Significance
Imaging flow cytometry is a powerful tool for analyzing every single cell in a large heterogeneous population but relies on fluorescent labeling, which comes with cytotoxicity, nonspecific binding, and interference with natural cellular functions. This paper presents label-free multicolor chemical imaging flow cytometry based on stimulated Raman scattering (SRS), a highly sensitive method of molecular vibrational spectroscopy. With the help of deep learning, it demonstrates high-precision characterization and classification of microalgal cells and cancer cells without the need for fluorescent labeling.
Keywords: imaging flow cytometry, stimulated Raman scattering, metabolite imaging, microalgae, cancer cells
Abstract
Combining the strength of flow cytometry with fluorescence imaging and digital image analysis, imaging flow cytometry is a powerful tool in diverse fields including cancer biology, immunology, drug discovery, microbiology, and metabolic engineering. It enables measurements and statistical analyses of chemical, structural, and morphological phenotypes of numerous living cells to provide systematic insights into biological processes. However, its utility is constrained by its requirement of fluorescent labeling for phenotyping. Here we present label-free chemical imaging flow cytometry to overcome the issue. It builds on a pulse pair-resolved wavelength-switchable Stokes laser for the fastest-to-date multicolor stimulated Raman scattering (SRS) microscopy of fast-flowing cells on a 3D acoustic focusing microfluidic chip, enabling an unprecedented throughput of up to ∼140 cells/s. To show its broad utility, we use the SRS imaging flow cytometry with the aid of deep learning to study the metabolic heterogeneity of microalgal cells and perform marker-free cancer detection in blood.
Measurements and statistical analysis of chemical, structural, and morphological phenotypes of numerous living cells are essential for providing systematic insights into biological processes in diverse fields including cancer biology (1, 2), immunology (3, 4), drug discovery (5, 6), microbiology (7, 8), and metabolic engineering (9, 10). By virtue of its ability to provide high-throughput (much higher than automated microscopy for high-content screening [11]) and high-information content simultaneously, imaging flow cytometry (7, 12–15) has become an established tool for addressing the demand. The availability of high-content cellular data (i.e., cell images) provided by imaging flow cytometry can form a useful basis for highly accurate identification, characterization, and classification of individual cells with emerging computational tools such as data mining and machine learning (16). Imaging flow cytometry has been shown effective for studying the intracellular localization of proteins (13), evaluating DNA damage and repair (14), and analyzing cell–cell interaction and cell cycle (15).
Despite its excellent capabilities, imaging flow cytometry has a fundamental drawback: its requirement of fluorescent labeling for phenotyping, i.e., discriminating between physically or chemically different cells. Fluorescent probes often come with cytotoxicity, nonspecific binding, and interference with natural cellular functions (17). Also, they are not available to all types of cells and molecules (17). Furthermore, fluorescent labeling is not applicable to cell types whose surface antigens often vary, such as circulating tumor cells (18, 19). Development of a high-throughput analytical technology for providing both the physically and chemically rich information of numerous single cells without the need for fluorescent labeling has been an important but challenging goal in the field of biotechnology and biomedical engineering. In fact, previously demonstrated high-throughput imaging flow cytometry based on nonspectroscopic measurements only meets this goal partly (20, 21).
In this article, we present label-free chemical imaging flow cytometry by high-speed multicolor stimulated Raman scattering (SRS) microscopy on a microfluidic platform to overcome the limitation. It is based on a fast pulse pair-resolved wavelength-switchable Stokes laser with a synchronized pump pulse laser and a galvanometric scanner for SRS imaging of acoustically 3D-focused single cells flowing in a microfluidic channel. It achieves motion artifact-free chemical imaging of fast-flowing cells at a record high flow speed of 2 cm/s and a record high image acquisition speed of 24 klines/s with a pixel dwell time of 0.2 µs for 4-color SRS signal acquisition, which is >10 times faster than previous >3-color SRS techniques (22–26). Specifically, this pixel acquisition time of 0.2 µs for 4 colors is much shorter than the previously reported shortest value of 5 µs in single-point multiplex SRS flow cytometry (26) and even faster than that of 0.5 µs in 2-color SRS signal acquisition (27). To show its biological and medical utility, we demonstrate SRS imaging flow cytometry of microalgal, hematological, and cancer cells in practical settings at an unprecedented throughput of up to 140 cells/s. Using the multicolor SRS images of each single cell in a large heterogeneous population (n = ∼10,000) with the help of deep learning, we achieved high-precision characterization and classification of the cells without the need for fluorescent labeling.
Results
Schematic.
