Short abstract
Cultivation of primary human hepatocytes (PHHs) often faces obstacles including failure of long-term in vitro culture, weak proliferation ability, rapid loss of liver-specific function and morphology, and tendency of fibrosis. Previous research focused on immortalization methods, such as telomerase and viral, to culture immortalized primary human hepatocytes, which may lose some of the normal properties. However, non-immortalized PHHs often fail to maintain long-term viability and functionality. These highlight the urgent need for developing new culture strategy for PHHs. In the present study, we isolated PHHs from fresh human liver tissues representing different liver diseases and age groups. We used conditional reprogramming, without permanent immortalization, for long-term in vitro primary human hepatocytes cultivation and characterization. For functional characterization, we assessed CYP3A4, 1A1 and 2C9 activities and measured the mRNA expression of albumin, s100a4, krt8, krt18, cyp1a1, cyp3a4, cyp2b6, cyp2c8, cyp2c9, and cyp2d6. Additionally, we compared the DNA fingerprint of the cells against their original liver tissues using short tandem repeat (STR) analysis. We found that PHHs-derived from young patients can survive for more than three months, while the lifespan of primary human hepatocytes derived from adult patients ranges from two to three months, which is longer than most commercial primary hepatocytes. Importantly, the cells at early passages retain strong CYP3A4, 1A1 and 2C9 activities and the DNA fingerprints are identical with their original tissues. Through conditional programming, we achieved, for the first time, a high level of success rate in the long-term in vitro cultivation of primary human hepatocytes-derived patients representing diverse liver disease. Moreover, the conditional programming cell culture technology reported in this paper requires neither co-culture with additive cells, nor complex and expensive components, such as collagen sandwich or spheroid culture. We thus believe that the patient-derived PHHs cultivation using conditional programming may provide a viable and valuable cell model to study liver disease-related mechanisms.
Impact statement
Commercially available primary human hepatocytes rapidly lose their proliferative ability and liver-specific functions over a few cultivation days. The demand for pharmaceutical toxicity screening and liver disease research requires the development of long-term primary hepatocyte culture methods. This manuscript addresses this challenge by introducing for the first time successful long-term in vitro cultivation of primary human hepatocytes from a range of liver transplantation patients using conditional reprogramming technique. The beauty of this technique is that it is not a permanent immortalization and does not require co-culture with additive cells. The primary human hepatocytes retain proliferative capacity, genetic stability, and hepatocyte-specific functions at early passages. In view of these, we believe that scientists and researchers will benefit from using these highly valuable cell models to study diverse liver diseases.
Keywords: Primary human hepatocytes, long-term culture, conditional reprogramming
Introduction
The liver is a vital organ, which is critically involved in metabolism, protein synthesis, detoxification, bile acid production, and many other important biological processes. Liver diseases, caused by a variety of genetic, unhealthy lifestyle, drug toxicity or environmental factors, represent a major threat to human health. It is estimated that five million adults are diagnosed with liver disease in the United States.1–3 Cirrhosis, liver cancer, and liver failure are serious conditions that cause death. Given the importance of maintaining appropriate liver function in human health, it is imperative to understand molecular mechanism underlying respective liver malfunctions for developing effective treatment. The extensive research on liver biology demands the use of an in vitro cell model of functional primary human hepatocytes (PHHs). In vitro cultured PHHs provide a valuable resource for studies in cell metabolism, inflammation, preclinical drug screening, toxicology screenings, and development of bioartificial liver devices.4 More importantly, establishing in vitro hepatocyte culture derived from a diversity liver patients allows assessment of drug effects at a cellular level on patients with different genetic backgrounds or disease conditions, supporting the development of personalized medicine.5,6 However, in vitro cultivation of PHHs often faces obstacles including failure of long-term in vitro culture, rapid loss of liver-specific function and morphology, tendency of fibrosis, and weak proliferation ability.7,8 Besides, the extensive use of cancerous liver cell lines, e.g. HepG2, limits the exploration of disease-specific molecular pathways in diverse liver patients. In order to overcome these issues, several diverse medium formulations have been used to culture PHH, such as modified DMED,9,10 modified MCM,8,11 Williams’ E medium,9 and hepatocyte medium.12 Co-cultivation of hepatocytes with non-parenchymal cells, such as endothelial cells and fibroblasts, is another commonly used technique to keep viability, hepatocyte morphology, and functions.13,14 3D technique has been reported to be beneficial for maintaining some PHH functions.15,16 However, the existing culture techniques face the loss of viability during early passages and/or contamination of irrelevant cell sources introduced by co-culture, resulting in the limited use of PHHs for long-term culture purpose and downstream biomedical analyses.
Conditional reprogramming has been used to establish patient-derived cell cultures from diverse human biospecimens, such as breast, skin, prostate and colon, with an indefinite proliferative state without the use of exogenous viral or genetic manipulation.17 This technique uses the combination of irradiated mouse fibroblast J2 cells (feeder cells) and the Rho-associated kinase (ROCK) inhibitor (Y-27632) to propagate human epithelial cells. Y-27632 is able to increase the viability of human bone marrow-derived mesenchymal stem cells18 and human keratinocyte stem cells19 and induce indefinite cell proliferation.20,21 Alterations of actin/myosin activity are the main consequence of ROCK inhibition, and this cytoskeletal network has a primary role in regulating cell proliferation. In addition, it has been reported that the use of ROCK Inhibitor may regulate p16/Rb pathway, which may also contribute to the cell immortalization.17,20 Feeder cells can be replaced by medium conditioned by irradiated J2 cells (conditional medium; CM) for cell culture.17 To our knowledge, the potential application of conditional programming cell technology for long-term cultivation of PHH has not been reported.
In the present study, we evaluated the use of conditional programming in in vitro culture of PHH derived from fresh liver tissues representing diverse liver patients. We report, for the first time, that this cell culture technology can be used in long-term in vitro PHH culture and maintaining important liver functions.
