Abstract
The mdm2 oncogene has recently been suggested to be a valuable target for cancer therapy and prevention. Overexpression of mdm2 is often seen in various human cancers and correlates with high-grade, late-stage, and more treatment-resistant tumors. The MDM2-p53 auto-regulatory loop has been extensively investigated and is an attractive cancer target, which indeed has been the main focus of anti-MDM2 drug discovery. Much effort has been expended in the development of small molecule MDM2 antagonists targeting the MDM2-p53 interaction, and a few of these have advanced into clinical trials. However, MDM2 exerts its oncogenic activity through both p53-dependent and -independent mechanisms. Recently, there is an increasing interest in identifying natural MDM2 inhibitors; some of them have been shown to decrease MDM2 expression and activity in vitro and in vivo. These identified natural MDM2 inhibitors include a plethora of diverse chemical frameworks, ranging from flavonoids, steroids, and sesquiterpenes to alkaloids. In addition to a brief review of synthetic MDM2 inhibitors, this review focuses on natural product MDM2 inhibitors, summarizing their biological activities in vitro and in vivo and the underlying molecular mechanisms of action, targeting MDM2 itself, regulators of MDM2, and/or the MDM2-p53 interaction. These MDM2 inhibitors can be used alone or in combination with conventional treatments, improving the prospects for cancer therapy and prevention. Their complex and unique molecular architectures may provide a stimulus for developing synthetic analogs in the future.
Keywords: MDM2, MDM2 inhibitors, MDM2-p53 interaction, mechanisms of action, natural product anticancer agents, p53-dependent, p53-independent, p53 wild-type, p53 mutant, small molecule inhibitors of MDM2
1. INTRODUCTION
Carcinogenesis and tumor progression are predominantly caused by alterations in oncogenes, tumor suppressor genes, and other genes regulating cell growth, cell cycle progression, and cell death. Although the loss of a tumor suppressor may or may not lead to oncogenesis, many human tumors possess at least one activated oncogene. The activation of one or more oncogenes, along with exposure to environmental stress or another cellular “hit”, is sufficient to initiate carcinogenesis. The products of oncogenes (i.e., oncoproteins) may be classified into six broad groups: transcription factors (e.g., c-myc), chromatin remodelers (e.g., MLL fusion proteins), growth factors (e.g., EGF), growth factor receptors (e.g., EGFR), signal transducers (e.g., Her2/neu), and apoptosis regulators (e.g., Bcl-2) [1]. Continuous expression of at least one oncogene is often necessary and sufficient for cells to attain unchecked growth and proliferation under conditions normally incompatible with survival. Inhibiting the expression or activity of a dominant oncogene leads to inhibition of tumor growth, demonstrating that overcoming the “oncogene addiction” of the affected cell(s) can stop cancer development and progression [2, 3]. Therefore, targeting oncogene has been suggested as a valid strategy for anticancer drug discovery and development. This review focuses on natural product MDM2 inhibitors, summarizing their biological activities in vitro and in vivo and the underlying molecular mechanisms of action, along with a discussion on MDM2 biology and development of MDM2 as a viable target for cancer therapy and prevention.
1.1. A Brief Historic Perspective of mdm2 Oncogene
The murine double minute 2 gene (mdm2) was originally identified along with two other genes (mdm1 and mdm3) which were overexpressed by more than 50-fold through amplification in a spontaneously transformed mouse BALB/c cell line (3T3-DM) [4, 5], These genes are located on small, acentromeric extrachromosomal nuclear bodies, called double minutes, and are retained in cells only if they provide a growth advantage. The gene product of mdm2 was later shown to be responsible for cell transformation when it was overexpressed [4, 5]. In 1992, Oliner et al. cloned the human mdm2 gene and mapped it to chromosome 12ql3–14 [6]. Both the mdm2 gene and its human counterpart, hdm2, consist of 12 exons that can generate many different MDM2 or HDM2 proteins (hereafter referred to as MDM2 for all species) [7]. Alternative splicing of the primary mRNA transcript can result in the generation of different MDM2 isoforms, some of which inhibit the activity of the wild-type protein in a dominant-negative fashion [8, 9], MDM2 possesses a nuclear localization signal (NLS), a nuclear export signal (NES), a central acidic domain, a C-terminal zinc-finger domain, and a RING finger domain possessing E3 ligase activity [10]. Under normal conditions, MDM2 is expressed in the nucleus, but it translocates to the cytoplasm to mediate the degradation of some of its targets by the proteasome [11–13].
Soon after the identification of the mdm2 gene, the reason for its transformation potential was discovered. MDM2 was shown to bind the tumor suppressor p53, and inhibit p53-mediated gene transactivation [11]. At the same time, MDM2 gene amplification was observed in over one-third of human sarcomas that retained wild-type p53 [6]. These exciting studies led investigators to hypothesize that the overexpression of MDM2 was another mechanism by which cells could inactivate p53 during the process of transformation. MDM2 affects the p53-mediated transcription of various genes by inhibiting p53-mediated transactivation [8, 11]. MDM2 uses multiple mechanisms to inactivate p53 and inhibit its transcriptional activity [14–16]; it targets p53 for ubiquitination and degradation by the proteasome [15], shuttles p53 out of the nucleus, prevents p53 from interacting with transcriptional coactivators, and recruits known transcriptional corepressors, such as hCtBP2, to p53 [15, 16].
MDM2 has an important role in modulation of several cell cycle proteins. The overexpression of MDM2 has been shown to negatively correlate with the expression of the cyclin-dependent kinase inhibitor, p21 [17, 18]. In breast cancer cells, MDM2 overexpression correlates with a lack of p21 expression [17]. MDM2 can also directly interact with p21 in a p53-independent manner and enhance its degradation [19], MDM2 also mediates the ubiquitin-independent proteasomal degradation of p21 [20]. In a recent study, Xu et al. proved that MDM2-p21 interaction resulted in a conformational change in p21 which depended on the central domain of MDM2; with the nuclear localization of both proteins being essential forp21 degradation [21]. MDM2 reverses the growth inhibition at G1 phase imposed by p53 and the retinoblastoma protein (pRb) [22, 23]. Overexpression of MDM2 can overcome the TGF-β-related growth inhibition via the pRb-E2F pathway [24]. Transgenic mouse experiments have shown that the expression of a BLG (β-lactoglobulin)/mdm2 transgene (BLG/mdm2) in the epithelial cells of the mouse mammary gland causes the mammary epithelial cells to undergo multiple rounds of DNA synthesis without cell division, resulting in polyploidy and tumor formation [25]. This effect of MDM2 on the S phase is independent of the pS3 status of the cells [25].
MDM2 also plays an important role in epidermal differentiation [26]. MDM2 overexpression in the granular layer disturbs the differentiation program [27]. In rhabdomyosarcoma, forced expression of MDM2 inhibits MyoD function and consequently inhibits muscle differentiation [28], MDM2 also ubiquitinates NUMB, a cell fate determinant protein, and influences cell differentiation and survival by inducing the translocation of NUMB to the nucleus and promoting its degradation [29], The loss of NUMB expression in breast cancer leads to decreased p53 expression and correlates with a poorer prognosis [29].
Additionally, MDM2 is involved in the ribosome biogenesis occurring in both the cell cytoplasm and in the nucleolus of eukaryotic cells. Several proteins contained in the large subunit of the ribosome such as L5 [30], L11 [31], and L23 [32], form a complex with MDM2 in response to ribosomal stress, such as exposure to actinomycin D. These proteins subsequently activate p53 by inhibiting its MDM2-mediated suppression, thereby causing an increase in G1 arrest [30–34]. Our group has discovered another novel MDM2-interacting ribosomal protein, S7, which is present in the small subunit of the ribosome [35]. This protein binds MDM2 in vitro and in vivo, forming a ternary complex of MDM2-p53-S7, which prevents p53 ubiquitination. The overexpression of S7 inhibits cell proliferation, induces apoptosis, and increases p53 transactivational activity [35]. A later study bv Zhu et al. demonstrated S7 itself as a substrate for MDM2 E3 ligase in addition to it being a regulator of MDM2 mediated p53 degradation [36].
There are an increasing number of molecules reported to have an interaction with MDM2, which play a role in regulation of MDM2 expression and activity and, in turn, modulate multiple molecular pathways important to the control of cell growth, proliferation and death. We discuss these interactions in the subsequent sections.
1.2. MDM2: A Ubiquitin Ligase
E3 ubiquitin ligases are a large family of proteins engaged in regulating the turnover and activity of many proteins [37]. The ubiquitination of proteins occurs through a complex series of steps that involve E1, E2, and E3 proteins. Together with the ubiquitin-activating enzyme, E1, and the ubiquitin-conjugating enzyme, E2, E3 ubiquitin ligases catalyze the ubiquitination of a variety of biologically significant protein substrates, leading to their degradation through the 26S proteasome [37]. MDM2 functions as an E3 ligase that ubiquitinates p53 at several lysine residues [38, 39]. It also has the ability to ubiquitinate itself [40] and various other substrates, such as NUMB [41], pRb [42], and MDMX [43], The RING motif is common in E3 ligases, and the C-terminal RING finger domain is responsible for the E3 ligase activity of MDM2 [40, 44]. E3 ligases are substrate-specific, and hence, play an important role in the ubiquitin-mediated proteolytic cascade [37]. Low levels of MDM2 activity induce the monoubiquitination and nuclear export of p53, whereas high levels promote the polyubiquitination and nuclear degradation of p53 [45]. MDM2 mediates monomeric p53 ubiquitination on multiple lysine residues [45]. Other proteins that co-operate with MDM2 in the polyubiquitination and degradation of p53 include p300/CBP [46]. Nevertheless, targeting MDM2 E3 ligase activity is currently one of the major strategies of developing MDM2 inhibitors for cancer therapy.
1.3. MDM2-p53 Interaction
The MDM2-p53 interaction has been the hottest focal point in research and discovery of MDM2 inhibitors for cancer therapy. The tumor suppressor p53 is a potent anti-proliferative and pro-apoptotic protein that can be detrimental to normal cells [47]. The cellular level of p53 must be tightly controlled in unstressed cells. It has been well established that MDM2 has a major role in this control. MDM2 and p53 form a sophisticated autoregulatory feedback loop in which the two proteins mutually control each other’s cellular level [47]. p53 binds to a promoter of the mdm2 gene (P1 promoter) and regulates its expression [8, 10]. As the level of MDM2 increases, it binds to and inactivates p53 by directly blocking the p53 transactivational domain and by targeting the p53 protein for ubiquitin-dependent degradation by the proteasome [12, 15]. It was initially reported that MDM2 and p53 bind to each other via their N-terminal domains [48]. Recently, Poyurovsky et al report that MDM2-p53 interaction is decreased upon deletion, mutation or acetylation of the p53 C terminus [49]. Using multiple approaches, they proved that a peptide fragment from the p53 C terminus directly binds the MDM2 N terminus in vitro; thus introducing a novel interface for MDM2-p53 interaction [49]. The MDM2 binding site on p53 partially overlaps with its transactivational domain, and thus, MDM2 effectively inhibits p53 transcriptional activity [12, 48]. In addition, as mentioned above, MDM2 serves as an E3 ubiquitin ligase for p53 proteolysis [15]. As a result, both p53 and MDM2 are kept at very low levels in unstressed cells. The crucial role of MDM2 in p53 regulation is strongly supported by the fact that targeted deletion of the mdm2 gene in mice is embryonic lethal, but mdm2−/− mice can be successfully rescued by a simultaneous deletion of the TP53 gene, which encodes p53 [50].
Although it has been demonstrated that MDM2 is the primary regulator of p53 stability and activity, accumulating evidence suggests that MDM2 is not the only protein involved in regulating p53. Recently, three additional proteins, PIRH2 (p53-induced protein with RING-H2 domain that can promote p53 ubiquitination and degradation independent of MDM2) [51], COP1 (constitutively photomorphogenic 1) [52], and ARF-BP1 (ARF-binding protein 1) [53], were discovered to bind p53 and act as p53 ubiquitin ligases. Currently, there is no evidence that any of these p53 ubiquitin ligases can be substitutes for MDM2 in the regulation of p53 stability, which is in agreement with the embryonic lethality of mdm2 knockout mice.
1.4. Interactions with Other Molecules
1.4.1. Affectors of MDM2
Many investigations have identified numerous MDM2 interacting molecules that regulate the expression of several key factors involved in cell proliferation, apoptosis, and tumor invasion and metastasis, independent of MDM2’s interaction with p53. However, these interactions may have an indirect effect on the degradation of p53. One of the first exemplary proteins discovered to interact with MDM2 was pl4ARF, an alternate reading frame protein expressed from the INK4a locus [54]. The ARF-MDM2 interaction blocks MDM2 shuttling between the nucleus and cytoplasm by sequestering MDM2 in the nucleolus [55]. The transport of p53 from the nucleus by MDM2 is required for the efficient degradation of p53. The sequestration of MDM2 in the nucleolus thus results in the indirect activation of p53 [55]. ARF deregulation results in increased nuclear MDM2 levels, thus decreasing p53, and leading to malignant transformation [56, 57]. Another example of the M DM2-interactive proteins, nucleophosmin (NPM, also known as B23, N038, or numatrin), stabilizes ARF, increasing its concentration in the nucleolus [58]. NPM also competes for the binding of MDM2 with p53, resulting in decreased p53 degradation.
In addition to p53, other proteins may affect MDM2 transcription and/or function. For example, PTEN decreases both MDM2 stability and transcription [59]. The mdm2 gene has two promoters, P1 and P2 [60]. PTEN disrupts the activation of the P1 promoter of mdm2, thereby decreasing MDM2 expression, independent of p53 [59]. On the other hand, the p72 RNA helicase can co-operate with both P/CAF and p300/CBP to enhance the transcription of MDM2 in both p53-dependent and -independent manners [61]. The Fli-1 Ets transcription factor increases MDM2 transcription; thus decreasing p53 stability and activity [62]. The Ras-Raf-MEK-MAPK pathway also increases MDM2 transcription, and MEK-mediated phosphorylation of MDM2 enhances its export from the nucleus, modulating its function [63].
The phosphorylation of MDM2 by kinases such as ATM kinase impairs the degradation and nuclear export of p53 by MDM2 [64–66]. The phosphorylation of MDM2 by ATM may inhibit HAUSP-mediated MDM2 de-ubiquitination, resulting in a decrease in MDM2 stability and a subsequent increase in p53 stability [67]. Phosphatidylinositol 3-OH-kinase (PI3-kinase) and its downstream target, Akt/PKB serine-threonine kinase, also appear to bind and phosphorylate MDM2 on serines 166 and 186 following mitogen-induced activation, helping to increase the trans location of MDM2 from the cytoplasm into the nucleus, decreasing the transcriptional activity of p53 [68–71]. Other enzymes, such as CK2, PI3K/Akt, and DNA-PK, as well as members of the Ras-Raf-MEK-MAPK pathway, also regulate MDM2 phosphorylation [72]. The phosphatase formed by cyclin G, a regulatory component of the active PP2A holoenzyme, is known to bind and activate MDM2 through dephosphorylation [73]. In fact, cells null for cyclin G expression show increased phosphorylation of MDM2.
Other mechanisms for posttranslational modifications of MDM2 include SUMOylation [74, 75] and acétylation [76]. MDM2 can undergo SUMOylation within its RING finger domain, leading to a decrease in MDM2 auto-ubiquitination, but to an increase in the ubiquitination of p53 [74]. In comparison, the ARF (p 14/p 19) protein upregulates MDM2 SUMOylation in a p53-independent manner [75]. While the SUMOylation of MDM2 by ARF does not appear to affect the MDM2-p53 loop, it may affect the p53-independent activities of MDM2 [75]. p300/CBP lead to acétylation of MDM2’s RING finger domain, decreasing its activity against both p53 and itself [76].
1.4.2. Targets Regulated by MDM2
MDM2 overexpression has been shown to increase the hypoxia inducible factor (HIF)-l expression levels in colon carcinoma cells, which is followed by a subsequent increase in the expression of vascular endothelial growth factor (VEGF) [77]. In addition, decreased p53 expression (which frequently occurs when MDM2 is overexpressed) results in increased HIF-1 and an increase in VEGF transcription [78]. However, studies have suggested that the effects of the HIF-1-MDM2-p53 interaction may be cell type- and condition-specific [77–81]. In addition, a G-protein-coupled receptor-activated regulator has been implicated in the MDM2-p53 axis [82]. The β-arrestins involved in this signaling pathway bind to MDM2, causing the translocation of MDM2 to the cytoplasm, thus downregulating p53 ubiquitination [82, 83]. β-arrestins have also been shown to regulate the ubiauitination of the mitoeenic IGF-1R through MDM2 thus serving as an interface for molecular crosstalk between GPCRs and receptor tyrosine kinases [83].
MDM2 has been shown to target pRB and the transcription factor E2F-1 and its binding partner, DP-1, among others [84]. A ternary complex comprising MDM2, pRb, and p53 inhibits the degradation of p53, and consequently rescues its pro-apoptotic functions [85]. The binding of pRb with MDM2 also regulates the activity of the Sp (specificity protein) transcription factor [86]. It has been reported that the RING domain of MDM2 binds to the androgen receptor (AR) in the nucleus and facilitates its degradation [87]. MDM2 also mediates the NEDDylation of the pro-apoptotic form of p73-TAp73, a process that promotes the translocation of the p73 protein to the cytoplasm and attenuates its p53-transactivational function [88]. The p73 protein, a structural homologue of p53, is also involved in cell cycle regulation and the induction of apoptosis.
