Significance
More than 50 years ago, Epstein–Barr virus (EBV) was found to infect resting human B lymphocytes and to induce their continuous proliferation in vitro. This seminal finding clearly mimics aspects of EBV’s oncogenic potential in vivo but how EBV activates primary human B lymphocytes and how lymphoblastoid cell lines (LCLs) evolve from the EBV-infected lymphocytes is uncertain. We studied this model and found that the EBV-infected naïve B lymphocytes undergo a germinal center-like activation and differentiation program within only a few days after infection to emerge with transcriptomic and phenotypic features resembling plasmablasts and early plasma cells. In vitro in established LCLs, EBV “locks” the infected B cells in a stage of differentiation that is only transient in vivo.
Keywords: Epstein–Barr virus, reprogramming, B cell, B lymphocyte
Abstract
Epstein–Barr virus (EBV) is a human tumor virus and a model of herpesviral latency. The virus efficiently infects resting human B lymphocytes and induces their continuous proliferation in vitro, which mimics certain aspects of EBV’s oncogenic potential in vivo. How lymphoblastoid cell lines (LCLs) evolve from the infected lymphocytes is uncertain. We conducted a systematic time-resolved longitudinal study of cellular functions and transcriptional profiles of newly infected naïve primary B lymphocytes. EBV reprograms the cells comprehensively and globally. Rapid and extensive transcriptional changes occur within 24 h and precede any metabolic and phenotypic changes. Within 72 h, the virus activates the cells, changes their phenotypes with respect to cell size, RNA, and protein content, and induces metabolic pathways to cope with the increased demand for energy, supporting an efficient cell cycle entry on day 3 postinfection. The transcriptional program that EBV initiates consists of 3 waves of clearly discernable clusters of cellular genes that peak on day 2, 3, or 4 and regulate RNA synthesis, metabolic pathways, and cell division, respectively. Upon onset of cell doublings on day 4, the cellular transcriptome appears to be completely reprogrammed to support the proliferating cells, but 3 additional clusters of EBV-regulated genes fine-tune cell signaling, migration, and immune response pathways, eventually. Our study reveals that more than 11,000 genes are regulated upon EBV infection as naïve B cells exit quiescence to enter a germinal center-like differentiation program, which culminates in immortalized, proliferating cells that partially resemble plasmablasts and early plasma cells.
Epstein–Barr virus (EBV) was discovered in 1964 in lymphoma biopsies of patients from sub-Saharan Africa (1, 2). In the following years, Pope (3) and Henle (4) independently reported that the newly termed virus induces the proliferation of immune cells from peripheral blood of any human donor. This seminal finding changed the view on the role of herpes viruses in general and provided a reliable assay for EBV’s transforming capacity mimicking its oncogenic potential. B cell transformation became the most important test to identify and investigate the contributing viral factors and to study the mechanisms leading to herpesviral latency. In the following years, EBV was found to be strongly associated with other lymphomas, but also gastric and nasopharyngeal carcinomas (5, 6). In vitro, EBV efficiently infects mature, resting B lymphocytes, activates them, and induces their continuous proliferation, leading to established lymphoblastoid cell lines (LCL). Many groups have contributed to the analysis of LCLs to understand the multiple viral contributions to B cell transformation and viral latency. We learned that EBV establishes the so-called latency III program in LCLs, which includes the expression of 6 EBV nuclear antigens (EBNAs), such as EBNA1, EBNA2, EBNA3A, EBNA3B, EBNA3C, and EBNA-LP, 3 latent membrane proteins (LMP1, LMP2A, and LMP2B), ncRNAs (EBER1, EBER2, snoRNA), and 44 miRNAs (7).
Recently, the early aspects of EBV infection during the so-called prelatent phase have gained more attention (8, 9) but, to our knowledge, a comprehensive analysis of the early events that lead to viral transformation of B cells is lacking. As a consequence, we do not know how the infected resting B lymphocyte eventually evolves to give rise to a proliferating LCL. During this initial phase of infection, the expression of certain latent genes such as EBNA2 is critical, but also lytic genes are temporarily expressed and seem to contribute to the early survival of the EBV-infected B cells promoting long-term persistence of EBV (10–12). In the prelatent phase, EBV has to overcome the initial antiviral responses of the host cell (and its organism) to be successful. Several viral processes are known that counteract these responses. The known viral processes encompass the mimicry of IL-10 by BCRF1 (13), repression of antigen presentation mediated by BNLF2a (14, 15), and certain viral miRNAs (16), as well as prevention of apoptosis by BALF1 and BHRF1 (10).
The success of infection lies in the complex strategy of EBV to reprogram the host B cell. One of the most important viral factors is EBNA2, which is supported by EBNA-LP, but several reports investigated other latent genes defining their roles immediately after infection, such as the EBNA3s (17), or revealing the functions of the delayed expression the LMPs (18–20). Initially after infection, EBNA2 plays a crucial role in cell activation and proliferation entry. EBNA2 together with EBNA-LP coregulate the expression of important cellular genes, including the proto-oncogene MYC (21) and transcription factors, such as PU.1 (22), EBF1 (23), IRF4 (24), and CBF1 (25). These cellular genes promote B cell activation directly or activate signaling cascades, which together support cell proliferation and virus persistence during latency or prepare the latently infected cells to support the productive lytic phase, including viral replication and de novo virus synthesis.
Our knowledge about EBV-mediated processes in newly infected B lymphocytes is incomplete. Many groups have investigated the regulation and function of specific cellular and individual viral genes and processes in stable, latently infected cell lines, but the initial events that are crucial and drive the early phase of EBV infection are less known. From the perspective of the virus, EBNA-LP and EBNA2 are the 2 viral genes that are expressed early, but a detailed and systematic analysis of viral gene expression and their impact on host cell transcription is not available.
The aim of this study was to examine the early interactions between EBV and its cellular host. We designed time-resolved infection experiments with a focus on the B cell biology and the dynamics of cellular and viral gene regulation. Our data reveal that naïve B lymphocytes infected with EBV undergo rapid and dramatic phenotypic changes involving cell size, content of macromolecules, metabolic and mitochondrial activities, and entry into the cell cycle. EBV imposes a strict timing of cellular genes supporting EBV’s prelatent phase and induces the global reprogramming of the transcriptome of the quiescent, resting B lymphocyte, which becomes an infected, activated, and cycling B blast with a transcriptional profile resembling plasmablasts and early plasma cells. Our analysis suggests that the most profound alterations at the level of the cellular transcriptome of the infected naïve B lymphocyte occur within the first 3 d, whereas phenotypic and metabolic features start changing from day 3 onwards. Later events seem to fine-tune the biology of the host cell preparing for the ensuing phase of stable latency. The data are a rich source of cell biology covering the early molecular steps of B cell transformation driven by the tightly controlled expression program of viral genes.
Results
EBV Infection Induces Fundamental Metabolic Alterations in Infected Cells.
We performed detailed time-course experiments to monitor basic metabolic parameters, such as mitochondrial activity and glucose uptake in uninfected and EBV-infected B cells during the first week of infection. B lymphocytes were obtained from adenoid tissue and purified by removing cells with other identities using a negative-depletion strategy (SI Appendix, Materials and Methods). Purified B cells were infected with the wild-type EBV strain B95-8 (wt/B95.8) and a multiplicity of infection that was found optimal for infectivity and cellular survival (26, 27). Equivalent numbers of uninfected cells and cells infected for the indicated times (Fig. 1 A and B) were stained with tetramethylrhodamine ethyl ester (TMRE), incubated with Annexin V, or with 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]-2-deoxy-d-glucose (2-NBDG), a glucose analog, to analyze mitochondrial activity, early apoptosis, and glucose uptake, respectively. TMRE is a cationic red-orange dye, which accumulates in active mitochondria reflecting their membrane potential, negatively charged matrix, and mitochondrial electron transport chain functions. Annexin V binds phosphatidylserine residues in the plasma membrane, indicating an early phase of cell apoptosis.
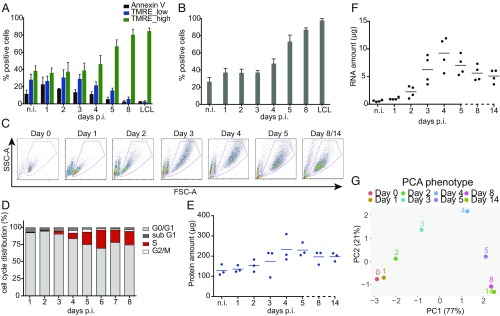
Fig. 1.