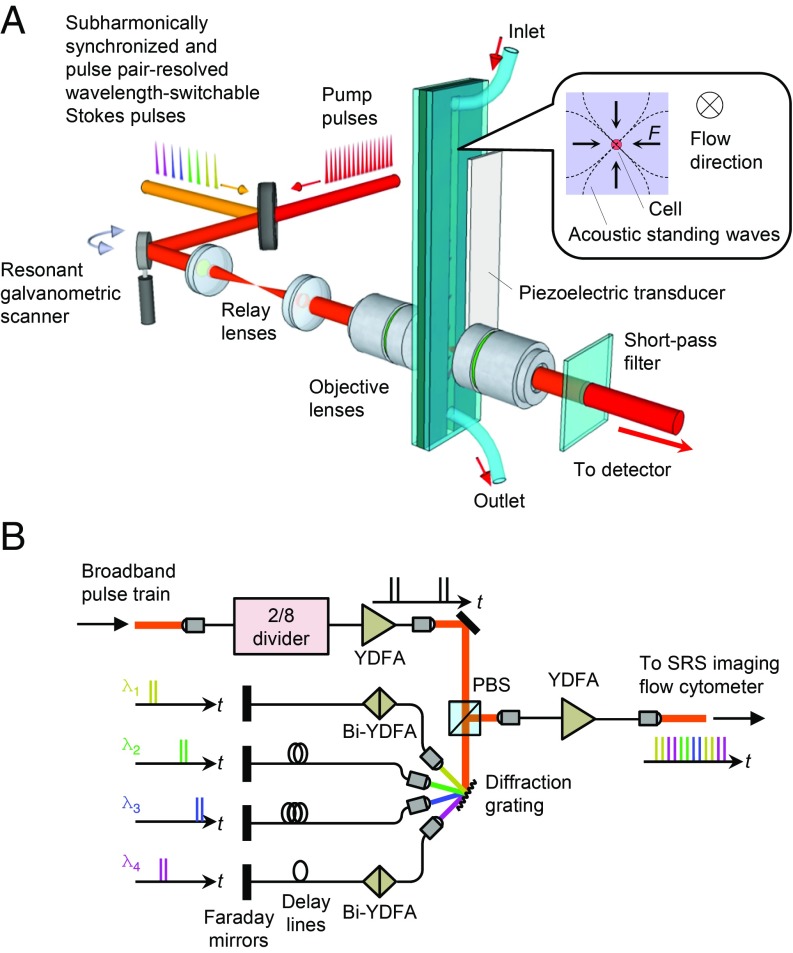
As schematically illustrated in Fig. 1A (see SI Appendix, Fig. S1, for a complete schematic), our SRS imaging flow cytometer consists of 1) pulse sources that generate the trains of picosecond pump laser pulses and subharmonically synchronized, pulse pair-resolved, wavelength-switched Stokes laser pulses for the acquisition of 4-color SRS signals; 2) a microfluidic chip (28) with a piezoelectric transducer for acoustic 3D focusing (29, 30) of fast-flowing cells; 3) focusing optics with a resonant galvanometric scanner for scanning the focal spot of the laser pulses in the direction perpendicular to the cell flow for 2D SRS imaging; and 4) a photodetector with a lock-in detection circuit for detecting the SRS signal by demodulating the pump intensity modulated by the SRS process. The cell flow is pumped by a syringe pump at a volume rate of ∼24 µL/min to produce a constant single stream of cells at a flow speed of 2 cm/s at the center of the microchannel. The resonant galvanometric scanner generates sinusoidal motion of the focal spot at 12 kHz, resulting in the horizontal scan rate of 24 kHz.
Fig. 1.
Schematic of the SRS imaging flow cytometer. (A) Schematic of the SRS imaging flow cytometer. It consists of 1) pulse sources that generate synchronized trains of pump pulses at a repetition rate of 76 MHz and subharmonically synchronized, pulse pair-resolved, wavelength-switchable Stokes pulses at a repetition rate of 38 MHz; 2) a microfluidic chip with a piezoelectric transducer for 2D acoustophoretic focusing of fast-flowing cells in a microchannel; and 3) a resonant galvanometric scanner, relay lenses, and objective lenses for scanning the focal spot in the direction perpendicular to the flow for 2D SRS image acquisition. The transmitted pump pulses are detected with a photodiode to measure the intensity modulation caused by SRS. (B) Schematic of the fast pulse pair-resolved wavelength-switchable laser. PBS, polarizing beam splitter; YDFA, ytterbium-doped fiber amplifier; Bi-YDFA, bidirectional YDFA.
The key component of the SRS imaging flow cytometer that enables the high-speed multicolor SRS image acquisition is the fast pulse pair-resolved wavelength-switchable laser, which is used as the Stokes pulse source (Fig. 1B). Specifically, the laser switches the wavelength of the pulses by every 2 pulses (every 52.5 ns) to achieve 4-color SRS signal acquisition at a period of 210 ns. The wavelength switching is realized by applying a time gate with an intensity modulator that picks up every 2 pulses in a sequence of 8 pulses from a homemade Yb fiber laser, separating the spectrum into 4 wavelength components with a diffraction grating, and applying time delays using fiber delay lines and Faraday mirrors. The time-delayed spectral components are partly amplified by bidirectional ytterbium-doped fiber amplifiers (Bi-YDFAs) and recombined to generate a 4-color pulse train. Compared with our previous galvanometric scanner-based wavelength tuning technique (31, 32), the present nonmechanical wavelength-switchable laser is 4 orders of magnitude faster, enabling multicolor SRS imaging of flowing cells.
Evaluation with Beads.
