Abstract
Intracellular delivery of nucleic acids to mammalian cells using polyplex nanoparticles remains a challenge both in vitro and in vivo, with transfections often suffering from variable efficacy. To improve reproducibility and efficacy of transfections in vitro using a next-generation polyplex transfection material poly(beta-amino ester)s (PBAEs), the influence of multiple variables in the preparation of these nanoparticles on their transfection efficacy was explored. The results indicate that the even though PBAE/pDNA polyplex nanoparticles are formed by self-assembly of polyelectrolytes, their transfection is not affected by the manner in which the components are mixed, facilitating self-assembly in a single step, but a timing for self-assembly of 5–20 min is optimal. In addition, even though the biomaterials are biodegradable in water, their efficacy is not affected by up to 8 freeze-thaw cycles of the polymer. It was found that there is greater stability of nucleic acid-complexed polymer as a polyplex nanoparticle compared to free polymer. Finally, by exploring multiple buffer systems, it was identified that utilization of divalent cation magnesium or calcium acetate buffers at pH 5.0 are optimal for transfection using these polymeric materials, boosting transfection several fold compared to monovalent cations. Together, these results can improve the reproducibility and efficacy of PBAE and similar polyplex nanoparticle transfections and improve the robustness of using these biomaterials for bioengineering and biotechnology applications.
Keywords: Polyplex nanoparticle, gene delivery, poly(beta-amino ester), transfection
1. Introduction:
Non-viral gene delivery is a routinely used approach in the laboratory to introduce genetic materials to mammalian cells. Nanoparticles (NPs) are often used to mediate effective cellular uptake, endosomal escape, and delivery to either the cytosol or the nucleus. Cationic polymeric materials, including polyethyleminine (PEI) (Boussif et al. 1995) and poly(beta-amino ester)s (PBAEs) (Akinc et al. 2003; Anderson et al. 2003; Lynn and Langer 2000), have been used extensively for gene delivery both in vitro and in vivo with promising results but to date have been slow to advance to the clinic due largely to insufficient efficacy of the materials (Yin et al. 2014).
In contrast to non-biodegradable PEI-based polymers, PBAEs are cationic polyesters that contain tertiary amines in the backbone of the polymer that facilitate rapid hydrolysis in aqueous solution, enabling PBAEs to be used with generally low cytotoxicity and effectively no risk of accumulation following repeat administration in vivo. PBAE NPs have been demonstrated to yield higher transfection efficacy than many commercial reagents in vitro and have been demonstrated to effectively deliver nucleic acids in vivo, even leading to improved survival outcomes in multiple tumor models with delivery of various plasmid DNA cargoes (Mangraviti et al. 2015; Min et al. 2018). Many researchers performing simple in vitro transfections may further benefit from the utilization of PBAEs for routine transfections for bioengineering and biotechnology applications due to their high efficacy, low cytotoxicity and rapid degradation rates that fully eliminate the polymers prior to peak gene expression approximately two days following transfection (Sunshine et al. 2012; Tzeng et al. 2011). The relative ease of synthesis of these materials and improvements in performance over many canonical transfection reagents makes PBAEs prime candidates for use as routine transfection reagents. Unlike many current commercial reagents, however, PBAEs are susceptible to degradation in aqueous solution, which may make them more sensitive to the manner in which these materials are stored and their NP derivatives are prepared.
To improve the reproducibility and efficacy of pre-clinical experiments using PBAE NPs and better enable high throughput methods for studying these materials, we performed assays to assess the influence of various experimental factors on transfection efficacy. To achieve this, we utilized two canonical, linear, end-capped PBAE polymers to transfect two well characterized cell lines with a reporter gene in vitro and quantified expression via flow cytometry. Multiple experimental parameters were explored with a focus on factors that can affect polyplex self-assembly including order of polyelectrolyte addition during polyplex self-assembly, volume of polyelectrolyte components during self-assembly, environmental conditions during polyplex self-assembly, polyplex self-assembly incubation time, and influence of hydrolysis of polymer. This approach enabled the discovery of parameters that produced optimized transfection as well as the determination of parameter ranges that ensure robust reproducibility.
2. Experimental Section:
2.1. Materials
Monomers for PBAE synthesis were purchased from vendors listed in Table S1. Stock solutions of 3 M sodium acetate (NaAc) (Sigma), 1 M HEPES (Quality biological), 1 M magnesium acetate (MgAc2) (Boston Bioproducts), 1 M calcium acetate (CaAc2) (VWR) were diluted to desired osmolarity and adjusted to set pH values. Buffer salts for 2-ethanesulfonic acid (MES), sodium citrate (Na2HCitr), sodium phosphate (Na2HPO4), magnesium citrate (MgHCitr) and calcium citrate (CaHCitr) were purchased from Sigma Aldrich. Branched PEI (BPEI, 25 kDa MW) was purchased from Sigma Aldrich. Plasmid eGFP-N1 (Addgene 2491) was used for assessment of transfection efficacy.
2.2. Transfection Assessment
All PBAE NPs were prepared in 25 mM NaAc, pH 5.0 at a 60 weight-weight (w/w) polymer/DNA ratio for all experiments and pipetting to mix as previously described unless otherwise noted (Tzeng et al. 2011). BPEI NPs were prepared in 150 mM NaCl by adding polymer to DNA in a 1:1 volume ratio at a 2 w/w ratio. Particles were added to HEK293T or MDA-MB-231 cells at DNA doses of 200 and 400 ng per well, respectively, in complete medium containing 10% serum and incubated for two hours, followed by a complete media change. Transfection efficacy was observed using a fluorescence microscope (Axiovert Observer A.1, Zeiss) and quantified using flow cytometry approximately 48 hours following transfection, using an Accuri C6 flow cytometer (BD Biosciences, Franklin Lakes, NJ) with Hypercyt high‐throughput sampler and reader (Intellicyt Corp., Albuquerque, NM). Transfection efficacy is generally reported as percent transfection in MDA-MB-231 cells and as normalized geometric mean expression in HEK293T cells, as the transfection was typically too high across all PBAE transfection conditions (>90% cells positive) in this cell type to differentiate between conditions using percent transfection. Figure S14 shows gating for flow cytometry analysis.
2.3. Mixing Volume ratio
PBAE NPs were prepared by pipet-mixing polymer and DNA at several volumetric ratios, with the polymer volume fractions varying from 0.15 to 0.95 in 25 mM NaAc buffer, pH 5.0. Follow-up experiments in which PBAE in DMSO was resuspended directly with dilute plasmid DNA were performed by aliquoting the PBAE in anhydrous DMSO (100 mg/mL) to tubes or round-bottom plates, then directly pipetting plasmid DNA pre-diluted in 25 mM NaAc, pH 5.0, to resuspend the PBAE for complexation into NPs.
2.4. Pre-transfection incubation time
To determine the influence of preparation time on transfection efficacy, NPs were prepared by pipet-mixing in a 1:1 volume ratio of dilute polymer and plasmid DNA at set times prior to addition of NPs to cells. For PBAE polymers, the additional step of dissolving polymer from anhydrous DMSO to aqueous buffer was performed two minutes prior to mixing dilute polymer solution with dilute plasmid DNA. Additional experiments were conducted in which polymer was dissolved in 25 mM NaAc by vortexing and pre-incubated for set amounts of time prior to complexation with plasmid DNA; after mixing with DNA, NPs were further incubated 5 minutes then added to cells. For all presented experiments, NPs were then added to HEK293T and MDA-MB-231 cells at low doses (200 ng and 400 ng DNA, respectively) and incubated with cells for two hours, after which media was aspirated and replaced with 100 μL/well of fresh, complete medium.