Metabolic and phenotypic parameters of B cells early after EBV infection. (A) The fraction of Annexin V+ and TMRE_low cells decreased in the course of EBV infection. The fraction of TMRE_high cells stayed constant until day 3 p.i., but increased starting on day 4 to amount to more than 80% 1 wk p.i. Mean and SDs from 4 independent biological replicates are shown. (B) During the first 3 d p.i., the fraction of cells that took up the glucose analog (2-NBDG) was constant. Starting on day 4 the fraction of glucose analog-positive cells increased up to 100%. Bars in the graph display mean and SD from 4 independent experiments. (C) Time-resolved FACS analysis of FSC-A (x axis) and SSC-A (y axis) criteria of EBV-infected cells. Uninfected B cells as well as newly infected cells on day 1 p.i. form a homogenous population of small cells with low granularity. The cells increase in size and reach maximal size and granularity 4 d p.i. Later, the cells adopt a more discrete population. On days 8 and 14 p.i. the cells are indistinguishable. (D) B cells after wt/EBV (2089) infection were incubated with BrdU for 1 h followed by intracellular staining with a BrdU-specific antibody. The cells were analyzed by FACS to determine their cell cycle distribution. Until day 3 the cells did not proliferate and stayed arrested in G0/G1 phase of the cell cycle. From day 3 p.i. the cells started to synthesize cellular DNA, as indicated by the presence of cells in the S-phase. Experiments were performed using 3 independent replicates. (E) Uninfected cells and cells infected with wt/EBV (2089) were collected daily and total protein lysates were obtained. The samples were measured using the Bradford assay. The graph shows the total amount of proteins (μg) obtained from 1 million cells. Measurements were performed every day after infection as indicated. The horizontal blue lines show the mean average from cells prepared from 3 different donors, individual samples are represented as single dots. (F) Amounts of total RNA (μg) obtained from 1 million cells were measured using a Shimadzu MultiNA instrument. Solid black lines represent the mean average from 4 different donors. Each donor is represented as a single dot. (G) The plot represents the PCA of the data in this figure and SI Appendix, Fig. S3 A and B (TMRE, 2-NBDG, cell diameter, DI, S-phase cell cycle distribution, protein content, and RNA content). The results from each experiment were normalized by the maximum value in each assay (SI Appendix, Materials and Methods). Next, PCA was performed and the first 2 components, PC1 (x axis) and PC2 (y axis), were plotted. The number in parentheses indicates the percentage of total variance explained by each PC. Colored dots represent samples infected with EBV at the depicted time points.
We compared the kinetics of TMRE staining and Annexin V binding at the indicated time points before and after EBV infection (Fig. 1A). Two distinct TMRE+ cell populations (TMRE_low and TMRE_high) were obvious in uninfected cells and cells infected for up to 3 d, which showed a similar and thus stable distribution (SI Appendix, Fig. S1). From day 4 onward, the TMRE_high population became more abundant (more than 80% positive cells), while the TMRE_low population disappeared on day 8 postinfection (p.i.). The binding of Annexin V indicated a substantial fraction of apoptotic cells during the first 4 d of infection, which decreased starting at day 5 p.i. (SI Appendix, Fig. S1).
The significant increase of mitochondrial activity could indicate a cellular requirement for glucose, the most important source of energy. To monitor the changes in glucose uptake we analyzed uninfected and infected cells with the aid of a fluorescently labeled (FITC) glucose analog (2-NBDG) at the indicated time points (Fig. 1B). Cells were transferred to low-glucose medium and incubated with 100 µM 2-NBDG for 1 h under optimal culture conditions. The accumulation of the 2-NBDG fluorescent signal in the cells was measured by FACS over time. Similar to the TMRE results, the uptake of glucose did not change in infected cells compared with uninfected B cells up to 3 d p.i., with levels of about 40% 2-NBDG+ cells (Fig. 1B and SI Appendix, Fig. S2). Four to 5 d p.i. the uptake of the glucose analog increased substantially and almost all cells were found to become 2-NBDG+, eventually.
Based on these results, we conclude that mitochondrial activity and glucose uptake increase in the time course of infection. Moreover, within the first 3 d p.i. the metabolism of the infected B cells is comparable with uninfected, resting B lymphocytes, but starting from day 4 onward, the infected cells increase their metabolic rate, which is maintained in established, stably proliferating LCLs.
Phenotypic Changes in the Host B Cells during Early Time Points after Virus Infection.
Next to the basic metabolic parameters, we studied additional cellular phenotypes of the EBV-infected cells. Quiescent naïve B lymphocytes (IgD+, CD38−) were purified from adenoid tissue by physical sorting. Visual daily inspection of the infected cells (Fig. 1C) demonstrated that they grew massively in size and gained granularity according to forward (FSC-A) and sideward scatter (SSC-A) FACS criteria, respectively. The population of cells started to become more homogenous on day 5 p.i. to reach a state that was maintained in established LCLs (Fig. 1C). We determined the cells’ diameter during EBV infection (SI Appendix, Fig. S3A). Initially, the diameter of the purified naïve B cells was found to be 5.5 µm. The cells grew constantly up to 9 µm on average within the first 4 d of infection, but their diameter shrank to 8 µm later and did not change further when a stable population of lymphoblastoid cells eventually emerged (SI Appendix, Fig. S3A).
EBV infection of the resting naïve B lymphocytes induced their delayed entry into the cell cycle. Naïve B cells were found arrested in G0/G1 for 2 d p.i. according to their DNA content and metabolic incorporation of BrdU (Fig. 1D). The first cells entered S-phase on day 3 p.i. and the fraction of S-phase cells increased on the following days. The B cells underwent substantial DNA synthesis as indicated by the fraction of cells in S-phase on days 5 and 6 p.i., exceeding this number in established latently infected lymphoblast populations. In parallel to the analysis of the cell cycle, we monitored the dynamics of cell divisions by loading the uninfected B lymphocytes with CellTrace violet and by following its distribution by FACS to calculate the division index (DI; i.e., the average number of divisions for all cells) (28) in the course of EBV infection (SI Appendix, Fig. S3B). Within the first 3 d the infected B cells did not proliferate. The first cell division was observed on day 4 p.i., immediately followed by a 2-d-long phase with a steep increase of the DI indicating a generation time of only 12 h. From day 7 p.i. onward, the cells decelerated and divided once within a period of 24 to 36 h.
The EBV-induced B cell growth and cellular proliferation resulted in global alterations of macromolecules in the infected cells. The contents of total cellular RNA and protein in 1 × 106 intact cells that had been physically sorted according to their forward and sideward scatter criteria were measured daily and compared with the lymphoblast cell population at 2 wk p.i. (Fig. 1 E and F). In cells infected with EBV for 4 d, the total protein content was about 2.5-fold higher compared with uninfected cells (Fig. 1E). At later time points, the protein content decreased slightly and became stable at a higher level than in resting B lymphocytes after about 1 wk p.i. Similarly, the total cellular RNA content increased substantially by a factor of 15 when it peaked at day 4 p.i. (Fig. 1F). A single noninfected B lymphocyte contained about 0.59 pg RNA on average, whereas an EBV-infected activated B cell contained about 9.2 pg on day 4 p.i. Starting from day 5, the amount of RNA in a single cell decreased constantly and stabilized at about 5.1 pg a fortnight postinfection (Fig. 1F).
The results from the metabolic and phenotypic assays were normalized (SI Appendix, Materials and Methods) and combined via principal component analysis (PCA) to visualize the global dynamics of the metabolic and phenotypic changes during EBV infection. The PCA shows that uninfected and infected cells were similar in terms of their metabolic functions and phenotypes during the first 2 d p.i. since they cluster close to each other (Fig. 1G). At days 3 and 4 p.i. the cells differed substantially from the initial cluster (Fig. 1G) due to their increased energy consumption, entry into the cell cycle, and first cell divisions (Fig. 1 A, B, and D and SI Appendix, Fig. S3 A and B). Infected cells on day 5 p.i. differed even more from cells during the previous days of infection, but they were closer to cells infected for 8 and 14 d, demonstrating their similar metabolic processes and phenotypes (Fig. 1G). In this PCA plot, the second principal component (PC2) is mainly driven by differences in total RNA content and cell diameter, while all of the other measurements give a larger contribution to the first principal component (PC1) (SI Appendix, Fig. S3 C–E). This broadly reflects the 2 dynamic patterns observed: either a marked peak at ∼4 d p.i. (total RNA content and cell diameter) or an approximate monotonic increase (all other measurements).
