Abstract
Inactivation of the tumor suppressor p53 and/or overexpression of the oncogene MDM2 frequently occur in human cancers, and are associated with poor prognosis, advanced forms of the disease, and chemoresistance. MDM2, the major negative regulator of p53, induces p53 degradation and inactivates its tumor suppressing activity. In turn, p53 regulates MDM2 expression. This MDM2-p53 negative feedback loop has been widely studied and presents an attractive target for cancer therapy, with a few of the inhibitors of this interaction already having advanced into clinical trials. Additionally, there is an increasing interest in understanding MDM2’s p53-independent activities in carcinogenesis and cancer progression, which may also have implications for cancer therapy. This review aims to highlight the various roles that the MDM2-p53 interaction plays in cancer, the p53 independent oncogenic activities of MDM2 and the various strategies that may be used to target MDM2 and the MDM2-p53 interaction. We will summarize the major preclinical and clinical evidences of MDM2 inhibitors for human cancer treatment and make suggestions to further improve efficacy and safety of this interesting class of cancer therapeutics.
Keywords: Drug target, MDM2, p53, MDM2-p53 interaction, small molecule inhibitor
1. INTRODUCTION
Human malignancies remain a leading cause of death in both women and men worldwide; there are still major challenges in understanding cancer etiology, pathogenesis, treatment and prevention. It is widely accepted that carcinogenesis and cancer progression are complex and multi-factorial processes with the emergence of cells possessing features such as sustained proliferation, evasion of growth suppressors, resistance to cell death and replicative mortality, increased angiogenesis, and activation of invasion and metastasis [1]. The cellular changes at genetic and epigenetic levels during carcinogenesis and cancer progression have been the center of cancer research for several decades. These genetic changes involve deletions, mutations, and rearrangements in a set of specific genes (typically oncogenes and tumor suppressors) that may affect their protein products. Oncogenes generally encode cell proliferation and apoptosis-controlling proteins; and their amplification or activation leads to development of the malignant phenotype [2–4]. In contrast, tumor suppressor genes activate anti-proliferative and pro-apoptotic pathways, thus halting cell cycle progression; and inactivation or loss of tumor suppressor genes also leads to the malignant phenotype.
Since its discovery more than thirty years ago, p53 has become the most widely studied tumor suppressor gene [5]. This short-lived protein has multiple functions in normal cells and is essential in preventing cancer onset and development. There are several outstanding reviews which document the functions and regulations of p53 [6–17]. As a transcription factor, p53 is responsible for maintaining genomic integrity and is activated in response to diverse stress signals, leading to DNA repair, cell cycle arrest, apoptosis, and senescence [9, 16]. The importance of p53 in cancer biology and etiology can be gauged by the fact that this protein is deleted or mutated in more than half of all human malignancies [15]; with the loss of its wild-type function negatively impacting prognosis and response to cancer therapy. These observations render the restoration of p53 functions in the cancerous cell as an attractive anticancer approach. However, after more than two-decades of intensive preclinical and clinical research, activation of p53, including p53 gene therapy, has not been proven to be a practical clinical approach to cancer therapy. One of the reasons for ineffective consequence of p53 therapy may be the inactivation of endogenous and exogenous p53 by its negative regulators such as MDM2 (murine double minute 2) [18].
MDM2 is a cellular phosphoprotein that forms a complex with p53 [18–22]. MDM2 has E3 ubiquitin ligase activity which plays a critical role in the degradation of p53 [23]. MDM2 is overexpressed in many human malignancies, indicating that it is a major mechanism utilized by cancer cells to escape p53 surveillance [24]. Indeed, the MDM2 gene is frequently amplified in most cancers; and alterations in the p53 and MDM2 genes and/or their protein products, separately or concomitantly, lead to poor prognosis and treatment failure in cancer patients [25, 26]. Therefore, considering the obstacles faced in p53-based cancer therapy [27–31], we [32, 33] and others [34] have proposed that MDM2 is a valuable molecular target for cancer therapy. In the last 15 years or so, the MDM2-p53 interaction has become a focal point of research in both academia and the industry to develop better targeted cancer therapeutics [28, 30, 33, 34]. In this review, we aim to critically evaluate the rationale and status of targeting MDM2 and advances made in the development of MDM2 inhibitors as novel cancer therapeutics and what the future holds for this class of novel cancer therapeutics.
2. MDM2, p53, AND CANCER
2.1. MDM2 As A Negative Regulator of p53
The p53 protein was first discovered in 1979 as a cellular partner of simian virus 40 large T-antigen; it was found to migrate as a 53-kD band in gel electrophoresis, thus imparting its name [6]. Subsequent studies showed that p53 maintains genomic integrity of the cell, prevents malignant transformation, induces cell cycle arrest and apoptosis in response to stress signals [16]. Disruption of this gene facilitates the oncogenic process leading to increased cancer risk [12, 13]. Indeed, p53 mutations are seen in more than 50% of all human cancers, being highly prevalent in both solid cancers as well as in leukemias and lymphomas; thus providing an insight into the genetic machinery that controls the carcinogenic process [15]. With the discovery of MDM2, a more complete picture of the p53 pathway emerged a decade after p53 discovery.
The mdm2 gene is located on chromosome 12q13–14 and encodes for a 491 amino acid protein [19]. Under normal conditions, MDM2 is expressed in the nucleus, but it trans-locates to the cytoplasm to mediate the degradation of its substrates. The first indication that MDM2 acts as an oncogene came from the observation that it could cause spontaneous transformation of an immortalized murine cell line, BALB/c 3T3 and that mdm2 overexpression rendered rodent fibroblasts tumorigenic in nude mice [20]. The direct evidence for the crucial role of MDM2 in negatively regulating p53 comes from the fact that targeted deletion of the mdm2 gene in mice is embryonic lethal due to p53-mediated apoptosis, whereas simultaneous deletion of the TP53 gene rescues the lethality [35, 36].
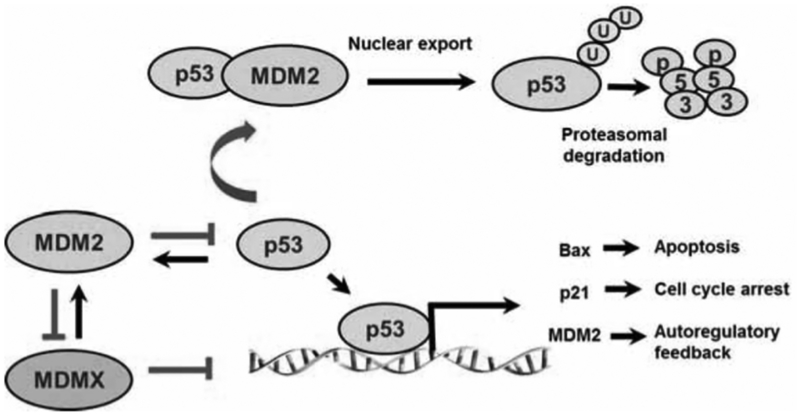
The auto-regulatory feedback loop between MDM2 and p53 may be the most important finding in this field. As illustrated in (Fig. 1), the MDM2 protein binds to the N-terminal transactivation domain of p53, inhibiting its transcriptional activity [37–39], promotes p53 export out of the nucleus [40, 41], prevents p53 from interacting with transcriptional co-activators [42], and targets p53 for ubiquitination and degradation by the proteasome [41, 43, 44]. On the other hand, p53 regulates the transcription of the mdm2 gene, thus forming a negative feedback loop that maintains cellular p53 at low level [45]. The MDM2-p53 interaction was initially thought to result from the mutual binding of MDM2 and p53 via their N-terminal domains, but recently the p53 C-terminus has also been shown to be involved in the MDM2-p53 interaction [46, 47]. The mechanisms responsible for the MDM2-induced p53 degradation have been intensively investigated. MDM2 serves as an E3 ubiquitin ligase via its C-terminal RING finger domain and ubiquitinates p53 at several lysine residues [23]. While low levels of MDM2 induce p53 mono ubiquitination and nuclear export, high levels of MDM2 cause polyubiquitination and degradation of p53 in the nucleus [48, 49]. MDM2 can also auto ubiquitinate itself, leading to self-degradation [23].
Fig. (1). The MDM2-p53 Interaction.
MDM2 binds to p53 and mediates its ubiquitination, leading to the nuclear export of p53 and proteasomal degradation. The MDM2-p53 interaction also masks the transactivation domain of p53, inhibiting its target gene expression. In turn, p53 induces the transcription of MDM2, forming a negative feedback loop that tightly maintain p53 protein at low level in normal cells.
Several studies demonstrate that the MDM2-p53 interaction is more complicated than previously thought. There have now been increasing evidences indicating that the MDM2-p53 feedback loop is regulated or modulated by a host of factors (as summarized in Table 1), including ribosomal proteins [50–63], proteasome activator (PA28γ) [64], polycomb protein RNF2 [65, 66] and RYBP [67], the ARF tumor suppressor [68–71], DNA damage response elements (14-3-3-σ, PML, and JMY) [72–74], and MDMX [75–77], among others. These molecules may bind to MDM2 and/or p53, altering their conformation, modification, interaction, and modulating the E3 ligase activity of MDM2 towards itself and p53. This complex interplay of cellular players adds multiple layers of regulation to the MDM2-p53 loop. The MDMX-MDM2-p53 interaction can serve as an excellent example of the complexity of p53 signaling pathways. MDMX (also referred as MDM4) possesses a high degree of homology to MDM2, especially in its N-terminal p53 binding domain [75, 77]. Similar to MDM2, it possesses, at its N-terminus, a p53 binding domain and at its C-terminus, a RING finger domain through which it forms hetero dimers with MDM2. MDMX is highly and/or abnormally expressed in human tumors and seems to promote carcinogenesis [77–79]. MDM2 and MDMX perform non-redundant functions to inactivate p53 during embryogenesis and throughout development [79–81]. MDM2 and MDMX are proposed to work independently to inhibit p53 activity, or alternatively, MDM2 and MDMX may form a complex that is more effective at inhibiting p53 transactivation or enhancing p53 turnover. MDM2 and MDMX are also postulated to form heterooligomers through their RING domains, whereby MDMX increases MDM2’s E3 ligase activity (as shown in Fig. 1). MDM2 can also directly ubiquitinate and degrade MDMX in response to DNA-damage stimuli [82–85].
Table 1.
Summary of Proteins Modulating the MDM2-p53 Loop
| MDM2-p53 Loop Modulator | Outcome | Mechanism | References |
|---|---|---|---|
| ARF | Stabilizes and/or activates p53 | Disrupts MDM2-p53 interaction, sequesters MDM2 to nucleolus, and promotes MDM2 degradation | [68, 69, 86] |
| NPM | Stabilizes and/or activates p53 | Competes with p53 for MDM2 binding | [87] |
| HIPK2 | Stabilizes and/or activates p53 | Prevents MDM2-mediated p53 nuclear export and ubiquitination | [88] |
| PML | Stabilizes and/or activates p53 | Protects p53 from MDM2-mediated inhibition and degradation by sequestering MDM2 into nucleus | [89–92] |
| 14–3-3-σ | Stabilizes and/or activates p53 | Disrupts MDM2-p53 interaction by translocating MDM2 into cytoplasm | [74] |
| PCAF | Stabilizes and/or activates p53 | Disrupts MDM2-p53 interaction and stimulates MDM2 auto-ubiquitination; Acetylates p53 in response to DNA damage | [93, 94] |
| SENP3 | Stabilizes and/or activates p53 | Binds to MDM2 and inhibits p53 ubiquitination | [95] |
| Tip60 | Stabilizes and/or activates p53 | Disrupts MDM2-p53 interaction by localizing p53 to PML bodies; Inhibits NEDDylation of p53 by MDM2 and promotes p53 acetylation | [96–98] |
| Caspase-2 | Stabilizes and/or activates p53 | Cleavage of MDM2 at Asp-367, leading to loss of C-terminal RING domain | [99] |
| pRb | Stabilizes and/or activates p53 | Forms trimeric complex with p53 and MDM2 to protect p53 pro-apoptotic activity | [100, 101] |
| RYBP | Stabilizes and/or activates p53 | Interacts with MDM2 to decrease MDM2-mediated p53 ubiquitination | [67] |
| TRIAD1 | Stabilizes and/or activates p53 | Binds to p53 C-terminus and facilitate its dissociation from MDM2 | [102, 103] |
| Aurora-A | Stabilizes and/or activates p53 | Prevents MDM2-p53 interaction by phosphorylating p53 at Ser-106 | [104] |
| Ribosomal proteins (L5/L11/L23/L26/S7/S14/S25/S27/S27L/S27a) | Stabilizes and/or activates p53 | Bind to the central acidic domain of MDM2 and inhibits its E3 ubiquitin ligase activity towards p53 | [50–63] |
| PAK1IP1 | Stabilizes and/or activates p53 | Interacts with MDM2 and inhibits p53 ubiquitination | [105] |
| DNAJC7 | Stabilizes and/or activates p53 | Dissociates MDM2 from p53 | [106] |
| Otubain-1 | Stabilizes and/or activates p53 | Binds to MDM2 cognate E2, UbcH5 and inhibits MDM2-mediated p53 ubiquitination | [107] |
| MDMX | Destabilizes and/or inactivates p53 | Hetero-oligomerization of MDM2 and MDMX via their RING domains enhances p53 ubiquitination | [75, 84, 108–110] |
| SENP2 | Destabilizes and/or inactivates p53 | Desumolyates MDM2 and permits its binding and ubiquitination of p53 | [111] |
| Enigma | Destabilizes and/or inactivates p53 | Binds to MDM2 and forms a ternary complex with p53; inhibits MDM2 self-ubiquitination and E3 ligase activity towards p53 | [112] |
| YY1 | Destabilizes and/or inactivates p53 | Promotes assembly of the MDM2-p53 complex, and blocks p300-dependent p53 acetylation and stabilization. | [113, 114] |
| RNF2 | Destabilizes and/or inactivates p53 | Binds with both p53 and MDM2 and promotes MDM2-mediated p53 ubiquitination | [65, 66] |
| PA28γ | Destabilizes and/or inactivates p53 | Binds with both p53 and MDM2 and increases MDM2-mediated p53 ubiquitination | [64] |
| HAUSP | Destabilizes and/or inactivates p53 | Deubiquitinates MDM2 leading to increased p53 degradation | [115] |
| SKI | Destabilizes and/or inactivates p53 | Increases MDM2 stability by stimulating sumoylation of MDM2 | [116] |
| Siva-1 | Destabilizes and/or inactivates p53 | Binds to both p53 and MDM2 and enhances MDM2-mediated p53 ubiquitination and degradation | [117] |
2.2. MDM2 As An Oncogene, Independent of p53
In addition to its role in the MDM2-p53 loop, recent findings indicate that MDM2 also has critical roles in carcinogenesis, independent of p53. For example, bladder cancer and sarcoma patients with increased MDM2 levels and mutant p53 present a worse prognosis than those with a single defect [26]. In animal studies, 33% lymphomas that arise in Eμ-myc transgenic mice with deleted or mutated p53 show overexpression of MDM2 [86, 118]. Studies with genetically engineered mice support the hypothesis that MDM2 has p53-independent functions that contribute to carcinogenesis. p53−/−mdm2+/−mice and p53−/−mice expressing an MDM2 trans-gene develop more sarcomas than p53−/−mice [119]. Many investigations have now identified numerous MDM2 interacting molecules that are involved in cell proliferation, apoptosis, and tumor invasion and metastasis [120–123].
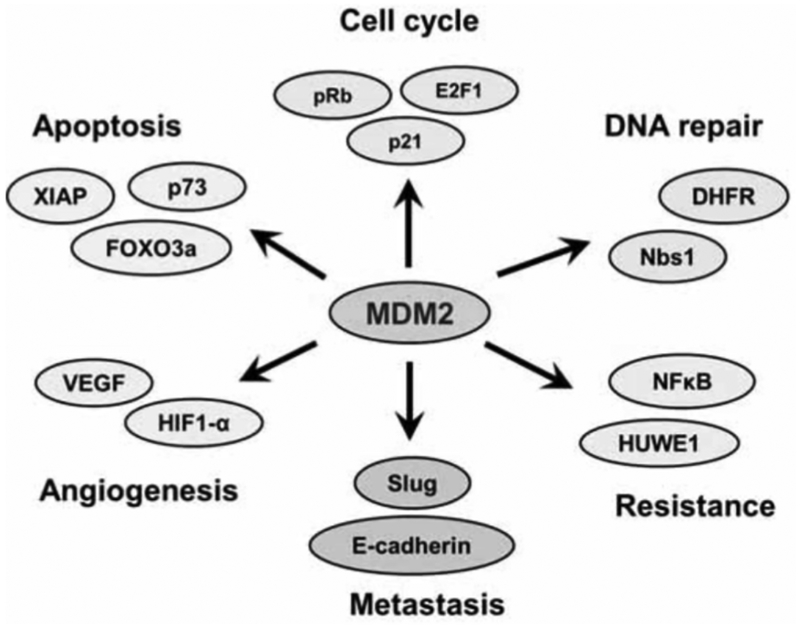
The representative p53-indpendent activities of MDM2 in carcinogenesis and cancer progression are depicted in (Fig. 2). In addition to the inhibition of p53, MDM2 overexpression in cancer cells influences various cellular signaling pathways and contributes to tumor development and progression. MDM2 binds and regulates multiple proteins that are involved in cell cycle control, apoptosis, DNA repair, cell migration and invasion, angiogenesis, and chemo resistance. These signaling pathways work together to ensure the integrity of genetic information, and as such MDM2 may be considered as a central meeting point regulating genome stability, cell transformation and ultimately tumor formation and progression.
Fig. (2). MDM2: p53-independent activities.
MDM2 regulates a variety of cellular proteins and contributes to multiple processes of carcinogenesis and cancer progression in a p53-independent manner (see text for more information).
The MDM2’s p53-independent activities have been intensively investigated, but remain not fully understood. MDM2 overexpression may contribute to genomic instability seen in cancer cells. For instance, MDM2 contributes to chromosomal breakage and failure of genomic integrity by direct interaction with proteins such as dihydrofolate reductase (DHFR), a key enzyme in folate metabolism [124]. MDM2 also plays an important role in DNA repair mechanisms by binding directly to the DNA repair protein Nbs1 [125]. MDM2 overexpression inhibits p21, the cyclin-dependent kinase inhibitor [126]. MDM2 directly interacts with p21, inducing its conformational change and ubiquitin-independent proteasomal degradation [126–128]. This may at least, in part, explain the MDM2’s p53-independent activity in promoting cell proliferation.
Furthermore, MDM2 overexpression drives cell cycle progression in the S-phase by binding with retinoblastoma (Rb) protein and E2F-1, members of the Rb-E2F tumor suppressor pathway [129–131]. MDM2 stabilizes E2F-1, inducing E2F-1 transcriptional activation and ultimately cell cycle progression, while inhibiting the ability of Rb to induce G1 arrest. MDM2 regulates Rb protein stability through both its ubiquitin ligase activity as well as by directly interacting with the C8 subunit of the 20S proteasome, thereby facilitating its destruction [129]. Interestingly, MDM2-dependent degradation of Rb increases DNA methyltransferase DNMT3A activity which is associated with silencing of tumor suppressor genes [132]. MDM2 overexpression also inhibits apoptosis induced by co-overexpression of E2F-1 and DP-1(E2F dimerization partner 1), in osteosarcoma cells lacking p53 and Rb [133].