Materials and methods
In vitro culture of patient-derived primary hepatocytes
The study has been approved by the Georgetown University Institutional Review Board (IRB). Informed consent was obtained from the majority of the research subjects that enrolled in a protocol (# 2005–206). For the PHH3 and 4 that participated in a protocol (# STUDY00000006), the requirement for informed consent was waived by the IRB. Fresh liver tissues were obtained from explant specimens upon liver transplantation or liver resection of diverse liver patients (Table 1). We isolated primary human hepatocytes (PHHs) from the respective liver tissues using collagenase/hyaluronidase digestion (STEMCELL Technologies, Vancouver, Canada). Cells were cultured in conditional reprogrammed F-Y medium (Table 2) in non-coated T75 flask (CellStar 658175). Cells were cultured in a 37°C incubator with 5% CO2. Medium was changed every other day or every three days. The cell concentration and viability were checked by incubation with trypan blue and then counted by a cell counter (Invitrogen, Waltham, USA). HepG2 was obtained from the Tissue Culture Shared Resource at Georgetown University. J2 and human fibroblast cells were obtained from the Center for Cell Reprogramming at Georgetown University.
Table 1.
Samples used to derive PHHs using conditional reprogramming.
| Patient identification | Age (years) | Gender | Diagnosis | Tissue size | Cell culture status |
|---|---|---|---|---|---|
| 1 (PHH 1) | 52 | F | Cirrhosis | 2 × 2×1 cm | Stopped growing at P5 |
| 2 | 56 | M | Hepatitis C virus-induced liver failure | 2p, each 1 × 1×1 cm | Got contaminated at P1 |
| 3 (PHH 2) | 66 | F | Cirrhosis | 0.5 × 0.5 × 1 cm | Stopped growing at P3 |
| 4 | 60 | M | Hepatocellular carcinoma | 2 × 2×2 cm | Got contaminated at P1 |
| 5 | 68 | F | Cirrhosis | 2.5 × 2.5 × 1 cm | Got contaminated at P1 |
| 6 | 55 | F | Hepatocellular carcinoma | 0.5 × 0.5 × 0.5 cm | Got contaminated at P1 |
| 7 | 61 | F | Acute liver failure | 2 × 2×2 cm | Got contaminated at P1 |
| 8 (PHH 3) | 3 | F | Maple syrup urine disease | 2 × 2×2 cm | Continued to grow to P10 until the experiment was terminated |
| 9 (PHH 4) | 9 | M | Citrullinemia type 1 disease | 2 × 2×2 cm | Continued to grow to P10 until the experiment was terminated |
| 10 (PHH 5) | 72 | M | Cirrhosis | 1.5 × 1.5 × 1 cm | Stopped growing at P3 |
| 11 (PHH 6) | 53 | M | Hepatitis C virus induced liver failure | 2 × 2×2 cm | Stopped growing at P4 |
PHH: primary human hepatocyte.
Table 2.
F-Y media formulation.
| Component | ≈ 500 mL |
|---|---|
| Complete DMEM | 373 mL |
| F12 nutrient mixture | 120 mL |
| Hydrocortisone/epidermal growth factor mixture | 0.5 mL |
| Insulin (5 mg/mL in distilled water) | 0.5 mL |
| Amphotericin B (250 µg/mL) | 0.5 mL |
| Gentamicin (10 mg/mL) | 0.5 mL |
| Cholera toxin (11.7 µM in distilled water) | 4.3 µL |
| ROCK inhibitor Y-27632 (10 mM in distilled water) | 0.5 mL |
| HEPES (1M) | 5 mL |
Note: Complete DMEM contains DMEM with 10% fetal bovine serum, 100 µg/mL glutamine, 100 µg/mL penicillin, and 100 µg/mL streptomycin.
Hydrocortisone/epidermal growth factor mixture: mix 1 mL of 0.5 mg/mL hydrocortisone in 100% ethanol with 19 mL DMEM containing 2.5 µg epidermal growth factor.
Conditioned medium (CM): irradiated 3T3-J2 cells were grown in F-Y medium for three days at a confluence of 90%. The cultured medium was concentrated to remove the cell debris and filtered through 0.2 µm filter systems.
Conditional Reprogramming F-Y media: combined the fresh F-Y media with CM in the presence of 10 µM Y-27632 with a ratio of 1:3.
Short tandem repeat analysis
Genomic DNA was isolated from cells at different passages and liver tissues using DNeasy Blood & Tissue kit (QIAGEN, Hilden, North Rhine-Westphalia, Germany). Short tandem repeat (STR) analysis was performed by Genetica DNA Laboratories (Burlington, Vermont, USA). We tested 15 STR markers: TH01, D21S11, D18S51, Penta E, D5S818, D13S317, D7S820, D16S539, CSF1PO, Penta D, vWA, D8S1179, TPOX, FGA, and Amelogenin. Data analysis and allele size determination were carried out using GeneMapper software.
Cytochromes P450 activity measurement
To characterize the cells, we performed cytochromes P450 (CYP450) 3A4, 1A1 and 2C9 assays following the protocol of the assay kits (Promega, Fitchburg, Massachusetts, USA) when we passaged the cells. Briefly, 20,000 cells were seeded into 96-well plates. On the following day, we replaced culture medium with 50 µL fresh medium containing corresponding luminogenic CYP substrate and incubated cells at 37°C for 1 h (for CYP3A4) or 3 h (for both CYP1A1 and CYP2C9). After incubation, we transferred 25 µL of culture medium from each well into a 96-well opaque white luminometer plate, and added 25 µL of luciferin detection reagent to initiate a luminescent reaction. After the plate was incubated at room temperature for 20 min, luminescence was read using a luminometer (Wallac Victor 1420, Conquer Scientific, San Diego, California, USA). After luminescence assay, 25 µL of medium remained in the wells into which an equal volume (25 µL) of the CellTiter-GloR reagent was added to determine cell viability. The contents were mixed for 2 min on an orbital shaker to induce cell lysis. The plate was then incubated at room temperature for 10 min to stabilize luminescent signal; 40 µL of cell lysate was transferred into a 96-well opaque white luminometer plate to record luminescence. We used HepG2 and human fibroblast cells as a positive control and a negative control, respectively.
