Abstract
Environmental stress such as genotoxic agents can cause DNA damage either indirectly through the generation of reactive oxygen species or directly by interactions with the DNA molecule. Damage to the genetic material may cause mutations and ultimately cancer. Genotoxic mutation can be prevented either by apoptosis or DNA repair. In response to DNA damage, cells have evolved DNA damage responses (DDR) to detect, signal, and repair DNA lesions. Epigenetic mechanisms play critically important roles in DDR, which requires changes in chromatin structure and dynamics to modulate DNA accessibility. Incorporation of histone variants into chromatin is considered as an epigenetic mechanism. Canonical histones can be replaced with variant histones that change chromatin structure, stability, and dynamics. Recent studies have demonstrated involvement of nearly all histone variants in environmental-stress-induced DNA damage repair through various mechanisms, including affecting nucleosome dynamics, carrying variant-specific modification, promoting transcriptional competence or silencing, mediating rearrangement of chromosomes, attracting specific repair proteins, among others. In this review, we will focus on the role of histone variants in DNA damage repair after exposure to environmental genotoxic agents. Understanding the mechanisms regulating environmental exposure-induced epigenetic changes, including replacement of canonical histones with histone variants, will promote the development of strategies to prevent or reverse these changes.
Keywords: Histone variant, DNA repair, Environmental exposure, Chromatin, Epigenetic change
Introduction
Epigenetics is the study of mitotically and/or meiotically heritable changes in gene expression that are not due to the alterations in the DNA sequence [1]. Epigenetic mechanisms include DNA methylation, post translational histone modifications, non-coding RNA expression, and incorporation of variant histones [2, 3].Epigenetic changes are critical for almost all DNA-templated processes such as transcription, DNA repair, replication and recombination [4]. Epigenetic changes can be induced by environmental factors at different times in life and are potentially reversible. Exposure-induced epigenetic alterations may play a direct or indirect role in disease occurrence and/or progression [5, 6]. For example, exposures to genotoxic agents can cause DNA damage either indirectly through the generations of reactive oxygen species (ROS) or directly by interactions with the DNA molecule [3, 7], which could cause disease through inducing mutation that leads to gain of function or loss of function of key genes associated with the disease. Recent studies have shown that epigenetic changes may contribute to environmental diseases through their roles in repair processes of DNA damage induced by environmental stress.
In this review, we will focus on the roles of histone variants in DNA damage repair after exposure to environmental genotoxic agents. Histones are small, basic, and highly conserved proteins that serve as structural scaffolds for DNA packaging. The nucleosome core particle is the first level of compaction, which consists of 147 base pairs of DNA wrapped around a histone octamer composed of two H2A–H2B dimers and a H3–H4 tetramer [8]. The linker histone H1 binds to the nucleosome core particle to form the chromatosome that are organized into a higher-order chromatin structure. Canonical histones in a nucleosome can be replaced with histone variants. Histone variants are different from canonical histones not only by their amino acid sequences, but also by the way they are deposited into chromatin. While canonical histones are assembled in a replication-coupled manner, variant histones are incorporated into chromatin in a replication-independent way. The replacement of their canonical counterparts with histone variants leads to changes in nucleosomes properties, i.e., structure, stability, dynamics, and ultimately DNA accessibility [9]. DNA accessibility in regions of DNA damage is a key factor for efficient DNA repair. In fact, numerous studies have implicated possible roles of histone variants in DNA damage repair. Among these studies, the roles of H3 variants in environmental exposure-induced chromatin dynamics and DNA damage responses are most well studied. Thus, in this review, we will mainly focus on the histone H3 family. The various histones and their variants are discussed below in numerical order.
1. Histone H1 Family
Until now, about 126 different members of the histone H1 family have been found in diverse species, which vary from 194 to 346 amino acids in their length (http://www.actrec.gov.in/histome/). In humans, histone H1 has eleven variants, coded by a single gene that exhibits either replication-dependent or replication-independent expression. The distribution of these eleven histone genes is highly conserved between the human, mouse and rat genomes. Three of the H1 variants are testis-specific (e.g. HIST1H1T, H1FNT, and H1LS1), one of them is oocyte-specific (H1oo), and the others are somatic variants [10]. HIST1H1T is expressed in spermatocytes [11], and both H1FNT and H1LS1 are expressed in spermatids [12, 13]. Linker histone H1 is involved in chromatin compaction and plays a role in the formation of higher-order chromatin structure and gene regulation [14, 15].