Other evidence supporting MDM2’s anti-apoptotic roles includes its interaction with the pro-apoptotic proteins p73 and FOXO3a [134–136]. MDM2 mediates the NED Dylation of the pro-apoptotic form of p73-TAp73, promoting its cytoplasmic translocation and mitigating its p53-transactivational activity [134]. MDM2 negatively regulates the stability of FOXO3a by mediating its ubiquitination and degradation [135]. FOXO3a is also responsible for regulating p27, which leads to MDM2-mediated control of cell cycle progression via oncogenic growth factor or Ras activation [137]. In addition, MDM2 positively regulates XIAP (X-linked inhibitor of apoptosis protein) by enhancing its translation and subsequent expression [138].
Apart from its roles in cell cycle and apoptosis control, MDM2 also regulates angiogenesis by increasing expression levels of transcription factors such as hypoxia inducible factor (HIF-1α), subsequently increasing the expression of vascular endothelial growth factor (VEGF) [139–141]. In addition, MDM2 has been identified in the epithelial-tomesenchymal transition process via its capability to target E-cadherin for proteasomal degradation [142], implicating its role in cancer metastasis. MDM2 also interacts with MTBP (MDM2 binding protein), a metastasis suppressor, and over-expression of MDM2 can reverse MTBP’s function, indicating a potential role for MDM2 in cancer metastasis [143]. MDM2 binds to and stabilizes Slug mRNA, increasing its expression and cellular invasiveness in p53-null cells [144]. MDM2 contributes to chemo resistance by degrading HUWE-1 which ubiquitinates the antiapoptotic protein Mcl-1 [145]. Further, MDM2 enhances p65-mediated transcriptional activity of NFκB in ALL cells, leading to resistance towards doxorubicin-induced apoptosis [146].
In brief, as discussed above, the p53-independent activities of MDM2 are important because more than half of all human cancers exhibit mutated and non-functional forms of p53. If MDM2 is to be considered as a valid and viable therapeutic target, the oncogenic activity of MDM2, independent of its role as a negative regulator of p53, must be understood in greater detail [147].
2.3. Clinical Relevance of Targeting MDM2 for Cancer Therapy and Prevention
Multiple lines of preclinical and clinical evidences suggest the existence of a direct relationship between MDM2 and cancer development and progression [26] (Table 2). MDM2 overexpression has been observed in many cancer types [148, 151, 156, 165, 179], with the overall frequency of gene amplification being 7% and the highest frequency being observed in soft tissue tumors [24]. These tumors often show a higher incidence of MDM2 amplification than p53 mutation. Overall, a negative correlation is seen between occurrence of p53 mutations and MDM2 amplification, thus supporting the hypothesis that MDM2 negatively regulates p53. In many cases, a higher frequency of MDM2 protein overexpression is observed in tumors with wild-type p53.
Table 2.
MDM2 Gene and/or Protein Alterations in Human Cancers
| Cancer Type | MDM2 Expression | Clinical Outcome | References |
|---|---|---|---|
| Breast | mdm2 gene amplification | Poor prognosis; reduced survival; advanced metastasis | [148, 149] |
| Prostate | MDM2 protein overexpression | Increased tumor growth and metastasis | [150–152] |
| Lung | mdm2 gene amplification MDM2 protein overexpression | Poor prognosis; decreased overall survival | [153–155] |
| Colorectal | MDM2 protein overexpression | Predictor of metastasis; correlation with advanced stage of the disease | [156–159] |
| Esophagus | mdm2 gene amplification | Shorter survival Increased chemo/radioresistance | [24, 26, 160, 161] |
| Osteosarcoma | mdm2 gene amplification | Advanced disease and metastasis | [24, 26, 162] |
| Glioblastoma | mdm2 gene amplification | Poor prognosis | [24, 26] |
| Liposarcoma | mdm2 gene amplification | Poor prognosis | [163] |
| Endometrial | SNP309 polymorphism mdm2 gene amplification | Increased risk | [24, 164] |
| Gastric | MDM2 protein overexpression | Poor prognosis; increased risk | [165] |
| Pancreatic | MDM2 protein overexpression | Increased risk | [166] |
| Ovarian | SNP309 polymorphism MDM2 protein overexpression | Poor prognosis; increased risk | [167, 168] |
| Bladder | SNP309 polymorphism MDM2 protein overexpression | Increased risk | [169, 170] |
| Leukemia | SNP309 polymorphism MDM2 protein overexpression | Poor prognosis | [171–173] |
| Melanoma | MDM2 protein overexpression mdm2 gene amplification SNP309 polymorphism | Poor prognosis; increased risk | [174–178] |
A well-documented mechanism for MDM2 overexpression is the single nucleotide polymorphism at nucleotide 309 (SNP309) in its promoter [180, 181]. The clinical significance of SNP309 remains to be clarified. Although the MDM2 SNP309 variant increases MDM2 expression and is associated with tumor formation [182–184], the SNP is not associated with increased risk or a prognostic factor in certain cancers such as glioblastoma [185]. MDM2 overexpression induced by TGF-β is observed in late stage metastatic breast cancer and correlates with poor prognosis [186]. Other mechanisms apart from gene amplification, such as increased transcription and translation also contribute to MDM2 over-expression [109]. High levels of both MDM2 and MDMX proteins without increased copy number have been observed in some cancer types, such as melanoma, Ewing’s sarcoma, colon carcinoma, and retinoblastoma [109].
In breast cancer cell lines, it appears that MDM2 expression is linked to the ER (estrogen receptor) status of the cells [187]. Higher MDM2 expression is observed in ER+ cells. Interestingly, estrogen-mediated increase in cell proliferation correlates with increased MDM2 levels, without a concomitant decrease in the p53 protein level [188]. These findings may have an implication in breast cancer diagnosis and treatment.
MDM2 is also linked to chemo resistance in several cancer types. Overexpression of MDM2 is an important event in regulating sensitivity to chemotherapy in childhood acute lymphoblastic leukemia (ALL) [189]. Functional inactivation of p53 by mutation or other mechanisms is common in relapsed neuroblastoma and associated with MDM2 gene amplification, contributing to chemo resistance [190]. Several studies have reported that either increased MDM2 activity or the SNP309G allele is associated with cellular resistance to topoisomerase II inhibitors [191].
3. STRATEGIES TO TARGET MDM2 FOR CANCER THERAPY
As aforementioned, in most situations, MDM2 overexpression is oncogenic and is associated with late stage disease, resistance to chemotherapy and radiotherapy, and poor prognosis, making it a valid and valuable target for developing cancer therapeutics. We and others first tested the value of MDM2 as a cancer target in preclinical models using antisense oligonucleotides (ASOs) and RNA interference (siRNA) to inhibit MDM2 expression. The results have validated that MDM2 inhibition in cells and mouse models of human cancer is indeed a viable approach to suppress tumor cell growth in vitro and in vivo [33, 192–197]. To date, investigations on the MDM2-p53 interaction has provided a basis for the design of novel small molecule therapeutics aiming at inhibiting MDM2 activity and eventually reactivating the wild-type p53 function [198–202]. In vivo studies have shown restoration of wild-type p53 function can lead to the suppression of soft tissue sarcoma, lymphomas, and liver cancers [202].
Studies on the crystal structure of the N-terminal domain of MDM2 reveal a deep hydrophobic cleft on which the p53 peptide binds as an amphipathic alpha helix, a structure also present in other pro-apoptotic proteins such as Bax [203]. In fact, three amino acids in p53 (Phe19, Trp23 and Leu26) are essential for the binding between p53 and MDM2, and they are inserted into the deep hydrophobic depression on the surface of the MDM2 molecule [203]. Various approaches followed in the discovery of novel small molecule inhibitors of MDM2-p53interaction have been recently developed. The major strategies that can be used to target MDM2 includes: (i) blocking MDM2 expression; (ii) inhibiting the E3 ubiquitin ligase activity of MDM2; (iii) inhibiting the MDM2-p53 interaction; and (iv) targeting the protein-protein complex in relation to MDM2 interactive proteins.
Cutting-edge technologies in structure biology, bioinformatics, computer-aided drug design, and high throughput screening, have led to the identification of numerous molecules belonging to different classes of chemical structures that have significant MDM2 inhibitory effects. The list of natural compounds and synthetic small molecules capable of selectively inhibiting MDM2 and/or the MDM2-p53 interaction keeps increasing rapidly. In the following sections, we will focus primarily on specific and selected molecules that have (or might have) a strong association at the clinical level.
3.1. Inhibiting MDM2 Expression
Several early studies by our group [32, 192–194, 197] and others [204, 205] using antisense oligonucleotides to inhibit MDM2 expression have established the proof of principle for this approach in cell and mouse models of human cancer. MDM2 down-regulation in various cancer cells results in p53 stabilization and activation of the p53 pathway in cancer cells in vitro, as well as in tumor xenografts in nude mice. Interestingly, not only p53 wild-type cells, but also cells that express mutant p53, respond to MDM2 inhibition [32, 197]. It has been suggested that the p53-independent stabilization of the cyclin-dependent kinase inhibitor, p21, due to MDM2 down-regulation might contribute to the antitumor activity of MDM2 antisense oligonucleotides and MDM2 inhibitors in these mutant cell lines [200]. MDM2 inhibition also results in chemosensitization and radiosensitization of cancer cells in various in vivo and in vitro models [33, 206].
Other gene targeting strategies include the use of ribozymes and RNA interference techniques. MDM2 ribozymes induce cell growth arrest and increase apoptosis in vitro [207]. RNAi-mediated MDM2 knockdown leads to similar effects on proliferation, survival, apoptosis, and cell cycle progression [208]. Also, aptamers targeting MDM2 provide another line of attack [209]. More recently, several groups have used novel approaches to deliver MDM2-siRNA for cancer therapy [210, 211].
In the past few years, we and others have investigated the value of MDM2 as a molecular target for natural product chemotherapeutic and chemopreventive agents [212–214]. Several well-known chemopreventive agents such as curcumin, genistein, and ginsenosides have been demonstrated to down-regulate MDM2 oncoprotein expression. These compounds were able to influence MDM2 levels in tumors with wild-type p53 as well as those with non-functional or mutant p53. For a comprehensive discussion on natural product MDM2 inhibitors, readers are directed towards a recent review [215].
3.2. Inhibiting MDM2-p53 Binding
MDM2 will be unable to down-regulate p53 if it is prevented from interacting with p53 protein. Therefore, inhibition of MDM2-p53 binding appears to be a desirable strategy for p53 stabilization and activation. However, targeting protein-protein interactions by small molecules is challenging. Generally, protein-protein interactions are difficult to interrupt by low molecular weight chemical entities as they involve large, flat interfaces with buried surfaces [216]. However, in the case of the MDM2-p53 interaction, it has been demonstrated that a limited number of amino acid residues (namely, Phe19, Trp23 and Leu26 in the N-terminal domain of p53) are crucial for the binding of the two proteins [203]. There exists a narrow, continuous, hydrophobic pocket on the MDM2 protein for p53 binding with the aforementioned amino acids critically regulating the interaction. Therefore, it is reasonable to expect that a synthetic molecule with three hydrophobic groups projecting into this pocket would essentially mimic the key amino acid residues of p53, thus competitively inhibiting the MDM2-p53 interaction. The nutlin class of MDM2 inhibitors developed by Roche Company from the screening of a large combinatorial library is based on this principle [198–200]. These inhibitors possess the capability to displace p53 from MDM2 in vitro with nanomolar potency (IC50 = 90 nM for nutlin-3a). Crystallographic studies demonstrate that nutlins bind to the p53-interacting domain of MDM2 in a way that closely resembles the molecular interactions of the crucial amino acid residues from p53 [198].
3.3. Modulating E3 Ubiquitin Ligase Activity of MDM2
The ubiquitin ligase activity of MDM2 is crucial for its negative regulation on p53 protein stability, indicating that this may be a valid drug target. Recently, small molecule inhibitors have been identified that specifically target the E3 ligase activity of MDM2. These compounds inhibit the ubiquitination of p53 in vitro, with IC50 values in the micromolar range [217–221]. They are able to activate p53 signaling, inducing apoptosis in a p53-dependent manner. The first report on MDM2 E3 ligase inhibitors dates back to 2002 with the arylsulfonamide, bisarylurea, and acylimidazolone compounds being discovered in an MDM2-mediated p53 ubiquitination chemical library screen [218]. They do not affect the physical interaction between MDM2 and p53, probably inhibiting MDM2 in an allosteric fashion by blocking a structural rearrangement of MDM2 necessary for p53 ubiquitination [218]. Selective inhibition of p53 ubiquitination is therapeutically desirable as opposed to total loss of MDM2 ubiquitin ligase activity (which will also inhibit MDM2 autoubiquitination, leading to enhanced MDM2 levels). In addition, several natural product MDM2 inhibitors have been shown to induce MDM2 autoubiquitination and degradation, although the exact mechanism of action remains to be revealed [215].
4. SMALL MOLECULE MDM2 INHIBITORS
4.1. SMIs Disrupting MDM2-p53 Interaction
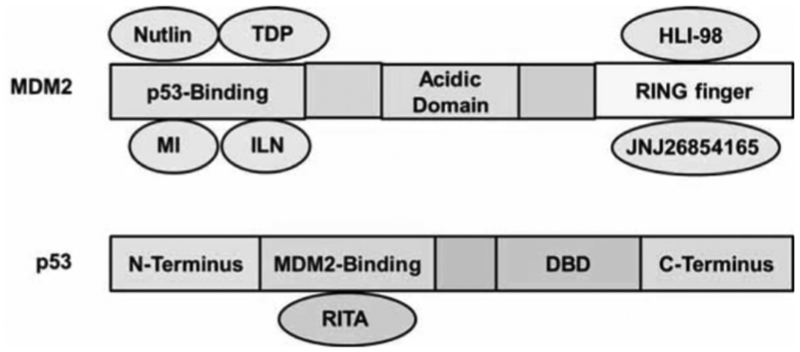
Advances in the understanding of the conformation and structure of MDM2 have sparked the rational design of small molecule MDM2 inhibitors. Selective binding of another molecule to the surface of MDM2 can prevent the MDM2-p53 interaction, leading to the accumulation of p53 in the nucleus, and the subsequent induction of pro-apoptotic and anti-proliferative p53 functions. Most new chemical entities developed as MDM2 inhibitors are based on this principle [222]. High throughput screening methods, combined with combinatorial library synthesis, have led to the development of a number of small molecule MDM2 inhibitors with diverse chemical structures. These include chalcones, piperazine-4-phenyl derivatives, nutlins (cis-imidazolines), spiro-oxindoles, sulfonamides, benzodiazepinediones, isoindolinones, and terphenyls among others [221, 223–225]. Their corresponding binding domains on MDM2 and/or p53 proteins are shown in (Fig. 3).
Fig. (3). Schematic representation of the MDM2 and p53 proteins, and the binding areas for small-molecule inhibitors.
Nutlin, cis-imidazoline; TDP, benzodiazepinedione; MI, spiro-oxindoles; ILN, isoindolinone; HLI-98, 5-deazaflavin; JNJ-26854165, tryptamine; RITA, thiophene; RING finger: E3 ubiquitin ligase domain; DBD: DNA binding domain. Binding of either HLI98 or JNJ-26854165 to MDM2 RING finger domain inhibits its E3 ubiquitin ligase activity.
4.1.1. SMIs Binding to MDM2
The imidazoline derivatives (better known as Nutlins) are one of the first small molecules for selective inhibition of MDM2-p53 binding [198]. They act by mimicking the three critical amino acid residues (Phe19, Trp23, Leu26) within the hydrophobic pocket of MDM2. Crystallographic studies demonstrate that the chlorophenyl moieties perfectly fit deeply into the Leu26 and Trp23 pockets while the isopropylphenyloxy moiety mimics Phe19 with the phenyl groups serving as a connector to place the phenyloxy group into the Phe19 pocket. Nutlins do not induce p53 phosphorylation on Ser15, suggesting that they are non-genotoxic, selective inhibitors. Studies also suggest that the 4-methoxy functionality in nutlin3 mimics p53 Leu22. Nutlin-3, the most potent analog of the series, induces p53 levels and enhances p53 transcriptional activity. It has been shown to be effective in various cancer models with wild-type p53, such as neuroblastoma, colon, mantle cell lymphoma, breast and osteosarcoma [202, 226–228]. Nutlin-3 also activates p53 and induces apoptosis and cellular senescence in myeloid and lymphoid leukemic cells [229, 230]. An interesting observation has been that androgen deprivation followed by two weeks of Nutlin-3 administration in LNCaP bearing nude mice leads to a greater tumor regression and dramatically increases survival, indicating cross talk between p53, MDM2, and the androgen receptor [231].
This class of compounds have been shown to have effects on other proteins as well. In the absence of functional p53, Nutlin-3 disrupts p73-MDM2 interaction and increases p73 transcriptional activity, leading to greater apoptosis and growth inhibition in hepatocellular carcinomas [232]. Interestingly, Nutlin-3a, a compound that was designed to inhibit MDM2-p53 binding, has been shown to interfere with the MDM2-E2F-1 interaction [233]. Simultaneous treatment of a p53 mutant peripheral nerve sheath tumor cell line with Nutlin-3a and cisplatin leads to an increase in E2F-1 levels and E2F-1-mediated apoptosis [234]. Furthermore, E2F-1 knockdown inhibits Nutlin-3-mediated apoptosis [234]. Nutlin-3 also inhibits VEGF via inhibition of HIF-MDM2 interaction [235]. Nutlin-3 has also been shown to inhibit the protein expression of NFκB target genes ICAM-1 and MCP-1 in a p53-dependent manner [236]. ICAM-1 and MCP-1 are involved in cell migration and metastasis, suggesting a role for Nutlin-3 in the treatment of advanced diseases. The nutlin series of compounds have been shown to cause cell death via interaction with proteins such as TRAIL, PUMA, and others [229]. Nutlin therapy has also been shown to sensitize different cell lines such as laryngeal, lung, and prostate cancer cells to ionizing radiation [237]. Nutlin-resistant cell lines have wild-type MDM2 but mutations in the p53 DNA binding and dimerization domains. This chemoresistance can be overcome by the p53 targeting agent RITA [238]. Interestingly, the nutlins do not bind the MDM2 homolog MDMX. Furthermore, despite the similarity between MDM2 and MDMX, MDM2 inhibitors such as Nutlin-3 are far less effective against MDM4. However, MDMX inhibitors not only can activate p53 and induce apoptosis in breast cancer cells, but also synergize with MDM2 inhibitors for p53 activation and induction of apoptosis [239]. Nutlins-3a and −3b (inactive enantiomer) have been demonstrated to be substrates of multi-drug resistance protein-1, and act to reverse multi-drug resistance by saturating the efflux capabilities of these transporters [240]. However, several nutlins developed in the early phases failed to proceed to the trials due to poor pharmacokinetic properties [241].
