Significance
The protein hormone florigen has dual functions in flowering plants: the systemic boosting of flowering in the apical meristems and growth attenuation in other vegetative meristems. Similar to global changes that occur during transition to maturity in mammals, the transition to flowering dictates a moderation of vegetative growth and a redistribution of physical loads, signals, and resources. Here we show that florigen triggers a flowering-independent developmental program, which culminates in the acceleration of secondary cell wall biogenesis in tomato stems, thereby promoting vascular maturation and adaptation of the shoot system to the emerging needs of the reproductive phase. By synchronizing mutually independent flowering and secondary cell wall biogenesis, the systemic florigen functions as a coordinator and great communicator of the reproductive phase.
Keywords: vegetative functions, SCWB, florigen, tomato, cellular targets
Abstract
Florigen, a proteinaceous hormone, functions as a universal long-range promoter of flowering and concurrently as a generic growth-attenuating hormone across leaf and stem meristems. In flowering plants, the transition from the vegetative phase to the reproductive phase entails the orchestration of new growth coordinates and a global redistribution of resources, signals, and mechanical loads among organs. However, the ultimate cellular processes governing the adaptation of the shoot system to reproduction remain unknown. We hypothesized that if the mechanism for floral induction is universal, then the cellular metabolic mechanisms underlying the conditioning of the shoot system for reproduction would also be universal and may be best regulated by florigen itself. To understand the cellular basis for the vegetative functions of florigen, we explored the radial expansion of tomato stems. RNA-Seq and complementary genetic and histological studies revealed that florigen of endogenous, mobile, or induced origins accelerates the transcription network navigating secondary cell wall biogenesis as a unit, promoting vascular maturation and thereby adapting the shoot system to the developmental needs of the ensuing reproductive phase it had originally set into motion. We then demonstrated that a remarkably stable and broadly distributed florigen promotes MADS and MIF genes, which in turn regulate the rate of vascular maturation and radial expansion of stems irrespective of flowering or florigen level. The dual acceleration of flowering and vascular maturation by florigen provides a paradigm for coordinated regulation of independent global developmental programs.
Nine decades ago, innovative grafting experiments firmly established a hypothetical signal, dubbed florigen, produced in leaves and transported to the apical meristems as a universal systemic inducer of flowering (1, 2). However, the discovery of flowering pathways in Arabidopsis, which predominantly converge on the gene FT (3, 4), practically sent the elusive florigen into oblivion. A surprising twist in the odyssey of florigen emerged with the establishment in tomato of a 1-to-1 genetic relationship between florigen and the SINGLE FLOWER TRUSS (SFT) gene, an ortholog of FT (5–8). This solitary genetic origin, unique among plant hormones, instituted florigen as a protein hormone universally encoded by FT orthologs, thus resolving the apparently conflicting paradigms.
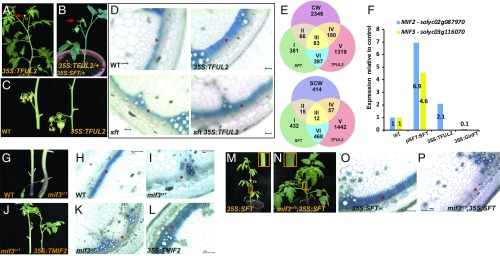
Floral induction, in addition to creating reproductive organs, transforms the shoot system from the vegetative phase to the reproductive phase. The reproductive phase entails the reprograming of metabolic networks, moderation of vegetative growth, and amendment of the global source–sink relationships. Florigen, in addition to flowering, promotes simplification of compound leaves (SI Appendix, Fig. S1) and radial contraction of stems (Fig. 1). Conversely, a lack of florigen promotes leaf complexity and radial expansion of stems (5, 9) (Fig. 1D and SI Appendix, Fig. S1). Significantly, in all its vegetative roles in tomato, florigen impacts the growth coordinates of organs but never their homeotic fate. In addition, in the shoot apical meristems (SAMs), florigen regulates the timing of flowering but does not alter the designated homeotic fate senso stricto. Other plant species behave similarly. Induction of florigenic genes or, alternatively, annulment of their antagonists was shown to regulate tuberization in potato, leaf size in Arabidopsis and tobacco, cluster shape in grapes, bud setting in aspen, and more (10, 11). Flowering in tomato is synonymous with termination (9). Since all florigen-dependent vegetative alterations may be similarly attributed to growth termination, instead of being designated as a flowering hormone, florigen is recognized as a generic regulator of growth and termination (9, 12) (Fig. 1 B–D). However, the links between flowering and reprogramming of the shoot system during floral transition, as well as the genetic and molecular bases for the growth termination functions of florigen, remain unknown.
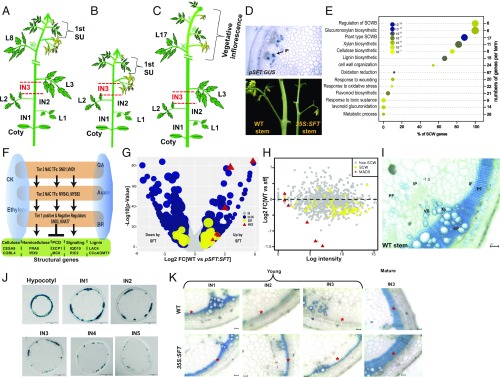
Fig. 1.
Florigen stimulates SCWB to restrict lateral expansion of tomato stems. (A–C) Schematic representations of major growth habits in tomato. (A) WT tomato plant. L, leaf; In, internode; SU, sympodial unit. (B) 35S:SFT induces premature flowering. (C) sft induces late flowering, suppresses sympodial growth, and forms single flower truss inflorescences. (D) pSFT:GUS (magnification: 10×) expressed in external and internal tomato stem phloem. (E) The SCWB transcription network is regulated amass by florigen. Shown is a dot plot classifying the top 14 GO terms (biological processes) enriched in pSFT:SFT vs. WT DEGs (Padj < 0.1; |FC >2|). The GO term order is proportional to the percentage of SCW genes in it. Circle size is proportional to fold change, color designates significance, and the y-axis on the right represents gene number per term. (F) Scheme of the SCWB transcription network consisting mainly of 3 or 4 interlocked tiers of NAC and MYB TFs and their downstream metabolic activities (15, 16, 24–26). All major hormones play important regulatory roles in formation of the SCW. (G) Florigen stimulates the SCWB gene network and MADS TFs. Shown is a volcano plot comparing WT and pSFT:SFT. A total of 56 SCWB genes were up-regulated, and 15 were down-regulated. The y-axis, which is proportional to significance, is plotted vs. fold change (log 2 scale). Circle size is proportional to expression level. Color-coded dots represent the classes of DEGs (Padj ≤ 0.1; |FC >2|). Blue, non-SCWB genes; yellow, SCWB genes; red triangles, MADS TFs. (H) The SCWB gene network is down-regulated in sft stems. The scatterplot shows the distribution of the 981 DEGs between WT and pSFT:SFT in the sft vs. WT contrast. Most of the 981 DEGs are scattered around zero, whereas MADS genes and SCWB genes show a significant down-regulation (P = 8.1*10–14, t test). (I) The principle vascular elements in a mature tomato stem. VB, vascular bundle; Xy, xylem; PF, phloem fibers; EP, external phloem; IP, internal phloem; IF, interfascicular xylem fibers. TBO staining for lignin. (Scale bars: 100 mm.) (J) GUS staining in cross-sections of 5-wk-old stems expressing the tomato pCESA4:GUS gene. An earlier developmental stage is shown for comparison in SI Appendix, Fig. S4. Note: At early stages the patterns of CESA activity and TBO staining for lignin overlap. During maturation, CESA activity diminishes, but lignin accumulates. (Scale bars: 2 mm, except for IN5, which is 1 mm.) (K) Florigen accelerates SG and IF xylem differentiation in tomato stems. Shown are cross-sections of 4-wk-old (young) and 40-d-old (mature) WT and 35S:SFT plants. TBO staining for lignin. (Scale bars: 100 mm.) *IF zones.