Among mutations in H1 variants in chicken cells, only H1.R mutants have increased sensitivity to the DNA damage agent methyl methanesulfonate (MMS) [16]. H1.R mutants exhibit reduced gene targeting, impaired sister chromatid exchange and an accumulation of ionizing radiation-induced chromosomal aberrations at the G2 phase, indicating H1.R plays a role in Rad54-mediated homologous recombination (HR) pathway [16]. However, the precise role of histone H1 variants in DNA repair in other species is still far from clear and is the least well studied. Studying the genomic distribution of H1 variants is also challenging due to the lack of H1 variant-specific antibodies [10].
2. Histone H2A Family
Histone H2A proteins are composed of ~130 amino acid residues. The H2A family is one of the most sequence-divergent families, including macroH2A, H2A.X, H2A.Z and H2A-Bbd. At least 265 different members of histone H2A have been discovered in various species (http://www.actrec.gov.in/histome/). In humans, nineteen variants of histone H2A encoded by 16 genes have been reported. Sequence diversity comes mainly from divergent C-terminal tails, but to date the biological relevance of this diversity remains unclear [17]. The H2A C-terminus is located at the DNA entry/exit site, making variations at this domain a powerful tool for differentiating nucleosomes by changing their stability and dynamics, binding to DNA and /or other interacting factors [18]. Canonical H2A proteins can differ in many more positions, especially in the C-terminal six amino acids [19]. However, no functional specialization of these canonical H2A isoforms has been demonstrated.
2.1 The Role of H2A.X at Double-Strand DNA Breaks
Variant H2A.X is most similar in sequence to the canonical H2A but has a divergent C-terminal tail [20]. Unlike canonical genes, the mRNA of H2A.X has either a polyA tail or the stem-loop structure in its 3’ end, suggesting that it undergoes both replication-dependent and independent transcription [21]. H2A.X can be ubiquitinated, acetylated or phosphorylated. In response to DNA double-strand breaks (DSBs), H2A.X in the vicinity of the DNA break is rapidly phosphorylated at serine 139, helping to recruit the DNA damage response proteins [22, 23]. Acetylation and ubiquitination of H2A.X also function in this process with acetylation at lysine 5 as a prerequisite for ubiquitination and subsequent release of H2A.X from the DNA damage sites [23].
When DSBs are detected, 53BP1 is rapidly recruited to the chromatin around the DSBs sites. This is regulated by a cascade signaling pathway, which is initiated by the ATM-mediated phosphorylation of H2AX (γH2A.X), followed by the accumulation of mediator of DNA damage checkpoint protein 1 (MDC1) and activation of RNF8-RNF168-dependent chromatin ubiquitylation at the DSBs sites [24]. The Mre11-Rad50-Nbs1 (MRN) complex binds to ends of the DSB and recruits auto-phosphorylated ATM monomers at the damaged chromatin sites to generate γH2A.X through ATM-dependent phosphorylation at Ser139 of H2A.X [25]. γH2A.X is then recognized by MDC1; phosphorylation of MDC1 leads to activation of a positive feedback loop by recruiting more MRN complexes and activated ATM around the damaged chromatin and the ATM phosphorylating proximal H2A.X. By these events, γH2A.X complexes can extend megabase pair distances from sites of DSBs on both sides of a break. Phosphorylated MDC1 recruits an RNF8-UBC13 complex to regulate ubiquitylation of H2A and H2A.X. Recent studies have demonstrated that RNF168 binds to ubiquitylated histones and facilitates the formation of polyubiquitilated histones, which further promotes recruitment of the BRCA1 complex and 53BP1 to DNA damage sites, leading to DSB repair and checkpoint arrest.
2.2 The Role of H2A.Z at Double-Strand DNA Breaks
Histone H2A.Z is highly conserved and is essential for development in higher eukaryotes [26]. Barski et al. have shown that H2A.Z localizes mainly in gene promoter regions, and in mammals it localizes to transcription start sites correlating with gene activation, heterochromatic silencing, and transcriptional memory [27]. Acetylation of H2A.Z has been shown to be related to its role in gene activation.