Derivatives structurally related to nutlins include pyrrolidine-2-carboxamide derivatives, and imidazothiazole derivatives (with a proline moiety) [223]. A series of compounds with 3-imidazolyl-indole scaffolds as dual inhibitors of MDM2/MDMX-p53 interaction have been identified [242]. Spiroxindole derivatives represent another class of MDM2-p53 inhibitors designed to mimic the Trp23 residue that is the most critical for binding of p53 to MDM2. MI-63, MI-219, MI-319, and MI-147 belong to this class. Oxindoles were synthesized using a de novo approach to mimic the indole ring and also the side chain of Trp23. Shangary and others then used a substructure search technique to identify natural products with an oxindole substructure [202], such as spirotryprostatin A and alstonisine. MI-219, the clinical grade compound, is shown to alter the functional activity of MDM2 through enhancing its auto ubiquitination and degradation [243]. MI-219 causes p53 activation by blocking the MDM2-p53 interaction in wild-type p53 cells but exhibits much lower activity in cells with mutant p53, which is consistent with its mechanism of action as a specific inhibitor of the MDM2-p53 protein-protein interaction [202]. However, in the studies, MI-219 failed to achieve tumor regression in various xenograft models of human cancer though it was able to completely inhibit tumor growth [244]. Recently, the group has reported the synthesis of a series of highly potent diastereomeric spiro-oxindoles, in which stereochemistry affects binding affinity to MDM2 greatly [245]. Optimization of the pharmacokinetic properties of one of the analogs reported therein has yielded MI-888 (Ki = 0.44 nM) with highly selective activity in inhibiting the growth of wild-type p53 cancer cell lines [296, 297]. MI-888 exhibits very good oral bioavailability in rats along with complete tumor regression in mice bearing osteosarcoma (SJSA-1) and acute leukemia (RS4;11) xenografts [297]. An analogue of MI-888 has been advanced into Phase I clinical development [246].
Other MDM2 binding SMIs include chalcone and the NAcylpolyamine derivatives. The latter class represents cyclic peptides in β-sheet confirmation which inhibit the association of both MDM2 and MDMX with p53 [247]. The chal-cone derivatives have been shown to have anticancer activities in a number of cancer cells. Several derivatives prepared from Licochalcone-A, obtained from licorice root, inhibit MDM2-p53 interaction by binding near the Trp23 pocket of MDM2 [248]. A second generation of chalcone derivatives bearing a boronic fragment has also been synthesized and it is believed that boronic acid analogs may form a stronger salt bridge with Lys51 than corresponding chalcone analogs of caxboxylic acid moiety [248–250]. Surprisingly, the DNA damaging agent 5-fluorouracil has been demonstrated to stabilize and activate p53 by blocking MDM2 feedback inhibition through ribosomal proteins [251]. This finding warrants further investigation into the mechanism of current chemotherapeutic agents to unravel if they activate p53 by inhibiting MDM2.
Recently, Amgen has developed a series of piperidinone based compounds [252]. These compounds also interact with the Trp23, Leu26, and Phe19 residues in MDM2 apart from ordering the N-terminal residues of MDM2, thus resulting in very high affinity for MDM2 binding (MDM2 IC50= 1.1 nM) [252]. In a mouse SJSA-1 tumor xenograft model, oral administration of AM-8553 at 200 mg/kg once daily resulted in partial tumor regression, demonstrating its excellent antitumor activity and clinical translational potential [252].
4.1.2. SMIs Binding to p53
The molecule RITA (Reactivation of p53 and Induction of Tumor cell Apoptosis) acts by binding to the N-terminal of p53 and reactivating p53 function [249]. It is shown that RITA acts not only on wild-type p53, but also on mutant p53. RITA may bind directly with mutant p53 or by disrupting the inhibitory complex that mutant p53 forms with p63 and smad2, or the complex with p73. However, some investigators have demonstrated that RITA does not block MDM2-p53 binding [253, 254]. Therefore, a better understanding of the binding mode of this class of compounds is required for further development of them as effective anti-cancer agents.
4.2. SMIs InhibitingMDM2 E3 Ubiquitin Ligase Activity
The 5-deazaflavin compounds inhibit the E3 ligase activity of MDM2 by targeting the RING finger domain of the oncoprotein. These compounds are also known as HDM2 ligase inhibitor (HLI series). Treatment of these compounds results in p53 activation, despite causing stabilization of both p53 and MDM2. This activation is speculated to result from the direct binding and inhibition of the RING finger domain, the functional domain necessary for MDM2’s E3 ligase activity. These compounds also affect the E2 ligase activity. However, at high concentrations, HLIs also cause cell death in p53 mutant cells [255].
Selective inhibitors of E3 ligase include three chemically distinct classes of compounds (namely, benzsulfonamides, ureas, and imidazolones). These are selective inhibitors of MDM2 E3 ligase, with little or no effect on auto ubiquitination of MDM2. They comprise of simple reversible inhibitors of MDM2 that bind to it in a manner noncompetitive with respect to the substrates, Ub-Ubc4 (E2) and p53. The lead candidate of tryptamine derivatives, JNJ26854165, blocks the degradation of p53 by inhibiting the binding of MDM2-p53 complex to proteasome. Treatment with this compound induced p53-mediated apoptosis in wild-type p53 cells, while in mutant p53 cells, JNJ26854165 induced S-phase cell cycle arrest and E2F1-mediated apoptosis. Notably, the compound is active against Nutlin-3a resistant samples, indicating involvement of a pathway other than MDM2-p53 inhibition [109, 220, 256].
Sempervirine, a natural indole alkaloid identified by a high throughput electrochemiluminescent screen that screened more than 100,000 natural compounds, has also been identified as an inhibitor of MDM2’s E3 ligase activity [257]. Sempervirine inhibits the MDM2-mediated ubiquiti-nation and degradation of p53, resulting in the accumulation of p53. This compound also preferentially induces apoptosis in wild-type p53 expressing cancer cells, meriting further investigation of its anticancer activity [257].
4.3. SMIs Inducing MDM2 Post-Translational Modifications
Based on the reported chemopreventive activities in several human cancers, we have demonstrated that the soy-derived isoflavone genistein directly down-regulates MDM2 at both the transcriptional and post-translational levels, independent of the p53 status [212]. Our studies have also shown that genistein down-regulates MDM2 and increases p21 levels, independent of tyrosine kinase inhibitory activity of the compound. Another common flavonoid, apigenin, has been shown to induce p53 via MDM2 attenuation and to inhibit the phosphorylation of MDM2 by Akt (which, in turn, increases MDM2 stability and p53 degradation) in ovarian cancer cells [258]. Similarly, the sesquiterpene lactone parthenolide, isolated from the European feverfew herb, Tanacetum parthenium, induces MDM2 ubiquitination and proteasomal degradation in an ATM-dependent manner, subsequently activating p53 and other MDM2-regulated tumor suppressor [259, 260]. Berberine, a natural isoquinoline alkaloid obtained from the herb Rhizoma coptidis and used in Traditional Chinese Medicine, has been widely investigated due to its myriad pharmacological effects, including anticancer, anti-inflammatory, and antibacterial activities. Berberine induces apoptosis in acute lymphoblastic leukemia (ALL) cells through downregulation of the MDM2 oncoprotein in wild-type p53 ALL cell lines [261]. Berberine also decreases DAXX transcription, and subsequently prevents the formation of the MDM2-DAXX-HAUSP complex, resulting in the persistent self-ubiquitination of MDM2 in ALL cells [261].
4.4. SMIs Inhibiting MDM2 Expression
Several natural products have shown inhibitory effects on MDM2 gene expression [215]. Ginsenoside saponins has been known to regulate multiple steps in cell proliferation, modulate the expression of tumor suppressors, oncogenes, growth factors, cell death mediators, pro-inflammatory molecules, and protein kinases [262]. These anticancer activities follow a well-defined structure-activity relationship. Over the past few years, our group has identified two new ginsenoside products, 20(S)-25-hydroxy-dammarane-3β,12β, 20-triol and its methoxyl derivative (25-OCH-PPD and 25-OCH3-PPD) which exhibit excellent anticancer activity against various human cancers, including prostate, pancreatic, lung, and breast cancers by decreasing MDM2 protein levels both in vitro and in vivo. 25-OCH3-PPD also inhibits the transcriptional activity of MDM2 in cancer cells [263–267].
Curcumin, a dietary polyphenol, also down-regulates MDM2 transcription through the PI3K/mTOR/ETS2 pathway and curcumin exposure sensitizes human cancer cells to chemotherapy and radiation via MDM2 [213]. Semisynthetic derivatives of the marine alkaloids such as the makaluvamines have been shown to decrease the levels of MDM2 oncoprotein, to cause apoptosis by the caspase-mediated pathway, and to inhibit the PI3k-Akt pathway [268–274].
5. CLINICAL DEVELOPMENT OF SMALL MOLECULE MDM2 INHIBITORS
5.1. Nutlins (Cis-Imidazolines)
A cis-imidazoline derivative belonging to the Nutlin family entered clinical trials in late 2007/early 2008. This compound codenamed RG-7112 molecule has just completed phase I clinical trials in patients with solid tumors, and hematologic neoplasia (ClinicalTrials.gov; identifiers: and ). This multicenter, open trial was conducted in the United States and France and designed to determine the maximum tolerated dose and the optimal dosing schedule of RG-7112, administered as monotherapy in patients with advanced solid tumors. A first cohort of patients received the starting dose of 20mg/m2/day, once daily for 10 days in each 28-day cycle. RG7112 treatment was seen to stabilize the p53 protein and induce p53 target genes such as CDKN1A, NOXA, PUMA, FAS and BAX. The compound exhibited good dose tolerance and positive effects in the treatment of acute or chronic leukemia and patients responded well to an escalation in dosage. It is reported that a single patient with AML who has been leukemia-free for 9 months subsequent to RG7112 treatment [275]. Reductions in lymph node and spleen size, as well as in circulating leukemia cells, were noticed in chronic lymphocytic leukemia (CLL) and small lymphocytic lymphoma (SLL) conditions [276–278].
RG-7112 has also been tested in patients with well-differentiated liposarcoma prior to debulking surgery (ClinicalTrials.gov; identifiers: ) [277]. The study conducted in France sought to study biomarker evidence of MDM2 and p53 pathway alterations in a clinical setup. The results of these initial proof of mechanism studies are preliminary since a very small number of patients (20 patients) were enrolled. Although, the data indicate that RG7112 does reach its target in a solid tumor and correlates with increased p53 and p21 levels, increased MIC-1 levels (induced by p53), and decreased proliferation, the data did not reach statistical significance. Also, as the tumors were not microdissected, it may be possible that the heterogeneous tumor tissue contained only a small fraction of cells responsive to RG7112 treatment. The most frequent adverse event seen was hematological toxicities. Thus, RG7112 may be a part of neoadjuvant therapy in combination with existing clinically approved non-genotoxic chemotherapeutics. This will help prevent aggravating the hematological toxicities that are a hallmark of standard DNA damaging drugs. Further information on the molecule’s pharmacokinetic profile and adverse effects are awaited. Currently, an ongoing clinical trial is recruiting participants for a multicenter, open-label, Phase Ib study for RO5045337 (the oral formulation of RG7112) that will evaluate the safety, pharmacokinetics and efficacy of this drug in combination with doxorubicin in patients with soft tissue sarcoma. Another small molecule MDM2 inhibitor, RO5503781, presumably with a similar structure, has been launched into clinical trials, presently recruiting participants for a multicenter, open label, dose-escalating study to evaluate the safety, pharmacokinetics, pharmacodynamics and efficacy in patients with advanced malignancies except leukemia. In the clinical trials, this drug is being tested as a single agent or in combination with cytarabine (ClinicalTrials.gov; identifiers: and ).
5.2. Spiroxindoles
SAR405838 (MI-888), an analogue of the MI series (MI-219: clinical prototype), discovered by Shangary et al. at the University of Michigan, was launched into phase I clinical trials by Sanofi S.A in 2012 (ClinicalTrials.gov; identifier: ). This study, as of date, is still recruiting participants and no results are available. The study aims to assess the drug’s efficacy in patients with dedifferentiated liposarcoma and determine safety and the maximum tolerated doses (MTD).
5.3. Tryptamine Derivative (JNJ-26854165/Serdemetan)
Another MDM2 inhibitor in clinical trials is the E3 ubiquitin ligase inhibitor JNJ-26854165 (ClinicalTrials.gov; identifier: ) [220, 279, 280]. The first observations from the clinical trials were presented in 2009. This orally bioavailable drug has just completed phase I clinical trials for the treatment of advanced stage or refractory solid tumors in USA [276]. The starting dose of JNJ-26854165 in the trial was 4 mg/day as a single oral dose. Side effects reported with the treatment of JNJ-26854165 included nausea, vomiting, fatigue, anorexia, insomnia, electrolyte imbalance, creatinine elevations, and asymptomatic QTc prolongation. No hematological or cardiovascular toxicities were observed. A single patient with higher dose exhibited a grade 3 QTcF prolongation which was reversed after discontinuation of the treatment. Linear pharmacokinetics was seen in 20 to 400 mg dose range, with the preclinically determined therapeutic concentration being achieved at doses above 300 mg [276]. The levels of p53 were upregulated in skin while MDM2 levels were enhanced in tumors (probably due to loss of auto ubiquitination function) in a dose dependent manner. Similarly, the levels of MIC-1, a plasma macrophage inhibitory cytokine, a member of the TGF-β super family and induced by p53, were also increased in dose-dependent manner. A dose of 350 mg was used on expanded cohort of patients to confirm maximum tolerated dose, and a separate trial was started with alternate dosing schedule (150 mg twice a day) to minimize QTc prolongation.
5.4. Other Compounds in Clinical Trials
A novel chemical moiety thioureidobutyronitrile (Kevetrin) is also being launched into Phase I dose escalation and safety clinical trials this year. In preclinical studies, Kevetr in activates p53, induces p21 and PUMA (p53 up-regulated modulator of apoptosis), a p53 activated proapoptotic protein (ClinicalTrials.gov; identifier: NCT 01664000). Recently, Novartis has launched an orally active MDM2-p53 interaction inhibitor, codenamed CGM097, into clinical trials for treatment of p53 wild-type tumors (ClinicalTrials.gov; identifier: ).
In addition to these MDM2 small molecule inhibitors, other compounds in clinical development include MK-8242 (or SCH 900242), which is being tested in two phase I clinical trials (ClinicalTrials.gov; identifier: and ). This compound is being tested as a single agent in patients with advanced solid tumors and in combination with cytarabine in acute myelogenous leukemia (AML) subjects. Lately, Daiichi Sankyo Inc. has commenced phase I clinical trial of DS-3032b (ClinicalTrials.gov; identifier: ) in patients with advanced solid tumors or lymphomas.
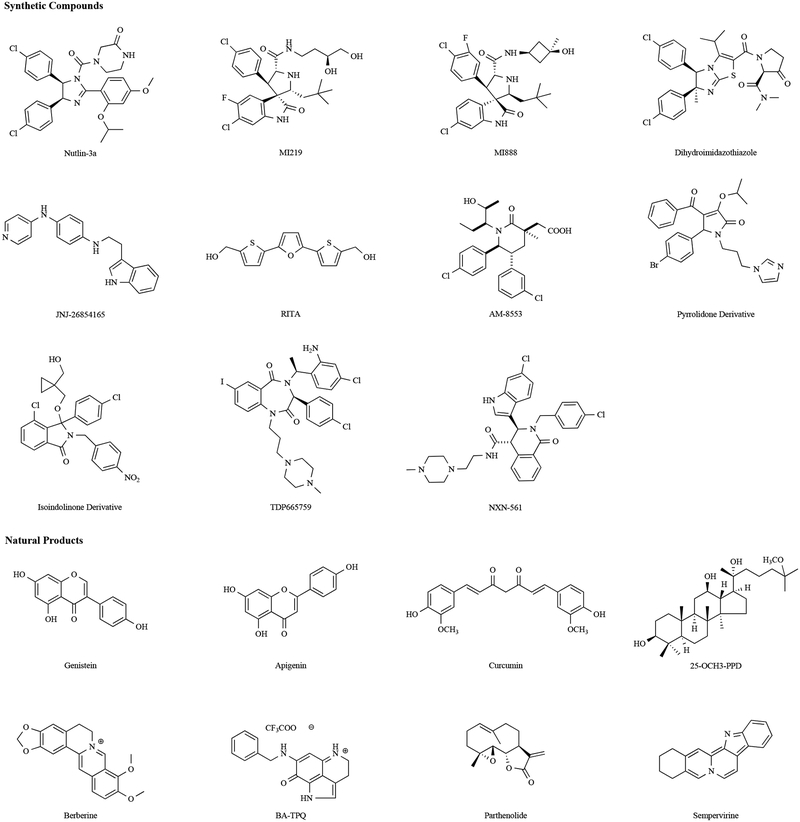
Of note, several of the naturally occurring compounds such as ginseng saponins and curcumin have been extensively studied for efficacy and safety in clinical setups, although they were not initially investigated as MDM2 inhibitors. They present a veritable treasure house of MDM2 inhibitors for future development. In (Fig. 4), we present the structures of several prototype MDM2 inhibitors both synthetic and natural origin, while in (Table 3) we summarize the mechanisms of action of these prototype synthetic and natural SMIs along with their clinical development statuses.
Fig. (4).
The chemical structures of representative synthetic small molecule and natural product MDM2 inhibitors.
Table 3.