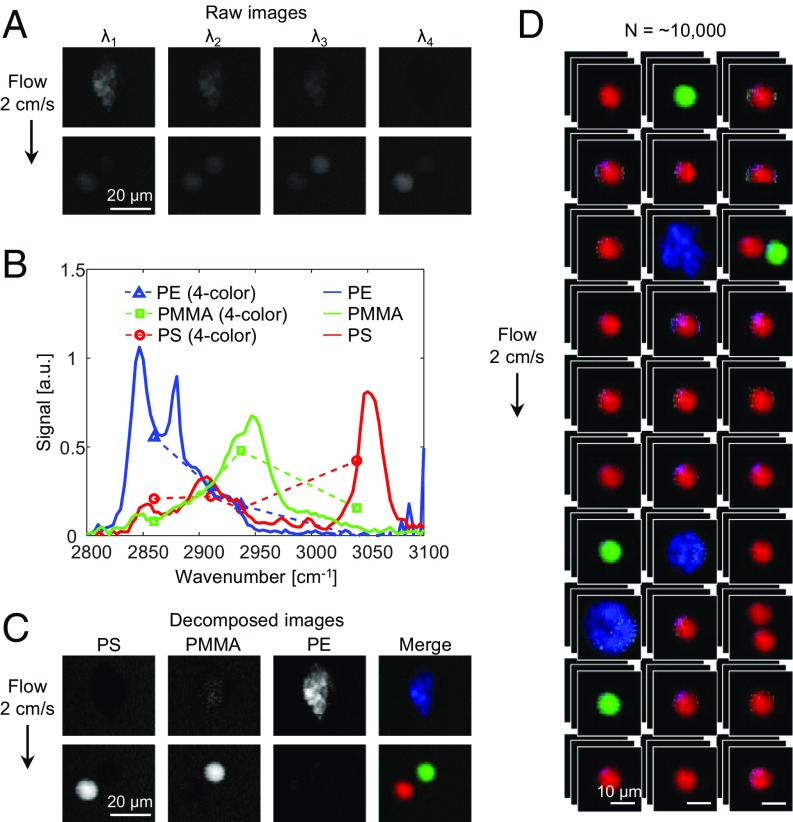
To validate the capability of multicolor SRS imaging in a high-speed flow, we characterized the SRS imaging flow cytometer with polymer beads made of polyethylene (PE), poly(methyl methacrylate) (PMMA), and polystyrene (PS) in the CH-stretching region between 2,800 and 3,100 cm−1 with an average throughput of ∼20 beads/s. As shown in Fig. 2A, SRS images of the beads at 4 Raman wavenumbers indicate their characteristic signatures at the different wavenumbers. These 4-color Raman spectra show a good agreement with those obtained by hyperspectral SRS microscopy (31, 32) (Fig. 2B). By linearly decomposing the multicolor SRS images with the 4-color Raman spectra (10), we obtained the decomposed images of the beads as shown in Fig. 2C, which show that the different types of beads are distinguishable. Fig. 2D shows an SRS image library of the beads, showing the ability to continuously acquire SRS images of flowing beads.
Fig. 2.
Characterization of the SRS imaging flow cytometer with polymer beads. (A) Four-color SRS images of PS, PMMA, and PE beads. The Raman shifts used are 2,860 cm−1 (λ1), 2,910 cm−1 (λ2), 2,937 cm−1 (λ3), and 3,040 cm−1 (λ4). (B) Four-color SRS spectra in comparison with the spectra between 2,800 and 3,100 cm−1 obtained by conventional SRS microscopy. (C) Linearly decomposed images of PS, PMMA, and PE beads shown in red, green, and blue, respectively. (D) SRS image library of polymer beads in a mixture flowing at a speed of 2 cm/s. The pixel dwell time for the 4-color SRS images is 210 ns.
SRS Imaging Flow Cytometry for Studying the Metabolic Heterogeneity of Microalgal Cells.
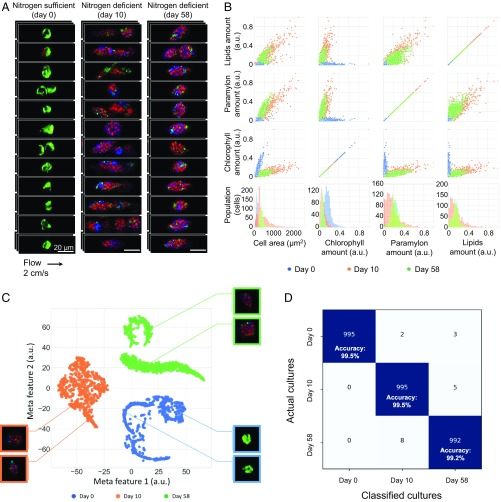
To demonstrate the diverse utility of the SRS imaging flow cytometer, we first used it to characterize the metabolic heterogeneity of microalgae as a practical application for highly efficient metabolic engineering (9, 10), which is an important research area for achieving the Sustainable Development Goals (SDGs) of the United Nations (33). Specifically, we evaluated the intracellular metabolites of each single cell of Euglena gracilis, which is a microalgal species of unicellular photosynthetic eukaryotes. E. gracilis is known to produce metabolites such as paramylon (a type of polysaccharides similar to starch) and lipids, which are useful for food supplements and biofuels, respectively (10). For an efficient production of these biomaterials, chemically specific analysis of cell-to-cell variations in a heterogeneous population of live E. gracilis cells is crucial for a better understanding of their biodiversity, which is more prominent under environmental perturbations (9). A careful examination and linear decomposition of the 4-color SRS images of E. gracilis cells (10) allowed us to identify 3 distinct spectra that correspond to paramylon, lipids, and chlorophyll (SI Appendix, Fig. S2). Fig. 3A shows an SRS image library of nitrogen-sufficient and nitrogen-deficient E. gracilis cells (n = 5,008 for nitrogen-sufficient cells, 60,854 for 10-d nitrogen-deficient cells, and 20,524 for 58-d nitrogen-deficient cells) imaged at a flow speed of 2 cm/s with average throughputs of 35 cells/s (for nitrogen-sufficient cells), 71 cells/s (for 10-d nitrogen-deficient cells), and 18 cells/s (for 58-d nitrogen-deficient cells). Here nitrogen deficiency was used as a stress condition to promote the intracellular accumulation of paramylon and lipids (34). The information-rich images show the intracellular distribution of the metabolites in each living cell as well as its morphological features. The numerous cell images also allowed us to quantify the chemical constituents with single-cell resolution in a matrix of scatterplots and histograms (Fig. 3B). It exhibits large cell-to-cell differences in the quantity of the chemical constituents, a decreased amount of chlorophyll and an increased amount of paramylon after 10 d of the nitrogen-deficiency stress, which agrees with previous reports (10, 34). After 58 d of the stress, the amounts of paramylon and lipids exhibit smaller variances, suggesting that the cells became slightly homogeneous. Furthermore, the availability of the large number of single-cell images enabled us to employ a convolutional neural network (CNN) to identify and classify the different cultures. Specifically, we employed the well-known neural