We hypothesized that if the mechanism for floral induction is universal, then the cellular metabolic changes underlying the global shift of vegetative organs to the reproduction phase would also be universal, similarly regulated long-range, and likely represented by the vegetative functions of florigen. We thus opted to search for entire cellular metabolic systems that, when modified by florigen, may eventually impose a tunable growth balance without changing homeotic fates. Specifically, we searched for cellular metabolic programs rather than transcription factors (TFs), because the latter are not the ultimate building blocks of cellular differentiation. We chose the stems of tomato as a model system because their age-dependent radial contraction is the single most robust pleiotropic effect of high florigen in tomato (Fig. 1 and ref. 9). Stems consist of a limited number of cell types, and their differentiating zones are entrenched in hierarchically demarcated developmental territories (13, 14). We identified the acceleration of the transcription network navigating secondary cell wall (SCW) biogenesis (SCWB) (15, 16) as an ultimate end metabolic network coopted by a plant hormone to regulate its fundamental morphogenetic function. SCWB encompasses a hierarchical transcription and metabolic network that integrates the biosynthesis of cellulose, lignin, and hemicellulose and guides their deposition onto the inner layers of the primary and SCWs (17–20) (Fig. 1F). This novel finding is accompanied by evidence showing that although flowering and SCWB are coordinately accelerated by florigen, they are not a consequence of each another.
Results
Florigen Targets the Rate of SCWB in Tomato Stems.
In tomato, high florigen levels stimulate precocious primary flowering, while the pace of flowering during the sympodial phase is only marginally affected. In contrast, the radial contraction of stems is accelerated with age in both primary and sympodial shoots, representing the single most robust pleiotropic effect of florigen (5, 9) (Fig. 1B). Florigen is produced exclusively in the companion cells of the phloem (2, 21, 22) and, as shown here, in both the external and internal phloem rings that characterize Solanaceae and Cucurbitaceae (23) (SI Appendix, Fig. S1). Importantly, SFT is the sole contributor of florigen in cultivated tomato (10). To unveil cellular systems targeted by florigen in stems, we conducted an exploratory RNA profiling from the third internodes of 5-wk-old WT, sft, and pSFT:SFT tomato plants, which provide a reproducible, easily accessible experimental resource. Comparison of the WT and pSFT:SFT transcriptomes (Padj < 0.1 and |FC >2|) revealed 981 differentially expressed genes (DEGs) (Dataset S1) significantly enriched for the metabolic and regulatory genes involved in SCWB (15, 16, 24–26) (Fig. 1 E and G). Consistent with this enrichment, 12 of the 69 DE TFs encode the tomato homologs of the MYB and NAC master regulators of SCWB (16, 27, 28). Surprisingly, 6 MADS genes, including 4 members of the FUL clade (Fig. 1G and SI Appendix, Fig. S1), known to mediate FT-dependent floral transition (29, 30), were also up-regulated in pSFT:SFT stems. Notably, other genes involved in flowering, such as orthologs of LEAFY or SEPALLATA, were not altered by florigen.
If SCWB is a bona fide target of florigen, then SCWB-related transcripts should be underrepresented in sft. While the expression of TFUL (tomato FUL homologs)-like genes, which were activated in pSFT:SFT stems, was reduced in sft compared with WT (SI Appendix, Table S1 and Dataset S1), only a weak impact on SCWB genes was detected. Nevertheless, the majority of SCWB genes up-regulated by pSFT:SFT were expressed in sft stems at levels below those seen in the WT. This broad response was consistent and significant (Fig. 1H).
Accelerated SCWB Is Correlated with Precocious Vascular Maturation.
To explore the anatomic manifestations of the enriched SCWB genes, we followed secondary growth (SG) and deposition of SCW along WT and SFT-overexpressing stems. The pattern of SCW deposition along tomato shoots follows the stereotypical basipetal gradient of eudicot plants (31), as demonstrated here by the activity of the tomato pCESA4 reporter (Fig. 1J). The bulk of the SCW is deposited onto the walls of the fascicular and interfascicular (IF) cells produced during SG by the cambium and are fated to form the vessels and fibers of the secondary xylem and the primary phloem fibers (18, 19) (Fig. 1I). In practice, we followed the formation and progressive lignification of the IF xylem fibers generated by the vascular cambium (Fig. 1I). At 25 d postgermination (DPG), the first lignified IF xylem layers in the basal internodes of WT stems had just differentiated, while several lignified IF layers had already formed in the same internodes of the SFT-overexpressing plants (Fig. 1K and SI Appendix, Fig. S2). These features correlated with the flowering status (30) of the tested plants: 9 leaves and stage 3 inflorescences in WT plants vs. 3 leaves and a fully blossomed inflorescence in SFT-overexpressing plants.
However, although vascular maturation correlates with flowering in WT plants, it can also proceed independently of flowering. The differentiation of lignified xylem layers in the late-flowering sft plants or in the never-flowering uf sft double-mutant genotype was only marginally different from that seen in WT plants. Moreover, at 20 DPG, no lignified IF xylem rings were evident in the basal internodes of SFTox plants bearing stage 10 primary inflorescences (Fig. 1 and SI Appendix, Fig. S3). The precocious activation of SG and SCWB by florigen may fulfill the need for a comprehensive reallocation of signals and metabolic resources prescribed by the shift to the reproductive phase.
Plant development dictates that all metabolic components for SCW deposition function as a single operating unit. The concerted activation of genes representing both regulatory and biosynthesis functions suggests that florigen regulates SCWB as a unit, directly or indirectly. To test this possibility, we assembled a core list of 498 SCW genes, 468 of which are expressed in tomato stems (Materials and Methods and Dataset S3) and found that 68 of them (P = 4.15 × 10−24, hypergeometric test) were regulated by florigen. Thus, if SCWB underlies florigen-dependent radial contraction of the stems, then it would be expected to respond as a unit to other florigen sources, be it graft-transmissible or transient induction.
Graft-Transmissible Florigen Is Sufficient to Promote SCWB in Recipient Stems.