Representative Synthetic and Natural-origin Small Molecule Inhibitors of MDM2
| Prototype Compound | Molecular Target | Mechanism of Action | Clinical Status | Ref. |
|---|---|---|---|---|
| 1. Synthetic Small Molecule Inhibitors | ||||
| Nutlin 3A (Cis-imidazolines) | MDM2 N-terminal p53-binding pocket | Disrupts MDM2-p53 interaction | Phase I clinical trial completed; Combination studies ongoing |
[109, 198–200, 275, 276] |
| JNJ-26854165 (Tryptamine derivative) | MDM2 RING finger domain | Inhibits MDM2 E3 ubiquitin ligase activity; p53-independent activity |
Phase I clinical trials | [109, 202, 276] |
| TDP665759 (Benzodiazepinedione derivatives) | MDM2 N-terminal p53-binding pocket | Disrupts MDM2-p53 interaction | No clinical translation as of yet | [281, 282] |
| Isoindolinone derivative | MDM2 N-terminal p53-binding pocket | Disrupts MDM2-p53 interaction | No in-vivo activity reported | [283] |
| MI-219/MI-888 (Spiro-oxindole derivatives) | MDM2 N-terminal p53-binding pocket | Disrupts MDM2-p53 interaction; Induces MDM2 autoubiquitination |
Analog-MI 773 (SAR SAR405838) is in Phase I clinical trials | [202, 241, 243] |
| NXN-561 (Isoquinoline derivative) | MDM2 N-terminal p53-binding pocket | Disrupts MDM2-p53 interaction | No in vivo studies reported | [284] |
| Pyrrolidone derivative | MDM2 N-terminal p53-binding pocket | Disrupts MDM2-p53 interaction | No clinical translation as of yet | [285] |
| HLI-198C (5-Deazaflavin derivatives) | MDM2 | Inhibits MDM2 E3 ubiquitin ligase activity; p53-independent activity |
Unknown/not reported | [255, 282] |
| AM-8553 (Piperidinone derivatives) | MDM2 N-terminal p53-binding pocket | Disrupts MDM2-p53 interaction | Preclinical studies in mouse SJSA-1 tumor xenograft; orally active |
[252] |
| 2. Natural Origin Small Molecule Inhibitors | ||||
| Parthenolide | MDM2 RING finger | Induces MDM2 ubiquitination in ATM dependent mannerresulting in p53 activation | Not clinically tested in cancer | [259] |
| Sempervirine | MDM2 RING finger | Inhibits MDM2 E3 ubiquitin ligase activity, preventing both MDM2 autoubiquitination as well as MDM2 mediated p53 ubiquitination. | No clinical translation as of yet | [257] |
| Genistein | MDM2 | Inhibits MDM2 protein expression; p53-independent activity |
Clinically tested | [212] |
| Apigenin | MDM2 | Inhibits MDM2 protein expression; Inhibits MDM2 phosphorylation by Akt |
Clinically tested | [258] |
| 25-OCH3-PPD | MDM2 | Inhibits MDM2 protein expression; p53-independent activity |
No clinical translation as of yet | [214, 263, 265–267] |
| Curcumin | MDM2 | Inhibits MDM2 transcription through PI3K/mTOR/ETS2 pathway; p53-independent activity |
Clinically tested | [213] |
| Berberine | MDM2 | Inhibits MDM2 protein expression; Induces MDM2 autoubiquitination |
Not clinically tested in cancer | [261] |
6. DISCUSSION AND FUTURE DIRECTIONS
Disruption of the MDM2-p53 interaction with small molecule inhibitors is an attractive cancer therapeutic strategy. However, drug discovery efforts in this area have been primarily focused on strategies to inhibit the binding of p53 to the N-terminal domain of MDM2 and several novel scaffolds (both from rational drug synthesis and natural sources) have been developed [286–293]. Some of these compounds such as the dihydroimidazothiazole derivatives synthesized by scientists at Daiichi employ structural modifications of the nutlins followed by subsequent optimization of potency and pharmacokinetic behavior [289]. We now know that other domains in MDM2 are also involved in the MDM2-p53 interaction and mutations in these domains are associated with cancer [294]. In fact, it has been reported that the ligands binding to the MDM2 acid domain cause p53-mediated inhibition of cell growth and induce apoptosis [295]. This highlights the importance of the acidic domain in addition to the N terminus as a potential target for small molecular MDM2 inhibitors. Directly inhibiting the MDM2-p53 interaction, as a means of restoring p53 wild-type functions, is potentially useful in the treatment of cancers which harbor wild-type p53, but there still exist concerns as to how viable this concept would be clinically: how efficient will a chemical moiety in inhibiting a highly specific protein–protein interaction? What will be the adverse effects of unleashing a potent pro-apoptotic molecule like p53 on healthy cells? And since p53 transcriptionally activates MDM2, will increased p53 lead to increased MDM2 levels due to the MDM2-p53 feedback loop? Present evidence is in favor of these inhibitors; with a number of them entering clinical trials, the ultimate proof of concept may be just around the corner. More than twenty different chemical classes have been claimed to inhibit the MDM2-p53 interaction, but the majority of studies have been directed toward three classes: benzodiazepinediones, spiro-oxindoles (the MI series of compounds), and cis-imidazolines (Nutlins). Further preclinical and clinical studies on other compounds may provide more information on the value of targeting MDM2 and MDM2-p53 interaction.
In order to critically evaluate the mechanism of action and therapeutic potential of a MDM2 inhibitor, it should have the following desirable “drug-like” properties: (i) high binding affinity and specificity towards MDM2, (ii) high cytotoxicity in cancer cells with wild-type p53, and (iii) a highly desirable pharmacokinetic (PK) profile [202]. The p53-binding site of MDM2 is highly hydrophobic, and therefore all the non-peptide drugs that have been developed are, necessarily, lipophilic, and, thus lack aqueous solubility. It is true that several potent MDM2 inhibitors have been tested in animal models of human cancer for their anticancer activity. However, some of these compounds such as Nutlin 3A and MI-219 were not able to achieve complete tumor regression and also showed variable activity in different cancer types (Nutlin-3 and MI-219 inhibit tumor growth completely in SJSA-1 derived xenografts but show minimal activity against HCT-116 colon cancer xenografts) [244]. These results were consistent with data obtained from in vitro cell experiments [244, 296]. It is noteworthy that the antitumor activity of these compounds (nutlins as well as the MI series) was achieved at doses that caused no visible toxicity to the animals, as evidenced by body weight and gross organ morphology at necropsy [198–200, 244, 246, 275, 276, 296, 297]. Since derivatives of nutlins (such as RG7112) with optimized pharmacological parameters (with respect to bioavailability) are able to achieve tumor regression (either partial or complete), it is evident that potent and highly optimized MDM2 inhibitors can achieve impressive anticancer activity in animal models of human cancers [275]. Indeed, researchers at University of Michigan have carried out structural modifications in their MI-series based on the stereo-chemical properties of these compounds alongwith optimization of the pharmacokinetic parameters (by adding more “biological friendly” side chains that increase bioavailability) [244–246]. These efforts have yielded MI-888 which exhibits excellent oral bioavailability alongwith complete tumor regression in two animal models of human cancer [246, 296, 297]. An analogue of MI-888 has also advanced into phase-I clinical trials [297]. An interesting study by Azmi et al. details the use of the essential trace element zinc along with the MDM2 antagonist, MI-219 [298]. Zinc is an important part of the p53 biochemistry, with p53 binding to DNA through a structurally complex domain stabilized by zinc atom [298]. The MDM2 protein also carries a C-terminal RING domain that coordinates two zinc atoms, which are responsible for p53 nuclear export and proteasomal degradation [298]. Zinc chloride supplemented MI-219 regimen suppresses the p53 feedback MDM2 activation, thus increasing its efficacy [298].
However, till date, two MDM2 inhibitors (compounds inhibiting the MDM2-p53 interaction), Nutlin-3 and MI-219, appear to meet enough “drug-like” criteria. In addition, the benzodiazepinedione compound TDP665759, with an IC50 value of 704 nM appears to be another suitable compound. On the contrary, other small molecule inhibitors that target the MDM2-p53 interaction have either exhibited modest cytotoxic activities in cells or do not possess high binding affinity and/or specificity towards MDM2. Moreover, information on their cellular mechanisms of action, in vivo anti-cancer activity in preclinical set-up, cellular specificity for cancerous cells versus normal cells has not yet been reported.
Inhibitors of the MDM2-p53 interaction are unique in the aspect that, unlike, several traditional chemotherapeutics, these compounds induce p53 accumulation and activation in a non-genotoxic manner (without inducing DNA damage or requiring ATM/ATR-dependent p53 phosphorylation). Thus, these compounds do not technically affect p53 post-translational modification pathways such as phosphorylation, acetylation and sumoylation that cause p53 activation in response to genotoxic stress.
Although these inhibitors are highly effective against tumors containing wild-type p53, tumors with mutated or nonfunctional p53 may not be responsive. In fact, mutant p53 is known to stabilize MDM2. Considering that MDM2 has multiple p53-independent oncogenic activities, compounds such as nutlins that have been developed to specifically inhibit MDM2-p53 interaction would display poor effects on p53-inactivated tumors. The development of resistance following MDM2 inhibitor treatment still needs to be further investigated. Moreover, the pharmacokinetic properties and bioavailability of synthetic MDM2 inhibitors need major improvements before clinical use can be permitted. For instance, several initial analogs in the nutlin series have inadequate bioavailability in vivo [202].
As aforementioned, MDM2 is one of the most important oncogenes in the entire process of cancer. In addition to being a negative regulator of p53, it interacts with numerous other proteins involved in diverse cellular functions ranging from cell proliferation, apoptosis, to metastasis and angiogenesis, indicating that these signaling pathways have a potential to be explored as therapeutic targets. Targeting these interactions may provide alternative or complementary approaches to targeting the MDM2-p53 interaction, especially for the treatment of cancers with mutant p53 or loss of p53 function. Given the p53-independent activities of MDM2 in cancer, it becomes necessary to focus on MDM2 itself as a drug target. The oncogenic activities of MDM2 that have potential to be translated into possible molecular targets for cancer therapy include: (i) binding and destabilization of p21, (ii) binding and inhibition of the p53 homologues, p63 and p73, (iii) positive modulation of HIF-1α transcription factor/VEGF [299], (iv) interaction with ribosomal proteins, (vi) inhibition of MDM2 by the tumor suppressor ARF, (vii) inhibition of pRb, (viii) its interaction with hormone receptors such as androgen receptor and estrogen receptor, and (iv) its role in epithelial-mesenchymal transition and metastasis. However, more studies are required to elucidate the role(s) of these interactions, and to define the circumstances under which these interaction(s) can be successfully targeted. The use of system biology and modern high throughput drug screening techniques, combined with an increasingly in-depth understanding of the biochemistry and molecular biology of MDM2 will help us to develop novel MDM2 inhibitors.
MDM2 expression is regulated at multiple levels through various cellular mechanisms. Therefore, it is imperative to understand these mechanisms to better develop therapeutic strategies for MDM2 inhibition. In addition to p53, the transcription of MDM2 is regulated by other transcription factors such as NF-κB [300], Fli-1 [301], ETS [213], AP1 [302], and NFAT1 [303]. Inhibition of these transcription factors may provide another strategy to overcome MDM2 deregulation in cancer cells. Indeed, we have seen that curcumin suppresses MDM2 though the inhibition of ETS transcription factor. Thus, we believe a therapeutic regimen combining an MDM2 transcriptional inhibitor may benefit those harboring high level of MDM2 and/or inactivated p53.
Another mechanism by which MDM2 can be prevented from destabilizing p53 is by inhibiting its E3 ubiquitin ligase activity. However, not much progress (in terms of number of new chemical entities generated) has been made on MDM2 E3 ubiquitin ligase inhibitors on account of the biological complexity of the ubiquitination process. It is hoped that a better understanding at the molecular level of how exactly MDM2 functions as an E3 ubiquitin ligase will facilitate structure-based drug design and discovery. Conceptually, E3 ligases are very attractive drug targets as they mediate a majority of protein destruction mechanisms. Moreover, targeting MDM2 E3 ubiquitin ligase may have a broader spectrum of activity in cancers exhibiting mutant and non-functional p53. In fact, a study employing a combinatorial regimen of bortezomib (a proteasome inhibitor) along with nutlin shows excellent activity against myeloma [304, 305].
At present, most of the available MDM2 inhibitors lack activity against MDMX. Although Nutlin 3a and MI-219 activate p53 in cancer cells with overexpressed MDM2, they do not have similar effects in cells with overexpressed MDMX. MDM2 inhibitors are not able to bind to MDMX although MDMX has similarities in the N-terminal p53-binding domains to that of MDM2. Subtle structural differences in the MDMX-p53 interaction pocket drastically reduce the binding affinity of both Nutlin 3a and MI-219 for MDMX. This observation is supported by in vitro binding studies which demonstrate that Nutlin 3a is 500-fold less potent against MDMX compared with MDM2 [306]. MDM2 and MDMX possess non-redundant roles in modulating p53 activity and are the major antagonists of p53 in vivo. Therefore, dual antagonists for both MDM2 and MDMX may effectively reactivate p53 in cancer. RO-5963 has been identified as a dual inhibitor of MDM2-p53 and MDMX-p53 interactions. Structural analyses suggest that the compound induces p53 activity via formation of MDM2-MDMX homodimers and heterodimers [306].
Finally, natural compounds present an attractive area for the development of MDM2 inhibitors as they have been shown to target both the p53-dependent and -independent activities of MDM2, exhibiting impressive activity even in p53 mutant or non-functional conditions, thus eliminating the need for p53-dependent mechanisms to exert their effects. On the other hand, most synthetic small molecule inhibitors of MDM2 are known to restore the pro-apoptotic and anti-proliferative functions of p53 by disrupting the MDM2-p53 interaction and, therefore, are less effective in p53 mutant tumors. Further research on the reasons for the effectiveness of naturally occurring compounds in a p53 mutant scenario may provide clues about the structural requirements of such p53-independent MDM2 inhibitory activity. The unique frameworks of the natural compounds can inspire to develop novel synthetic and semi-synthetic derivatives with improved efficacy, pharmacokinetic, and bioavail-ability profiles, thus opening new avenues in MDM2-p53 research.
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (NIH) grants R01 CA112029 and R01 CA121211 and a Susan G Komen Foundation grant BCTR0707731 (to R.Z.). We are grateful to all the current and former members of our laboratories for their excellent contributions to the research projects that were cited in this article. We apologize for not being able to cite all of the publications in the field due to the limitations of the length of the review article. We thank Dr. J.-J. Qin for critical review and comments on this manuscript.
Footnotes
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflicts of interest.
REFERENCES
- [1].Hanahan D; Weinberg RA Hallmarks of cancer: the next generation. Cell, 2011, 144(5), 646–674. [DOI] [PubMed] [Google Scholar]
- [2].Weinstein IB; Joe A Oncogene addiction. Cancer Res, 2008, 68(9), 3077–3080; discussion 3080. [DOI] [PubMed] [Google Scholar]
- [3].Letai AG Diagnosing and exploiting cancer’s addiction to blocks in apoptosis. Nat. Rev. Cancer, 2008, 8(2), 121–132. [DOI] [PubMed] [Google Scholar]
- [4].Zhang Z; Li M; Rayburn ER; Hill DL; Zhang R; Wang H Oncogenes as novel targets for cancer therapy (part I): growth factors and protein tyrosine kinases. Am. J. Pharmacogenomics, 2005, 5(3), 173–190. [DOI] [PubMed] [Google Scholar]
- [5].Zilfou JT; Lowe SW Tumor suppressive functions of p53. Cold Spring Harb. Perspect. Biol, 2009, 1(5), a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Levine AJ; Oren M The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer, 2009, 9(10), 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Meek DW Tumour suppression by p53: a role for the DNA damage response? Nat. Rev. Cancer, 2009, 9(10), 714–723. [DOI] [PubMed] [Google Scholar]
- [8].Teodoro JG; Evans SK; Green MR Inhibition of tumor angiogenesis by p53: a new role for the guardian of the genome. J. Mol. Med, 2007, 85(11), 1175–1186. [DOI] [PubMed] [Google Scholar]
- [9].Vousden KH; Lu X Live or let die: the cell’s response to p53. Nat. Rev. Cancer, 2002, 2(8), 594–604. [DOI] [PubMed] [Google Scholar]
- [10].Joerger AC; Fersht AR Structural biology of the tumor suppressor p53. Ann. Rev. Biochem, 2008, 77, 557–582. [DOI] [PubMed] [Google Scholar]