H2A.Z has been found to be enriched at sites of DNA DSBs [28], but this enrichment could be detectable only at late time points after damage and in poorly transcribed chromatin regions, which originally display low H2A.Z content [28, 29]. The accumulation of H2A.Z at DSBs sites occurs by exchanging p400 motor ATPase into chromatin. p400 is a component of the Tip60 (Tat-interacting protein of 60kDa) chromatin-remodeling complex. Tip60 complex acetylates H2A.Z and H4 to create a more open chromatin structure. H2A.Z exchange is important for ubiquitylation of the chromatin through RNF8 (RING finger protein 8, acts as an ubiquitin ligase (E3) in the ubiquitination of certain nuclear proteins), and also for recruiting downstream components of both NHEJ and HR repair pathways, such as 53BP1 and RIF1 [28, 30].
2.3 The Role of macroH2A at Double-Strand DNA Breaks
The histone H2A variant macroH2A is characterized by a large 'macro domain' at the C-terminal [31]. MacroH2A is generally considered as transcriptionally repressive because of its association with condensed chromatin, such as the inactive X chromosome [32, 33]. There are two paralogous genes encoding macroH2A1 and macroH2A2, and macroH2A1 has two splicing variants, macroH2A1.1 and macroH2A1.2 [34].
One of the early events in DSBs repair is the activation of Poly (ADP-ribose) polymerase 1 (PARP1) binding to the DNA DSB ends. PARP1 is an enzyme, that adds ADP-ribose moieties (ADPR) from NAD+ to form PAR [35]. The accumulation of macroH2A1.1 on damaged chromatin contributes to DSB repair by HR and local chromatin compaction [36, 37]. In particular, the macrodomain-containing form of macroH2A—macroH2A1.1 but not macroH2A1.2 and macroH2A2—associates with damaged chromatin in a PAR-dependent manner [37, 38]. Unlike other variants behaving as a bona fide histone protein, macroH2A1.1 acts as a chromatin-associated factor. Moreover, macroH2A1 can accumulate at DSBs sites after being transiently depleted from damaged chromatin [36].
2.4 The Role of H2A.Bbd in DNA Damage Response
Histone H2A.Bdb (Barr body deficient) was first identified about one decade ago [39]. H2A.Bbd is encoded by a polyadenylated mRNA. On the protein level, H2A.Bbd lacks the C-terminal tail and part of the docking domain. H2A.Bbd is not present in all tissues, but it is strongly expressed in testis and brain [40].
H2A.Bbd has been found to be enriched at sites of UVA laser damage repair [41]. In mouse embryonic fibroblasts, GFP-tagged H2A.Bbd, which wraps less DNA than canonical H2A, was transiently localized to sites of DNA synthesis during S-phase and during DNA repair [41]. However, the general functional relevance to cells of this ectopic expression is uncertain, because it needs the ectopic expression of H2A.Bbd in mouse embryonic fibroblasts, which is normally expressed only in testis and brain cells [36].
3. Histone H2B Family
The histone H2B family contains 214 different members described from diverse species (http://www.actrec.gov.in/histome/). Histone H2B forms a dimer with histone H2A in nucleosome cores. Histone H2B has 17 isoforms encoded by 25 genes in humans, the majority of which are assembled in cluster 1 [19]. Compared with other core histones, there are not many posttranslational modifications (PTMs) identified to date among the amino acid resides of histone H2B [42]. Some studies have shown that H2B ubiquitination can contribute to DSB damage responses in mammals [43]. After IR exposure, RNF20, which can ubiquitinate H2B at K119, formed nuclear foci that accumulated at the DSB sites [44].
4. Histone H3 Family
Histone H3 consists of ~136 amino acids. The histone H3 family contains 216 different members from various species (http://www.actrec.gov.in/histome/). In humans, 20 genes encode 8 variants of histone H3, most of which are located on chromosome 6 [19]. Histone H3 is the most extensively post-translationally modified among the five histones. Three main variants of H3 are surprisingly similar in sequence: H3.2 only differs from H3.1 in Cysteine 96 to Serine 96, and H3.3 differs from H3.1 by only 5 residues [20]. However, these variants have great differences in their expression, localization in chromatin and modification state. The canonical variants (H3.1 and H3.2) are only expressed during S phase, while H3.3 is expressed in a replication-independent manner [45]. H3.3 is usually localized to active promoters and gene bodies, regulatory regions, and pericentric and telomere regions. H3.3 is enriched for histone modifications that are associated with gene activation. In contrast, H3.1 and H3.2 do not show specific regions of localization in the genome [46, 47]. Different histone chaperones recognize and assemble H3.1 and H3.3 into nucleosomes in a replication-dependent and independent manner, respectively [48].
