Abstract
Purpose:
An ever-growing number of predictive models used to inform clinical decision making have included quantitative, computer-extracted imaging biomarkers, or “radiomic features.” Broadly generalizable validity of radiomics-assisted models may be impeded by concerns about reproducibility. We offer a qualitative synthesis of 41 studies that specifically investigated the repeatability and reproducibility of radiomic features, derived from a systematic review of published peer-reviewed literature.
Methods and Materials:
The PubMed electronic database was searched using combinations of the broad Haynes and Ingui filters along with a set of text words specific to cancer, radiomics (including texture analyses), reproducibility, and repeatability. This review has been reported in compliance with Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines. From each full-text article, information was extracted regarding cancer type, class of radiomic feature examined, reporting quality of key processing steps, and statistical metric used to segregate stable features.
Results:
Among 624 unique records, 41 full-text articles were subjected to review. The studies primarily addressed non-small cell lung cancer and oropharyngeal cancer. Only 7 studies addressed in detail every methodologic aspect related to image acquisition, preprocessing, and feature extraction. The repeatability and reproducibility of radiomic features are sensitive at various degrees to processing details such as image acquisition settings, image reconstruction algorithm, digital image preprocessing, and software used to extract radiomic features. First-order features were overall more reproducible than shape metrics and textural features. Entropy was consistently reported as one of the most stable first-order features. There was no emergent consensus regarding either shape metrics or textural features; however, coarseness and contrast appeared among the least reproducible.
Conclusions:
Investigations of feature repeatability and reproducibility are currently limited to a small number of cancer types. Reporting quality could be improved regarding details of feature extraction software, digital image manipulation (preprocessing), and the cutoff value used to distinguish stable features.
Summary
We offer a qualitative synthesis of 41 studies that specifically investigated the repeatability and reproducibility of radiomic features. The repeatability and reproducibility of radiomic features are sensitive at various degrees to image quality and to software used to extract radiomic features. Investigations of feature repeatability and reproducibility are currently limited to a small number of cancer types. No consensus was found regarding the most repeatable and reproducible features with respect to different settings.
Introduction
Medical imaging is widespread, and its value is firmly established in routine oncologic practice. Image-based biomarkers are used during screening, staging, stratifying, and intervention planning (surgery and radiation therapy) (1–3) and for predicting treatment outcomes (4). In current practice, a radiologist semantically annotates only a small number of radiologic features as having some clinical significance during manual assessment of the images (ie, with the unaided human eye). These few features may include Response Evaluation Criteria in Solid Tumors (5) and World Health Organization criteria (6) for treatment response; a change in the mean apparent diffusion coefficient (7); morphologic descriptors (eg, spiculation) (8); or the number of voxels exceeding a threshold for selective uptake of a radioactive tracer (9).
Tumor phenotypes, as they are manifest in medical images, may contain more information than can be readily processed by the unaided human eye. Recent studies have suggested that complex shape metrics and the nonuniform appearance of the tumor mass in images (ie, texture) also provide information about the likely outcome of the disease (3, 10–12).
Radiomics is the computerized extraction of quantitative features from medical images, beyond the level of detail accessible to an unaided human eye, with the intent to automatically label clinically significant tumor phenotypes (13). Radiomics entails large-scale batch processing (14) via high-throughput computational “pipelines” that integrate some or all of the following steps: image pre-processing, tumor segmentation, feature extraction, feature selection, machine learning—based predictive modeling, and model validation (15).
A systematic review of false discovery rates in textural analysis of medical images (16) clearly showed that optimal cutoff selection for tuning machine-learning predictive models (17) in combination with a large number of candidate image features (approximately 100) leads to an increased risk of type I error. Furthermore, related sets of image-derived features tend to be strongly correlated with each other, and this increases the risk of falsely significant associations. There are strong interclass correlations for features derived from similar matrix operations (18), and there may be correlations to the absolute tumor volume (19).
There are ways to reduce the risk of a false-positive association. First, only features with high repeatability and high reproducibility should be used for training the predictive models. “Repeatability” refers to features that remain the same when imaged multiple times in the same subject, be that a human person or a suitable phantom (14, 20). “Reproducibility” refers to features that remain the same when imaged using different equipment, different software, different image acquisition settings, or different operators (eg, other clinics), be that in the same subject or in different subjects (14, 20). Second, estimates of predictive performance in single-institution cohorts should include multiple-folded repeated cross validation to minimize the risk of overfitting (21). Last, assessment of predictive models based on radiomic features should be based on independent external validation in multi-institutional settings (22).
The purpose of this systematic review was to determine which broadly generic type of radiomic features has been shown to be repeatable and/or reproducible in peer-reviewed studies and, if applicable, what degree of repeatability and reproducibility might be achievable. It was out of the scope of the reviewers performing this study to provide any subjective evaluation concerning the goodness of a study. For example, when evaluating the quality of reporting of a study, we only retrieved objective information from the article, without assigning a score aiming at judging the overall quality of the article.
Methods and Materials
Eligibility criteria
We conducted this systematic review during March and April 2017. Reporting of this review complies with the PRISMA-P Preferred Reporting Items for Systematic Reviews and Meta-Analyses statement (23). The included articles met all of the eligibility criteria given in the subsequent paragraphs.
Report design
We included only peer-reviewed full-text reports published in journals that presented full results of repeatability and/or reproducibility tests on radiomic features. With full results, we intend all the articles that matching the inclusion criteria defined in material and methods, presented a statistical analysis of radiomics reproducibility and repeatability. Only articles that included (in their titles or abstracts) at least 1 of the search words specified in the search string were identified.
Population
With regard to the population reported in the study, we included either (1) studies of human persons diagnosed with 1 (or more) known and clearly stated primary solid tumor where medical imaging in the form of computed tomography (CT), positron emission tomography (PET), and/or magnetic resonance imaging (MRI) was used or (2) studies consisting of radiologic phantoms where medical imaging in the form of CT, PET, and/or MRI was used. We excluded studies consisting of animal subjects, studies using biological samples taken from the human body, nonclinical imaging studies, or studies in which the type of primary tumor was not objectively known.
Outcomes
The primary criterion for inclusion was an assessment of the repeatability and/or reproducibility of any number of radiomic features with respect to any equipment-, scan-, subject-, or observer-related cause. Included studies also had to report at least 1 of the following quantitative outcomes of interest: variability of radiomic features with respect to image acquisition parameters, imaging modalities examined, or effect of preprocessing steps applied to the images from which features were extracted.
Language
Only full-text reports in the English language were included in this review.
Information sources
The Cochrane Database of Systematic Reviews was screened for any previous systematic reviews addressing repeatability and/or reproducibility of radiomic features. An electronic search was conducted in PubMed (MEDLINE citations had been previously merged into the PubMed repository). For all articles for which the full text was obtained for data extraction, the bibliographic references within them were also screened for potentially eligible studies. No search was made in gray-literature sources for unpublished material or conference proceedings.
Search strategy
A search of PubMed citations was performed using the broad Haynes (24) and Ingui (25) filters in combination with the modifications proposed by Geersing et al (26) (each combined using “OR”). For the final database search, additional criteria of “cancer” (Medical Subject Headings major topic) and text terms that were each related to reproducibility, repeatability (fundamental to include test-retest studies), variability, and radiomics (including textural analyses) were also included. All PubMed search results were admitted up to and including the second week of April 2017.
Study records
Data management
Electronic full-text articles were downloaded using university library subscriptions. A review-specific SharePoint (Microsoft Corporation, USA) page was set up to handle document collection, data extraction forms, and dissemination of reviewer findings.
Selection process
Two reviewers worked independently throughout all phases of the study selection process (abstract screening, eligibility, and inclusion for full-text evaluation). They compared the titles and abstracts against the inclusion criteria. Each reported whether an abstract was eligible for evaluation in full. Disagreements were resolved by consensus. All of the articles deemed eligible were successfully downloaded.
Two reviewers independently evaluated whether the full-text reports were suitable for inclusion and synthesis. Dis-agreements were again resolved by consensus. A third reviewer was available if disagreements could not be resolved, but this option was not exercised. Reasons for excluding a specific full-text article were documented.
Data extraction: data items
We extracted information about the population used in the studies, including the sample size and type of primary tumor for human studies and the phantom details for phantom studies. The inclusion of metastatic, secondary, or synchronous tumors was noted, as was any case in which the nonprimary tumor was used in the derivation of radiomic features. The study design and image modality used were noted, including any image acquisition parameters explicitly stated in the text. We noted the total number of radiomic features tested and grouped these features according to (1) shape features (defining the 2-dimensional or 3-dimensional [3D] properties of the tumor, eg, volume or surface area); (2) first-order statistics (derived from statistical moments of the image intensity histogram); and (3) higher-order textural features (describing spatial patterns of voxel intensities) (27).
