected with a Leica HCX PL APO CS2 20.0 × 0.70 IMM UV water objective upon illumination of the sample with a 514-nm argon laser line with an emission band of 520–560 nm for the YFP or with a 458-nm argon laser line with an emission band of 463–490 nm for the CFP. Detection were performed using Leica HyD detectors. The specificity of the YFP signal was systematically checked by determining the fluorescence emission spectrum between 525 and 600 nm with a 10-nm window and under an excitation at 514 nm. Images were processed using Leica LASX and Adobe Photoshop software.
Pull-Downs.
Three pull-downs on Arabidopsis cell suspension culture expressing N-terminally GSrhino tagged PANS1 were performed as described (49). On-bead digested samples were analyzed on a Q Exactive mass spectrometer (Thermo Fisher Scientific), and copurified proteins were identified using standard procedures (27). After identification, the protein list was filtered versus a list of nonspecific proteins, assembled similarly as described (27). True interactors that might have been filtered out due to their presence in the list of nonspecific proteins were selected by means of semiquantitative analysis using the average normalized spectral abundance factors (NSAF) of the identified proteins in the PANS1 pull-downs. Proteins identified with at least two peptides in at least two experiments, showing high (at least 10-fold) and significant [−log10(P value (t test)) ≥ 10] enrichment compared with calculated average NSAF values from a large dataset of pull-downs with nonrelated bait proteins, were selected.
Protein Sequence Analyses.
Searches for homologs of AtPANS1 (PANS1_ARATH) were performed using PSI-BLAST (50) against the nr database using e values thresholds from 1e-3 to 1e-5. Multiple sequence alignments were calculated using MAFFT (51) and represented using Jalview (52). PSI-BLAST searches starting from AtPANS1 established the homologous relationship with OsRSS1 (XP_015627424.1) after four iterations, but further iterations were not successful in identifying plant homologs in clades other than monocots and dicots. Repeating the search from OsRSS1 on plant sequences excluding dicots helped identify homologs in mosses. These sequences were used in turn as inputs to search for the most likely homologs of AtPANS1 in green algae and one sequence in O. lucimarinus was detected (XP_001422400.1) as a potential candidate. Reciprocal PSI-BLAST analyses starting from the O. lucimarinus protein against the nr sequence database restricted to the Viridiplantae clade recovered OsRSS1 as a significant match after seven iterations and the Arabidopsis PATRONUS1 and 2 after 13 iterations.
PSI-BLAST analyses using (XP_001422400.1) also detected as significant homologs several sequences from bivalves such as M. yessoensis (XP_021355525.1) after 10 iterations without apparent divergence of the sequence profile. PSI-BLAST from M. yessoensis (XP_021355525.1) against metazoan sequences integrated homologs of securin in vertebrates after four iterations including those of Homo sapiens, establishing a transitivity link from Arabidopsis PANS1 to human securin passing through green algae and bivalve homologs. The sequence accession numbers retrieved after the 10 iterations of PSI-BLAST from the O. lucimarinus sequence are listed in SI Appendix, Table S3.
Supplementary Material
Acknowledgments
We thank Mathilde Grelon, Christine Mezard, and Eric Jenczewski for critical reading of the manuscript. The Institute Jean-Pierre Bourgin benefits from the support of the LabEx Saclay Plant Sciences-SPS Grant ANR-10-LABX-0040-SPS. This work was financially supported by a European Research Council Grant ERC 2011 StG 281659 (MeioSight) (to R.M.), the Fondation Simone et Cino Del Duca (to R.M.), and the CEFIPRA project SMOKY (to R.M.). R.G. is supported by French Infrastructure for Integrated Structural Biology Grant ANR-10-INSB-05-01 and CHIPSET Grant ANR-15-CE11-0008-01. This work was supported by the European Union Marie-Curie “COMREC” network Grant FP7 ITN-606956 (to M.A.P. and A.S.). In addition, core funding from the University of Hamburg to A.S. is gratefully acknowledged.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906237116/-/DCSupplemental.