4.1 UV Radiation and Histone H3 Variants
UV radiation exposure through sunlight and artificial sources is a primary risk factor for the development of melanoma. Chronic exposure of the skin of mice to UVB radiation or of human keratinocytes in culture to UVA radiation induces changes in chromatin and DNA methylation at specific gene promoters [49, 50]. Exposure of skin to UV radiation induces oxidative stress, inflammation, and DNA damage in the form of DNA photoproducts such as cyclobutane pyrimidine dimers [51].
The role of histone H3 variants in DNA damage responses has been investigated mainly in human cells exposed to ultraviolet (UVC) irradiation or laser micro-irradiation, which results in nucleotide excision repair [52]. The first evidence for de novo histone incorporation at DNA damage sites was found for H3.1. With the development of the SNAP-tag technology, tracking newly synthesized histones in vivo was instrumental for visualizing the de novo deposition of more histone variants at UV sites [53, 54]. A number of studies have demonstrated that the deposition of newly synthesized H3.1 in UVC damaged chromatin is promoted by histone chaperone Chromatin Assembly Factor 1 (CAF-1), which recruits both the H3.1 and H3.2 coupled to late repair steps [52, 55]. In contrast, after UVC irradiation, newly synthesized H3.3 variants deposited by the histone chaperone HIRA (histone regulator A) to sites of UVC irradiation upon detection of damage prior to repair that primes damaged chromatin for later reactivation of transcription [56, 57]. Beyond restoring nucleosome structure, newly synthesized histone deposition at DNA damage sites also has critical functional consequences. Although neither H3.1 nor H3.3 deposition seems to affect UVC damage repair in human cells [52, 56], H3.3 plays a critical role in replication fork progression after DNA damage in chicken cells [58]. Moreover, histone chaperone HIRA specific for H3.3 deposition is required for transcription recovery upon repair completion in human cells [56, 59].
4.2 Arsenic Exposure and Histone H3 Variants
Chromatin structure and dynamics regulates DNA accessibility and DNA repair process. Environmental toxins may modulate chromatin organization in regions of DNA damage and repair by interfering with deposition of histone variants. Arsenic is a well-established human carcinogen. Even today, hundreds of millions of people are exposed to dangerous levels of naturally occurring arsenic in drinking water throughout the world [60]. While the ROS produced by arsenic exposure are considered as a key factors in arsenic carcinogenicity [61], it has been demonstrated that arsenic can alter histone modifications and DNA methylation at gene promoters, leading to abnormal gene expression [62–64].
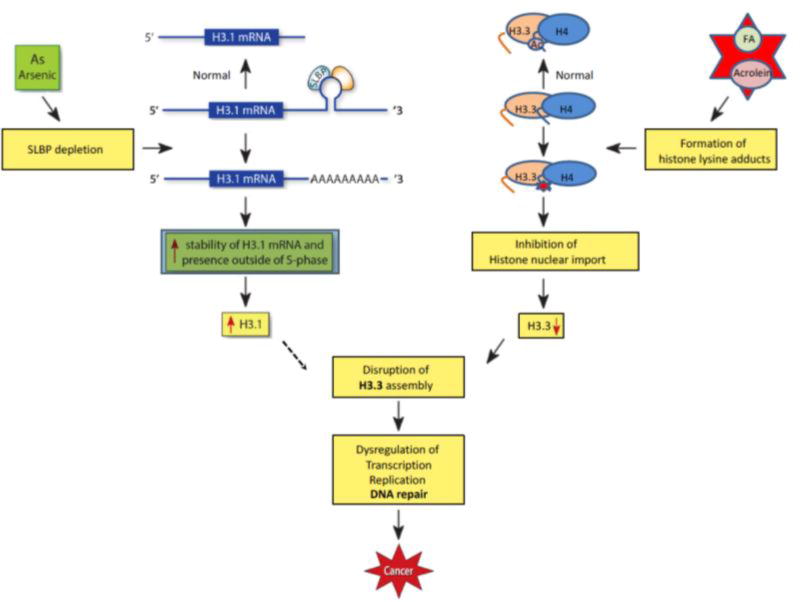
The histone H3 genes can be classified into three distinct groups. One group contains replication-dependent genes encoding the canonical histones, whose expression is largely limited to the S phase of the cell cycle (e.g., H3.1). Another group contains replication-independent genes encoding replacement histones that are expressed throughout the cell cycle (H3.3). A third group of H3 genes encodes tissue-specific isotopes (H3t genes)[65]. Canonical histone H3 mRNAs are unique–they do not have poly(A) tail in their 3’ ends, but instead possess a stem-loop structure. Stem-loop binding protein (SLBP) binds to this stem-loop structure and regulates histone pre-mRNA processing. We found that arsenic exposure induces polyadenylation of the canonical histone H3.1 mRNA, which was accompanied by a depletion of SLBP mRNA and protein [57]. In addition, the expression of other genes that are known to play a role in canonical histone mRNA processing (i.e. LSM10, CPSF2, CPSF3, etc.) was not altered by arsenic [66], suggesting that