In addition, we noted the names and versions of the software used to quantitatively extract radiomic features, including whether any particular preprocessing steps were applied to the images before feature extraction. Finally, we noted the statistical methods used as a metric of repeatability and/or reproducibility of the studied features.
Outcomes and prioritizations
Primary outcome
The primary outcome of interest in this review synthesis was the degree of repeatability or reproducibility of radiomic features, along with any independent validation used to test repeatability and reproducibility at an external institution.
Secondary outcomes
The secondary outcomes were the impact of image acquisition settings on the reproducibly of features and the effect of preprocessing imaging filters applied before feature extraction.
Additional outcomes
Additional outcomes were the statistics and metrics used for reporting robustness and reliability and the investigation of the impact of different segmentation methods used to define the regions of interest (ROIs).
Risk of bias in individual studies
To assess the risk of bias in each study, 2 reviewers independently extracted detailed information from the reports in the following specific domains:
Characteristics of the cohort used to perform the study in the case of human studies or characteristics of the phantom used to perform the study in the case of phantom studies
Description of the software used to compute the features
Image acquisition parameters reported in the study
Filtering and/or image preprocessing operation(s) performed on the original scan before the radiomic features underwent computation
Segmentation method(s) used to derive an ROI
Use of either cross validation or independent validation to show that features are repeatable and/or reproducible after folding or in separate data sets
Threshold (cutoff) values used in repeatability and/or reproducibility metric(s) to segregate features
Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis classifications were not applicable here because tests of repeatability and reproducibility of radiomic features do not strictly link to diagnostic verification or predictive performance. The impact of undocumented (or inadequately reported) steps on the potential repeatability or reproducibility of features was noted. Discrepancies in data extraction between the 2 reviewers were resolved by consensus after discussion. A third reviewer was available to resolve a deadlock, but this option was not needed.
Data synthesis
The included studies were not uniform by way of reported metrics, and we could not attempt a quantitative meta-analysis of pooled metrics. A systematic qualitative synthesis is given in this publication, with details presented in text and tables to summarize our findings about the included studies.
Subgroup analyses
The summary findings on repeatability and reproducibility of features were grouped by the following: disease type (lung cancer, head and neck cancer, and other anatomic sites) or phantom study and type of imaging modality (CT, PET, and MRI).
Results
Literature search results
The Preferred Reporting Items for Systematic Reviews and Meta-Analyses flow diagram is shown in Fig. 1. The PubMed search yielded 624 abstracts for screening against our selection criteria, reduced to 623 after elimination of duplicates. The full text was retrieved for 52 abstracts deemed suitable for in-depth evaluation, including 5 that were located in the references of retrieved studies and 2 previously known studies. After full-text evaluation, 11 studies were excluded because they did not meet the aforementioned eligibility criteria. A qualitative synthesis was derived from 41 studies, of which 35 were performed in human subjects and 6 were exclusively performed on radiologic phantoms. A detailed checklist is provided in Appendix E1 (available online at https://doi.org/10.1016/j.ijrobp.2018.05.053).
Fig. 1.
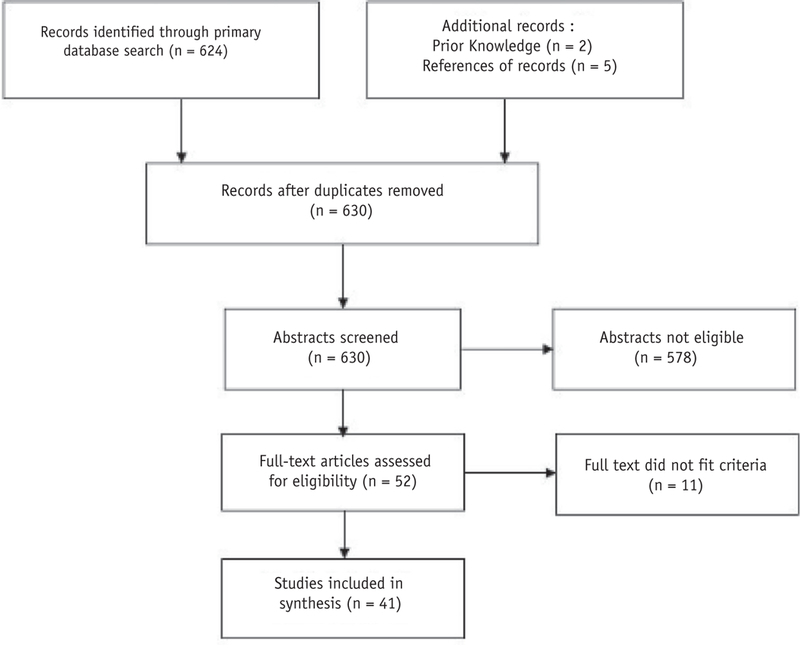
Preferred Reporting Items for Systematic Reviews and Meta-Analyses flow diagram. The primary PubMed search returned 624 records. A further 5 records were added from references in full-text articles. Two records were added owing to prior knowledge. After screening and full-text assessment, a total of 41 studies were included in the qualitative synthesis.
Human study characteristics
Table 1 summarizes the general characteristics of the human studies. The vast majority of studies addressed lung cancer (25 of 35 studies), of which 21 specifically addressed non-small cell lung cancer (NSCLC). There were 3 studies each on head and neck cancer, esophageal cancer, and rectal cancer. There was only 1 study each on breast cancer and cervical cancer. In 2 studies, multiple cancer types were combined, but details within the subgroups of cancer types were not specified (59, 60). Two studies combined features from multiple specified cancers (19, 57).
Table 1.
Summary of human studies included in analysis
| Reference | Disease | Modality investigated |
No. of patients |
Primary set made public |
Total NO. Of features |
Class of features |
Statistical analysis |
Type of study |
|---|---|---|---|---|---|---|---|---|
| Aerts et al (4), 2014 | NSCLC | CT | 52 | Yes | 440 | FO, SM, TA | ICC | Repeatability, reproducibility |
| Balagurunathan et al (28), 2014 | NSCLC | CT | 32 | Yes | 330 | FO, SM, TA | CCC | Reproducibility |
| Cheng et al (29), 2016 | NSCLC | PET | 56 | No | 12 | TA | ICC | Reproducibility |
| Coroller et al (30), 2016 | NSCLC | CT | 32 | Yes | 1603 | FO, SM, TA | ICC | Repeatability |
| Desseroit et al (31), 2017 | NSCLC | PET, CT | 73 | No | 49 | FO, TA | ICC | Repeatability |
| Desseroit et al (32), 2016 | NSCLC | CT | 32 | Yes | 34 | FO, SM, TA | Mean standard deviation | Repeatability |
| Fave et al (33), 2015 | NSCLC | CBCT | 10 | No | 68 | FO, TA | CCC | Reproducibility |
| Fave et al (34), 2015 | NSCLC | CT | 40 | No | 20 | FO, SM, TA | Spearman correlation | Reproducibility |
| Fave et al (35), 2016 | NSCLC | CT | 134 | No | 55 | FO, TA | Spearman correlation | Reproducibility |
| Fried et al (36), 2014 | NSCLC | CT | 91 | No | 30 | FO, TA | CCC | Reproducibility |
| Huynh et al (37), 2017 | NSCLC | CT | 32 | Yes | 644 | FO, SM, TA | ICC | Repeatability |
| Kalpathy-Cramer et al (38), 2016 | NSCLC | CT | 40 | Yes | 830 | FO, SM, TA | CCC | Reproducibility |
| Leijenaar et al (39), 2013 | NSCLC | PET | 34 | No | 108 | FO, SM, TA | ICC | Repeatability |
| Mackin et al (40), 2015 | NSCLC | CT | 20 | No | 10 | FO, TA | Mean standard deviation | Reproducibility |
| Oliver et al (41), 2015 | NSCLC | PET | 23 | No | 56 | FO, SM, TA | Average percentage difference | Reproducibility |
| Parmar et al (42), 2014 | NSCLC | PET | 20 | Yes | 56 | FO, SM, TA | ICC | Reproducibility |
| Van Velden et al (43), 2016 | NSCLC | PET | 11 | No | 105 | FO, SM, TA | ICC | Reproducibility |
| Yan et al (44), 2015 | NSCLC | PET | 17 | No | 64 | FO, TA | Mean standard deviation | Reproducibility |
| Yip et al (45), 2014 | NSCLC | PET | 26 | No | 4 | TA | Relative difference | Reproducibility |
| Zhao et al (46), 2016 | NSCLC | CT | 32 | No | 89 | FO, SM, TA | CCC | Repeatability, reproducibility |
| Grootjans et al (47), 2016 | Lung cancer | PET | 60 | No | 4 | TA | Mean standard deviation | Reproducibility |
| Koo et al (48), 2017 | Lung cancer | CT | 194 | No | 9 | FO, SM | ICC | Reproducibility |
| Lasnon et al (49), 2016 | Lung cancer | PET | 60 | No | 5 | TA | Mean standard deviation | Reproducibility |
| Bagher-Ebadian et al (50), 2017 | Oropharyngeal cancer | CT, CBCT | 18 | No | 165 | FO, TA | Mean absolute percentage change | Reproducibility |