2.5. Freeze-Thaw Cycles and Water Content
Individual PBAE aliquots at 100 mg/mL in anhydrous DMSO were subjected to repeated freeze-thaw cycles performed on subsequent days. For thaw only cycles, polymer aliquots were thawed at room temperature for one hour in a container with desiccant then refrozen at −20°C a set number of times. Under matched conditions for open-air thaw conditions, polymer aliquots were thawed at room temperature with desiccant for one hour, then opened to the laboratory atmosphere for 5 minutes, capped, and refrozen to simulate controlled multiple uses from a single polymer aliquot. After freeze-thaw cycles were completed, all polymer aliquots were stored at −20°C for one month then used for assessment of transfection efficacy as described above. For water fraction experiments, PBAE 4-4-6 in anhydrous DMSO at 100 μg/μL was diluted to 50 μg/μL with additional solvent so that the final aliquot contained between 0–50% water by volume. Aliquots were then frozen at −20°C for two months, thawed, and held at room temperature for set amounts of time prior to be used for transfection efficacy assessment as described above.
2.6. Polymer degradation:
Two replicates each of PBAE (free polymer) and PBAE/DNA (60 w/w NPs) were incubated in either PBS (pH 7.4), citrate buffer (pH 6), or sodium acetate buffer (pH 5.2) at 37°C. At each specified time point, samples were frozen and lyophilized to remove aqueous solvent. Polymer was then dissolved in THF, filtered, and analyzed by gel permeation chromatography (GPC) as previously described (Bishop et al. 2013). Number average (Mn) and weight average (Mw) molecular weights were determined against linear polystyrene standards at each time point. One-phase decay plots were fit to degradation plots to determine ester bond half-life.
2.7. Buffer system
For the initial experiment, buffer solutions of sodium acetate (NaAc), 2-(N-morpholino)ethanesulfonic acid (MES), sodium phosphate (NaH2PO4), sodium citrate (Na2HCitr), potassium acetate (KAc), and magnesium acetate (MgAc2) were prepared as 1 M stocks in ultrapure water and sterile-filtered. Each stock buffer was split into four groups, diluted to 25 mM in ultrapure water, and adjusted to pH 4, 5, 6, or 7 assessed with a Mettler Toledo SevenEasy pH meter. PBAE NPs were formed in each buffer by resuspending PBAE polymer at 100 mg/mL in DMSO in a 96-well round-bottom plate, followed by mixing with dilute plasmid DNA using a multichannel pipette. NPs were then added to HEK293T and MDA-MB-231 cells at low doses (200 ng and 400 ng DNA, respectively) and incubated with cells for two hours, after which media was aspirated and replaced with 100 μL/well of fresh, complete medium. Following results from the first experiment, additional buffers of calcium acetate (CaAc2) magnesium citrate (MgHCitr), and calcium citrate (CaHCitr) were prepared at 25 mM concentrations and pH 5.0. Divalent cation citrate buffers were marginally soluble and used at their solubility limits, estimated to be approximately 4 mM. Composite buffers mimicking calcium phosphate transfection conditions were also utilized (Kingston et al. 2003). Uptake experiments were performed with 20% of the plasmid DNA labeled with Cy5 as previously described (Wilson et al. 2017b).
2.8. Data analysis, statistics and Figures:
FlowJo (FlowJo, LLC) was used for flow cytometry analysis. Servier Medical Art (CC license) was used for illustrations. Prism 6 (Graphpad, La Jolla, CA) was used for all statistical analyses and curve plotting. For multiple comparisons between all tested conditions, ordinary one-way ANOVA with Tukey corrected multiple comparisons were performed. Similarly, for multiple comparisons tests against a specific control condition one-way ANOVA with Dunnett corrected multiple comparisons was performed. Following one-way ANOVA, a Brown-Forsythe test of equal variance was performed to assess if specific conditions possessed significantly different standard deviations. Unless otherwise specified, statistical tests were performed with a global alpha value of 0.05, and experiments were repeated at least twice with representative results shown. Unless otherwise stated, absence of statistical significance markings where a test was stated to have been performed signifies no statistical significance. Statistical significance is denoted as follows: *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
3. Results and Discussion
We sought to determine if incubation time influences transfection efficacy with these materials since PBAE polymers undergo hydrolysis following dissolution in aqueous solutions, with ester bond half-life usually between 2–6 hours (Sunshine et al. 2012; Tzeng and Green 2013). The experiments revealed that the optimal aqueous incubation time for PBAE polymer prior to and after mixing with plasmid DNA are <10 minutes and between 5–20 minutes, respectively. Further exploring the influence of hydrolysis in stored aliquots of PBAE on NP efficacy, our results indicated that polymer aliquots stored at −20°C were stable over up to eight freeze-thaw cycles, making these materials promising for use as general-use transfection reagents where a single polymer aliquot can be used multiple times without concern of reduction in efficacy.
We further explored the influence of volumetric ratio of PBAE to DNA during NP mixing to show that, as long as NPs are sufficiently mixed, the volume of each solution prior to mixing made no difference in transfection efficacy. These results were further expanded to show that PBAE polymer in DMSO could be directly resuspended with dilute plasmid DNA in buffer to give equally effective NPs, which has implications for making NPs at high DNA concentrations where PBAE polymer solubility would otherwise limit the dose. Finally, we explored alternative buffer systems to the traditional pH 5 sodium acetate buffer used and discovered that divalent cation magnesium and calcium acetate buffers improve transfection efficacy. Here, we explored the influence of assembly parameters on only two relatively hydrophilic PBAE structures which are representative of the most commonly used PBAE materials for gene delivery in the literature. PBAEs possessing more hydrophilic (such as PEG-diacrylate based) or hydrophobic (such as PBAE utilizing alkyl side chains (Eltoukhy et al. 2013)) may perform differently.
Polymer synthesis and characterization
PBAE polymers were synthesized as previously described (Tzeng et al. 2011; Wilson et al. 2017a). PBAE 4-4-6 used in this study was characterized via GPC to have Mn 5,170 Da, Mw 10,220 Da, and dispersity (D) 1.98, while PBAE 4-5-39 was characterized to have Mn 4,580 Da, Mw14,290 Da, and dispersity (D) 3.12. PBAE 4-5-39 is a new chemical entity using end-cap E39, which is similar to our previously reported E7 end-cap but is slightly more hydrophilic, possessing a secondary amine in the piperazine ring instead of a tertiary amine. Together, these two PBAE structures are representative of relatively hydrophilic PBAE structures (via the use of PBAE 4-4-6) and more hydrophobic structures similar to C32 (via 4-5-39) {Anderson, 2004 #902}.