depletion of SLBP is likely the main reason for arsenic-induced polyadenylation of H3.1 mRNA. We further demonstrated that the observed increase in canonical histone gene expression was at least in part due to the aberrant polyadenylation of canonical histone mRNAs in a number of human cell lines, including epithelial lung cells (BEAS2B and A549), lymphoma B cell line (BL41), as well as in As-transformed clones (BEAS2B) that no longer had any arsenic exposure [66]. Interestingly, polyadenylation of canonical histone H3.1 mRNA stabilized the transcripts, which in turn greatly increased the level of H3.1 in the M cell cycle phase [67]. In normal condition, only H3.3 should exists during M phase. The significant increase of H3.1 in M phase following arsenic exposure might compete with H3.3 for incorporation into the chromatin that could change the chromatin structure and dynamics (Fig. 1). This is currently under investigation.
Figure 1. Disruption of chromatin structure by environmental exposures.
In normal condition, the stem-loop binding protein (SLBP) binds to the stem-loop structure on 3’ end of canonical histone H3.1 mRNA, generating the histone mRNA without poly(A) tail by cleaving downstream of the stem-loop. SLBP is depleted following arsenic exposure. The loss of SLBP results in binding of less SLBP to the stem-loop structure, adding a poly(A) tail via a downstream poly(A) signal sequence. The polyadenylation increases stability of H3.1 mRNA and subsequently H3.1 protein levels outside of S-phase, which may interfere with assembly of histone variant H3.3. On the other hand, aldehyde chemicals such as acrolein and formaldehyde (FA) form adducts with lysine residues on newly synthesized histones, including H3.3 and H4, inhibiting their acetylation and subsequently compromising histone nuclear import and assembly into chromatin. Defective chromatin assembly due to H3.3 reduction may dysregulate gene expression, DNA replication and repair thereby facilitating carcinogenesis.
4.3 The Role of CENP-A in DNA Repair
CENP-A is the centromere-specific histone H3 variant. Vertebrate cenH3 (CENP-A) has been shown to be recruited transiently to some DNA damage sites, such as DSBs [68]. Expression of GFP-tagged CENP-A resulted in a rapid localization of CENP-A to sites of the DSBs in human and mouse cells, while in a study using SNAP-tag technology, new CENP-A was not observed at DNA damage sites [56]. In another study, endogenous CENP-A was not detected at laser damage sites, whereas GFP-CENP-A was only weakly recruited in a limited number of cells [69]. Perpelescu M. et al. also failed to detect CENP-A by immunofluorescence at IR damage sites, although an interaction between CENP-A and ATM, which is dependent on IR and remodeling and spacing factor 1 (RSF1), could be detected after crosslinking. RSF1 has an IR-dependent association with ATM, which is required for DSBs repair [69, 70]. RSF1 also affects the establishment of CENP-A at centromeres [71] and its interaction with CENP-A is independent of IR [70], suggesting that some CENP-A deposition might be depend on other histone chaperones such as HJURP (Holliday junction recognition protein). HJURP is a CENP-A chaperone originally reported to be involved in DSBs repair through interaction with the MRN complex [72].
5. Histone H4 Family
Histone H4 contains only 103 amino acids and constitutes a heterodimer (H3–H4) or heterotetramer (H3–H4)2 with histone H3. The histone H4 family has 116 members discovered in a variety of organisms. Surprisingly, in humans the histone H4 protein is encoded by 14 distinct H4 genes, and most of them are clustered on chromosome 6 [19]. Interestingly, no H4 variants have been found in higher eukaryotes [73]. Yet, histone H4 may have impacts on DNA repair process by changing the dynamics of other histone variants.