| Bogowicz et al (51), 2016 | Oropharyngeal cancer | CT | 22 | No | 315 | FO, TA | ICC | Reproducibility |
| Lu et al (52), 2016 | Oropharyngeal cancer | PET | 40 | No | 88 | FO, SM, TA | ICC | Reproducibility |
| Doumou et al (53), 2015 | Esophageal cancer | PET | 64 | No | 57 | TA | Spearman correlation | Reproducibility |
| Hatt et al (54), 2013 | Esophageal cancer | PET | 50 | No | 10 | FO, TA | Mean standard deviation | Reproducibility |
| Tixier et al (55), 2012 | Esophageal cancer | PET | 16 | No | 25 | FO, TA | ICC | Reproducibility |
| Hu et al (56), 2016 | Rectal cancer | CT | 40 | No | 775 | TA | ICC | Repeatability |
| Orlhac et al (19), 2014 | Rectal cancer, NSCLC, breast cancer | PET | 28, 24, 54 | No | 41 | FO, TA | Spearman correlation | Reproducibility |
| Van Timmeren et al (57), 2016 | Rectal cancer, lung cancer | CT | 40, 40 | No | 542 | FO, SM, TA | CCC | Repeatability |
| Guan et al (58), 2016 | Cervical cancer | MR | 51 | No | 8 | FO, TA | ICC | Reproducibility |
| Galavis et al (59), 2010 | Various solid tumors | PET | 20 | No | 50 | FO, TA | Average percentage difference | Reproducibility |
| Hatt et al (60), 2015 | Various solid tumors | PET | 555 | No | TA | Spearman correlation | Reproducibility | |
Abbreviations: CBCT = cone beam computed tomography; CCC = concordance correlation coefficient; CT = computed tomography; FO = first order; ICC = intraclass correlation coefficient; MR = magnetic resonance; NSCLC = non-small cell lung cancer; PET = positron emission to-mography; SM = shape metric; TA = textural analysis.
The number of patients reported in the retrieved studies ranged from 10 (33) to 555 (54), and only 1 study was prospective (56). Few studies (7 of 41) specifically referred to a publicly available image set.
The imaging modalities mentioned in the human lung studies were PET (17 of 35), CT (17 of 35), MRI (1 of 35), and cone beam CT (CBCT) (2 of 35). Two studies used multiple imaging modalities. Six studies exclusively investigated feature repeatability; all others examined either reproducibility alone or both reproducibility and repeatability.
The number of investigated radiomic features in the studies ranged from 4 (45) to 830 (38). The latter was a multi-institutional study, so it was unclear whether the number included repeated instances of some of the features. All the studies included textural analysis; the majority (28 of 35) also evaluated first-order features, but less than half (15 of 35) evaluated shape metrics. Fourteen studies investigated all categories of features.
Phantom study characteristics
Table 2 shows the main characteristics of 6 studies exclusively concerning radiologic phantoms. Among these, CT was the most common image modality (5 of 6), and PET was investigated in only 1 study. We did not locate any phantom study of repeatable and/or reproducible features from MRI. All of these studies investigated feature reproducibility, and only 1 phantom study was prospective (65).
Table 2.
Summary of pure phantom studies included in analysis
| Reference | Phantom used | Modality investigated |
Primary set made public |
Total no. of features |
Class of features |
Statistical analysis |
Type of study |
|---|---|---|---|---|---|---|---|
| Buch et al (61), 2017 | Nonanatomic in-house phantom | CT | No | 42 | FO, TA | Student t test | Reproducibility |
| Forgacs et al (62), 2016 | NEMA-IQ phantom | PET | No | 26 | TA | Coefficient of variation | Reproducibility |
| Kim et al (63), 2015 | AAPM CT performance phantom model 76-410-4130 | CT | No | 5 | TA | Unclear | Reproducibility |
| Lo et al (64), 2016 | Water phantom | CT | No | 26 | FO, TA | Mean standard deviation | Reproducibility |
| Shafiq-Ul-Hassan et al (65), 2017 | Credence Cartridge Radiomics phantom | CT | No | 213 | FO, SM, TA | Coefficient of variation | Reproducibility |
| Zhao et al (66), 2014 | Anthropomorphic thorax phantom | CT | No | 15 | SM, TA | Multilinear regression | Reproducibility |
Abbreviations: AAPM = American Association of Physicists in Medicine; CT = computed tomography; FO = first order; NEMA-IQ = National Electrical Manufacturers Association-Image Quality; PET = positron emission tomography; SM = shape metric; TA = textural analysis.
The number of investigated radiomic features in the phantom studies ranged from 5 (63) to 213 (65). All studies included textural analysis, 3 evaluated first-order features, and 2 evaluated shape metrics. Only 1 study investigated all categories of features (65).
Quality of reporting in included studies
Human studies
Table 3 gives a summary of methodology and reporting quality for human studies. In general, methodologic aspects were adequately documented. However, only 7 of 35 studies reported detailed information in every one of the aforementioned quality domains (28, 33, 42, 51–53, 56). In 3 quality aspects, the overall standard of reporting was lower: (1) providing details of the software implementation to extract radiomic features, (2) providing details pertaining to image preprocessing before extracting radiomic features, and (3) stating the cutoff value for discriminating a subset of repeatable and/or reproducible features.
Table 3.
Quality of reporting analysis for human studies
| Reference | Cohort details stated |
Software details stated |
Image acquisition settings provided |
Preprocessing details provided |
Segmentation details provided |
Metric thresholds stated |
|---|---|---|---|---|---|---|
| Aerts et al (4), 2014 | Yes | Yes | Yes | No | Yes | No |
| Bagher-Ebadian et al (50), 2017 | Yes | Yes | Yes | Yes | Yes | No |
| Balagurunathan et al (28), 2014 | Yes | Yes | Yes | Yes | Yes | Yes |
| Bogowicz et al (51), 2016 | Yes | Yes | Yes | Yes | Yes | Yes |
| Cheng et al (29), 2016 | Yes | Yes | Yes | No | Yes | No |
| Coroller et al (30), 2016 | Yes | No | Yes | Yes | Yes | Yes |
| Desseroit et al (31), 2017 | Yes | No | No | Yes | Yes | No |
| Desseroit et al (32), 2016 | Yes | No | Yes | No | No | No |
| Doumou et al (53), 2015 | Yes | Yes | Yes | Yes | Yes | Yes |
| Fave et al (33), 2015 | Yes | Yes | Yes | Yes | Yes | Yes |
| Fave et al (34), 2015 | Yes | Yes | Yes | Yes | Yes | No |
| Fave et al (35), 2016 | Yes | Yes | Yes | Yes | No | Yes |
| Fried et al (36), 2014 | Yes | Yes | Yes | Yes | Yes | Yes |
| Galavis et al (59), 2010 | No | Yes | Yes | Yes | Yes | Yes |
| Guan et al (58), 2016 | Yes | No | Yes | Yes | Yes | Yes |
| Grootjans et al (47), 2016 | Yes | No | Yes | Yes | Yes | No |
| Hatt et al (54), 2013 | Yes | No | Yes | Yes | Yes | No |
| Hatt et al (60), 2015 | Yes | No | Yes | Yes | Yes | No |
| Hu et al (56), 2016 | Yes | Yes | Yes | Yes | Yes | Yes |
| Huynh et al (37), 2017 | Yes | Yes | Yes | No | No | Yes |
| Kalpathy-Cramer et al (38), 2016 | Yes | Yes | Yes | No | Yes | Yes |
| Koo et al (48), 2017 | Yes | No | Yes | Yes | Yes | No |
| Lasnon et al (49), 2016 | Yes | No | Yes | Yes | Yes | No |
| Leijenaar et al (39), 2013 | Yes | Yes | Yes | No | Yes | Yes |
| Lu et al (52), 2016 | Yes | Yes | Yes | Yes | Yes | Yes |
| Mackin et al (40), 2015 | Yes | Yes | Yes | Yes | No | No |
| Oliver et al (41), 2015 | Yes | No | Yes | No | Yes | Yes |
| Orlhac et al (19), 2014 | Yes | No | Yes | Yes | Yes | No |
| Parmar et al (42), 2014 | Yes | Yes | Yes | Yes | Yes | Yes |
| Tixier et al (55), 2012 | Yes | No | Yes | Yes | Yes | No |
| Van Velden et al (43), 2016 | Yes | No | Yes | Yes | Yes | Yes |
| Van Timmeren et al (57), 2016 | Yes | No | Yes | Yes | No | Yes |
| Yan et al (44), 2015 | Yes | No | Yes | Yes | Yes | Yes |
| Yip et al (45), 2014 | Yes | Yes | Yes | Yes | No | No |
| Zhao et al (46), 2016 | Yes | No | Yes | No | Yes | Yes |
Each cell indicates whether it was possible to evince sufficient information from the text to re-create the experiment (yes) or not (no).