The acceleration of SCWB in the stems by high SFT reflects the combined impact of endogenous, stem-borne florigen and of a mobile florigen (m-florigen) imported from the leaves. To identify cellular systems regulated by the m-florigen alone, we studied 5 graft combinations that report the response of sft and SFT recipient genotypes to m-florigen contributed by WT and 35S:SFT donors (Fig. 2 A and B and Materials and Methods). The inherent variation associated with the graft assemblies, the cumulative and pleiotropic functions of florigen, the efficiency of mobility, and the dynamics of vascular maturation (26, 29, 31) delineated low transcriptional responses and modest expectations. A group of 593 genes, differing significantly (Padj ≤ 0.1; |FC ≥2|) between the WT and sft recipients of the homografts and the corresponding 3 recipients of the heterografts were identified (Fig. 2C and Dataset S2). Enriched Gene Ontology (GO) terms for the 593 DEGs exclusively represented SCW functions (SI Appendix, Fig. S2), and hierarchical clustering linked almost all DE SCWB genes into 1 cluster (red in Fig. 3C). The clustering of the master regulators of SCWB with their target metabolic genes and of the “floral” MADS genes with the SCW network (Fig. 2D) suggests that m-florigen impacts SCWB as a network, and that this role, as in flowering (32), is mediated in part by the TFUL genes (Fig. 2 and SI Appendix, Fig. S2). Analysis of the relationship between the absolute expression levels of the representative genes depicted in Fig. 2D suggests that all genes are regulated in all genotypes, but that m-florigen is significantly more effective in a functional SFT background.
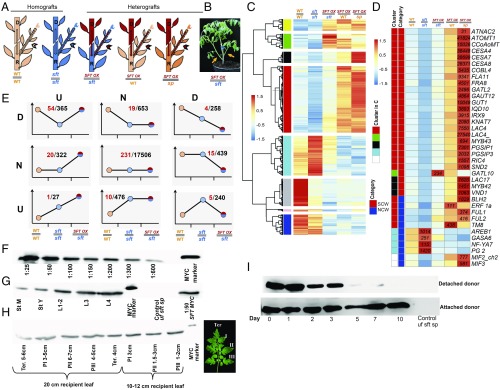
Fig. 2.
Graft-transmissible florigen enhances the SCWB network in sft and WT recipients. (A) Graft combinations: donor scions (upper part) and recipient shoots (lower part). Stems of lateral recipient shoots (arrowheads) were harvested at 25 d postgrafting. WT, light orange; sp, orange; sft, blue; 35S:SFT, red. (B) A live 35S:SFT//sft graft. Here 5-wk-old plants were grafted with shoots of 35S:SFT donors of the same age. (C) Hierarchical clustering of 593 genes regulated by m-florigen (Padj ≤ 0.1; |FC ≥2|). Almost all SCW genes are clustered in red. (D) Heat map of selected m-florigen-responsive SCW and non- CW genes (from C) and their absolute expression levels in 5 recipient genotypes. Genes are color-coded in accordance with the hierarchical clustering in C and with respect to the categories CW and non-CW. (E) PG1. All expressed genes in the recipients of the leftmost 3 grafts in A were sorted into 9 cohorts based on comparisons of WT//WT vs. sft//sft and SFT//sft vs. sft//sft and subsequently by 3 possible response categories—up (U), down (D) and no change (N)—demanding |FC >1.5| as the cutoff (Materials and Methods). Remarkably, cohort DU, with genes down-regulated in sft of the sft//sft homograft but up-regulated by at least 1.5-fold in comparison with sft of the 35S SFT//sft heterograft, is enriched for the florigen-regulated genes analyzed in C (101 of 593; P < 5.4*10–70 hypergeometric) and for SCWB genes (54 of 365; P < 4.4*10–26). The color-coding of the circles in the line graphs corresponds to the recipient genotypes as in A. Red numbers indicate SCWB genes. (F–I) Distribution and stability of the m-florigen protein in tomato. (F) A dilution series for the SFT-3XMYC protein from donor 35S:SFT-3XMYC leaves (Materials and Methods). (G) M-florigen is accumulated in young and old recipient leaves and stems at approximately 1% of donor leaf levels. L1, old leaves; St M, mature stems; St Y, young stems. (H) MYC tagged m-florigen is distributed among all leaflets of mature recipient leaves. P, a pair of leaflets; Ter, most mature terminal leaflet (on the right). (I) The MYC-tagged, graft-transmissible florigen had a half-life of ∼4 d. (Top) At 20 d postgrafting, the donor 35S:SFT scions were removed, and the presence of mobile SFT-MYC in recipient leaves was recorded daily. (Bottom) Control mobile SFT-MYC from recipient shoots of intact grafts.
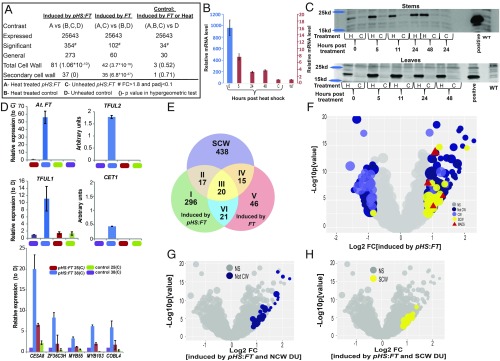
Fig. 3.
Transient heat-shock induction of AtFT significantly stimulated SCWB genes in tomato stems. (A) Identification of heat-shock–induced FT-responsive genes. A 1-factor design was used to compare the RNA profiles of heat-treated pHS:FT stems with the combined control treatments (Padj < 0.1; |FC ≥1.8|) (first column, A vs. B, C, and D). Comparisons to find FT leaky genes (A and C vs. B and D) and heat-induced genes (A, B, and C vs. D) were also made. (B) Time course activation and degradation of HS-induced FT RNA in tomato stems. The initial ∼100-fold expression level (blue y-axis) of FT mRNA was at the 3- to 5-fold level after 5 h (red y-axis). (C) HS-induced SFT-FLAG protein persisted for at least 48 h in stems (Top) and leaves (Bottom) in tomato. (D) qRT-PCR validation of 9 genes activated by heat-induced FT in tomato stems. (E) Venn diagram showing significant overlap of FT-induced genes with SCW genes in tomato. (F) Volcano plot showing the comparison between pHS:FT-induced and all other control conditions. DEG (Padj ≤ 0.1; |FC 1.8|). (G and H) pHS:FT induced genes overlap with 55 non-CW genes (G) and 23 SCW genes (H) from cohort DU of PG1, which are also up-regulated.
The observed enrichment of SCWB genes hinges on their functional classification. In an alternative analytical approach, we performed multiple tripartite comparisons between recipient genotypes. All 20,286 expressed genes were ranked solely by their expression trajectories and then divided into 9 categories visualized in what we refer to as Punnett grids (PGs). In the most instructive of these grids, PG1 (Fig. 2E), we compared the transcription profiles of sft recipients in the 2 bipartite contrasts: WT//WT vs. sft//sft and 35S:SFT//sft vs. sft//sft. Cohort DU of PG1 includes 365 genes that were down-regulated (D) in the recipient sft stems in the first bipartite contrast but were “rescued” (Up-U) by m-florigen in the recipient sft stems of the second bipartite contrast. Functional annotation of these 365 genes (Dataset S2) revealed an exclusive enrichment of SCWB genes (Fig. 2 and SI Appendix, Fig. S3). Significantly, 54 of the 359 expressed SCW genes (P = 5.9 × 10−33, hypergeometric test) were assigned to cohort DU, with no preferential presentation in any other cohort (Fig. 2E, red numbers in PG1). Thus, while moving along the phloem track, florigen emits lateral signals, or perhaps moves itself, enhancing the SCWB metabolic network in the xylem stems.
An Unmodified and Extremely Stable m-Florigen Is Produced in, and Imported by, all Aerial Organs.