Several proteasome-associated proteins other than MDM2 also associate with p53, affecting the MDM2-p53 interaction. For example, gankyrin, a seven-repeat protein commonly overexpressed in early hepatocarcinogenesis and associated with the 19S regulatory complex of the 26S proteasome, facilitates the MDM2-p53 interaction by binding to MDM2 [89], This results in increased ubiquitination and degradation of p53. Even in the absence of p53, however, the protein enhances the auto-ubiquitination of MDM2 [89]. A de-ubiquitinating protein, HAUSP (herpes virus-associated ubiquitin-specific protease, also known as USP7; ubiquitin specific protease 7), was originally thought to just cleave ubiquitin from p53, thus stabilizing it [90]. However, it was later found to bind to MDM2 as well [91]. The binding of HAUSP to either p53 or MDM2 leads to their de-ubiquitination, and its binding to MDM2 leads to p53 destabilization due to increased MDM2 stability [91]. The proteasome activator PA28γ also regulates the MDM2-p53 interaction (independent of its proteasome-activator function) and serves as a necessary co-factor for p53 degradation [92]. Further, PA28γ binds p21 to regulate its degradation in an ubiquitin-independent manner; it also binds pi 4/p 19 ARF and pi 6 (INK4A) [92]. These observations suggest that the MDM2-interactive proteins, such as PA28γ, p21, and pl4ARF, may form a complex to enhance the proteasomal degradation of the various proteins.
Recently, it has been shown that the phosphorylated form of Foxo3A is a target for MDM2-dependent ubiquitination and degradation [93, 94]. Foxo3A is responsible for regulating p27, which leads to MDM2-mediated control of cell cycle progression in response to oncogenic growth factor signaling or Ras activation [93, 94]. MDM2 has also been shown to regulate the expression of the anti-apoptotic protein, XIAP, by a novel mechanism [95]. MDM2 binds to XIAP mRNA and enhances its translation, leading to the increased expression of XIAP. This leads to an MDM2-dependent inactivation of caspase-mediated apoptosis [95].
MDM2 has also recently been shown to target E-cadherin for degradation via the 26S proteasome [96]. E-cadherin has a well-characterized role in the epithelial-to-mesenchymal transition, a critical step for the metastatic progression of solid tumors of epithelial origin. MDM2 may also confer TGF-β resistance (overcoming G1 arrest) by directly interacting with TGF-β1-activated SMA- and MAD3 (Smad3/4) transcription factors, consequently increasing MDM2 protein expression and destabilization of p53 in human cancer cell lines [24]. Additionally, TGF-β1 expression leads to Smad3 activation and MDM2 expression in murine mammary epithelial cells during the epithelial-to-mesenchymal transition (EMT). These facts suggest that MDM2 may play a critical role in cell migration and metastasis [97].
MDMX, another RING finger ubiquitin ligase, possesses a high degree of homology to MDM2, especially in its N-terminal p53 binding domain [98]. However, although MDMX possesses a RING domain, it is unable to ubiquitinate and degrade p53. MDMX possesses a p53 binding domain at its N-terminus and a RING finger domain at its C-terminus through which it heterodimerizes with MDM2 [98, 99]. Because of its sequence similarity with MDM2 and its ability to inhibit p53-induced transcription when overexpressed, MDMX has been hypothesized to act as a negative regulator of p53 through physical binding [100]. Despite its inability to facilitate p53 degradation, MDMX seems to be essential for mouse development, because mdmx−/− mice are embryonic lethal, and similar to themdm2−/− mice, can be rescued by the deletion of TP53 [101]. Similar to MDM2, the overexpression of aberrant forms of MDMX has been observed in human tumors and seems to facilitate tumorigenesis [102]. MDMX also has been shown to promote the proteasomal turnover of p21 in cooperation with MDM2 at the G1 phase [103].
We summarize some of the important MDM2 interactive proteins in (Table 1). An exhaustive analysis of all MDM2 interactions is beyond the scope of this review. However, interested readers are directed to several other reviews that provide an excellent discussion on the signaling pathways and interactions of MDM2 with other intracellular proteins and/or receptors [129–135].
Table 1.
MDM2-interactive Proteins and their Effects
| Protein Names | Effect of Interaction with Protein on MDM2 | Effcct of Interaction with Protein on p53 | Effect of MDM2 on Protein | Ref. |
|---|---|---|---|---|
| 14–3-3-σ | MDM2 stability decreased, translocation to cytoplasm | Increased Stability | No Direct Effect | [104] |
| TGF-β | Not Well Understood | No Significant Effect | Not well understood, MDM2 may mediate TGF-β resistance | [24,97] |
| pl4(ARf) | MDM2 Activity Decreased, MDM2 localized to nucleolus | Increased Stability | MDM2 decreases its transcriptional activity and increases degradation | [55] |
| Nucleophosmin (NPM, B23) | Inhibits Binding of p53 with MDM2 | Increased Stability | Not Well Understood | [58] |
| Gankyrin (PSM10) | E3 ligase activity of MDM2 increased | Enhanced ubiquitination and degradation | Not Well Understood | [89] |
| MTBP | Stability Increased | Ubiquitination and degradation of p53 increased | Not Well Understood | [105] |
| PML | Decreases Ubiquitinating Ability | Protects p53 from Mdm2-mediated inhibition and degradation. | Not Well Understood | [106] |
| Merlin | Induces MDM2 degradation | Not Well Understood | Not Well Understood | [107] |
| PCAF | Inhibits binding of MDM2 with p53; stimulates auto-ubiquitination | Decreased Acetylation | MDM2 increases its proteasomal degradation | [108] |
| Tip60 | Localization to PML bodies | Decreased MDM2-mediated NEDDylation; acetylation | MDM2 increases its proteasomal degradation | [109] |
| JMY | Not Well Understood | Decreased Transcriptional Activity | MDM2 increases proteasomal degradation | [110] |
| TSG-101 | Inhibits Ubiquitination; increased stabilization | Decreased Activity | MDM2 increases its proteolysis | [111] |
| YYI | Promotes the assembly of the MDM2-p53 complex. | Disrupts the interaction between p53 and the coactivator p300, blocks p300-dependent acetylation and stabilization of p53. | Not Well Understood | [112] |
| IGFR | Not Well Understood | Increased Stability | MDM2 increases its proteasomal degradation | [113] |
| Estrogen Receptor α | Increased transcription following estrogen ligation by the ER | Increased stability and activity | Interaction with MDM2 decreases its stability; it also increases its activity when p53 is absent | [114] |
| NFκB | Not Well Understood | Not Well Understood | Pharmacologic inhibition of Mdm2 results in activation-induced apoptosis. MDM2 overexpression leads to p65 subunit induction and chemoresistance | [115,116] |
| PSD-95 | Not Well Understood | Not Well Understood | Promotes ubiquitination and proteasomal degradation | [117] |
| ENIGMA | Inhibits MDM2 self-ubiquitination | Decreased stability; Increases MDM2 ubiquitin ligase activity toward p53 | Not Well Understood | [118] |
| Siva-1 | Not Well Understood | Increases Mdm2-mediated p53 degradation. | Not Well Understood | [119] |
| elf4e | Increases MDM2 expression | Not Well Understood | Not Well Understood | [120] |
| VEGF | Not Well Understood | Not Well Understood | MDM2 Positively regulates VEGF mRNA stabilization | [121] |
| Nucleostomin | Nucleoplasmic mobilization of nucleostemin stabilizes MDM2 | Decreases p53 transcriptional activity | Not Well Understood | [122] |
| Caspase-2 | Cleaves MDM2 at asp 367 leading to loss of C-terminal RING domain | Stability Increased | Not Well Understood | [123] |
| MYCN | MDM2 positively regulated by MYCN | Not Well Understood | Not Well Understood | [124] |
| S14 | Decreased ubiquitination of p53 | Increased stability and activity | Not Well Understood | [125] |
| S100 | Not Well Understood | Disrupt extent of MDM2 mediated p53 ubiquitination | Not Well Understood | [126] |
| HIPK2 | Not Well Understood | HIPK2 and p53 colocalize with PML-3 into the nuclear bodies and cooperate in the activation of p53-dependent transcription and induction of apoptosis | MDM2 causes degradation of HIPK2 | [127] |
| HAUSP/USP7 | MDM2 stability increased due to de-ubiquitination | p53 stability decreased due to increased MDM2-mediated ubiquitination | Not Well Understood | [67] |
| MDMX | Stability Increased | Stability increased; NEDDylation | MDM2 increases its ubiquitination | [98] |
| NUMB | No Known Effect | p53 Activity increased | MDM2 increases its degradation | [29] |
| p2l | Expression Decreased | Decreased Expression of p53 | MDM2 induces conformational change in p53 leading to increased proteasomal degradation | [17,18] |
| p73 | Not Well Understood | Increased stability and transcription; NEDDylation | Interaction with MDM2 decreases its transcriptional activity | [88] |
| PA28γ | Increased Stability | Increased Ubiquitination | Not Well Understood | [92] |
| pRb | Decreased E3 ligase activity toward p53 | Increases p53-mediated apoptosis | MDM2 binding decreases activity | [42] |
| L5 | Decreased Ubiquitination of p53 | Increased stability and activity | Not Well Understood | [30] |
| L11 | Decreased Ubiquitination of p53 | Increased stability and activity | Not Well Understood | [31] |
| L23 | Decreased Ubiquitination of p53 | Increased synthesis and protection form MDM2-mediated ubiquitination | Not Well Understood | [32] |
| S7 | Not Well Understood | Increased stability and activity | Not Well Understood | [36] |
| S25 | Inhibit E3 ligase activity of MDM2 | Increased stability and activity, decrease p53 ubiquitination by MDM2 | Not Well Understood | [128] |
| β-arrestins | Cytoplasmic localization, decreased E3 ligase activity (β-A2) | Increased protein stability and activity (β-arrestin 2) | Not well understood (β-arrestin 2), increased ubiquitination (β-arrestin 1) | [82,83] |
| HIF-1α | Increased activity toward p53 | Decreased Stability | MDM2 may increase its stability | [78] |
| Androgen Receptor | Not Well Understood | Not Well Understood | MDM2 increases its ubiquitination and proteolysis. MDM2 also decreases its transcriptional activity | [87] |
| E2F1 | Not Well Understood | Not Well Understood | p53 represses its activation/function; MDM2 increases its stability and stimulates activity | [42] |
1.5. MDM2 As a Cancer Target
It has become clear that MDM2 is an important orchestrator of various steps in the oncogenic process. The liberation of p53 from MDM2, which stabilizes the tumor suppressor and activates the p53-mediated pathways, leading to cell cycle arrest and apoptosis, has emerged as an important paradigm for cancer therapy and prevention. Different approaches have been exploited to release p53 from the control of MDM2, including methods to inhibit the MDM2-p53 interaction, MDM2 expression, and MDM2 ubiquitin ligase activity. In addition, inhibiting MDM2’s p53-independent activity also becomes one of the major strategies in developing MDM2 inhibitors for cancer therapy and prevention.
1.5.1. Blocking MDM2 Expression (p53-Dependent and -Independent Pathways)
Several studies using antisense oligonucleotides to inhibit MDM2 expression have established the proof-of-concept for this approach in cells and mouse models of human cancer. MDM2 downregulation by antisense oligonucleotides results in p53 stabilization and activation of the p53 pathway in cancer cells in vitro, as well as in tumor xenografts in nude mice [136–139]. Interestingly, not only p53 wild-type cells, but also cells that express mutant p53, respond to MDM2 inhibition. Studies have suggested that the p53-independent stabilization of the cyclin-dependent kinase inhibitor, p21, as a result of MDM2 downregulation might contribute to the antitumor activity of MDM2 antisense oligonucleotides and MDM2 inhibitors in these mutant cell lines [140]. Several naturally derived compounds also have been shown to exert their anticancer activities by inhibiting MDM2 interaction with other molecules, independent of p53, such as berberine (disruption of MDM2-DAXX-HAUSP complex) [141]. A comprehensive review of natural products blocking MDM2 expression in a p53-denpendent or -independent manner is discussed below.
1.5.2. Modulating MDM2’s E3 Ubiquitin Ligase Activity or/and Blocking MDM2-Mediated Ubiquitination of p53
Because the ubiquitination and subsequent degradation of p53 are the major ways in which MDM2 negatively regulates p53, targeting the ubiquitin ligase activity of MDM2 has been pursued as a p53-activating strategy [142, 143], Recently, small-molecule inhibitors have been identified that specifically target the E3 ligase activity of MDM2 [144]. Several compounds from this class have been shown to inhibit the ubiquitination of p53 in vitro, with IC50 values in the 20–50 μM range. In cancer cells, they activate p53 signaling and induce apoptosis in a p53-dependent manner. However, these compounds have low potency and selectivity. Further optimization of these inhibitors to increase their potency, to increase their E3 ligase specificity, and to eliminate their p53-independent off-target activities is necessary before the potential of this p53-activating strategy can be adequately assessed [144].
1.5.3. Inhibiting MDM2-p53 Binding
MDM2 is unable to downregulate p53 if it is prevented from interacting with the tumor suppressor. Therefore, inhibition of MDM2-p53 binding is a desirable strategy for p53 stabilization and activation. However, targeting protein-protein interactions by small molecules is challenging. Protein-protein interactions usually involve large and flat surfaces that are difficult to interrupt by low molecular weight compounds [145, 146], However, in the case of the p53-MDM2 interaction, it has been demonstrated that a limited number of amino acid residues are crucial for the binding of the two proteins. In fact, just three amino acids in p53 - Phel9, Trp23 and Leu26 - are essential for the binding between the two proteins, and they are inserted into a deep hydrophobic pocket on the surface of the MDM2 molecule [10, 147]. Thus, one can design small molecules that mimic this interaction.
1.6. Synthetic Small Molecule MDM2 Inhibitors
Advances in the understanding of the conformation and structure of MDM2 have sparked the rational development of synthetic small molecule MDM2 inhibitors [148]. Molecular modeling approaches, such as molecular dynamics, pharmacophore-based, and molecular docking, have helped in this structure-based design. The hypothesis that a small-molecule could be developed to bind to MDM2, and thereby inhibit the MDM2-p53 interaction, resulted from the resolution of the crystal structure of MDM2 bound to a peptide from the transactivation domain of p53, which revealed that MDM2 possesses a relatively deep hydrophobic pocket that is filled primarily by three side chains from the helical region of the peptide [147]. Also, the discovery that MDM2-p53 binding is dependent on only three p53 amino acid residues interacting with a discrete MDM2 pocket stimulated efforts to identify potential small molecule inhibitors [10, 147]. Selective binding of another molecule to the surface of MDM2 can prevent the MDM2-p53 interaction, leading to the accumulation of p53 in the nucleus, and the subsequent induction of p53 responsive genes and activation of the p53 signaling pathway [148]. High throughput screening methods, combined with combinatorial library synthesis, have led to the development of a number of small molecule MDM2 inhibitors with diverse chemical structures. These include chalcones, piperazine-4-phenyl derivatives, nutlins (cis-imidazolines), spiro-oxindoles, sulfonamides, benzodiazepinediones, isoindolinones, terphenyls, and many more [142, 148]. There have been concerns whether it is possible to inhibit a protein-protein interaction with a drug-like molecule, and what the effects of unregulated p53 would be on healthy cells. However, experiments with a number of small molecules have proven that it is possible to inhibit this interaction, and a few of these molecules have already progressed to advanced preclinical development or early phase clinical trials [149]. Nearly 20 different chemical classes have been claimed to inhibit the MDM2-p53 interaction, but the majority of studies have been directed toward three classes: benzodiazepinediones, spiro-oxindoles, and cis-imidazolines [149]. The spiro-oxindole core structure was discovered using a structure-based de novo design strategy to develop entities that could mimic the interaction of Trp 23 in p53 with MDM2 [150].
Despite their different structural motifs, all synthetic inhibitors adhere to the basic hydrophobic pharmacophore model of MDM2. Potent inhibitors belonging to the three major classes of inhibitors have been obtained by the introduction of halide-substituted aromatic groups to improve the pharmacokinetic properties of the molecules [151]. The development of MDM2 E3 ubiquitin ligase inhibitors is less advanced, but it has been pointed out that E3 ligases are, in general, conceptually attractive drug targets. Table 2 provides the structures and describes the major findings of the most potent compounds of the different classes of inhibitors [144, 148–157].
Table 2.
Synthetic MDM2 Inhibitors and their Potential Targets
| Name and Structure | Mechanism of Action | In vitro Evidence | In vivo Evidence | Current Status | Ref. |
|---|---|---|---|---|---|
 |
Inhibits the MDM2-p53 interaction | IC50 = 17 nM for the most potent compound, but not RG7112 (data not disclosed yet) | Not reported for RG7112 | Phase I clinical trial for the treatment of solid tumors and hematological neoplasms, and treatment of leukemia | [141,142,144) |
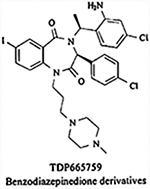 |
Inhibits the MDM2-p53 interaction | IC50= 0.7 μM | Inhibited tumor growth in A375 melanoma xenografts in female nude mice (synergism with Doxorubicin) | Preclinical development | [145] |
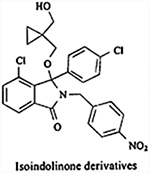 |
Inhibits the MDM2-p53 interaction | IC50 = 0.17 ± 0.2 μM | Not reported | Unknown/not reported | [146] |
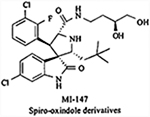 |
Inhibits the MDM2-p53 interaction | KI = 0.6 nM | Inhibited tumor growth in SJSA-1 and LNCaP tumor xenografts in nude mice. | Preclinical development | [143,147] |
| Inhibits the MDM2-p53 interaction by binding to p53 | KD=1.4 nM | Inhibited tumor growth in HCT116 and HCTll6p53−/− xenografts in SCID mice | Unknown/not reported | [148] | |
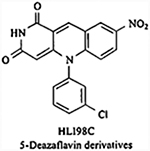 |
Inhibits the E3 ligase | IC50= 20 μM | Not reported | Unknown/not reported | [137] |
 |
Inhibits the E3 ligase | Not disclosed | Inhibited tumor growth in NCI-H1373 non-small cell lung cancer xenografts, PC-3M orthotopic prostate and p53 mutant HT-29 colon xenografts, and U87 glioblastoma xenografts | Phase I clinical trials for the treatment of advanced stage or refractory solid tumors in the USA and non-small cell lung cancer and prostate cancer in Europe. | [137,149,150] |
IC50, Inhibition of MDM2 binding to pS3, or inhibition οf E3 ubiquitin ligase activity; Ki or KD. Affinity constants.