network structure, VGG-16 (35), with 5 convolution segments to extract 4,096 features from each image, followed by a fully connected classifier to provide the classification result (see SI Appendix, Deep Learning with CNNs, for details). A scatterplot of the cells shown in Fig. 3C generated by the extracted features through t-distributed stochastic neighbor embedding (t-SNE) indicates a clear separation between the different cultures. Furthermore, SRS images of each cluster shown in Fig. 3 C, Inset, resemble those shown in Fig. 3A. The classification result shown in the confusion matrix (Fig. 3D) exhibits an excellent accuracy of >99% for all of the cultures. These results can help us develop and optimize culture or genetic transformation conditions under which the cells change their metabolism by modifying their biosynthetic pathways toward the formulation and accumulation of their intracellular metabolites.
Fig. 3.
SRS imaging flow cytometry and large-scale single-cell analysis of E. gracilis cells. (A) SRS images of flowing E. gracilis cells cultured under 3 different conditions: nitrogen-sufficient culture (day 0), 10 d of nitrogen-deficient culture, and 58 d of nitrogen-deficient culture. Green, chlorophyll; red, paramylon; blue, lipids. The pixel dwell time for the 4-color SRS images is 210 ns. (B) Scatterplots and histograms of the cultures in the amounts of intracellular chlorophyll, paramylon, and lipids. The histograms are projections of the scatterplots. (C) t-SNE plot of the cells after the CNN is applied to the acquired SRS images of the cells (n = 10,000 per culture for training; n = 1,000 per culture for analysis). Insets show SRS images of typical cells in each culture condition. (D) Confusion matrix of the cultures. The CNN identifies and classifies the different cultures with an excellent classification accuracy of >99%.
SRS Imaging Flow Cytometry for Marker-Free Detection of Cancer Cells in Blood.
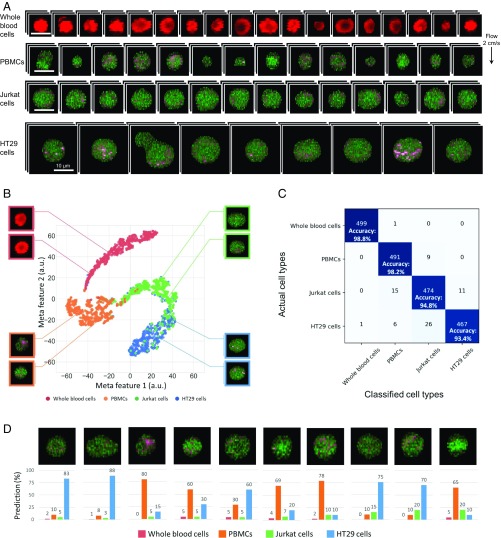
To show another practical application of the SRS imaging flow cytometer, we employed it to demonstrate simulated marker-free detection of circulating tumor cells (CTCs) in human blood. CTCs are cells that have entered blood vessels from a primary tumor and circulate in the body and are considered to be precursors to cancer metastasis (18). Detection of CTCs is effective for monitoring of cancer progression and determining a therapeutic strategy. Although the expression of the cell surface marker, epithelial cell adhesion molecule (EpCAM), is usually assayed in the detection of CTCs, this antigen can disappear through the epithelial-to-mesenchymal transition (36). Accordingly, it is important to accurately identify and enumerate CTCs without fluorescent labeling in liquid biopsy. In fact, a previous report shows promising results that coherent Raman microscopy can detect lipid-rich CTCs with the aid of preenrichment (19) but not with high throughput. For this purpose, we first used the SRS imaging flow cytometer to obtain numerous images of human whole blood cells (mostly red blood cells) (n = 12,438), peripheral blood mononuclear cells (PBMCs) (n = 18,385), human colon cancer (HT29) cells (n = 1,035), and human T lymphoma (Jurkat) cells (n = 1,437) separately to produce an image database with average throughputs of 123 cells/s (for whole blood cells), 142 cells/s (PBMCs), 8 cells/s (HT29 cells), and 113 cells/s (Jurkat cells). HT29 cells are used as a model of CTCs, while Jurkat cells are considered as a model of blood cancer cells. Fig. 4A shows an SRS image library of chemically decomposed images of these cell types (SI Appendix, Fig. S3), exhibiting a significant amount of lipid droplets (highlighted in pink) in HT29 cells but not in whole blood cells, PBMCs, and Jurkat cells. While cell areas and 4-color SRS signals of these types of cells have significant overlaps (SI Appendix, Fig. S4), a t-SNE plot (Fig. 4B) shows clusters of the 4 types of cells with their SRS images in the insets which are similar to those shown in Fig. 4A. Furthermore, a confusion matrix of these cells based on a constructed and trained CNN show the SRS imaging flow cytometer’s ability to identify and classify these cell types with an excellent classification accuracy of >98% for whole blood cells, >98% for PBMCs, >94% for Jurkat cells, and >93% for HT29 cells without the need for fluorescent