Pre-Transfection Incubation Time
We performed experiments to test if the incubation time of PBAE NPs in buffer prior to adding to cells influenced transfection efficacy. Many transfection reagents recommend rapidly pipetting or vortexing aqueous solutions of transfection reagent and plasmid DNA followed by an undisturbed incubation period ranging from 5–30 minutes for DNA complexation. We hypothesize that the rapid degradation of PBAE polymers in aqueous solution might influence the optimal incubation time of these materials prior to adding them to cells. To explore this variable, we tested both the pre-incubation time effects of free PBAE polymer and the incubation time effects of complexed PBAE/DNA NPs (Fig. 1A). First, the pre-incubation time of PBAE polymer alone in aqueous solution was tested, showing decreases in efficacy following pre-incubation times greater than 10–20 minutes (Fig. 1B,C). This result was consistent across cell lines and with two PBAE polymers of varying hydrophobicity (Fig. S2A,B). Following these results, we tested the effect of incubating PBAE/DNA NPs following NP formation to determine optimal incubation time to allow for effective polymer/DNA self-assembly and found that an incubation time of between 5–20 minutes was optimal (Fig. 1D,E and S2C,D). While long incubation times showed decreases transfection efficacy presumably due to degradation, short incubation times of less than one minute following mixing of plasmid DNA and PBAE polymer also showed statistically lower transfection efficacy across polymers and cell types.
Figure 1.
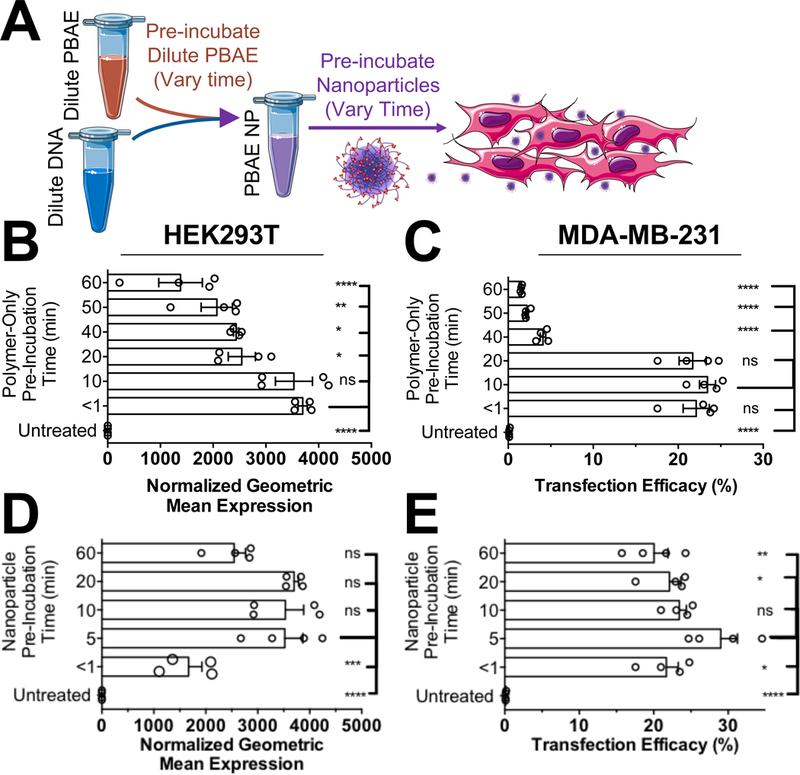
Effect of pre-incubation time of free PBAE polymer and of formed PBAE/DNA nanoparticles on transfection. A) Either PBAE 4–4-6 nanoparticles were formed and pre-incubated for set timepoints before adding to cells or PBAE 4–4-6 was pre-dissolved in 25 mM NaAc buffer and pre-incubated for set timepoints before forming nanoparticles that were then added to cells. PBAE polymer pre-incubated for 10 minutes or less prior to nanoparticle formation was most effective for transfection in (B) HEK293T and (C) MDA-MB-231 cells. Nanoparticles that were pre-incubated 5–20 minutes before adding to cells were most effective for transfection of (D) HEK293T and (E) MDA-MB-231 cells.
To identify why PBAE NPs were susceptible to optimal incubation times, we assessed the diameter of PBAE 4-4-6 free polymer and NPs over the time span of one hour in 25 mM NaAc buffer, pH 5. Supporting the transfection results, PBAE/DNA NPs were stable over one hour (Fig. S3), indicating that PBAE/DNA NPs were effectively stable in acidic buffer over time. We also measured the degradation rate of free PBAE polymer and PBAE polymer/DNA complexes in aqueous buffer at different pH values (Fig. S4). At pH 5.2 in NaAc buffer, the ester bond half-life of free PBAE polymer MN was only 1.6 hours, while PBAE/pDNA polyplexes was 3.5 hours (Table S2). This difference in degradation rate for free PBAE polymer compared to PBAE/pDNA complexes was statistically significant as assessed by a one-phase decay model at pH 5.2 (p<0.0001 for both MN and MW). Free PBAE polymer was further shown to degrade faster at pH 6 and pH 7.4 than pH 5.2, though there were no statistical differences between free PBAE polymer and PBAE/pDNA polyplexes in degradation rate at higher pH values. This result is consistent with ours and others’ previously published results showing that degradation of this class of polymers is more rapid at neutral pH (Lynn and Langer 2000; Tzeng and Green 2013; Zhong et al. 2005) than at weakly acidic conditions. We hypothesize that when many of the amines in the PBAE backbone are not protonated (i.e., >pH 6), they behave as bases and can locally generate -OH species near the ester backbone. The -OH species is then free to act as a nucleophile and degrade the ester bonds. At lower pH values (<pH 6), when more amines in the PBAE backbone are protonated, the amines no longer generate as much hydroxide, and degradation of the polymer backbone is slowed, although this degradation is still significantly faster than the degradation of other polyesters such as PLGA, which degrade with a half-life on the order of weeks (Anderson and Shive 1997).
Together, these results support limiting the residence time of free PBAE polymer in aqueous solution to <20 minutes and keeping the PBAE polymer/DNA complex incubation time between 5–20 minutes for optimal transfection. In contrast, we performed a similar assay with BPEI/DNA and found that pre-incubation times between 10–60 minutes were not statistically different in transfection efficacy for this non-degradable cationic polymer (Fig. S5). We hypothesize that the differences observed in the optimal pre-incubation time between PBAE NPs and BPEI NPs are primarily attributable to degradation associated with PBAEs.
Mixing Volume Ratio
Polyplex NPs of pDNA interacting with PBAEs and BPEI prepared by pipet mixing have traditionally been mixed in a 1:1 volume ratio of dilute nucleic acid to dilute polymer for simplicity (Anderson et al. 2004; Boussif et al. 1995). Other strategies for forming polyplex NPs, including microfluidic mixing (Lu et al. 2014) and flash nanoprecipitation (Santos et al. 2016), vary from this 1:1 mixing volume ratio only minimally, with nucleic acids accounting for at least one third of the total volume of solution. For many assays, including semi-automated high throughput screening situations, and for in vivo administration, however, the ability to vary the volume fraction of the polymer solution in forming polyplex NPs would be beneficial. For PBAEs, their amphiphilic nature makes them less water-soluble than many other polyplex NP systems with a solubility limit generally approximately 20 mg/mL in acidic buffer.