Here we have shown that the stem vasculature is a bona fide target for the phloem-mobile hormone and not merely a conduit for a rectilinear passage from leaves to the apical meristems (Figs. 1 and 2C). To further understand the mechanisms underlying the systemic functions of florigen, we performed mass spectrometry analysis of affinity-enriched SFT-MYC from recipient leaves (Materials and Methods), which demonstrated that florigen moves free of major posttranslational modifications. In contrast, the antagonistic SP-MYC protein was phosphorylated in Serine 157, which is lacking in SFT (Fig. 2 and SI Appendix, Fig. S4). We next recorded the tagged m-florigen in recipient organs and found it in the stems and in every single leaflet of mature and young leaves at ∼1% of its level in the donor leaves (Fig. 2 G and H). Depending solely on the quantification of the dilution series, we recorded a relatively elevated level of the protein in recipient tissues, considering the low expression level in the donor (SI Appendix, Fig. S2E). Note that a single donor leaf is sufficient to promote lasting flowering in sft recipient plants (5), and that the expression level of SFT in 35S:SFT plants never exceeds 5% of the promoter potential (i.e., ∼1,000 vs. 25,000 normalized read counts for other genes). This, together with the “self-propagation” of florigenic signals (2) and the quantitative nature of floral stimulation, led us to examine the stability of the mobile hormone. By recording the mobile SFT-MYC in recipient leaves daily following removal of the donor scions, we determined that the tomato florigen has an exceptionally long half-life of ∼4 d (33) (Fig. 2I). The long-range mobility and indiscriminate distribution of the florigen protein among all vegetative organs are consistent with the regulation of leaf and stem morphogenesis via grafting and the autoregulatory model for the production and distribution of florigen (9). The unusual extended stability of florigen protein may underlie the quantitative regulation of flowering.
Transient Heat Induction of the Arabidopsis FT Gene Up-Regulates SCW Genes after Only 24 h.
Our hypothesis that florigen guides the dynamics of SCWB is contingent on steady-state transcription profiles. To determine whether florigen impacts SCWB on a shorter time scale, we used the pHS:FT transgene (pHEAT SHOCK:FT, a gift from O. Nilsson) to create sft pHS:FT plants bearing a heat-inducible florigen. The experimental scheme comprised heat-treated sft pHS:FT plants (A) and 3 control treatments: untreated sft pHS:FT (B), heat-treated sft (C), and untreated sft (D) plants (Fig. 3A). The 5-wk-old plants were exposed for 90 min to 38° C hot air, and stem segments from their third internodes were harvested 24 h later (Materials and Methods) to allow for the recovery of normal transcription and accumulation of florigen (34).
To investigate the fate of heat-induced transcripts and polypeptides, we next created pHS:SFT-FLAG plants. Although >95% of the SFT-FLAG transcripts present at the end of the 90-min heat treatment vanished 5 h later, their levels remained 3-fold above control levels at 48 h post-heat treatment (Fig. 3B). However, at 5 h post-heat treatment, the SFT-FLAG protein level was only 2-fold higher in both stems and leaves, for at least 48 h (Fig. 3C).
In response to heat induction of FT, we identified 354 DEGs (Fig. 3A and Dataset S4), including 37 SCW-related genes, 44 cell wall (CW)-related genes, and 273 non-CW genes, including TFUL1 and TFUL2. A quantitative validation of 9 FT-induced SCW genes is shown in Fig. 3D. Genes induced by heat treatment or by basal expression in pHS:FT plants significantly overlapped with the core list of tomato SCW genes (Fig. 3E), and all DE FT-induced SCW genes, as well as the 2 MADS genes, were up-regulated (volcano plot in Fig. 3F). Significantly, 23 of the 37 FT SCW-related regulated genes and 55 of the 273 total genes activated by FT were part of cohort DU of PG1 (Fig. 3 G and H) and thus may represent unknown SCWB genes. Therefore, florigen can trigger up-regulation of the SCWB network in tomato stems before any anatomic changes are evident.
Ginkgo biloba FT: A Mobile Antagonist of Flowering and a Suppressor of SCWB Provides a Molecular Portrait of an Antiflorigenic Syndrome.
In its role as a floral enhancer, florigen is checked by systemic antagonistic systems (2, 11, 35). If florigen deploys similar mechanisms in boosting SCWB in stems, then its floral antagonists would be expected to suppress SCWB. Gymnosperm FT-like genes have been reported to suppress flowering in Arabidopsis (36, 37), and we found that 35S:GinFT (Ginkgo biloba) generates an antiflorigenic syndrome in tomato including leafy inflorescences and, most pertinent here, extreme radial swelling and twisting of the stems and leaf petioles (Fig. 4 A–E).
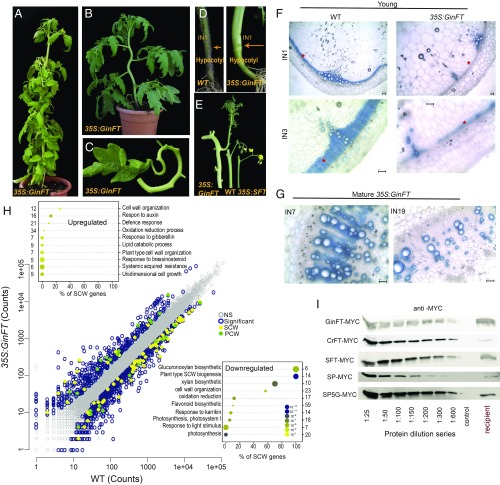
Fig. 4.
The gymnosperm GinFT gene antagonizes florigen, suppresses SCWB, and prolongs vascular maturation. (A–E) Characterization of the GinFT developmental syndrome in tomato. (A) A mature 35S:GinFT-MYC transgenic plant. The inward curling of stems and leaves is age-dependent. (B) A convoluted stem from a 4-wk-old 35S:GinFT plant. (C) The petiole and rachis of an adult 35S:GinFT leaf. Lateral leaflets have been removed for demonstration. (D) Hypocotyls of 35S:GinFT plants undergo differential radial constriction. Arrows mark the hypocotyl stem junctions. (E) Size comparisons of inflorescence-bearing stems. (Left) 35S:GinFT; (Center) WT; (Right) 35S:SFT. (F and G) High expression levels of GinFT delays lignification of IF fibers in young (F) and mature (G) stems. (F) 30-d-old WT and 35S:GinFT plants. (Upper) Internode 1. (Lower) Internode 3 at higher magnification. (G) 75-d-old 35S:GinFT plants, internodes 7 and 19. Note the accumulation of nonlignified xylem fibers. TBO staining for lignin. (H) SCWB genes are significantly down-regulated in 35S:GinFT stems. The scatterplot shows expression levels of 35S:GinFT vs. WT. Colored circles represent 1,122 DEGs (Padj < 0.1; |FC ≥2|). (Insets) Top 10 GO terms in up-regulated (Top) and down-regulated (Bottom) DEGs. (I) CETS proteins of gymnosperms conserve mobility potential. Protein blots show a dilution series of MYC-tagged proteins from the indicated CETS donor leaves, along with the undiluted mobile CETS proteins from the leaves of the corresponding recipient shoots. *IF zones. (Scale bars: 100 mm.)