- [11].Yoshida K; Miki Y The cell death machinery governed by the p53 tumor suppressor in response to DNA damage. Cancer Sci, 2010, 101(4), 831–835. [DOI] [PubMed] [Google Scholar]
- [12].Vazquez A; Bond EE; Levine AJ; Bond GL The genetics of the p53 pathway, apoptosis and cancer therapy. Nature reviews. Drug Discov, 2008, 7(12), 979–987. [DOI] [PubMed] [Google Scholar]
- [13].Riley T; Sontag E; Chen P; Levine A Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell. Biol, 2008, 9(5), 402–412. [DOI] [PubMed] [Google Scholar]
- [14].Hainaut P; Hollstein M p53 and human cancer: the first ten thousand mutations. Adv. Cancer Res, 2000, 77, 81–137. [DOI] [PubMed] [Google Scholar]
- [15].Rivlin N; Brosh R; Oren M; Rotter V Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes & cancer, 2011, 2(4), 466–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Vousden KH; Prives C Blinded by the Light: The Growing Complexity of p53. Cell, 2009, 137(3), 413–431. [DOI] [PubMed] [Google Scholar]
- [17].Menendez D; Inga A; Resnick MA The expanding universe of p53 targets. Nat. Rev. Cancer, 2009, 9(10), 724–737. [DOI] [PubMed] [Google Scholar]
- [18].Momand J; Zambetti GP; Olson DC; George D; Levine AJ The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell, 1992, 69(7), 1237–1245. [DOI] [PubMed] [Google Scholar]
- [19].Cahilly-Snyder L; Yang-Feng T; Francke U; George DL Molecular analysis and chromosomal mapping of amplified genes isolated from a transformed mouse 3T3 cell line. Somat. Cell Mol. Genet, 1987, 13(3), 235–244. [DOI] [PubMed] [Google Scholar]
- [20].Fakharzadeh SS; Trusko SP; George DL Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBOJ, 1991, 10(6), 1565–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Oliner JD; Kinzler KW; Meltzer PS; George DL; Vogel-stein B Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature, 1992, 358(6381), 80–83. [DOI] [PubMed] [Google Scholar]
- [22].Olson DC; Marechal V; Momand J; Chen J; Romocki C; Levine AJ Identification and characterization of multiple mdm-2 proteins and mdm-2-p53 protein complexes. Oncogene, 1993, 8(9), 2353–2360. [PubMed] [Google Scholar]
- [23].Fang S; Jensen JP; Ludwig RL; Vousden KH; Weissman AM Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem, 2000, 275(12), 8945–8951. [DOI] [PubMed] [Google Scholar]
- [24].Momand J; Jung D; Wilczynski S; Niland J The MDM2 gene amplification database. Nucleic Acids Res, 1998, 26(15), 3453–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Onel K; Cordon-Cardo C MDM2 and prognosis. Mol. Cancer Res, 2004, 2(1), 1–8. [PubMed] [Google Scholar]
- [26].Rayburn E; Zhang R; He J; Wang H MDM2 and human malignancies: expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr. Cancer Drug Targets, 2005, 5(1), 27–41. [DOI] [PubMed] [Google Scholar]
- [27].Beaudry GA; Bertelsen AH; Sherman MI Therapeutic targeting of the p53 tumor suppressor gene. Curr. Opin. Biotechnol, 1996, 7(6), 592–600. [DOI] [PubMed] [Google Scholar]
- [28].Mandinova A; Lee SW The p53 pathway as a target in cancer therapeutics: obstacles and promise. Sci. Transl. Med, 2011, 3(64), 64rv61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ventura A; Kirsch DG; McLaughlin ME; Tuveson DA; Grimm J; Lintault L; Newman J; Reczek EE; Weissleder R; Jacks T Restoration of p53 function leads to tumour regression in vivo. Nature, 2007, 445(7128), 661–665. [DOI] [PubMed] [Google Scholar]
- [30].Martins CP; Brown-Swigart L; Evan GI Modeling the therapeutic efficacy of p53 restoration in tumors. Cell, 2006, 127(7), 1323–1334. [DOI] [PubMed] [Google Scholar]
- [31].Kastan MB Wild-type p53: tumors can’t stand it. Cell, 2007, 128(5), 837–840. [DOI] [PubMed] [Google Scholar]
- [32].Wang H; Nan L; Yu D; Lindsey JR; Agrawal S; Zhang R Anti-tumor efficacy of a novel antisense anti-MDM2 mixed-backbone oligonucleotide in human colon cancer models: p53-dependent and p53-independent mechanisms. Mol. Med, 2002, 8(4), 185–199. [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang; Wang H MDM2 oncogene as a novel target for human cancer therapy. Curr. Pharmaceut. Des, 2000, 6(4), 393–416. [DOI] [PubMed] [Google Scholar]
- [34].Chene P Inhibiting the p53-MDM2 interaction: an important target for cancer therapy. Nat. Rev. Cancer, 2003, 3(2), 102–109. [DOI] [PubMed] [Google Scholar]
- [35].Montes de Oca Luna R; Wagner DS; Lozano G Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature, 1995, 378(6553), 203–206. [DOI] [PubMed] [Google Scholar]
- [36].de Rozieres S; Maya R; Oren M; Lozano G The loss of mdm2 induces p53-mediated apoptosis. Oncogene, 2000, 19(13), 1691–1697. [DOI] [PubMed] [Google Scholar]
- [37].Oliner JD; Pietenpol JA; Thiagalingam S; Gyuris J; Kinzler KW; Vogelstein B Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature, 1993, 362(6423), 857–860. [DOI] [PubMed] [Google Scholar]
- [38].Cross B; Chen L; Cheng Q; Li B; Yuan ZM; Chen J Inhibition of p53 DNA binding function by the MDM2 protein acidic domain. J. Biol. Chem, 2011, 286(18), 16018–16029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Thut CJ; Goodrich JA; Tjian R Repression of p53-mediated transcription by MDM2: a dual mechanism. Genes Dev, 1997, 11(15), 1974–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bottger A; Bottger V; Garcia-Echeverria C; Chene P; Hochkeppel HK; Sampson W; Ang K; Howard SF; Picksley SM; Lane DP Molecular characterization of the hdm2-p53 interaction. J. Mol. Biol, 1997, 269(5), 744–756. [DOI] [PubMed] [Google Scholar]
- [41].Kubbutat MH; Jones SN; Vousden KH Regulation of p53 stability by Mdm2. Nature, 1997, 387(6630), 299–303. [DOI] [PubMed] [Google Scholar]
- [42].Wsierska-Gadek J; Horky M How the nucleolar sequestration of p53 protein or its interplayers contributes to its (re)-activation. Ann. N. Y. Acad. Sci, 2003, 1010, 266–272. [DOI] [PubMed] [Google Scholar]
- [43].Haupt Y; Maya R; Kazaz A; Oren M Mdm2 promotes the rapid degradation of p53. Nature, 1997, 387(6630), 296–299. [DOI] [PubMed] [Google Scholar]
- [44].Honda R; Tanaka H; Yasuda H Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS. Lett, 1997, 420(1), 25–27. [DOI] [PubMed] [Google Scholar]
- [45].Wu X; Bayle JH; Olson D; Levine AJ The p53-mdm-2 autoregulatory feedback loop. Genes Dev, 1993, 7(7A), 1126–1132. [DOI] [PubMed] [Google Scholar]
- [46].Chi SW; Lee SH; Kim DH; Ahn MJ; Kim JS; Woo JY; Torizawa T; Kainosho M; Han KH Structural details on mdm2-p53 interaction. J. Biol. Chem, 2005, 280(46), 38795–38802. [DOI] [PubMed] [Google Scholar]
- [47].Poyurovsky MV; Katz C; Laptenko O; Beckerman R; Lok-shin M; Ahn J; Byeon IJ; Gabizon R; Mattia M; Zupnick A; Brown LM; Friedler A; Prives C The C terminus of p53 binds the N-terminal domain of MDM2. Nat. Struct. Mol. Biol, 2010, 17(8), 982–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Nakamura S; Roth JA; Mukhopadhyay T Multiple lysine mutations in the C-terminal domain of p53 interfere with MDM2-dependent protein degradation and ubiquitination. Mol. Cell. Biol, 2000, 20(24), 9391–9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Rodriguez MS; Desterro JM; Lain S; Lane DP; Hay RT Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol. Cell. Biol, 2000, 20(22), 8458–8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhang Y; Lu H Signaling to p53: ribosomal proteins find their way. Cancer Cell, 2009, 16(5), 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Dai MS; Lu H Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem, 2004, 279(43), 44475–44482. [DOI] [PubMed] [Google Scholar]
- [52].Zhang Y; Wolf GW; Bhat K; Jin A; Allio T; Burkhart WA; Xiong Y Ribosomal protein L11 negatively regulates onco-protein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol, 2003, 23(23), 8902–8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Dai MS; Shi D; Jin Y; Sun XX; Zhang Y; Grossman SR; Lu H Regulation of the MDM2-p53 pathway by ribosomal protein L11 involves a post-ubiquitination mechanism. J. Biol. Chem, 2006, 281(34), 24304–24313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Jin A; Itahana K; O’Keefe K; Zhang Y Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell. Biol, 2004, 24(17), 7669–7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Dai MS; Zeng SX; Jin Y; Sun XX; David L; Lu H Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell. Biol, 2004, 24(17), 7654–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zhang Y; Wang J; Yuan Y; Zhang W; Guan W; Wu Z; Jin C; Chen H; Zhang L; Yang X; He F Negative regulation of HDM2 to attenuate p53 degradation by ribosomal protein L26. Nucleic Acids Res, 2010, 38(19), 6544–6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Chen D; Zhang Z; Li M; Wang W; Li Y; Rayburn ER; Hill DL; Wang H; Zhang R Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene, 2007, 26(35), 5029–5037. [DOI] [PubMed] [Google Scholar]
- [58].Zhu Y; Poyurovsky MV; Li Y; Biderman L; Stahl J; Jacq X; Prives C Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol. Cell, 2009, 35(3), 316–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zhou X; Hao Q; Liao J; Zhang Q; Lu H Ribosomal protein S14 unties the MDM2-p53 loop upon ribosomal stress. Oncogene, 2013, 32(3), 388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Zhang X; Wang W; Wang H; Wang MH; Xu W; Zhang R Identification of ribosomal protein S25 (RPS25)-MDM2-p53 regulatory feedback loop. Oncogene, 2013, 32(22), 2782–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Sun XX; DeVine T; Challagundla KB; Dai MS Interplay between ribosomal protein S27a and MDM2 protein in p53 activation in response to ribosomal stress. J. Biol. Chem, 2011, 286(26), 22730–22741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Xiong X; Zhao Y; He H; Sun Y, Ribosomal protein S27-like and S27 interplay with p53-MDM2 axis as a target, a substrate and a regulator. Oncogene, 2011, 30(15), 1798–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lohrum MA; Ludwig RL; Kubbutat MH; Hanlon M; Vousden KH Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell, 2003, 3(6), 577–587. [DOI] [PubMed] [Google Scholar]
- [64].Zhang Z; Zhang R Proteasome activator PA28 gamma regulates p53 by enhancing its MDM2-mediated degradation. EMBOJ, 2008, 27(6), 852–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wen W; Peng C; Kim MO; Ho Jeong C; Zhu F; Yao K; Zykova T; Ma W; Carper A; Langfald A; Bode AM; Dong Z Knockdown of RNF2 induces apoptosis by regulating MDM2 and p53 stability. Oncogene, 2013. doi: 10.1038/onc.2012.605. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Su WJ; Fang JS; Cheng F; Liu C; Zhou F; Zhang J RNF2/Ring1b negatively regulates p53 expression in selective cancer cell types to promote tumor development. Proc. Natl. Acad. Sci. U S A, 2013, 110(5), 1720–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Chen D; Zhang J; Li M; Rayburn ER; Wang H; Zhang R RYBP stabilizes p53 by modulating MDM2. EMBO Rep, 2009, 10(2), 166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kamijo T; Weber JD; Zambetti G; Zindy F; Roussel MF; Sherr CJ Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl. Acad. Sci. U S A, 1998, 95(14), 8292–8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Honda R; Yasuda H Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBOJ, 1999, 18(1), 22–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Weber JD; Kuo ML; Bothner B; DiGiammarino EL; Kriwacki RW; Roussel MF; Sherr CJ Cooperative signals governing ARF-mdm2 interaction and nucleolar localization of the complex. Mol. Cell. Biol, 2000, 20(7), 2517–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Chen D; Kon N; Li M; Zhang W; Qin J; Gu W ARFBP1/Mule is a critical mediator of the ARF tumor suppressor. Cell, 2005, 121(7), 1071–1083. [DOI] [PubMed] [Google Scholar]
- [72].Louria-Hayon I; Grossman T; Sionov RV; Alsheich O; Pandolfi PP; Haupt Y The promyelocytic leukemia protein protects p53 from Mdm2-mediated inhibition and degradation. J. Biol. Chem, 2003, 278(35), 33134–33141. [DOI] [PubMed] [Google Scholar]
- [73].Coutts AS; Boulahbel H; Graham A; La Thangue NB Mdm2 targets the p53 transcription cofactor JMY for degradation. EMBO Rep, 2007, 8(1), 84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Yang HY; Wen YY; Chen CH; Lozano G; Lee MH 14–3-3 sigma positively regulates p53 and suppresses tumor growth. Mol. Cell. Biol, 2003, 23(20), 7096–7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Shvarts A; Steegenga WT; Riteco N; van Laar T; Dekker P; Bazuine M; van Ham RC; van der Houven van Oordt W; Hate-Hateboer G; van der Eb AJ; Jochemsen AG MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J, 1996, 15(19), 5349–5357. [PMC free article] [PubMed] [Google Scholar]
- [76].Zdzalik M; Pustelny K; Kedracka-Krok S; Huben K; Pecak A; Wladyka B; Jankowski S; Dubin A; Potempa J; Dubin G Interaction of regulators Mdm2 and Mdmx with transcription factors p53, p63 and p73. Cell Cycle, 2010, 9(22), 4584–4591. [DOI] [PubMed] [Google Scholar]
- [77].Danovi D; Meulmeester E; Pasini D; Migliorini D; Capra M; Frenk R; de Graaf P; Francoz S; Gasparini P; Gobbi A; Helin K; Pelicci PG; Jochemsen AG; Marine JC Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol. Cell. Biol, 2004, 24(13), 5835–5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Prodosmo A; Giglio S; Moretti S; Mancini F; Barbi F; Avenia N; Di Conza G; Schunemann HJ; Pistola L; Ludovini V; Sacchi A; Pontecorvi A; Puxeddu E; Moretti F Analysis of human MDM4 variants in papillary thyroid carcinomas reveals new potential markers of cancer properties. J. Mol. Med, 2008, 86(5), 585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Huang L; Yan Z; Liao X; Li Y; Yang J; Wang ZG; Zuo Y; Kawai H; Shadfan M; Ganapathy S; Yuan ZM The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc. Natl. Acad. Sci. U S A, 2011, 108(29), 12001–12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Francoz S; Froment P; Bogaerts S; De Clercq S; Maetens M; Doumont G; Bellefroid E; Marine JC Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc. Natl. Acad. Sci. U S A, 2006, 103(9), 3232–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Tanimura S; Ohtsuka S; Mitsui K; Shirouzu K; Yoshimura A; Ohtsubo M MDM2 interacts with MDMX through their RING finger domains. FEBS Lett, 1999, 447(1), 5–9. [DOI] [PubMed] [Google Scholar]
- [82].Shadfan M; Lopez-Pajares V; Yuan ZM MDM2 and MDMX: Alone and together in regulation of p53. Transl. Cancer Res, 2012, 1(2), 88–89. [PMC free article] [PubMed] [Google Scholar]
- [83].Marine JC; Jochemsen AG Mdmx as an essential regulator of p53 activity. Biochem. Biophys. Res. Commun, 2005, 331(3), 750–760. [DOI] [PubMed] [Google Scholar]
- [84].Marine JC; Jochemsen AG Mdmx and Mdm2: brothers in arms? Cell Cycle, 2004, 3(7), 900–904. [PubMed] [Google Scholar]
- [85].Pazgier M; Liu M; Zou G; Yuan W; Li C; Li C; Li J; Monbo J; Zella D; Tarasov SG; Lu W Structural basis for high-affinity peptide inhibition of p53 interactions with MDM2 and MDMX. Proc. Natl. Acad. Sci. U S A, 2009, 106(12), 4665–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Eischen CM; Weber JD; Roussel MF; Sherr CJ; Cleveland JL Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev, 1999, 13(20), 2658–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Kurki S; Peltonen K; Latonen L; Kiviharju TM; Ojala PM; Meek D; Laiho M Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell, 2004, 5(5), 465–475. [DOI] [PubMed] [Google Scholar]
- [88].Di Stefano V; Blandino G; Sacchi A; Soddu S; D’Orazi G HIPK2 neutralizes MDM2 inhibition rescuing p53 transcriptional activity and apoptotic function. Oncogene, 2004, 23(30), 5185–5192. [DOI] [PubMed] [Google Scholar]
- [89].Alsheich-Bartok O; Haupt S; Alkalay-Snir I; Saito S; Appella E; Haupt Y PML enhances the regulation of p53 by CK1 in response to DNA damage. Oncogene, 2008, 27(26), 3653–3661. [DOI] [PubMed] [Google Scholar]
- [90].Bernardi R; Scaglioni PP; Bergmann S; Horn HF; Vousden KH; Pandolfi PP PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat. Cell Biol, 2004, 6(7), 665–672. [DOI] [PubMed] [Google Scholar]
- [91].Kurki S; Latonen L; Laiho M Cellular stress and DNA damage invoke temporally distinct Mdm2, p53 and PML complexes and damage-specific nuclear relocalization. J. Cell. Sci, 2003, 116(Pt 19), 3917–3925. [DOI] [PubMed] [Google Scholar]
- [92].Gottifredi V; Prives C P53 and PML: new partners in tumor suppression. Trend. Cell Biol, 2001, 11(5), 184–187. [DOI] [PubMed] [Google Scholar]
- [93].Linares LK; Kiernan R; Triboulet R; Chable-Bessia C; Latreille D; Cuvier O; Lacroix M; Le Cam L; Coux O; Benkirane M Intrinsic ubiquitination activity of PCAF controls the stability of the oncoprotein Hdm2. Nat. Cell Biol, 2007, 9(3), 331–338. [DOI] [PubMed] [Google Scholar]
- [94].Jin Y; Zeng SX; Dai MS; Yang XJ; Lu H MDM2 inhibits PCAF (p300/CREB-binding protein-associated factor)-mediated p53 acetylation. J. Biol. Chem, 2002, 277(34), 30838–30843. [DOI] [PubMed] [Google Scholar]
- [95].Nishida T; Yamada Y The nucleolar SUMO-specific protease SMT3IP1/SENP3 attenuates Mdm2-mediated p53 ubiquitination and degradation. Biochem. Biophys. Res. Commun, 2011, 406(2), 285–291. [DOI] [PubMed] [Google Scholar]
- [96].Kim JW; Song PI; Jeong MH; An JH; Lee SY; Jang SM; Song KH; Armstrong CA; Choi KH TIP60 represses transcriptional activity of p73beta via an MDM2-bridged ternary complex. J. Biol. Chem, 2008, 283(29), 20077–20086. [DOI] [PubMed] [Google Scholar]
- [97].Tang Y; Luo J; Zhang W; Gu W Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell, 2006, 24(6), 827–839. [DOI] [PubMed] [Google Scholar]