For this purpose, we sought to test if the volume fraction of mixing dilute PBAE polymer with dilute plasmid DNA influenced the transfection efficacy of the resulting NPs (Fig. 2A). Testing PBAE polymer volume fractions of 0.15–0.95 demonstrated that the mixing volume ratio did not have a statistically significant influence on the transfection efficacy in either cell line with PBAE 4-4-6 (Fig. 2B,C). These results were confirmed with PBAE 4-5-39 as well (Fig S6), where no statistically significant difference in transfection efficacy was noted among the different polymer volume fractions. The standard deviations of wells transfected with high polymer volume fractions were statistically higher as assessed by the Brown-Forsythe test of equal variance, but only for HEK293T cells transfected with PBAE 4-4-6.
Figure 2.
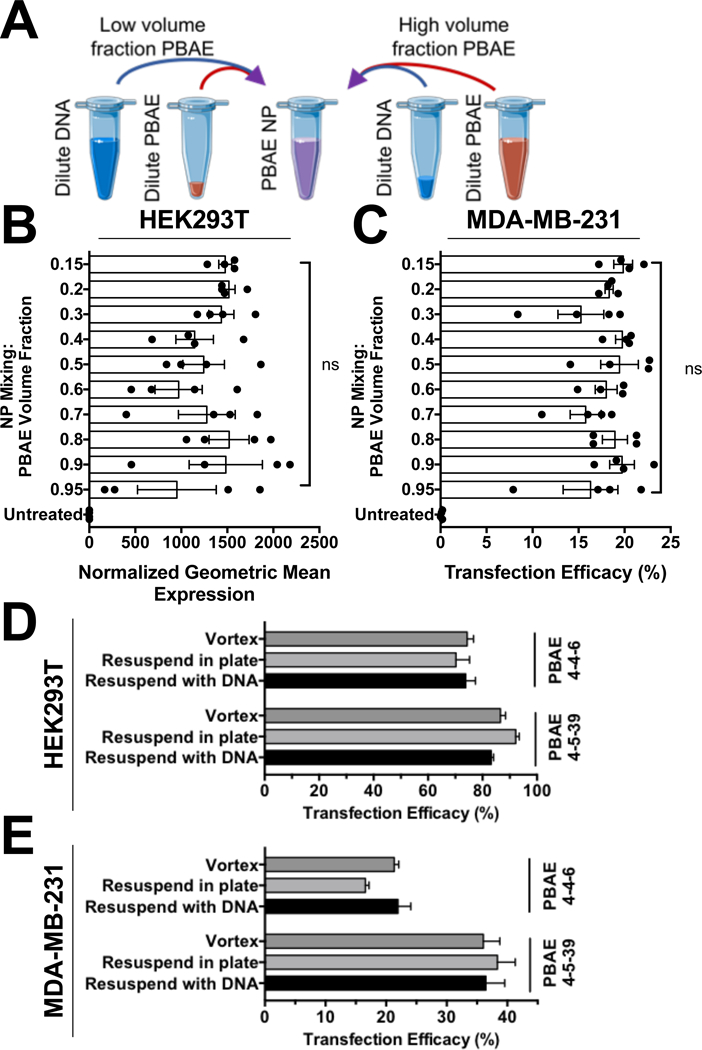
(A) PBAE NPs are not highly sensitive to the mixing volume ratio between dilute polymer and plasmid DNA. There were no statistical differences (one-way ANOVA with Tukey corrected multiple comparisons) between any transfected groups for (B) HEK293T or (C) MDA-MB-231 cells transfected with PBAE 4–4-6. The method of resuspending PBAE polymer in aqueous solution likewise did not influence the efficacy of the nanoparticles in (D) HEK293T or (E) MDA-MB-231 cells. Bars show mean ± SEM of four wells.
To facilitate high-throughput screening of polymeric NPs, we further sought to test if resuspending PBAE polymer by vortexing, resuspending by pipetting in a plate with buffer, or resuspending by pipetting in a plate with pre-diluted pDNA in buffer influenced the resulting transfection efficacy. The ability to resuspend PBAE polymer and form NPs in a single step simply by simply adding pre-diluted pDNA in buffer to a set volume of stock polymer at 100 mg/mL in DMSO would considerably facilitate rapid testing of these materials by skipping the polymer dilution step and enable parallelization in preparation of NPs. Surprisingly, we found no difference in transfection efficacy resulting from resuspending PBAE polymers in a round-bottom plate by forming the NPs in two steps or by direct resuspension to NPs in a single step by pipetting dilute pDNA in buffer to resuspend the polymer (Fig. 2D,E). Together, these results regarding methods of self-assembly are promising for using PBAEs in parallel semi-automated screening techniques and for their use as routine transfection reagents, robust to mixing technique variability among users.
Freeze/Thaw Cycles
PBAE polymers are notable for their rapid degradation in aqueous solution compared to traditional polyesters such as poly(lactide-co-glycolide) (PLGA) or polycaprolactone (PCL) (Akinc et al. 2003; Lynn and Langer 2000). The degradation half-life of the ester bond in the backbone of PBAE polymers has been measured to typically range from 2 to 6 hours depending on hydrophobicity (Lynn and Langer 2000; Sunshine et al. 2012) and local pH (Lynn and Langer 2000; Sunshine et al. 2012; Tzeng and Green 2013; Zhong et al. 2005). To avoid the early degradation that would be expected to reduce efficacy of the polymers, PBAEs are traditionally stored in anhydrous DMSO at −20°C with silica desiccant, but the use of molecular sieves or other stronger desiccants is not needed. Utilization of PBAE polymers as routine in vitro transfection reagents or for clinical formulations requires sufficient stability during storage to ensure the polymers have the same efficacy after storage.
To address these concerns, we performed thawed PBAE aliquots in anhydrous DMSO repeatedly on subsequent days either under anhydrous conditions or under open-atmosphere laboratory conditions to simulate multiple opening-closing events for individual aliquots of polymer. After a set number of freeze-thaw cycles on subsequent days, polymer aliquots were stored at −20°C for one month, then thawed a final time and used for transfection. We hypothesized that the repeated freeze-thaw process or water absorption from the atmosphere would result in sufficient polymer degradation to reduce efficacy of the materials following multiple freeze-thaw cycles. DMSO is a highly hygroscopic solvent, and water absorption from the atmosphere can be a major limiting factor for high throughput compound library screening when compounds are stored as DMSO stocks (Ellson et al. 2005). As reported, open-air exposure of DMSO to typical laboratory air for one hour can result in absorption of 6% water by volume (Ellson et al. 2005).
Transfections were conducted on two cell lines with two PBAE structures (Fig. 3 and S7) and surprisingly demonstrated no statistical change in transfection efficacy of these materials over eight freeze-thaw cycles for the PBAE aliquots exposed to anhydrous or open-atmosphere conditions compared to the control aliquot of polymer thawed only once. While the hygroscopic nature of DMSO was a concern, the amount of time the polymers spent at room temperature and open to laboratory air did not seem to appreciably influence the stability of the polymers stored at −20°C.
Figure 3.