Histological sections of 35S:GinFT stems revealed that, unlike in WT plants, the basal internodes of 30-d-old 35S:GinFT stems were still devoid of concentric lignified IF fibers (Fig. 4F). At 75 DPG, while fully blossoming, lignification of secondary xylem fibers in the basal internodes of the stem were still significantly delayed (Fig. 4G). To investigate whether GinFT attenuates SCWB as a unit, we profiled RNA from the first internodes of 35-d-old WT and 35S:GinFT plants. GO analysis of the 1,122 DEGs (|FC ≥2|) revealed significant enrichment of SCW genes; however, this time the majority of the SCWB genes were down-regulated rather than up-regulated, providing a molecular portrait of an antiflorigenic syndrome (Fig. 4H and Dataset S5).
The GinFT gene fulfilled all functions of a florigen antagonist, including binding of the Arabidopsis FD and its tomato homolog SPGB (38) (SI Appendix, Fig. S4). Thus, we hypothesized that CETS genes may already have the potential for long-range mobility in gymnosperms. Consistent with this speculation, the MYC-tagged polypeptides produced by 35S:GinFT-MYC and 35:CrFT-MYC transgenic plants were as mobile as tomato CETS proteins encoded by the SFT, SP, or SP5G genes (Fig. 4I).
High TFUL2 Accelerates SCWB in Stems Independent of Flowering.
TFUL2 is consistently activated by all forms of florigen (Figs. 1–3). If activation of TFUL2 is involved in mediating florigen in stems, then high TFUL2 expression would be expected to mimic the phenotypic expression and GO of florigen in stems, at least partly independent of florigen. Indeed, overexpression of TFUL2 in tomato induces extremely slender and rigid stems (Fig. 5 A–C). This is observed for 4 other members of the clade and in 3 different WT backgrounds (SI Appendix, Fig. S5A). We monitored SG and lignification in stems of 35S:TFUL2, WT, sft 35S:TFUL2, and sft plants, which form a nonoverlapping graded series of 5–6, 9, 11, and 13 leaves to flowering, respectively. At 4 wk postgermination, 2- to 3-layer-thick lignified IF fibers populated the 2 basal internodes of WT and sft plants, but 10–11 layers were prematurely produced in the same internodes of 35S:TFUL2 and sft 35S:TFUL2 plants. Remarkably, the extremely late flowering sft 35S:TFUL2 stems were nearly as slender and prematurely lignified as stems from the early flowering 35S:TFUL2 plants (Fig. 5D).
Fig. 5.
TFUL2, TMIF2, and TMIF3 induce premature, florigen-independent SCW deposition. (A) Early primary flowering (5 leaves) in 35S:TFUL2 plants. First inflorescence differentiated after 5 leaves instead of in 2 or 3 leaves as in 35S:SFT plants and 8 or 9 leaves as in WT plants. (B) Additive effects on flowering time and stem girth in 35S:TFUL2/+ 35S:TFUL2/+ double-heterozygous plants. (C) Slender stems and beaked fruits in 35S:TFUL2 plants. All genes belonging to the TFUL clan showed similar phenotypes when overexpressed in 3 different WT backgrounds (SI Appendix, Fig. S1A). (D) 35S:TFUL2 induces flowering-independent precocious SG and SCW deposition. Shown is a cross-section of the first internodes of 30-d-old plants. (E) Venn diagrams showing overlap of DEGs (Padj ≤ 0.1; |FC ≥2|) of 35S:SFT or 35S:TFUL2 with CW (Top) or SCW (Bottom) genes in the first internode. (F) Relative expression of the tomato MIF2 and MIF3 genes in lines overexpressing SFT, TFUL2, and GinFT. (G) Radial expansion of 6-wk-old mif3cr1 stems. (H and I) Vascular maturation is arrested in stem internodes of mif3cr1 mutant plants. (J–L) Slender stems (J) and precocious vascular differentiation (L) in 35S:MIF2-FLAG plants. Compare with mif3cr1 (K). (M–P) Flowering time and vascular differentiation are uncoupled. (M and N) 35S:SFT and mif3cr1 35S:SFT plants flower after 3 leaves, but the radial size of the latter (Inset) is similar to that of mif3cr1 plants. (O and P) Differentiation of IF xylem layers in the first internode of the 35S:SFT and mif3cr1 35S:SFT plants shown in M and N. *IF zones. (Scale bars: 100 mm.)
Since florigen activates TFUL2 in stems and TFUL2 accelerates secondary xylem differentiation, we studied differences and commonalities in the ways in which TFUL2 and SFT affect the activity of the SCWB network during vascular maturation. To this end, we compared the transcription profiles of the first internodes of 35S:TFUL2 and 35S:SFT plants with those of WT plants. We identified DEGs for each overexpressing genotype and calculated the overlap of genes regulated by SFT and TFUL2. We then superimposed this overlap with our reference lists of CW and SCW genes, generating the classification shown in Fig. 5E, SI Appendix, Fig. S5B, and Dataset S6. A significant number of CW and non-CW genes were differentially regulated by either SFT or TFUL2. SFT itself is up-regulated by 3-fold in 35S: TFUL2 stems, duplicating the regulatory module used in stimulating flowering. The GO analysis of all classes of DE genes comprising Fig. 5E indicated that lignin metabolism is differentially sensitive to TFUL2 (SI Appendix, Fig. S5B), and that 80 of the 83 CW genes regulated by both genes (Fig. 5E, class III, top diagram) responded in the same direction. Non-CW genes regulated by SFT or TFUL2 (classes V, VI, and I) may represent auxiliary activities consequential to the acceleration of SCWB, as well as functions not previously associated with SCWB.
Directly or indirectly, the activation of MADS genes and their impact on SCWB and radial contraction of the stems validate the discovery of SCWB as a bona fide target of florigen. It is expected that accelerating the rate of an existing metabolic system may require different mechanisms than those in initial activation. Nevertheless, genes of the FUL clade are a common relay partner in 2 pivotal systems: boosting floral transition in apical meristems (32) and, as shown here, vascular maturation. It is possible that the same module may play a similar role in other context-specific pleiotropic functions of florigen.
MIF2 and MIF3 Mediate Vascular Maturation Downstream of and Independent from Florigen.
Our RNA-Seq data revealed SCWB as a target for florigen. For this conclusion to be valid, some genes not previously associated with SCWB but inferred from the same dataset as being regulated by florigen may be involved in SCWB, and importantly, in a flowering independent manner. To test this assumption, we used CRISPR-editing (39) to mutate several genes that are modified in stems. Here we focused on 2 MIF genes, MINI ZINC FINGERS (40–43), which are activated by SFT and TFUL2 but, conversely, silenced in GinFT stems (Fig. 5F). In tomato, the MIF genes are expressed at moderate levels in all organs, and their 5 regulatory regions contain potential MADS-responsive elements (40, 43). We could not find a severe defect in mif2cr1 stems, but plants with mif3cr1 single or mif2cr1 mif3cr1 double mutants (SI Appendix, Figs. S2–S5 and Materials and Methods) had significantly swollen stems (Fig. 5G), as well as delayed lignification of secondary xylem fibers (Fig. 5 I and K). These defects were reminiscent of 35S:GinFT plants but with important differences; the overall stature, flowering time, and fertility of mif2cr1 and mif3cr1plants, as well as the double mutant mif2cr1 mif3cr1 plants, were essentially normal.