The acetylation of lysines 5 and 12 on histone H4 plays an important role in chromatin assembly regulation [74]. H4K5&K12ac, a diacetylation catalyzed by histone acetyltransferase 1 (HAT1), is detectable on newly synthesized histone H4 from yeast to humans as an early modification occurring on H3–H4 [74–78]. The H4K5&K12 double mutants are imported into the nucleus less efficiently than wild-type histones [75, 79]. Moreover, HAT1 and H4K5&K12 regulate the association of H3–H4 with the histone translocator protein importin 4 and the histone chaperone ASF1, indicating that H4K5&K12 might regulate chromatin assembly pathways by regulating H3–H4 nuclear import and assembly [75, 78–80]. We have demonstrated that the potential carcinogen acrolein, which is abundant in cigarette smoke and cooking fumes, reacts with lysines 5 and 12 on cytosolic histone H4 in vitro and in cells, preventing these sites from being physiologically acetylated by histone acetyltransferases including HAT1 [71, 72]. Reduction of H4K5&12ac and/or other histone modifications following acrolein exposure disrupted the association of H3–H4 with importin 4 as well as Asf1, leading to inhibition of histone nuclear import and assembly into chromatin [81, 82]. To examine if this could be a common characteristic of other type of aldehydes, we further investigated how established carcinogen formaldehyde, a simplest aldehyde, regulates histone modifications and chromatin assembly. We found that formaldehyde exposure is also able to reduce the level of cytosolic H4K5&12ac add the global level of nucleosomal histone H4 as compared with the untreated controls, a suggestive of inhibition of chromatin assembly [83]. The outcome of defective chromatin assembly is the lack of histone supplies. Since histone variant H3.3 localizes primarily to active regions of the genome with high rates of histone turnover, the impact of lack of histone supplies should be greatest in these H3.3 regions. In addition, while the assembly of canonical H3.1 and H3.2 is replication-coupled, the assembly of H3.3 is not dependent on DNA replication. Therefore, we made use of chromatin immunoprecipitation assays (CHIP) to test the level of H3.3 at several genomic loci, since the assembly of H3.3 should be affected first by lack of histone supplies. Indeed, ChIP results showed that incorporation of histone variant H3.3 was decreased following formaldehyde exposure in the majority of sites we tested [83]. As mentioned above, H3.3 seems to be important for DNA repair process. Thus, it is possible that environmental toxins such as aldehydes may affect DNA repair processed by perturbing chromatin (e.g., H3.3) dynamics, at least in part through changing the modification status of histone H4 (Fig. 1).
Concluding remarks and future directions
The revolutionary developments of new molecular chromatin analysis technologies have allowed us to make high-resolution genomic maps of histone variants and modified nucleosomes. These maps have disclosed where, and to what extent, different histone variants are enriched on DNA damage sites. Nearly all histone variants seem to be involved in environmental-stress-induced DNA damage repair through various mechanisms. Further work still needs to be carried out to reveal detailed mechanisms by which environmental exposures induce changes in histone variant deposition and dynamics. Understanding these mechanisms will promote the development of new biomarkers for identifying environmental exposure, and will perhaps also contribute to mitigated damage via enhanced DNA repair, in order to protect the human health.
Acknowledgments
Funding
This work was supported by the National Institutes of Health [1R01ES026138-01 to C.J.]; NIEHS Center of Excellence Pilot Project Program and Career Development [5P30ES000260 to C.J.].