One study did not provide detailed information about the disease groups used in the analysis, apart from stating that different types of solid cancers were included (59). The practice of pooling heterogeneous tumors is questionable because there is no a priori reason to assume that any arbitrary feature that may be stable in one disease site will also prove to be stable in others.
Software details (application framework used for analysis, programming language, and version) were not reported in detail in 16 studies. Standards for radiomic features have not yet been universally adopted; therefore, software should be described because differences due to feature extraction are likely to influence the apparent stability of features.
All but 2 studies (31, 32) provided detailed tables describing image acquisition settings including information about scanners (manufacturer, model, reconstruction package, and software version) and scan protocols. Eight studies lacked detailed descriptions regarding preprocessing steps (if any) applied to the original images (4, 29, 37–39, 41, 46). Digital image manipulations (eg, voxel size resampling, de-noising, and sharpening) are known to drastically alter the extracted values, and this is likely to hamper reproducibility across data sets.
Six studies did not provide sufficient information regarding the segmentation procedures to define an ROI (32, 35, 37, 40, 45, 57). Differences in segmentation methods are likely to bias the stability of shape metrics and perhaps textural and first-order features.
Fifteen studies did not document the cutoff value used in their statistical metrics to discriminate between reproducible and irreproducible features. One of these selected only the top-ranking feature from each of 4 feature groups (first order, shape metric, texture, and wavelet filtered) (4). The subset of stable radiomic features selected in a given study obviously depends on arbitrary threshold values of the repeatability or reproducibility metric; therefore, the cutoff criterion should be clearly stated.
Two studies validated feature stability in texture phantoms combined with publicly available clinical images (33, 40). One study assessed feature reproducibility across 7 institutions (38).
Seven studies made their primary data set of clinical images publicly available to other researchers (4, 28, 30, 32, 37, 38, 42), and some of these studies used the same publicly available data set.
Phantom studies
Table 4 provides an overview of methodologic aspects and reporting quality for the phantom studies. All studies reported information about the phantom used in the analysis. All provided detailed tables describing image acquisition settings, including information about scanners (manufacturer, model, reconstruction package, and software version) and scan protocols. However, no studies reported detailed information in every one of the aforementioned quality domains. In 2 quality aspects, the overall standard of reporting was lower: (1) providing details of the software implementation to extract radiomic features and (2) stating the cutoff value for discriminating within a subset of reproducible features.
Table 4.
Quality of reporting analysis for phantom studies
| Reference | Phantom details stated |
Software details stated |
Image acquisition settings provided |
Preprocessing details provided |
Segmentation details provided |
Metric thresholds stated |
|---|---|---|---|---|---|---|
| Buch et al (61), 2017 | Yes | Yes | Yes | Yes | Yes | No |
| Forgacs et al (62), 2016 | Yes | No | Yes | No | No | Yes |
| Kim et al (63), 2015 | Yes | Yes | Yes | Yes | Yes | No |
| Lo et al (64), 2016 | Yes | No | Yes | Yes | Yes | No |
| Shafiq-ul-Hassan et al (65), 2017 | Yes | No | Yes | Yes | Yes | No |
| Zhao et al (66), 2014 | Yes | No | Yes | Yes | Yes | No |
Each cell indicates whether it was possible to evince sufficient information from the text to re-create the experiment (yes) or not (no).
Four studies made use of commercially available phantoms originally designed for scanner calibration or image quality checks, whereas 2 studies used an in-house texture phantom (61, 65). Software details (application framework used for analysis, programming language, and version) were not reported in the majority of studies. Five studies described their in-house software as based on MATLAB (The MathWorks) (61, 62, 64–66), and Kim et al (63) developed a plug-in for the open-source software ImageJ (National Institutes of Health), but none of these studies made their code accessible.
Only Forgacs et al (62) lacked a detailed description regarding preprocessing steps (if any) that were applied to the original images. One study did not provide sufficient information regarding the segmentation procedures to define an ROI (62). Three studies relied on manual segmentations (61, 63, 65), and the remainder used semi-automated segmentations while also specifying the algorithms used.
Five studies did not document the cutoff values used in their statistical metrics to discriminate between reproducible and irreproducible features (61, 63, 64, 65, 66). The statistical analysis was unclear in the text in the study by Kim et al (63).
Radiomic features according to cancer diagnosis
Lung cancers
The imaging modalities among lung cancer studies were CT (14 studies), PET (11 studies), and CBCT (1 study). Desseroit et al (31) investigated PET and CT at the same time. All of the studies investigating PET acquired these images using a combined PET-CT scanner. Twenty-one studies used NSCLC data sets, and 4 studies used a combination of different lung cancers (47–49, 57).
Three studies evaluated the reproducibility of radiomic features with respect to multiple manual segmentations in the same patient (interobserver sensitivity) by use of PET (4, 39, 43). Each showed that interobserver differences in delineations affected feature reproducibility to some degree. Interobserver differences were amplified in textural features. Leijenaar et al (39) found that features with high test-retest repeatability were also less affected by interobserver differences. Parmar et al (42) studied interobserver variability with respect to manual versus semiautomated segmentation (3D Slicer) and concluded that semiautomated methods improved feature reproducibility in PET. Orlhac et al (19) studied textural feature reproducibility with respect to 2 different semiautomated segmentation algorithms: an adaptive method (67) and a more conventional thresholding method based on 40% of the maximum standardized uptake value. Homogeneity, contrast, dissimilarity, and coarseness were found to be the most reproducible features.
The only multicenter CT study found that among shape metrics, 3 variants—local shape descriptors, global shape descriptors, and textural features—had the highest variation with respect to segmentations, whereas size measures had the least variation resulting from segmentation (38). First-order features were highly reproducible across participating centers, and there were strong internal correlations within each class of features.
Four studies examined the impact of different PET image reconstruction algorithms or image processing filters (19, 43, 44, 49). Grid size had a larger impact on feature reproducibility than did simple Gaussian filters applied inside the image reconstruction algorithms; the latter affected reproducibility in only shape and textural features. Gray-level resampling sensitively affects textural feature reproducibility, whereas first-order features are less affected. Differences in reconstruction algorithms strongly affect feature reproducibility, with the exception of first-order entropy. Entropy was reproducible for both image preprocessing and several reconstruction algorithms.
Three studies compared features using free-breathing PET versus respiratory-gated PET to evaluate the impact of motion on reproducibility but showed conflicting results. In the study by Oliver et al (41), spatial blurring effects due to respiratory motion and intrinsic noise during acquisition were major factors leading to irreproducibility. Similar results were seen when textural features on 3D versus 4-dimensional (4D) PET were compared (45). The latter study concluded that 4D imaging reduced motion artifacts, producing less blurred images and potentially more reproducible textural features. However, in the study by Grootjans et al (47), the differences between features derived from 3D and 4D imaging were not statistically significant.
Zhao et al (46) evaluated the combined effect of 3 different CT slice thicknesses and 2 different CT reconstruction algorithms. Many features that had been repeatable under test-retest conditions became irreproducible with respect to altered slice thickness and image reconstruction settings, with first-order features and shape metrics being less sensitive than textural features.
Fave et al (35) tested the effect of different image preprocessing filters, such as bit-depth resampling and smoothing filters, on CT radiomic features. Correlation to tumor volume and use of preprocessing filters increased the chance of features being significant on univariate analysis against outcome, but whether these retain their predictive value in an independent validation set remains unclear.