- [98].Legube G; Linares LK; Lemercier C; Scheffner M; Khochbin S; Trouche D Tip60 is targeted to proteasome-mediated degradation by Mdm2 and accumulates after UV irradiation. EMBOJ, 2002, 21(7), 1704–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Oliver TG; Meylan E; Chang GP; Xue W; Burke JR; Humpton TJ; Hubbard D; Bhutkar A; Jacks T Caspase-2-mediated cleavage of Mdm2 creates a p53-induced positive feedback loop. Mol. Cell, 2011, 43(1), 57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Pan W; Issaq S; Zhang Y, The in vivo role of the RP-Mdm2-p53 pathway in signaling oncogenic stress induced by pRb inactivation and Ras overexpression. Plos One, 2011, 6(6), e21625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Mathew R; Arora S; Khanna R; Mathur M; Shukla NK; Ralhan R Alterations in p53 and pRb pathways and their prognostic significance in oesophageal cancer. Eur. J. Cancer, 2002, 38(6), 832–841. [DOI] [PubMed] [Google Scholar]
- [102].Bae S; Jung JH; An IS; Kim OY; Lee MJ; Lee JH; Park IC; Lee SJ; An S TRIAD1 is negatively regulated by the MDM2 E3 ligase. Oncology reports, 2012, 28, (5), 1924–1928. [DOI] [PubMed] [Google Scholar]
- [103].Bae S; Jung JH; Kim K; An IS; Kim SY; Lee JH; Park IC; Jin YW; Lee SJ; An S, TRIAD1 inhibits MDM2-mediated p53 ubiquitination and degradation. FEBS. Lett, 2012, 586(19), 3057–3063. [DOI] [PubMed] [Google Scholar]
- [104].Hsueh KW; Fu SL; Chang CB; Chang YL; Lin CH A novel Aurora-A-mediated phosphorylation of p53 inhibits its interaction with MDM2. Biochim. Biophys. Acta, 2013, 1834(2), 508–515. [DOI] [PubMed] [Google Scholar]
- [105].Yu W; Qiu Z; Gao N; Wang L; Cui H; Qian Y; Jiang L; Luo J; Yi Z; Lu H; Li D; Liu M PAK1IP1, a ribosomal stress-induced nucleolar protein, regulates cell proliferation via the p53-MDM2 loop. Nucleic Acids Res, 2011, 39(6), 2234–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kubo N; Wu D; Yoshihara Y; Sang M; Nakagawara A; Ozaki T Co-chaperon DnaJC7/TPR2 enhances p53 stability and activity through blocking the complex formation between p53 and MDM2. Biochem. Biophys. Res. Commun, 2013, 430(3), 1034–1039. [DOI] [PubMed] [Google Scholar]
- [107].Sun XX; Challagundla KB; Dai MS Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBOJ, 2012, 31(3), 576–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].ElSawy KM; Verma CS; Joseph TL; Lane DP; Twarock R; Caves LS On the interaction mechanisms of a p53 peptide and nutlin with the MDM2 and MDMX proteins: a Brownian dynamics study. Cell Cycle, 2013, 12(3), 394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Wade M; Li YC; Wahl GM MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer, 2013, 13(2), 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Xia M; Knezevic D; Tovar C; Huang B; Heimbrook DC; Vassilev LT Elevated MDM2 boosts the apoptotic activity of p53-MDM2 binding inhibitors by facilitating MDMX degradation. Cell Cycle, 2008, 7(11), 1604–1612. [DOI] [PubMed] [Google Scholar]
- [111].Jiang M; Chiu SY; Hsu W SUMO-specific protease 2 in Mdm2-mediated regulation of p53. Cell Death Differ, 2011, 18(6), 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Jung CR; Lim JH; Choi Y; Kim DG; Kang KJ; Noh SM; Im DS Enigma negatively regulates p53 through MDM2 and promotes tumor cell survival in mice. J. Clin. Invest, 2010, 120(12), 4493–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Gronroos E; Terentiev AA; Punga T; Ericsson J YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc. Natl. Acad. Sci. U S A, 2004, 101(33), 12165–12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Sui G; Affar el B; Shi Y; Brignone C; Wall NR; Yin P; Donohoe M; Luke MP; Calvo D; Grossman SR; Shi Y Yin Yang 1 is a negative regulator of p53. Cell, 2004, 117(7), 859–872. [DOI] [PubMed] [Google Scholar]
- [115].Cummins JM; Vogelstein B HAUSP is required for p53 destabilization. Cell Cycle, 2004, 3(6), 689–692. [PubMed] [Google Scholar]
- [116].Ding B; Sun Y; Huang J Overexpression of SKI oncoprotein leads to p53 degradation through regulation of MDM2 protein sumoylation. J. Biol. Chem, 2012, 287(18), 14621–14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Du W; Jiang P; Li N; Mei Y; Wang X; Wen L; Yang X; Wu M Suppression of p53 activity by Siva1. Cell Death Dffer, 2009, 16(11), 1493–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Alt JR; Greiner TC; Cleveland JL; Eischen CM Mdm2 haplo-insufficiency profoundly inhibits Myc-induced lymphoma-genesis. EMBOJ, 2003, 22(6), 1442–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Jones SN; Hancock AR; Vogel H; Donehower LA; Bradley A Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc. Natl. Acad. Sci. U S A, 1998, 95(26), 15608–15612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Zhang Z; Wang H; Li M; Rayburn E; Agrawal S; Zhang R Novel MDM2 p53-independent functions identified through RNA silencing technologies. Ann. N. Y. Acad. Sci, 2005, 1058, 205–214. [DOI] [PubMed] [Google Scholar]
- [121].Manfredi JJ The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev, 2010, 24(15), 1580–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Ganguli G; Wasylyk B p53-independent functions of MDM2. Mol. Cancer Res, 2003, 1(14), 1027–1035. [PubMed] [Google Scholar]
- [123].Bouska A; Eischen CM Murine double minute 2: p53-independent roads lead to genome instability or death. Trend. Biochem Sci, 2009, 34(6), 279–286. [DOI] [PubMed] [Google Scholar]
- [124].Maguire M; Nield PC; Devling T; Jenkins RE; Park BK; Polanski R; Vlatkovic N; Boyd MT MDM2 regulates dihydrofolate reductase activity through monoubiquitination. Cancer Res, 2008, 68(9), 3232–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Alt JR; Bouska A; Fernandez MR; Cerny RL; Xiao H; Eischen CM Mdm2 binds to Nbs1 at sites of DNA damage and regulates double strand break repair. J. Biol. Chem, 2005, 280(19), 18771–18781. [DOI] [PubMed] [Google Scholar]
- [126].Zhang Z; Wang H; Li M; Agrawal S; Chen X; Zhang R MDM2 is a negative regulator of p21WAF1/CIP1, independent of p53. J. Biol. Chem, 2004, 279(16), 16000–16006. [DOI] [PubMed] [Google Scholar]
- [127].Jin Y; Lee H; Zeng SX; Dai MS; Lu H MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBOJ, 2003, 22(23), 6365–6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Xu H; Zhang Z; Li M; Zhang R MDM2 promotes proteasomal degradation of p21Waf1 via a conformation change. J. Biol. Chem, 2010, 285(24), 18407–18414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Xiao ZX; Chen J; Levine AJ; Modjtahedi N; Xing J; Sellers WR; Livingston DM Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature, 1995, 375(6533), 694–698. [DOI] [PubMed] [Google Scholar]
- [130].Sun P; Dong P; Dai K; Hannon GJ; Beach D p53-independent role of MDM2 in TGF-beta1 resistance. Science, 1998, 282(5397), 2270–2272. [DOI] [PubMed] [Google Scholar]
- [131].Lundgren K; Montes de Oca Luna R; McNeill YB; Emerick EP; Spencer B; Barfield CR; Lozano G; Rosenberg MP; Finlay CA Targeted expression of MDM2 uncouples S phase from mitosis and inhibits mammary gland development independent of p53. Genes Dev, 1997, 11(6), 714–725. [DOI] [PubMed] [Google Scholar]
- [132].Tang YA; Lin RK; Tsai YT; Hsu HS; Yang YC; Chen CY; Wang YC MDM2 overexpression deregulates the transcriptional control of RB/E2F leading to DNA methyltransferase 3A overexpression in lung cancer. Clin. Cancer Res, 2012, 18(16), 4325–4333. [DOI] [PubMed] [Google Scholar]
- [133].Loughran O; La Thangue NB Apoptotic and growth-promoting activity of E2F modulated by MDM2. Mol. Cell. Biol, 2000, 20(6), 2186–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Malaguarnera R; Vella V; Pandini G; Sanfilippo M; Pezzino V; Vigneri R; Frasca F TAp73 alpha increases p53 tumor suppressor activity in thyroid cancer cells via the inhibition of Mdm2-mediated degradation. Mol. Cancer Res, 2008, 6(1), 64–77. [DOI] [PubMed] [Google Scholar]
- [135].Fu W; Ma Q; Chen L; Li P; Zhang M; Ramamoorthy S; Nawaz Z; Shimojima T; Wang H; Yang Y; Shen Z; Zhang Y; Zhang X; Nicosia SV; Zhang Y; Pledger JW; Chen J; Bai W MDM2 acts downstream of p53 as an E3 ligase to promote FOXO ubiquitination and degradation. J. Biol. Chem, 2009, 284(21), 13987–14000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Zeng X; Chen L; Jost CA; Maya R; Keller D; Wang X; Kaelin WG Jr.; Oren M; Chen J; Lu H MDM2 suppresses p73 function without promoting p73 degradation. Mol. Cell. Biol, 1999, 19(5), 3257–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Yang JY; Zong CS; Xia W; Yamaguchi H; Ding Q; Xie X; Lang JY; Lai CC; Chang CJ; Huang WC; Huang H; Kuo HP; Lee DF; Li LY; Lien HC; Cheng X; Chang KJ; Hsiao CD; Tsai FJ; Tsai CH; Sahin AA; Muller WJ; Mills GB; Yu D; Hortobagyi GN; Hung MC ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat. Cell. Biol, 2008, 10(2), 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Gu L; Zhu N; Zhang H; Durden DL; Feng Y; Zhou M Regulation of XIAP translation and induction by MDM2 following irradiation. Cancer Cell, 2009, 15(5), 363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Nieminen AL; Qanungo S; Schneider EA; Jiang BH; Agani FH Mdm2 and HIF-1alpha interaction in tumor cells during hypoxia. J. Cell. Physiol, 2005, 204(2), 364–369. [DOI] [PubMed] [Google Scholar]
- [140].Ravi R; Mookerjee B; Bhujwalla ZM; Sutter CH; Artemov D; Zeng Q; Dillehay LE; Madan A; Semenza GL; Bedi A Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes Dev, 2000, 14(1), 34–44. [PMC free article] [PubMed] [Google Scholar]
- [141].Carroll VA; Ashcroft M Regulation of angiogenic factors by HDM2 in renal cell carcinoma. Cancer Res, 2008, 68(2), 545–552. [DOI] [PubMed] [Google Scholar]
- [142].Yang JY; Zong CS; Xia W; Wei Y; Ali-Seyed M; Li Z; Broglio K; Berry DA; Hung MC MDM2 promotes cell motility and invasiveness by regulating E-cadherin degradation. Mol. Cell. Biol, 2006, 26(19), 7269–7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Iwakuma T; Agarwal N MDM2 binding protein, a novel metastasis suppressor. Cancer Metas. Rev, 2012, 31(3–4), 633–640. [DOI] [PubMed] [Google Scholar]
- [144].Jung CH; Kim J; Park JK; Hwang SG; Moon SK; Kim WJ; Um HD Mdm2 increases cellular invasiveness by binding to and stabilizing the Slug mRNA. Cancer Lett, 2013, 335(2), 270–277. [DOI] [PubMed] [Google Scholar]
- [145].Kurokawa M; Kim J; Geradts J; Matsuura K; Liu L; Ran X; Xia W; Ribar TJ; Henao R; Dewhirst MW; Kim WJ; Lucas JE; Wang S; Spector NL; Kornbluth S A Network of Substrates of the E3 Ubiquitin Ligases MDM2 and HUWE1 Control Apoptosis Independently of p53. Sci. Signal, 2013, 6(274), ra32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Gu L; Findley HW; Zhou M MDM2 induces NF-kappaB/p65 expression transcriptionally through Sp1-binding sites: a novel, p53-independent role of MDM2 in doxorubicin resistance in acute lymphoblastic leukemia. Blood, 2002, 99(9), 3367–3375. [DOI] [PubMed] [Google Scholar]
- [147].Rayburn ER; Ezell SJ; Zhang R Recent advances in validating MDM2 as a cancer target. Anti-Cancer Agent. Med. Chem, 2009, 9(8), 882–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Lukas J; Gao DQ; Keshmeshian M; Wen WH; Tsao-Wei D; Rosenberg S; Press MF Alternative and aberrant messenger RNA splicing of the mdm2 oncogene in invasive breast cancer. Cancer Res, 2001, 61(7), 3212–3219. [PubMed] [Google Scholar]
- [149].Sjostrom J; Blomqvist C; Heikkila P; Boguslawski KV; Raisanen-Sokolowski A; Bengtsson NO; Mjaaland I; Malmstrom P; Ostenstadt B; Bergh J; Wist E; Valvere V; Saksela E Predictive value of p53, mdm-2, p21, and mib-1 for chemotherapy response in advanced breast cancer. Clin. Cancer Res, 2000, 6(8), 3103–3110. [PubMed] [Google Scholar]
- [150].Osman I; Drobnjak M; Fazzari M; Ferrara J; Scher HI; Cordon-Cardo C Inactivation of the p53 pathway in prostate cancer: impact on tumor progression. Clin. Cancer Res, 1999, 5(8), 2082–2088. [PubMed] [Google Scholar]
- [151].Leite KR; Franco MF; Srougi M; Nesrallah LJ; Nesrallah A; Bevilacqua RG; Darini E; Carvalho CM; Meirelles MI; Santana I; Camara-Lopes LH Abnormal expression of MDM2 in prostate carcinoma. Mod. Pathol, 2001, 14(5), 428–436. [DOI] [PubMed] [Google Scholar]
- [152].Khor LY; Desilvio M; Al-Saleem T; Hammond ME; Grignon DJ; Sause W; Pilepich M; Okunieff P; Sandler H; Pollack A; Radiation Therapy Oncology G MDM2 as a predictor of prostate carcinoma outcome: an analysis of Radiation Therapy Oncology Group Protocol 8610. Cancer, 2005, 104(5), 962–967. [DOI] [PubMed] [Google Scholar]
- [153].Dworakowska D; Jassem E; Jassem J; Karmolinski A; Dworakowski R; Wirth T; Gruchala M; Rynkiewicz A; Skokowski J; Yla-Herttuala S; Jaskiewicz K; Czestochowska E Clinical significance of apoptotic index in non-small cell lung cancer: correlation with p53, mdm2, pRb and p21WAF1/CIP1 protein expression. J. Cancer Res. Clin. Oncol, 2005, 131(9), 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Mori S; Ito G; Usami N; Yoshioka H; Ueda Y; Kodama Y; Takahashi M; Fong KM; Shimokata K; Sekido Y p53 apoptotic pathway molecules are frequently and simultaneously altered in nonsmall cell lung carcinoma. Cancer, 2004, 100(8), 1673–1682. [DOI] [PubMed] [Google Scholar]
- [155].Higashiyama M; Doi O; Kodama K; Yokouchi H; Kasugai T; Ishiguro S; Takami K; Nakayama T; Nishisho I MDM2 gene amplification and expression in non-small-cell lung cancer: immunohistochemical expression of its protein is a favourable prognostic marker in patients without p53 protein accumulation. Brit. J. Cancer, 1997, 75(9), 1302–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Abdel-Fattah G; Yoffe B; Krishnan B; Khaoustov V; Itani K MDM2/p53 protein expression in the development of colorectal adenocarcinoma. J. Gastrointest. Surg, 2000, 4(1), 109–114. [DOI] [PubMed] [Google Scholar]
- [157].Elkablawy MA; Maxwell P; Williamson K; Anderson N; Hamilton PW Apoptosis and cell-cycle regulatory proteins in colorectal carcinoma: relationship to tumour stage and patient survival. J. Pathol, 2001, 194(4), 436–443. [DOI] [PubMed] [Google Scholar]
- [158].Takata O; Kawamura YJ; Konishi F; Sasaki J; Kai T; Miyakura Y; Nagai H; Tsukamoto T cDNA array analysis for prediction of hepatic metastasis of colorectal carcinoma. Surgery Today, 2006, 36(7), 608–614. [DOI] [PubMed] [Google Scholar]
- [159].Paradiso A; Ranieri G; Simone G; Silvestris N; Costa A; De Lena M; Leone A; Vallejo C; Lacava J mdm2-p53 Interaction: lack of correlation with the response to 5-fluorouracil in advanced colorectal cancer. Oncology, 2002, 62(3), 278–285. [DOI] [PubMed] [Google Scholar]
- [160].Cescon DW; Bradbury PA; Asomaning K; Hopkins J; Zhai R; Zhou W; Wang Z; Kulke M; Su L; Ma C; Xu W; Marshall AL; Heist RS; Wain JC; Lynch TJ Jr.; Christiani DC; Liu G p53 Arg72Pro and MDM2 T309G polymorphisms, histology, and esophageal cancer prognosis. Clin. Cancer Res, 2009, 15(9), 3103–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Okamoto H; Fujishima F; Nakamura Y; Zuguchi M; Miyata G; Kamei T; Nakano T; Katsura K; Abe S; Taniyama Y; Teshima J; Watanabe M; Sato A; Ohuchi N; Sasano H Murine double minute 2 and its association with chemoradioresistance of esophageal squamous cell carcinoma. Anticancer Res, 2013, 33(4), 1463–1471. [PubMed] [Google Scholar]
- [162].Ladanyi M; Cha C; Lewis R; Jhanwar SC; Huvos AG; Healey JH MDM2 gene amplification in metastatic osteosarcoma. Cancer res, 1993, 53(1), 16–18. [PubMed] [Google Scholar]
- [163].Ito M; Barys L; O’Reilly T; Young S; Gorbatcheva B; Mona-han J; Zumstein-Mecker S; Choong PF; Dickinson I; Crowe P; Hemmings C; Desai J; Thomas DM; Lisztwan J Comprehensive mapping of p53 pathway alterations reveals an apparent role for both SNP309 and MDM2 amplification in sarcomagenesis. Clin. Cancer Res, 2011, 17(3), 416–426. [DOI] [PubMed] [Google Scholar]
- [164].Terry K; McGrath M; Lee IM; Buring J; De Vivo I MDM2 SNP309 is associated with endometrial cancer risk. Cancer Epidemiol. Biomarkers Prev, 2008, 17(4), 983–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Ye Y; Li X; Yang J; Miao S; Wang S; Chen Y; Xia X; Wu X; Zhang J; Zhou Y; He S; Tan Y; Qiang F; Li G; Roe OD; Zhou J MDM2 is a useful prognostic biomarker for resectable gastric cancer. Cancer Sci, 2013, 104(5), 590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Asomaning K; Reid AE; Zhou W; Heist RS; Zhai R; Su L; Kwak EL; Blaszkowsky L; Zhu AX; Ryan DP; Christiani DC; Liu G MDM2 promoter polymorphism and pancreatic cancer risk and prognosis. Clin. Cancer Res, 2008, 14(12), 4010–4015. [DOI] [PubMed] [Google Scholar]
- [167].Baekelandt M; Kristensen GB; Nesland JM; Trope CG; Holm R Clinical significance of apoptosis-related factors p53, Mdm2, and Bcl-2 in advanced ovarian cancer. J. Clin. Oncol, 1999, 17(7), 2061. [DOI] [PubMed] [Google Scholar]
- [168].Ma YY; Guan TP; Yao HB; Yu S; Chen LG; Xia YJ; He XJ; Wang HJ; Jiang XT; Tao HQ The MDM2 309T>G polymorphism and ovarian cancer risk: a meta-analysis of 1534 cases and 2211 controls. Plos One, 2013, 8(1), e55019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Onat OE; Tez M; Ozcelik T; Toruner GA MDM2 T309G polymorphism is associated with bladder cancer. Anticancer Res, 2006, 26(5A), 3473–3475. [PubMed] [Google Scholar]
- [170].Lianes P; Orlow I; Zhang ZF; Oliva MR; Sarkis AS; Reuter VE; Cordon-Cardo C Altered patterns of MDM2 and TP53 expression in human bladder cancer. J. Natl. Cancer Inst, 1994, 86(17), 1325–1330. [DOI] [PubMed] [Google Scholar]
- [171].Watanabe T; Ichikawa A; Saito H; Hotta T Overexpression of the MDM2 oncogene in leukemia and lymphoma. Leuk. Lymphoma, 1996, 21(5–6), 391–397, color plates XVI following 395. [DOI] [PubMed] [Google Scholar]