All of the compounds described above target only the MDM2-p53 feedback loop and attempt to reactivate or stabilize p53 in the cell. However, a concern that remains is the fact that p53 activation requires not only stabilization and accumulation of the protein, but also post-translational modifications, such as phosphorylation, acetylation and SUMOylation, in response to genotoxic stress. Although small molecule antagonists of MDM2 prevent p53 from binding to MDM2, they do not technically affect post-translational modifications [151]. Thus, while they are likely to be highly effective against tumors containing wild-type p53, tumors with mutated or non-functional p53 may not be responsive. The possibility of chemoresistance in such tumors and the development of resistance following MDM2 inhibitor treatment need to be investigated. Secondly, it is debatable if the signaling pathways downstream of p53 are intact in tumorous cells. Although defects in the signaling upstream of p53 can be compensated by other molecules, such as HAUSP and COP-1 (which also stabilize p53 and p73), there are few compensatory mechanistic alternatives for defects downstream of p53; small molecular inhibitors such as nutlins that are specifically designed only to target the MDM2-p53 interaction may be rendered ineffective in the latter context. Furthermore, the pharmacokinetic properties and bioavailability of these synthetic compounds still need major improvements before they can be used in the clinical setting.
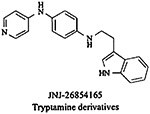
Among the small molecule MDM2 inhibitors, drugs currently under clinical trials include RG7112 and JNJ-2684165. RG7112 is an imidazoline derivative (belonging to the nutlin family) developed by Hoffman-La Roche and is currently under phase I clinical trial for the treatment of solid tumors [158], hematologic neoplasms and leukaemia [159]. This compound inhibits the MDM2-p53 interaction in a highly selective manner. JNJ-26854165, a tryptamine derivative developed by Johnson & Johnson Company is currently under phase I clinical trial for the treatment of advanced stage or refractory solid tumors in USA [157], non-small cell lung cancer and prostate cancer in Europe [149]. It blocks the degradation of p53 by inhibiting the binding of MDM2-p53 complex to proteasome. Side effects reported with the treatment of JNJ-26854165 include nausea, vomiting, fatigue, anorexia, insomnia, electrolyte imbalance, creatinine elevations, and asymptomatic QTc prolongation, mostly grade 1–2. A patient with higher dose exhibited grade 3 QTcF prolongation, which recovered after stopping the treatment [160]. Since, most of the compounds are in early clinical trials, there are not many reports regarding their adverse effects and efficacy in the clinic. These challenges have led researchers to explore other avenues for the development ofMDM2 inhibitors.
2. NATURAL PRODUCT MDM2 INHIBITORS
Natural products with their diverse structures and complicated carbon skeletons have always played an invaluable role in development of anti-cancer compounds. Indeed, almost 50% of the antitumor agents approved in the last 50 years of the 20th century are either compounds derived from natural sources or (semi-) synthetic analogs of these products [161–165]. Chemical entities of natural origin may be classified into three major categories, including original natural products, semisynthetic derivatives of natural products, and total synthetics based upon naturally occurring skeletons or with pharmacophores identified from natural products [163–165]. In this review, we also followed the three major categories to summarize the MDM2 inhibitors of natural origin, which could provide both broader and more in-depth insights into the discovery and development of MDM2 inhibitors.
The rationale for developing natural products as MDM2 inhibitors is based on two major factors: that several natural compounds exhibit MDM2 inhibition irrespective of p53 status of cell, and that natural products possess immense chemical diversity (yielding different kinds of candidate compounds) and often have good “drug-like” properties. The recognized and potential advantages of developing natural products as MDM2 inhibitors over synthetic compounds are discussed below.
In addition to their diversity and abundance, one of the most attractive properties of natural products is their chemical stability. Although a number of synthetic compounds have been designed based on the ciystal structure of the MDM2-p53 complex [129, 166, 167], with impressive activity profiles, this design principle has also resulted in some unavoidable drawbacks, such as high molecular weight and chemically unstable compounds. Natural products usually show greater stability than synthetic compounds due to their more stable stereochemical structures. Recently, a homogeneous time-resolved fluorescence (HTRF)-based high throughput screening study helped Allen et al. identify chromenotriazolopyrimidine as a potent inhibitor of the MDM2-p53 interaction, with a low micromolar IC50 (3.88 ± 1.48 μM) [168]. Unfortunately, this compound was shown to be chemically unstable, and it had poor solubility in DMSO. Even though structural optimization has been carried out based upon cocrystallization studies with MDM2, the poor solubility of the derivatives of the compound can also be predicted owing to the relatively high molecular weight of the compounds.
Among the other appealing properties of natural products is the fact that they are often less toxic than synthetic compounds, because many natural products are isolated from dietary vegetables, fruits, or medicinal herbs. As a result they have a long history of use either as food or as folk medicine. A classic example would be curcumin which has been an integral part of the South Asian diet for centuries and has, recently, been investigated for its anti-cancer properties [169]. In turn, none of the synthetics designed as anticancer agents are known to have low toxicity, which has led to a significantly higher rate of failure for such compounds during development. Additionally, natural products often show longer-lasting target effects and fewer side effects compared with synthetic compounds. Recently, a number of reports have demonstrated that several natural products in different classes possess considerable MDM2 binding affinity at micromolar concentrations, which further prompted biologists to screen and develop MDM2 inhibitors from natural resources. Below, we review the natural product MDM2 inhibitors that can be broadly classified into three categories based upon the main strategies that have been used to discover and evaluate them: natural products that inhibit MDM2 expression, those blocking the MDM2-p53 interaction, and those that modulate the E3 ubiquitin ligase activity and/or block the MDM2-mediated ubiquitination of p53.
2.1. Natural Products that Inhibit MDM2 Expression
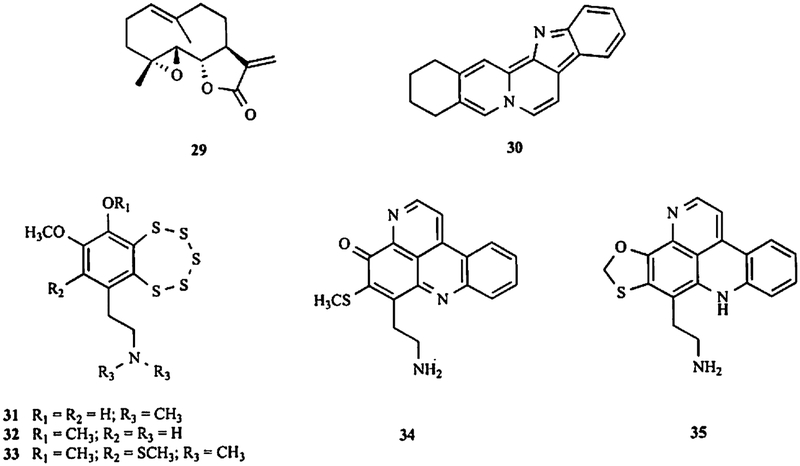
In this section, we review the natural products that inhibit MDM2 expression to emphasize their potential, grouping them according to their carbon frameworks (Fig. (1) and Table 3).
Fig. (1). The chemical structures of natural products that inhibit MDM2 expression.
(1) Genistein; (2) apigenin; (3) oroxylin A; (4) 25-OCH3-PPD; (5) 25-OH-PPD; (6) 25-OH-PPT; (7) berberine; (8) FBA-TPQ; (9) BA-TPQ; (10) gambogic acid; (11) triptolide; (12) curcumin; (13) l-(4-hydroxy-3-methoxyphenyl)-7-(3,4-dihydroxyphenyl)-4E-en-3-heptanone.
Table 3.
Natural Product MDM2 Inhibitors and their Potential Targets
| No. | Compounds | Origin | Chemical Class | In vitro Effects | In vivo Effects | Target(s) | Ref. |
|---|---|---|---|---|---|---|---|
| 1 | Gcnistein | N | Isoflavonoid | Reduces MDM2 protein and mRNA levels in human cell lines of breast, colon, and prostate cancer; primary fibroblasts; and breast epithelial cells | Inhibits MDM2 expression and the growth of PC3 xenograft tumors | MDM2 Expression | [159] |
| 2 | Apigenin | N | Flavonoid | Downregulates MDM2 and inhibits the phosphorylation of MDM2 in ovarian cancer cells | Not Reported | MDM2 Expression | [163] |
| 3 | Oroxylin A | N | Flavonoid | Downregulates MDM2 expression and interferes with the MDM2-modulated proteasome-related p53 degradation | Not Reported | MDM2 Expression | [164] |
| 4 | 25-OCH3-PPD | N | Ginsenoside | Decreases MDM2 protein levels in cell lines of prostate, pancreatic, and lung cancers; Inhibits MDM2 transcription in pancreatic cancer cell lines | Inhibits the growth of PC3, Panc-1, and A549 xenograft tumors | MDM2 Expression | [166–169] |
| 5 | 25-OH-PPD | N | Ginsenoside | Inhibits MDM2 expression in pancreatic and prostate cancer cell lines. | Inhibits the growth of Panc-1 and PC3 xenograft tumors | MDM2 Expression | [170,171] |
| 6 | 25-OH-PPT | N | Ginsenoside | Inhibits MDM2 expression in prostate cancer cell lines | Not Reported | MDM2 Expression | [170] |
| 7 | Berberine | N | Isoquinoline Alkaloid | Downregulates the MDM2 protein level and subsequently activates p53, but does not inhibit MDM2 mRNA expression | Not Reported | MDM2 Expression | [134] |
| 8 | FBA-TPQ | SN | Makaluvamine | Inhibits MDM2 expression in breast, colon, and ovarian cancer cell lines | Inhibits MDM2 expression and growth in MCF7 and OVCAR-3 xenograft tumors | MDM2 Expression | [177,178] |
| 9 | BA-TPQ | SN | Makaluvamine | Inhibits MDM2 expression in breast cancer cell lines | Inhibits MDM2 expression and growth in MCF7 and MDA-MB-468 xenograft tumors | MDM2 Expression | [179] |
| 10 | Gambogic Acid | N | Xanthonoid | Downregulates MDM2 at both the mRNA and protein levels | Inhibits the growth of HepG2 and H1299 xenograft tumors | MDM2 Expression | [181,182] |
| 11 | Triptolide | N | Diterpenoid | Decreases the levels of MDM2 and p21 proteins; Inhibits MDM2 phosphorylation, and increases p53 expression in a dose-dependent manner; Stabilizes p53 and activates p53 signaling | Not Reported | MDM2 Expression | [188–190] |
| 12 | Curcumin | N | Diary lheptanoid | Reduces MDM2 protein and mRNA and enhances p21wafl/CIPI protein expression | Inhibits the growth of PC3 xenograft tumors | MDM2 Expression | [192] |
| 13 | 1-(4-hydroxy-3-methoxyphenyl)-7-(3,4-dihydroxyphenyl)-4E-en-3-heptanone | N | Diarylheptanoid | Inhibits MDM2 expression | Not Reported | MDM2 Expression | [195] |
| 14 | No Name Reported | ND | Chalcone Derivative | IC50 = 49 μM; KD = 90 μM | Not Reported | MDM2-p53 Interaction | [196] |
| 15 | No Name Reported | ND | Chalcone Derivative | IC50= 117 μM; KD = 150 μM | Not Reported | MDM2-p53 Interaction | [196] |
| 16 | No Name Reported | ND | Chalcone Derivative | IC50 = 206 μM; KD = 220 μM | Not Reported | MDM2-p53 Interaction | [196] |
| 17 | No Name Reported | ND | Chalcone Derivative | IC50 = 250 μM; KD = 244 μM | Not Reported | MDM2-p53 Interaction | [196] |
| 18 | AMI 14 | Chalcone Derivative | No significant inhibition of the MDM2-p53 interaction | Not Reported | MDM2-p53 Interaction | [197,198] | |
| 19 | Hexylitaconic Acid | N | Hexylitaconic Acid | IC50 = 250 μg/mL | Not Reported | MDM2-p53 Interaction | [199] |
| 20 | No Name Reported | ND | Hexylitaconic Acid Derivative | No significant inhibition of the MDM2-p53 interaction | Not Reported | MDM2-p53 Interaction | [199] |
| 21 | No Name Reported | ND | Hexylitaconic Acid Derivative | No significant inhibition of the MDM2-p53 interaction | Not Reported | MDM2-p53 Interaction | [199] |
| 22 | No Name Reported | ND | Hexylitaconic Acid Derivative | No significant inhibition of the MDM2-p53 interaction | Not Reported | MDM2-p53 Interaction | [199] |
| 23 | Chlorofusin | N | Cyclic Peptide | lC50 = 4.6 μM; KD = 4.7 μM | Not reported | MDM2-p53 Interaction | [201,202] |
| 24 | (4R,8S,9S)-Ch]orofusin | ND | Cyclic Peptide | IC50 = 8 μM | Not Reported | MDM2-p53 Interaction | [209] |
| 25 | (4S,8R,9S-Chlorofusin | ND | Cyclic Peptide | IC50 = 8 μM | Not Reported | MDM2-p53 Interaction | [209] |
| 26 | (4S,8S,9R-Chlorofusin | ND | Cyclic Peptide | IC50 = 8 μM | Not Reported | MDM2-p53 Interaction | [209] |
| 27 | Hoiamide D | N | Hoiamide | EC50 = 4.5 μM | Not Reported | MDM2-p53 Interaction | [210] |
| 28 | Hoiamide C | ND | Hoiamide | None reported | Not Reported | MDM2-p53 Interaction | [210] |
| 29 | Parthenolide | N | Sesquiterpene Lactone | Induces the dissociation of p53-boirnd HDAC1, ubiquitination of p53-bound MDM2, and p53 activation. | Not Reported | E3 Ubiquitin Ligase | [212] |
| 30 | Sempervirine | N | Indole Alkaloid | Inhibits the ubiquitination and degradation of p53 by MDM2, and leads to accumulation of p53 | Not Reported | E3 Ubiquitin Ligase | [214] |
| 31 | Isolissoclinotoxin B | N | Pyridoacridine Alkaloid | lC50b = 58.6 ± 4 μM | Not Reported | E3 Ubiquitin Ligase | [215] |
| 32 | Varacin | N | Pyridoacridine Alkaloid | IC50b > 295 μM | Not Reported | E3 Ubiquitin Ligase | [215] |
| 33 | N,N-dimethyl-5-methylvaracin | N | Pyridoacridine Alkaloid | IC50b = 120.8 ± 9 μM | Not Reported | E3 Ubiquitin Ligase | [215] |
| 34 | Diplamine B | N | Pyridoacridine Alkaloid | IC50b =101.3 ± 4 μM | Not Reported | E3 Ubiquitin Ligase | [215] |
| 35 | Lissoclinidine B | N | Pyridoacridine Alkaloid | IC50b = 98.1 ± 6 μM | Not Reported | E3 Ubiquitin Ligase | [215] |
N, Natural product; ND, Natural product derivate; SN, Synthetic compound with a pharmacophore from a natural product; IC50. Inhibition of MDM2 binding to p53 measured by ELISA, DELFIA-modificd ELISA assay, or Scarccy assay; KD. Affinity constants between compounds and the N-lerminal domain of MDM2 measured by NMR titration experiments or surface plasmon resonance; EC50, Inhibition of the MDM2-p53 interaction measured by HTR-FRET-based competition assay; IC50b, Inhibition of the ubiquitin ligase activity measured by an electrochemiluminescence assay.
2.1.1. Flavonoids and Isoflavonoids
To the best of our knowledge, genistein (1), a soy-derived isoflavone, was one of the first compounds demonstrated to downregulate MDM2 at both the transcriptional and posttranslational levels [170]. Based on the reported chemopreventive activities of genistein in several human cancers, Li et al. further demonstrate that the compound directly decreases the MDM2 oncoprotein and mdm2 oncogene expression levels, independent of the p53 status of the cells. Additional studies have shown that genistein downregulates MDM2 without requiring the known tyrosine kinase inhibitory activity of the compound, and it consequently increases the p21 level. A subsequent in vivo investigation confirmed that genistein’s antitumor effects are linked to its inhibitory effects on MDM2 expression [170]. Correlatively, synthetic genistein glycosides and 7-O-modified genistein derivatives have been shown to have remarkable cytotoxicity in in vitro screening studies, and may be promising for the development of new anticancer chemotherapeutics [171–173]. However, it is currently unknown if these genistein derivatives have MDM2 inhibitory effects.
Apigenin (2), a common dietary flavonoid found in many fruits and vegetables, has been reported to induce p53 through the downregulation of MDM2 and to inhibit the phosphorylation of MDM2 by AKT in ovarian cancer cells [174]. Another flavonoid, oroxylin A (3), a naturally occurring derivative extracted from Scutellariae radix, has been shown to induce apoptosis in HepG2 hepatocellular carcinoma cells [175]. During their study, Wu and coworkers observed that oroxylin A stabilized p53 expression at the posttranslational level and consequently induced apoptosis. These changes were brought via downregulating MDM2 expression and interfering with MDM2-moduIated proteasome-related p53 degradation [175].