Three studies focused exclusively on repeatability using the same test-retest images (31, 37, 57). They consistently found shape metrics and first-order features to be highly repeatable, but there was no consensus on repeatable textural features.
Two studies investigated feature reproducibility across different scanning equipment using a specially constructed texture phantom: Fave et al (33) using CBCT and Mackin et al (40) using CT. Both investigations went on to validate their phantom results using clinical images. Feature reproducibility using CBCT was adversely affected by motion and scattered radiation, whereas interscanner CT differences were found to be of the same magnitude as interpatient feature differences.
Head and neck cancers
Three studies were concerned with head and neck cancers; all primary tumors were located in the oropharynx. Modalities investigated were CT (51), CT and CBCT (50), and PET (52). Each of these investigated some aspect of image resampling filters or other preprocessing regarding feature reproducibility.
Bagher-Ebadian et al (50) applied different smoothing, sharpening, and noise filters to CBCT and CT images and found that feature reproducibility in both modalities was most strongly affected by high-pass filters and logarithmic filters. Smoothing filters and Gaussian noise kernels had a similar but smaller impact on reproducibility. The authors found no major differences in reproducibility between CT and CBCT.
The effect of gray-level discretization was discussed in 2 studies: those by Bogowicz et al (51) using CT and Lu et al (52) using PET. The first found that bin size strongly affected reproducibility on perfusion CT, but the second found a qualitatively similar though less severe impact on reproducibility in PET.
Lu et al (52) also compared different PET segmentation methods (manual, semiautomated, and fully automated) and found that more than half of the radiomic features were reproducible. In addition, the sensitivity of features due to segmentation differences was less than that due to voxel dimension resampling.
Esophageal cancers
Three studies were concerned with esophageal cancer. All 3 investigated PET modalities. Tixier et al (55) investigated the effect of different PET reconstruction algorithms in esophageal cancer. The most reproducible tumor heterogeneity markers were entropy, homogeneity, and dissimilarity (for local characterization) and variability in the size and intensity of homogeneous tumor regions (for regional characterization). The other 2 studies investigated how different thresholding-based semiautomated segmentation algorithms affect feature reproducibility. The impact was less marked with first-order features than with textural features. Entropy was the most reproducible first-order feature, and homogeneity was the most reproducible textural feature. Segmentation affected reproducibility more than either smoothing or filtering.
Rectal cancers
Three studies were concerned with rectal cancers. Modalities investigated included CT (2 studies) and PET (1 study). Orlhac et al (19) studied textural feature reproducibility using PET with respect to different segmentation algorithms and gray-level resampling. Only a few features (homogeneity, contrast, dissimilarity, and coarseness) were found to be highly reproducible with respect to segmentation and resampling. Two studies investigated feature repeatability using CT (56, 57). Shape features were again found to be the most repeatable, and higher-order textural features were the least reproducible. Normalizing the extracted values by ROI volume generally improved the overall repeatability of features.
Other cancers
Studies were limited for other cancers. Guan et al (58) investigated feature reproducibility in the apparent diffusion coefficient from MRI of cervical cancer with respect to interobserver and intraobserver variability. All entropy measures were highly reproducible independent of observer effects. Orlhac et al (19) investigated textural feature reproducibility using PET in breast cancer. Only a few features (contrast, coarseness, and high gray-level run emphasis) were reproducible with respect to the number of gray levels used for resampling.
Radiomic features according to imaging modality
Positron emission tomography
PET was the second most common imaging modality overall and the most common in lung cancer. First-order statistics derived from a standard uptake value (SUV) histogram, such as mean SUV and maximum SUV, were consistently among the most repeatable and reproducible. Interclass correlation coefficients of these features were consistently higher than 0.95. First-order PET features were generally robust with respect to segmentation, but textural features consistently showed greater sensitivity to segmentation differences (4, 19, 39, 42, 43, 53). The choice of image reconstruction algorithm had a greater effect on reproducibility of shape metrics and textural features relative to first-order features (43, 44, 49).
Computed tomography
Studies using CT were most common in head and neck cancer (2 studies) (50, 51), and CT was the second most common modality in lung cancer (14 studies). First-order and shape CT features were generally more repeatable than textural features (32, 56, 57). Slice thickness resampling and different reconstruction algorithms strongly degraded feature reproducibility (35, 46, 50, 51). The magnitude of degradation was greater for textural features than for first-order features.
Cone beam CT
CBCT was used in 1 NSCLC study (33) and 1 oropharyngeal cancer study (50). Radiomic feature reproducibility on CBCT appeared to be adversely affected by scattered X rays and specifics of the imaging device. Low-amplitude noise and smoothing did not appear to affect the correlation of CBCT features to planning CT.
Radiomic features according to phantom studies
All the studies that investigated reproducibility on CT agreed that voxel size resampling strongly affected feature reproducibility. Zhao et al (66) demonstrated substantial differences in reproducibility when comparing 1.25- and 5-mm slices. Volume, homogeneity, and energy (gray-level co-occurrence matrix) were more reproducible for the finer slice thickness. This study recommended using images with a slice thickness between 1 and 2.5 mm for radiomic analysis. Studies confirmed that other CT acquisition parameters, such as tube voltage or tube current, had no influence on feature reproducibility (61, 62).
Predictive or prognostic power of reproducible or repeatable features
Of the 35 articles investigating feature reproducibility and repeatability in human studies, only 11 also addressed the prognostic or predictive value of computed features (Table E1, available online at https://doi.org/10.1016/jAjrobp.2018.05.053). Ten studies used NSCLC data sets, and only 1 study was based on esophageal cancers. Five studies investigated clinical outcome and patients’ overall survival, 2 investigated the role of features in stratifying patients according to poor or good prognosis (based on mean overall survival), 3 investigated the pathologic response to treatment, and 1 investigated tumor recurrence. All studies agreed that models including quantitative imaging features have better performance than models including only clinical features. The majority of the studies found textural analysis features to be predictive or prognostic. Unfortunately, there is no consensus on most predictive textural analysis features. In addition, some studies found some first-order features to be predictive, but unfortunately, it was not possible to find a consensus. We suggest that authors clearly document the procedure adopted for feature selection for their models, possibly by making use of workflow figures.
Methodologic issues identified in review
Accessibility of software for feature extraction and of image collections
The included studies used a wide range of software to process images and extract features. Fourteen studies specifically identified MATLAB as the framework for their feature extraction algorithms (4, 28, 29, 36, 37, 39, 42, 45, 50, 52, 53, 56, 59, 61). Software in the studies by Bogowicz et al (51) and Kim et al (63) were based on in-house code written for Python and ImageJ, respectively. Twenty studies did not report any details about the software used. Only 1 of the aforementioned studies has made its source code available in a GitHub repository (52).
Among MATLAB users, only Aerts et al (4) and Balagurunathan et al (28) have made their image sets publicly accessible online. Kalpathy-Cramer et al (38) and Oliver et al (41) also used in-house created software, but neither provided additional details or made the software publicly accessible. Kalpathy-Cramer et al (38) provided open access to images and structure sets (for 40 patients and 1 phantom) via The Cancer Imaging Archive (TCIA). Four studies used the IBEX open-source radiomics package (68), developed by MD Anderson Cancer Center, but their image collections are not publicly accessible online (33, 34, 35, 40).
Among phantom studies, only 2 reported the software used to extract radiomic features. Buch et al (61) used MATLAB as the framework application for their feature extraction algorithms, and Kim et al (63) developed a dedicated plug-in for ImageJ. However, none of the phantom studies had publicly released their image sets.
It would be difficult to compare, for consistency and standardization, the radiomic features extracted by different software implementations if values for a canonical set of features were not openly accessible. Furthermore, feature stability and predictive performance of radiomic features cannot easily be externally validated unless other researchers have access to either the extraction software or the medical images (or both).
Heterogeneity in statistical metric and cutoff values
The human subject studies in this synthesis were highly heterogeneous regarding statistical metrics for repeatability and/ or reproducibility. The metrics encountered were the intra-class correlation coefficient (ICC) in 14 studies, concordance correlation coefficient (CCC) in 7 studies, Spearman rank correlation in 5 studies, and various descriptive measures of difference among the remaining 9 studies. Some studies reported more than a single metric. However, the specific cutoff values used to segregate stable from unstable features were not always stated. When stated, the threshold values were highly study dependent. This led to differences in the individual features that were deemed repeatable or reproducible, and there was no universal consensus.
