Abstract
CCCTC-binding factor (CTCF) is a highly conserved, ubiquitously expressed zinc finger protein. CTCF is a multifunctional protein, associated with a number of vital cellular processes such as transcriptional activation, repression, insulation, imprinting and genome organization. Emerging evidence indicates that CTCF is also involved in DNA damage response. In this review, we focus on this newly identified role of CTCF in facilitating DNA double-strand break repair. Due to the large number of cellular processes in which CTCF is involved, factors that functionally affect CTCF could have serious implications on genomic stability. It is becoming increasingly clear that exposure to environmental toxicants could have adverse effects on CTCF functions. Here we discuss the various ways that the environmental toxicants could impact CTCF functions and the potential consequences on DNA damage response.
Keywords: CTCF, γH2AX, cohesin, DNA double-strand breaks, environmental toxicants, nickel
1. Introduction
Cells are constantly exposed to both endogenous and exogenous DNA damaging agents. Endogenous DNA damage is caused by spontaneous hydroxylation, deamination, DNA replication misincorporation and DNA topoisomerase errors [1–3]. In addition, reactive oxygen species (ROS) generated by normal physiological processes could also damage DNA [4, 5]. Types of DNA damage induced by endogenous sources include base modifications [6], single-strand breaks (SSBs) and double-strand breaks (DSBs) [5, 7]. Exogenous DNA damage can be caused by physical and chemical sources. Physical sources of DNA damage include ultraviolet (UV) light, which causes pyrimidine-pyrimidone (6-4) photoproducts, as well as ionizing radiation (IR), which causes DSBs [8–12]. Environmental toxicants such as heavy metals, pesticides, dioxins and particulate air pollutants are a major category of chemical agents that damage DNA. They induce several types of DNA damage including base modifications, DNA fragmentation, SSBs and DSBs [13–21].
Consequences of DNA damage can be diverse. If left unchecked, DNA damage could lead to increased risk of a number of diseases including cancer and hereditary disorders [3, 22–24]. To repair DNA, the cells have evolved a number of mechanisms termed DNA damage response (DDR) [25]. Depending on the type of DNA damage, specific repair pathways are activated. SSBs and nucleotide errors are repaired by base excision repair (BER), nucleotide excision repair (NER), or mismatch repair (MMR) [26–30]. DSBs are repaired by two main mechanisms, non-homologous end-joining (NHEJ) and homology-directed repair or homologous recombination (HR) [31]. NHEJ, which involves direct ligation of broken or damaged DNA strands, does not require homology between DNA strands and therefore, is error-prone. This pathway is active through all stages of the cell cycle. HR requires homologous pairing of the sister chromatids and is active during S and G2 phase [31]. Since HR relies on the homology between two DNA strands, it is generally considered error-free [31, 32].
DDR is a complex process involving a number of enzymatic activities. For efficient DNA repair, recruitment of repair factors to the damaged sites needs to be temporally regulated. In addition, it is important that the repair process is spatially constrained [33]. Access of repair factors to inappropriate regions can affect genomic integrity. In eukaryotes spatial constraining of DNA is achieved through its organization into higher-order chromatin structures. Recent genome-wide studies have identified such spatially constrained higher-order chromatin structures such as heterochromatin domains [34, 35]. Furthermore, the advent of chromosome conformation capture (3C) and 3C based technologies such as chromosome conformation capture-on-chip (4C), chromosome conformation capture carbon copy (5C) and Hi-C have been instrumental in identifying chromatin organization and interactions in three-dimensional (3D) space [36–38]. Recently, 5C and Hi-C technologies have enabled the discovery of local interacting megabase sized regions of the genome called topologically associated domains (TADs) [39–47]. TADs are structural as well as functional components of chromatin. While interactions occur at high frequency within a given TAD, the interaction frequencies between TADs are low [39].
Insulator binding proteins, which mediate long-range interactions via their ability to loop DNA through protein-protein interactions are essential for the establishment of higher order chromatin structures including heterochromatin domains and TADs [39, 41, 43, 48]. A number of insulator binding proteins have been characterized in Saccharomyces cerevisiae and Drosophila melanogaster [49–52]. However, CCCTC-binding factor (CTCF) is the only insulator binding protein identified in vertebrates [53, 54]. Recent studies have uncovered a major role for CTCF in DNA double strand break response [55–57]. In this review, we discuss emerging evidence that have begun to shed light on the role of CTCF in DNA repair. We also evaluate the deleterious effects of environmental toxicants on CTCF, which could potentially impact DNA repair processes.
2. CTCF is a multifunctional protein
CTCF is a highly conserved, ubiquitously expressed protein. It consists of an N-terminal domain, a C-terminal domain and a central DNA binding domain. CTCF binds to thousands of sites across the genome [34, 58], and its binding is largely invariant among various cell-types. CTCF DNA binding domain contains 11 zinc fingers (ZFs): 10 Cys2His2 (C2H2) ZFs and 1 Cys2His-Cys (C3H1) ZF. Studies on CTCF ZF mutants show that mutation in any of the 11 ZFs lower DNA binding affinity. This suggests that all the 11 ZFs contribute to DNA binding. Furthermore mutations at the core ZFs (ZFs 4–7) impacted the DNA binding of CTCF considerably more than the peripheral ZFs [59, 60]. Deletion of N- and C- terminal regions outside the ZF domain did not impact CTCF’s DNA binding ability [61]. CTCF binding to DNA occurs sequence specifically and the core consensus CTCF DNA binding sequence is well characterized [34, 58, 59]. However, CTCF is known to recognize a diverse set of sequences through combinatorial use of its ZFs [59].
CTCF is a versatile protein with a wide array of functions. Originally identified as a transcriptional repressor, CTCF is now known to be involved in several processes including transcriptional activation, X-chromosome inactivation, V(D)J recombination, RNA polymerase II (Pol II) pausing and imprinting [60, 62, 63]. Of the many functions of CTCF, it is most well-characterized for its role in chromatin insulation [53, 64, 65]. Insulators are DNA sequence elements that play a key role in preventing inappropriate interactions between adjacent regions of the genome [53, 64, 65]. The insulator function is mediated by architectural proteins that bind the insulator sequences. While a number of insulator-binding proteins are known in yeast and flies, CTCF is the only known insulator-binding protein in vertebrates [49–54]. CTCF can function both as an enhancer-blocking insulator and domain-barrier insulator [53, 54, 64, 65]. Through its enhancer-blocking function, CTCF prevents communication between enhancers and inappropriate promoters. Imprinting at the Igf2/H19 locus is a classic example of the enhancer blocking insulation function of CTCF [66–68]. The domain barrier function of CTCF enables organization of the genome into transcriptionally active and silent regions, which is necessary to prevent spreading of condensed chromatin, into the neighboring active regions [35]. CTCF also contributes to the formation of TADs by binding their boundaries and functioning as barriers. CTCF enables long-range interactions through looping of DNA [43, 44, 69]. This ability of CTCF to mediate long-range interactions and organize genome in three-dimensional (3D) nuclear space forms the basis for a number of its functions [43, 44]. Through recent investigations, it is becoming increasingly clear that CTCF plays a major role in DSB repair [55–57].