2.1.2. Ginsenosides
Ginsenosides, a major class of steroid compounds extracted from Panax species, are considered to be responsible for the diverse pharmacological activities of Panax plants, including their anti-inflammatory, anti-diabetes, and anticancer effects [176]. 20(S)-25-methoxyl-dammarane-3/β,12β,20-triol (25-OCH3-PPD, 4), a novel ginsenoside isolated from the dried leaves of Panax notoginseng, has been shown to have considerable cytotoxicity against 12 human cancer cell lines and demonstrated potential as a novel anticancer drug [177]. The anticancer activities of 25-OCH3-PPD have been shown in various in vitro and in vivo models of human prostate [178], pancreatic [179], and lung [180] cancers. The downregulation of the MDM2 protein levels is observed in all of the tested cancer cell lines, which is considered to be at least partially responsible for the anticancer effects of the compound. It is found that 25-OCH3-PPD reduces MDM2 transcription in a dose-dependent manner in cancer cells. Additionally, since the greatest effect occurs with the shortest promoter construct (−132 to +33) for MDM2 promoter, it is likely that at least one of the transcription factor binding sites (ETS, AP1, MEF2, or NFAT) within this section is affected by the compound [179].
A total of 11 ginsenosides, including 20(R)-dammarane-3β,12β,20,25-tetrol (25-OH-PPD, 5) and 20(R)-dammarane-3β,6α,12β,20,25-pentol (25-OH-PPT, 6) have recently been isolated and identified from the fruits of P. ginseng [181]. One of the compounds, 25-OH-PPD, has remarkable cytotoxicity to cancer cells with low micromolar IC50 values (10–60 μM) and dose-dependent effects on apoptosis, proliferation, and cell cycle progression [181]. The downregulation of the MDM2 protein levels is demonstrated, following exposure to both 25-OH-PPD and 25-OH-PPT [182], Further investigations are needed to clarify the importance of MDM2 inhibition in their anticancer effects.
2.1.3. Natural Isoquinoline Alkaloids
Berberine (7), a natural isoquinoline alkaloid from the Chinese medicinal herb Rhizoma coptidis, has recently attracted attention due to its diverse pharmacological effects, including anticancer, anti-inflammatory, and antibacterial activities. Zhang et al. recently report that berberine could induce apoptosis in acute lymphoblastic leukemia (ALL) cells through downregulation of the MDM2 oncoprotein [141]. Their results suggest that both the high MDM2 expression levels and wild-type p53 status of ALL cell lines are necessary for the proapoptotic effects of berberine, and that the compound persistently downregulates the MDM2 level to subsequently activate p53 in a steady-state manner, but does not inhibit MDM2 mRNA expression. It is further discovered that berberine also decreases DAXX expression at the transcriptional level, and subsequently prevents the formation of the MDM2-DAXX-HAUSP complex, resulting in the persistent self-ubiquitination of MDM2 in ALL cells [141]. In addition, numerous analogs and derivatives of berberine, such as palmatine, coralyne, and sanguinarine, have been reported to possess remarkable cytotoxicity to cancer cells, and are also expected to be natural product MDM2 inhibitors [183].
2.1.4. Makaluvamine Analogs
The makaluvamines, a class of marine pyrroloiminoquinone alkaloids isolated from sponges of the genus Zyzzya, have been demonstrated to have considerable in vitro and in vivo anticancer activities, which are initially thought to result from the inhibition of topoisomerase II [184]. Further studies indicate that the cytotoxicity of the makaluvamines could not be attributed solely to the inhibition of topoisomerase II, but also resulted from the direct DNA damage that occurs under the reductive activation conditions caused by the makaluvamines [185].
In order to further explore the novel mechanisms of action of makaluvamines, more than 40 makaluvamine analogs have been synthesized and evaluated for their anticancer activities in our laboratories [186, 187]. Their notable activities prompted the further consideration of four novel makaluvamine analogs, including 7-(4-fluorobenzylamino)-1,3,4,8-tetrahydropyrroIo[4,3,2-de]quinolin-8(1H)-one (FBA-TPQ, 8), 7-(phenethylamino)-l,3,4,8-tetrahydropyrrolo[4,3,2-de]quinolin-8(1H)-one (PEA-TPQ), 7-(3,4-methylenedioxyphenethylamino)-l, 3,4,8-tetrahydropyrrolo[4,3,2-de]quinolin-8(1H)-one (MPA-TPQ), and 7-(3,4-dimethoxy phenethylamino)-l,3,4,8-tetrahydropyrrolo[4,3,2-de]quinolin-8(lH)-one (DPA-TPQ). As expected, the compounds have broad-spectrum anticancer activities, with nanomolar IC50 values against thirteen human cancer cell lines [188]. Further studies have shown an increase in p53/p-p53 and a decrease in the MDM2 expression level in both in vitro and in vivo models of breast cancer [188]. A follow-up study using ovarian cancer models have also confirmed that the activation of the MDM2-p53 feedback loop is largely responsible for the anticancer activities of FBA-TPQ [189].
Another synthetic makaluvamine analog, 7-(benzylamino)-1,3,4,8-tetrahydropyrrolo [4,3,2-de]quinolin-8(1H)-one (BA-TPQ, 9) has been shown to dose-dependently inhibit cell growth, induce apoptosis and cell cycle arrest, and decrease the growth of xenograft tumors, all of which have been partially attributed to the action of BA-TPQ on the p53/MDM2 interaction [190]. We have also found the downregulation of the MDM2, cyclin D1, Cdk2, Cdk4, Cdk6, and E2F1 protein levels and the upregulation of the p53 protein level in MCF-7 cells. Interestingly, similar effects on these cell cycle/proliferation-related proteins also are observed in MCF-7 p53 KD cells and MDA-MB-468 (p53 mutant) cells, suggesting that BA-TPQ has a p53-independent mechanism of action [190]. MDM2 appears to be a major target of BA-TPQ, and p53-independent oncogenic effects of MDM2 may be inhibited in these cells. Reports about natural makaluvamines, makaluvamine derivatives, and synthetic makaluvamine analogs are still being published [191], and the data suggest that this class of compounds may yield a number of potential therapeutic agents for cancer, some of which may exert their effects by inhibiting MDM2.
2.1.5. Xanthonoids
Gambogic acid (GA, 10), a polyprenylated xanthone, is the major active ingredient of gamboges, which is derived from Garcinia hanburyi, which grows in Southeast Asia [192]. It has a long history of use as folk medicine due to its broad spectrum of activities. Recent pharmacological studies have shown that GA has potent anti-tumor activity, and that the compound exerts its effects by acting as an apoptosis inducer in cancer cell lines. Kasibhatla et al. report that G A targets the transferrin receptor (TfR or CD71) [192]. However, Gu et al. demonstrate that G A exerts its potent antitumor activity by modulating the MDM2-p53 pathway [193]. G A up-regulates p53 protein expression, but has no influence on p53 mRNA level. A subsequent study suggests that the downregulation of MDM2 at both the transcriptional and posttranscriptional levels be responsible for the p53 activation, subsequently triggering cancer cell apoptosis. in addition, a xenograft study also validates that GA inhibits the growth of p53 wild-type tumors, but not p53-null tumors [193]. Further research by Rong et al. confirms the downregulation of MDM2 at both the transcriptional and posttranscriptional levels by GA [194]. GA reduces the MDM2 expression in a concentration- and time-dependent manner, regardless of the p53 status of the cells. In addition, both the P1 and P2 promoters of mdm2 are responsive to GA, resulting in a decrease in the mdm2 mRNA expression level. GA also increases the protein level of p21, regardless of the p53 status of the cells, probably due to the alleviation of the binding of MDM2 to p21 and the stabilization of p21. A further in vivo investigation shows that GA inhibits tumor growth in a mouse xenograft model, possibly by downregulating MDM2 [194]. Furthermore, several GA derivatives, such as gambogoic acid [195], N-(2-ethoxyethyl)gambogamide [196], and N-(carboxy-methyl)gambogamide [197], have been reported to have antitumor activities in vitro and in vivo. However, it remains to be determined if their anticancer activities can be attributed to MDM2 inhibition.
2.1.6. Diterpenes
Several reports have highlighted the anticancer efficacy of natural diterpenes, of which taxol is the best-known, as mitotic inhibitors that can be used for cancer chemotherapy [198]. In recent years, several natural diterpenes have been reported to exert their anticancer activities by modulating the p53/MDM2 pathway. Kivihaiju et al. have demonstrated that low concentrations of triptolide (11), a diterpene triepoxide from Tripterygium wilfordii Hook, f., inhibit cell proliferation and induce cell senescence, and at high concentrations induce apoptosis, which is partly attributed to the nuclear accumulation of p53 [199]. Further studies show that triptolide exposure decreases the levels of both the MDM2 and p21 proteins [199]. Yang et al. report that triptolide inhibits MDM2 phosphorylation and increases p53 expression in a dose-dependent manner [200]. In addition, Carter et al. also report that triptolide is more effective for stabilizing p53 and activating p53 signaling than nutlin3a, an MDM2 inhibitor, and that it decreases the levels of MDM2 and XIAP [201]. Li et al. report a novel synthetic triptolide analog, (14s)-14,21-epoxytriptolide, has significant selective in vivo anticancer activity, especially against human prostate (PC-3) and ovarian (SK-OV-3) cancers, and has high efficacy against multidrug resistant cancer cell models [202]. (14s)-14,21-epoxytriptolide could be a promising candidate for further development of novel MDM2 inhibitors because of its chemical relationship with triptolide [202].
2.1.7. Diarylheptanoids
Curcumin (diferuloylmethane, 12), a dietary polyphenol, is the major active ingredient of Curcuma longa (known as the spice turmeric). Increasing evidence has suggested that curcumin can affect numerous molecular targets and influence diverse biochemical and molecular cascades [169]. We have demonstrated that curcumin exerts anticancer activity by downregulating MDM2 transcription through the PI3K/mTOR/ETS2 pathway [203]. The p53-independent downregulation of MDM2 expression is seen in both human normal and cancer cell lines after treatment with curcumin, resulting in p21 upregulation. Further investigations in PC3 cells, including control (parental) PC3 cells, and PC3 cells with stable knockdown or overexpression of MDM2, have demonstrated that MDM2 expression is at least partially responsible for the p53-independent anticancer activity of curcumin, and that curcumin exposure also contributes to the sensitization of human cancer cells to chemotherapy and radiation through MDM2, regardless of the p53 status of the cells [203]. Moreover, the biological significance of curcumin has also prompted medicinal chemists to synthesize curcumin derivatives, which has led to several potent new candidate chemotherapeutic agents [204, 205]. A more thorough elucidation of the molecular mechanisms for these derivatives is still needed, considering that MDM2 expression may also be a target for these compounds. Much attention has been paid to various analogs of curcumin, such as diaryheptanoids [206]. The most potent diaryheptanoid, 1-(4-hydroxy-3-methoxyphenyl)-7-(3,4-dihydroxyphenyl)-4E-en-3-heptanone (13), induces S phase arrest and apoptosis by upregulating ATF3 and stabilizing p53 and downregulation of MDM2 [206].
2.2. Natural Products Blocking the MDM2-p53 Interaction
As a result of the elucidation of the crystal structure of the MDM2-p53 complex, especially the identification of the three crucial amino acids in p53 (Phe19, Trp23, and Leu26) required for the binding of these two proteins, hundreds of peptides, synthetic compounds, and natural products have been designed, synthesized, or screened for their ability to target the MDM2-p53 interaction, preventing MDM2 binding to p53, stabilizing 53 protein and activating p53 functions. This strategy has resulted in several classes of MDM2 inhibitors under development The examples of natural products that are currently known to block the MDM2-p53 interaction are listed in Fig. (2) and Table 3.
Fig. (2). The chemical structures of natural products that block the MDM2-p53 interaction.
(14–17) Chalcone derivatives (No names reported); (18) AMI 14; (19) hexylitaconic acid; (20–22) hexylitaconic acid derivatives (No names reported); (23) chlorofusin; (24) (4R,8S,9S)-chlorofusin; (25) (4R,8S,9S)-chlorofusin (26); (4S,8S,9R)-chlorofusin; (27) hoiamide D; (28) hoiamide C.
2.2.1. Chalcone Derivatives
Chalcones and their derivatives have exhibited broad spectrum and potent anticancer activities. Recently, this class of natural compounds has been revealed to inhibit the MDM2-p53 interaction by a two-site ELISA and multidimensional NMR spectrometry [207]. The ELISA data show that compounds 14 and 15 have the best inhibitory activities, with IC50 values of 49 and 117 μM, respectively, and that compounds 16 and 17 have modest activities, with IC50 values of 206 and 250 μM, respectively. Accordingly, NMR titration experiments also have demonstrated that these chalcone derivatives are bound in or near the tryptophan (Trp23) binding pocket of MDM2, and that compounds 14 (KD = 90 μM) and 15 (kD = 150 μM) have better activities than compounds16 (KD= 220 μM) and 17 (KD = 244 μM). Their activities are attributed to the presence of the chlorophenyl group and augmented by increasing the number of chloro-substitutions [207]. In addition, among several newly synthesized chalcone derivatives tested for their anticancer activities is a boronic-chalcone derivative, 3,5-bis-(4-boronic acid-benzylidene)-1-methyl-piperidin-4-one (AM 114, 18), that has the most potent inhibitory activity [208]. AM1 14 treatment leads to p53 and p21 accumulation and apoptosis, but does not significantly disrupt the MDM2-p53 interaction [209].
2.2.2. Hexylitaconic Acid and its Derivatives
Hexylitaconic acid (19) is an itaconic acid derivative isolated from a culture of the marine-derived Arthrinium sp fungi, which was initially separated from a marine sponge from Toyama Bay in the Sea of Japan [210, 211]. The compound has been identified as a new inhibitor of the MDM2-p53 interaction by an ELISA assay, blocking MDM2-p53 interaction in a dose-dependent manner, with an IC50 value of 50 μg/mL [210]. However, a SAR study measuring the inhibitory activities of three derivatives and two analogs of hexylitaconic acid, including a monomethyl ester (20), a dihydro derivative (21), a dihydro derivative of monomethyl ester (22), itaconic acid, and succinic acid, reveals that no significant inhibitory activities at the concentration of 50 μg/mL [210].
2.2.3. Chlorofusin
Chlorofusin (23), isolated from the extract of the fungal strain Microdochium caespitosum, is considered to be among the first MDM2-p53 interaction inhibitor derived from a natural source [212]. Duncan et al. first reported its inhibitory effects (IC50 = 4.6 μM) on the MDM2-p53 interaction by using the DELFIA-modified ELISA assay [212]. The results of that study prompted Duncan et al. to continue their investigation of the interaction between chlorofusin and the N-terminal domain of MDM2 by surface plasmon resonance. They found that it had an affinity constant (KD = 2.11 × 105 M−1 or 4.7 μM) that was consistent with the IC50 obtained in their previous study [213]. As a result of the increasing interest in this exciting lead compound, total synthetic studies of this natural product have been carried out, leading to the reassignment of the chromophore-relative stereochemistry, assignment of the absolute stereochemistry, and a series of analogs and structural moieties have been constructed [214–220], Clark et al. have evaluated the inhibition of MDM2-p53 binding by chlorofusin, its seven chromophore diastereomers, and several key analogs and partial moieties by the Searcey assay, showing that all of the tested chromophore diastereomers are effective inhibitors of the MDM2-p53 interaction, and three of them, (4R, 8S,9S)-chlorofusin (24), (4S,8S,9S)-chloroftisin (25), and (4S,8S,9R)-chlorofusin (26), exhibit equipotent effects (IC50 = 8 μM) with chlorofusin [220]. Two of these have unnatural 4S configurations and one of them is the enantiomer of the natural product. However, the cyclic peptide moiety and chromophore moiety of chlorofusin and the simple derivatives synthesized using these features do not show significant inhibitory activities on the protein-protein binding. Based on these findings, it is concluded that neither the chromophore nor the absolute stereochemistry is essential for inhibiting the MDM2-p53 interaction.
2.2.4. Hoiamide and its Derivatives
A bioassay-guided isolation of two cyanobacterial extracts from Papua New Guinea has led to the purification of hoiamide D (27) and C (28), two polyketide synthase/non-ribosomal peptide synthetase-derived natural products [221]. Hoiamide D is obtained in both its carboxylic acid and conjugate base forms, and hoiamide C is considered to be an artifact that formed during the extraction process. The inhibitory activity of hoiamide D in the carboxylic acid form on the MDM2-p53 interaction is examined by a HTR-FRET-based competition assay. The carboxylate anion exhibits significant inhibitory effects on MDM2-p53 binding, with an EC50 value of 4.5 μM. However, this compound has minimal cytotoxicity to mammalian H460 cells at a concentration of 40 μM. Even though there is no direct proof that hoiamide C inhibits MDM2-p53 binding, this compound may also be a potential inhibitor due to the structural relationships between the two hoiamides [221].
2.3. Natural Products that Modulate the E3 Ubiquitin Ligase Activity and/or Block the MDM2-Mediated Ubiquitination of p53
As the major negative regulator of p53, MDM2 controls p53 ubiquitination and subsequent degradation by its ubiquitin ligase (E3) activity, for which the C-terminal RING finger domain is required. An increasing number of studies have demonstrated that many small molecule inhibitors can specifically inhibit the E3 ligase activity of MDM2, preventing the degradation of p53, thereby stabilizing it. In this section, we review the natural product inhibitors falling into this category, and classify them based on their chemical structures (Fig. (3) and Table 3).
Fig. (3).
The chemical structures of natural products that modulate MDM2’s E3 ubiquitin ligase activity and/or block the MDM2-mediated ubiquitination of p53. (29) Parthenolide; (30) sempervirine; (31) isolissoclinotoxin B; (32) varacin; (33) N,N-dimethyl-5-methylvaracin; (34) diplamine B; (35) lissoclinidine B.