The ICC metric (69) is appropriate where one expects strong correlation within a given class but weak correlation between classes, and it was most commonly reported in reproducibility experiments. Five of these ICC-based studies failed to report the threshold value used to consider a feature as reproducible. The others defined a feature as highly reproducible if ICC was >0.9 (30, 43, 51); if ICC was >0.81 (58); if ICC was >0.8 (37, 39); and finally, if ICC was ≥0.8 (42, 52).
The CCC metric (70) assumed each observation was independent and was commonly reported in both repeatability and reproducibility studies. In the studies by Kalpathy-Cramer et al (38) and Zhao et al (46), the cutoff was set at CCC ≥0.75. Other reported cutoffs were CCC ≥0.8 (33, 56), CCC >0.85 (57), and CCC >0.9 (28, 36). Spearman rank correlation (71) measures the ordinal correlation between features in 2 experiments and was reported in 5 studies.
Human studies also tended to stratify features into ordinal groups (eg, poor, medium, or high reproducibility or repeatability) according to the statistical metric. No study made available its calculated metrics at the level of the individual feature. We did not attempt a meta-analysis of summary statistics in this review.
The phantom studies also used diverging statistical metrics. In the study by Kim et al (63), the metric was ambiguous. Two studies used the coefficient of variation (62, 65); one study used the mean standard deviation (64); one study used a multilinear regression method (66); and Buch et al (61) used a t test. Only Forgacs et al (62) reported the cutoff used to select reproducible features. For phantom studies, lack of consensus also excluded quantitative meta-analysis of the results.
Reporting of digital image manipulations before feature extraction
Radiomic feature values appeared to be sensitive to preprocessing filters applied to the original image. There was some consensus that first-order features were not as sensitive to image preprocessing as were textural features. Because the latter class of features is highly sensitive to perturbations in local intensity distribution and short-range correlations, the use of prefilters might have enhanced certain details and eliminated information from others.
However, the aforementioned preprocessing steps (if any) were embedded within each software implementation and were seldom explicitly documented. Discrepancies between studies using the same image modalities and the same software may be partly due to undocumented differences in preprocessing, but it would be impossible to rule out differences resulting from image reconstruction algorithm or image acquisition settings.
Qualitative synthesis
Lung studies generally agreed that ROI segmentation affects the reproducibility of radiomic features for both PET and CT modalities, especially among the shape metrics and textural features. Image reconstruction algorithms revealed a difference between filtration and voxel sampling. The former had more impact on reproducibility of textural features, but the latter reduced the reproducibility of all features. Respiratory motion appears to have had a significant adverse impact on reproducibility of PET and CBCT features. Feature values were correlated to ROI volume in some software implementations, which may lead to a confounding association with certain outcomes.
In general, the head and neck cancer studies agreed that either modifying voxel size or applying intensity discretization influenced feature reproducibility for both CT and PET, but PET seemed overall less sensitive with respect to differences in segmentation. This review did not find any studies addressing differences in image acquisition settings, reconstruction algorithms, or scanners for head and neck cancers.
In PET human subject studies, first-order entropy was one of the most stable features across multiple settings. There were mixed findings for reproducibility of skewness and kurtosis. Shape metrics were also reproducible using PET but were less reproducible using CT, likely because of the manual delineation sensitivity of the latter. Coarseness and contrast appeared to be least stable among the textural features. There was no overall pattern for stable textural features, nor were any significant differences noted due to different isotopes (52) (ie, 18F and 11C).
First-order entropy emerged as among the most consistently reproducible features on CT for both oropharyngeal and lung cancers. Single-institution studies concluded that CT shape metrics were highly reproducible, but the only multi-institutional study concluded that shape descriptors (ie, flatness and sphericity) and textural features were the least reproducible (38). Kalpathy-Cramer et al (38) also showed that first-order CT features were highly reproducible across participating centers, even for skewness and kurtosis. There was consensus that certain texture features, such as coarseness and contrast, were poorly reproducible. No emergent pattern regarding reproducible PET texture features was found.
No overall trend emerged regarding repeatable and reproducible CBCT textural features. Among first-order features, entropy was one of the most stable features, whereas kurtosis was the least stable.
In general, all phantom studies on CT consistently reported that first-order features such as histogram mean and entropy were the most reproducible features. Similar results for entropy were observed on PET radiomic analysis (62) when examining reproducibility with respect to different acquisition time intervals and reconstruction settings. The aforementioned qualitative synthesis across all included studies has been summarized in Fig. 2, indicating which process steps are most likely, probable, or least likely to affect the repeatability and reproducibility of radiomic features.
Fig. 2.
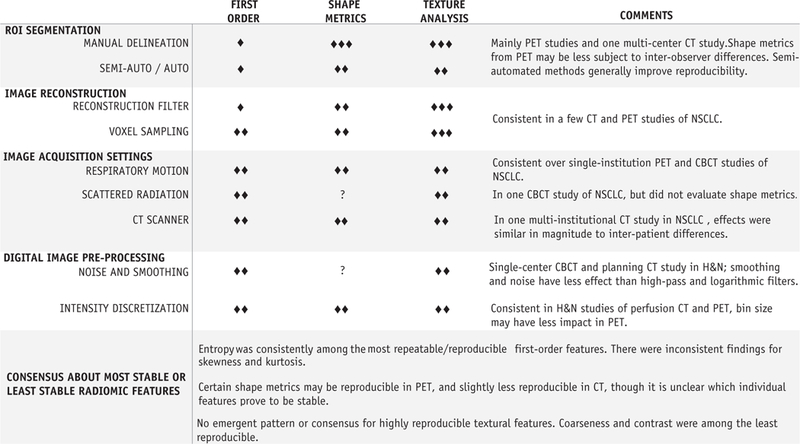
Qualitative synthesis of radiomic feature classes, indicating processing steps that are either highly likely (3 diamonds), probable (2 diamonds), or less likely (1 diamond) to exert an adverse effect on repeatability and reproducibility for each class of radiomic features. Feature classes for which no information was available are marked as unknown (question mark). Abbreviations: CBCT = cone beam computed tomography; CT = computed tomography; H&N = head and neck cancer; NSCLC = non-small cell lung cancer; PET = positron emission tomography; ROI = region of interest.
Discussion
The total number of published predictive modeling studies using image-based quantitative features has been rapidly rising, but global consensus about features that are repeatable and reproducible has not yet emerged. Lack of unified synthesis could potentially undermine future discussions about clinical applicability and prospective multi-institutional external-validation trials. The primary objective of this review was to identify radiomic features that were shown to be repeatable and reproducible through an electronic search of peer-reviewed journal publications. We also evaluated the methodologic details provided in each of the studies. Summaries of our findings have been presented in tables.
We located a number of general reviews focusing on the process and challenges of radiomic studies (15, 72). The previous work has drawn particular attention to the lack of standardization (73) and need for calibration of imaging settings. At the time of this writing, there has been no systematic review focusing on repeatability and/or reproducibility studies of radiomic features.
General recommendations for radiomic research
To homogenize radiomic reproducibility and repeatability studies, we suggest that the community perform bench-marking studies on common, shared, and publicly available data sets. In particular, this concept has already been proposed within the Image Biomarker Standardization Initiative, where different institutions computed features on a common data set. However, to expand this effort, we have been working on (1) providing users with a common repository with shared data sets for feature benchmarking; (2) providing a computational infrastructure, which directly connects to the repository; and (3) suggesting a standardized way of reporting and collecting computational results.
With regard to point 1, we believe that common data sets should include both phantom and human studies. Because features could be dependent on several acquisition parameters (eg, slice thicknesses and different scanning protocols and/or scanner manufacturers), our recommendations are as follows:
Include benchmarking data sets collected by different institutions to guarantee the maximum heterogeneity in terms of the aforementioned parameters.
Include different data sets for most common modalities and diseases because, as we have shown in the review, feature reproducibility results can be different when considering different diseases and/or different modalities.
With regard to point 2, we are currently working on developing an infrastructure, based on workflow programming language, that allows users to connect to the mentioned repository and run their feature extraction software. This infrastructure can be expanded by introducing in the workflow a “benchmarking module,” where users can easily “test” their software on common data sets, directly choosing them according to the modality and/or disease population of interest. In such a module, computational results can then automatically be uploaded, and a “sanity” report of feature reproducibility, compared with values already obtained by other institutions (bench-marking), is returned to the user. We believe that such an infrastructure not only will stimulate users to perform benchmarking calculations but also will help them in terms of debugging their software in case of possible errors.