3. H2AX phosphorylation
Phosphorylation of histone H2A variant H2AX, at serine 139, termed γH2AX, is one of the first events in DNA damage response [70]. γH2AX plays a key role in DDR. It is widely used as a surrogate marker of DSBs and as a biomarker for cancer therapy evaluation [71]. Upon induction of DSB, H2AX is phosphorylated by the phosphoinositide-3-kinase-related protein kinase (PIKK) family, which includes ataxia telangiectasia mutated (ATM), DNA-PKcs and ATM and RAD3-related (ATR) [71]. Formation of γH2AX initiates within 30 seconds of DNA damage and reaches maximal levels at 10–30 min [70]. γH2AX spreads into megabase regions (0.5–2 Mb) surrounding DNA lesions and forms DNA repair foci [72]. One γH2AX focus exists for each DSB.γH2AX foci function as a signal for the recruitment of DDR proteins. γH2AX also decondenses chromatin at the damage sites, thereby increasing accessibility, which enables assembly and retention of DNA repair factors [73]. Upon repair of DSBs, the γH2AX foci decline and disappear. The half-life of γH2AX foci ranges between 2 and 7 h [74, 75].
3.1 CTCF delimits γH2AX domains
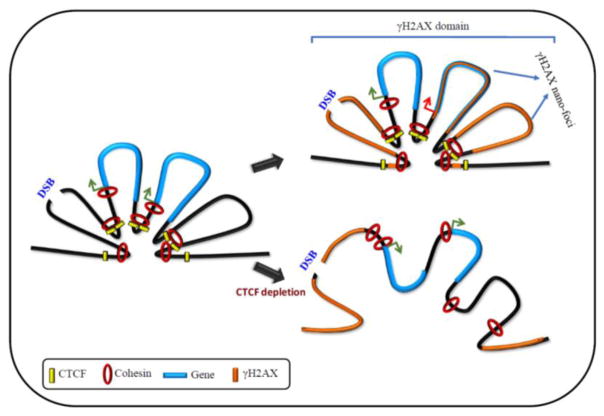
CTCF is rapidly recruited to DSBs induced by several sources including γ-irradiation, IR and laser micro irradiation [55–57, 76]. Recruitment of CTCF occurring within 30 seconds of DSB induction suggests this to be an early event in DDR [56]. Analysis of CTCF mutants show that the zinc finger domain is important for the recruitment of CTCF to DSBs [56]. Interestingly, the timing of CTCF recruitment to DSBs coincides with the phosphorylation of H2AX. This suggests that CTCF is recruited to DNA after DSB induction. However, analysis of CTCF binding sites that are conserved among different cell-types showed that the γH2AX domains were flanked by CTCF before and during DDR [57]. Together, these studies suggest that upon DNA damage, both newly recruited, as well as constitutively DNA bound CTCF flank γH2AX domains. Whether this happens in a site-specific manner remains to be seen. CTCF ChIP-Seq signals at the boundaries of γH2AX domains are stronger compared to those within the domains [57]. Given the role of CTCF in delimiting heterochromatin domains and TADs by binding their boundaries, it is reasonable to speculate that CTCF could be involved in confining γH2AX foci to DSB sites. Supporting this notion, γH2AX domains increase in size upon siRNA-mediated depletion of CTCF [56]. This suggests that γH2AX can spread outside the domain boundaries and expand the DDR region in the absence of CTCF. In addition, CTCF depletion increased both the number of IR-induced γH2AX foci (IRIF) in each cell as well as the number of foci-positive cells [55, 56], and CTCF depleted cells display hypersensitiveness to IR [55, 56]. Full-length CTCF could restore the IRIF formation [56]. This suggests that CTCF plays a key role in constraining γH2AX domains by binding its boundaries and limiting its distribution to the vicinity of DSBs, thereby restricting the area of DDR (Fig. 1).
Fig. 1. CTCF establishes γH2AX domains during DSB repair.
Upon DNA damage, CTCF binds DSBs and facilitates γH2AX domains formation by functioning as domain barrier. Cohesin likely interacts with CTCF during γH2AX foci and nano-foci establishment. Cohesin binding at the promoters of active genes within domains inhibit γH2AX establishment, thereby protecting transcription. Loss of cohesin binding at gene promoters cause γH2AX spreading and gene silencing. CTCF depletion causes loss of higher-order chromatin structures resulting in spreading of γH2AX foci and impaired DNA repair. Green arrow: active gene; red arrow: repressed gene
3.2 CTCF aids formation of γH2AX nano-foci
Super-resolution microscopy imaging has shown that a single confocal microscopy identified γH2AX focus can be further resolved into spatially clustered substructures [57]. These 200 nm diameter substructures named γH2AX nano-foci have been suggested as the smallest γH2AX modified chromatin regions [57]. ChIP-chip studies by Iacovoni et al have revealed similar substructures within γH2AX domains. This study showed that the γH2AX domains are discontinuous and contains holes (no-γH2AX regions) and peaks (high-γH2AX regions) [72]. By profiling histone H3 using ChIP-chip, Iacovoni et al demonstrate normal histone occupancy in the holes. Therefore, the γH2AX-depleted holes are formed by reduction in the levels of phosphorylation [72] rather than decreased nucleosome occupancy [72]. Promoters and transcription start sites (TSS) within γH2AX domains show reduction in γH2AX levels [72]. Furthermore, RNA polymerase II (Pol II) is enriched at the holes, indicating active gene promoters. Moreover, highly expressed genes within γH2AX domains were less enriched for γH2AX [72]. This could suggest that γH2AX depletion at active gene promoters is necessary to maintain gene transcription within γH2AX foci during DDR.
Occurrence of γH2AX as spatially proximal clusters at DSBs suggests higher-order chromatin organization. CTCF mediated long-range interactions form the basis of higher-order chromatin organization in vertebrates. Therefore, long-range interactions mediated by CTCF likely bring the edges of the DNA damaged regions into close proximity thereby restricting DDR to the damaged sites. Spreading of γH2AX domains caused by CTCF depletion is indicative of the DDR regions spreading beyond the damages sites. Moreover, CTCF depletion resulted in reduction in the number and size of γH2AX nano-foci [57]. This suggests that CTCF, through its ability to organize 3D structure of the genome, functions as a delimiter of γH2AX domains and an organizer of γH2AX nano-foci.