Real-time PCR
Total RNA was extracted from cells using Direct-zol RNA MinipPrep kit (Zymo Research, Irvine, California, USA) and treated with DNase (Zymo Research, Irvine, California, USA) according to the manufacturers’ instructions. First-strand cDNA was synthesized with an equal amount of RNA using High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, California, USA). Real-time PCR was performed in a QuantStudio 12K Flex system (Thermo Fisher, Waltham, MA, USA) using Applied Biosystems™ Power SYBR™ Green PCR Master Mix (Applied Biosystems, Foster City, California, USA). The relative quantities of mRNA were analyzed using the 2−ΔΔCT method.22 The primers used in the quantitative PCR were as follows: gapdh (NM_002046.7, as a housekeeping gene): (F) 5´-ACATCGCTCAGACACCATG-3´ and (R) 5´-TGTAGTTGAGGTCAATGAAGGG-3´; albumin (NM_000477.7): (F) 5´-CCTGATTACTCTGTCGTGCTG-3´ and (R) 5´-ATTCTGAGGCTCTTCCACAAG-3´; s100a4 (NM_002961.3): (F) 5´-AGTCAGAACTAAAGGAGCTGC-3´ and (R) 5´-GACACAGTACTCTTGGAAGTC C-3´; krt8 (NM_002273.3): (F) 5´-CAGGAGAAGGAGCAGATCAAG-3´ and (R) 5´-GTTGTCCATGTTGCTTCGAG -3´; krt18 (NM_000224.3): (F) 5´-CAGAGACTGGAGCCATTACTTC -3´ and (R) 5´-GCCAGCTCTGTCTCATACTTG-3´; cyp1a1 (NM_000499.4): (F) 5´-CCCAACCCTTCCCTGAATG -3´ and (R) 5´-TTCTCCTGACAGTGCTCAATC -3´; cyp3a4 (NM_017460.5): (F) 5´-ACCAGTGGAAAACTCAAGGAG -3´ and (R) 5´- TGATCACATCCATGCTGTAGG-3´; cyp2b6 (NM_000767.4): (F) 5´-GCGATTCTCTGTGACCACTATG -3´ and (R) 5´-GTAATGGACTGGAAGAGGAAGG -3´; cyp2c8 (NM_000770.3): (F) 5´-CTT GCGGAATTTTGGGATGG -3´ and (R) 5´-AGATCACATTGCAGGGAGC -3´; cyp2c9 (NM_000771.3): (F) 5´- GATATGCTCTCCTTCTCCTGC -3´ and (R) 5´- TCACACGTTCAATCTCTTCCTG -3´; cyp2d6 (NM_000106.5): (F) 5´- ACTGCCGTGATTCATGAGG -3´ and (R) 5´- ATCCTTCAGCACCGATGA C -3´
Results
Cell culture
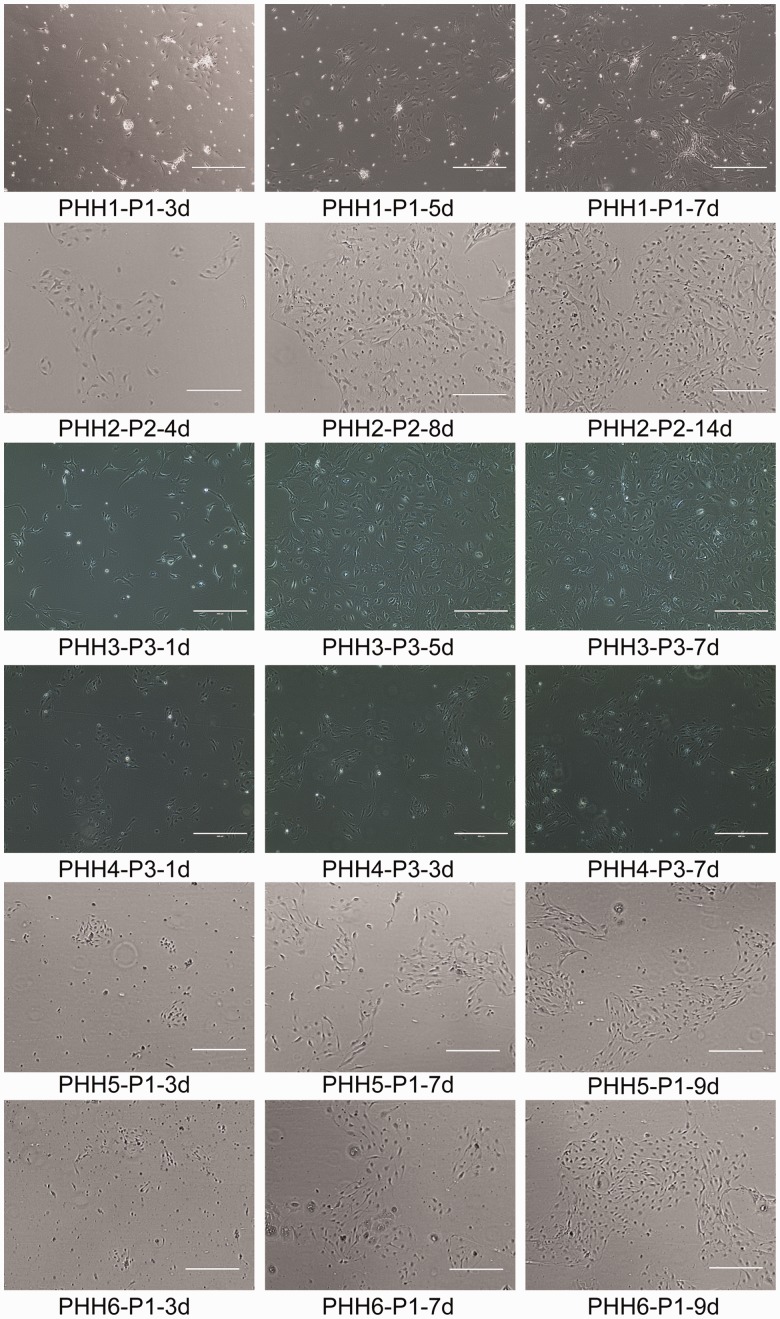
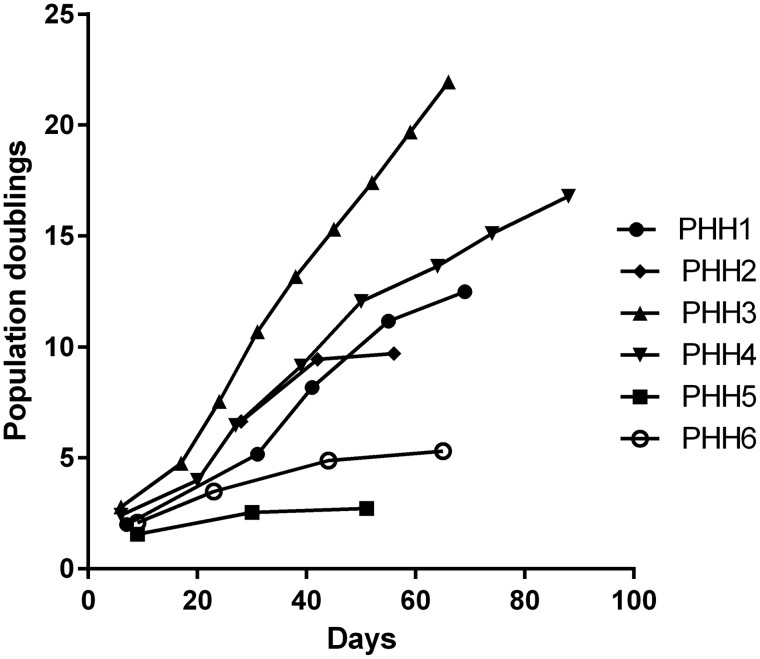