- [172].Gustafsson B; Axelsson B; Gustafsson B; Christensson B; Winiarski J MDM2 and p53 in childhood acute lymphoblastic leukemia: higher expression in childhood leukemias with poor prognosis compared to long-term survivors. Pediatr. Hematol. Oncol, 2001, 18(8), 497–508. [DOI] [PubMed] [Google Scholar]
- [173].Gryshchenko I; Hofbauer S; Stoecher M; Daniel PT; Steurer M; Gaiger A; Eigenberger K; Greil R; Tinhofer I MDM2 SNP309 is associated with poor outcome in B-cell chronic lymphocytic leukemia. J. Clin. Oncol, 2008, 26(14), 2252–2257. [DOI] [PubMed] [Google Scholar]
- [174].Muthusamy V; Hobbs C; Nogueira C; Cordon-Cardo C; McKee PH; Chin L; Bosenberg MW Amplification of CDK4 and MDM2 in malignant melanoma. Genes Chromosomes Cancer, 2006, 45(5), 447–454. [DOI] [PubMed] [Google Scholar]
- [175].Polsky D; Bastian BC; Hazan C; Melzer K; Pack J; Houghton A; Busam K; Cordon-Cardo C; Osman I HDM2 protein overexpression, but not gene amplification, is related to tumori-genesis of cutaneous melanoma. Cancer res, 2001, 61(20), 7642–7646. [PubMed] [Google Scholar]
- [176].Rajabi P; Karimian P; Heidarpour M The relationship between MDM2 expression and tumor thickness and invasion in primary cutaneous malignant melanoma. J. Res. Med. Sci, 2012, 17(5), 452–455. [PMC free article] [PubMed] [Google Scholar]
- [177].Firoz EF; Warycha M; Zakrzewski J; Pollens D; Wang G; Shapiro R; Berman R; Pavlick A; Manga P; Ostrer H; Celebi JT; Kamino H; Darvishian F; Rolnitzky L; Goldberg JD; Osman I; Polsky D Association of MDM2 SNP309, age of onset, and gender in cutaneous melanoma. Clin. Cancer Res, 2009, 15(7), 2573–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Coupland SE; Anastassiou G; Stang A; Schilling H; Anagnostopoulos I; Bornfeld N; Stein H The prognostic value of cyclin D1, p53, and MDM2 protein expression in uveal melanoma. J. Pathol, 2000, 191(2), 120–126. [DOI] [PubMed] [Google Scholar]
- [179].Dworakowska D; Jassem E; Jassem J; Peters B; Dziadziuszko R; Zylicz M; Jakobkiewicz-Banecka J; Kobierska-Gulida G; Szymanowska A; Skokowski J; Roessner A; Schneider-Stock R MDM2 gene amplification: a new independent factor of adverse prognosis in non-small cell lung cancer (NSCLC). Lung Cancer, 2004, 43(3), 285–295. [DOI] [PubMed] [Google Scholar]
- [180].Grochola LF; Zeron-Medina J; Meriaux S; Bond GL Single-nucleotide polymorphisms in the p53 signaling pathway. Cold Spring Harb. Perspect. Biol, 2010, 2(5), a001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Bond GL; Hu W; Bond EE; Robins H; Lutzker SG; Arva NC; Bargonetti J; Bartel F; Taubert H; Wuerl P; Onel K; Yip L; Hwang SJ; Strong LC; Lozano G; Levine AJ A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell, 2004, 119(5), 591–602. [DOI] [PubMed] [Google Scholar]
- [182].Bond GL; Hirshfield KM; Kirchhoff T; Alexe G; Bond EE; Robins H; Bartel F; Taubert H; Wuerl P; Hait W; Toppmeyer D; Offit K; Levine AJ MDM2 SNP309 accelerates tumor formation in a gender-specific and hormone-dependent manner. Cancer Res, 2006, 66(10), 5104–5110. [DOI] [PubMed] [Google Scholar]
- [183].Wan Y; Wu W; Yin Z; Guan P; Zhou B MDM2 SNP309, gene-gene interaction, and tumor susceptibility: an updated meta-analysis. BMC Cancer, 2011, 11, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Wilkening S; Bermejo JL; Hemminki K MDM2 SNP309 and cancer risk: a combined analysis. Carcinogenesis, 2007, 28(11), 2262–2267. [DOI] [PubMed] [Google Scholar]
- [185].El Hallani S; Marie Y; Idbaih A; Rodero M; Boisselier B; Laigle-Donadey F; Ducray F; Delattre JY; Sanson M No association of MDM2 SNP309 with risk of glioblastoma and prognosis. J. Neurooncol, 2007, 85(3), 241–244. [DOI] [PubMed] [Google Scholar]
- [186].Araki S; Eitel JA; Batuello CN; Bijangi-Vishehsaraei K; Xie XJ; Danielpour D; Pollok KE; Boothman DA; Mayo LD TGF-beta1-induced expression of human Mdm2 correlates with late-stage metastatic breast cancer. J. Clin. Invest, 2010, 120(1), 290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Phelps M; Darley M; Primrose JN; Blaydes JP p53-independent activation of the hdm2-P2 promoter through multiple transcription factor response elements results in elevated hdm2 expression in estrogen receptor alpha-positive breast cancer cells. Cancer Res, 2003, 63(10), 2616–2623. [PubMed] [Google Scholar]
- [188].Brekman A; Singh KE; Polotskaia A; Kundu N; Bargonetti J A p53-independent role of Mdm2 in estrogen-mediated activation of breast cancer cell proliferation. Breast Cancer Res, 2011, 13(1), R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Zhou M; Gu L; Findley HW; Jiang R; Woods WG PTEN reverses MDM2-mediated chemotherapy resistance by interacting with p53 in acute lymphoblastic leukemia cells. Cancer Res, 2003, 63(19), 6357–6362. [PubMed] [Google Scholar]
- [190].Carr J; Bell E; Pearson AD; Kees UR; Beris H; Lunec J; Tweddle DA Increased frequency of aberrations in the p53/MDM2/p14(ARF) pathway in neuroblastoma cell lines established at relapse. Cancer Res, 2006, 66(4), 2138–2145. [DOI] [PubMed] [Google Scholar]
- [191].Liu W; He L; Ramirez J; Ratain MJ Interactions between MDM2 and TP53 Genetic Alterations, and Their Impact on Response to MDM2 Inhibitors and Other Chemotherapeutic Drugs in Cancer Cells. Clin. Cancer Res, 2009, 15(24), 7602–7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Zhang R; Wang H; Agrawal S Novel antisense anti-MDM2 mixed-backbone oligonucleotides: proof of principle, in vitro and in vivo activities, and mechanisms. Curr. Cancer Drug Targets, 2005, 5(1), 43–49. [DOI] [PubMed] [Google Scholar]
- [193].Wang H; Nan L; Yu D; Agrawal S; Zhang R Antisense anti-MDM2 oligonucleotides as a novel therapeutic approach to human breast cancer: in vitro and in vivo activities and mechanisms. Clin. Cancer Res, 2001, 7(11), 3613–3624. [PubMed] [Google Scholar]
- [194].Zhang Z; Li M; Wang H; Agrawal S; Zhang R Antisense therapy targeting MDM2 oncogene in prostate cancer: Effects on proliferation, apoptosis, multiple gene expression, and chemotherapy. Proc. Natl. Acad. Sci. U S A, 2003, 100(20), 11636–11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Zhang Z; Wang H; Prasad G; Li M; Yu D; Bonner JA; Agrawal S; Zhang R Radiosensitization by antisense anti-MDM2 mixed-backbone oligonucleotide in in vitro and in vivo human cancer models. Clin. Cancer Res, 2004, 10(4), 1263–1273. [DOI] [PubMed] [Google Scholar]
- [196].Zhang R; Wang H Antisense oligonucleotide inhibitors of MDM2 oncogene expression. Met. Mol. Med, 2003, 85, 205–222. [DOI] [PubMed] [Google Scholar]
- [197].Wang H; Zeng X; Oliver P; Le LP; Chen J; Chen L; Zhou W; Agrawal S; Zhang R MDM2 oncogene as a target for cancer therapy: An antisense approach. Int. J. Oncol, 1999, 15(4), 653–660. [PubMed] [Google Scholar]
- [198].Vassilev LT; Vu BT; Graves B; Carvajal D; Podlaski F; Filipovic Z; Kong N; Kammlott U; Lukacs C; Klein C; Fotouhi N; Liu EA In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science, 2004, 303(5659), 844–848. [DOI] [PubMed] [Google Scholar]
- [199].Fry DC; Vassilev LT Targeting protein-protein interactions for cancer therapy. J. Mol. Med, 2005, 83(12), 955–963. [DOI] [PubMed] [Google Scholar]
- [200].Vassilev LT MDM2 inhibitors for cancer therapy. Trends Mol. Med, 2007, 13(1), 23–31. [DOI] [PubMed] [Google Scholar]
- [201].Chen J; Wang J; Xu B; Zhu W; Li G Insight into mechanism of small molecule inhibitors of the MDM2-p53 interaction: molecular dynamics simulation and free energy analysis. J. Mol. Graphics Modell, 2011, 30, 46–53. [DOI] [PubMed] [Google Scholar]
- [202].Shangary S; Wang S Small-molecule inhibitors of the MDM2-p53 protein-protein interaction to reactivate p53 function: a novel approach for cancer therapy. Ann. Rev. Pharmacol. Toxicol, 2009, 49, 223–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Kussie PH; Gorina S; Marechal V; Elenbaas B; Moreau J; Levine AJ; Pavletich NP Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science, 1996, 274(5289), 948–953. [DOI] [PubMed] [Google Scholar]
- [204].Chen L; Agrawal S; Zhou W; Zhang R; Chen J Synergistic activation of p53 by inhibition of MDM2 expression and DNA damage. Proc. Natl. Acad. Sci. U S A, 1998, 95(1), 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Tortora G; Caputo R; Damiano V; Bianco R; Chen J; Agrawal S; Bianco AR; Ciardiello F A novel MDM2 anti-sense oligonucleotide has anti-tumor activity and potentiates cytotoxic drugs acting by different mechanisms in human colon cancer. Int. J. Cancer, 2000, 88(5), 804–809. [DOI] [PubMed] [Google Scholar]
- [206].Wang H; Oliver P; Zhang Z; Agrawal S; Zhang R Chemosensitization and radiosensitization of human cancer by antisense anti-MDM2 oligonucleotides: in vitro and in vivo activities and mechanisms. Ann. N. Y. Acad. Sci, 2003, 1002, 217–235. [DOI] [PubMed] [Google Scholar]
- [207].Castanotto D; Li JR; Michienzi A; Langlois MA; Lee NS; Puymirat J; Rossi JJ Intracellular ribozyme applications. Biochem. Soc. Trans, 2002, 30(Pt 6), 1140–1145. [DOI] [PubMed] [Google Scholar]
- [208].Brummelkamp TR; Fabius AW; Mullenders J; Madiredjo M; Velds A; Kerkhoven RM; Bernards R; Beijersbergen RL An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat. Chem. Biol, 2006, 2(4), 202–206. [DOI] [PubMed] [Google Scholar]
- [209].Brown CJ; Dastidar SG; See HY; Coomber DW; Ortiz-Lombardia M; Verma C; Lane DP Rational design and biophysical characterization of thioredoxin-based aptamers: insights into peptide grafting. J. Mol. Biol, 2010, 395(4), 871–883. [DOI] [PubMed] [Google Scholar]
- [210].Kenjo E; Asai T; Yonenaga N; Ando H; Ishii T; Hatanaka K; Shimizu K; Urita Y; Dewa T; Nango M; Tsukada H; Oku N Systemic delivery of small interfering RNA by use of targeted polycation liposomes for cancer therapy. Biol. Pharm. Bull, 2013, 36(2), 287–291. [DOI] [PubMed] [Google Scholar]
- [211].Yu H; Zou Y; Jiang L; Yin Q; He X; Chen L; Zhang Z; Gu W; Li Y Induction of apoptosis in non-small cell lung cancer by downregulation of MDM2 using pH-responsive PMPC-b-PDPA/siRNA complex nanoparticles. Biomaterials, 2013, 34(11), 2738–2747. [DOI] [PubMed] [Google Scholar]
- [212].Li M; Zhang Z; Hill DL; Chen X; Wang H; Zhang R Genistein, a dietary isoflavone, down-regulates the MDM2 oncogene at both transcriptional and posttranslational levels. Cancer Res, 2005, 65(18), 8200–8208. [DOI] [PubMed] [Google Scholar]
- [213].Li M; Zhang Z; Hill DL; Wang H; Zhang R Curcumin, a dietary component, has anticancer, chemosensitization, and radio-sensitization effects by down-regulating the MDM2 oncogene through the PI3K/mTOR/ETS2 pathway. Cancer Res, 2007, 67(5), 1988–1996. [DOI] [PubMed] [Google Scholar]
- [214].Wang W; Zhao Y; Rayburn ER; Hill DL; Wang H; Zhang R In vitro anti-cancer activity and structure-activity relationships of natural products isolated from fruits of Panax ginseng. Cancer Chemoth. Pharm, 2007, 59(5), 589–601. [DOI] [PubMed] [Google Scholar]
- [215].Qin JJ; Nag S; Voruganti S; Wang W; Zhang R Natural product MDM2 inhibitors: anticancer activity and mechanisms of action. Curr. Med. Chem, 2012, 19(33), 5705–5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Lauria A; Tutone M; Ippolito M; Pantano L; Almerico AM Molecular modeling approaches in the discovery of new drugs for anti-cancer therapy: the investigation of p53-MDM2 interaction and its inhibition by small molecules. Curr. Med. Chem, 2010, 17(28), 3142–3154. [DOI] [PubMed] [Google Scholar]
- [217].Sun Y, Targeting E3 ubiquitin ligases for cancer therapy. Cancer Biol. Ther, 2003, 2(6), 623–629. [PubMed] [Google Scholar]
- [218].Yang Y; Ludwig RL; Jensen JP; Pierre SA; Medaglia MV; Davydov IV; Safiran YJ; Oberoi P; Kenten JH; Phillips AC; Weissman AM; Vousden KH Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell, 2005, 7(6), 547–559. [DOI] [PubMed] [Google Scholar]
- [219].Kitagaki J; Agama KK; Pommier Y; Yang Y; Weissman AM Targeting tumor cells expressing p53 with a water-soluble inhibitor of Hdm2. Mol. Cancer Ther, 2008, 7(8), 2445–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Kojima K; Burks JK; Arts J; Andreeff M The novel tryptamine derivative JNJ-26854165 induces wild-type p53- and E2F1-mediated apoptosis in acute myeloid and lymphoid leukemias. Mol. Cancer Ther, 2010, 9(9), 2545–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Wang W; Hu Y Small molecule agents targeting the p53-MDM2 pathway for cancer therapy. Med. Res. Rev, 2012, 32(6), 1159–1196. [DOI] [PubMed] [Google Scholar]
- [222].Arkin MR; Wells JA Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat. Rev. Drug Disco, 2004, 3(4), 301–317. [DOI] [PubMed] [Google Scholar]
- [223].Kamal A; Mohammed AA; Shaik TB p53-Mdm2 inhibitors: patent review (2009 – 2010). Exp. Opin. Ther. Patents, 2012, 22(2), 95–105. [DOI] [PubMed] [Google Scholar]
- [224].Millard M; Pathania D; Grande F; Xu S; Neamati N Small-molecule inhibitors of p53-MDM2 interaction: the 2006–2010 update. Curr. Pharm. Des, 2011, 17(6), 536–559. [DOI] [PubMed] [Google Scholar]
- [225].Zak K; Pecak A; Rys B; Wladyka B; Domling A; Weber L; Holak TA; Dubin G Mdm2 and MdmX inhibitors for the treatment of cancer: a patent review (2011-present). Exp. Opin. Ther. Patents, 2013, 23(4), 425–448. [DOI] [PubMed] [Google Scholar]
- [226].Tovar C; Rosinski J; Filipovic Z; Higgins B; Kolinsky K; Hilton H; Zhao X; Vu BT; Qing W; Packman K; Myklebost O; Heimbrook DC; Vassilev LT Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc. Natl. Acad. Sci. U S A, 2006, 103(6), 1888–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Van Maerken T; Ferdinande L; Taildeman J; Lambertz I; Yigit N; Vercruysse L; Rihani A; Michaelis M; Cinatl J Jr.; Cuvelier CA; Marine JC; De Paepe A; Bracke M; Speleman F; Vandesompele J Antitumor activity of the selective MDM2 antagonist nutlin-3 against chemoresistant neuroblastoma with wild-type p53. J. Natl. Cancer Inst, 2009, 101(22), 1562–1574. [DOI] [PubMed] [Google Scholar]
- [228].Tabe Y; Sebasigari D; Jin L; Rudelius M; Davies-Hill T; Miyake K; Miida T; Pittaluga S; Raffeld M MDM2 antagonist nutlin-3 displays antiproliferative and proapoptotic activity in mantle cell lymphoma. Clin. Cancer Res, 2009, 15(3), 933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Ohnstad HO; Paulsen EB; Noordhuis P; Berg M; Lothe RA; Vassilev LT; Myklebost O MDM2 antagonist Nutlin-3a potentiates antitumour activity of cytotoxic drugs in sarcoma cell lines. BMC Cancer, 2011, 11, 211–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Hasegawa H; Yamada Y; Iha H; Tsukasaki K; Nagai K; Atogami S; Sugahara K; Tsuruda K; Ishizaki A; Kamihira S Activation of p53 by Nutlin-3a, an antagonist of MDM2, induces apoptosis and cellular senescence in adult T-cell leukemia cells. Leukemia, 2009, 23(11), 2090–2101. [DOI] [PubMed] [Google Scholar]
- [231].Tovar C; Higgins B; Kolinsky K; Xia M; Packman K; Heim-brook DC; Vassilev LT MDM2 antagonists boost antitumor effect of androgen withdrawal: implications for therapy of prostate cancer. Mol. Cancer, 2011, 10, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Wang J; Zheng T; Chen X; Song X; Meng X; Bhatta N; Pan S; Jiang H; Liu L MDM2 antagonist can inhibit tumor growth in hepatocellular carcinoma with different types of p53 in vitro. J. Gastroenterol. Hepatol, 2011, 26(2), 371–377. [DOI] [PubMed] [Google Scholar]
- [233].Kitagawa M; Aonuma M; Lee SH; Fukutake S; McCormick F E2F-1 transcriptional activity is a critical determinant of Mdm2 antagonist-induced apoptosis in human tumor cell lines. Oncogene, 2008, 27(40), 5303–5314. [DOI] [PubMed] [Google Scholar]
- [234].Ambrosini G; Sambol EB; Carvajal D; Vassilev LT; Singer S; Schwartz GK Mouse double minute antagonist Nutlin-3a enhances chemotherapy-induced apoptosis in cancer cells with mutant p53 by activating E2F1. Oncogene, 2007, 26(24), 3473–3481. [DOI] [PubMed] [Google Scholar]
- [235].LaRusch GA; Jackson MW; Dunbar JD; Warren RS; Donner DB; Mayo LD Nutlin3 blocks vascular endothelial growth factor induction by preventing the interaction between hypoxia inducible factor 1alpha and Hdm2. Cancer Res, 2007, 67(2), 450–454. [DOI] [PubMed] [Google Scholar]
- [236].Dey A; Wong ET; Bist P; Tergaonkar V; Lane DP Nutlin-3 inhibits the NFkappaB pathway in a p53-dependent manner: implications in lung cancer therapy. Cell Cycle, 2007, 6(17), 2178–2185. [DOI] [PubMed] [Google Scholar]
- [237].Impicciatore G; Sancilio S; Miscia S; Di Pietro R Nutlins and ionizing radiation in cancer therapy. Curr. Pharm. Des, 2010, 16(12), 1427–1442. [DOI] [PubMed] [Google Scholar]
- [238].Jones RJ; Bjorklund CC; Baladandayuthapani V; Kuhn DJ; Orlowski RZ Drug resistance to inhibitors of the human double minute-2 E3 ligase is mediated by point mutations of p53, but can be overcome with the p53 targeting agent RITA. Mol. Cancer Ther, 2012, 11(10), 2243–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Wang H; Ma X; Ren S; Buolamwini JK; Yan C A small-molecule inhibitor of MDMX activates p53 and induces apoptosis. Mol. Cancer Ther, 2011, 10(1), 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Michaelis M; Rothweiler F; Klassert D; von Deimling A; Weber K; Fehse B; Kammerer B; Doerr HW; Cinatl J Jr. Reversal of P-glycoprotein-mediated multidrug resistance by the murine double minute 2 antagonist nutlin-3. Cancer Res, 2009, 69(2), 416–421. [DOI] [PubMed] [Google Scholar]
- [241].Yu S; Qin D; Shangary S; Chen J; Wang G; Ding K; McEachern D; Qiu S; Nikolovska-Coleska Z; Miller R; Kang S; Yang D; Wang S Potent and orally active small-molecule inhibitors of the MDM2-p53 interaction. J. Med. Chem, 2009, 52(24), 7970–7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Popowicz GM; Czarna A; Wolf S; Wang K; Wang W; Domling A; Holak TA Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery. Cell Cycle, 2010, 9(6), 1104–1111. [DOI] [PubMed] [Google Scholar]