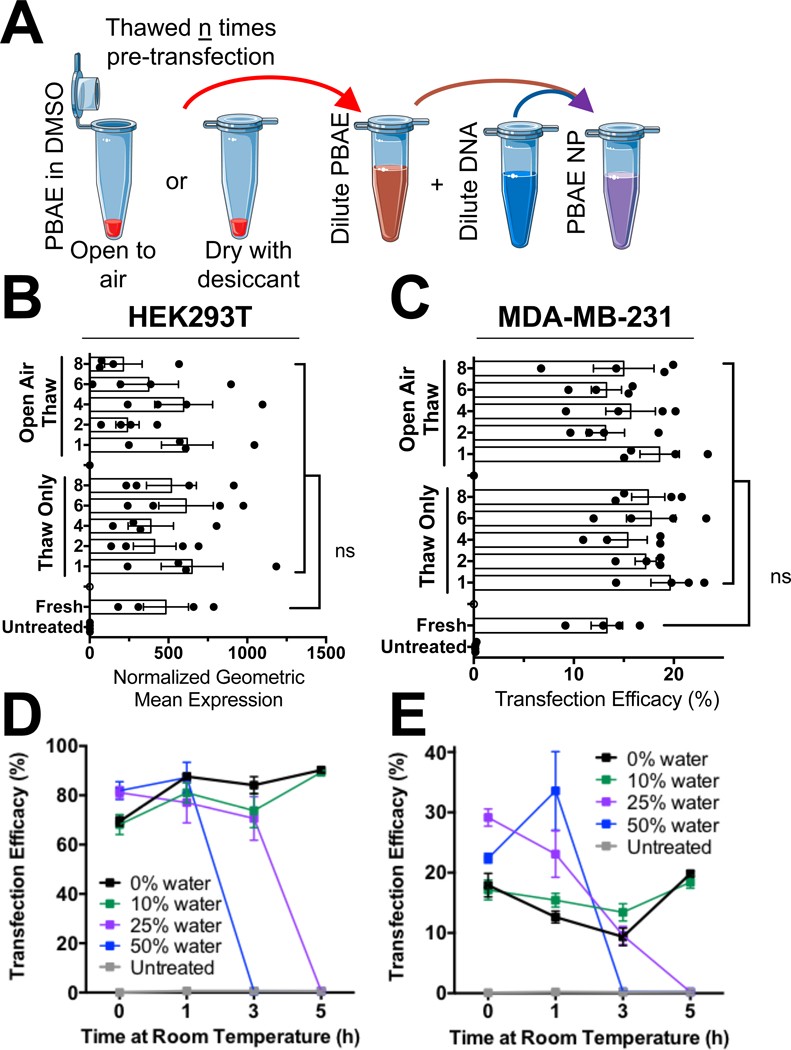
PBAE polymers are not prone to freeze-thaw induced reductions in efficacy. (A) Schematic of experimental outline for testing effect of freeze-thaw cycles on polymer efficacy. (B) HEK293T and (C) MDA-MB-231 cell transfection efficacy were not statistically different even with PBAE 4–4-6 having undergone eight freeze-thaw cycles (one-way ANOVA with multiple comparisons to the fresh polymer). Similarly, PBAE 4–4-6 aliquots pre-mixed to 50 mg/mL prior to storage with varying degrees of hydrated DMSO showed no loss in efficacy when stored at −20°C for two weeks and then used within an hour after thawing in (D) HEK293T or (E) MDA-MB-231 cells.
We next sought to determine if water absorption, specifically in anhydrous DMSO, and subsequent degradation when stored at −20°C has the potential to reduce efficacy of these materials over time. For this experiment, polymer aliquots were prepared with defined water content ranging from 0.25% to 50%, frozen at −20°C for two months, then thawed and used for transfection. Our results surprisingly showed that PBAE 4-4-6 lost no efficacy over two months when stored at −20°C and used immediately after thawing, even with 50% water solutions (Fig. S8). The amount of time that polymer spent in hydrated DMSO at room temperature did, however, affect transfection efficacy. Aliquots stored with 25% and 50% water by volume became completely ineffective at three and five hours of room temperature incubation respectively, while aliquots with 10% water by volume maintained equal efficacy to anhydrous polymer aliquots even after five hours of room temperature incubation (Fig. 3D,E). These results were consistent between both HEK293T and MDA-MB-231 cells, which we hypothesize is attributable to appreciable polymer degradation in hydrated DMSO solutions. Given the water absorption rate of DMSO, we recommend limiting room temperature incubation of these polymers to less than five hours and open-air exposure of the DMSO stocks to less than an hour to keep absorbed water content below 10%. With these recommendations, PBAEs are unlikely to suffer degradation induced reductions in efficacy. These results are largely consistent with previous reports of storing lyophilized PBAE NPs at 4°C and −20°C, demonstrating that the minute amounts of water absorbed from the air are effectively inconsequential as long as the materials are stored at −20°C and the amount of time at room temperature is limited to during use for transfection (Tzeng et al. 2015).
Buffer System
PBAE NPs have traditionally been prepared in low salt (25 or 50 mM), acidic, pH 5 sodium acetate (NaAc) buffer, which ensures tertiary amines in the backbone of the polymer are charged, facilitating complexation with anionic DNA (Anderson et al. 2003). Due to the amphiphilic and pH-sensitive nature of PBAE polymers, buffer pH as well as osmolarity and valence were all expected to influence NP formation conditions by affecting the cationicity and hydrophobicity of the PBAE polymer as well as potentially influencing DNA structure. At pH 5, most tertiary amines of linear PBAE structures are protonated, whereas, at neutral pH, tertiary amines in the polymer backbone remain largely unprotonated (Sunshine et al. 2012). This increase in cationicity likewise influences the solubility of PBAE structures, as more highly charged cationic structures are able to undergo hydrogen bonding at low pH but are less soluble above pH 6.0.
The sodium acetate buffer system is notably monovalent for both ions, which are expected to participate minimally in interactions between cationic polymer and anionic DNA. To explore the influence of buffer valence, we evaluated buffer systems with both divalent cations and anions, which were anticipated to participate in binding interactions with either the cationic polymer or anionic DNA. Among anions, citrate buffers have long been used when forming liposomes and lipid NPs, typically at pH 4 (Chen et al. 2012; Dahlman et al. 2014; Whitehead et al. 2014). Similarly, the zwitterionic buffer HEPES has often been used to buffer BPEI in 150 mM NaCl, but due to its limited neutral buffering range, we substituted MES as a single-component zwitterion buffer (Ogris et al. 2003). Phosphate buffers have likewise been used in transfection, although primarily as a means to precipitate calcium phosphate DNA particles for low-efficiency calcium phosphate-based transfection (Kingston et al. 2003).
We initially tested six biological buffer systems at 25 mM concentration and pH values of 4, 5, 6 and 7 with PBAE 4-4-6/pDNA polyplex NPs. Comparing by pH across all buffer systems, PBAE 4-4-6 yielded statistically higher transfection efficacy in both cell lines when using acidic buffers of pH of 4 or 5 compared to pH 7 buffers (Fig. 4A,B), although this result was not universal across all buffer systems and was more pronounced in HEK293T cells. No buffers resulted in dramatic differences in measured cell viability (Fig. 4C,D), and all buffer systems yielded some level of transfection. Magnesium acetate and sodium citrate buffers at pH 5 repeatedly yielded the highest transfection efficacy and were further explored in subsequent transfection screens in which we also included a calcium acetate (CaAc2) buffer (pH 5, 25 mM).
Figure 4.