Point 3 is strictly related to point 2. In fact, bench-marking intrinsically brings the concept of comparisons. For this reason, a standardized way of reporting should be preferred. As already pointed out in this article, users should report not only the obtained features’ raw values but also the configurations and/or parameters used to perform computations, together with details of the software used for computations. To facilitate this process, we are working on providing users with standard template tables that need to be filled in by the users and that include all the information mentioned earlier. In our view, this represents the first step toward homogenizing and increasing the quality of reporting. However, to facilitate feature comparison, we recommend using ontology techniques combined with Semantic Web to transform template tables into semantically linked data that can easily be queried by means of universal concepts defined by the ontology.
Finally, to increase the general validity of a radiomics-based model, we believe that external validation of the developed model should be performed, and only reproducible and repeatable features should be included in the model. To achieve this goal, we suggest using a distributed learning approach. In fact, in a distributed learning environment, models are “learned” and validated in different centers to increase their general validity. In addition, we recommend that authors describe in detail the procedure adopted for selecting features in the model. We suggest the following possible workflow:
Perform a reproducibility experiment and rank features from most reproducible to least reproducible.
Start scanning the list, picking up the most reproducible features; train the model; and validate the model externally to investigate the predictive or prognostic power of the features.
As a final point, we recognize the need to create a community of radiomics users, sharing common methodology in terms of both feature computation and methodology. In addition, we believe that this community should be guided by findable, accessible, interoperable, and reusable (FAIR) principles for a standardized, reliable, and reproducible use of radiomics.
Limitations of review
We are not able to rule out possible publication bias toward favorable results among the included studies. We did not generate a funnel plot because of the relatively low number of eligible studies and because of the specific exclusion of unpublished reports and conference proceedings. Every published study included in our review identified at least 1 radiomic feature that was repeatable or reproducible.
This systematic review was limited to only 2 reviewers; though a third reviewer was available to resolve disagreements, this option was not exercised. No disagreements were found after discussion, when comparing the results of the quality of reporting and the qualitative synthesis.
Furthermore, our search was limited to only 1 literature repository (PubMed) after its incorporation of MEDLINE and Embase citations. We did not permit conference proceedings, non—peer-reviewed publications, or other sources of gray literature in this review, which may have limited the number of studies located.
As a fundamental final point, this review would have benefited from a quantitative synthesis of the analyzed articles. Unfortunately, as already mentioned, the reviewed studies applied arbitrary cutoffs during the statistical analysis of reproducible and repeatable features; in some cases, as we documented in our article, thresholds used to define a feature as “reproducible” were not reported. We have performed the analysis that was amenable to us at this time. However, we strongly support consensus toward a standardized metric to quantitatively evaluate reporting of radiomic studies that will be useful for the community. No such consensus presently exists, and our review was the first attempt to describe what has been reported in the reviewed literature. We did not specifically propose a metric to evaluate the quality of reporting because this needs to be a consensus effort by our community. This can be seen as one of our studys limitations.
An anticipated update of the current review is proposed for April 2019. We hope by that date to have agreed on a common quantitative evaluation metric within the research community so that we will be able to update the review, in terms of not only up-to-date publications but also the inclusion of a quantitative analysis.
Supplementary Material
Footnotes
Supplementary material for this article can be found at https://doi.org/10.1016/j.ijrobp.2018.05.053.
Conflict of interest: none.
References
- 1.Emaminejad N, Qian W, Guan Y, et al. Fusion of quantitative image and genomic biomarkers to improve prognosis assessment of early stage lung cancer patients. IEEE Trans Biomed Eng 2016;63:1034–1043. [DOI] [PubMed] [Google Scholar]
- 2.Popovici V, Budinská E, Dusek L, et al. Image-based surrogate biomarkers for molecular subtypes of colorectal cancer. Bioinformatics 2017;33:2002–2009. [DOI] [PubMed] [Google Scholar]
- 3.Scalco E, Rizzo G. Texture analysis of medical images for radio-therapy applications. Br J Radiol 2017;90:20160642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–247. [DOI] [PubMed] [Google Scholar]
- 6.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 2000;92:205–216. [DOI] [PubMed] [Google Scholar]
- 7.Bulakbasi N, Guvenc I, Onguru O, et al. The added value of the apparent diffusion coefficient calculation to magnetic resonance imaging in the differentiation and grading of malignant brain tumors. J Comput Assist Tomogr 2004;28:735–746. [DOI] [PubMed] [Google Scholar]
- 8.Zwirewich CV, Vedal S, Miller RR, et al. Solitary pulmonary nodule: High-resolution CT and radiologic-pathologic correlation. Radiology 1991;179:469–476. [DOI] [PubMed] [Google Scholar]
- 9.Thie JA. Understanding the standardized uptake value, its methods, and implications for usage. J Nucl Med 2004;45:1431–1434. [PubMed] [Google Scholar]
- 10.Alobaidli S, McQuaid S, South C, et al. The role of texture analysis in imaging as an outcome predictor and potential tool in radiotherapy treatment planning. Br J Radiol 2014;87:20140369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chicklore S, Goh V, Siddique M, et al. Quantifying tumour heterogeneity in 18F-FDG PET/CT imaging by texture analysis. Eur J Nucl Med Mol Imaging 2013;40:133–140. [DOI] [PubMed] [Google Scholar]
- 12.Miles KA, Ganeshan B, Hayball MP. CT texture analysis using the filtration-histogram method: What do the measurements mean? Cancer Imaging 2013;13:400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar V, Gu Y, Basu S, et al. Radiomics: The process and the challenges. Magn Reson Imaging 2012;30:1234–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larue RTHM, Defraene G, De Ruysscher D, et al. Quantitative radiomics studies for tissue characterization: A review of technology and methodological procedures. Br J Radiol 2017;90:20160665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chalkidou A, O’Doherty MJ, Marsden PK. False discovery rates in PET and CT studies with texture features: A systematic review. PloS One 2015;10:e0124165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altman DG, Lausen B, Sauerbrei W, et al. Dangers of using ‘optimal’ cutpoints in the evaluation of prognostic factors. J Natl Cancer Inst 1994;86:829–835. [DOI] [PubMed] [Google Scholar]
- 18.Bagci U, Yao J, Miller-Jaster K, et al. Predicting future morphological changes of lesions from radiotracer uptake in 18F-FDG-PET images. PloS One 2013;8:e57105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orlhac F, Soussan M, Maisonobe JA, et al. Tumor texture analysis in 18F-FDG PET: Relationships between texture parameters, histogram indices, standardized uptake values, metabolic volumes, and total lesion glycolysis. J Nucl Med 2014;55:414–422. [DOI] [PubMed] [Google Scholar]
- 20.O’Connor JPB, Aboagye EO, Adams JE, et al. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol 2017;14:169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baumann K Cross-validation as the objective function for variable-selection techniques. TrAC Trends Anal Chem 2003;22:395–406. [Google Scholar]
- 22.Collins GS, Reitsma JB, Altman DG, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD statement. BMJ 2015;350:g7594. [DOI] [PubMed] [Google Scholar]
- 23.Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med 2009;6:e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haynes RB, McKibbon KA, Wilczynski NL, et al. Optimal search strategies for retrieving scientifically strong studies of treatment from Medline: Analytical survey. BMJ 2005;330:1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingui BJ, Rogers MA. Searching for clinical prediction rules in MEDLINE. J Am Med Inform Assoc 2001;8:391–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geersing GJ, Bouwmeester W, Zuithoff P, et al. Search filters for finding prognostic and diagnostic prediction studies in Medline to enhance systematic reviews. PloS One 2012;7:e32844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Depeursinge A, Foncubierta-Rodriguez A, Van De Ville D, et al. Three-dimensional solid texture analysis in biomedical imaging: Review and opportunities. Med Image Anal 2014;18:176–196. [DOI] [PubMed] [Google Scholar]