2.3.1. Sesquiterpene Lactone
Sesquiterpene lactones have been considered to be a promising natural resource for developing anticancer drugs, such as artemisinin and its derivatives, which mainly exert their anticancer activities by inducing apoptosis [222]. Several sesquiterpene lactones are reported to exert their anticancer activities by modulating the MDM2-p53 feedback loop. Gopal et αl report that parthenolide (29), isolated from the European feverfew herb, Tanacetum parthenium, induces MDM2 ubiquitination and proteasomal degradation in an ATM-dependent manner, subsequently activating p53 and other MDM2-regulated tumor suppressors [223]. Their results demonstrate that parthenolide treatment induces an increase in p53 and MDM2 modification in a time- and dose-dependent manner and that the change in MDM2 coincides with the parthenolide-dependent HDAC1 depletion. Parthenolide induces the dissociation of p53-bound HDAC1 and the ubiquitination of p53-bound MDM2, resulting in p53 activation. In addition, the protein levels of PUMA and p21 are increased after parthenolide exposure, further confirming the activation of p53 [223]. In order to improve the water solubility of parthenolide (29), Nasim and Crooks have designed and synthesized a series of aminoparthenolide analogs through diastereoselective conjugate addition, by which several primary and secondary amines are added to the α-methylene γ-butyrolactone moiety of parthenolide [224]. The anticancer activities of these 17 derivatives are evaluated in NCI 60 cancer cell lines. A tyramine derivative shows cytostatic effects and cytotoxicity against lymphoblastic leukemia cells, and a (R)-(l, 2,3,4-tetrahydro-1-naphthyl)amino derivative exhibits cytostatic effects in human anaplastic large T-cell lymphoma cells at nanomolar concentrations [224]. Further studies on this class of natural product derivatives are needed, especially to determine their mechanism of action, and to elucidate whether the MDM2-p53 pathway is one of the major targets, since these derivatives are closely related to parthenolide.
2.3.2. Sempervirine (An Indole Alkaloid)
Sempervirine (30), a natural indole alkaloid, has been identified as an inhibitor of MDM2’s E3 ligase activity from more than 144,000 natural product extracts evaluated by a high-throughput electrochemiluminescent screen [225]. Sempervirine inhibits the MDM2-mediated ubiquitination and degradation of p53, resulting in the accumulation of p53. This compound also preferentially induces apoptosis in transformed cells and cancer cells expressing wild-type p53, warranting further studies of its anticancer activity [225].
2.3.3. Pyridoacridine Alkaloids
Using the same high throughput screening method described above, an extract of the ascidian Lissoclmum cf. badium from the coast of Papua New Guinea has been identified as an inhibitor of MDM2’s E3 ligase activity [226]. A subsequent study has led to the purification and identification of five novel pyridoacridine alkaloids, including isolissoclinotoxin B (31), varacin (32), NJV-dimethy 1–5-methylvaracin (33), diplamine B (34), and lissoclinidine B (35). The MDM2-inhibitory activity of these five compounds are evaluated in a MDM2 electrochemiluminescence assay, demonstrating that isolissoclinotoxin B (31) has the best activity, with an IC50 value of 58.6 μM, and lissoclinidine B (35) displays weaker activity, with an IC50 value of 98.1 μM. However, a farther study suggested that lissoclinidine B (35) inhibits the ubiquitination and degradation of p53, selectively kills transformed cells expressing wild-type p53, and could be a promising lead candidate for the development of new anti-MDM2 chemotherapeutic agents [226].
In summary, we see a variety of natural compounds obtained from diverse sources, which possessing highly dissimilar carbon skeletons have the capability of inhibiting the MDM2 oncoprotein. However, in most cases, we do not know if MDM2 inhibition is indeed a major anticancer mechanism for those compounds. This sort of information would be highly valuable while designing combination therapy and for devising methods to counter drug resistance. Studies involving elucidating the effect of a particular compound in presence or absence of MDM2 (i.e. involving MDM2 overexpressing stably transfected or inducible cell lines, MDM2 knockout systems) should be performed to understand the actual importance of MDM2 inhibition in mediating the anticancer effects of the drug.
3. FUTURE DIRECTIONS AND CONCLUSIONS
As discussed in the previous sections, MDM2 is one of the most important factors controlling the oncogenic process. In addition to being a negative regulator of p53, it interacts with several other proteins involved in cell cycle progression and growth. The disruption of the MDM2-p53 interaction with small molecule inhibitors has been validated as a potentially viable and attractive cancer therapeutic strategy. Current drug discovery efforts in this area are focused on the binding of p53 to the N-terminal domain of MDM2, which has been extensively characterized at the molecular level [148]. Less work has been done on the development of molecules to target other regions of MDM2, such as its RING finger domain. As detailed in this review [148] and other reviews [117, 129, 130, 227–230], there are numerous proteins in addition to p53 that interact with MDM2. Many of these interactions enhance MDM2’s ability to downregulate p53, and their interaction with MDM2 should be explored as the basis for novel cancer therapeutic strategies. Targeting these interactions may provide alternative or complementary approaches to targeting the MDM2-p53 interaction. It is also necessary to focus on MDM2 itself as a drug target and not limit studies to its role in the MDM2-p53 feedback loop. The p53-independent oncogenic activities of MDM2 that can be investigated include, but are not limited to (i) its binding to, and destabilization of, p21, (ii) its binding to the p53 homologue, p73, (iii) its destabilization of NUMB, (iv) its positive modulation of the HIF-1 transcription factor, (v) its modulation of ribosomal proteins, (vi) role of MDM2 in modulation of apoptotic proteins, and (vii) interaction with androgen receptor (may be important in prostate cancer management).
Another area that needs a close attention is the role of MDM2 in cell migration and metastasis. Until now, most research on MDM2 has focused on its role in the degradation of p53 or its effects on the cell cycle and apoptosis. Studies by Yang el al and Araki el al proved that MDM2’s influence spreads much farther and wider than was previously thought [96, 97]. Given that research into the epithelial-mesenchymal transition is still in its early stages, it is possible that MDM2 may be identified as a molecule that can be targeted to regulate this process as well.
Although MDM2 overexpression is oncogenic and is associated with a poorer prognosis in most situations, in certain scenarios overexpression of MDM2 may be associated with a good prognosis. For example, MDM2 overexpression correlates with a favorable prognosis in estrogen receptor alpha positive breast cancer [231]. Also, in malignant melanoma IGF-1R, an oncogenic receptor tyrosine kinase, can be downregulated by MDM2-mediated ubiquitination, targeting it for proteasomal degradation [113]. Thus, while there may be some stipulations to using MDM2 inhibitors for cancer therapy, in most scenarios, MDM2 inhibition appears to have potential as a novel and effective anticancer approach.
Further, MDM2 also inactivates the tumor suppressor pRb, and stimulates the E2F1/DP1 transcription factors, resulting in the promotion of the G1 to S phase transition [42, 85]. It has also been shown to confer a growth advantage to cells, even in the absence of p53 and pRb, causing them to overcome the G1 cell cycle blockade induced by p107 [232]. These examples are given to emphasize the crucial role that this molecule plays in the cell cycle machinery, and the numerous opportunities still remaining that could be investigated for anticancer drug discovery targeting MDM2.
In addition, MDM2 has several domains that can be additional targets for therapy, including its NLS, NES, and its C-terminal RING finger domain [10]. To date, most drugs have been investigated for their ability to inhibit the N-terminal domain-mediated interaction with p53 [142, 151]. Altering the posttranslational modifications, including phosphorylation and SUMOylation, may also represent novel approaches for inhibiting MDM2 [72]. The p53-binding domain has been extensively targeted, and a few agents have focused on the E3 ligase activity of the RING finger domain [143, 144, 148]. However, the other regions of MDM2, including the binding domains for other proteins, as well as the other functional domains, have not been considered. Also, recently, other domains in p53 that bind to MDM2 (such as the C-terminus of p53) have been identified [49]. This information may also be potentially used to develop a new generation of small molecule compounds targeting these new sites. Further, the numerous MDM2-interactive (p53-dependent and -independent) proteins can also be targeted to improve the therapeutic response to cancers [129].
A recent study shows that the proteasome inhibitor velcade (also known as bortezomib), when combined with nutlin, exhibited synergistic activity against myeloma [233]. These results and other studies employing similar combinations, provide the framework for further development of MDM2 inhibitors that can be used either alone or in combination with conventional and novel therapies to increase the anticancer efficacy and improve the patient outcome.
Natural compounds present an attractive area for the development of MDM2 inhibitors. Unlike synthetic drug development, which can cost billions of dollars to develop from the bench to the bedside, many natural compounds have known safety and pharmacokinetic profiles; having been used as part of traditional medicines or in supplements. Their chemical diversity and complex structures can also serve as elegant progenitors for novel synthetic compounds. The natural product inhibitors of MDM2 have been shown to target both the p53-dependent and -independent activities of MDM2. Importantly, many of them have shown impressive activity in p53 mutant or non-functional tumor environments, thus eliminating the need for p53 dependent mechanisms to exert their effects. On the other hand, synthetic small molecule inhibitors (SMIs) of MDM2 are known to restore the apoptotic and cell cycle regulatory functions of p53 by disrupting the MDM2-p53 interaction. Therefore, in principle, these SMIs are ineffective against p53 mutant tumors, which is a hallmark of most cancers. Although their mechanisms need to be more clearly defined before definitive conclusions can be made, many of the naturally derived compounds appear to decrease MDM2 expression and/or activity in animal models, suggesting that they could be effective cancer therapeutic or preventive agents. However, what is lacking is a systematic study of chemically-related natural MDM2 inhibitors in established, well characterized cancer models to ascertain the relationships between the structures and inhibitory activities of these compounds. Such data could be used to develop novel semi-synthetic derivatives with improved efficacy, pharmacokinetic, and bioavailability profiles.
Targeting individual interactive molecules or the interaction of MDM2 with specific co-factors or regulators (dependent or independent of p53) can provide an effective therapeutic strategy. However, more research is needed to elucidate the role(s) of each of these interactions, and to define the circumstances under which the interaction(s) can be successfully targeted. The use of modern combinatorial libraries and high throughput screening techniques, coupled with an increasingly in-depth understanding of the biochemistry and molecular biology of MDM2 will help us to develop novel MDM2 inhibitors.
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (NIH) grants RO1 CA112029 and RO1 CA121211 and a Susan G Komen Foundation grant BCTR0707731 (to R.Z.). We thank the current and former members of our laboratory for their excellent contributions to the research projects that were cited in this review. We apologize for not being able to cite all of the publications in the field due to the limitations of the length of the review.
Footnotes
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflicts of interest.
REFERENCES
- [1].Croce CM Oncogenes and cancer. N. Engl. J. Med, 2008,358,502–511. [DOI] [PubMed] [Google Scholar]
- [2].Weinstein IB; Joe AK. Mechanisms of disease: oncogene addiction – a rationale for molecular targeting in cancer therapy. Nat. Clin. Pract. Oncol, 2006,5,448–457. [DOI] [PubMed] [Google Scholar]
- [3].Letai AG Diagnosing and exploiting cancer’s addiction to blocks in apoptosis. Nat. Rev. Cancer; 2008,8,121–132. [DOI] [PubMed] [Google Scholar]
- [4].Cahilly-Snyder L; Yang-Feng T; Francke U; George DL Molecular analysis and chromosomal mapping of amplified genes isolated from a transformed mouse 3T3 cell line. Somat. Cell Mol. Genet, 1987, 13, 235–244. [DOI] [PubMed] [Google Scholar]
- [5].Fakharzadeh SS; Trusko SP; George DL Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBOJ, 1991,10,1565–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Oliner JD; Kinzler KW; Meitzer PS; George DL; Vogelstein B Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature, 1992,358, 80–83. [DOI] [PubMed] [Google Scholar]
- [7].Olson DC; Marechal V; Momand J; Chen J; Romocki C; Levine AJ Identification and characterization of multiple mdm-2 proteins and mdm-2-p53 protein complexes. Oncogene, 1993,8,2353–2360. [PubMed] [Google Scholar]
- [8].Perry ME; Mendrysa SM; Saucedo LJ; Tannous P; Holubar M p76(MDM2) inhibits the ability of p90(MDM2) to destabilize p53. J. Biol. Chem, 2000,275,5733–5738. [DOI] [PubMed] [Google Scholar]
- [9].Evans SC; Viswanathan M; Grier JD; Narayana M; EI-Naggar AK; Lozano G An alternatively spliced HDM2 product increases p53 activity by inhibiting HDM2. Oncogene, 2001,20,4041–4049. [DOI] [PubMed] [Google Scholar]
- [10].Chen J; Marechal V; Levine AJ Mapping of the p53 and mdm-2 interaction domains. Mol. Cell. Biol, 1993,13,4107–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Momand J; Zambetti GP; Olson DC; George D; Levine AJ The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell, 1992, 69, 1237–1245. [DOI] [PubMed] [Google Scholar]
- [12].Böttger A; Böttger V; Garcia-Echeverria C; Chène P; Hochkeppel HK; Sampson W; Ang K; Howard SF; Picksley SM; Lane DP Molecular characterization of the hdm2-p53 interaction. J. Mol. Biol, 1997, 269,744–756. [DOI] [PubMed] [Google Scholar]
- [13].Wsierska-Gadek J; Horky M How the nucleolar sequestration of p53 protein or its interplayers contributes to its (re)-activation. Ann. N. Y. Acad. Sei, 2003, 1010, 266–272. [DOI] [PubMed] [Google Scholar]
- [14].Haupt Y; Maya R; Kazaz A; Oren M Mdm2 promotes the rapid degradation of p53. Nature, 1997,387,296–299. [DOI] [PubMed] [Google Scholar]
- [15].Honda R; Tanaka H; Yasuda H Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett, 1997,420,25–27. [DOI] [PubMed] [Google Scholar]
- [16].Kubbutat MH; Jones SN; Vousden KH Regulation of p53 stability by Mdm2. Nature, 1997,387,299–303. [DOI] [PubMed] [Google Scholar]