References
- 1.Kamenz J., Hauf S., Time to split up: Dynamics of chromosome separation. Trends Cell Biol. 27, 42–54 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Hornig N. C. D., Knowles P. P., McDonald N. Q., Uhlmann F., The dual mechanism of separase regulation by securin. Curr. Biol. 12, 973–982 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Hauf S., Waizenegger I. C., Peters J. M., Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science 293, 1320–1323 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Luo S., Tong L., Molecular mechanism for the regulation of yeast separase by securin. Nature 542, 255–259 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boland A., et al. , Cryo-EM structure of a metazoan separase-securin complex at near-atomic resolution. Nat. Struct. Mol. Biol. 24, 414–418 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou H., McGarry T. J., Bernal T., Kirschner M. W., Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 285, 418–421 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Luo S., Tong L., Structural biology of the separase-securin complex with crucial roles in chromosome segregation. Curr. Opin. Struct. Biol. 49, 114–122 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitajima T. S., Kawashima S. A., Watanabe Y., The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 427, 510–517 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Miyazaki S., Kim J., Sakuno T., Watanabe Y., Hierarchical regulation of centromeric cohesion protection by meikin and shugoshin during meiosis I. Cold Spring Harb. Symp. Quant. Biol. 82, 259–266 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Liu Z., Makaroff C. A., Arabidopsis separase AESP is essential for embryo development and the release of cohesin during meiosis. Plant Cell 18, 1213–1225 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moschou P. N., Bozhkov P. V., Separases: Biochemistry and function. Physiol. Plant. 145, 67–76 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Fülöp K., et al. , Arabidopsis anaphase-promoting complexes: Multiple activators and wide range of substrates might keep APC perpetually busy. Cell Cycle 4, 1084–1092 (2005). [PubMed] [Google Scholar]
- 13.Kevei Z., et al. , Conserved CDC20 cell cycle functions are carried out by two of the five isoforms in Arabidopsis thaliana. PLoS One 6, e20618 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cromer L., et al. , Centromeric cohesion is protected twice at meiosis, by SHUGOSHINs at anaphase I and by PATRONUS at interkinesis. Curr. Biol. 23, 2090–2099 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Zamariola L., et al. , SGO1 but not SGO2 is required for maintenance of centromere cohesion in Arabidopsis thaliana meiosis. Plant Reprod. 26, 197–208 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Yuan G., et al. , PROTEIN PHOSHATASE 2A B′α and β maintain centromeric sister chromatid cohesion during meiosis in Arabidopsis. Plant Physiol. 178, 317–328 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zamariola L., et al. , SHUGOSHINs and PATRONUS protect meiotic centromere cohesion in Arabidopsis thaliana. Plant J. 77, 782–794 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Singh D. K., Spillane C., Siddiqi I., PATRONUS1 is expressed in meiotic prophase I to regulate centromeric cohesion in Arabidopsis and shows synthetic lethality with OSD1. BMC Plant Biol. 15, 201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juraniec M., et al. , Arabidopsis COPPER MODIFIED RESISTANCE1/PATRONUS1 is essential for growth adaptation to stress and required for mitotic onset control. New Phytol. 209, 177–191 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Ogawa D., et al. , RSS1 regulates the cell cycle and maintains meristematic activity under stress conditions in rice. Nat. Commun. 2, 278 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prusicki M. A., et al. , Live cell imaging of meiosis in Arabidopsis thaliana. eLife 8, e42834 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grelon M., Vezon D., Gendrot G., Pelletier G., AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20, 589–600 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen-Fix O., Peters J. M., Kirschner M. W., Koshland D., Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10, 3081–3093 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Funabiki H., et al. , Cut2 proteolysis required for sister-chromatid seperation in fission yeast. Nature 381, 438–441 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Clift D., Marston A. L., The role of shugoshin in meiotic chromosome segregation. Cytogenet. Genome Res. 133, 234–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bontinck M., et al. , Recent trends in plant protein complex analysis in a developmental context. Front. Plant Sci. 9, 640 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Leene J., et al. , An improved toolbox to unravel the plant cellular machinery by tandem affinity purification of Arabidopsis protein complexes. Nat. Protoc. 