We grew PHHs derived from 11 liver samples from liver transplant patients with diverse diseases and ages using conditional programming (Table 1). While five of them got contaminated due to technical issues, six PHHs survived (Table 1). Figure 1 depicts the phase contrast images of the six PHHs at different passages. Briefly, the isolated cells attached to the surface of the cell culture flask and started to grow within 24 h after incubation, reaching confluency within 10 days (for PHH 1, 3, 4, 5 and 6) or 25 days (for PHH 2). At the beginning of seeding, there were several types of cells attached in the flask. After several days of cultivation, only the cells with hepatocyte appearance remained and other types of cells floated into the medium and were discarded. In addition, we found that PHHs derived from younger patients have higher proliferative activity than those from adults (Figure 2). We selected PHH 1, PHH 2, PHH 3, and PHH 4 for further observation and characterization because of the lack of viability of PHH 5 and PHH 6. PHH 3 and 4, which are derived from a three-years-old and a nine-years-old patient, respectively, could be passaged for more than 10 passages (at least 80 days and 15 population doublings) until the experiment was terminated (Figure 2). On the other hand, PHH 1, 2, 5, and 6, which are derived from 53, 57, 73, and 54-years-old patient, respectively, could only be cultured for 3–5 passages (50–70 days). We observed that the proliferative activity of PHH 1 and 2 was high during the first 30 days and was gradually decreased (Figure 2). STR analysis indicated that all of the four PHHs at different passages are identical with their corresponding original tissues (Table 3).
Figure 1.
The representative phase contrast images of PHHs at different passages across a time course. Magnification, 10×; bar, 400 µm. (A color version of this figure is available in the online journal)
Figure 2.
The proliferative activity of the four PHHs was displayed in different fashions. Cells were cultured in conditional reprogramming F-Y media. The cell number and viability were recorded at each passage and a plot of population doublings versus time (days) was constructed using Graphpad Prism 6.
Table 3.
STR analysis of PHHs at different passages and their respectively original tissues.