- [17].Jiang M; Shao ZM; Wu J; Lu JS; Yu LM; Yuan JD; Han QX; Shen ZZ; Fontana JA p21/wafl/cip1 and mdm-2 expression in breast carcinoma patients as related to prognosis. Int. J. Cancer, 1997, 74,529–534. [DOI] [PubMed] [Google Scholar]
- [18].Ng IO; Lam KY; Ng M; Regezi JA Expression of p21/wafl in oral squamous cell carcinomas--correlation with p53 and mdm2 and cellular proliferation index. Oral Oncol, 1999,35,63–69. [DOI] [PubMed] [Google Scholar]
- [19].Zhang Z; Wang H; Li M; Agrawal S; Chen X; Zhang R MDM2 is a negative regulator of p21 WAFl/CIPl, independent of p53. J. Biol. Chem, 2004,279, 16000–16006. [DOI] [PubMed] [Google Scholar]
- [20].Jin Y; Lee H; Zeng SX; Dai MS; Lu H MDM2 promotes p21WAFl/CIPl proteasomal turnover independently of ubiquitylation. EMBO J, 2003, 22, 6365–6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Xu H; Zhang Z; Li M; Zhang R MDM2 promotes proteasomal degradation of p21Wafl via a conformation change. J. Biol. Chem, 2010,285, 18407–18414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen J; Wu X; Lin J; Levine AJ mdm-2 inhibits the G1 arrest and apoptosis functions of the p53 tumor suppressor protein. Mol. Cell. Biol, 1996,16,2445–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Xiao ZX; Chen J; Levine AJ; Modjtahedi N; Xing J; Sellers WR; Livingston DM Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature, 1995,375,694–698. [DOI] [PubMed] [Google Scholar]
- [24].Sun P; Dong P; Dai K; Hannon GJ; Beach D p53-independent role of MDM2 inTGF-pl resistance. Science, 1998,282,2270–2272. [DOI] [PubMed] [Google Scholar]
- [25].Lundgren K; Montes de Oca Luna R; McNeill YB; Emerick EP; Spencer B; Barfield CR; Lozano G; Rosenberg MP; Finlay CA Targeted expression of MDM2 uncouples S phase from mitosis and inhibits mammary gland development independent of p53. Genes Dev, 1997, II, 714–725. [DOI] [PubMed] [Google Scholar]
- [26].Dazard JE; Augias D; Neel H; Mils V; Maréchal V; Basset-Seguin N; Piette J MDM-2 protein is expressed in different layers of normal human skin. Oncogene, 1997, 14,1123–1128. [DOI] [PubMed] [Google Scholar]
- [27].Alkhalaf M; Ganguli G; Messaddeq N; Le Meur M; Wasylyk B MDM2 overexpression generates a skin phenotype in both wild type and p53 null mice. Oncogene, 1999,18,1419–1434. [DOI] [PubMed] [Google Scholar]
- [28].Fiddler TA; Smith L; Tapscott SJ; Thayer MJ Amplification of MDM2 inhibits MyoD-mediated myogenesis. Mol. Cell. Biol, 1996, 16, 5048–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Juven-Gershon T; Shifman O; Unger T; Elkeles A; Haupt Y; Oren M The Mdm2 oncoprotein interacts with the cell fate regulator Numb. Mol. Cell Biol, 1998, 18,3974–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Marechal V; Elenbaas B; Piette J; Nicolas JC; Levine AJ. The ribosoma) L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol. Cell Biol, 1994,14,7414–7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhang Y; Wolf GW; Bhat K; Jin A; Allio T; Burkhart WA; Xiong Y Ribosomal protein LU negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell. Biol, 2003, 23, 8902–8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dai MS; Zeng SX; Jin Y; Sun XX; David L; Lu H Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol, 2004,24,7654–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lohrum MA; Ashcroft M; Kubbutat MH; Vousden KH Identification of a cryptic nucleolar-localization signal in MDM2. Nat. Cell Biol, 2000, 2, 179–181. [DOI] [PubMed] [Google Scholar]
- [34].Zhang Y; Lu H Signaling to p53: ribosomal proteins find their way. Cancer Cell 2009,16,369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chen D; Zhang Z; Li M; Wang W; Li Y; Rayburn ER; Hill DL; Wang H; Zhang R Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene, 2007,26,5029–5037. [DOI] [PubMed] [Google Scholar]
- [36].Zhu Y; Poyurovsky MV; Li Y; Biderman L; Stahl J; Jacq X; Prives C Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell, 2009,35, 316–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Deshaies RJ; Joazeiro CA RING domain E3 ubiquitin ligases. Annu. Rev. Biochem, 2009, 78,399–434. [DOI] [PubMed] [Google Scholar]
- [38].Nakamura S; Roth JA; Mukhopadhyay T Multiple lysine mutations in the C-terminal domain of p53 interfere with MDM2-dependent protein degradation and ubiquitination. Mol Cell Biol, 2000,20,9391–9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Rodriguez MS; Desterro JM; Lain S; Lane DP; Hay RT Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol Cell Biol, 2000,20, 8458–8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Fang S; Jensen JP; Ludwig RL; Vousden KH; Weissman AM Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol Chem, 2000,275, 8945–8951. [DOI] [PubMed] [Google Scholar]
- [41].Yogosawa S; Miyauchi Y; Honda R; Tanaka H; Yasuda H Mammalian Numb is a target protein of Mdm2, ubiquitin ligase. Biochem. Biophys. Res. Commun, 2003,302, 869–872. [DOI] [PubMed] [Google Scholar]
- [42].Uchida C; Miwa S; Kitagawa K; Hattori T; Isobe T; Otani S; Oda T; Sugimura H; Kamijo T;Ookawa K; Yasuda H; Kitagawa M Enhanced Mdm2 activity inhibits pRB function via ubiquitin-dependent degradation. EMBOJ, 2005,24,160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pan Y; Chen J MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol, 2003,23, 5113–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lee JT; Gu W The multiple levels of regulation by p53 ubiquitination. Cell Death Differ, 2010,17, 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lai Z; Ferry KV; Diamond M; Wee K; Kim YB; Ma J; Yang T; Benfield PA; Copeland RA; Auger KR Human Mdm2 mediates multiple mono-ubiquitination of p53 by a mechanism requiring enzyme isomerization. JBiol. Chem, 2001,276,31357–31367. [DOI] [PubMed] [Google Scholar]
- [46].Ferreon JC; Lee CW; Arai M; Martinez-Yamout MA; Dyson HJ; Wright PE Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300and HDM2. Proc. Natl Acad. Sei. USA, 2009,106,6591–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Brady CA; Attardi LD p53 at a glance. J. Cell Sei, 2010, 123, 2527–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Chi SW; Lee SH; Kim DH; Ahn MJ; Kim JS; Woo JY; Torizawa T; Kainosho M; Han KH Structural details on mdm2-p53 interaction. J. Biol Chem, 2005,280, 38795–38802. [DOI] [PubMed] [Google Scholar]
- [49].Poyurovsky MV; Katz C; Laptenko O; Beckerman R; Lokshin M; Ahn J; Byeon IJ; Gabizon R; Mattia M; Zupnick A; Brown LM; Friedler A; Prives C The C terminus of p53 binds the N-terminal domain of MDM2. Nat. Struct. Mol Biol, 2010, 17,982–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Montes de Oca Luna R; Wagner DS; Lozano G Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature, 1995,378,203–206. [DOI] [PubMed] [Google Scholar]
- [51].Wang Z; Yang B; Dong L; Peng B; He X; Liu W A novel oncoprotein Pirh2: rising from the shadow of MDM2. Cancer Sei, 2011, 102,909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wang L; He G; Zhang P; Wang X; Jiang M; Yu L Interplay between MDM2, MDMX, Pirh2 and COP1: the negative regulators of p53. Mol Biol. Rep, 2011,38,229–236. [DOI] [PubMed] [Google Scholar]
- [53].Chen D; Kon N; Li M; Zhang W; Qin J; Gu W ARF-BPI/Mule is a critical mediator of the ARF tumor suppressor. Cell, 2005, 121, 1071–1083. [DOI] [PubMed] [Google Scholar]
- [54].Quelle DE; Zindy F; Ashmun RA; Sherr CJ Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated protein capable of inducing cell cycle arrest. Cell, 1995,83,993–1000. [DOI] [PubMed] [Google Scholar]
- [55].Weber JD; Kuo ML; Bothner B; DiGiammarino EL; Kriwacki RW; Roussel MF; Sherr CJ Cooperative signals governing ARF-mdm2 interaction and nucleolar localization of complex. Mol Cell Biol, 2000, 20, 2517–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kamijo T; Weber JD; Zambetti G; Zindy F; Roussel MF; Sherr CJ Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl. Acad. Sei. USA, 1998,95, 8292–8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Honda R; Yasuda H Association of pl9(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBOJ, 1999, 18,22–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Korgaonkar C; Hagen J; Tompkins V; Frazier AA; Allamargot C; Quelle FW; Quelle DE Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol Cell Biol, 2005,25,1258–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chang CJ; Freeman DJ; Wu H PTEN regulates Mdm2 expression through the P1promoter. J. Biol Chem, 2004,279,29841–29848. [DOI] [PubMed] [Google Scholar]
- [60].Barak Y; Gottlieb E; Juven-Gershon T; Oren M Regulation of mdm2 expression by p53: alternative promoters produce transcripts with nonidentical translation potential. Genes Dev, 1994,8, 1739–1749. [DOI] [PubMed] [Google Scholar]
- [61].Shin S; Janknecht RJ Concerted activation of the Mdm2 promoter by p72 RNA helicase and the coactivators p300 and P/CAF. J. Cell Biochem, 2007, 101, 1252–65. [DOI] [PubMed] [Google Scholar]
- [62].Truong AH; Cervi D; Lee J; Ben-David Y Direct transcriptional regulation of MDM2 by Fli-1. Oncogene, 2005,24,962–969. [DOI] [PubMed] [Google Scholar]
- [63].Ries S; Biederer C; Woods D; Shifman O; Shirasawa S; Sasazuki T; McMahon M; Oren M; McCormick F Opposing effects of Ras on p53: transcriptional activation of mdm2 and induction of pl9ARF. Cell, 2000,103, 321–323. [DOI] [PubMed] [Google Scholar]
- [64].Khosravi R; Maya R; Gottlieb T; Oren M; Shiloh Y; Shkedy D Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc. Natl. Acad. Sei. USA, 1999, 96, 14973–14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].De Toledo SM; Azzam EI; Dahlberg WK; Gooding TB; Little JB ATM complexes with HDM2 and promotes its rapid phosphorylation in a p53-independent manner in normal and tumor human cells exposed to ionizing radiation. Oncogene, 2000,19,6185–6193. [DOI] [PubMed] [Google Scholar]
- [66].Maya R; Baiass M; Kim ST; Shkedy D; Leal JF; Shifman O; Moas M; Buschmann T; Ronai Z; Shiloh Y; Kastan MB; Katzir E; Oren M ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev, 2001,15,1067–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Meulmeester E; Pereg Y; Shiloh Y; Jochemsen AG ATM-mediated phosphorylations inhibit Mdmx/Mdm2 stabilization by HAUSP in favor of p53 activation. Cell Cycle, 2005,4,1166–1170. [DOI] [PubMed] [Google Scholar]
- [68].Zhou BP; Liao Y; Xia W; Zou Y; Spohn B; Hung MC HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat. Cell Biol, 2001, 3, 973–982. [DOI] [PubMed] [Google Scholar]
- [69].Ogawara Y; Kishishita S; Obata T; Isazawa Y; Suzuki T; Tanaka K; Masuyama N; Gotoh Y Akt enhances Mdm2-mediated ubiquitination and degradation of p53. J. Biol Chem, 2002,277,21843–21850. [DOI] [PubMed] [Google Scholar]
- [70].Ashcroft M; Ludwig RL; Woods DB; Copetand TD; Weber HO; MacRae EJ; Vousden KH Phosphorylation of HDM2 by Akt. Oncogene, 2002,21,1955–1962. [DOI] [PubMed] [Google Scholar]
- [71].Mayo LD; Donner DB A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc. Natl Acad. Sei. USA, 2001,98,11598–11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Meek DW; Knippschild U Posttranslational modification of MDM2. Mol Cancer Res, 2003, 1, 1017–1026. [PubMed] [Google Scholar]
- [73].Okamoto K; Li H; Jensen MR; Zhang T; Taya Y; Thoigeirsson SS; Prives C Cyclin G recruits PP2A to dephosphorylate Mdm2. Mol Cell, 2002, 9,761–771. [DOI] [PubMed] [Google Scholar]
- [74].Buschmann T; Fuchs SY; Lee CG; Pan ZQ; Ronai Z SUMO-1 modification of Mdm2 prevents its self-ubiquitination and increases Mdm2 ability to ubiquitinate p53. Cell, 2000,101,753–762. [DOI] [PubMed] [Google Scholar]
- [75].Tago K; Chiocca S; Sherr CJ Sumoylation induced by the Arf tumor suppressor: a p53-independent function. Proc. Natl Acad. Sei. USA, 2005, 102,7689–7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Ito A; Lai CH; Zhao X; Saito S; Hamilton MH; Appella E; Yao TP p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBOJ, 2001,20,1331–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Nieminen AL; Qanungo S; Schneider EA; Jiang BH; Agani FH Mdm2 and HlF-la interaction in tumor cells during hypoxia. J. Cell Physiol, 2005,204, 364–369. [DOI] [PubMed] [Google Scholar]
- [78].Ravi R; Mookeijee B; Bhujwalla ZM; Sutter CH; Artemov D; Zeng Q; Diltehay LE; Madan A; Semenza GL; Bedi A Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor-la. Genes Dev, 2000,14,34–44. [PMC free article] [PubMed] [Google Scholar]
- [79].Carroll VA; Ashcroft M Regulation of angiogenic factors by HDM2 in renal cell carcinoma. Cancer Res, 2008, 68,545–552. [DOI] [PubMed] [Google Scholar]
- [80].Rempe DA; Lelli KM; Vangeison G; Johnson RS; Federoff HJ In cultured astrocytes, p53 and MDM2 do not alter hypoxia-inducible factor-1 a function regardless of the presence of DNA damage. J. Biol Chem, 2007, 282, 16187–16201. [DOI] [PubMed] [Google Scholar]
- [81].Suzuki H; Tomida A; Tsuruo T Dephosphorylated hypoxia-inducible factor la as a mediator of p53-dependent apoptosis during hypoxia. Oncogene, 2001,20, 5779–5788. [DOI] [PubMed] [Google Scholar]
- [82].Wang P; Gao H; Ni Y; Wang B; Wu Y; Ji L; Qin L; Ma L; Pei G β-arrestin 2 functions as a G-protein-coupled receptor-activated regulator of oncoprotein Mdm2. J. Biol Chem, 2003,278,6363–6370. [DOI] [PubMed] [Google Scholar]
- [83].Gimita L; Shenoy SK; Sehat B; Vasilcanu R; Gimita A; Lefkowitz RJ; Larsson O β-Arrestin is crucial for ubiquitination and down-regulation of the insulin-like growth factor-1 receptor by acting as adaptor for the MDM2 E3 ligase. J. Biol Chem, 2005,280,24412–24419. [DOI] [PubMed] [Google Scholar]
- [84].Sdek P; Ying H; Zheng H; Margulis A; Tang X; Tian K; Xiao ZX The central acidic domain of MDM2 is critical in inhibition of retinoblastoma-mediated suppression of E2Fand cell growth. J. Biol. Chem, 2004,279,53317–53122. [DOI] [PubMed] [Google Scholar]
- [85].Hsieh JK; Chan FS; O’Connor DJ; Mittnacht S; Zhong S; Lu X RB regulates the stability and the apoptotic function of p53 via MDM2. Mol. Cell, 1999,3, 181–193. [DOI] [PubMed] [Google Scholar]
- [86].Guo CS; Degnin C; Fiddler TA; Stauffer D; Thayer MJ Regulation of MyoD activity and muscle cell differentiation by MDM2, pRb, and Spl. J. Biol. Chem, 2003,278, 22615–22622. [DOI] [PubMed] [Google Scholar]
- [87].Gaughan L; Logan IR; Neal DE; Robson CN Regulation of androgen receptor and histone deacetylase 1 by Mdm2-mcdiated ubiquitylation. Nucleic Acids Res, 2005,33,13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Malaguamera R; Vella V; Pandini G; Sanftlippo M; Pezzino V; Vigneri R; Frasca F TAp73α increases p53 tumor suppressor activity in thyroid cancer cells via the inhibition of Mdm2-mediated degradation. Mol. Cancer Res, 2008, 6,64–77. [DOI] [PubMed] [Google Scholar]
- [89].Higashitsuji H; Higashitsuji H; Itoh K; Sakurai T; Nagao T; Sumitomo Y; Masuda T; Dawson S; Shimada Y; Mayer RJ; Fujita J The oncoprotein gankyrin binds to MDM2/HDM2, enhancing ubiquitylation and degradation of p53. Cancer Cell 2005, 8,75–87. [DOI] [PubMed] [Google Scholar]
- [90].Li M; Chen D; Shiloh A; Luo J; Nikolaev AY; Qin J; Gu W Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature, 2002, 416,648–653. [DOI] [PubMed] [Google Scholar]
- [91].Cummins JM; Vogelstein B HAUSP is required for p53 destabilization. Cell Cycle 2004,3,689–692. [PubMed] [Google Scholar]