Overall, this analysis revealed that the changes in metabolic and phenotypic features are very limited within the first 2 d of infection, whereas they massively change between days 2 and 5 p.i. Already on day 8 p.i. the cells reached a stable, latent cell phenotype, which reflects their established and final cellular fate.
EBV Infection Causes Global Alterations in the B Cell Transcriptome.
The prominent phenotypic changes of naïve B cells early after EBV infection indicated comprehensive alterations in the entire B cell biology. To analyze the detailed dynamics of gene expression in the course of infection, we performed sequential, time-resolved RNA-sequencing (RNA-seq) experiments with noninfected primary B lymphocytes and with cells infected with EBV from day 1 to day 5, daily, and on day 8 and day 14 p.i. Sorted, quiescent naïve B cells (IgD+/CD38−) from 3 different donors were infected with the wt/B95.8 (2089) EBV strain (29) with an optimal multiplicity of infection (26, 27). Each day postinfection 106 living, physically intact cells were collected by FACS sorting according to their FSC and SSC criteria. We extracted their RNA and established polyA enriched libraries to allow subsequent whole-transcriptome sequencing on an Illumina HiSeq 4000 instrument with a high coverage. After gene counting but before any downstream analyses, we performed quality controls to ensure that all libraries were of acceptable quality. All but 1 sample from day 8 p.i. passed the quality controls (SI Appendix, Fig. S4). SI Appendix, Fig. S5 provides the chart of the entire bioinformatic workflow and detailed information can be found in SI Appendix, Materials and Methods.
Initially, we focused on the global transcriptional changes during infection (Fig. 2). A PCA was performed on the log-transformed gene-expression matrix. The first 2 principal components, representing the 2 directions of largest variance (PC1 and PC2) are shown in Fig. 2A and the corresponding scree plot is provided in SI Appendix, Fig. S3F. The expression levels of the top 100 genes that contribute the most to PC1 and PC2 are displayed in heat maps in SI Appendix, Fig. S6, while lists of the top 1,000 genes are in Datasets S1 and S2.
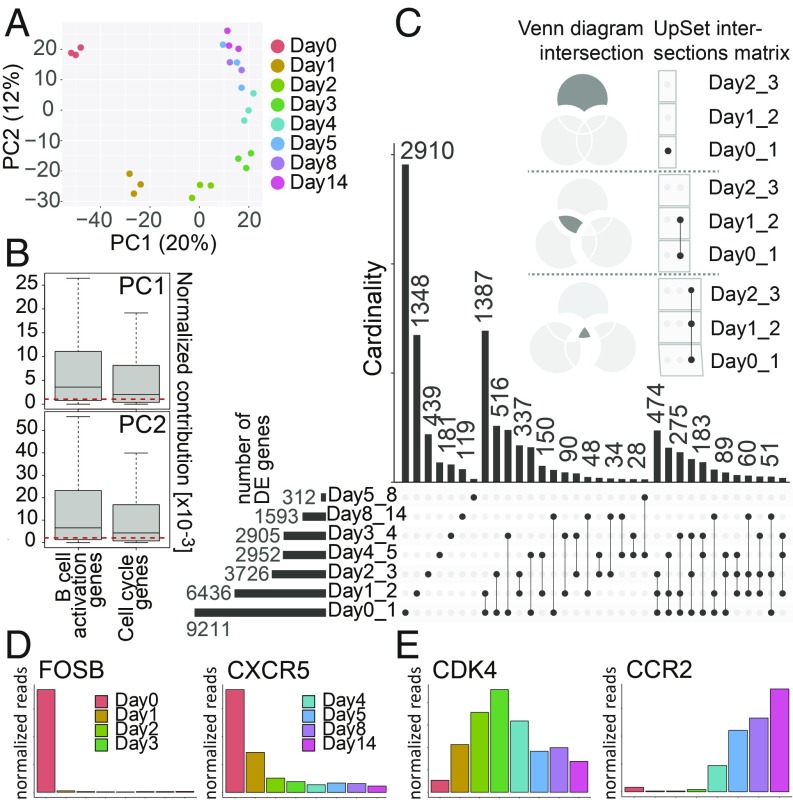
Fig. 2.
Analysis of the time-resolved RNA-seq data. (A) PCA of the samples from sequencing libraries at the given time points postinfection. The daily samples are represented by 3 biological replicates with the exception of day 8 (n = 2). The samples are shown as a function of PC1 and PC2. The x and y axes show the percentages of variance explained by PC1 and PC2. (B) The normalized contributions of cell cycle annotated genes and genes marking any of the stages of B cell activation (Fig. 5A) to PC1 and PC2, as plotted in A, are shown. The genes that are annotated as both cell cycle and B cell activation genes were removed from the analysis. The red dashed lines indicate the median normalized contributions calculated on all genes. The contributions of both cell cycle genes and B cell activation genes are statistically significantly higher than expected (Wilcoxon rank-sum test; P values for cell cycle genes are 0.0002 and 8 × 10−11 for PC1 and PC2, respectively; P values for B cell activation genes are <2.2 × 10−16 for both PC1 and PC2). The y axes provide the scaled values (×10−3) of their normalized contributions. (C) The UpSet plot visualizes intersecting sets of DE genes at different time points after EBV infection. In the upper part of this panel, 3 Venn diagrams serve as examples to illustrate the syntax of the UpSet diagram. The single dot and dots with interconnecting vertical lines form the intersection matrix. Paired intersections are depicted as black dots; gray dots indicate the sets that are not part of the intersection. Black lines connecting 2 or more black dots indicate which sets form the intersections. In the lower part, groups of DE genes, obtained from the pairwise comparison (performed by DESeq2) (SI Appendix, Materials and Methods) as indicated (Day0_1 = day 0 vs. day 1, etc.) form 7 sets in the UpSet matrix, which are depicted as black horizontal bars with “Number of DE genes” to indicate the set sizes (ranging from 312 to 9,211). Each row represents a matrix set. The columns show the intersections depicted as areas in the schematic Venn diagram as explained in upper part of this panel. The heights of the black columns indicate the cardinality (number of elements) in the different intersections. (D) The bar plots show the time-resolved expression of 2 selected genes, which displayed high expression in noninfected cells or only in cells very early after EBV infection. These genes are among those with the highest loadings on PC1 and/or PC2. The x axis displays days postinfection, whereas the y axis depicts normalized read counts averaged over different biological replicates. (E) Mean expression of 2 genes that contributed to cell cycle progression and B cell activation as function of time after infection. The indicated genes were barely expressed in noninfected cells but became activated at different time points after EBV infection.
First, this analysis shows that the 3 biological replicates of each time point have a very low variability, since they are very close to each other in the PCA plot, while there is a clear separation between samples collected during different days postinfection. As expected, the cell cycle is a major driver of the transcriptional changes, as the average contributions of cell cycle annotated genes to PC1 and PC2 are statistically significantly larger than expected by chance (Fig. 2B). Genes associated with B cell activation and differentiation (as defined in ref. 30) (see SI Appendix, Materials and Methods for details) contribute even more to both PC1 and PC2 (Fig. 2B and Dataset S5), suggesting that the signal seen in the PCA plot also stems from processes intrinsic to the specific biology of the activated B cells.
Furthermore, the PCA separates more the samples obtained from uninfected cells and infected cells of the first 3 d, which formed 4 separate groups (Fig. 2A) compared with samples from day 4 onward, which are close together. This observation suggests that the profoundest variations in gene expression occurred already during the first 3 d of infection in contrast to the phenotypic and metabolic changes that primarily occurred on day 3 and later (Fig. 1). After day 3 the transcriptional changes were more limited and probably restricted to specific cellular processes.
This observation was confirmed when looking at the differentially expressed (DE) genes between consecutive days after EBV infection, which we visualized using the UpSet package (31) (Fig. 2C). The most abundant group of DE genes (n = 9,211) was identified comparing uninfected B cells and cells infected for 24 h, and the second most abundant group of DE genes (n = 6,436) was found when comparing Day1 with Day2. The pairwise comparison between the following days demonstrated a constant decrease in the numbers of DE genes over time (Fig. 2C and SI Appendix, Fig. S7). Similarly, the set analysis also uncovered a gradual time-dependent reduction in the sizes of the sets (i.e., the cardinality or set size describing the number of genes per set) that belonged to unique subsets or set interactions (Fig. 2C). This finding indicates that the number of DE genes decreased substantially at later time points (e.g., Day4_5, Day5_8, and Day8_14), when latency was established. For example, 2,910 genes changed specifically between day 0 and day 1, while only 119 genes were differentially expressed comparing the last 2 time points (day 8 and day 14).