| Sample | D3S1358 | TH01 | D21S11 | D18S51 | Penta E | D5S818 | D13S317 | D7S820 | D16S539 | CSF1PO | Penta D | vWA | D8S1179 | TPOX | FGA | AMEL | Mouse |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PHH 1-tissue | 15, 17 | 6, 9 | 28, 31.2 | 14, 15 | 11, 12 | 12, 13 | 8, 12 | 11 | 9, 10 | 11, 13 | 9 | 16, 17 | 13 | 8, 11 | 20, 22 | x | Negative |
| PHH 1-P2 | 15, 17 | 6, 9 | 28, 31.2 | 14, 15 | 11, 12 | 12, 13 | 8, 12 | 11 | 9, 10 | 11, 13 | 9 | 16, 17 | 13 | 8, 11 | 20, 22 | x | Negative |
| PHH 2-tissue | 16 | 6, 9 | 30 | 15 | 8, 11 | 10, 13 | 13 | 11, 13 | 9 | 11, 12 | 11, 12 | 16 | 10, 13 | 8, 12 | 24, 25 | x | Negative |
| PHH 2-P2 | 16 | 6, 9 | 30 | 15 | 8, 11 | 10, 13 | 13 | 11, 13 | 9 | 11, 12 | 11, 12 | 16 | 10, 13 | 8, 12 | 24, 25 | x | Negative |
| PHH 3-tissue | 14, 16 | 6 | 29, 32.2 | 12, 14 | 11, 19 | 11 | 13, 14 | 11, 12 | 10, 11 | 12, 13 | 9, 11 | 14 | 10, 13 | 9, 12 | 21 | x | Negative |
| PHH 3-P6 | 14, 16 | 6 | 29, 32.2 | 12, 14 | 11, 19 | 11 | 13, 14 | 11, 12 | 10, 11 | 12, 13 | 9, 11 | 14 | 10, 13 | 9, 12 | 21 | x | Negative |
| PHH 3-P9 | 14, 16 | 6 | 29, 32.2 | 12, 14 | 11, 19 | 11 | 13, 14 | 11, 12 | 10, 11 | 12, 13 | 9, 11 | 14 | 10, 13 | 9, 12 | 21 | x | Negative |
| PHH 4-tissue | 15 | 6 | 28, 30 | 14, 18 | 10, 12 | 13 | 8, 10 | 9, 10 | 9, 10 | 11, 13 | 12 | 16, 17 | 8, 14 | 8 | 20, 22 | X, Y | Negative |
| PHH 4-P9 | 15 | 6 | 28, 30 | 14, 18 | 10, 12 | 13 | 8, 10 | 9, 10 | 9, 10 | 11, 13 | 12 | 16, 17 | 8, 14 | 8 | 20, 22 | X, Y | Negative |
| PHH 4-P11 | 15 | 6 | 28, 30 | 14, 18 | 10, 12 | 13 | 8, 10 | 9, 10 | 9, 10 | 11, 13 | 12 | 16, 17 | 8, 14 | 8 | 20, 22 | X, Y | Negative |
PHH: primary human hepatocyte; STR: short tandem repeat.
PHH culture is not overwhelmed by fibroblast overgrowth
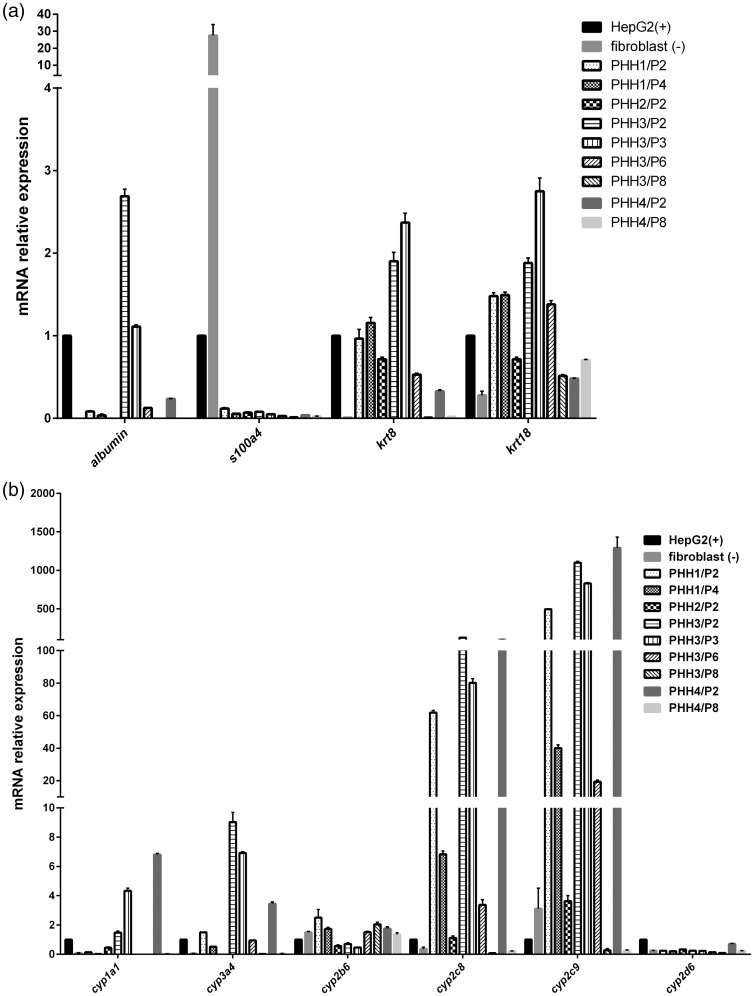
Primary cell growth is often gradually overwhelmed by fibroblast overgrowth, making it difficult to establish primary cell culture.23,24 Epithelial-fibroblast transition is one type of epithelial-mesenchymal transition (EMT). Keratin 8 and keratin 18 are two kinds of cytokeratin, which are markers used to identify epithelial cells to fibroblast in kidney, liver, and lung.25 We measured the mRNA expression of keratin 8 and keratin 18 of all cells in different passages to assess presence of fibroblast overgrowth during the cell culture. We found that the mRNA expression of keratin 8 and keratin 18 of HepG2 (positive control) is significantly higher than that in human fibroblast cells (negative control) (Figure 3(a)). PHH 1 at passages 2 and 4 expressed high amount of keratin 8 and keratin 18. PHH 2 at passage 2 expressed higher amounts of keratin 8 and keratin 18 than human fibroblast cells (Figure 3(a)). PHH 3 and 4, in early passages (up to passages 6 and 2, respectively) expressed significantly higher amount of keratin 8 and keratin 18 (Figure 3(a)).
Figure 3.
mRNA expression of krt8, krt18, s100a4, albumin (a), cyp1a1, 3a4, 2b6, 2c8, 2c9, and 2d6 (b) in four PHHs at different passages, human fibroblast cells (negative control), and HepG2 (positive control). The mRNA levels of corresponding genes relative to gapdh (a house keeping gene) were determined by the quantitative real-time PCR. The results were obtained from three replicate wells.