labeling (Fig. 4C). Next, we applied the validated CNN to a mixed sample of HT29 cells spiked in PBMCs (with a mixing ratio of HT29/PBMCs = 1 ± 0.15/16 and an average throughput of 63 cells/s) that simulated lysed blood containing CTCs. Fig. 4D shows the CNN-calculated probability of identifying each cell in its SRS image as a red blood cell, PBMC, Jurkat cell, or HT29 cell. The CNN-predicted classification ratio of HT29/PBMCs = 222/3,387 = 1.05/16 is in good agreement with the mixing ratio of the mixed sample, demonstrating the SRS imaging flow cytometer’s potential capability of identifying CTCs in human blood. In practice, preenrichment techniques (e.g., density gradient centrifugation, lysis, and mechanical filtration) are required to reduce the total time duration of screening.
Fig. 4.
SRS imaging flow cytometry and label-free cancer cell detection in liquid biopsy. (A) SRS images of whole blood cells, PBMCs, Jurkat cells, and HT29 cells. Green, protein; pink, lipids; red, hemoglobin. The pixel dwell time for the 4-color SRS images is 210 ns. (B) t-SNE plot of the cells after the CNN is applied to the acquired SRS images of the cells (n = 1,000 for Jurkat cells, 600 for HT29 cells, 18,000 for PBMCs, and 12,500 for whole blood cells for training; n = 500 per cell type for analysis). Insets show typical SRS images of each type of cells. (C) Confusion matrix of the cells. The CNN identifies and classifies the different cell types with an excellent classification accuracy of >98% for white blood cells, >98% for PBMCs, >94% for Jurkat cells, and >93% for HT29 cells. (D) SRS images of cells classified by the CNN as HT29 cells and PBMCs with prediction probabilities in the sample of HT29 cells spiked in PBMCs.
Discussion
In this article, we have demonstrated label-free chemical imaging flow cytometry on a microfluidic platform with a high throughput of up to ∼140 cells/s enabled by high-speed 4-color SRS microscopy and acoustic 3D focusing. Its ability to provide numerous motion artifact-free and blur-free SRS images allows the accurate classification of cells based on the spatial distribution of intracellular molecular vibrational signatures. To show the diverse applicability of our SRS imaging flow cytometry, we employed it to study the metabolic heterogeneity of microalgal cells and conduct the marker-free detection of cancer cells in blood. Our results indicate that the SRS imaging flow cytometry holds promise for microbiology, cancer biology, and the SDGs.
The multicolor molecular vibrational image contrast provided by our SRS imaging flow cytometry offers various possibilities as follows. First, label-free phenotyping with SRS imaging will be useful for the quality control of stem cells for regenerative medicine (37, 38), where fluorescent labeling is not applicable due to its potential cytotoxicity and interference with their differentiation. Second, the molecular vibrational contrast allows for analyzing certain types of biomolecules such as unsaturated fatty acids (39) and polysaccharides (10), which are difficult to detect with fluorescent labeling. Third, SRS imaging is compatible with stable isotope labeling based on deuterium (40) or 13C (41) and with small Raman tags based on alkynes (42, 43), which are effective for metabolic analysis of small biomolecules. Fourth, SRS can be combined with labeling methods using supermultiplex (>10 colors) Raman probes (44, 45), which paves the way toward detailed analysis of intracellular constituents.
The capabilities of the SRS imaging flow cytometry can further be improved in multiple directions. First, the number of colors (i.e., molecular vibrational frequencies) is not limited to 4 and can be increased by adding more optical fibers and/or optical modulators to the pulse pair-resolved wavelength-switchable laser shown in Fig. 1B. Second, the wavelength-switched multicolor SRS imaging method is compatible with line-focusing geometry (46), which will further increase the SRS image acquisition speed. Third, a cell sorting capability can be added to the SRS imaging flow cytometry platform to isolate target cells from heterogeneous populations (7). With these improvements, the SRS imaging flow cytometry is expected to become a much more versatile tool and find numerous applications in diverse fields in biology and medicine.
Materials and Methods
SRS Imaging Flow Cytometer.