4. Role of cohesin in DSB repair
A number of functions mediated by CTCF involve interaction with its numerous protein partners. One of the major CTCF-interacting proteins is the cohesin complex. Cohesin is important for CTCF-mediated chromatin looping. Cohesin is a multi-subunit protein complex, which plays an essential role in DNA replication and sister chromatid cohesion. Cohesin complex consists of SMC1, SMC3, SCC1 (Mcd1 in yeast and Rad21 in humans) and SCC3 (IRR/SCC3 in yeast and SA1 and SA2 in humans). Cohesin is recruited to DNA upon DSB induction [72, 77–79]. The extent of cohesin recruitment around DSBs is species specific. While in yeast cohesin binds 50–100 kb around DSBs, its distribution is limited to 5 kb in humans. During HR, cohesin tethers sister chromatids, facilitating DNA repair [80–82]. Cohesin binding to DSBs is essential for HR and the cohesin subunit SA2 has actually been shown to antagonize the error-prone NHEJ and favour HR [78]. SA2 deletion on the other hand, increased DNA end joining and promoted NHEJ [78].
4.1 Cohesin maintains H2AX profiles within domains
Recent studies have revealed an important role for cohesin in γH2AX distribution within the domains. Distribution of γH2AX within the domains is discontinuous and cohesin binding is enriched at the γH2AX-depleted regions [83]. Depletion of cohesin subunit SCC1 resulted in γH2AX spreading to the previously cohesin bound regions [83]. This suggests a role for cohesin in the organization of γH2AX within domains by preventing γH2AX establishment at specific loci. The cohesin enriched/γH2AX depleted regions correlated with the promoters of active genes. SCC1 depletion caused downregulation of these genes [83]. Interestingly, ChIP-Seq experiments before and after IR-induced DNA damage demonstrated that upon DNA damage increase in cohesin binding occurred at the regions already bound by cohesin prior to damage [79]. This suggests that cohesin recruitment to active gene promoters occurs constitutively, and is not dependent on DSB induction. Upon DSB induction, levels of cohesin at the active gene promoters are further enriched, which inhibits γH2AX establishment and helps maintain transcription [83]. Upon cohesin depletion, γH2AX depleted regions at the TSS of genes within the γH2AX domains (holes) were undetectable [83]. Moreover, cohesin depletion increased γH2AX levels on cohesin-bound genes resulting in decreased transcription after DSB induction [83]. This suggests that cohesin at active gene promoters function as a protector of transcription in the event of DNA damage, by preventing γH2AX establishment (Fig. 1).
In addition to spreading γH2AX within the domains, SCC1 depletion also induces γH2AX spreading outside the domains [83], suggesting domain barrier function for cohesin. However, not all of the γH2AX domains that spread upon cohesin depletion were associated with cohesin binding [83]. It has been suggested that γH2AX spreading could merely be an outcome of SCC1 depletion-induced increase in the total levels of γH2AX [83]. Therefore, while the role of cohesin in protecting transcription within γH2AX domains during DSB repair is clear, its role in constraining the γH2AX domains remains inconclusive.
4.2 Cohesin cooperates with CTCF during establishment of long-range interactions
Cohesin shares the core consensus DNA binding sequence with CTCF [84, 85]. Genome-wide studies show extensive colocalization of cohesin and CTCF [84, 85]. Moreover, CTCF deletion abrogated cohesin binding to specific sites showing that CTCF is involved in its recruitment [86]. Therefore, it is likely that CTCF is required for cohesin recruitment at DSBs. Moreover, CTCF and cohesin cooperate during the organization of higher-order chromatin structures [86]. Therefore, it is plausible that CTCF and cohesin interact during the establishment and maintenance of γH2AX foci, which requires higher order chromatin organization. Regulation of γH2AX domains by CTCF and cohesin is reminiscent of TADs. CTCF depletion result sin increased inter-TAD interactions, suggesting its role as barriers of TADs. On the other hand, cohesin depletion inhibited intra-TAD interactions, suggesting its role in chromatin organization within the TAD domains [86]. Similarly, CTCF borders γH2AX domains and depletion of CTCF results in the spreading of γH2AX into the surrounding regions. However, cohesin depletion, while not always associated with γH2AX domain spreading, is clearly associated with the increase in γH2AX within the domain [83]. This suggests that while CTCF primarily functions as a γH2AX domain barrier, cohesin functions in the maintenance of the nano-foci patterns within γH2AX domains (Fig. 1).
5. CTCF recruits BRCA2 to DSBs during HR
Hilmi et al recently demonstrated a direct role of the CTCF in HR through its ability to recruit BRCA2 to DSBs [55]. BRCA2 is a tumour suppressor gene. Mutations in BRCA2 are associated with increased risk of breast and ovarian cancers. BRCA2 deleted cells as well as mice are hypersensitive to radiation and genotoxic agents, and exhibit spontaneous chromosomal aberrations [87, 88]. Moreover, BRCA2 deficient cells display chromosomal instability and defects in homologous recombination [87, 88]. Upon DNA damage, BRCA2 is recruited to DNA, which in turn recruits RAD51. RAD51 filament formation, which allows strand invasion and homologous pairing [89–91], is an initial step in the process of HR.
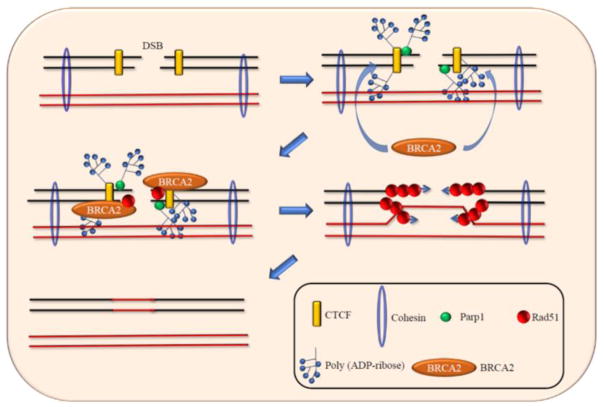
Co-immunoprecipitation experiments demonstrate that exposure to multiple DNA damaging agents favour interaction between CTCF and BRCA2 [55]. Furthermore, BRCA2 recruitment to DSBs is diminished in CTCF-depleted cells, although BRCA2 protein levels remained unaltered [55]. This shows that BRCA2 is recruited to DSBs through its interaction with CTCF (Fig. 2). Interestingly, CTCF-mediated recruitment of BRCA2 to DSBs is dependent on poly(ADP-ribosyl)ation (PARlation) of CTCF. PARP1 inhibitors decrease the interaction between CTCF and BRCA2. Moreover, CTCF PARlation-defective mutant cannot recruit BRCA2 to DSBs although it can bind DNA efficiently [55]. Therefore, although PARlation of CTCF is not required for its DNA binding, it is essential for the recruitment of BRCA2 to DSBs (Fig. 2). Interestingly, CTCF knockdown did not diminish the binding of NHEJ associated protein 53BP1 to DSBs [55].