S100A4 is a biomarker to characterize the mesenchymal products caused by EMT that occur during the formation of fibrosis in various organs.25 The RTPCR results revealed that compared to HepG2, human fibroblast cells expressed excessively high mRNA of s100A4. All the PHHs at any passage did not express s100A4 (Figure 3(a)).
Albumin expression is a specific biomarker of liver function in hepatocyte cultivation. We found that PHH 1 and 2 did not express mRNA of albumin (Figure 3(a)). PHH 3 at passages 2 and 3 expressed high level of albumin, but lost the function from passage 6 (Figure 3(a)). PHH 4 at passage 2 slightly expressed mRNA of albumin, but failed to express it at passage 8 (Figure 3(a)). These results indicate that PHHs can maintain some hepatic functions at early passages without fibroblastic overgrowth.
PHH possesses cytochromes P450 enzymatic activities and express mRNA of cytochromes P450 enzymes
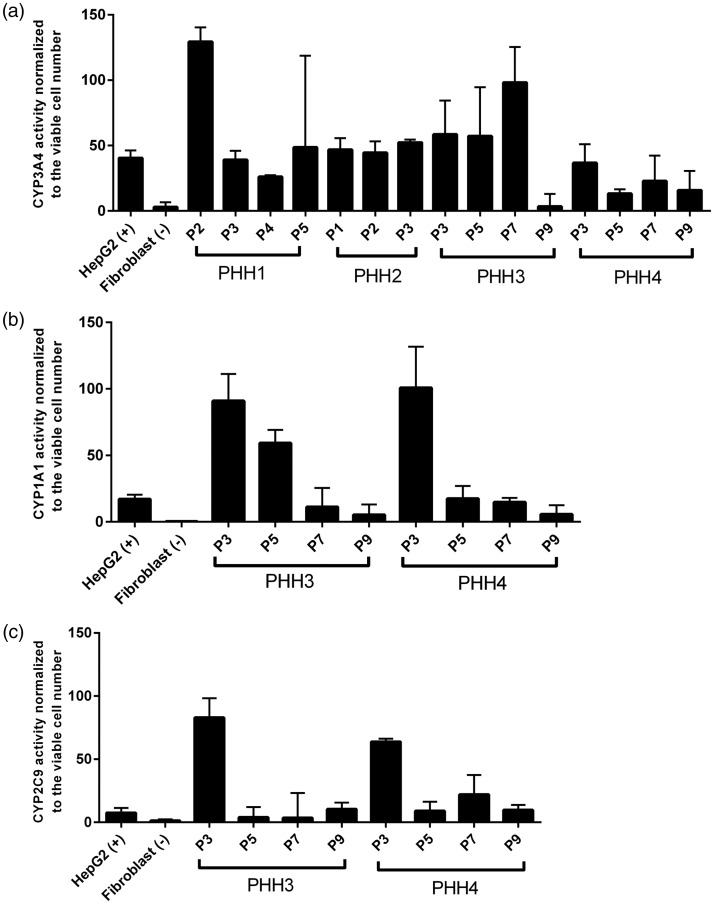
Human cytochromes P450 enzymes superfamily predominantly exist in liver organ to metabolize drugs and other xenobiotics. CYP3A4 is the member with the highest abundance and it metabolizes the majority of drugs with known metabolic pathways.26 CYP1A1, a member of the CYP1 family, is responsible for detoxification of drugs and metabolic activation of polycyclic aromatic hydrocarbons and aromatic amines.27 CYP2C9 typically metabolizes nonsteroidal anti-inflammatory drugs and hypoglycemic.26 We measured the enzymatic activities of CYP1A1, CYP3A4, and CYP2C9 when we passaged the cells. We found that PHH 3, in early passages (up to passage 7), exhibited strong enzymatic activities of CYP1A1 and CYP3A4 (Figure 4(a) and (b)). Only in passage 3, PHH 3 displayed stronger CYP2C9 activity than HepG2 did (Figure 4(c)). Similar to PHH 3, PHH 4, up to passage 7, showed strong activities of CYP1A1 and CYP3A4 (Figure 4(a) and (b)). PHH 4 displayed stronger CYP2C9 activity, until passage 9, than HepG2 did (Figure 4(c)). Both PHH 1 and 2 exhibited strong CYP3A4 activity throughout their lifespan (Figure 4(a)). Due to lack of viable cells, we did not test CYP1A1 and 2C9 activities of PHH1 and 2. We further determined mRNA expression of cyp1a1, cyp3a4, cyp2b6, cyp2c8, cyp2c9, and cyp2d6. Compared to HepG2, PHH 3 expressed significantly higher mRNA level of cyp2c8 and cyp2c9 until passage 6 and higher mRNA level of cyp1a1, cyp3a4 until passage 3 (Figure 3(b)). PHH 4, at passage 2, expressed significantly higher mRNA level of cyp1a1, cyp3a4, cyp2c8, and cyp2c9 (Figure 3(b)). However, the cyp2b6 and cyp2d6 mRNA expression between PHH, HepG2, and fibroblast did not exhibit differences (Figure 3(b)). Both PHH 3 and PHH 4 failed to express high mRNA level of CYP450 members and displayed high CYP450 enzymatic activities at late passages (Figure 3(b)). PHH 1 and 2 displayed a different fashion compared to PHH 3 and 4 (Figure 3(b)). Compared to HepG2, PHH 1 expressed significantly higher mRNA level of cyp3a4, cyp2c8, and cyp2c9 and PHH 2 only expressed significantly higher mRNA level of cyp2c9 (Figure 3(b)).
Figure 4.
CYP 3A4 (a), 1A1 (b) and 2C9 (c) activities of four PHHs. The assays were carried out following the protocol of P450-Glo CYP3A4, 1A1 and 2C9 assay kits. The enzymatic activities were normalized to cell viability. HepG2 and human fibroblast cell were positive control and negative control, respectively. The results were obtained from three replicate readings.