Having identified sets, subsets, and their multiple intersections in a time-resolved approach, we analyzed the expression kinetics of potentially interesting genes selected among those that explain most of the variation along the first 2 PCs (SI Appendix, Fig. S6) and are expected to be functionally relevant in the course of viral infection (Fig. 2 D and E).
CXCR5, a homing receptor, and the transcription factors FOSB were highly expressed in uninfected cells but their expression was lost within 24 to 48 h p.i. CXCR5 orchestrates lymphocyte migration to secondary lymphoid organs and is known to be strongly down-regulated during plasma cell differentiation (32) and EBV infection (33, 34), probably interfering with the localization of activated B cells. FOSB is a proto-oncogene and G0/G1 switch protein, which is induced upon B cell receptor induction (35), but not in our infection model (Fig. 2D). In contrast, the expression of a large group of genes gradually increased during the progression of EBV infection, which is exemplified by CDK4 and CCR2 (Fig. 2E). CDK4 is the catalytic subunit of the cyclin D/CDK4 complex that phosphorylates and inhibits retinoblastoma protein family members and regulates the cell-cycle during the G1/S transition. Therefore, CDK4 is expected to be expressed when the infected cells start to enter the cell cycle on day 3 p.i. (Fig. 1D). CCR2, a C-C chemokine receptor 2, was already reported to be induced in B cells upon EBV infection and in established LCLs, confirming our findings (36). The expression of these 2 genes was induced about 3 to 4 d p.i.
In summary, this analysis revealed global changes in transcription that EBV induces in naïve primary B lymphocytes. The most dramatic transcriptional changes occurred during the first 3 d of infection, affecting the entire biology of the host cell and paving the way to the profound phenotypic and metabolic changes we observed starting from day 3 onwards (Fig. 1).
Gene-Expression Cluster Analysis Identifies Multiple and Specific Biological Processes Regulated by EBV Infection.
Next, we wanted to exploit our time-resolved dataset to classify genes according to their expression dynamics, aiming to pinpoint specific biological processes that EBV drives at the different time points postinfection. To this aim, we followed the computational strategy illustrated in Fig. 3. First, we selected all genes with a dynamic behavior, defined as the union of all genes significantly DE (false-discovery rate < 0.1) with a fold-change greater than 2 between at least 2 time points (SI Appendix, Materials and Methods). We identified 11,178 genes that satisfy these criteria, representing 86% of all genes tested (n = 13,007). On the other hand, only 241 genes (1.85% of all tested genes) did not show any significant change in their expression levels during the course of EBV infection. This finding suggests that EBV globally affects the entire transcriptome in the prelatent phase of infection.
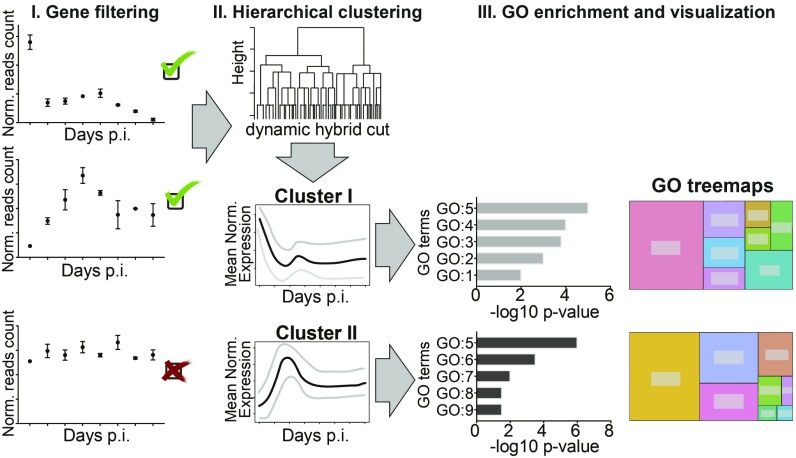
Fig. 3.
Workflow of the bioinformatic analysis and gene clustering. The schematic overview represents the 3 main steps of the bioinformatic analysis: gene filtering, hierarchical clustering, and GO enrichment and visualization performed to identify specific clusters of DE genes and related biological processes. The SI Appendix, Materials and Methods provides the details to this analysis.
The significant DE genes were classified into 6 robust clusters (SI Appendix, Fig. S8), each characterized by different expression dynamics (Fig. 4; see SI Appendix, Materials and Methods for details on gene filtering and clustering). Finally, a gene ontology (GO) enrichment analysis identified biological processes within each cluster of genes (Fig. 4 and SI Appendix, Fig. S9).
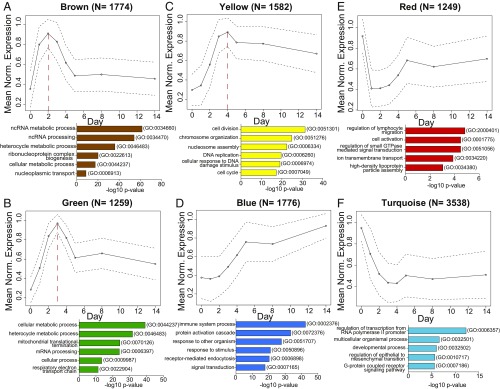
Fig. 4.
Cluster analyses of DE genes in the course of EBV infection define 6 clusters of genes with specific expression dynamics and the corresponding biological processes. Six clusters of genes are shown with their arbitrarily assigned color name codes and mean expression levels. The y axis represents mean normalized expression as a function of time (days postinfection) on the x axis. The numbers on the top of the plots (N) indicate the numbers of the genes that form the cluster. The circles indicate average expression levels, while the dashed lines delineate the SDs. The red, dashed vertical lines mark the expression peak of 3 clusters. All genes within a given specific cluster were used to perform a GO analysis that is summarized in a bar plot below the depicted graphs. The plots contain the top 5 or 6 enriched GO terms with their GO specific IDs (on the right) according to their −log10 P value (x axis). The color of the bars relates to the color name of the clusters. (A) The graph depicts the expression pathway of the brown cluster. This group contains genes that peaked on day 2 p.i., decreased until day 5, and showed stable levels thereafter. The GO analysis identified the most significant biological processes involved in ncRNAs metabolism and processing, heterocycle metabolism, and ribonucleoprotein complex biogenesis. (B) The green cluster peaked on day 3 p.i. Within this cluster, the most frequently found GO terms were cellular metabolic process and mitochondrial biogenesis, such as mitochondrial translational termination or mitochondrial processes linked to energy production as the respiratory electron transport chain. (C) Genes in the yellow cluster peaked on day 4 after EBV infection and decreased slightly thereafter. The analysis pointed to the enrichment of biological processes categorized into cell division, chromosome organization, DNA replication, and cell cycle processes. (D) The blue cluster encompasses genes that constantly but slowly increased during infection. The majority of these identified genes belong to the immune system process, protein activation cascade, or are genes that are involved in response to external stimulus. (E) The group of genes in the red cluster is characterized by an initial loss of expression on the first day postinfection and a rapid recovery until day 5. The GO analysis of this cluster highlighted biological processes related to lymphocyte migration, cell activation, or regulation of small GTPase-mediated signal transduction. (F) The graph displays pathways present in the turquoise cluster. It encompasses genes that are highly expressed in uninfected cells but down-regulated in infected cells until day 4 p.i., when the genes adapted a reduced but stable expression level. GO terms enriched in this group of genes indicated processes of cell development and multicellular organismal process. In this group of genes, very broad GO terms were found.
In particular, we distinguished 2 groups of clusters: 3 clusters had well-defined peaks on day 2, 3, or 4 p.i. (Fig. 4 A–C). In contrast, 3 clusters had peaks either in noninfected cells or 2 wk p.i. (Fig. 4 D–F).