- [92].Zhang Z; Zhang R Proteasome activator PA28γ regulates p53 by enhancing its MDM2-mediated degradation. EMBOJ, 2008,27,852–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Yang JY; Zong CS; Xia W; Yamaguchi H; Ding Q; Xie X; Lang JY; Lai CC; Chang CJ; Huang WC; Huang H; Kuo HP; Lee DF; Li LY; Lien HC; Cheng X; Chang KJ; Hsiao CD; Tsai FJ; Tsai CH; Sahin AA; Muller WJ; Mills GB; Yu D; Hortobagyi GN; Hung MC ERK promotes tumorigenesis by inhibiting F0X03a via MDM2-mediated degradation. Nat. Cell Biol, 2008,10, 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Fu W; Ma Q; Chen L; Li P; Zhang M; Ramamoorthy S; Nawaz Z; Shimojima T; Wang H; Yang Y; Shen Z; Zhang Y; Zhang X; Nicosia SV; Zhang Y; Pledger JW; Chen J; Bai W MDM2 acts downstream of p53 as an E3 ligase to promote FOXO ubiquitination and degradation. J. Biol. Chem,2009, 284, 13987–14000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Gu L; Zhu N; Zhang H; Durden DL; Feng Y; Zhou M Regulation of XIAP translation and induction by MDM2 following irradiation. Cancer Cell, 2009, 15,363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Yang JY; Zong CS; Xia W; Wei Y; Ali-Seyed M; Li Z; Broglio K; Berry DA; Hung MC MDM2 promotes cell motility and invasiveness by regulating E-cadherin degradation. Mol. Cell. Biol, 2006,26,7269–7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Araki S; Eitel JA; Batuello CN; Bijangi-Vishehsaraei K; Xie XJ; Danielpour D; Pollok KE; Boothman DA; Mayo LD TGF-βl-induced expression of human Mdm2 correlates with late-stage metastatic breast cancer. J. Clin. Invest, 2010,120,290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Shvarts A; Steegenga WT; Riteco N; van Laar T; Dekker P; Bazuine M; van Ham RC; van der Houven van Oordt W; Hateboer G; van der Eb AJ; Jochemsen AG MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBOJ, 1996, 15, 5349–5357. [PMC free article] [PubMed] [Google Scholar]
- [99].Zdzalik M; Pustelny K; Kedracka-Krok S; Huben K; Pecak A; Wladyka B; Jankowski S; Dubin A; Potempa J; Dubin G Interaction of regulators Mdm2 and Mdmx with transcription factors p53, p63 and p73. Cell Cycle, 2010, 9,4584–4591. [DOI] [PubMed] [Google Scholar]
- [100].Danovi D; Meulmeester E; Pasini D; Migliorini D; Capra M; Frenk R; de Graaf P; Francoz S; Gasparini P; Gobbi A; Helin K; Pelicci PG; Jochemsen AG; Marine JC Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol. Cell. Biol, 2004,24, 5835–5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Huang L; Yan Z; Liao X; Li Y; Yang J; Wang ZG; Zuo Y; Kawai H; Shadfan M; Ganapathy S; Yuan ZM The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc. Natl. Acad. Sei. USA, 2011,108,12001–12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Prodosmo A; Giglio S; Moretti S; Mancini F; Barbi F; Avenia N; Di Conza G; SchOnemann HJ; Pistola L; Ludovini V; Sacchi A; Pontecorvi A; Puxeddu E; Moretti F Analysis of human MDM4 variants in papillaiy thyroid carcinomas reveals new potential markers of cancer properties. J Mol. Med. (Beri), 2008, 86,585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Jin Y; Zeng SX; Sun XX; Lee H; Blattner C; Xiao Z; Lu H MDMX promotes proteasomal turnover of p21 at GI and early S phases independently of, but in cooperation with, MDM2. Mol. Cell. Biol, 2008,28, 1218–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Yang HY; Wen YY; Chen CH; Lozano G; Lee MH 14-3-3 sigma positively regulates p53 and suppresses tumor growth. Mol. Cell. Biol, 2003, 23,7096–7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Brady M; Vlatkovic N; Boyd MT Regulation of p53 and MDM2 activity by MTBP. Mol. Cell Biol, 2005,25,545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Louria-Hayon I; Grossman T; Sionov RV; Alsheich O; Pandolfi PP; Haupt Y The promyelocytic leukemia protein protects p53 from Mdm2-mediated inhibition and degradation. J. Biol. Chem, 2003, 278, 33134–33141. [DOI] [PubMed] [Google Scholar]
- [107].Kim H; Kwak NJ; Lee JY; Choi BH; Lim Y; Ko YJ; Kim YH; Huh PW; Lee KH; Rha HK; Wang YP Merlin neutralizes the inhibitory effect of Mdm2 on p53. J. Biol. Chem, 2004,279,7812–7818. [DOI] [PubMed] [Google Scholar]
- [108].Linares LK; Kieman R; Triboulet R; Chable-Bessia C; Latreille D; Cuvier O; Lacroix M; Le Cam L; Coux O; Benkirane M Intrinsic ubiquitination activity of PCAF controls the stability of the oncoprotein Hdm2. Nat. Cell Biol, 2007,9,331–338. [DOI] [PubMed] [Google Scholar]
- [109].Sapountzi V; Logan IR; Robson CN Cellular functions of TIP60. Int. J. Biochem. Cell Biol, 2006,38, 1496–1509. [DOI] [PubMed] [Google Scholar]
- [110].Coutts AS; Boulahbel H; Graham A; La Thangue NB Mdm2 targets the p53 transcription cofactor JMY for degradation. EMBO Rep, 2007, 8, 84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Li L; Cohen SN Tsg101: a novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell, 1996,85,319–329. [DOI] [PubMed] [Google Scholar]
- [112].Sui G; Affar EIB; Shi Y; Brignone C; Wall NR; Yin P; Donohoe M; Luke MP; Calvo D; Grossman SR; Shi Y Yin Yang 1 is a negative regulator of p53. Cell, 2004,117, 859–872. [DOI] [PubMed] [Google Scholar]
- [113].Gimita L; Gimita A; Larsson O Mdm2-dependent ubiquitination and degradation of the insulin-like growth factor 1 receptor. Proc. Natl. Acad. Sei. USA, 2003,100, 8247–8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Sengupta S; Wasylyk B Ligand-dependent interaction of the glucocorticoid receptor with p53 enhances their degradation by Hdm2. Genes Dev, 2001, 15,2367–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Busuttil V; Drain N; McCormick L; Bemassola F; Candi E; Melino G; Green DR NF-κB inhibits T-ccll activation-induced, p73-dependent cell death by induction of MDM2. Proc. Natl. Acad. Sei. USA, 2010, 107, 18061–18066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Gu L; Findley HW; Zhou M MDM2 induces NF-kappaB/p65 expression transcriptionally through Spl-binding sites: a novel, p53-independent role of MDM2 in doxorubicin resistance in acute lymphoblastic leukemia. Blood, 2002, 99, 3367–3375. [DOI] [PubMed] [Google Scholar]
- [117].Raybum E; Zhang R; He J; Wang H MDM2 and human malignancies: expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr. Cancer Drug Targets, 2005,5,27–41. [DOI] [PubMed] [Google Scholar]
- [118].Jung CR; Lim JH; Choi Y; Kim DG; Kang KJ; Noh SM; Im DS Enigma negatively regulates p53 through MDM2 and promotes tumor cell survival in mice. J. Clin. Invest, 2010,120,4493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Ray RM; Bhattacharya S; Johnson LR Mdm2 inhibition induces apoptosis in p53 deficient human colon cancer cells by activating p73- and E2F1-mediated expression of PUMA and Siva-1. Apoptosis, 2011,16,35–44. [DOI] [PubMed] [Google Scholar]
- [120].Hsu HS; Chen HW; Kao CL; Wu ML; Li AF; Cheng T MDM2 is overexpressed and regulated by the eukaryotic translation initiation factor 4E (eIF4E) in human squamous cell carcinoma of esophagus. Ann. Surg. Oncol, 2011,18, 1469–1477. [DOI] [PubMed] [Google Scholar]
- [121].Zhou S; Gu L; He J; Zhang H; Zhou M MDM2 regulates vascular endothelial growth factor mRNA stabilization in hypoxia. Mol. Cell. Biol, 2011,31,4928–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Lo D; Lu H Nucleostemin: Another nucleolar “Twister” of the p53-MDM2 loop. Cell Cycle, 2010, 9,3227–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Oliver TG; Meylan E; Chang GP; Xue W; Burke JR; Humpton TJ; Hubbard D; Bhutkar A; Jacks T Caspase-2-mediated cleavage of Mdm2 creates a p53-induced positive feedback loop. Mol. Cell, 2011,43,57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Gu L; Zhang H; He J; Li J; Huang M; Zhou M MDM2 regulates MYCN mRNA stabilization and translation in human neuroblastoma cells. Oncogene, 2012,31, 1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Zhou X; Hao Q; Liao J; Zhang Q; Lu H Ribosomal protein S14 unties the MDM2-p53 loop upon ribosomal stress. Oncogene, 2012, doi: 10.1038/onc.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].van Dteck J; Lum JK; Teufel DP; Fersht AR SI00 proteins interact with the N-terminal domain of MDM2. FEBS Lett, 2010,584,3269–3274. [DOI] [PubMed] [Google Scholar]
- [127].Rinaldo C; Prodosmo A; Mancini F; Iacovelli S; Sacchi A; Moretti F; Soddu S MDM2-reguIated degradation of HIPK2 prevents p53Ser46 phosphorylation and DNA damage-induced apoptosis. Mol. Cell, 2007, 25, 739–750. [DOI] [PubMed] [Google Scholar]
- [128].Zhang X; Wang W; Wang H; Wang MH; Xu W; Zhang R Identification of ribosomal protein S25 (RP25)-MDM2-p53 regulatory feedback loop. Oncogene, 2012, DOI: 10.1038/onc.2012.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Raybum ER; Ezell SJ; Zhang R Recent advances in validating MDM2 as a cancer target. Anticancer Agents Med. Chem, 2009, 9, 882–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Manfredi JJ The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressors. Genes Dev, 2010,24,1580–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Bartel F; Taubert H; Harris LC Alternative and aberrant splicing of MDM2 mRNA in human cancer. Cancer Cell, 2002,2,9–15. [DOI] [PubMed] [Google Scholar]
- [132].Cheok CF; Verma CS; Baselga J; Lane DP Translating p53 into the clinic. Nat. Rev. Clin. Oncol, 2011,8,25–37. [DOI] [PubMed] [Google Scholar]
- [133].Brown CJ; Lain S; Verma CS; Fersht AR; Lane DP Awakening guardian angels: drugging the p53 pathway. Nat. Rev. Cancer, 2009, 9, 862–873. [DOI] [PubMed] [Google Scholar]
- [134].Macchiaruto A; Giacchè N; Mancini F; Puxeddu E; Moretti F; Pellicciari R Alternative strategies for targeting mouse double minute 2 activity with small molecules: novel patents on the horizon? Expert Opin. Ther. Pat, 2011,21,287–294. [DOI] [PubMed] [Google Scholar]
- [135].Di J; Zhang Y; Zheng J Reactivation of p53 by inhibiting Mdm2 E3 ligase: a novel antitumor approach. Curr. Cancer Drug Targets, 2011, 11, 987–994. [DOI] [PubMed] [Google Scholar]
- [136].Chen L; Agrawal S; Zhou W; Zhang R; Chen J Synergistic activation of p53 by inhibition of MDM2 expression and DNA damage. Proc. Natl. Acad. Sei. USA, 1998, 95, 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Tortora G; Caputo R; Damiano V; Bianco R; Chen J; Agrawal S;Bianco AR; Ciardiello F A novel MDM2 anti-sense oligonucleotide has anti-tumor activity and potentiates cytotoxic drugs acting by different mechanisms in human colon cancer. Int. J. Cancer, 2000,88, 804–809. [DOI] [PubMed] [Google Scholar]
- [138].Wang H; Prasad G; Buolamwini JK; Zhang R Antisense anticancer oligonucleotide therapeutics. Curr. Cancer Drug Targets, 2001,1, 177–196. [DOI] [PubMed] [Google Scholar]
- [139].Zhang R; Wang H; Agrawal S Novel antisense anti-MDM2 mixed-backbone oligonucleotides: proof of principle, in vitro and in vivo activities, and mechanisms. Curr. Cancer Drug Targets, 2005,5,43–49. [DOI] [PubMed] [Google Scholar]
- [140].Zhang Z; Wang H; Li M; Rayburn E; Agrawal S; Zhang R Novel MDM2 p53-independent functions identified through RNA silencing technologies. Ann. N. Y. Acad. Sei, 2005,1058,205–214. [DOI] [PubMed] [Google Scholar]
- [141].Zhang X; Gu L; Li J; Shah N; He J; Yang L; Hu Q; Zhou M Degradation of MDM2 by the interaction between berberine and DAXX leads to potent apoptosis in MDM2-overexpressing cancer cells. Cancer Res, 2010, 70: 9895–9904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Buolamwini JK; Addo J; Kamath S; Patil S; Mason D; Ores M; Small molecule antagonists of the MDM2 oncoprotein as anticancer agents. Curr. Cancer Drug Targets, 2005,5,57–68. [DOI] [PubMed] [Google Scholar]
- [143].Sun Y Targeting E3 ubiquitin ligases for cancer therapy. Cancer Biol. Ther, 2003,2,623–629. [PubMed] [Google Scholar]
- [144].Yang Y; Ludwig RL; Jensen JP; Pierre SA; Medaglia MV; Davydov IV; Safiran YJ; Oberoi P; Kenten JH; Phillips AC; Weissman AM; Vousden KH Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell, 2005,7,547–559. [DOI] [PubMed] [Google Scholar]
- [145].Arkin MR; Wells JA Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat. Rev. Drug Discov, 2004,3, 301–317. [DOI] [PubMed] [Google Scholar]
- [146].Fry DC; Vassilev LT Targeting protein-protein interactions for cancer therapy. J. Mol. Med, 2005,83,955–963. [DOI] [PubMed] [Google Scholar]
- [147].Kussie PH; Gorina S; Maréchal V; Elenbaas B; Moreau J; Levine AJ; Pavletich NP Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science, 1996,274,948–953. [DOI] [PubMed] [Google Scholar]
- [148].Lauria A; Tutone M; Ippolito M; Pantano L; Almerico AM Molecular modeling approaches in the discovery of new drugs for anti-cancer therapy: the investigation of p53-MDM2 interaction and its inhibition by small molecules. Curr. Med. Chem, 2010,17,3142–3154. [DOI] [PubMed] [Google Scholar]
- [149].Kamal A; Mohammed AA; Shaik TB p53-Mdm2 inhibitors: patent review (2009–2010). Expert Opin. Ther. Pat, 2012,22,95–105. [DOI] [PubMed] [Google Scholar]
- [150].Shangary S; Qin D; McEachem D; Liu M; Miller RS; Qiu S; Nikolovska-Coleska Z; Ding K; Wang G; Chen J; Bernard D; Zhang J; Lu Y; Gu Q; Shah RB; Pienta KJ; Ling X; Kang S; Guo M; Sun Y; Yang D; Wang S Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc. Nati Acad. Sei. USA, 2008,105,3933–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Vassilev LT; Vu BT; Graves B; Carvajal D; Podlaski F; Filipovic Z; Kong N; Kammlott U; Lukacs C; Klein C; Fotouhi N; Liu EA In vivo activation of the p53 pathway by small-molecule antagonists of mdm2.Science, 2004,303,844–848. [DOI] [PubMed] [Google Scholar]
- [152].Koblish HK; Zhao S; Franks CF; Donatelli RR; Tominovich RM; LaFrance LV; Leonard KA; Gushue JM; Parks DJ; Calvo RR;Milkiewicz KL; Marugan JJ; Raboisson P; Cummings MD; Grasberger BL; Johnson DL; Lu T; Molloy CJ; Maroney AC Benzodiazepinedione inhibitors of the Hdm2: p53 complex suppress human tumor cell proliferation in vitro and sensitize tumors to doxorubicin in vivo. Mol. Cancer Ther, 2006,5,160–169. [DOI] [PubMed] [Google Scholar]
- [153].Hardcastle IR; Liu J; Valeur E; Watson A; Ahmed SU; Blackburn TJ; Bennaceur K; Clegg W; Drummond C; Endicott JA; Golding BT; Griffin RJ; Gruber J; Haggerty K; Harrington RW; Hutton C; Kemp S; Lu X; McDonnell JM; Newell DR; Noble ME; Payne SL; Reviil CH; Riedinger C; Xu Q; Lunec J Isoindolinone inhibitors of the murine double minute 2 (MDM2)-p53 protein-protein interaction: structure-activity studies leading to improved potency. J. Med. Chem, 2011,54, 1233–1243. [DOI] [PubMed] [Google Scholar]
- [154].Yu S; Qin D; Shangary S; Chen J; Wang G; Ding K; McEachem D;Qiu S; Nikolovska-Coleska Z; Miller R; Kang S; Yang D; Wang S Potent and orally active small-molecule inhibitors of the MDM2-p53 interaction. J. Med. Chem, 2009,52,7970–7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Issaeva N; Bozko P; Enge M; Protopopova M; Verhoef LG; Masucci M; Pramanik A; Selivanova G Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat. Med,; 2004,70, 1321–1328. [DOI] [PubMed] [Google Scholar]
- [156].Jones RJ; Arts J; Orlowski RZ Inhibition of the human double minute (HDM)-2 E3 ubiquitin ligase activates different programmed cell death (PCD) pathways in models of Non-Hodgkin lymphoma (NHL) with wild type (wt) and mutant (mut) p53 In: 50th ASH Annual Meeting and Exposition; San Francisco; December 6–9,2008. [Google Scholar]
- [157].Clinical trials.gov US NIH. A phase I study to determine the safety, pharmacotogy, and pharmacodynamics of JNJ26854165 in subjects with advanced stage and/or refractory solid tumors http://www.healthetreatment.com/clinical-trial/NCT00676910/ (Accessed February 29,2012).
- [158].Clinicaltrials.gov US NIH. A study of R05045337 [RG7112] in patients with advanced solid tumors http://www.clinicaltrials.gov/ct2/show/NCT00559533 (Accessed May 24,2012).
- [159].Clinicaltrials.gov US NIH. A study of R05045337 [RG7112] in patients with hematologic neoplasms http://www.clinicaltrials.gov/ct2/show/NCT00623870 (Accessed May 24,2012).