A detailed schematic of the SRS imaging flow cytometer is shown in SI Appendix, Fig. S1. It is composed of a mode-locked Ti:sapphire pulse laser (Coherent, Mira 900D) and a home-built mode-locked Yb fiber pulse laser to generate synchronized pump and Stokes pulse trains at repetition rates of 76 and 38 MHz, respectively. The pump laser has a center wavelength of 790 nm and a spectral width of 0.2 nm, while the Yb fiber laser generates broadband pulses at a wavelength of 1,030 nm with a spectral width of 20 nm. The latter is used to generate 4-color wavelength-switched Stokes pulses as described above. The pump laser and the Yb fiber laser are synchronized by using a feedback loop. Specifically, the intensity cross-correlation between the pump pulses and Yb fiber laser pulses is detected by a GaAsP photodiode (Hamamatsu, G1115) via 2-photon absorption, such that its signal is used to control the intracavity electro-optic modulator and a piezoelectric transducer in the Yb fiber laser. The 2 laser beams are overlapped both spatially and temporally and are focused inside the microchannel via the first objective lens (50×, NA = 0.65). The optical power in the sample plane is <140 mW for both the pump and Stokes laser beams. The SRS process occurs in the focused volume of the combined beam inside the target sample in the microchannel, resulting in the intensity modulation of the pump beam at the repetition rate of the Stokes pulses. The transmitted pump beam is detected by a Si photodetector (Hamamatsu, S3399) via the second objective lens (50×, NA = 0.65) while the transmitted Stokes beam is removed by an optical short-pass filter. The photodetector is equipped with homemade electrical band-pass filters with a center frequency of 38 MHz, band elimination filters at 76 MHz, and electrical amplifiers so as to extract the intensity modulation of the pump pulses and to avoid unwanted saturation of electrical amplifiers. The photodetector signal is demodulated by a homemade lock-in amplifier at 38 MHz to obtain the SRS signal. The SRS signal is digitized with an 8-bit analog–digital converter (Analog Devices, AD9287), whose output digital data are processed by a FPGA (Xilinx, Spartan-6) to continuously send the data via Gigabit Ethernet to a server computer. To perform SRS imaging of flowing beads or cells, we prepared a solution of biological cells or polymer beads with a concentration of ∼5 × 105 mL−1 and injected the solution by a syringe pump (Harvard Apparatus) into the microchannel whose cross-sectional area is 200 × 200 μm2. To align the beads or cells at the center of the microchannel, a piezoelectric transducer attached to the surface of the microfluidic chip is used to generate acoustic standing waves inside the microchannel. To perform SRS imaging flow cytometry, the focal spot is scanned in the direction perpendicular to the direction of the flow by a resonant galvanometric scanner operating at 12 kHz such that 2D SRS images are obtained by stacking the 1D SRS signals. The pixel dwell time for 4-color SRS images is 210 ns with no pixel averaging used for image production. By changing the wavelength of the Stokes laser with the fast wavelength-switched pulse laser and by digitally mapping the Raman signal into a 3D matrix (x, y, λ), 4-color SRS images are produced. The 4-color SRS images are used to generate cell mask patterns as described in the next section and are decomposed into images of several constituents by using a Moore–Penrose pseudoinverse matrix. The decomposed images are then combined to produce multicolor images, to which cell masks are applied to highlight cell images. The concentration of the cells or beads was adjusted so that the average spacing was ∼150 µm (which determined the throughput) to avoid doublet formation, the probability of which is given as the ratio between the cell/bead size and their average spacing according to Poisson statistics. The precision of the flow rate was estimated to be ∼10% from the ellipticity of the bead images.
Generation of Cell Masks.
To generate the cell masks for E. gracilis cells, we first calculated the inner product between each pixel’s 4-color SRS spectrum dn, where n represents the index of the image pixels, and the spectral basis si (i = 1, 2, 3) of lipid, paramylon, and chlorophyll, given by . For each nth pixel, we chose the largest value of among different i values. Using the generated cell image of the maximum values among different spectra, we obtained a binary cell image by applying a threshold value and then iteratively applied dilation and erosion operations to connect the segmented areas within a cell. We then removed unnaturally small objects in the binarized images to generate the cell masks. To generate the cell masks for blood cells, we first applied a moving average with a 2D Gaussian window to the λ3 images. Note that we used only λ3 (= 2,937 cm−1) images to generate cell masks because blood cells have strong protein signals at this wavenumber. After the moving average, we generated the cell masks by binarizing the image using a threshold value, followed by an iterative use of dilation and erosion operations to connect the segmented areas within a cell and removal of unnaturally small objects in the binarized images.
Deep Learning with CNNs.
To analyze the images of E. gracilis and blood cells acquired by our SRS imaging flow cytometer, we used the CNN structure (VGG-16) to classify different cell types. For the training process in the E. gracilis screening experiment, we randomly picked 10,000 cell images as the training and validation dataset and 1,000 cell images as the test dataset from each E. gracilis culture. As for the cancer detection experiment, we randomly picked 1,000 Jurkat cell images, 600 HT29 cell images, 18,000 PBMC images, and 12,500 whole blood cell images as the training and validation dataset and 500 cell images as the test dataset for each cell type. For each cell image, a total number of 4,096 features were extracted from the 4-channel (λ1−λ4 signals) input image through 5 convolution segments. Each convolution segment consists of 2 or 3 convolutional layers for feature extraction and a maximum pooling layer for the reduction of data volume. At last, a fully connected classifier was used to downsize the 4,096 features to 1 dimension (probability values) to provide a classification result. The CNN was trained for 100 epochs using categorical cross-entropy as the loss function and stochastic gradient descent with standard parameters: 0.9 momentum, an initial learning rate of 0.0008, and a slightly regularizing weight decay of 0.000001 as the optimizer. Training took around 2 h and was stopped automatically by inspecting the validation accuracy.
t-SNE Plots.