The genes whose expression levels peaked on day 2, 3, or 4 were strongly enriched for very specific GO terms (Fig. 4 A–C and SI Appendix, Fig. S9). In particular, the brown cluster (Fig. 4A) displayed the peak of expression on day 2 p.i. and is enriched with genes involved in mostly noncoding RNA (ncRNA) metabolism and processing, ribonucleoprotein complex biogenesis, or nucleoplasmic transport. The green cluster, with its peak of gene expression on day 3 p.i. (Fig. 4B), included genes that are involved in mitochondrial translation termination, the respiratory electron transport chain, and cellular metabolism, in agreement with our metabolic data in Fig. 1 A and B. Third in the group, the yellow cluster (Fig. 4C), had its peak of expression on day 4 p.i., when the infected cells start to proliferate and undergo the first cell divisions (Fig. 1D and SI Appendix, Fig. S3B). This cluster, with GO terms related to chromosome organization, cell cycle process, cell division, and DNA replication faithfully reflects the phenotypic changes in cell biology we recorded in our initial set of experiments (Fig. 1).
The second group of clusters (Fig. 4 D–F) included the blue cluster (Fig. 4D), which showed a constant increase in gene expression during the course of EBV infection. This cluster is enriched for discrete biological GO processes, including the immune system process, the protein activation cascade, which consists of a sequential series of enzymatic modifications (proteolysis, covalent modifications, binding events, and so forth), and defense responses to other organisms, including viruses. In contrast, the red cluster (Fig. 4E) was characterized by an immediate decrease of cellular gene expression followed by a rapid increase in expression levels and included genes mostly involved in lymphocyte migration, cellular activation, and regulation of GTPase activity. Additionally, the turquoise cluster (Fig. 4F) displayed an immediate loss early after infection and a significant stable reduction of gene expression throughout the observation period. This group of genes was enriched in GO categories, including transcription from RNA polymerase II promoters as well as developmental processes, which might reflect the final status of lymphoblast cells that do not further differentiate in vitro.
The cluster analysis defined very specific dynamic patterns of a plethora of biological processes that are regulated during the ongoing EBV infection. Some of these processes are related to the metabolic and phenotypic changes that we observed, confirming the results from experiments shown in Fig. 1 and providing a list of specific genes associated with those changes (green and yellow clusters). Moreover, additional processes not previously linked to EBV infection (e.g., ncRNA processing, mitochondrial activity, or a group of genes that decrease their expression immediately after infection) were identified and their dynamics characterized. The RNA-seq data were confirmed by testing 7 candidates for their protein-expression dynamics (SI Appendix, Fig. S11A), which mostly followed the kinetics of transcript levels. This is in contrast to noninfected cells, which were cultured in full medium for 3 consecutive days and did not display substantial changes in protein expression. This indicates that the very rapid and dramatic changes observed in the infected cells are not artifacts due to the experimental conditions but are caused by the infection with EBV (SI Appendix, Fig. S11A).
Within a very strict and well-defined time window, the virus completely changes the transcriptome and, consequently, the proteome of resting, naïve B cells, which is followed by specific metabolic and phenotypic changes (Fig. 1). The most prominent transcriptional alterations take place very early during the first 3 d of infection, when many cellular processes are induced or down-regulated. Later during infection cellular genes undergo relatively minor changes to adopt the stable gene-expression profile of the activated and EBV transformed, immortalized B cells.
Regulation of Viral Gene Expression in the Early Phase of EBV Infection.
In parallel with cellular genes, viral genes are expected to show a very dynamic but regulated expression during the prelatent early phase of EBV infection. We knew from published work that initially after infection with EBV, latent genes are induced (9, 12, 18), but also certain lytic genes are expressed, albeit only temporarily. Some of the latter have essential or auxiliary roles supporting the early survival of the infected B cells (10, 37, 38).
We found that the relative abundance of viral genes with respect to cellular genes peaked immediately after infection but started to decline within 48 h (SI Appendix, Fig. S10A). Viral gene expression at the early time points after infection also implicated differences defined with PCA of viral genes (SI Appendix, Fig. S10 B and C). The samples from days 1 and 2 differed the most, and starting from day 3 the samples formed a more homogenous group, indicating their similarities in terms of viral gene expression. However, different viral genes showed different dynamics. In our RNA-seq experiments followed by hierarchical clustering, we identified 3 clusters with viral genes that showed specific gene-expression dynamics and kinetics (SI Appendix, Fig. S10 D and E). Certain viral genes in cluster I, including EBNA2/BYRF1, EBNA-LP, and BHRF1, are characterized by an extraordinarily high expression, especially during the first 2 d p.i. The expression levels of viral genes in cluster II peak around days 2 and 3, including genes such as EBNA3C or BKRF1/EBNA1, which are known to follow the initial EBNA2 expression. In contrast to the majority of DE viral genes, cluster III contained viral latent genes, such as BNLF1/LMP1 and LMP2A, which accelerated their expression starting from day 2 after EBV infection reaching modest, often variable levels of expression 2 wk p.i.
The most dynamic regulation of EBV gene expression took place during the first 2 d p.i. (SI Appendix, Fig. S10B), pointing to a very stringent expression program in the prelatent phase. The specific expression patterns of viral genes strongly correlated with the most prominent changes in the host B cells (Figs. 1 and 2A), consistent with viral genes being the main trigger of B cell activation and transformation in vitro.
Genes in the B Cell Activation Program Are Dynamically Regulated during EBV Infection.
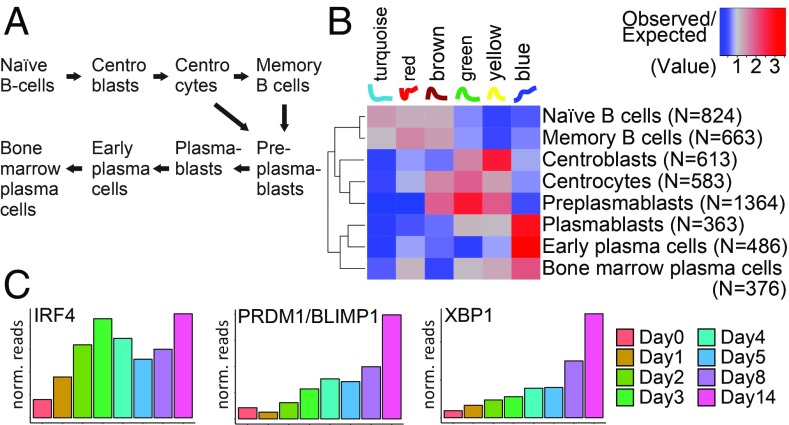
We noticed that a number of DE genes had functions related to the biology of B cells (Fig. 2B). Thus, we exploited our gene clusters (Fig. 4) to classify the expression dynamics of a list of genes that mark the various stages of B cell differentiation (30), from naïve B cells to plasma cells (Fig. 5A). We found that the differentiation markers of any of the B cell types are distributed nonrandomly across the clusters (χ2 tests, P < 2.2 × 10−16 for any group of genes), and each shows enrichments/depletions in clusters corresponding to specific expression dynamics. In particular, as the heatmap in Fig. 5B illustrates, the genes associated with naïve and memory B cells are enriched in the clusters corresponding to early-peaking genes; the genes that characterize the germinal center reaction and entry into plasma cell differentiation (centroblasts, centrocytes, and preplasmablasts) are more likely to reach their maximum between days 2 and 4 p.i.; and finally, the genes expressed in plasmablasts and early plasma cells typically reach their maximum at the end of the infection trajectory. Markers of bone marrow plasma cells are also enriched in the blue cluster (i.e., the cluster of genes peaking at the end of the trajectory), although not as strongly as markers of plasmablasts and early plasma cells (Fig. 5B).
Fig. 5.
Expression dynamics of genes during EBV infection and associated differentiation of mature B cells. (A) Schematic representation of the different stages of mature B cells. For our analysis, we took a list of genes that are most specific to each of the individual stages as reported previously (30). (B) This heatmap shows the ratio between the observed and the expected frequencies of genes marking the stages of B cell differentiation (in rows; number of genes indicated in parentheses) in each of the clusters we identified (in columns; names and corresponding expression patterns at the top). Red indicates enrichment, blue depletion. Markers of naïve and memory B cells tend to peak at the beginning of the infection; genes associated to centroblasts, centrocytes, and preplasmablasts typically peak between day 2 and day 4; markers of plasmablasts and early plasma cells mostly reach their maximum at the end of the infection trajectory, but markers characteristic of bone marrow plasma cells are less prominent at this stable stage. (C) The bar plots represent gene expression of 3 selected genes as normalized read counts averaged over 3 biological replicates (y axis) during the course of EBV infection (x axis). The genes were selected for their known roles in later stages of B cell differentiation. IRF4, PRDM1/BLIMP1, and XBP1 reach maximal levels in preplasmablasts, plasmablasts, and early plasmablast, but are substantially (IRF4) or moderately (PRDM1/BLIMP1, XBP1) down-regulated in bone marrow plasma cells (30).