If MIFs are required for the normal course of SCWB in stems as legitimate components of the florigen-SCWB developmental program, then their overexpression would be expected to enhance it. Indeed, 35S:TMIF2-FLAG and 35S:TMIF3 plants, in addition to the already reported growth retardation and fruit development (40, 43), feature rigid slender stems with an extensive, precociously lignified secondary xylem (Fig. 5 J and L) but a normal flowering time.
To determine the MIF–florigen relationship, we bred mif2cr1 mif3cr1 35S:SFT plants. Like 35S:SFT plants, mif2cr1 mif3cr1 35S:SFT flowered after 3 leaves, but their stems remained as swollen as those of mif2cr1 mif3cr1 plants (Fig. 5 M and N), and lignification of the IF cambial cells was similarly delayed (Fig. 5 O and P and SI Appendix, Fig. S5C). These plants provide conclusive evidence that SCW deposition and xylem differentiation are not dependent on flowering time, and that the lateral expansion of stems can be uncoupled from precocious flowering. The 2 MIF genes form a regulatory tier linking florigen and SCWB in tomato stems.
The final girth and stiffness of the stems reflect a regulated balance between SG and SCW deposition, although the precise mechanism regulating this balance is unknown. High florigen and TFUL2 precociously activate radial proliferation of the secondary cambium and at the same time differentially enhance the deposition of SCWs. This new balance results in the termination of radial growth and rigid slender stems. Conversely, because SCWB is radically delayed in 35S:GinFT or mif stems, radial proliferation proceeds unchecked, resulting in swollen and fragile stems.
The Florigen-SCWB Developmental Link Is Valid in Photoperiodic Wild Tomato.
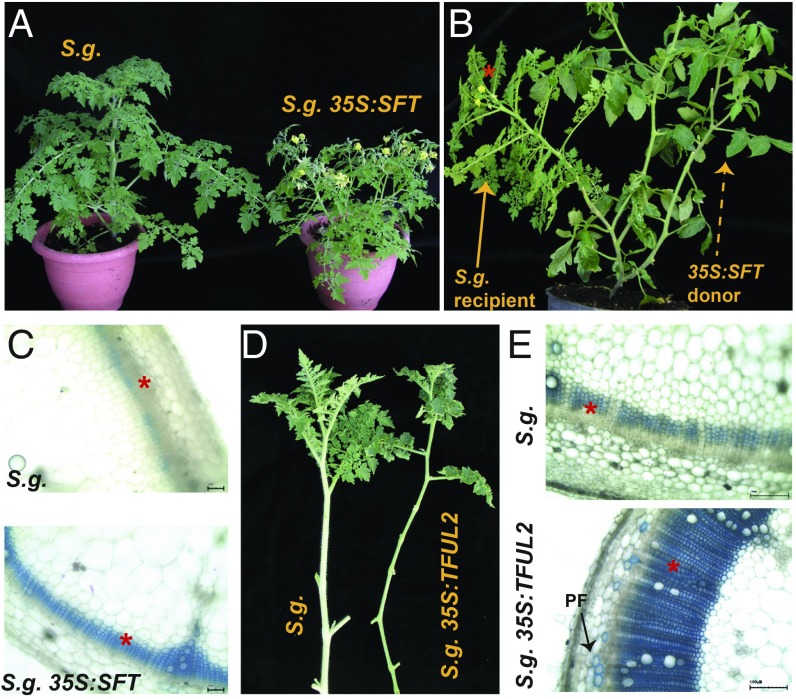
Cultivated tomato is a day-neutral plant. To determine whether the florigen-SCWB link is valid in photoperiodic plants, we created Solanum galapagense (S.g.) plants, a short-day, wild relative of tomato that overexpresses SFT or TFUL2. When grown under noninductive long-day conditions, S.g. plants overexpressing SFT or S.g. recipient shoots grafted by 35S:SFT donors displayed extensive flowering (Fig. 6 A and B). Under the same conditions, the nonflowering S.g. plants showed a protracted delay in vascular maturation, but S.g. 35S:SFT plants developed slender stems and enhanced xylem differentiation (Fig. 6C). Similarly, 2-mo-old S.g. 35S:TFUL2 plants grown under long days and completely devoid of floral primordia formed extremely slender stems (Fig. 6D) with fully differentiated IF fiber layers (Fig. 6E). Taken together, these findings show that the rate of secondary xylem differentiation in both day-neutral and short-day plants is correlated primarily with the expression levels of TFUL2 and not with flowering per se.
Fig. 6.
The SFT-TFUL2 module is conserved in the short-day wild tomato S. galapaganese (S.g.). (A) 35S:SFT induces flowering in S.g. under nonpermissive long days. (Left) A nonflowering 6-wk-old S.g. plant. (Right ) A flowering 6-wk-old 35S:SFT S.g. plant. (B) Graft-transmissible florigen stimulates flowering in the short-day species S.g. Donor tomato 35S:SFT scions (arrow) rescue flowering (arrow and asterisks) in WT S.g. recipients under nonpermissive long-day conditions. (C) 35S:SFT induces precocious SCW deposition and vascular maturation in the short–day S.g. grown under long-day conditions; cross-sections are from the first internodes of the plants shown in A. TBO staining for lignin. (D and E) TFUL2 enhances vascular differentiation independent of flowering in S.g. stems under long-day conditions. (D) Shoots of nonflowering WT and S.g. 35S:TFUL2 plants grown under long-day conditions. Both plants were completely devoid of inflorescence primordia. (E) Maturation status of the first internodes from the stems shown alongside in D. The S.g. 35S:TFUL2 plants formed approximately 30 concentric layers of lignified IF xylem fibers, but only 5 such layers evolved in WT plants. Note that the external phloem fibers (PF) in S.g. 35S:TFUL2 are already lignified, indicating complete differentiation. *IF zones. (Scale bars: 100 mm.)
Discussion
Flowering-Independent Developmental Differentiation by Florigen.
SCWB has emerged as an ultimate metabolic target of florigen in tomato stems. The significant overlap of the cohorts of genes involved in SCWB in 4 independent experimental platforms substantiates the hypothesis that florigen enhances SCWB as a gene regulatory network and, mechanistically, independent of flowering. Florigen, in its endogenous and mobile forms, can trigger up-regulation of the SCWB network in the tomato stem before any anatomic changes are evident, as early as 24 h after FT induction (Fig. 3). The genes downstream of florigen dramatically illustrate the uncoupling of vascular differentiation and SCWB from flowering: mif3cr1 35S:SFT plants flower as early as 35S:SFT plants alone. However, their stems swell and vascular maturation is attenuated as in mif3cr1 plants. Therefore, in tomato, flowering is not a condition for xylem differentiation in stems, as has been suggested for Arabidopsis hypocotyls (44). The function of florigen in the SAMs is universal, and we predict that its link with SCWB will prove to be universal as well. In support of this proposal, flowering and independent maturation of the vasculature in the short-day S.g. are enhanced by TFUL2 (Fig. 6), SFT arrests stem expansion in cotton (45), and leaf growth in Arabidopsis, tomato, and roses is restricted in preparation for flowering (46).
While Florigen accelerates vascular maturation, it is not essential for normal vascular development. Vegetative SAMs develop normally without florigen, and mutant sft plants in tomato or ft tsf plants in Arabidopsis will eventually flower without florigen (47). Overlooked is the fact that among all the potent TFs involved in leaf shape (48), florigen is most effective suppressor of leaf complexity in tomato (9) and is completely epistatic to the dominant KN2 gene (SI Appendix, Fig. S1D). Thus, the flowering-independent regulation of vascular maturation by the florigen-TFUL module may offer an additional regulatory tier for leaf size and shape.