To better understand the features learned by the VGG-16 model, we used the t-SNE implementation in Scikit-learn (47) to visualize the activations in the final dense layer. The results were learned in an unsupervised way, meaning that there was no label information on cell types in the 4,096 features learned by the VGG-16 model. We identified clear clusters in the 2D t-SNE plots as shown in Figs. 3C and 4B, suggesting that the VGG-16 model effectively learned the important information about the cell images but not the artificial noise.
Preparation of Jurkat Cells.
Jurkat cells (ATCC, TIB-152) were obtained and cultured in a medium with l-glutamine and sodium bicarbonate (Sigma-Aldrich, R8758-500), 10% FBS (Biowest, S1780-500), 100 units/mL penicillin and 100 µg/mL streptomycin (Wako, 168-23191) at 37 °C, and 5% CO2. The cells were placed into 50-mL suspension culture flasks (Greiner Bio-One, 690195) and allowed to spread for 3–7 d.
Preparation of HT29 Cells.
HT29 (human colorectal cancer, ATCC, HTB-38) cells were cultured in Dulbecco’s Modified Eagle Medium (Gibco, 11995-065) supplemented with 10% FBS (Gibco, 10437-028), 100 units/mL penicillin, and 100 µg/mL streptomycin (Wako, 168-23191) in a 100-mm tissue culture dish (Iwaki, 3020-100) until the cells became nearly confluent. The cells were detached from the dish by incubating with 2 mL of 0.05% (W/V) trypsin-EDTA (Gibco, 15400-054) for 10 min at 37 °C, collected by centrifugation (Hitachi, CT6D) for 3 min, and resuspended in DMEM containing 10% FBS at 1 × 105 cells/mL.
Preparation of Whole Blood Cells and PBMCs.
The blood samples were taken from healthy adult volunteers. One milliliter of whole blood was incubated with 5 mL of the hemolytic agent (Beckman Coulter, Optilyse C) for 10 min and additional 15 min after adding 10 mL of CellWash (BD Biosciences). Then, the samples were centrifuged at 430 g for 5 min. After the removal of the supernatants, the pellets were suspended with 5 mL of CellWash and used as PBMCs. PBMCs and whole blood cells were diluted with phosphate buffer saline to adjust a cell count of 107 cells/mL and subjected to the SRS imaging flow cytometer within 5 h after the sample preparation. This procedure was performed at room temperature (18–25 °C). This study was approved by the Institutional Ethics Committee of Faculty of Medicine, the University of Tokyo (No. 11049-5) and conducted according to the Declaration of Helsinki. All volunteers provided written informed consent.
Data Availability.
All data generated and analyzed during the current study are available on Zenodo (DOI: 10.5281/zenodo.3269336).
Code Availability.
All codes used in the data analysis are available on Zenodo (DOI: 10.5281/zenodo.3269336).
Supplementary Material
Acknowledgments
This work was funded mainly by the ImPACT Program of the Council for Science, Technology and Innovation (Cabinet Office, Government of Japan).
Footnotes
Conflict of interest statement: Y.O., Y.S., Y.W., and K.G. are inventors of a patent application covering SRS imaging with a wavelength-switched laser. All other authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.F.L. is a guest editor invited by the Editorial Board.
Data deposition: All data generated and analyzed during the current study, as well as the codes used in the data analysis, are available on Zenodo (DOI: 10.5281/zenodo.3269336).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902322116/-/DCSupplemental.
References
- 1.McGranahan N., Swanton C., Clonal heterogeneity and tumor evolution: Past, present, and the future. Cell 168, 613–628 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Prasetyanti P. R., Medema J. P., Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer 16, 41 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Satija R., Shalek A. K., Heterogeneity in immune responses: From populations to single cells. Trends Immunol. 35, 219–229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stubbington M. J. T., Rozenblatt-Rosen O., Regev A., Teichmann S. A., Single-cell transcriptomics to explore the immune system in health and disease. Science 358, 58–63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perlman Z. E., et al. , Multidimensional drug profiling by automated microscopy. Science 306, 1194–1198 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Heath J. R., Ribas A., Mischel P. S., Single-cell analysis tools for drug discovery and development. Nat. Rev. Drug Discov. 15, 204–216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nitta N., et al. , Intelligent image-activated cell sorting. Cell 175, 266–276.e13 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Ackermann M., A functional perspective on phenotypic heterogeneity in microorganisms. Nat. Rev. Microbiol. 13, 497–508 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Georgianna D. R., Mayfield S. P., Exploiting diversity and synthetic biology for the production of algal biofuels. Nature 488, 329–335 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Wakisaka Y., et al. , Probing the metabolic heterogeneity of live Euglena gracilis with stimulated Raman scattering microscopy. Nat. Microbiol. 1, 16124 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Boutros M., Heigwer F., Laufer C., Microscopy-based high-content screening. Cell 163, 1314–1325 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Basiji D. A., Ortyn W. E., Liang L., Venkatachalam V., Morrissey P., Cellular image analysis and imaging by flow cytometry. Clin. Lab. Med. 27, 653–670, viii (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borah S., Nichols L. A., Hassman L. M., Kedes D. H., Steitz J. A., Tracking expression and subcellular localization of RNA and protein species using high-throughput single cell imaging flow cytometry. RNA 18, 1573–1579 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parris C. N., et al. , Enhanced γ-H2AX DNA damage foci detection using multimagnification and extended depth of field in imaging flow cytometry. Cytometry A 87, 717–723 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blasi T., et al. , Label-free cell cycle analysis for high-throughput imaging flow cytometry. Nat. Commun. 