We thank Cathy Klein for critical review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Riggs AD, Porter TN. Overview of epigenetic mechanisms. Cold Spring Harbor monograph series. 1996;32:29–46. [Google Scholar]
- 2.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nature reviews. Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klaunig JE, Wang Z, Pu X, Zhou S. Oxidative stress and oxidative damage in chemical carcinogenesis. Toxicology and applied pharmacology. 2011;254:86–99. doi: 10.1016/j.taap.2009.11.028. [DOI] [PubMed] [Google Scholar]
- 4.MacAlpine DM, Almouzni G. Chromatin and DNA replication. Cold Spring Harb Perspect Biol. 2013;5:a010207. doi: 10.1101/cshperspect.a010207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cortessis VK, Thomas DC, Levine AJ, Breton CV, Mack TM, Siegmund KD, Haile RW, Laird PW. Environmental epigenetics: prospects for studying epigenetic mediation of exposure-response relationships. Human genetics. 2012;131:1565–1589. doi: 10.1007/s00439-012-1189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou L, Zhang X, Wang D, Baccarelli A. Environmental chemical exposures and human epigenetics. International journal of epidemiology. 2012;41:79–105. doi: 10.1093/ije/dyr154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poirier MC. Chemical-induced DNA damage and human cancer risk. Discovery medicine. 2012;14:283–288. [PMC free article] [PubMed] [Google Scholar]
- 8.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 9.Weber CM, Henikoff S. Histone variants: dynamic punctuation in transcription. Genes Dev. 2014;28:672–682. doi: 10.1101/gad.238873.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Izzo A, Kamieniarz-Gdula K, Ramirez F, Noureen N, Kind J, Manke T, van Steensel B, Schneider R. The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells. Cell Rep. 2013;3:2142–2154. doi: 10.1016/j.celrep.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Drabent B, Franke K, Bode C, Kosciessa U, Bouterfa H, Hameister H, Doenecke D. Isolation of two murine H1 histone genes and chromosomal mapping of the H1 gene complement. Mamm Genome. 1995;6:505–511. doi: 10.1007/BF00356166. [DOI] [PubMed] [Google Scholar]
- 12.Martianov I, Brancorsini S, Catena R, Gansmuller A, Kotaja N, Parvinen M, Sassone-Corsi P, Davidson I. Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc Natl Acad Sci U S A. 2005;102:2808–2813. doi: 10.1073/pnas.0406060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan W, Ma L, Burns KH, Matzuk MM. HILS1 is a spermatid-specific linker histone H1-like protein implicated in chromatin remodeling during mammalian spermiogenesis. Proc Natl Acad Sci U S A. 2003;100:10546–10551. doi: 10.1073/pnas.1837812100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Millan-Arino L, Islam AB, Izquierdo-Bouldstridge A, Mayor R, Terme JM, Luque N, Sancho M, Lopez-Bigas N, Jordan A. Mapping of six somatic linker histone H1 variants in human breast cancer cells uncovers specific features of H1.2. Nucleic Acids Res. 2014;42:4474–4493. doi: 10.1093/nar/gku079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Over RS, Michaels SD. Open and closed: the roles of linker histones in plants and animals. Molecular plant. 2014;7:481–491. doi: 10.1093/mp/sst164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashimoto H, Sonoda E, Takami Y, Kimura H, Nakayama T, Tachibana M, Takeda S, Shinkai Y. Histone H1 variant, H1R is involved in DNA damage response. DNA repair. 2007;6:1584–1595. doi: 10.1016/j.dnarep.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Marino-Ramirez L, Kann MG, Shoemaker BA, Landsman D. Histone structure and nucleosome stability. Expert Rev Proteomics. 2005;2:719–729. doi: 10.1586/14789450.2.5.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ausio J, Abbott DW. The many tales of a tail: carboxyl-terminal tail heterogeneity specializes histone H2A variants for defined chromatin function. Biochemistry. 2002;41:5945–5949. doi: 10.1021/bi020059d. [DOI] [PubMed] [Google Scholar]
- 19.Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- 20.West MH, Bonner WM. Histone 2A, a heteromorphous family of eight protein species. Biochemistry. 1980;19:3238–3245. doi: 10.1021/bi00555a022. [DOI] [PubMed] [Google Scholar]
- 21.Mannironi C, Bonner WM, Hatch CL. H2A.X. a histone isoprotein with a conserved C-terminal sequence, is encoded by a novel mRNA with both DNA replication type and polyA 3' processing signals. Nucleic Acids Res. 1989;17:9113–9126. doi: 10.1093/nar/17.22.9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. The Journal of biological chemistry. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 23.Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, Kimura H, Ikura M, Nishikubo S, Ito T, Muto A, Miyagawa K, Takeda S, Fishel R, Igarashi K, Kamiya K. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Molecular and cellular biology. 2007;27:7028–7040. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nature cell biology. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- 25.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nature reviews. Molecular cell biology. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 26.Draker R, Cheung P. Transcriptional and epigenetic functions of histone variant H2A.Z. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2009;87:19–25. doi: 10.1139/O08-117. [DOI] [PubMed] [Google Scholar]