Fig. 2. CTCF recruits BRCA2 to DSB during HR.
During DSB repair, CTCF recruits BRCA2 to DSBs in a PARlation dependent manner. BRCA2 in turn recruits RAD 51, which form filaments to allow strand invasion and homologous recombination.
6. Effects of environmental toxicants on CTCF
Given the key role of CTCF in DSB repair, factors that can affect the expression and function of CTCF could have major implications in genomic stability. Here we discuss some of the factors that can adversely affect CTCF functions and potentially impact DNA repair processes.
6.1 Ni-exposure lowers CTCF DNA binding affinity
Exposure of human lung epithelial cells to nickel (Ni), an environmental carcinogen associated with lung and nasal cancers in humans, diminishes the DNA binding ability of CTCF [35]. The DNA binding affinity of proteins is dependent on the underlying sequences, with the binding being strongest to the core consensus sequences. Variants of the consensus sequences, which are considered weaker binding sequences, bind the protein with lower affinities [92]. Ni inhibited CTCF DNA binding in a dose dependent manner. At low concentrations, Ni disrupted CTCF binding only at the weakest sites. With increasing concentrations of Ni, binding was affected even at the stronger binding sites. Therefore, Ni impacts CTCF DNA binding affinity in a dose as well as binding sequence dependent manner [35]. CTCF functions as a repressive domain barrier by binding the boundaries of domains marked by H3K27me3 and H3K9me2 [34, 35, 58]. Ni-exposure-induced impairment of CTCF binding caused disruption of H3K9me2 domain boundaries resulting in spreading of H3K9me2 into active chromatin regions causing gene silencing [35]. Therefore, it is clear that Ni-induced loss of CTCF binding has functional consequences in terms of cellular regulation.
6.2 Zn2+ substitution by metal ions impairs DNA binding of ZF-proteins
Although the mechanisms underlying Ni-induced reduction in the DNA binding affinity of CTCF is still not fully understood, extensive studies on the impact of toxic metals on the DNA binding of ZF-transcription factors, XPA and SP1, offer some clues. The DNA binding domains of XPA and SP1 contain 1 C4-type ZF and 3 C2H2-type ZF, respectively [93]. Zn2+ in the ZFs of XPA and SP1 can be substituted by Ni2+ [94, 95]. This substitution dramatically alters the structure of these proteins, impacting their DNA binding affinities [94, 95]. In addition, As3+, a major environmental carcinogen prevalent in air and drinking water can also substitute Zn2+ in the ZFs of XPA, thus interfering with its DNA binding ability [96, 97]. In addition, Co2+ and Cd2+ have also been shown to inhibit XPA activity by altering its ZF structure. It has been suggested that alteration in the ZF structure due to Zn2+ substitution by toxic metals could change the DNA binding sequence preference of these proteins, thus functionally affecting them [94, 95, 98]. Furthermore, exposure to certain toxic metals has also been associated with reduction in the Zn content of several proteins. For instance, As3+ exposure caused decrease in the Zn content of PARP1 (C3H3-type ZF-protein) and XPA [96, 99]. MMA3+ exposure decreased the Zn content in APTX (C2H2-type ZF-protein), PARP1 and XPA [96]. Based on these studies, it is clear that ZFs are major targets of several toxic metals.
Since ZFs are clearly important targets of several toxic metals, it is plausible that the DNA binding affinity of CTCF, a ZF-protein, could be affected by environmental toxicants via similar mechanisms (Fig. 3). DSB repair studies in the presence or absence of Ni showed that HR was inhibited in cells exposed to 500 μM NiCl2 [19]. However, DSB repair was not impacted at lower doses of Ni, suggesting dose dependent effect of Ni on DNA repair processes. Moreover, all the tested doses of AsO3 inhibited HR pathway and favored the error-prone NHEJ [19]. It is conceivable that inhibition of DNA repair processes due to exposure to toxic metals could be at least in part due to the impact of metal ions on CTCF DNA binding affinity.
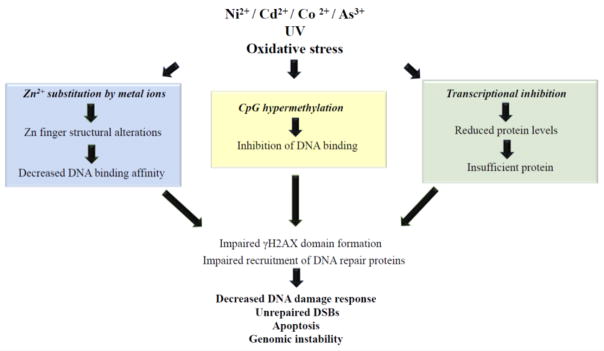
Fig. 3.
Environmental exposures could functionally impact CTCF in multiple ways and potentially impair DNA repair function.
6.3. CTCF DNA binding is CpG methylation sensitive
CTCF core consensus DNA binding sequence contains CpG, which can be methylated. A large number of studies have shown that CTCF cannot bind CpG methylated DNA [60, 66, 67, 100]. Negative correlation between DNA methylation and CTCF binding is in fact an important gene regulatory mechanism. Although CTCF binding sites are largely invariant between various cell-types, a subset of CTCF binding sites have been shown to be cell-type specific. Interestingly, 41% of these cell-type specific sites exhibit differential DNA methylation profiles [101]. Therefore, DNA methylation is a key factor in determining the target specificity of CTCF. Imprinting at the Igf2/H19 locus is one of the most well-understood processes involving interplay between DNA methylation and CTCF binding [66, 67]. Maternally inherited imprinting control region (ICR) at the Igf2/H19 is hypomethylated, which favors CTCF binding [66, 67]. This insulates the IGF2 promoter from the distal enhancer thus ensuring its silencing. On the other hand, the paternally inherited ICR is methylated, which prevents CTCF binding. This promotes interaction of IGF2 with the distal enhancers enabling its transcription.
In addition to being a normal process, interplay between CTCF and DNA methylation has also been implicated in pathogenic processes [102]. Expression of BCL6 oncogene is associated with B cell lymphoma. Lai et al showed that in lymphomas, BCL6 is hypermethylated within its first intron, which prevented CTCF binding. Lack of CTCF binding abrogated the CTCF-mediated silencing of BCL6 and resulted in its increased expression [103]. Similarly, increased expression of PDGFRA, a glioma oncogene has also been shown to be caused by the CpG methylation-induced loss of CTCF binding [104]. Hypermethylation at a TAD boundary, which causes loss of CTCF binding, allows activation of PDGFRA by enabling interaction with a previously blocked constitutive enhancer [104]. Collectively, these studies demonstrate CpG methylation as one of the most important factors associated with CTCF function.