Discussion
Studies that involve human hepatocyte culture are limited due to the lack of reliable techniques for PHH cultivation to generate a large number of viable hepatocyte. Commercially available human hepatocytes are costly and can only survive for several days. Recently developed methods to culture hepatocyte include coculture with endothelial cells or 3T3-J2 fibroblasts, collagen double-gel configuration, and hepatocyte spheroids.7 The cocultivation of human hepatocyte with endothelial cells or Swiss 3T3-J2 mouse fibroblasts could long-term maintain the liver-specific functions through heterotypic interactions and production of extracellular materials.28,29 However, the co-culture technique of PHHs always leads to contamination with other cells. It has been reported that direct physical contact between feeder cells and keratinocyte epithelial is not required for the maintenance of indefinite proliferative state of epithelial cells.30 Collagen sandwich configuration and spheroid culture provide 3D-like environment for hepatocyte and could promote the lifespan and hepatic functions of human hepatocyte. However, they cannot generate a large amount of cells.7,31
In the present study, we demonstrate, for the first time, that conditional programming is applicable to long-term expansion of PHH derived from liver patients with broad pathological conditions, including cirrhosis, liver failure, and metabolic diseases. The technology enables physical separation between PHH and feeder cells, thus providing pure PHH, as compared to traditionally used co-culture methods. All PHHs survived more than one month using conditional programming excluding the five PHHs which got contaminated with fungi or bacteria during the first several days after in vitro cultivation. The potential use of the extended culture time of PHH may allow long-term maintenance of the cells for characterization of their physiological or pathophysiological features. Additionally, the method enables researchers to perform experiments with sufficient biological replicates using the same human hepatocyte sample. We found that the PHHs derived from young patients (PHH 3, 4, from 3- and 9-years-old patients, respectively) survived longer and possessed higher proliferative activity than the PHHs derived from adult patients (PHH 1, 2, 5 and 6, from 66-, 68-, 72- and 53-years-old patients, respectively) (Figure 2). Comparing albumin mRNA expression between the two groups, we observed that a decrease in albumin mRNA expression coincides with an increase in age (Figure 3(a)). The differences in lifespan and albumin mRNA expression between the two groups likely suggest that the patient’s age may play an important role in the long-term proliferation of PHH in vitro, which is in agreement with previous studies.32,33 It should be also noted that the nature of liver disease may have influence on the capacity of the PHH being long-term cultured in vitro, giving rise to the difference in the passaging time. Strikingly, PHH 1, 2, 3, and 4 at early passages maintain high CYP450 activities and mRNA expression (Figures 3(b) and 4(a) to (c)). This implies that conditional programming could generate large number of viable cells with hepatic functions to predict drug toxicity, side effects, and metabolism. These observations may also suggest that cells from different patients have somewhat different growth condition requirements. Although the medium may not be optimal for growth of all PHH, it appears that conditional programming is generally applicable. This is evident from a high level of success rate we achieved in the long-term in vitro cultivation of PHH from a range of liver disease patients. The technology is a simple, inexpensive yet efficient cultivation technique that can be potentially applied to establish in vitro pure hepatocyte cultivation to offer a large amount of cell source.
We conclude that conditional programming is of considerable advantage as compared to other culture systems. Optimizations of culture medium are currently ongoing to prolong the lifespan and enhance hepatic functions. We believe the finding in this study will enable our team and other researchers in this challenging field to enhance future studies on long-term cultivation of PHHs.
Authors’ contributions
Su S, Roy R, Liu X and Ressom WH designed the study. Cui W and Kroemer A provided fresh liver tissues. Su S and Di Poto C conducted the experiments. Su S analyzed data and wrote the manuscript. Ressom WH, Roy R, Liu X, Kroemer A and Cui W revised the manuscript. All authors read and approved the final manuscript.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The work presented in this paper is supported by NIH grant U01CA185188 awarded to HWR.
References
- 1.Karlsen T, Lammert F, Thompson R. Genetics of liver disease: from pathophysiology to clinical practice. J Hepatol 2015; 62:S6–14 [DOI] [PubMed] [Google Scholar]
- 2.Kaplowitz N. Drug-induced liver injury. Clin Infect Dis 2004; 38:S44–8 [DOI] [PubMed] [Google Scholar]
- 3.Farazi P, DePinho R. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer 2006; 6:674–87 [DOI] [PubMed] [Google Scholar]
- 4.Guillouzo A. Liver cell models in in vitro toxicology. Environ Health Perspect 1998; 106:511–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitra A, Mishra L, Li S. Technologies for deriving primary tumor cells for use in personalized cancer therapy. Trends Biotechnol 2013; 31:347–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fiegel H, Kaufmann P, Bruns H, Kluth D, Horch R, Vacanti J, Kneser U. Hepatic tissue engineering: from transplantation to customized cell-based liver directed therapies from the laboratory. J Cell Mol Med 2008; 12:56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shulman M, Nahmias Y. Long-term culture and coculture of primary rat and human hepatocytes. Epithelial cell culture protocols 2nd ed. Berlin: Spinger, 2013, pp.287–302. [DOI] [PMC free article] [PubMed]