The acceleration of SCWB as an end target for florigen does not imply direct molecular relations between the 2 ends of the process. Defining the mediating links is problematic, because florigen accelerates an already existing program rather than activating a new one. Consistent with this premise, genes involved in the specification or patterning of SG and SCWB, such as MP, class III HD-ZIPs, KANADY, or APL (20, 49, 50), are not differentially regulated by florigen. Taylor-Teeples et al. (26) identified E2c as a positive master regulator of tier 3 SCWB (Fig. 1F) in Arabidopsis roots, but such a relationship is not supported in tomato stems. All major plant hormones are involved in regulating SG and xylem differentiation (Fig. 1). Gibberellic acid (GA) is essential for xylem differentiation in Solanaceae (51) but, unlike in grasses and Arabidopsis, in tomato GA is required for floral maturation but not for floral induction (52). Several GA metabolic genes are regulated by SFT and TFUL2 (Fig. 5 and SI Appendix, Fig. S5) but thus far they have not consolidated to form a regulatory tier for SCWB.
Florigen as an Organizer of the Reproductive Shoot.
The central role of the florigen-SCWB developmental link is echoed in its conservation and universality. The developmental choice to adjust global growth by coordinating floral induction with vascular maturation is logical. The continuous growth of plant organs hinges on cell divisions and cell expansion, both of which are contingent on the composition of the cell wall (53, 54). An accelerated vascular maturation meets the anticipated needs of the commencing reproductive phase for mechanical support and altered redistribution of resources and signals. The selection of a CETS gene for the systemic induction of flowering and the concomitant adjustment of vascular differentiation implies that such genes were already endowed with related/parallel potentials in nonflowering plants. As shown here, Ginkgo FT is equipped with the potential to interact with SPGB (tomato FD) and FD, antagonize florigen, attenuate SCWB, and move systemically along the phloem track of tomato. The SPGB is expressed in apical meristems (55) as well as stems. It is tempting to speculate that the link between CETS genes and SCWB underlies the annual growth cycles in the trunks of perennial trees. This speculation is consistent with the induction of xylem differentiation by the phloem-translocated florigen and by the unexpected stability of the tomato florigen protein (Fig. 2). It is possible that other “flowering genes,” such as LEAFY (46), carried originally vegetative functions and were recruited to regulate different aspects of the newly emerging flowering system.
Florigen functions as a versatile molecular adaptor of TFs (38, 56). Unlike classic plant hormones, florigen performs identical developmental functions across species and has no metabolic intermediates, and its positive roles are restricted to the reproductive phase. By coordinating vascular maturation with the transition to flowering, florigen harmonizes the progression of 2 global developmental processes that may otherwise proceed independently and are not the consequence of one another. In this role, florigen functions as a “peripheral” regulator of the reproductive shoot system, as in peripheral to the basic tenets of the plants, not in reference to spatial operation or importance.
Materials and Methods
Plant Material.
Plants were grown in a greenhouse under natural conditions. The WT cultivars NY, M82 (sp background), and Money Maker (MM) were obtained from the C. M. Rick Tomato Genetics Resource Center at University of California Davis. Other lines mentioned in the text were bred for this work. Transgenic plants were generated as described previously (57).
Grafting Procedure.
In this procedure, 5- to 6 wk-old shoots of 35S:CETS scions containing apices and 4 expanded leaves were grafted onto 5-wk-old WT or sft recipients pruned to have 3 to 4 expanded leaves and their associated dormant axillary buds. In such a configuration, the protein has to enter the recipient stem, move 1 or 2 internodes (5–10 cm), and cross 1 or 2 nodes and a leaf-bud node junction before entering the stems and leaves of the released recipient shoots roughly 20 cm away. The tagged protein was then recorded at 3 wk postgrafting.
Creating the Core List of CW Genes in Tomato.
A list of 2,675 tomato SCW and primary CW (PCW) genes was compiled from previous reports (16, 28) and curated (Dataset S3). We next screened the list of 180 TFs presented by Taylor-Teeples et al. (26) to bind regulatory sequences of SCW genes in Arabidopsis roots and added to our list 65 TFs that showed differential expression in at least 1 of our RNA-Seq experiments. The resulting complete list of 498 tomato genes represents 265 Arabidopsis genes. We categorized the genes as SCW, PCW, or both based on the aforementioned sources. The numbers of genes in each category are listed as total and those expressed in our stem RNA-Seq data, respectively: CW, 2,675 and 2,440; PCW, 273 and 238; PCW + SCW, 24 and 24; SCW, 498 and 468 (Dataset S3).
Next-generation sequencing analysis and other experimental procedures are described in detail in SI Appendix. Sequencing data were deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE132280).
Supplementary Material
Acknowledgments
We thank Ilana Nepomniashchi and Ziva Amesellem for generating transgenic plants and Tom Harel for planning and creating CRISPR constructs. We also thank Idan Efroni, Benny Podbilewicz, Zach Lippman, Sarah F. Prewitt, and Sigal Savaldi-Goldstein for valuable comments on the manuscript and Haim Bar (University of Connecticut) for validating our statistical tests. We thank Tal Katz-Ezov and the staff at the Technion Life Sciences and Engineering Infrastructure Center and Technion Genome Center centers for NGS analysis, and Tamar Ziv and the staff at the Smoler Proteomic center for their support of the protein experiments. This work was continuously supported by Russell Berrie Nanotechnology Institute grants to E.L. for NGS (data generating and analysis) and imaging, by an Israel Science Foundation (ISF) research grant to E.L., by ISF and US–Israel Binational Agricultural Research and Development Fund grants to Y.E., and by a joint ISF (Bikura) grant (to E.L. and Y.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All RNA-Seq data have been deposited to the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE132280).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906405116/-/DCSupplemental.