6.4 DNA methylation can be altered by environmental agents
DNA methylation status is thus a major factor that can influence DNA binding of CTCF. Therefore, exposure to environmental chemicals that can alter DNA methylation profiles could potentially affect the CTCF DNA binding and impair DNA repair outcome (Fig. 3). Incidentally, DNA methylation is the most extensively studied epigenetic mark in cells and animals exposed to a multitude of environmental toxicants [105–107]. A large number of studies have shown that a plethora of environmental carcinogens such as Ni, As, Cr, Pb, bisphenol A and persistent organic pollutants (POPs) alter DNA methylation profiles [105, 107]. For instance, Ni exposure causes extensive DNA hypermethylation [108]. Silencing of p16 by DNA hypermethylation has been observed in tumors of mice exposed to NiS [108]. In Pb2+ exposed individuals dose dependent p16 hypermethylation has been observed [109]. Moreover, extensive DNA hypermethylation has been observed in mammalian cells and in workers with chromate-induced lung cancers [110]. Oxidative stress induced by exposure to a number of environmental toxicants could also potentially play a major role in altering DNA methylation profiles.
6.5 Environmental toxicants affect CTCF transcription
In addition to impaired DNA binding, another way that CTCF could be affected by environmental agents is through transcriptional regulation (Fig. 3). Cells exposed to inorganic arsenic (iAs3+) show decreased levels of CTCF [111]. Human corneal epithelial and human hematopoietic myeloid cells exposed to UV display decrease in the mRNA and protein levels of CTCF [112]. In PPC-1 prostate cancer cells, oxidative stress induced by H2O2 resulted in increased binding of NF-kB to CTCF promoter causing its repression [113]. Consequently, loss of imprinting was observed at the IGF2 locus [113].
7. Conclusion
Several years of intensive investigations on CTCF have identified a multitude of processes in which it is involved. Although the association of CTCF with DSB repair has been detected only recently, it is already clear that it plays a critical role in this process. CTCF is a genome organizer. It has the ability to mediate long-range interactions and establish higher-order chromatin structures. This ability of CTCF to organize the genome through long-range interactions underlies most of its well-known functions such as transcriptional activation, insulation and imprinting. Indeed, the genome organization function of CTCF appears to play a major role in influencing DNA repair processes as well. CTCF likely constrains the DNA repair sites by functioning as a barrier, thus enabling spatial restriction of DNA repair associated processes such as H2AX phosphorylation, chromatin structural changes and assembly of repair factors. In addition, CTCF could also potentially be involved in restricting DNA damage-induced loss of torsional stress to a single domain [114, 115]. Cellular processes such as replication and transcription generate DNA supercoiling and torsional stress within a topological domain [114, 115]. Increase in torsional stress is inhibitory to transcription [116]. Single or double strand break could release this torsional stress [117, 118]. Although DNA damage is generally considered a harmful process, programmed DNA damage and subsequent repair is currently being recognized as a facilitator of transcription due its role in relieving torsional stress [117, 119]. Whether CTCF has any role in restricting loss of topological stress to a single domain during programmed DNA damage needs to be investigated. However, CTCF’s association with DNA repair processes may not be limited to its genome organization function. It could facilitate recruitment of DNA repair proteins to their target sites, as exemplified by its role in recruiting BRCA2 to DSBs.
Due to the large number of processes in which CTCF in involved, factors that functionally affect CTCF function could have catastrophic consequences on genomic stability. Harmful effects of environmental toxicants on CTCF are currently beginning to be understood. CTCF is particularly vulnerable to environmental factors due to the number of ways that it could be affected: alterations to its ZF structure due to Zn2+ substitution by metal ions could affect its DNA binding affinity; CpG hypermethylation due to exposure to a multitude of environmental toxicants could also inhibit its DNA binding; finally, environmental toxicants could decrease its levels through transcriptional inhibition. Since CTCF’s DNA binding ability is critical for its functions, any CTCF dependent process including DNA repair could be impaired by environmental toxicants (Fig. 3). Environmental toxicants that impair CTCFs DNA repair function could particularly be deleterious to genomic stability since many of these toxicants could also induce DNA damage, thereby leaving the cell with damaged DNA and weakened DDR.
The mechanistic details of how CTCF regulates DSB repair is still not fully understood. Moreover, how environmental agents impact CTCF function in terms of DNA repair and the long-term consequences of environmental exposures on DNA repair needs to be investigated in much greater detail.