This result suggests that EBV hijacks the intrinsic B cell activation program and induces an autonomous, germinal center-like reaction in naïve B cells during the first days of infection to generate immortalized lymphoblastoid cells that carry it. However, certain genes that are highly expressed in centroblasts and centrocytes—such as BCL6, LMO2, POLH, and AID/AICDA—are not up-regulated in EBV-infected cells during the early days of EBV’s prelatent phase (http://ebv-b.helmholtz-muenchen.de/), suggesting that EBV infection bypasses certain steps of the genuine germinal center program and minimizes the risk of losing the cells’ B cell receptor expression inherent to somatic hypermutation. At day 14 the transcriptional profile of the cells resembles plasmablasts and early plasma cells that proliferate in vivo, as suggested by the up-regulation of IRF4, PRDM1/BLIMP1, and XBP1 (Fig. 5C) (30), but these cells still express, for example, CD20/MS4A1 and PAX5 (http://ebv-b.helmholtz-muenchen.de/), indicating that they are not terminally differentiated plasma cells. Our protein analyses in SI Appendix, Fig. S11B support this view as certain activation markers—such as CD23, CD27, CD38, or the adhesion molecule CD62L—are induced by viral infection. Noninfected B cells do not show this up-regulation, as expected. The surface marker analysis also revealed that the infected cells do not acquire substantial levels of CD138, a typical plasma cell marker.
More in general, this analysis shows how our dataset can be combined with existing data to generate or test new hypotheses.
Discussion
A Detailed, Time-Resolved Characterization of the Transcriptome of Infected B Cells.
The dramatic metabolic and phenotypical changes we saw in our experiments (Fig. 1 and SI Appendix, Figs. S1–S3) raised questions about the transcriptional events driving them. To address these questions, we performed a time-resolved RNA-seq experiment at an unprecedented resolution: we collected and processed samples daily during the first 5 d of infection, and then at day 8 and day 14 p.i. After removing technical (e.g., sequencing depth) as well as biological (e.g., RNA content) confounding effects through data normalization, we devised and applied a computational strategy consisting of several steps (Fig. 3 and SI Appendix, Fig. S5) that allowed us to: (i) detect genes that change their expression levels during EBV infection; (ii) define clusters of genes sharing similar expression patterns; and (iii) characterize the different steps of the B cell reprogramming by identifying the biological processes involved and their dynamics.
We observed rapid and extensive transcriptional changes that occurred already within 24 h of infection and preceded any metabolic and phenotypical changes. Moreover, we found processes (e.g., ncRNA processing) that have only recently been linked to EBV infection (39). We also analyzed the dynamics of viral genes that are dominated by the immediate maximal expression of 3 viral genes (EBNA2, EBNA-LP, and BHRF1) already on day 1 p.i.
Our time-resolved transcriptional dataset represents an important resource for the entire community. Hence, to facilitate access to it, we created a website (http://ebv-b.helmholtz-muenchen.de/) where all our processed data, including the lists of genes in each cluster, can be downloaded and explored. Via this user-friendly web-based interface, the dynamics of individual cellular and viral genes can also be tracked starting from uninfected B cells, through the prelatent phase of EBV infection, until the establishment of LCLs 2 wk p.i. Moreover, all raw data are deposited and publicly available (SI Appendix, Materials and Methods).
Overall, our data indicate that EBV triggers a plethora of substantial changes in the biology of the entire host cell. In a well-defined and short interval of time, the virus drastically overturns the transcriptional program of resting and quiescent B cells and reprograms a multitude of different biological processes and pathways.
B and T Cells Differ in Their Activation Kinetics.
Alterations in the metabolism of immune cells, collectively termed immunometabolism (40), trigger important changes in lymphocytes, including their activation, proliferation, and eventual differentiation into different subtypes of multipotent precursor, effector, or memory cells. After encountering antigens, naïve and quiescent T cells undergo activation and start cell divisions within 24 h, which is in stark contrast to B lymphocytes infected with EBV, as demonstrated in this paper. The differences between the 2 classes of B and T lymphocytes might stem from the fact that T cells are programmed to react immediately upon encountering antigens, become activated within minutes, and start dividing every 4 to 6 h (41). To support this acute process, the lymphocytes have to adapt their metabolism in a very short time to fulfill the needs of increased energy and biomolecules in different microenvironments and different parts of the entire organism. These dynamic changes in basic bioenergetic processes enable the cells to modulate their catalytic and anabolic reactions in a very rapid and effective manner (42).
Upon activation, the naïve T cells rely mostly on oxidative phosphorylation production of energy to realize the basic need of energy to promote cell survival. The activated T cells are characterized by an elevated glycolysis and glutaminolysis, which provide the cells not only with ATP but also with the most important precursors for newly made biomolecules, such as nucleic acids, amino acids, lipids, and so forth (43–45). Glucose as a primary fuel has an important role during T cell activation, as well as subsequent differentiation since blocking of glucose catabolism inhibits T effector cell function in vitro and in vivo. Activated CD8+ T cells shift from fatty acid oxidation to aerobic glycolysis and glutamine oxidation to support their differentiation into cytotoxic T cells and generation of memory CD8+ T cells (46).
Similarly, upon CD40/IL-4 stimulation, B cells synthesize and accumulate macromolecules and gain in volume. B cell receptor (BCR) cross-linking, as well as IL-4 and Toll-like receptor-mediated activation, increase the uptake of glucose and aerobic glycolysis concomitant with the up-regulation of glycolytic enzymes (47, 48). Infection of primary B cells with EBV leads to cell activation, which mimics the physiological signaling, such as BCR signaling pathway mediated by LMP2A (49) or CD40 stimulation supported by LMP1 (50). Our results confirm that EBV infection also causes substantial changes in cell growth, cell cycle, and cellular content of macromolecules, such as protein and RNA (Fig. 1). Activating adjustments in the cellular phenotype increase the demand for energy and biomolecules to support cellular functions and cell divisions. The uptake of glucose increased after a 3-d-long latency period and started on day 4 p.i. (Fig. 1B), indicating the up-regulation of glycolysis only before the highly proliferative stage of the infected B cells on days 5 and 6 p.i. Thus, the increase of glucose uptake and the activation of immunometabolic functions are late events in B cells induced by EBV infection, which might not differ from physiological activating signals, such as antigen encounter of these immune cells in vivo. Similarly, mitochondrial activity increased starting from day 4 p.i. (Fig. 1A), when the cells enter the cell cycle, replicate DNA, and start dividing.
T cell activation induces many different signaling pathways that control diverse metabolism-associated factors, such as Myc, HIF-1α, mammalian target of rapamycin, or Gsk3. The proto-oncogene Myc is known to up-regulate glucose, glutamine, and polyamine metabolism during T cell activation, similar to the up-regulated metabolic pathways in cancer cells (51–53). Antigen-specific immune responses by B cells also affect their metabolic program and activate HIF-1α and Myc. Myc is especially crucial for the induction of the glucose transporter (GLUT1) during B cell activation (46) and enhances the expression of hexokinase 2 (HK2) and pyruvate dehydrogenase kinase-1 in Burkitt lymphoma cells (54). Oncogenic Myc levels increase the mitochondrial mass and oxygen consumption since several Myc targets are involved in mitochondrial biogenesis and functions (54, 55). The main driver of B cell transformation during EBV infection is EBNA2, which directly targets MYC and promotes its expression (21, 56). In fact, transcript levels of EBNA2 reach maximal levels 1 d after EBV infection, followed by extreme levels of Myc on the next day (this study and ref. 26). Thus, the very early high levels of Myc seem to be the main trigger for B cell activation and reprogramming in the prelatent phase of EBV-infected cells.
Dynamic and Comprehensive Adjustments in the Host Cell Transcriptome Early after Infection.