7, 10256 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eliceiri K. W., et al. , Biological imaging software tools. Nat. Methods 9, 697–710 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen E. C., Use of fluorescent probes: Their effect on cell biology and limitations. Anat. Rec. (Hoboken) 295, 2031–2036 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Yu M., Stott S., Toner M., Maheswaran S., Haber D. A., Circulating tumor cells: Approaches to isolation and characterization. J. Cell Biol. 192, 373–382 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitra R., Chao O., Urasaki Y., Goodman O. B., Le T. T., Detection of lipid-rich prostate circulating tumour cells with coherent anti-stokes Raman scattering microscopy. BMC Cancer 12, 540 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lei C., et al. , High-throughput label-free image cytometry and image-based classification of live Euglena gracilis. Biomed. Opt. Express 7, 2703–2708 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y., et al. , Label-free detection of aggregated platelets in blood by machine-learning-aided optofluidic time-stretch microscopy. Lab Chip 17, 2426–2434 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Liao C. S., et al. , Stimulated Raman spectroscopic imaging by microsecond delay-line tuning. Optica 3, 1377–1380 (2016). [Google Scholar]
- 23.He R., et al. , Stimulated Raman scattering microscopy and spectroscopy with a rapid scanning optical delay line. Opt. Lett. 42, 659–662 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Liao C. S., et al. , Microsecond scale vibrational spectroscopic imaging by multiplex stimulated Raman scattering microscopy. Light Sci. Appl. 4, e265 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao C. S., et al. , Spectrometer-free vibrational imaging by retrieving stimulated Raman signal from highly scattered photons. Sci. Adv. 1, e1500738 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang C., et al. , Stimulated Raman scattering flow cytometry for label-free single-particle analysis. Optica 4, 103–109 (2017). [Google Scholar]
- 27.He R., et al. , Dual-phase stimulated Raman scattering microscopy for real-time two-color imaging. Optica 4, 44–47 (2017). [Google Scholar]
- 28.Sakuma S., Kasai Y., Hayakawa T., Arai F., On-chip cell sorting by high-speed local-flow control using dual membrane pumps. Lab Chip 17, 2760–2767 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Thalhammer G., et al. , Combined acoustic and optical trapping. Biomed. Opt. Express 2, 2859–2870 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park J. W., et al. , Acoustofluidic harvesting of microalgae on a single chip. Biomicrofluidics 10, 034119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozeki Y., et al. , High-speed molecular spectral imaging of tissue with stimulated Raman scattering. Nat. Photonics 6, 845–851 (2012). [Google Scholar]
- 32.Ozeki Y., Asai T., Shou J., Yoshimi H., Multicolor stimulated Raman scattering microscopy with fast wavelength-tunable Yb fiber laser. IEEE J. Sel. Top. Quantum Electron. 25, 7100211 (2018). [Google Scholar]
- 33.Griggs D., et al. , Policy: Sustainable development goals for people and planet. Nature 495, 305–307 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Coleman L. W., Rosen B. H., Schwartzbach S. D., Environmental control of carbohydrate and lipid synthesis in Euglena. Plant Cell Physiol. 29, 423–432 (1988). [Google Scholar]
- 35.Simonyan K., Zisserman A., Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556 (4 September 2014).
- 36.Hyun K. A., et al. , Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. Oncotarget 7, 24677–24687 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ember K. J. I., et al. , Raman spectroscopy and regenerative medicine: A review. NPJ Regen. Med. 2, 12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Downes A., Mouras R., Bagnaninchi P., Elfick A., Raman spectroscopy and CARS microscopy of stem cells and their derivatives. J. Raman Spectrosc. 42, 1864–1870 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J., et al. , Lipid desaturation is a metabolic marker and therapeutic target of ovarian cancer stem cells. Cell Stem Cell 20, 303–314.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei L., Yu Y., Shen Y., Wang M. C., Min W., Vibrational imaging of newly synthesized proteins in live cells by stimulated Raman scattering microscopy. Proc. Natl. Acad. Sci. U.S.A. 110, 11226–11231 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen Y., Xu F., Wei L., Hu F., Min W., Live-cell quantitative imaging of proteome degradation by stimulated Raman scattering. Angew. Chem. Int. Ed. Engl. 53, 5596–5599 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamakoshi H., et al. , Imaging of EdU, an alkyne-tagged cell proliferation probe, by Raman microscopy. J. Am. Chem. Soc. 133, 6102–6105 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Wei L., et al. , Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering. Nat. Methods 11, 410–412 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei L., et al. , Super-multiplex vibrational imaging. Nature 544, 465–470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu F., et al. , Supermultiplexed optical imaging and barcoding with engineered polyynes. Nat. Methods 15, 194–200 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z., Zheng W., Huang Z., Lock-in-detection-free line-scan stimulated Raman scattering microscopy for near video-rate Raman imaging. Opt. Lett. 41, 3960–3963 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Pedregosa F., et al. , Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and analyzed during the current study are available on Zenodo (DOI: 10.5281/zenodo.3269336).