Acknowledgments
This work was supported by National Institutes of Health (NIH), National Institute of Environmental Health Sciences Grants (NIEHS) R01ES023174, R01ES024727 to SC
Footnotes
Conflicts of Interest statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 2.Jackson AL, Loeb LA. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat Res. 2001;477:7–21. doi: 10.1016/s0027-5107(01)00091-4. [DOI] [PubMed] [Google Scholar]
- 3.Vilenchik MM, Knudson AG. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc Natl Acad Sci U S A. 2003;100:12871–12876. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiseman H, Halliwell B. Damage to DNA by reactive oxygen and nitrogen species: role in inflammatory disease and progression to cancer. Biochem J. 1996;313(Pt 1):17–29. doi: 10.1042/bj3130017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 6.Sonneborn JS, Gottsch H, Cubin E, Oeltgen P, Thomas P. Alternative strategy for stress tolerance: opioids. J Gerontol A Biol Sci Med Sci. 2004;59:433–440. doi: 10.1093/gerona/59.5.b433. [DOI] [PubMed] [Google Scholar]
- 7.Woodbine L, Brunton H, Goodarzi AA, Shibata A, Jeggo PA. Endogenously induced DNA double strand breaks arise in heterochromatic DNA regions and require ataxia telangiectasia mutated and Artemis for their repair. Nucleic Acids Res. 2011;39:6986–6997. doi: 10.1093/nar/gkr331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brash DE, Haseltine WA. UV-induced mutation hotspots occur at DNA damage hotspots. Nature. 1982;298:189–192. doi: 10.1038/298189a0. [DOI] [PubMed] [Google Scholar]
- 9.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward JF. The yield of DNA double-strand breaks produced intracellularly by ionizing radiation: a review. Int J Radiat Biol. 1990;57:1141–1150. doi: 10.1080/09553009014551251. [DOI] [PubMed] [Google Scholar]
- 11.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vignard J, Mirey G, Salles B. Ionizing-radiation induced DNA double-strand breaks: a direct and indirect lighting up. Radiother Oncol. 2013;108:362–369. doi: 10.1016/j.radonc.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 13.Garaj-Vrhovac V, Zeljezic D. Evaluation of DNA damage in workers occupationally exposed to pesticides using single-cell gel electrophoresis (SCGE) assay. Pesticide genotoxicity revealed by comet assay. Mutat Res. 2000;469:279–285. doi: 10.1016/s1383-5718(00)00092-9. [DOI] [PubMed] [Google Scholar]
- 14.Nagy E, Adachi S, Takamura-Enya T, Zeisig M, Moller L. DNA damage and acute toxicity caused by the urban air pollutant 3-nitrobenzanthrone in rats: characterization of DNA adducts in eight different tissues and organs with synthesized standards. Environ Mol Mutagen. 2006;47:541–552. doi: 10.1002/em.20227. [DOI] [PubMed] [Google Scholar]
- 15.Huang HB, Lai CH, Chen GW, Lin YY, Jaakkola JJ, Liou SH, Wang SL. Traffic-related air pollution and DNA damage: a longitudinal study in Taiwanese traffic conductors. PLoS One. 2012;7:e37412. doi: 10.1371/journal.pone.0037412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin PH, Lin CH, Huang CC, Fang JP, Chuang MC. 2,3,7,8-Tetrachlorodibenzo-p-dioxin modulates the induction of DNA strand breaks and poly(ADP-ribose) polymerase-1 activation by 17beta-estradiol in human breast carcinoma cells through alteration of CYP1A1 and CYP1B1 expression. Chem Res Toxicol. 2008;21:1337–1347. doi: 10.1021/tx700396d. [DOI] [PubMed] [Google Scholar]
- 17.Hassoun EA, Wilt SC, Devito MJ, Van Birgelen A, Alsharif NZ, Birnbaum LS, Stohs SJ. Induction of oxidative stress in brain tissues of mice after subchronic exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Sci. 1998;42:23–27. doi: 10.1006/toxs.1997.2411. [DOI] [PubMed] [Google Scholar]
- 18.Hengstler JG, Bolm-Audorff U, Faldum A, Janssen K, Reifenrath M, Gotte W, Jung D, Mayer-Popken O, Fuchs J, Gebhard S, Bienfait HG, Schlink K, Dietrich C, Faust D, Epe B, Oesch F. Occupational exposure to heavy metals: DNA damage induction and DNA repair inhibition prove co-exposures to cadmium, cobalt and lead as more dangerous than hitherto expected. Carcinogenesis. 2003;24:63–73. doi: 10.1093/carcin/24.1.63. [DOI] [PubMed] [Google Scholar]
- 19.Morales ME, Derbes RS, Ade CM, Ortego JC, Stark J, Deininger PL, Roy-Engel AM. Heavy Metal Exposure Influences Double Strand Break DNA Repair Outcomes. PLoS One. 2016;11:e0151367. doi: 10.1371/journal.pone.0151367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wise SS, Holmes AL, Wise JP., Sr Hexavalent chromium-induced DNA damage and repair mechanisms. Rev Environ Health. 2008;23:39–57. doi: 10.1515/reveh.2008.23.1.39. [DOI] [PubMed] [Google Scholar]
- 21.Jia J, Chen J. Chronic nickel-induced DNA damage and cell death: the protection role of ascorbic acid. Environ Toxicol. 2008;23:401–406. doi: 10.1002/tox.20346. [DOI] [PubMed] [Google Scholar]
- 22.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 23.Wiesmuller L, Ford JM, Schiestl RH. DNA Damage, Repair, and Diseases. J Biomed Biotechnol. 2002;2:45. doi: 10.1155/S1110724302001985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moriwaki S. Hereditary Disorders with Defective Repair of UV-Induced DNA Damage. Jpn Clin Med. 2013;4:29–35. doi: 10.4137/JCM.S10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giglia-Mari G, Zotter A, Vermeulen W. DNA damage response. Cold Spring Harb Perspect Biol. 2011;3:a000745. doi: 10.1101/cshperspect.a000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parsons JL, Dianov GL. Co-ordination of base excision repair and genome stability. DNA Repair (Amst) 2013;12:326–333. doi: 10.1016/j.dnarep.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Krokan HE, Bjoras M. Base excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 29.Scharer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5:a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamileri I, Karakasilioti I, Garinis GA. Nucleotide excision repair: new tricks with old bricks. Trends Genet. 2012;28:566–573. doi: 10.1016/j.tig.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 31.Altieri F, Grillo C, Maceroni M, Chichiarelli S. DNA damage and repair: from molecular mechanisms to health implications. Antioxid Redox Signal. 2008;10:891–937. doi: 10.1089/ars.2007.1830. [DOI] [PubMed] [Google Scholar]
- 32.Guirouilh-Barbat J, Lambert S, Bertrand P, Lopez BS. Is homologous recombination really an error-free process? Front Genet. 2014;5:175. doi: 10.3389/fgene.2014.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jose CC, Xu B, Jagannathan L, Trac C, Mallela RK, Hattori T, Lai D, Koide S, Schones DE, Cuddapah S. Epigenetic dysregulation by nickel through repressive chromatin domain disruption. Proc Natl Acad Sci U S A. 2014;111:14631–14636. doi: 10.1073/pnas.1406923111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sati S, Cavalli G. Chromosome conformation capture technologies and their impact in understanding genome function. Chromosoma. 2017;126:33–44. doi: 10.1007/s00412-016-0593-6. [DOI] [PubMed] [Google Scholar]