- 8.Godoy P, Hewitt N, Albrecht U, Anderson M, Ansari N, Bhattacharya S, Bode J, Bolleyn J, Borner C, Böttger J, Braeuning A, Budinsky R, Burkhardt B, Cameron N, Camussi G. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol 2013; 87:1315–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryan C, Carter E, Jenkins R, Sterling L, Yarmush M, Malt R, Tompkins R. Isolation and long-term culture of human hepatocytes. Surgery 1993; 113:48–54 [PubMed] [Google Scholar]
- 10.Ploss A, Khetani S, Jones C, Syder A, Trehan K, Gaysinskaya V, Mu K, Ritola K, Rice C, Bhatia S. Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proc Natl Acad Sci USA 2010; 107:3141–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lecluyse E, Alexandre E. Isolation and culture of primary hepatocytes from resected human liver tissue. Methods Mol Biol 2010; 640:57–82 [DOI] [PubMed] [Google Scholar]
- 12.LeCluyse E. Human hepatocyte culture systems for the in vitro evaluation of cytochrome P450 expression and regulation. Eur J Pharm Sci 2001; 13:343–68 [DOI] [PubMed] [Google Scholar]
- 13.Bhatia S, Yarmush M, Toner M. Controlling cell interactions by micropatterning in co-cultures: hepatocytes and 3T3 fibroblasts. J Biomed Mater Res 1997; 34:189–99 [DOI] [PubMed] [Google Scholar]
- 14.Mitaka T, Sato F, Mizuguchi T, Yokono T, Mochizuki Y. Reconstruction of hepatic organoid by rat small hepatocytes and hepatic nonparenchymal cells. Hepatology 1999; 29:111–25 [DOI] [PubMed] [Google Scholar]
- 15.Goral V, Hsieh Y, Petzold O, Clark J, Yuen P, Faris R. Perfusion-based microfluidic device for three-dimensional dynamic primary human hepatocyte cell culture in the absence of biological or synthetic matrices or coagulants. Lab Chip 2010; 10:3380–6 [DOI] [PubMed] [Google Scholar]
- 16.Meng Q. Three-dimensional culture of hepatocytes for prediction of drug-induced hepatotoxicity. Expert Opin Drug Metab Toxicol 2010; 6:733–46 [DOI] [PubMed] [Google Scholar]
- 17.Liu X, Krawczyk E, Suprynowicz F, Palechor-Ceron N, Yuan H, Dakic A, Simic V, Zheng Y, Sripadhan P, Chen C, Lu J, Hou T, Choudhury S, Kallakury B, Tang D, Darling T, Thangapazham R, Timofeeva O, Dritschilo A, Randell S, Albanese C, Agarwal S, Schlegel R. Conditional reprogramming and long-term expansion of normal and tumor cells from human biospecimens. Nat Protoc 2017; 12:439–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heng B. Effect of Rho-associated kinase (ROCK) inhibitor Y-27632 on the post-thaw viability of cryopreserved human bone marrow-derived mesenchymal stem cells. Tissue Cell 2009; 41:376–80 [DOI] [PubMed] [Google Scholar]
- 19.Terunuma A, Limgala R, Park C, Choudhary I, Vogel J. Efficient procurement of epithelial stem cells from human tissue specimens using a Rho-associated protein kinase inhibitor Y-27632. Tissue Eng Part A 2010; 16:1363–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, Timofeeva O, Nealon C, Dakic A, Simic V, Haddad B, Rhim J, Dritschilo A, Riegel A, McBride A, Schlegel R. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol 2012; 180:599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapman S, Liu X, Meyers C, Schlegel R, McBride A. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. J Clin Invest 2010; 120:2619–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25:402–8 [DOI] [PubMed] [Google Scholar]
- 23.Murakami H, Masui H. Hormonal control of human colon carcinoma cell growth in serum-free medium. Proc Natl Acad Sci USA 1980; 77:3464–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terano A, Ivey K, Stachura J, Sekhon S, Hosojima H, Mckenzie W, Jr, Krause W, Wyche J. Cell culture of rat gastric fundic mucosa. Gastroenterology 1982; 83:1280–91 [PubMed] [Google Scholar]
- 25.Kalluri R, Weinberg R. The basics of epithelial-mesenchymal transition. J Clin Invest 2009; 119:1420–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anzenbacher P, Anzenbacherova E. Cytochromes P450 and metabolism of xenobiotics. Cell Mol Life Sci 2001; 58:737–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelkonen O, Turpeinen M, Hakkola J, Honkakoski P, Hukkanen J, Raunio H. Inhibition and induction of human cytochrome P450 enzymes: current status. Arch Toxicol 2008; 82:667–715 [DOI] [PubMed] [Google Scholar]
- 28.Salerno S, Campana C, Morelli S, Drioli E, De Bartolo L. Human hepatocytes and endothelial cells in organotypic membrane systems. Biomaterials 2011; 32:8848–59 [DOI] [PubMed] [Google Scholar]
- 29.Clement B, Guguen-Guillouzo C, Campion J, Glaise D, Bourel M, Guillouzo A. Long-term co-cultures of adult human hepatocytes with rat liver epithelial cells: modulation of albumin secretion and accumulation of extracellular material. Hepatology 1984; 4:373–80 [DOI] [PubMed] [Google Scholar]
- 30.Palechor-Ceron N, Suprynowicz F, Upadhyay G, Dakic A, Minas T, Simic V, Johnson M, Albanese C, Schlegel R, Liu X. Radiation induces diffusible feeder cell factor(s) that cooperate with ROCK inhibitor to conditionally reprogram and immortalize epithelial cells. Am J Pathol 2013; 183:1862–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunn J, Tompkins R, Yarmush M. Long-term in vitro function of adult hepatocytes in a collagen sandwich configuration. Biotechnol Prog 1991; 7:237–45 [DOI] [PubMed] [Google Scholar]
- 32.Vondran F, Katenz E, Schwartlander R, Morgul M, Raschzok N, Gong X, Cheng X, Kehr D, Sauer I. Isolation of primary human hepatocytes after partial hepatectomy: criteria for identification of the most promising liver specimen. Artif Organs 2008; 32:205–13 [DOI] [PubMed] [Google Scholar]
- 33.Lloyd T, Orr S, Patel R, Crees G, Chavda S, Vadyar H, Berry D, Sherlock D, Dennison A. Effect of patient, operative and isolation factors on subsequent yield and viability of human hepatocytes for research use. Cell Tissue Bank 2004; 5:81–7 [DOI] [PubMed] [Google Scholar]