To our knowledge, only a few publications describe global cellular changes after viral infection, focusing on the expression and regulation of protein-coding as well as ncRNA genes. A recent report studied the host transcriptome and proteome during Kaposi sarcoma-associated herpesvirus (KSHV) infection and identified global changes in the cellular networks that are essential for virus latency (57). The authors identified alterations in ∼17% of the host transcriptome and 13% of the cellular proteome that also includes dramatic changes in the phosphoproteome 48 h after KSHV infection. EBV is infecting a cell not yet in the proliferation cycle, whereas Sychev et al. (57), in their KSHV infection model, used Tert-immortalized, proliferating microvascular endothelial cells. Similar to KSHV, EBV establishes a latent infection, but the impact on the host cells is much more dramatic. EBV reprograms the B cells and we found statistically significant changes for all but 241 cellular genes corresponding to 1.85% of all 13,007 tested genes. In other words, our analysis suggests that a large proportion of cellular genes are affected by EBV.
This view is supported by our time-resolved PCA (Fig. 2A). It documents a well-defined status of the quiescent naïve B cells, which immediately changes upon EBV infection as a direct response to virus-mediated activating signals. Noninfected B cells express a characteristic set of genes that probably make up the signature of these quiescent naïve B lymphocytes in vivo. This signature may support the cells in G0 and minimize their demand for energy, but upon infection many cellular genes show specific, time-regulated changes in their expression. Our bioinformatic work flow identified 6 distinct clusters (Fig. 4), which group them according to their expression pattern during infection and confirm our initial observations regarding cellular phenotypes and immunometabolism of the infected cells (Fig. 1).
The UpSet tool made it possible to identify and visualize the number of genes contained in unique as well as intersecting sets of DE genes between different days postinfection. Using this tool (Fig. 2C), we identified the most dynamic changes in host gene expression to occur very early after infection within 24 h, followed by considerable additional alterations within the next 48 h, whereas later adjustments of the cells’ transcriptomes were relatively minor. Noninfected cells and cells infected for 24 h formed a unique set of DE genes that does not intersect with any other set later during infection. Overall, our RNA-seq data analyses revealed that the most dynamic adjustments of gene expression occurred within the first 4 d p.i. When the infection progresses further EBV might need only to fine-tune the expression profile of the host cell to adjust the cell for the ensuing latent phase of infection.
Viral Gene Expression Precedes the Time-Controlled Alterations in the Cellular Transcriptome.
Scattered reports in the literature describe single viral genes and their early expression in B lymphocytes (ref. 18 and references therein). The results from our study mostly confirm the known or presumed kinetics of in vitro EBV gene expression in the prelatent phase (SI Appendix, Fig. S10). We identified the immediate and maximal induction of EBNA2, EBNA-LP, and BHRF1 on day 1 p.i., followed by a considerable reduction of their expression levels later. The expression of the EBNA3s peaks at day 2 p.i., whereas viral genes, including LMP1 and LMP2s, displayed a gradual increase in expression starting from day 3 p.i. We also noticed that several viral genes, defined as lytic genes, are moderately expressed immediately after infection, as expected (12), but become rapidly down-regulated starting already on the day 2 p.i. (SI Appendix, Fig. S10 D and E).
The profile of viral gene expression strongly correlates with the most prominent changes in the host B cell but highlights EBNA2 as the main driver, together with EBNA-LP and BHRF1. EBNA2’s immediate and high expression induces MYC, which presumably governs most subsequent alterations in the host B cell. This hypothesis is supported by an accompanying paper (26). In a systematic genetic approach with individual viral genes, we identify EBNA2 to be the only viral gene that is indispensable for B cell activation, cell cycle entry, and proliferation. We also noticed that not all naïve B lymphocytes uniformly respond to EBV infection, suggesting that EBV might specifically target nonidentified B cell subpopulations or is incapable of activating all cells because viral or cellular gene expression is inconsistent. These open issues can only be addressed by single-cell experiments (e.g., single-cell RNA-seq), which will likely unveil how EBV reprograms individual target cells.
EBV Infection of Naïve B Cells Induces a Germinal Center-Like Differentiation Program to Yield Lymphoblastoid Cells Resembling Plasma Blasts and Early Plasma Cells.
The emergence of LCLs from primary and quiescent B lymphocytes after EBV infection is a fascinating phenomenon and has been studied by many, but only recently with a focus on EBV’s prelatent phase (17, 18; see ref. 39 for a recent review). In Fig. 5 we used our newly acquired transcriptomic data to investigate the expression dynamics of a list of genes that mark the various stages of mature B cells (30). The heatmap in Fig. 5B suggests that EBV infection induces a transient differentiation program in naïve B lymphocytes in vitro, with transcriptomic features partially reminiscent of centroblasts, centrocytes, and preplasmablasts within a few days postinfection. Subsequently, cells emerge that resemble plasmablasts and early plasma cells (Fig. 5B). In fact, established lymphoblastoid cells release substantial amounts of immunoglobulins (58), which is in line with our correlative study here. It is noteworthy that at least in vitro EBV does not permit the terminal differentiation of these cells to become bone marrow plasma cells, nor does it allow their escape to adopt a nonproliferative state reminiscent of memory B cells, as indicated by the heat map in Fig. 5B. It thus appears as if EBV’s latency III program “locks” the infected B cells in a stage of B cell differentiation in vitro that is only transient in nature in vivo.
Materials and Methods
Detailed descriptions of the materials and methods are provided in SI Appendix and include the following: eukaryotic cell lines; virus supernatants from the HEK293 2089 cell line; collection and quantitation of virus supernatant from the B95-8 cell line; preparation of B cells from adenoid tissue; purification of B cells on MACS columns; primary B cells infection with EBV; TMRE and Annexin V detection; glucose uptake using 2-NBDG; FACS sorting for naïve B cells; analysis of the cells’ diameter; cell cycle analysis; calculation of the cell DI; Western blot protein quantification with the Wes platform technology; FACS staining for surface B cell markers; preparation of cell lysates; sample collection for time-course RNA-seq experiments; RNA isolation; library preparation and sequencing; and bioinformatic analysis, including transcript quantification by Salmon, quality control and data normalization, identification of DE genes, gene clustering, GO analysis, heatmaps of log10-transformed expression data, PCA of phenotypic data, PCA with viral genes, clustering of viral genes, and data availability.
Supplementary Material
Acknowledgments
We thank Christine Göbel and Stefanie Voigt (Munich) for precious experimental support; Ursula Zimber-Strobl (Munich) for essential advice on human B cell differentiation; and Hannah Busen (Munich) for advice and support during the initial phase of bioinformatic analyses. This work was financially supported by Deutsche Forschungsgemeinschaft Grants SFB1064/TP A13 and SFB-TR36/TP A04 (to W.H.); Deutsche Krebshilfe Grant 70112875 (to W.H.); and National Cancer Institute Grant CA70723 (to W.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The raw RNA-seq data have been deposited in the European Bioinformatics Institute, ArrayExpress database, https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-7805/ (accession no. E-MTAB-7805) Processed RNA-seq data can be downloaded from the following website http://ebv-b.helmholtz-muenchen.de/, which also provides a user-friendly R Shiny App to visualize and explore the data and individual genes.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901314116/-/DCSupplemental.