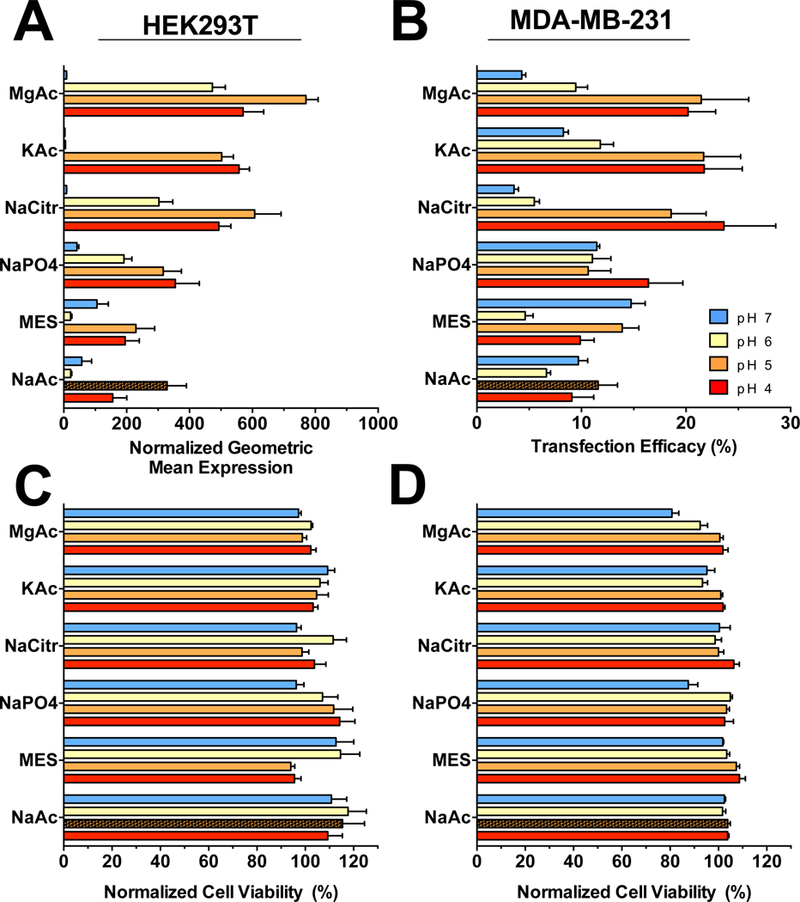
Buffer effect on PBAE polyplex NP transfection efficacy. Buffers from six salts were prepared at 25 mM osmolarity and pH values of 4, 5, 6 and 7 and used to make PBAE 4–4-6 nanoparticles compared against the historical buffer of pH 5 sodium acetate (NaAc) shown as the hatched bar. Several buffers showed promising increases in transfection efficacy in (A) HEK293T and (B) MDA-MB-231 cells with effectively no difference in cytotoxicity (C,D). To assess the influence of buffer pH on transfection, we performed a Tukey-corrected matched comparisons one-way ANOVA across all buffer conditions with respect to pH to demonstrate that pH 4 and pH 5 buffers were statistically the most effective buffers among all groups with significantly greater transfection than pH 7 buffers. Bars show mean ± SEM of four wells.
Divalent cation MgAc2 and CaAc2 buffers at pH 5 and 25 mM concentrations were observed to statistically improve transfection efficacy in all tested cases with PBAE NPs 4-4-6 and 4-5-39 in both HEK293T and MDA-MB-231 cells (Fig. 5 and S9). The improvement in transfection efficacy was notable in both cell types as assessed by either geometric mean expression, where calcium acetate yielded five-fold higher expression in either cell type, and percent transfection, where calcium acetate yielded 20% and 50% higher number of cells detectably expressing GFP in HEK293T and MDA-MB-231 cells respectively. To identify why divalent cation buffers improve transfection efficacy, we measured the uptake of PBAE 4-4-6 NPs formed in different acetate buffers but found no statistically significant differences in the level of NP uptake in HEK293T cells (Fig. S10). MDA-MB-231 cell uptake was statistically but only modestly higher with divalent cation buffers (Fig. S10,C,D). We also measured the particle diameter and zeta-potential of NPs formed in NaAc and MgAc2 buffers, which were observed to have the no statistically significant difference between particles prepared in either buffer (Fig. S11). These results indicated that the presence of divalent cations was not leading to aggregation of the PBAE NPs or a change in their diameters that would bias them towards greater uptake or transfection efficacy exclusively due to size differences.
Figure 5.
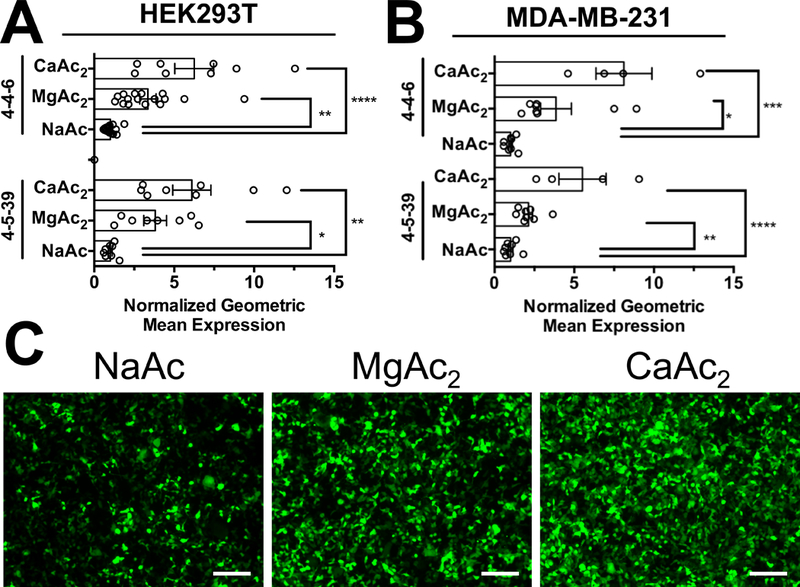
Acetate anion buffers with divalent magnesium or calcium cations improve transfection efficacy over monovalent sodium acetate buffer. PBAEs 4–4-6 and 4–5-39 had statistically higher transfection efficacy with MgAc2 and CaAc2 buffers than NaAc in (A) HEK293T cells and (B) MDA-MB-231 cells, shown as normalized to the level of transfection efficacy in pH 5, 25 mM NaAc buffer. (C) Microscopy showed increase in geometric mean expression of HEK293T cells using divalent cation acetate buffers. Results show multiple experiments with four replicates each, normalized to NaAc buffer transfection efficacy for that experiment. Scale bars 200 μm.
To further examine how divalent cation buffers might be affecting transfection efficacy, we measured the degree of pDNA binding with PBAE polymer using a variety of assays. Gel electrophoresis retention assays with a PBAE w/w titration and heparin sulfate competition binding assay both showed slightly reduced binding efficacy between PBAE polymer and pDNA in MgAc2 buffer compared to NaAc buffers (Fig. S12). Competition binding assays performed using the DNA-binding carbocyanine nucleic acid dyes YO-PRO-1 and YOYO-1 were further consistent with gel retention assay results, indicating slightly stronger binding of PBAE polymers to pDNA in NaAc buffer (Fig. S12,F,G). The presence of magnesium ions notably affected the fluorescence of YOYO-1 and YO-PRO-1 even in the absence of PBAE polymer, indicating that the concentration of divalent cation salts used were likely influencing DNA structure (Fig. S12,C–E). YOYO-1 and YO-PRO-1 are known to primarily interact with DNA via intercalation, resulting in approximately 1000-fold higher fluorescence upon binding double stranded nucleic acids (Armitage 2005). In the absence of PBAE polymer, we observed a dramatic increase of over 40% in the fluorescence of YOYO-1 binding to pDNA in MgAc2 buffer compared to that in NaAc, whereas YO-PRO-1 fluorescence decreased by 85% in MgAc2 buffer compared to that NaAc buffer, which is consistent with the magnesium ion’s influencing pDNA structure via electrostatic interactions.