10, 169–187 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Viadiu H., Stemmann O., Kirschner M. W., Walz T., Domain structure of separase and its binding to securin as determined by EM. Nat. Struct. Mol. Biol. 12, 552–553 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Hilioti Z., Chung Y. S., Mochizuki Y., Hardy C. F. J., Cohen-Fix O., The anaphase inhibitor Pds1 binds to the APC/C-associated protein Cdc20 in a destruction box-dependent manner. Curr. Biol. 11, 1347–1352 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Kitagawa R., Law E., Tang L., Rose A. M., The Cdc20 homolog, FZY-1, and its interacting protein, IFY-1, are required for proper chromosome segregation in Caenorhabditis elegans. Curr. Biol. 12, 2118–2123 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Harashima H., Dissmeyer N., Schnittger A., Cell cycle control across the eukaryotic kingdom. Trends Cell Biol. 23, 345–356 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Nagao K., Yanagida M., Securin can have a separase cleavage site by substitution mutations in the domain required for stabilization and inhibition of separase. Genes Cells 11, 247–260 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Lin Z., Luo X., Yu H., Structural basis of cohesin cleavage by separase. Nature 532, 131–134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klepikova A. V., Kasianov A. S., Gerasimov E. S., Logacheva M. D., Penin A. A., A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 88, 1058–1070 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Ogawa D., Morita H., Hattori T., Takeda S., Molecular characterization of the rice protein RSS1 required for meristematic activity under stressful conditions. Plant Physiol. Biochem. 61, 54–60 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Ebel C., Hanin M., Maintenance of meristem activity under stress: Is there an interplay of RSS1-like proteins with the RBR pathway? Plant Biol (Stuttg) 18, 167–170 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Afanasyeva A., Bockwoldt M., Cooney C. R., Heiland I., Gossmann T. I., Human long intrinsically disordered protein regions are frequent targets of positive selection. Genome Res. 28, 975–982 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nilsson J., Grahn M., Wright A. P. H., Proteome-wide evidence for enhanced positive Darwinian selection within intrinsically disordered regions in proteins. Genome Biol. 12, R65 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clough S. J., Bent A. F., Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Azimzadeh J., et al. , Arabidopsis TONNEAU1 proteins are essential for preprophase band formation and interact with centrin. Plant Cell 20, 2146–2159 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Girard C., et al. , FANCM-associated proteins MHF1 and MHF2, but not the other Fanconi anemia factors, limit meiotic crossovers. Nucleic Acids Res. 42, 9087–9095 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ross K. J., Fransz P., Jones G. H., A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res. 4, 507–516 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Siaud N., et al. , Brca2 is involved in meiosis in Arabidopsis thaliana as suggested by its interaction with Dmc1. EMBO J. 23, 1392–1401 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakagawa T., et al. , Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Cifuentes M., et al. , TDM1 regulation determines the number of meiotic divisions. PLoS Genet. 12, e1005856 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu C.-D., Chinenov Y., Kerppola T. K., Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9, 789–798 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Martin K., et al. , Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 59, 150–162 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Schwarz-Sommer Z., et al. , Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: Evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 11, 251–263 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Leene J., et al. , Capturing the phosphorylation and protein interaction landscape of the plant TOR kinase. Nat. Plants 5, 316–327 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Altschul S. F., et al. , Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M., Barton G. J., Jalview Version 2–A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crooks G. E., Hon G., Chandonia J.-M., Brenner S. E., WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.