References
- 1.Chailakhyan M. K., About the mechanism of the photoperiodic response (in Russian). Dokl. Akad. Nauk SSSR 1, 85–89 (1936). [Google Scholar]
- 2.Zeevaart J. A. D., Physiology of flower formation. Annu. Rev. Plant Physiol. 27, 321–348 (1976). [Google Scholar]
- 3.Koornneef M., Hanhart C. J., van der Veen J. H., A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66 (1991). [DOI] [PubMed] [Google Scholar]
- 4.Simpson G. G., Dean C., Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Lifschitz E., et al. , The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. U.S.A. 103, 6398–6403 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sparks E., Wachsman G., Benfey P. N., Spatiotemporal signalling in plant development. Nat. Rev. Genet. 14, 631–644 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y., Klejnot J., Yu X., Liu X., Lin C., Florigen (II): It is a mobile protein. J. Integr. Plant Biol. 49, 1665–1669 (2007). [Google Scholar]
- 8.Kobayashi Y., Weigel D., Move on up, it’s time for change: Mobile signals controlling photoperiod-dependent flowering. Genes Dev. 21, 2371–2384 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Shalit A., et al. , The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. U.S.A. 106, 8392–8397 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navarro C., et al. , Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478, 119–122 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Lifschitz E., Ayre B. G., Eshed Y., Florigen and anti-florigen: A systemic mechanism for coordinating growth and termination in flowering plants. Front. Plant Sci. 5, 465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbull C., Long-distance regulation of flowering time. J. Exp. Bot. 62, 4399–4413 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Taiz L., Zeiger E., Plant Physiology (Sinauer Associates, Sunderland, MA, ed. 5, 2010). [Google Scholar]
- 14.Petricka J. J., Benfey P. N., Root layers: Complex regulation of developmental patterning. Curr. Opin. Genet. Dev. 18, 354–361 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong R., Ye Z. H., Secondary cell walls: Biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol. 56, 195–214 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Hussey S. G., Mizrachi E., Creux N. M., Myburg A. A., Navigating the transcriptional roadmap regulating plant secondary cell wall deposition. Front. Plant Sci. 4, 325 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar M., Turner S., Plant cellulose synthesis: CESA proteins crossing kingdoms. Phytochemistry 112, 91–99 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Aloni R., Foliar and axial aspects of vascular differentiation: Hypotheses and evidence. J. Plant Growth Regul. 20, 22–34 (2001). [Google Scholar]
- 19.Chaffey N., Cholewa E., Regan S., Sundberg B., Secondary xylem development in Arabidopsis: A model for wood formation. Physiol. Plant. 114, 594–600 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Scarpella E., Helariutta Y., Vascular pattern formation in plants. Curr. Top. Dev. Biol. 91, 221–265 (2010). [DOI] [PubMed] [Google Scholar]
- 21.An H., et al. , CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131, 3615–3626 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Chen Q., et al. , FLOWERING LOCUS T mRNA is synthesized in specialized companion cells in Arabidopsis and Maryland Mammoth tobacco leaf veins. Proc. Natl. Acad. Sci. U.S.A. 115, 2830–2835 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turnbull C. G. N., Lopez-Cobollo R. M., Heavy traffic in the fast lane: Long-distance signalling by macromolecules. New Phytol. 198, 33–51 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Kubo M., et al. , Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oda Y., Fukuda H., Secondary cell wall patterning during xylem differentiation. Curr. Opin. Plant Biol. 15, 38–44 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Taylor-Teeples M., et al. , An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–575 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hao Z., Mohnen D., A review of xylan and lignin biosynthesis: Foundation for studying Arabidopsis irregular xylem mutants with pleiotropic phenotypes. Crit. Rev. Biochem. Mol. Biol. 49, 212–241 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Caffall K. H., Mohnen D., The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 344, 1879–1900 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Brady S. M., et al. , A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Brukhin V., Hernould M., Gonzalez N., Chevalier C., Flower development schedule in tomato Lycopersicon esculentum cv. sweet cherry. Sex. Plant Reprod. 15, 311–320 (2003). [Google Scholar]
- 31.Hall H., Ellis B., Developmentally equivalent tissue sampling based on growth kinematic profiling of Arabidopsis inflorescence stems. New Phytol. 194, 287–296 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Fornara F., de Montaigu A., Coupland G., SnapShot: Control of flowering in Arabidopsis. Cell 141, 550, 550.e1-e2 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Kim S. J., et al. , Post-translational regulation of FLOWERING LOCUS T protein in Arabidopsis. Mol. Plant 9, 308–311 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Swindell W. R., Huebner M., Weber A. P., Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8, 125 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeevaart J. A., Florigen coming of age after 70 years. Plant Cell 18, 1783–1789 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klintenäs M., Pin P. A., Benlloch R., Ingvarsson P. K., Nilsson O., Analysis of conifer FLOWERING LOCUS T/TERMINAL FLOWER1-like genes provides evidence for dramatic biochemical evolution in the angiosperm FT lineage. New Phytol. 196, 1260–1273 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Liu Y. Y., Yang K. Z., Wei X. X., Wang X. Q., Revisiting the phosphatidylethanolamine-binding protein (PEBP) gene family reveals cryptic FLOWERING LOCUS T gene homologs in gymnosperms and sheds new light on functional evolution. New Phytol. 212, 730–744 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Pnueli L., et al. , Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell 13, 2687–2702 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soyk S., et al. , Bypassing negative epistasis on yield in tomato imposed by a domestication gene. Cell 169, 1142–1155.e12 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Bollier N., et al. , At-MINI ZINC FINGER2 and Sl-INHIBITOR OF MERISTEM ACTIVITY, a conserved missing link in the regulation of floral meristem termination in Arabidopsis and tomato. Plant Cell 30, 83–100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han M., et al. , A mini zinc-finger protein (MIF) from Gerbera hybrida activates the GASA protein family gene, GEG, to inhibit ray petal elongation. Front. Plant Sci. 8, 1649 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu W., Ma H., Characterization of a novel putative zinc finger gene MIF1: Involvement in multiple hormonal regulation of Arabidopsis development. Plant J. 45, 399–422 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Sicard A., Petit J., Mouras A., Chevalier C., Hernould M., Meristem activity during flower and ovule development in tomato is controlled by the mini zinc finger gene INHIBITOR OF MERISTEM ACTIVITY. Plant J. 55, 415–427 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Sibout R., Plantegenet S., Hardtke C. S., Flowering as a condition for xylem expansion in Arabidopsis hypocotyl and root. Curr. Biol. 18, 458–463 (2008). [DOI] [PubMed] [Google Scholar]
- 45.McGarry R. C., et al. , Monopodial and sympodial branching architecture in cotton is differentially regulated by the Gossypium hirsutum SINGLE FLOWER TRUSS and SELF-PRUNING orthologs. New Phytol. 212, 244–258 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Efroni I., Eshed Y., Lifschitz E., Morphogenesis of simple and compound leaves: A critical review. Plant Cell 22, 1019–1032 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamaguchi A., Kobayashi Y., Goto K., Abe M., Araki T., TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 46, 1175–1189 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Hay A., Tsiantis M., KNOX genes: Versatile regulators of plant development and diversity. Development 137, 3153–3165 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Caño-Delgado A., Lee J.-Y., Demura T., Regulatory mechanisms for specification and patterning of plant vascular tissues. Annu. Rev. Cell Dev. Biol. 26, 605–637 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Lucas W. J., et al. , The plant vascular system: Evolution, development and functions. J. Integr. Plant Biol. 55, 294–388 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Dayan J., et al. , Leaf-induced gibberellin signaling is essential for internode elongation, cambial activity, and fiber differentiation in tobacco stems. Plant Cell 24, 66–79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rappaport L., Effect of gibberellin on growth, flowering and fruiting of the earlypak tomato, Lycopersicum esculentum. Plant Physiol. 32, 440–444 (1957). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cosgrove D. J., Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Yang W., et al. , Regulation of meristem morphogenesis by cell wall synthases in Arabidopsis. Curr. Biol. 26, 1404–1415 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lifschitz E., Eshed Y., Universal florigenic signals triggered by FT homologues regulate growth and flowering cycles in perennial day-neutral tomato. J. Exp. Bot. 57, 3405–3414 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Taoka K., Ohki I., Tsuji H., Kojima C., Shimamoto K., Structure and function of florigen and the receptor complex. Trends Plant Sci. 18, 287–294 (2013). [DOI] [PubMed] [Google Scholar]
- 57.McCormick S., Transformation of tomato with Agrobacterium tumifaciens. Plant Tissue Cult. Manual B6, 1–9 (1991). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.