- [243].Sosin AM; Burger AM; Siddiqi A; Abrams J; Mohammad RM; Al-Katib AM HDM2 antagonist MI-219 (spiro-oxindole), but not Nutlin-3 (cis-imidazoline), regulates p53 through enhanced HDM2 autoubiquitination and degradation in human malignant B-cell lymphomas. J. Hematol. Oncol, 2012, 5, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Shangary S; Qin D; McEachern D; Liu M; Miller RS; Qiu S; Nikolovska-Coleska Z; Ding K; Wang G; Chen J; Bernard D; Zhang J; Lu Y; Gu Q; Shah RB; Pienta KJ; Ling X; Kang S; Guo M; Sun Y; Yang D; Wang S Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc. Natl. Acad. Sci. U S A, 2008, 105(10), 3933–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Zhao Y; Liu L; Sun W; Lu J; McEachern D; Li X; Yu S; Bernard D; Ochsenbein P; Ferey V; Carry JC; Deschamps JR; Sun D; Wang S Diastereomeric Spirooxindoles as Highly Potent and Efficacious MDM2 Inhibitors. J. Am. Chem. Soc, 2013, 135(19), 7223–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Zhao Y; Yu S; Sun W; Liu L; Lu J; McEachern D; Shar-gary S; Bernard D; Li X; Zhao T; Zou P; Sun D; Wang S A Potent Small-Molecule Inhibitor of the MDM2-p53 Interaction (MI-888) Achieved Complete and Durable Tumor Regression in Mice. J. Med. Chem, 2013, June 20 {Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Hayashi R; Wang D; Hara T; Iera JA; Durell SR; Appella DH N-acylpolyamine inhibitors of HDM2 and HDMX binding to p53. Bioorg. Med. Chem, 2009, 17(23), 7884–7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Stoll R; Renner C; Hansen S; Palme S; Klein C; Belling A; Zeslawski W; Kamionka M; Rehm T; Muhlhahn P; Schumacher R; Hesse F; Kaluza B; Voelter W; Engh RA; Holak TA Chalcone derivatives antagonize interactions between the human oncoprotein MDM2 and p53. Biochemistry, 2001, 40(2), 336–344. [DOI] [PubMed] [Google Scholar]
- [249].Kumar SK; Hager E; Pettit C; Gurulingappa H; Davidson NE; Khan SR Design, synthesis, and evaluation of novel boronic-chalcone derivatives as antitumor agents. J. Med. Chem, 2003, 46(14), 2813–2815. [DOI] [PubMed] [Google Scholar]
- [250].Achanta G; Modzelewska A; Feng L; Khan SR; Huang P A boronic-chalcone derivative exhibits potent anticancer activity through inhibition of the proteasome. Mol. Pharmacol, 2006, 70(1), 426–433. [DOI] [PubMed] [Google Scholar]
- [251].Sun XX; Dai MS; Lu H 5-fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. J. Biol. Chem, 2007, 282(11), 8052–8059. [DOI] [PubMed] [Google Scholar]
- [252].Rew Y; Sun D; Gonzalez-Lopez De Turiso F; Bartberger MD; Beck HP; Canon J; Chen A; Chow D; Deignan J; Fox BM; Gustin D; Huang X; Jiang M; Jiao X; Jin L; Kayser F; Kopecky DJ; Li Y; Lo MC; Long AM; Michelsen K; Oliner JD; Osgood T; Ragains M; Saiki AY; Schneider S; Toteva M; Yakowec P; Yan X; Ye Q; Yu D; Zhao X; Zhou J; Medina JC; Olson SH Structure-based design of novel inhibitors of the MDM2-p53 interaction. J. Med. Chem, 2012, 55(11), 4936–4954. [DOI] [PubMed] [Google Scholar]
- [253].Issaeva N; Bozko P; Enge M; Protopopova M; Verhoef LG; Masucci M; Pramanik A; Selivanova G Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat. Med, 2004, 10(12), 1321–1328. [DOI] [PubMed] [Google Scholar]
- [254].Krajewski M; Ozdowy P; D’Silva L; Rothweiler U; Holak TA NMR indicates that the small molecule RITA does not block p53-MDM2 binding in vitro. Nat. Med, 2005, 11(11), 1135–1136; author reply 1136–1137. [DOI] [PubMed] [Google Scholar]
- [255].Roxburgh P; Hock AK; Dickens MP; Mezna M; Fischer PM; Vousden KH Small molecules that bind the Mdm2 RING stabilize and activate p53. Carcinogenesis, 2012, 33(4), 791–798. [DOI] [PubMed] [Google Scholar]
- [256].Lane DP; Cheok CF; Lain S p53-based cancer therapy. Cold Spring Harb. Perspect. Biol, 2010, 2(9), a001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Sasiela CA; Stewart DH; Kitagaki J; Safiran YJ; Yang Y; Weissman AM; Oberoi P; Davydov IV; Goncharova E; Beutler JA; McMahon JB; O’Keefe BR Identification of inhibitors for MDM2 ubiquitin ligase activity from natural product extracts by a novel high-throughput electrochemiluminescent screen. J. Biomol. Screen, 2008, 13(3), 229–237. [DOI] [PubMed] [Google Scholar]
- [258].Fang J; Xia C; Cao Z; Zheng JZ; Reed E; Jiang BH Apigenin inhibits VEGF and HIF-1 expression via PI3K/AKT/p70S6K1 and HDM2/p53 pathways. FASEB J, 2005, 19(3), 342–353. [DOI] [PubMed] [Google Scholar]
- [259].Gopal YN; Chanchorn E; Van Dyke MW Parthenolide promotes the ubiquitination of MDM2 and activates p53 cellular functions. Mol. Cancer Ther, 2009, 8(3), 552–562. [DOI] [PubMed] [Google Scholar]
- [260].Nasim S; Crooks PA Antileukemic activity of aminoparthenolide analogs. Bioorg. Med. Chem. Lett, 2008, 18(14), 3870–3873. [DOI] [PubMed] [Google Scholar]
- [261].Zhang X; Gu L; Li J; Shah N; He J; Yang L; Hu Q; Zhou M Degradation of MDM2 by the interaction between berberine and DAXX leads to potent apoptosis in MDM2-overexpressing cancer cells. Cancer Res, 2010, 70(23), 9895–9904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Nag SA; Qin JJ; Wang W; Wang MH; Wang H; Zhang R Ginsenosides as Anticancer Agents: In vitro and in vivo Activities, Structure-Activity Relationships, and Molecular Mechanisms of Action. Front. Pharmacol, 2012, 3, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Zhao Y; Wang W; Han L; Rayburn ER; Hill DL; Wang H; Zhang R Isolation, structural determination, and evaluation of the biological activity of 20(S)-25-methoxyl-dammarane-3beta, 12beta, 20-triol [20(S)-25-OCH3-PPD], a novel natural product from Panax notoginseng. Med. Chem, 2007, 3(1), 51–60. [DOI] [PubMed] [Google Scholar]
- [264].Wang W; Wang H; Rayburn ER; Zhao Y; Hill DL; Zhang R 20(S)-25-methoxyl-dammarane-3beta, 12beta, 20-triol, a novel natural product for prostate cancer therapy: activity in vitro and in vivo and mechanisms of action. Brit. J. Cancer, 2008, 98(4), 792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Wang W; Rayburn ER; Zhao Y; Wang H; Zhang R Novel ginsenosides 25-OH-PPD and 25-OCH3-PPD as experimental therapy for pancreatic cancer: anticancer activity and mechanisms of action. Cancer Lett, 2009, 278(2), 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Wang W; Rayburn ER; Hang J; Zhao Y; Wang H; Zhang R Anti-lung cancer effects of novel ginsenoside 25-OCH(3)-PPD. Lung Cancer, 2009, 65(3), 306–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Wang W; Rayburn ER; Hao M; Zhao Y; Hill DL; Zhang R; Wang H Experimental therapy of prostate cancer with novel natural product anti-cancer ginsenosides. Prostate, 2008, 68(8), 809–819. [DOI] [PubMed] [Google Scholar]
- [268].Shinkre BA; Raisch KP; Fan L; Velu SE Analogs of the marine alkaloid makaluvamines: synthesis, topoisomerase II inhibition, and anticancer activity. Bioorg. Med. Chem. Lett, 2007, 17(10), 2890–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [269].Shinkre BA; Raisch KP; Fan L; Velu SE Synthesis and antiproliferative activity of benzyl and phenethyl analogs of makaluvamines. Bioorg. Med. Chem, 2008, 16(5), 2541–2549. [DOI] [PubMed] [Google Scholar]
- [270].Wang W; Rayburn ER; Velu SE; Nadkarni DH; Murugesan S; Zhang R In vitro and in vivo anticancer activity of novel synthetic makaluvamine analogues. Clin. Cancer Res, 2009, 15(10), 3511–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Chen T; Xu Y; Guo H; Liu Y; Hu P; Yang X; Li X; Ge S; Velu SE; Nadkarni DH; Wang W; Zhang R; Wang H Experimental therapy of ovarian cancer with synthetic makaluvamine analog: in vitro and in vivo anticancer activity and molecular mechanisms of action. Plos One, 2011, 6(6), e20729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [272].Wang W; Rayburn ER; Velu SE; Chen D; Nadkarni DH; Murugesan S; Chen D; Zhang R, A novel synthetic iminoqui-none, BA-TPQ, as an anti-breast cancer agent: in vitro and in vivo activity and mechanisms of action. Breast Cancer Res. Treat, 2010, 123(2), 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Nadkarni DH; Wang F; Wang W; Rayburn ER; Ezell SJ; Murugesan S; Velu SE; Zhang R Synthesis and in vitro anti-lung cancer activity of novel 1, 3, 4, 8-tetrahydropyrrolo [4, 3, 2-de]quinolin-8(1H)-one alkaloid analogs. Med. Chem, 2009, 5(3), 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Zhang X; Xu H; Zhang X; Voruganti S; Murugesan S; Nadkarni DH; Velu SE; Wang MH; Wang W; Zhang R Pre-clinical evaluation of anticancer efficacy and pharmacological properties of FBA-TPQ, a novel synthetic makaluvamine analog. Marine Drug, 2012, 10(5), 1138–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Secchiero P; Bosco R; Celeghini C; Zauli G Recent advances in the therapeutic perspectives of Nutlin-3. Curr. Pharm. Des, 2011, 17(6), 569–577. [DOI] [PubMed] [Google Scholar]
- [276].Yuan Y; Liao YM; Hsueh CT; Mirshahidi HR Novel targeted therapeutics: inhibitors of MDM2, ALK and PARP. J. Hematol. Oncol, 2011, 4, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [277].Ray-Coquard I; Blay JY; Italiano A; Le Cesne A; Penel N; Zhi J; Heil F; Rueger R; Graves B; Ding M; Geho D; Middleton SA; Vassilev LT; Nichols GL; Bui BN Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. Lancet Oncol, 2012, 13(11), 1133–1140. [DOI] [PubMed] [Google Scholar]
- [278].Carol H; Reynolds CP; Kang MH; Keir ST; Maris JM; Gorlick R; Kolb EA; Billups CA; Geier B; Kurmasheva RT; Houghton PJ; Smith MA; Lock RB Initial testing of the MDM2 inhibitor RG7112 by the Pediatric Preclinical Testing Program. Pediatr. Blood Cancer, 2013, 60(4), 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Chargari C; Leteur C; Angevin E; Bashir T; Schoentjes B; Arts J; Janicot M; Bourhis J; Deutsch E Preclinical assessment of JNJ-26854165 (Serdemetan), a novel tryptamine compound with radiosensitizing activity in vitro and in tumor xenografts. Cancer Lett, 2011, 312(2), 209–218. [DOI] [PubMed] [Google Scholar]
- [280].Smith MA; Gorlick R; Kolb EA; Lock R; Carol H; Maris JM; Keir ST; Morton CL; Reynolds CP; Kang MH; Arts J; Bashir T; Janicot M; Kurmasheva RT; Houghton PJ Initial testing of JNJ-26854165 (Serdemetan) by the pediatric preclinical testing program. Pediatr. Blood Cancer, 2012, 59(2), 329–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [281].Koblish HK; Zhao S; Franks CF; Donatelli RR; Tominovich RM; LaFrance LV; Leonard KA; Gushue JM; Parks DJ; Calvo RR; Milkiewicz KL; Marugan JJ; Raboisson P; Cummings MD; Grasberger BL; Johnson DL; Lu T; Molloy CJ; Maroney AC Benzodiazepinedione inhibitors of the Hdm2:p53 complex suppress human tumor cell proliferation in vitro and sensitize tumors to doxorubicin in vivo. Mol. Cancer Ther, 2006, 5(1), 160–169. [DOI] [PubMed] [Google Scholar]
- [282].Wilson JM; Henderson G; Black F; Sutherland A; Ludwig RL; Vousden KH; Robins DJ Synthesis of 5-deazaflavin derivatives and their activation of p53 in cells. Bioorg. Med. Chem, 2007, 15(1), 77–86. [DOI] [PubMed] [Google Scholar]
- [283].Hardcastle IR; Liu J; Valeur E; Watson A; Ahmed SU; Blackburn TJ; Bennaceur K; Clegg W; Drummond C; Endicott JA; Golding BT; Griffin RJ; Gruber J; Haggerty K; Harrington RW; Hutton C; Kemp S; Lu X; McDonnell JM; Newell DR; Noble ME; Payne SL; Revill CH; Riedinger C; Xu Q; Lunec J Isoindolinone inhibitors of the murine double minute 2 (MDM2)-p53 protein-protein interaction: structure-activity studies leading to improved potency. J. Med. Chem, 2011, 54(5), 1233–1243. [DOI] [PubMed] [Google Scholar]
- [284].Rothweiler U; Czarna A; Krajewski M; Ciombor J; Kalinski C; Khazak V; Ross G; Skobeleva N; Weber L; Holak TA Isoquinolin-1-one inhibitors of the MDM2-p53 interaction. ChemMedChem, 2008, 3(7), 1118–1128. [DOI] [PubMed] [Google Scholar]
- [285].Zhuang C; Miao Z; Zhu L; Dong G; Guo Z; Wang S; Zhang Y; Wu Y; Yao J; Sheng C; Zhang W Discovery, synthesis, and biological evaluation of orally active pyrrolidone derivatives as novel inhibitors of p53-MDM2 protein-protein interaction. J. Med. Chem, 2012, 55(22), 9630–9642. [DOI] [PubMed] [Google Scholar]
- [286].Leao M; Pereira C; Bisio A; Ciribilli Y; Paiva AM; Machado N; Palmeira A; Fernandes MX; Sousa E; Pinto M; Inga A; Saraiva L Discovery of a new small-molecule inhibitor of p53-MDM2 interaction using a yeast-based approach. Biochem. Pharmacol, 2013, 85(9), 1234–1245. [DOI] [PubMed] [Google Scholar]
- [287].Lucas BS; Fisher B; McGee LR; Olson SH; Medina JC; Cheung E An expeditious synthesis of the MDM2-p53 inhibitor AM-8553. J. Am. Chem. Soc, 2012, 134(30), 12855–12860. [DOI] [PubMed] [Google Scholar]
- [288].Zhang R; Mayhood T; Lipari P; Wang Y; Durkin J; Syto R; Gesell J; McNemar C; Windsor W Fluorescence polarization assay and inhibitor design for MDM2/p53 interaction. Analytical Biochem, 2004, 331(1), 138–146. [DOI] [PubMed] [Google Scholar]
- [289].Miyazaki M; Naito H; Sugimoto Y; Yoshida K; Kawato H; Okayama T; Shimizu H; Miyazaki M; Kitagawa M; Seki T; Fukutake S; Shiose Y; Aonuma M; Soga T Synthesis and evaluation of novel orally active p53-MDM2 interaction inhibitors. Bioorg. Med. Chem, 2013, 21(14), 4319–4331. [DOI] [PubMed] [Google Scholar]
- [290].Wang W; Lv D; Qiu N; Zhang L; Hu C; Hu Y Design, synthesis and biological evaluation of novel 3,4,5-trisubstituted aminothiophenes as inhibitors of p53-MDM2 interaction. Part 2. Bioorg. Med. Chem, 2013, 21(11), 2886–2894. [DOI] [PubMed] [Google Scholar]
- [291].Wang W; Shangguan S; Qiu N; Hu C; Zhang L; Hu Y Design, synthesis and biological evaluation of novel 3,4,5-trisubstituted aminothiophenes as inhibitors of p53-MDM2 interaction. Part 1. Bioorg. Med. Chem, 2013, 21(11), 2879–2885. [DOI] [PubMed] [Google Scholar]
- [292].Gonzalez-Lopez de Turiso F; Sun D; Rew Y; Bartberger MD; Beck HP; Canon J; Chen A; Chow D; Correll TL; Huang X; Julian LD; Kayser F; Lo MC; Long AM; McMinn D; Oliner JD; Osgood T; Powers JP; Saiki AY; Schneider S; Shaffer P; Xiao SH; Yakowec P; Yan X; Ye Q; Yu D; Zhao X; Zhou J; Medina JC; Olson SH Rational design and binding mode duality of MDM2-p53 inhibitors. J. Med. Chem, 2013, 56(10), 4053–4070. [DOI] [PubMed] [Google Scholar]
- [293].Hu C; Li X; Wang W; Zhang L; Tao L; Dong X; Sheng R; Yang B; Hu Y Design, synthesis, and biological evaluation of imidazoline derivatives as p53-MDM2 binding inhibitors. Bioorg. Med. Chem, 2011, 19(18), 5454–5461. [DOI] [PubMed] [Google Scholar]
- [294].Lindstrom MS; Jin A; Deisenroth C; White Wolf G; Zhang Y Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and attenuate MDM2-induced p53 degradation. Mol. Cell. Biol, 2007, 27(3), 1056–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [295].Sczaniecka M; Gladstone K; Pettersson S; McLaren L; Huart AS; Wallace M MDM2 protein-mediated ubiquitination of numb protein: identification of a second physiological substrate of MDM2 that employs a dual-site docking mechanism. J. Biol. Chem, 2012, 287(17), 14052–14068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [296].Wang S; Zhao Y; Bernard D; Aguilar A; Kumar S Targeting the MDM2-p53 Protein-Protein Interaction for New Cancer Therapeutics. In Topics in Medicinal Chemistry ed; Springer Berlin Heidelberg: 2012; Vol. 8. [Google Scholar]
- [297].Zhao Y; Bernard D; Wang S Small molecule inhibitors of MDM2-p53 and MDMX-p53 interactions as new cancer therapeutics. BioDiscovery, 2013, 8, 4. [Google Scholar]
- [298].Azmi AS; Philip PA; Beck FW; Wang Z; Banerjee S; Wang S; Yang D; Sarkar FH; Mohammad RM MI-219-zinc combination: a new paradigm in MDM2 inhibitor-based therapy. Oncogene, 2011, 30(1), 117–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [299].Xiong J; Yang Q; Li J; Zhou S Effects of MDM2 inhibitors on vascular endothelial growth factor-mediated tumor angiogenesis in human breast cancer. Angiogenesis, 2013, August 2 [Epub ahead of Print]. [DOI] [PubMed] [Google Scholar]
- [300].Busuttil V; Droin N; McCormick L; Bernassola F; Candi E; Melino G; Green DR NF-kappaB inhibits T-cell activation-induced, p73-dependent cell death by induction of MDM2. Proc. Natl. Acad. Sci. U S A, 2010, 107(42), 18061–18066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [301].Truong AH; Cervi D; Lee J; Ben-David Y Direct transcriptional regulation of MDM2 by Fli-1. Oncogene, 2005, 24(6), 962–969. [DOI] [PubMed] [Google Scholar]
- [302].Ries S; Biederer C; Woods D; Shifman O; Shirasawa S; Sasazuki T; McMahon M; Oren M; McCormick F Opposing effects of Ras on p53: transcriptional activation of mdm2 and induction of p19ARF. Cell, 2000, 103(2), 321–330. [DOI] [PubMed] [Google Scholar]
- [303].Zhang X; Zhang Z; Cheng J; Li M; Wang W; Xu W; Wang H; Zhang R Transcription factor NFAT1 activates the mdm2 oncogene independent of p53. J. Biol. Chem, 2012, 287(36), 30468–30476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [304].Saha MN; Jiang H; Jayakar J; Reece D; Branch DR; Chang H MDM2 antagonist nutlin plus proteasome inhibitor velcade combination displays a synergistic anti-myeloma activity. Cancer Biol. Ther, 2010, 9(11), 936–944. [DOI] [PubMed] [Google Scholar]
- [305].Jin L; Tabe Y; Kojima K; Zhou Y; Pittaluga S; Konopleva M; Miida T; Raffeld M MDM2 antagonist Nutlin-3 enhances bortezomib-mediated mitochondrial apoptosis in TP53-mutated mantle cell lymphoma. Cancer Lett, 2010, 299(2), 161–170. [DOI] [PubMed] [Google Scholar]
- [306].Graves B; Thompson T; Xia M; Janson C; Lukacs C; Deo D; Di Lello P; Fry D; Garvie C; Huang KS; Gao L; Tovar C; Lovey A; Wanner J; Vassilev LT Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization. Proc. Natl. Acad. Sci. U S A, 2012, 109(29), 11788–11793. [DOI] [PMC free article] [PubMed] [Google Scholar]