- 37.de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou C, Li L, Qin ZS, Corces VG. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell. 2012;48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, Gribnau J, Barillot E, Bluthgen N, Dekker J, Heard E. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Bonev B, Cavalli G. Organization and function of the 3D genome. Nat Rev Genet. 2016;17:661–678. doi: 10.1038/nrg.2016.112. [DOI] [PubMed] [Google Scholar]
- 44.Gomez-Diaz E, Corces VG. Architectural proteins: regulators of 3D genome organization in cell fate. Trends Cell Biol. 2014;24:703–711. doi: 10.1016/j.tcb.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ciabrelli F, Cavalli G. Chromatin-driven behavior of topologically associating domains. J Mol Biol. 2015;427:608–625. doi: 10.1016/j.jmb.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 46.Sexton T, Cavalli G. The role of chromosome domains in shaping the functional genome. Cell. 2015;160:1049–1059. doi: 10.1016/j.cell.2015.02.040. [DOI] [PubMed] [Google Scholar]
- 47.Dekker J, Heard E. Structural and functional diversity of Topologically Associating Domains. FEBS Lett. 2015;589:2877–2884. doi: 10.1016/j.febslet.2015.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sequeira-Mendes J, Gutierrez C. Genome architecture: from linear organisation of chromatin to the 3D assembly in the nucleus. Chromosoma. 2016;125:455–469. doi: 10.1007/s00412-015-0538-5. [DOI] [PubMed] [Google Scholar]
- 49.West AG, Gaszner M, Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- 50.Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, Stein L, Henikoff S, Kellis M, White KP. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 52.Kellum R, Elgin SC. Chromatin boundaries: punctuating the genome. Curr Biol. 1998;8:R521–524. doi: 10.1016/s0960-9822(07)00337-5. [DOI] [PubMed] [Google Scholar]
- 53.Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, Ohlsson R, Lobanenkov VV. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 54.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 55.Hilmi K, Jangal M, Marques M, Zhao T, Saad A, Zhang C, Luo VM, Syme A, Rejon C, Yu Z, Krum A, Fabian MR, Richard S, Alaoui-Jamali M, Orthwein A, McCaffrey L, Witcher M. CTCF facilitates DNA double-strand break repair by enhancing homologous recombination repair. Sci Adv. 2017;3:e1601898. doi: 10.1126/sciadv.1601898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han D, Chen Q, Shi J, Zhang F, Yu X. CTCF participates in DNA damage response via poly(ADP-ribosyl)ation. Sci Rep. 2017;7:43530. doi: 10.1038/srep43530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Natale F, Rapp A, Yu W, Maiser A, Harz H, Scholl A, Grulich S, Anton T, Horl D, Chen W, Durante M, Taucher-Scholz G, Leonhardt H, Cardoso MC. Identification of the elementary structural units of the DNA damage response. Nat Commun. 2017;8:15760. doi: 10.1038/ncomms15760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 59.Nakahashi H, Kieffer Kwon KR, Resch W, Vian L, Dose M, Stavreva D, Hakim O, Pruett N, Nelson S, Yamane A, Qian J, Dubois W, Welsh S, Phair RD, Pugh BF, Lobanenkov V, Hager GL, Casellas R. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 2013;3:1678–1689. doi: 10.1016/j.celrep.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Renda M, Baglivo I, Burgess-Beusse B, Esposito S, Fattorusso R, Felsenfeld G, Pedone PV. Critical DNA binding interactions of the insulator protein CTCF: a small number of zinc fingers mediate strong binding, and a single finger-DNA interaction controls binding at imprinted loci. J Biol Chem. 2007;282:33336–33345. doi: 10.1074/jbc.M706213200. [DOI] [PubMed] [Google Scholar]
- 61.Quitschke WW, Taheny MJ, Fochtmann LJ, Vostrov AA. Differential effect of zinc finger deletions on the binding of CTCF to the promoter of the amyloid precursor protein gene. Nucleic Acids Res. 2000;28:3370–3378. doi: 10.1093/nar/28.17.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 65.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 66.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 67.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 68.Schoenherr CJ, Levorse JM, Tilghman SM. CTCF maintains differential methylation at the Igf2/H19 locus. Nat Genet. 2003;33:66–69. doi: 10.1038/ng1057. [DOI] [PubMed] [Google Scholar]
- 69.Phillips-Cremins JE, Corces VG. Chromatin insulators: linking genome organization to cellular function. Mol Cell. 2013;50:461–474. doi: 10.1016/j.molcel.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 71.Mah LJ, El-Osta A, Karagiannis TC. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24:679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 72.Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Turinetto V, Giachino C. Multiple facets of histone variant H2AX: a DNA double-strand-break marker with several biological functions. Nucleic Acids Res. 2015;43:2489–2498. doi: 10.1093/nar/gkv061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sanchez-Flores M, Pasaro E, Bonassi S, Laffon B, Valdiglesias V. gammaH2AX assay as DNA damage biomarker for human population studies: defining experimental conditions. Toxicol Sci. 2015;144:406–413. doi: 10.1093/toxsci/kfv011. [DOI] [PubMed] [Google Scholar]
- 75.Bouquet F, Muller C, Salles B. The loss of gammaH2AX signal is a marker of DNA double strand breaks repair only at low levels of DNA damage. Cell Cycle. 2006;5:1116–1122. doi: 10.4161/cc.5.10.2799. [DOI] [PubMed] [Google Scholar]
- 76.Izhar L, Adamson B, Ciccia A, Lewis J, Pontano-Vaites L, Leng Y, Liang AC, Westbrook TF, Harper JW, Elledge SJ. A Systematic Analysis of Factors Localized to Damaged Chromatin Reveals PARP-Dependent Recruitment of Transcription Factors. Cell Rep. 2015;11:1486–1500. doi: 10.1016/j.celrep.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim JS, Krasieva TB, LaMorte V, Taylor AM, Yokomori K. Specific recruitment of human cohesin to laser-induced DNA damage. J Biol Chem. 2002;277:45149–45153. doi: 10.1074/jbc.M209123200. [DOI] [PubMed] [Google Scholar]
- 78.Kong X, Ball AR, Jr, Pham HX, Zeng W, Chen HY, Schmiesing JA, Kim JS, Berns M, Yokomori K. Distinct functions of human cohesin-SA1 and cohesin-SA2 in double-strand break repair. Mol Cell Biol. 2014;34:685–698. doi: 10.1128/MCB.01503-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim BJ, Li Y, Zhang J, Xi Y, Li Y, Yang T, Jung SY, Pan X, Chen R, Li W, Wang Y, Qin J. Genome-wide reinforcement of cohesin binding at pre-existing cohesin sites in response to ionizing radiation in human cells. J Biol Chem. 2010;285:22784–22792. doi: 10.1074/jbc.M110.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Uhlmann F, Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr Biol. 1998;8:1095–1101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- 81.Potts PR, Porteus MH, Yu H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 2006;25:3377–3388. doi: 10.1038/sj.emboj.7601218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McAleenan A, Clemente-Blanco A, Cordon-Preciado V, Sen N, Esteras M, Jarmuz A, Aragon L. Post-replicative repair involves separase-dependent removal of the kleisin subunit of cohesin. Nature. 2013;493:250–254. doi: 10.1038/nature11630. [DOI] [PubMed] [Google Scholar]