The presence of magnesium, calcium and other divalent cations in solution has long been known to affect DNA structure, as hexahydrated magnesium ions bind the major groove of double stranded DNA and are involved in stabilization of single-stranded RNAs (Guéroult et al. 2012; Robinson et al. 2000). Magnesium or calcium ions are further required to facilitate the DNA binding activity of many enzymes and affect the melting temperature of oligonucleotides in predictable fashions. To determine if the presence of magnesium and calcium improved transfection by changing NP properties or by changing cell phenotype due to their presence in media during transfection, we added extra divalent cation salts to cell culture media directly either before or after transfection with PBAE NPs formed in NaAc buffer (Fig. 6A). Interestingly, the increase in transfection efficacy associated with the use of divalent cation buffers only occurred when PBAE NPs were prepared directly with divalent cation acetate buffers and not when divalent cation salts were simply added to media (Fig. 6B,C). Magnesium sulfate and calcium chloride are present at 0.8 and 1.8 mM concentrations in DMEM, respectively, meaning that the addition of NPs formed in 25 mM divalent cation acetate buffer increased the media concentration of magnesium or calcium only by 5.2 and 2.3 fold respectively. This short duration of exposure to higher divalent cation salt concentrations was not expected to change cell phenotype directly in vitro. Magnesium and calcium ions are largely considered biocompatible with the normal range of blood plasma levels of magnesium and calcium between 0.6–1.1 and 2.1–2.6 mM respectively. The fact that divalent cation buffers improved transfection only when used to form PBAE NPs and not when simply added to cell culture medium indicates that these buffers may likewise improve in vivo transfection efficacy.
Figure 6.
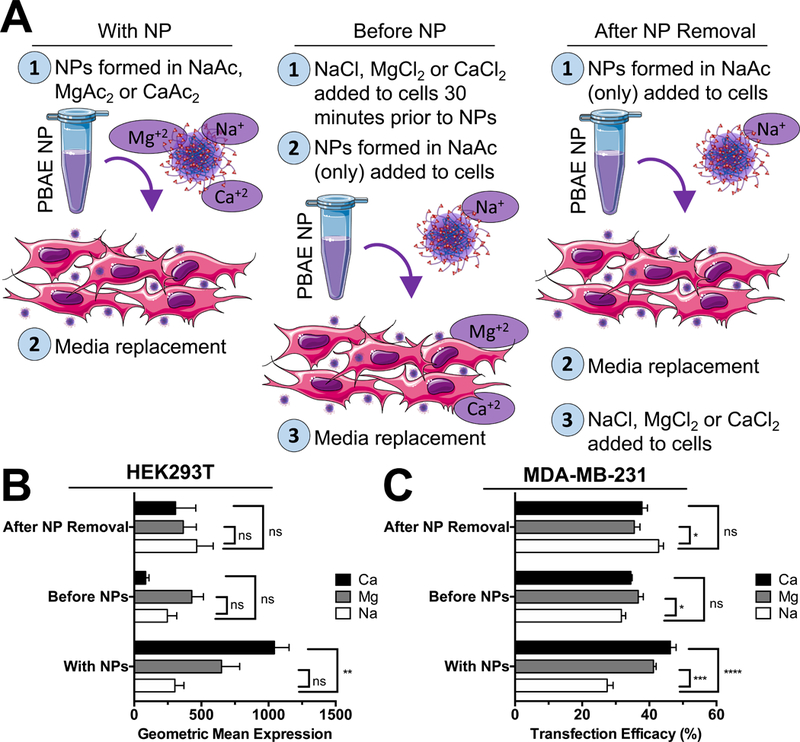
Divalent cation buffers only improve transfection when used directly to prepare NPs instead of simply by being added to media. (A) To identify if divalent cations were affecting NPs or cells directly, NaCl, MgCl2 or CaCl2 were added at concentrations to mimic presence when using MgAc2 or CaAc2 buffers and added to cells either prior to or after removal of NPs prepared in NaAc buffers only. In parallel, NPs formed with divalent cations were complexed in NaAc, MgAc2 or CaAc2 buffers and added to cells. Transfection efficacy with PBAE 4–4-6 was improved only for NPs prepared in divalent cation acetate buffers in both (B) HEK293T and (C) MDA-MB-231 cells and not by addition of divalent cation salts to media. Bars show mean ± SEM of four well replicates.
Inspired by historical transfection systems using divalent calcium and phosphate buffers to co-precipitate plasmid DNA, we also explored using composite buffer systems that, once mixed, resulted in insoluble or marginally soluble composites designed to purposefully precipitate NPs of polymer and plasmid DNA (Fig. S13). While calcium phosphate transfection is no longer commonly used due to its inconsistency and low transfection efficacy (usually <10% cells), we hypothesized that the ability to selectively enrich the media in direct contact with cells with NPs containing plasmid DNA may prove useful (Kingston et al. 2003). Our results indicated that these buffer systems were either generally ineffective, unreliable in the distribution of transfected cells or highly cytotoxic at the doses explored. Divalent calcium and magnesium acetate buffers outperformed all other buffer combinations, with notably higher cell viability and consistency of transfection throughout each well.
4. Conclusions:
The transfection studies demonstrate the importance of multiple experimental variables involved in the reproducible utilization of polyplex poly(beta-amino ester)/pDNA NPs for transfection. Importantly, these results demonstrate that PBAE polymers are largely insensitive to changes in the ratio of mixing between polymer and pDNA and are able to be resuspended directly from DMSO stocks using dilute DNA solutions to self-assemble into nanoparticles in a single step. These polymers are further insensitive to freeze-thaw cycles and contamination by water as long as they are stored at −20°C and are not kept thawed at room temperature for extended times. While consistent when used within 20 min, the polyplex NPs formed with these polymers become sensitive to degradation in aqueous buffers beyond 20 min of complexation time. Finally, our exploration of buffer systems other than sodium acetate identified divalent cation acetate buffers, such as magnesium acetate and calcium acetate, as optimal for transfection efficacy in multiple PBAE structures and cell lines in vitro, boosting transfection by severalfold. Together, these results demonstrate PBAE polymers a promising general-purpose transfection reagent for in vitro transfections and high-throughput screening applications.
Supplementary Material
Acknowledgements
The authors thank the following sources for funding support: NSF Graduate Research Fellowships DGE-0707427 to DRW; the Bloomberg~Kimmel Institute for Cancer Immunotherapy; the NIH (R01EB016721, R01EB022148, R01CA195503, the Wilmer Core Grant P30 EY001765 and T32 GM7300); and a Research to Prevent Blindness / Dr. H. James and Carole Free Catalyst Award for Innovative Research Approaches for Age-Related Macular Degeneration to JJG.