- [160].Tabemero J; Dirix L; Schoffski P; Cervantes A; Capdevila J; Baselga J; van Beijsterveldt L; Winkler H; Kraljević S; Zhuang SH Phase I pharmacokinetic (PK) and pharmacodynamic (PD) study of HDM-2 antagonist JNJ-26854165 in patients with advanced refractory solid tumors. J. Clin. Oncol (Meeting Abstracts), 2009,27(15S), 3514. [Google Scholar]
- [161].Li JWH; Vederas JC Drug discovery and natural products: End of an era or an endless frontier? Science, 2009,325,161–165. [DOI] [PubMed] [Google Scholar]
- [162].Newman DJ; Cragg GM; Snader KM The influence of natural products upon drug discovery. Nat. Prod. Rep, 2000, 17,215–234. [DOI] [PubMed] [Google Scholar]
- [163].Cragg GM; Newman DJ; Snader KM Natural products in drug discovery and development. J. Nat. Prod, 1997,60,52–60. [DOI] [PubMed] [Google Scholar]
- [164].Newman DJ; Cragg GM; Snader KM Natural products as sources of new drugs over the period 1981–2002. J. Nat. Prod, 2003,66,1022–1037. [DOI] [PubMed] [Google Scholar]
- [165].Newman DJ; Cragg GM Natural products as sources of new drugs over the last 25 years. J. Nat. Prod, 2007, 70,461–477. [DOI] [PubMed] [Google Scholar]
- [166].Vassilev LT MDM2 inhibitors for cancer therapy. Trends Mol. Med, 2007, 13,23–31. [DOI] [PubMed] [Google Scholar]
- [167].Hu CQ; Hu YZ Small molecular inhibitors of the p53-MDM2. Curr. Med. Chem, 2008,15, 1720–1730. [DOI] [PubMed] [Google Scholar]
- [168].Allen JG; Bourbeau MP; Wohlhieter GE; Bartberger MD; Michelsen K; Hungate R; Gadwood RC; Gaston RD; Evans B; Mann LW; Matison ME; Schneider S; Huang X; Yu D; Andrews PS; Reichelt A; Long AM; Yakowec P; Yang EY; Lee TA; Oliner JD Discovery and optimization of chromenotriazolopyrimidines as potent inhibitors of the mouse double minute 2-tumor protein 53 protein-protein interaction. J. Med. Chem, 2009,52,7044–7053. [DOI] [PubMed] [Google Scholar]
- [169].Bar-Sela G; Epelbaum R; Schaffer M Curcumin as an anti-cancer agent: review of the gap between basic and clinical applications. Curr. Med. Chem, 2010,17,190–197. [DOI] [PubMed] [Google Scholar]
- [170].Li M; Zhang Z; Hill DL; Chen X; Wang H; Zhang R Genistein, a dietary isoflavone, down-rcgulates the MDM2 oncogene at both transcriptional and posttranslational levels. Cancer Res, 2005, 65, 8200–8208. [DOI] [PubMed] [Google Scholar]
- [171].Polkowski K; Popiolkiewicz J; Krzeczyftski P; Ramza J; Pucko W; Zegrocka-Stendel O; Boryski J; Skierski JS,; Mazurek AP; Grynkiewicz G Cytostatic and cytotoxic activity of synthetic genistein glycosides against human cancer cell lines. Cancer Lett, 2004,203,59–69. [DOI] [PubMed] [Google Scholar]
- [172].Popiolkiewicz J; Polkowski K; Skierski JS; Mazurek AP In vitro toxicity evaluation in the development of new anticancer drugs-genistein glycosides. Cancer Lett, 2005,229,67–75. [DOI] [PubMed] [Google Scholar]
- [173].Zhang LN; Xiao ZP; Ding H; Ge HM; Xu C; Zhu HL; Tan RX Synthesis and cytotoxic evaluation of novel 7-O-modified genistein derivatives. Chem. Biodivers, 2007,4,248–255. [DOI] [PubMed] [Google Scholar]
- [174].Fang J; Xia C; Cao Z; Zheng JZ; Reed E; Jiang BH Apigenin inhibits VEGF and HIFI expression via PI3K/AKT/p70S6KI and HDM2/p53 pathways. FASEBJ, 2005,19,342–53. [DOI] [PubMed] [Google Scholar]
- [175].Mu R; Qi Q; Gu H; Wang J; Yang Y; Rong J; Liu W; Lu N; You Q; Guo Q Involvement of p53 in oroxylin A-induced apoptosis in cancer cells. Mol. Carcinog, 2009,48,1159–1169. [DOI] [PubMed] [Google Scholar]
- [176].Nag SA; Qin JJ; Wang W; Wang MH; Wang H; Zhang R Ginsenosides as anticancer agents: In vitro and in vivo activities, structure-activity relationships, and molecular mechanisms of action. Front. Pharmacol, 2012,3,25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Zhao Y; Wang W; Han L; Rayburn ER; Hill DL; Wang H; Zhang R Isolation, structural determination, and evaluation of the biological activity of 20(S)-25-methoxyl-dammarane-3β, 12β, 20-triol [20(S)-25-OCH3-PPD], a novel natural product from Panax notoginseng. Med. Chem,2007,3,51–60. [DOI] [PubMed] [Google Scholar]
- [178].Wang W; Wang H; Rayburn ER; Zhao Y; Hill DL; Zhang R 20(S)-25-methoxyl-dammarane-3ß, 12ß, 20-triol, a novel natural product for prostate cancer therapy: activity in vitro and in vivo and mechanisms of action. Br. J. Cancer, 2008,98, 792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Wang W; Rayburn ER; Zhao Y; Wang H; Zhang R Novel ginsenosides 25-OH-PPD and 25-OCHj-PPD as experimental therapy for pancreatic cancer: anticancer activity and mechanisms of action. Cancer Lett, 2009,278,241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Wang W; Rayburn ER; Hang J; Zhao Y; Wang H; Zhang R Antilung cancer effects of novel ginsenoside 25-OCH3-PPD. Lung Cancer, 2009, 65,306–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Wang W; Zhao Y; Rayburn ER; Hill DL; Wang H; Zhang R In vitro anti-cancer activity and structure-activity relationships of natural products isolated from fruits of Panax ginseng. Cancer Chemother. Pharmacol, 2007, 59,589–601. [DOI] [PubMed] [Google Scholar]
- [182].Wang W; Rayburn ER; Hao M; Zhao Y; Hill DL; Zhang R; Wang H Experimental therapy of prostate cancer with novel natural product anti-cancer ginsenosides. Prostate, 2008,68, 809–819. [DOI] [PubMed] [Google Scholar]
- [183].Iwasa K; Moriyasu M; Yamori T; Turuo T; Lee DU; Wiegrebe W ln vitro cytotoxicity of the protoberberine-type alkaloids. J. Nat. Prod, 2001, 64,896–898. [DOI] [PubMed] [Google Scholar]
- [184].Barrows LR; Radisky DC; Copp BR; Swaffar DS; Kramer RA; Warters RL; Ireland CM Makaluvamines, marine natural products, are active anti-cancer agents and DNA topo II inhibitors. Anticancer Drug Des,1993,8, 333–347. [PubMed] [Google Scholar]
- [185].Dijoux MG; Schnabel PC; Hallock YF; Boswell JL; Johnson TR; Wilson JA; Ireland CM; van Soest R; Boyd MR; Barrows LR; Cardellina JH Antitumor activity and distribution of pyrroloiminoquinones in the sponge genus Zyzzya. Bioorg. Med. Chem 2005,13,6035–6044. [DOI] [PubMed] [Google Scholar]
- [186].Shinkre BA; Raisch KP; Fan L; Velu SE Analogs of the marine alkaloid makaluvamines: synthesis, topoisomerase II inhibition, and anticancer activity. Bioorg. Med. Chem. Lett, 2007,17,2890–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Shinkre BA; Raisch KP; Fan L; Velu SE Synthesis and antiproliferative activity of benzyl and phenethyl analogs of makaluvamines. Bioorg. Med. Chem, 2008,16,2541–2549. [DOI] [PubMed] [Google Scholar]
- [188].Wang W; Rayburn ER; Velu SE; Nadkami DH; Murugesan S; Zhang R, In vitro and in vivo anticancer activity of novel synthetic makaluvamine analogues. Clin. Cancer Res, 2009,15,3511–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Chen T; Xu Y; Guo H; Liu Y; Hu P; Yang X; Li X; Ge S; Velu SE; Nadkami DH; Wang W; Zhang R; Wang H Experimental therapy of ovarian cancer with synthetic makaluvamine analog: in vitro and in vivo anticancer activity and molecular mechanisms of action. PLoS One, 2011, 6, e20729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Wang W; Rayburn ER; Velu SE; Chen D; Nadkami DH; Murugesan S; Chen D; Zhang R A novel synthetic iminoquinone, BA-TPQ, as an anti-breast cancer agent: in vitro and in vivo activity and mechanisms of action. Breast Cancer Res. Treat, 2010, 123,321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Nadkami DH; Wang F; Wang W; Rayburn ER; Ezell SJ; Murugesan S; Velu SE; Zhang R Synthesis and in vitro anti-lung cancer activity of novel 1, 3, 4, 8-tetrahydropyrrolo [4, 3, 2-de]quinolin-8(IH)-one alkaloid analogs. Med. Chem, 2009,5,227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Kasibhatla S; Jessen KA; Maliartchouk S; Wang JY; English NM; Drewe J; Qiu L; Archer SP; Ponce AE; Sirisoma N; Jiang S; Zhang HZ; Gehlsen KR; Cai SX; Green DR; Tseng B A role for transferrin receptor in triggering apoptosis when targeted with gambogic acid. Proc. Natl. Acad. Set. USA, 2005,102, 12095–12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Gu H; Wang X; Rao S; Wang J; Zhao J; Ren FL; Mu R; Yang Y;Qi Q; Liu W; Lu N; Ling H; You Q; Guo Q Gambogic acid mediates apoptosis as a p53 inducer through down-regulation of mdm2 in wild-type p53-expressing cancer cells. Mol. Cancer Ther, 2008,7,3298–3305. [DOI] [PubMed] [Google Scholar]
- [194].Rong JJ; Hu R; Qi Q; Gu HY; Zhao Q; Wang J; Mu R; You QD;Guo QL Gambogic acid down-regulates MDM2 oncogene and induces p21(Wafl/CIPl) expression independent of p53. Cancer Lett, 2009, 284, 102–112. [DOI] [PubMed] [Google Scholar]
- [195].Han QB; Cheung S; Tai J; Qiao CF; Song JZ; Xu HX Stability and cytotoxicity of gambogic acid and its derivative, gambogoic acid. Biol. Pharm. Bull, 2005,28,2335–2337. [DOI] [PubMed] [Google Scholar]
- [196].Tao Z; Zhou Y; Lu J; Duan W; Qin Y; He X; Lin L; Ding J Caspase-8 preferentially senses the apoptosis-inducing action of NG-18, a Gambogic acid derivative, in human leukemia HL-60 cells. Cancer Biol. Ther, 2007,6,691–696. [DOI] [PubMed] [Google Scholar]
- [197].Xie H; Qin YX; Zhou YL; Tong LJ; Lin LP; Geng MY; Duan WH; Ding J GA3, a new gambogic acid derivative, exhibits potent antitumor activities in vitro via apoptosis-involved mechanisms. Acta. Pharmacol. Sin, 2009,30, 346–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Wall ME; Wani MC Camptothecin and taxol: discovery to clinic-thirteenth Bruce F. Cain Memorial Award Lecture. Cancer Res, 1995, 55,753–760. [PubMed] [Google Scholar]
- [199].Kiviharju TM; Lecane PS; Sellers RG; Peehl DM Antiproliferative and proapoptotic activities of triptolide (PG490), a natural product entering clinical trials, on primary cultures of human prostatic epithelial cells. Clin. Cancer Res, 2002,8,2666–2674. [PubMed] [Google Scholar]
- [200].Yang M; Huang J; Pan HZ; Jin J Triptolide overcomes dexamethasone resistance and enhanced PS-341-induced apoptosis via PI3k/Akt/NF-kappaB pathways in human multiple myeloma cells. Int. J. Mol. Med, 2008, 22,489–496. [PubMed] [Google Scholar]
- [201].Carter BZ; Mak DH; Schober WD; Dietrich MF; Pinilla C;Vassilev LT; Reed JC; Andreeff M Triptolide sensitizes AML cells to TRAIL-induced apoptosis via decrease of XIAP and p53-mediated increase of DR5. Blood, 2008, 111, 3742–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Li Z; Zhou ZL; Miao ZH; Lin LP; Feng HJ; Tong LJ; Ding J; Li YC Design and synthesis of novel Cl 4-hydroxyl substituted triptolide derivatives as potential selective antitumor agents. J. Med. Chem, 2009, 52, 5115–5123. [DOI] [PubMed] [Google Scholar]
- [203].Li M; Zhang Z; Hill DL; Wang H; Zhang R Curcumin, a dietary component, has anticancer, chemosensitization, and radiosensitization effects by down-regulating the MDM2 oncogene through the P13K/mTOR/ETS2 pathway. Cancer Res, 2007, 67,1988–1996. [DOI] [PubMed] [Google Scholar]
- [204].Anuchapreeda S; Tima S; Duangrat C; Limtrakul P Effect of pure curcumin, demethoxycurcumin, and bisdemethoxycurcumin on WT1 gene expression in leukemic cell lines. Cancer Chemother. Pharmacol, 2008, 62,585–594. [DOI] [PubMed] [Google Scholar]
- [205].Subramaniam D; May R; Sureban SM; Leem KB; Georgem R; Kuppusamy P; Ramanujamm RP; Hideg K; Dieckgraefe BK; Houchen CW; Anant S Diphenyl difluoroketone: a curcumin derivative with potent in vivo anticancer activity. Cancer Res, 2008, 68,1962–1969. [DOI] [PubMed] [Google Scholar]
- [206].Tian Z; An N; Zhou B; Xiao P; Kohane IS; Wu E Cytotoxic diarylheptanoid induces cell cycle arrest and apoptosis via increasing ATF3 and stabilizing p53 in SH-SY5Y cells. Cancer Chemother. Pharmacol 2009, 63,1131–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Stoll R; Renner C; Hansen S; Palme S; Klein C; Belling A; Zeslawski W; Kamionka M; Rehm T; Mühlhahn P; Schumacher R; Hesse F; Kaluza B; Voelter W; Engh RA; Holak TA Chalcone derivatives antagonize interactions between the human oncoprotein MDM2 and p53. Biochemistry, 2001,40, 336–344. [DOI] [PubMed] [Google Scholar]
- [208].Kumar SK; Hager E; Pettit C; Gurulingappa H; Davidson NE; Khan SR Design, synthesis, and evaluation of novel bononic-chalcone derivatives as antitumor agents. J. Med. Chem, 2003,46,2813–2815. [DOI] [PubMed] [Google Scholar]
- [209].Achanta G; Modzelewska A; Feng L; Khan SR; Huang P A boronic-chalcone derivative exhibits potent anticancer activity through inhibition of the proteasome. Mol. Pharmacol, 2006, 70,426–433. [DOI] [PubMed] [Google Scholar]
- [210].Tsukamoto S; Yoshida T; Hosono H; Ohta T; Yokosawa H Hexylitaconic acid: a new inhibitor of p53-HDM2 interaction isolated from a marine-derived fungus, Arthrinium sp. Bioorg. Med. Chem. Lett, 2006, 16, 69–71. [DOI] [PubMed] [Google Scholar]
- [211].Nakahashi A; Miura N; Monde K; Tsukamoto S Stereochemical studies of hexylitaconic acid, an inhibitor of p53-HDM2 interaction. Bioorg. Med. Chem. Lett, 2009,19,3027–3030. [DOI] [PubMed] [Google Scholar]
- [212].Duncan SJ; Grüschow S; Williams DH; McNicholas C; Purewal R; Hajek M; Gerlitz M; Martin S; Wrigley SK; Moore M Isolation and structure elucidation of Chlorofusin, a novel p53-MDM2 antagonist from a Fusarium sp. J. Am. Chem. Soc, 2001,123,554–560. [DOI] [PubMed] [Google Scholar]
- [213].Duncan SJ; Cooper MA; Williams DH Binding of an inhibitor of the p53/MDM2 interaction to MDM2. Chem. Commun, 2003,3,316–317. [DOI] [PubMed] [Google Scholar]
- [214].Malkinson JP; Zloh M; Kadom M; Errington R; Smith PJ; Searcey M Solid-phase synthesisof the cyclic peptide portion of chlorofusin, an inhibitor of p53-MDM2 interactions. OrgLett, 2003,5,5051–5054. [DOI] [PubMed] [Google Scholar]
- [215].Woon EC; Arcieri M; Wilderspin AF; Malkinson JP; Searcey M; Solid-phase synthesis of chlorofusin analogues. J. Org. Chem, 2007, 72, 5146–5151. [DOI] [PubMed] [Google Scholar]
- [216].Clark RC; Lee SY; Boger DL Total synthesis of chlorofusin, its seven chromophore diastereomers, and key partial structures. J. Am. Chem. Soc, 2008,130, 12355–12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Lee SY; Clark RC; Boger DL Total synthesis, stereochemical reassignment, and absotute configuration of chlorofusin. J. Am. Chem. Soc, 2007,129,9860–9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Lee SY; Boger DL Synthesis of the chlorofusin cyclic peptide. Tetrahedron, 2009,65,3281–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Clark RC; Lee SY; Searcey M; Boger DL The isolation, total synthesis and structure elucidation of chlorofusin, a natural product inhibitor of the p53-MDM2 protein-protein interaction. Nat. Prod. Rep, 2009, 26, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Clark RC; Lee SY; Hwang L; Searcey M; Boger DL Evaluation of chlorofusin, its seven chromophore diastereomers, and key analogues. Tetrahedron Lett, 2009,50,3151–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Malloy KL; Choi H; Fiorilia C; Valeriote FA; Matainaho T; Gerwick WH Hoiamide D, a marine cyanobacteria-derived inhibitor of p53/MDM2 interaction. Bioorg. Med. Chem. Lett, 2012,22,683–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Hou J; Wang D; Zhang R; Wang H Experimental therapy of hepatoma with artemisinin and its derivatives: in vitro and in vivo activity, chemosensitization, and mechanisms of action. Clin. Cancer Res, 2008,14, 5519–5530. [DOI] [PubMed] [Google Scholar]
- [223].Gopal YN; Chanchom E; Van Dyke MW Parthenolide promotes the ubiquitination of MDM2 and activates p53 cellular functions. Mol. Cancer Ther, 2009,8,552–562. [DOI] [PubMed] [Google Scholar]
- [224].Nasim S; Crooks PA Antileukemic activity of aminoparthenolide analogs. Bioorg. Med. Chem. Lett, 2008,18,3870–3873. [DOI] [PubMed] [Google Scholar]
- [225].Sasiela CA; Stewart DH; Kitagaki J; Safiran YJ; Yang Y; Weissman AM; Oberoi P; Davydov IV.; Goncharova E; Beutler JA; McMahon JB; O’Keefe BR Identification of inhibitors for MDM2 ubiquitin ligase activity from natural product extracts by a novel high-throughput electrochemiluminescent screen. J. Biomol. Screen, 2008,13,229–237. [DOI] [PubMed] [Google Scholar]
- [226].Clement JA; Kitagaki J; Yang Y; Saucedo CJ; O’Keefe BR; Weissman AM; McKee TC; McMahon JB Discovery of new pyridoacridine alkaloids from Lissoclinum cf. badium that inhibit the ubiquitin ligase activity of Hdm2 and stabilize p53. Bioorg. Med. Chem,2008,16,10022–10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Eischen CM; Lozano G p53 and MDM2: antagonists or partners in crime? Cancer Cell, 2009, 15,161–162. [DOI] [PubMed] [Google Scholar]
- [228].Bouska A; Eischen CM Mdm2 affects genome stability independent of p53. Cancer Res, 2009,69, 1697–1701. [DOI] [PubMed] [Google Scholar]
- [229].Deisenroth C; Zhang Y Ribosome biogenesis surveillance: probing the ribosomal protcin-Mdm2-p53 pathway. Oncogene, 2010,29,4253–4260. [DOI] [PubMed] [Google Scholar]
- [230].Lipkowitz S; Weissman AM RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat. Rev. Cancer, 2011, 11, 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Hori M; Shimazaki J; Inagawa S; Itabashi M; Hori M Overexpression of MDM2 oncoprotein correlates with possession of estrogen receptor alpha and lack of MDM2 mRNA splice variants in human breast cancer. Breast Cancer Res. Treat, 2002, 71,77–83. [DOI] [PubMed] [Google Scholar]
- [232].Dubs-Poterszman C; Tocquc B; Wasylyk B MDM2 transformation in the absence of p53 and abrogation of the pl07 cell-cycle arrest Oncogene, 1995, 11, 2445–2449. [PubMed] [Google Scholar]
- [233].Saha MN; Jiang H; Jayakar J; Reece D; Branch DR; Chang H MDM2 antagonist nutlin plus proteasome inhibitor velcade combination displays a synergistic anti-myeloma activity. Cancer Biol. Ther, 2010, 9, 936–944. [DOI] [PubMed] [Google Scholar]