- 83.Caron P, Aymard F, Iacovoni JS, Briois S, Canitrot Y, Bugler B, Massip L, Losada A, Legube G. Cohesin protects genes against gammaH2AX Induced by DNA double-strand breaks. PLoS Genet. 2012;8:e1002460. doi: 10.1371/journal.pgen.1002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 85.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, van de Corput MP, van de Werken HJ, Knoch TA, van IWF, Grosveld FG, Ren B, Wendt KS. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 88.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 89.Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 91.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 92.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Asahina H, Kuraoka I, Shirakawa M, Morita EH, Miura N, Miyamoto I, Ohtsuka E, Okada Y, Tanaka K. The XPA protein is a zinc metalloprotein with an ability to recognize various kinds of DNA damage. Mutat Res. 1994;315:229–237. doi: 10.1016/0921-8777(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 94.Bal W, Schwerdtle T, Hartwig A. Mechanism of nickel assault on the zinc finger of DNA repair protein XPA. Chem Res Toxicol. 2003;16:242–248. doi: 10.1021/tx025639q. [DOI] [PubMed] [Google Scholar]
- 95.Nagaoka M, Kuwahara J, Sugiura Y. Alteration of DNA binding specificity by nickel (II) substitution in three zinc (II) fingers of transcription factor Sp1. Biochem Biophys Res Commun. 1993;194:1515–1520. doi: 10.1006/bbrc.1993.1996. [DOI] [PubMed] [Google Scholar]
- 96.Zhou X, Sun X, Mobarak C, Gandolfi AJ, Burchiel SW, Hudson LG, Liu KJ. Differential binding of monomethylarsonous acid compared to arsenite and arsenic trioxide with zinc finger peptides and proteins. Chem Res Toxicol. 2014;27:690–698. doi: 10.1021/tx500022j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chung JY, Yu SD, Hong YS. Environmental source of arsenic exposure. J Prev Med Public Health. 2014;47:253–257. doi: 10.3961/jpmph.14.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hartwig A. Zinc finger proteins as potential targets for toxic metal ions: differential effects on structure and function. Antioxid Redox Signal. 2001;3:625–634. doi: 10.1089/15230860152542970. [DOI] [PubMed] [Google Scholar]
- 99.Huestis J, Zhou X, Chen L, Feng C, Hudson LG, Liu KJ. Kinetics and thermodynamics of zinc(II) and arsenic(III) binding to XPA and PARP-1 zinc finger peptides. J Inorg Biochem. 2016;163:45–52. doi: 10.1016/j.jinorgbio.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ghirlando R, Felsenfeld G. CTCF: making the right connections. Genes Dev. 2016;30:881–891. doi: 10.1101/gad.277863.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R, Thurman RE, Kaul R, Myers RM, Stamatoyannopoulos JA. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bergman Y, Cedar H. DNA methylation dynamics in health and disease. Nat Struct Mol Biol. 2013;20:274–281. doi: 10.1038/nsmb.2518. [DOI] [PubMed] [Google Scholar]
- 103.Lai AY, Fatemi M, Dhasarathy A, Malone C, Sobol SE, Geigerman C, Jaye DL, Mav D, Shah R, Li L, Wade PA. DNA methylation prevents CTCF-mediated silencing of the oncogene BCL6 in B cell lymphomas. J Exp Med. 2010;207:1939–1950. doi: 10.1084/jem.20100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suva ML, Bernstein BE. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hou L, Zhang X, Wang D, Baccarelli A. Environmental chemical exposures and human epigenetics. Int J Epidemiol. 2012;41:79–105. doi: 10.1093/ije/dyr154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr Opin Pediatr. 2009;21:243–251. doi: 10.1097/mop.0b013e32832925cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ruiz-Hernandez A, Kuo CC, Rentero-Garrido P, Tang WY, Redon J, Ordovas JM, Navas-Acien A, Tellez-Plaza M. Environmental chemicals and DNA methylation in adults: a systematic review of the epidemiologic evidence. Clin Epigenetics. 2015;7:55. doi: 10.1186/s13148-015-0055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Govindarajan B, Klafter R, Miller MS, Mansur C, Mizesko M, Bai X, LaMontagne K, Jr, Arbiser JL. Reactive oxygen-induced carcinogenesis causes hypermethylation of p16(Ink4a) and activation of MAP kinase. Mol Med. 2002;8:1–8. [PMC free article] [PubMed] [Google Scholar]
- 109.Kovatsi L, Georgiou E, Ioannou A, Haitoglou C, Tzimagiorgis G, Tsoukali H, Kouidou S. p16 promoter methylation in Pb2+-exposed individuals. Clin Toxicol (Phila) 2010;48:124–128. doi: 10.3109/15563650903567091. [DOI] [PubMed] [Google Scholar]
- 110.Kondo Y, Shen L, Issa JP. Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol Cell Biol. 2003;23:206–215. doi: 10.1128/MCB.23.1.206-215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rea M, Eckstein M, Eleazer R, Smith C, Fondufe-Mittendorf YN. Genome-wide DNA methylation reprogramming in response to inorganic arsenic links inhibition of CTCF binding, DNMT expression and cellular transformation. Sci Rep. 2017;7:41474. doi: 10.1038/srep41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lu L, Wang L, Li T, Wang J. NF-kappaB subtypes regulate CCCTC binding factor affecting corneal epithelial cell fate. J Biol Chem. 2010;285:9373–9382. doi: 10.1074/jbc.M109.094425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yang B, Wagner J, Damaschke N, Yao T, Wuerzberger-Davis SM, Lee MH, Svaren J, Miyamoto S, Jarrard DF. A novel pathway links oxidative stress to loss of insulin growth factor-2 (IGF2) imprinting through NF-kappaB activation. PLoS One. 2014;9:e88052. doi: 10.1371/journal.pone.0088052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gilbert N, Allan J. Supercoiling in DNA and chromatin. Curr Opin Genet Dev. 2014;25:15–21. doi: 10.1016/j.gde.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kouzine F, Levens D, Baranello L. DNA topology and transcription. Nucleus. 2014;5:195–202. doi: 10.4161/nucl.28909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Naughton C, Avlonitis N, Corless S, Prendergast JG, Mati IK, Eijk PP, Cockroft SL, Bradley M, Ylstra B, Gilbert N. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol. 2013;20:387–395. doi: 10.1038/nsmb.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Madabhushi R, Gao F, Pfenning AR, Pan L, Yamakawa S, Seo J, Rueda R, Phan TX, Yamakawa H, Pao PC, Stott RT, Gjoneska E, Nott A, Cho S, Kellis M, Tsai LH. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell. 2015;161:1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.El-Khamisy SF, Caldecott KW. TDP1-dependent DNA single-strand break repair and neurodegeneration. Mutagenesis. 2006;21:219–224. doi: 10.1093/mutage/gel024. [DOI] [PubMed] [Google Scholar]
- 119.Puc J, Aggarwal AK, Rosenfeld MG. Physiological functions of programmed DNA breaks in signal-induced transcription. Nat Rev Mol Cell Biol. 2017;18:471–476. doi: 10.1038/nrm.2017.43. [DOI] [PMC free article] [PubMed] [Google Scholar]