References
- 1.Epstein M. A., Achong B. G., Barr Y. M., Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet 1, 702–703 (1964). [DOI] [PubMed] [Google Scholar]
- 2.Pulvertaft J. V., Cytology of Burkitt’s tumour (African Lymphoma). Lancet 1, 238–240 (1964). [DOI] [PubMed] [Google Scholar]
- 3.Pope J. H., Establishment of cell lines from peripheral leucocytes in infectious mononucleosis. Nature 216, 810–811 (1967). [DOI] [PubMed] [Google Scholar]
- 4.Henle W., Evidence for viruses in acute leukemia and Burkitt’s tumor. Cancer 21, 580–586 (1968). [DOI] [PubMed] [Google Scholar]
- 5.Shannon-Lowe C., Rickinson A. B., Bell A. I., Epstein-Barr virus-associated lymphomas. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young L. S., Yap L. F., Murray P. G., Epstein-Barr virus: More than 50 years old and still providing surprises. Nat. Rev. Cancer 16, 789–802 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Young L. S., Rickinson A. B., Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4, 757–768 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Woellmer A., Hammerschmidt W., Epstein-Barr virus and host cell methylation: Regulation of latency, replication and virus reactivation. Curr. Opin. Virol. 3, 260–265 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jochum S., Ruiss R., Moosmann A., Hammerschmidt W., Zeidler R., RNAs in Epstein-Barr virions control early steps of infection. Proc. Natl. Acad. Sci. U.S.A. 109, E1396–E1404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altmann M., Hammerschmidt W., Epstein-Barr virus provides a new paradigm: A requirement for the immediate inhibition of apoptosis. PLoS Biol. 3, e404 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalla M., Göbel C., Hammerschmidt W., The lytic phase of Epstein-Barr virus requires a viral genome with 5-methylcytosine residues in CpG sites. J. Virol. 86, 447–458 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalla M., Hammerschmidt W., Human B cells on their route to latent infection—Early but transient expression of lytic genes of Epstein-Barr virus. Eur. J. Cell Biol. 91, 65–69 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Zeidler R., et al. , Downregulation of TAP1 in B lymphocytes by cellular and Epstein-Barr virus-encoded interleukin-10. Blood 90, 2390–2397 (1997). [PubMed] [Google Scholar]
- 14.Hislop A. D., et al. , A CD8+ T cell immune evasion protein specific to Epstein-Barr virus and its close relatives in Old World primates. J. Exp. Med. 204, 1863–1873 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jochum S., Moosmann A., Lang S., Hammerschmidt W., Zeidler R., The EBV immunoevasins vIL-10 and BNLF2a protect newly infected B cells from immune recognition and elimination. PLoS Pathog. 8, e1002704 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albanese M., Tagawa T., Buschle A., Hammerschmidt W., MicroRNAs of Epstein-Barr virus control innate and adaptive anti-viral immunity. J. Virol. 91, e01667-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikitin P. A., et al. , An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe 8, 510–522 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price A. M., Luftig M. A., Dynamic Epstein-Barr virus gene expression on the path to B-cell transformation. Adv. Virus Res. 88, 279–313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price A. M., et al. , Analysis of Epstein-Barr virus-regulated host gene expression changes through primary B-cell outgrowth reveals delayed kinetics of latent membrane protein 1-mediated NF-κB activation. J. Virol. 86, 11096–11106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitzsimmons L., Kelly G. L., EBV and apoptosis: The viral master regulator of cell fate? Viruses 9, E339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaiser C., et al. , The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J. Virol. 73, 4481–4484 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao B., et al. , Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc. Natl. Acad. Sci. U.S.A. 108, 14902–14907 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu F., et al. , EBNA2 drives formation of new chromosome binding sites and target genes for B-cell master regulatory transcription factors RBP-jκ and EBF1. PLoS Pathog. 12, e1005339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang S., et al. , Epstein-Barr virus nuclear antigen 3C binds to BATF/IRF4 or SPI1/IRF4 composite sites and recruits Sin3A to repress CDKN2A. Proc. Natl. Acad. Sci. U.S.A. 111, 421–426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strobl L. J., et al. , Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP-J kappa. Immunobiology 198, 299–306 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Pich D., et al. , The first days in the life of naïve human B-lymphocytes infected with Epstein-Barr virus. biorXiv: 10.1101/666297v1 (11 June 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinbrück L., et al. , K1 and K15 of Kaposi’s sarcoma-associated herpesvirus are partial functional homologues of latent membrane protein 2A of Epstein-Barr virus. J. Virol. 89, 7248–7261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roederer M., Interpretation of cellular proliferation data: Avoid the panglossian. Cytometry A 79, 95–101 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Delecluse H. J., Hilsendegen T., Pich D., Zeidler R., Hammerschmidt W., Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. U.S.A. 95, 8245–8250 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kassambara A., et al. , GenomicScape: An easy-to-use web tool for gene expression data analysis. Application to investigate the molecular events in the differentiation of B cells into plasma cells. PLoS Comput. Biol. 11, e1004077 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lex A., Gehlenborg N., Strobelt H., Vuillemot R., Pfister H., UpSet: Visualization of intersecting sets. IEEE Trans. Vis. Comput. Graph. 20, 1983–1992 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jourdan M., et al. , An in vitro model of differentiation of memory B cells into plasmablasts and plasma cells including detailed phenotypic and molecular characterization. Blood 114, 5173–5181 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakayama T., et al. , Human B cells immortalized with Epstein-Barr virus upregulate CCR6 and CCR10 and downregulate CXCR4 and CXCR5. J. Virol. 76, 3072–3077 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehlin-Henriksson B., et al. , Changes in chemokines and chemokine receptor expression on tonsillar B cells upon Epstein-Barr virus infection. Immunology 127, 549–557 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin Q., Wang X., McBride J., Fewell C., Flemington E., B-cell receptor activation induces BIC/miR-155 expression through a conserved AP-1 element. J. Biol. Chem. 283, 2654–2662 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kholodnyuk I., Rudevica Z., Leonciks A., Ehlin-Henriksson B., Kashuba E., Expression of the chemokine receptors CCR1 and CCR2B is up-regulated in peripheral blood B cells upon EBV infection and in established lymphoblastoid cell lines. Virology 512, 1–7 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Wen W., et al. , Epstein-Barr virus BZLF1 gene, a switch from latency to lytic infection, is expressed as an immediate-early gene after primary infection of B lymphocytes. J. Virol. 81, 1037–1042 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalla M., Schmeinck A., Bergbauer M., Pich D., Hammerschmidt W., AP-1 homolog BZLF1 of Epstein-Barr virus has two essential functions dependent on the epigenetic state of the viral genome. Proc. Natl. Acad. Sci. U.S.A. 107, 850–855 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C., et al. , RNA sequencing analyses of gene expression during Epstein-Barr virus infection of primary B lymphocytes. J. Virol. 93, e00226-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murray P. J., Rathmell J., Pearce E., SnapShot: Immunometabolism. Cell Metab. 22, 190–190.e1 (2015). [DOI] [PubMed] [Google Scholar]
- 41.van Stipdonk M. J., et al. , Dynamic programming of CD8+ T lymphocyte responses. Nat. Immunol. 4, 361–365 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Slack M., Wang T., Wang R., T cell metabolic reprogramming and plasticity. Mol. Immunol. 68, 507–512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang R., Green D. R., Metabolic checkpoints in activated T cells. Nat. Immunol. 13, 907–915 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Buck M. D., O’Sullivan D., Pearce E. L., T cell metabolism drives immunity. J. Exp. Med. 212, 1345–1360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Almeida L., Lochner M., Berod L., Sparwasser T., Metabolic pathways in T cell activation and lineage differentiation. Semin. Immunol. 28, 514–524 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Cao Y., Rathmell J. C., Macintyre A. N., Metabolic reprogramming towards aerobic glycolysis correlates with greater proliferative ability and resistance to metabolic inhibition in CD8 versus CD4 T cells. PLoS One 9, e104104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doughty C. A., et al. , Antigen receptor-mediated changes in glucose metabolism in B lymphocytes: Role of phosphatidylinositol 3-kinase signaling in the glycolytic control of growth. Blood 107, 4458–4465 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dufort F. J., et al. , Cutting edge: IL-4-mediated protection of primary B lymphocytes from apoptosis via Stat6-dependent regulation of glycolytic metabolism. J. Immunol. 179, 4953–4957 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Mancao C., Hammerschmidt W., Epstein-Barr virus latent membrane protein 2A is a B-cell receptor mimic and essential for B-cell survival. Blood 110, 3715–3721 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kieser A., Sterz K. R., The latent membrane protein 1 (LMP1). Curr. Top. Microbiol. Immunol. 391, 119–149 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Rathmell J. C., T cell Myc-tabolism. Immunity 35, 845–846 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang R., et al. , The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stine Z. E., Walton Z. E., Altman B. J., Hsieh A. L., Dang C. V., MYC, metabolism, and cancer. Cancer Discov. 5, 1024–1039 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim J., Lee J. H., Iyer V. R., Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS One 3, e1798 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scarpulla R. C., Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 88, 611–638 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Liang J., et al. , Epstein-Barr virus super-enhancer eRNAs are essential for MYC oncogene expression and lymphoblast proliferation. Proc. Natl. Acad. Sci. U.S.A. 113, 14121–14126 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sychev Z. E., et al. , Integrated systems biology analysis of KSHV latent infection reveals viral induction and reliance on peroxisome mediated lipid metabolism. PLoS Pathog. 13, e1006256 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee D. Y., Sugden B., The LMP1 oncogene of EBV activates PERK and the unfolded protein response to drive its own synthesis. Blood 111, 2280–2289 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.