References:
- Akinc A, Anderson DG, Lynn DM, Langer R. 2003. Synthesis of poly (β-amino ester) s optimized for highly effective gene delivery. Bioconjugate chemistry 14(5):979–988. [DOI] [PubMed] [Google Scholar]
- Anderson DG, Lynn DM, Langer R. 2003. Semi-automated synthesis and screening of a large library of degradable cationic polymers for gene delivery. Angewandte Chemie (International ed. in English) 42(27):3153–8. [DOI] [PubMed] [Google Scholar]
- Anderson DG, Peng W, Akinc A, Hossain N, Kohn A, Padera R, Langer R, Sawicki JA. 2004. A polymer library approach to suicide gene therapy for cancer. Proceedings of the National Academy of Sciences of the United States of America 101(45):16028–16033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JM, Shive MS. 1997. Biodegradation and biocompatibility of PLA and PLGA microspheres. Advanced drug delivery reviews 28(1):5–24. [DOI] [PubMed] [Google Scholar]
- Armitage BA. 2005. Cyanine dye–DNA interactions: intercalation, groove binding, and aggregation DNA binders and related subjects: Springer; p 55–76. [Google Scholar]
- Bishop CJ, Ketola T-m, Tzeng SY, Sunshine JC, Urtti A, Lemmetyinen H, Vuorimaa-Laukkanen E, Yliperttula M, Green JJ. 2013. The effect and role of carbon atoms in poly(β-amino ester)s for DNA binding and gene delivery. Journal of the American Chemical Society 135(18):6951–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussif O, Lezoualc’h F, Zanta Ma, Mergny MD, Scherman D, Demeneix B, Behr JP. 1995. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proceedings of the National Academy of Sciences of the United States of America 92(16):7297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Love KT, Chen Y, Eltoukhy AA, Kastrup C, Sahay G, Jeon A, Dong Y, Whitehead KA, Anderson DG. 2012. Rapid Discovery of Potent siRNA-Containing Lipid Nanoparticles Enabled by Controlled Micro fl uidic Formulation.8–11. [DOI] [PubMed] [Google Scholar]
- Dahlman JE, Barnes C, Khan OF, Thiriot A, Jhunjunwala S, Shaw TE, Xing Y, Sager HB, Sahay G, Speciner L and others. 2014. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nature nanotechnology 9(May). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellson R, Stearns R, Mutz M, Brown C, Browning B, Harris D, Qureshi S, Shieh J, Wold D. 2005. In situ DMSO hydration measurements of HTS compound libraries. Combinatorial chemistry & high throughput screening 8(6):489–498. [DOI] [PubMed] [Google Scholar]
- Eltoukhy AA, Chen D, Alabi CA, Langer R, Anderson DG. 2013. Degradable terpolymers with alkyl side chains demonstrate enhanced gene delivery potency and nanoparticle stability. Advanced Materials 25(10):1487–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guéroult M, Boittin O, Mauffret O, Etchebest C, Hartmann B. 2012. Mg2+ in the major groove modulates B-DNA structure and dynamics. PloS one 7(7):e41704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston RE, Chen CA, Rose JK. 2003. Calcium phosphate transfection. Current protocols in molecular biology:9–1. [DOI] [PubMed] [Google Scholar]
- Lu M, Ho Y-P, Grigsby CL, Nawaz AA, Leong KW, Huang TJ. 2014. Three-dimensional hydrodynamic focusing method for polyplex synthesis. ACS nano 8(1):332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn DM, Langer R. 2000. Degradable Poly(β-amino esters): Synthesis, Characterization, and Self-Assembly with Plasmid DNA. Journal of the American Chemical Society 122(44):10761–10768. [Google Scholar]
- Mangraviti A, Tzeng SY, Kozielski KL, Wang Y, Jin Y, Gullotti D, Pedone M, Buaron N, Liu A, Wilson DR and others. 2015. Polymeric Nanoparticles for Nonviral Gene Therapy Extend Brain Tumor Survival in Vivo. ACS Nano 9(2):1236–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S, Jin Y, Hou CY, Kim J, Green JJ, Kang TJ, Cho SW. 2018. Bacterial tRNase–Based Gene Therapy with Poly (β‐Amino Ester) Nanoparticles for Suppressing Melanoma Tumor Growth and Relapse. Advanced healthcare materials:1800052. [DOI] [PubMed] [Google Scholar]
- Ogris M, Walker G, Blessing T, Kircheis R, Wolschek M, Wagner E. 2003. Tumor-targeted gene therapy: Strategies for the preparation of ligand-polyethylene glycol-polyethylenimine/DNA complexes. Journal of Controlled Release 91:173–181. [DOI] [PubMed] [Google Scholar]
- Robinson H, Gao Y-G, Sanishvili R, Joachimiak A, Wang AHJ. 2000. Hexahydrated magnesium ions bind in the deep major groove and at the outer mouth of A-form nucleic acid duplexes. Nucleic Acids Research 28(8):1760–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JL, Ren Y, Vandermark J, Archang MM, Williford JM, Liu HW, Lee J, Wang TH, Mao HQ. 2016. Continuous Production of Discrete Plasmid DNA‐Polycation Nanoparticles Using Flash Nanocomplexation. Small 12(45):6214–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunshine JC, Peng DY, Green JJ. 2012. Uptake and transfection with polymeric nanoparticles are dependent on polymer end-group structure, but largely independent of nanoparticle physical and chemical properties. Molecular pharmaceutics 9(11):3375–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng SY, Green JJ. 2013. Subtle changes to polymer structure and degradation mechanism enable highly effective nanoparticles for siRNA and DNA delivery to human brain cancer. Advanced healthcare materials 2(3):468–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng SY, Guerrero-Cázares H, Martinez EE, Sunshine JC, Quiñones-Hinojosa A, Green JJ. 2011. Non-viral gene delivery nanoparticles based on poly(β-amino esters) for treatment of glioblastoma. Biomaterials 32(23):5402–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng SY, Young NP, Abutaleb AO, Guerrero-ca H. 2015. Biodegradable Polymeric Nanoparticles Show High Efficacy and Specificity at DNA Delivery to Human Glioblastoma in Vitro and in Vivo. (5):5141–5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead KA, Dorkin JR, Vegas AJ, Chang PH, Veiseh O, Matthews J, Fenton OS, Zhang Y, Olejnik KT, Yesilyurt V. 2014. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nature communications 5:4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DR, Mosenia A, Suprenant MP, Upadhya R, Routkevitch D, Meyer RA, Quinones-Hinojosa A, Green JJ. 2017a. Continuous microfluidic assembly of biodegradable poly(beta-amino ester)/DNA nanoparticles for enhanced gene delivery. J Biomed Mater Res A 105(6):1813–1825. [DOI] [PubMed] [Google Scholar]
- Wilson DR, Routkevitch D, Rui Y, Mosenia A, Wahlin KJ, Quinones-Hinojosa A, Zack DJ, Green JJ. 2017b. A Triple-Fluorophore-Labeled Nucleic Acid pH Nanosensor to Investigate Non-viral Gene Delivery. Molecular Therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. 2014. Non-viral vectors for gene-based therapy. Nat Rev Genet 15(8):541–555. [DOI] [PubMed] [Google Scholar]
- Zhong Z, Song Y, Engbersen JFJ, Lok MC, Hennink WE, Feijen J. 2005. A versatile family of degradable non-viral gene carriers based on hyperbranched poly (ester amine) s. Journal of controlled release 109(1–3):317–329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