- 28.Balagurunathan Y, Gu Y, Wang H, et al. Reproducibility and prognosis of quantitative features extracted from CT images. Transl Oncol 2014;7:72–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng N-M, Fang Y-HD, Tsan D-L, et al. Respiration-averaged CT for attenuation correction of PET images—Impact on PET texture features in non-small cell lung cancer patients. PloS One 2016;11: e0150509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coroller TP, Agrawal V, Narayan V, et al. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother Oncol 2016;119:480–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desseroit M-C, Tixier F, Weber WA, et al. Reliability of PET/CT shape and heterogeneity features in functional and morphologic components of non-small cell lung cancer tumors: A repeatability analysis in a prospective multicenter cohort. J Nucl Med 2017;58: 406–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Desseroit M-C, Visvikis D, Tixier F, et al. Development of a nomogram combining clinical staging with (18)F-FDG PET/CT image features in non-small-cell lung cancer stage I-III. Eur J Nucl Med Mol Imaging 2016;43:1477–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fave X, Mackin D, Yang J, et al. Can radiomics features be reproducibly measured from CBCT images for patients with non-small cell lung cancer? Med Phys 2015;42:6784–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fave X, Cook M, Frederick A, et al. Preliminary investigation into sources of uncertainty in quantitative imaging features. Comput Med Imaging Graph 2015;44:54–61. [DOI] [PubMed] [Google Scholar]
- 35.Fave X, Zhang L, Yang J, et al. Impact of image preprocessing on the volume dependence and prognostic potential of radiomics features in non-small cell lung cancer. Transl Cancer Res 2016;5:349–363. [Google Scholar]
- 36.Fried DV, Tucker SL, Zhou S, et al. Prognostic value and reproducibility of pretreatment CT texture features in stage III non-small cell lung cancer. Int J Radiat Oncol Biol Phys 2014;90:834–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huynh E, Coroller TP, Narayan V, et al. Associations of radiomic data extracted from static and respiratory-gated CT scans with disease recurrence in lung cancer patients treated with SBRT. PloS One 2017;12:e0169172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalpathy-Cramer J, Mamomov A, Zhao B, et al. Radiomics of lung nodules: A multi-institutional study of robustness and agreement of quantitative imaging features. Tomography 2016;2:430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leijenaar RTH, Carvalho S, Velazquez ER, et al. Stability of FDG-PET radiomics features: An integrated analysis of test-retest and inter-observer variability. Acta Oncol 2013;52:1391–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mackin D, Fave X, Zhang L, et al. Measuring computed tomography scanner variability of radiomics features. Invest Radiol 2015;50:757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oliver JA, Budzevich M, Zhang GG, et al. Variability of image features computed from conventional and respiratory-gated PET/CT images of lung cancer. Transl Oncol 2015;8:524–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parmar C, Rios Velazquez E, Leijenaar R, et al. Robust radiomics feature quantification using semiautomatic volumetric segmentation. PloS One 2014;9:e102107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Velden FHP, Kramer GM, Frings V, et al. Repeatability of radiomic features in non-small-cell lung cancer [(18)F]FDG-PET/CT studies: Impact of reconstruction and delineation. Mol Imaging Biol 2016;18:788–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan J, Chu-Stern JL, Loi HY, et al. Impact of image reconstruction settings on texture features in 18F-FDG PET. J Nucl Med 2015;56: 1667–1673. [DOI] [PubMed] [Google Scholar]
- 45.Yip S, McCall K, Aristophanous M, et al. Comparison of texture features derived from static and respiratory-gated PET images in non-small cell lung cancer. PloS One 2014;9:e115510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao B, Tan Y, Tsai WY, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep 2016;6:23428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grootjans W, Tixier F, van der Vos CS, et al. The impact of optimal respiratory gating and image noise on evaluation of intratumor heterogeneity on 18F-FDG PET imaging of lung cancer. J Nucl Med 2016;57:1692–1698. [DOI] [PubMed] [Google Scholar]
- 48.Koo HJ, Sung YS, Shim WH, et al. Quantitative computed tomography features for predicting tumor recurrence in patients with surgically resected adenocarcinoma of the lung. PloS One 2017;12:e0167955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lasnon C, Majdoub M, Lavigne B, et al. 18F-FDG PET/CT heterogeneity quantification through textural features in the era of harmonisation programs: A focus on lung cancer. Eur J Nucl Med Mol Imaging 2016;43:2324–2335. [DOI] [PubMed] [Google Scholar]
- 50.Bagher-Ebadian H, Siddiqui F, Liu C, et al. On the impact of smoothing and noise on robustness of CT and CBCT radiomics features for patients with head and neck cancers. Med Phys 2017;44: 1755–1770. [DOI] [PubMed] [Google Scholar]
- 51.Bogowicz M, Riesterer O, Bundschuh RA, et al. Stability of radiomic features in CT perfusion maps. Phys Med Biol 2016;61:8736–8749. [DOI] [PubMed] [Google Scholar]
- 52.Lu L, Lv W, Jiang J, et al. Robustness of radiomic features in [(11)C] choline and [(18)F]FDG PET/CT imaging of nasopharyngeal carcinoma: Impact of segmentation and discretization. Mol Imaging Biol 2016;18:935–945. [DOI] [PubMed] [Google Scholar]
- 53.Doumou G, Siddique M, Tsoumpas C, et al. The precision of textural analysis in (18)F-FDG-PET scans of oesophageal cancer. Eur Radiol 2015;25:2805–2812. [DOI] [PubMed] [Google Scholar]
- 54.Hatt M, Tixier F, Cheze Le Rest C, et al. Robustness of intratumour 18F-FDG PET uptake heterogeneity quantification for therapy response prediction in oesophageal carcinoma. Eur J Nucl Med Mol Imaging 2013;40:1662–1671. [DOI] [PubMed] [Google Scholar]
- 55.Tixier F, Hatt M, Le Rest CC, et al. Reproducibility of tumor uptake heterogeneity characterization through textural feature analysis in 18F-FDG PET. J Nucl Med 2012;53:693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu P, Wang J, Zhong H, et al. Reproducibility with repeat CT in radiomics study for rectal cancer. Oncotarget 2016;7:71440–71446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Timmeren JE, Leijenaar RTH, van Elmpt W, et al. Test—retest data for radiomics feature stability analysis: Generalizable or study-specific? Tomography 2016;2:361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guan Y, Li W, Jiang Z, et al. Whole-lesion apparent diffusion coefficient-based entropy-related parameters for characterizing cervical cancers: Initial findings. Acad Radiol 2016;23:1559–1567. [DOI] [PubMed] [Google Scholar]
- 59.Galavis PE, Hollensen C, Jallow N, et al. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncol 2010;49:1012–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hatt M, Majdoub M, Vallieres M, et al. 18F-FDG PET uptake characterization through texture analysis: Investigating the complementary nature of heterogeneity and functional tumor volume in a multi-cancer site patient cohort. J Nucl Med 2015;56:38–44. [DOI] [PubMed] [Google Scholar]
- 61.Buch K, Li B, Qureshi MM, et al. Quantitative assessment of variation in CT parameters on texture features: Pilot study using a nonanatomic phantom. AJNR Am J Neuroradiol 2017;38:981–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Forgacs A, Pall Jonsson H, Dahlbom M, et al. A study on the basic criteria for selecting heterogeneity parameters of F18-FDG PET images. PloS One 2016;11:e0164113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim HG, Chung YE, Lee YH, et al. Quantitative analysis of the effect of iterative reconstruction using a phantom: Determining the appropriate blending percentage. Yonsei Med J 2015;56:253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lo P, Young S, Kim HJ, et al. Variability in CT lung-nodule quantification: Effects of dose reduction and reconstruction methods on density and texture based features. Med Phys 2016;43:4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shafiq-Ul-Hassan M, Zhang GG, Latifi K, et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys 2017;44:1050–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao B, Tan Y, Tsai WY, et al. Exploring variability in CT characterization of tumors: A preliminary phantom study. Transl Oncol 2014;7:88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nestle U, Kremp S, Schaefer-Schuler A, et al. Comparison of different methods for delineation of 18F-FDG PET-positive tissue for target volume definition in radiotherapy of patients with non-small cell lung cancer. J Nucl Med 2005;46:1342–1348. [PubMed] [Google Scholar]
- 68.Zhang L, Fried DV, Fave XJ, et al. IBEX: An open infrastructure software platform to facilitate collaborative work in radiomics. Med Phys 2015;42:1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weir JP. Quantifying test-retest reliability using the intraclass correlation coefficient and the SEM. J Strength Cond Res 2005;19:231–240. [DOI] [PubMed] [Google Scholar]
- 70.Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics 1989;45:255–268. [PubMed] [Google Scholar]
- 71.Myers L, Sirois MJ. Spearman correlation coefficients, differences between, Encyclopedia of Statistical Sciences. Hoboken, NJ: John Wiley & Sons; 2004. [Google Scholar]
- 72.Sollini M, Cozzi L, Antunovic L, et al. PET radiomics in NSCLC: State of the art and a proposal for harmonization of methodology. Sci Rep 2017;7:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zwanenburg A, Leger S, Valliéres M, et al. Image biomarker standardisation initiative. ArXiv161207003 Cs. 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