- 27.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 28.Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Molecular cell. 2012;48:723–733. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taty-Taty GC, Courilleau C, Quaranta M, Carayon A, Chailleux C, Aymard F, Trouche D, Canitrot Y. H2A.Z depletion impairs proliferation and viability but not DNA double-strand breaks repair in human immortalized and tumoral cell lines. Cell cycle (Georgetown, Tex.) 2014;13:399–407. doi: 10.4161/cc.27143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daley JM, Sung P. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Molecular and cellular biology. 2014;34:1380–1388. doi: 10.1128/MCB.01639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pehrson JR, Fried VA. MacroH2A, a core histone containing a large nonhistone region. Science (New York, N.Y.) 1992;257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- 32.Ratnakumar K, Duarte LF, LeRoy G, Hasson D, Smeets D, Vardabasso C, Bonisch C, Zeng T, Xiang B, Zhang DY, Li H, Wang X, Hake SB, Schermelleh L, Garcia BA, Bernstein E. ATRX-mediated chromatin association of histone variant macroH2A1 regulates alpha-globin expression. Genes Dev. 2012;26:433–438. doi: 10.1101/gad.179416.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costanzi C, Stein P, Worrad DM, Schultz RM, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation mouse embryos. Development (Cambridge, England) 2000;127:2283–2289. doi: 10.1242/dev.127.11.2283. [DOI] [PubMed] [Google Scholar]
- 34.Costanzi C, Pehrson JR. MACROH2A2, a new member of the MARCOH2A core histone family. The Journal of biological chemistry. 2001;276:21776–21784. doi: 10.1074/jbc.M010919200. [DOI] [PubMed] [Google Scholar]
- 35.Langelier MF, Planck JL, Roy S, Pascal JM. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science (New York, N.Y.) 2012;336:728–732. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khurana S, Kruhlak MJ, Kim J, Tran AD, Liu J, Nyswaner K, Shi L, Jailwala P, Sung MH, Hakim O, Oberdoerffer P. A macrohistone variant links dynamic chromatin compaction to BRCA1-dependent genome maintenance. Cell Rep. 2014;8:1049–1062. doi: 10.1016/j.celrep.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, Hottiger MO, Ladurner AG. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nature structural & molecular biology. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 38.Mehrotra PV, Ahel D, Ryan DP, Weston R, Wiechens N, Kraehenbuehl R, Owen-Hughes T, Ahel I. DNA repair factor APLF is a histone chaperone. Molecular cell. 2011;41:46–55. doi: 10.1016/j.molcel.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chadwick BP, Willard HF. A novel chromatin protein, distantly related to histone H2A, is largely excluded from the inactive X chromosome. The Journal of cell biology. 2001;152:375–384. doi: 10.1083/jcb.152.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soboleva TA, Nekrasov M, Pahwa A, Williams R, Huttley GA, Tremethick DJ. A unique H2A histone variant occupies the transcriptional start site of active genes. Nature structural & molecular biology. 2011;19:25–30. doi: 10.1038/nsmb.2161. [DOI] [PubMed] [Google Scholar]
- 41.Sansoni V, Casas-Delucchi CS, Rajan M, Schmidt A, Bonisch C, Thomae AW, Staege MS, Hake SB, Cardoso MC, Imhof A. The histone variant H2A.Bbd is enriched at sites of DNA synthesis. Nucleic Acids Res. 2014;42:6405–6420. doi: 10.1093/nar/gku303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamakaka RT, Biggins S. Histone variants: deviants? Genes Dev. 2005;19:295–310. doi: 10.1101/gad.1272805. [DOI] [PubMed] [Google Scholar]
- 43.Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang SY, Eppink B, Chung YM, Shalev G, Shema E, Shkedy D, Smorodinsky NI, van Vliet N, Kuster B, Mann M, Ciechanover A, Dahm-Daphi J, Kanaar R, Hu MC, Chen DJ, Oren M, Shiloh Y. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Molecular cell. 2011;41:529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DV, Shimada M, Tauchi H, Suzuki H, Tashiro S, Zou L, Komatsu K. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Molecular cell. 2011;41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 45.Hereford LM. In: All about histone genes. Histone genes: Structure, organization, and regulation. Stein GS, Stein JL, Marzluff WF, editors. John Wiley and Sons; New York, Chichester: 1984. p. 483. £78.70, $105.75, BioEssays, 2 (1985) 43–43. [Google Scholar]
- 46.McKittrick E, Gafken PR, Ahmad K, Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc Natl Acad Sci U S A. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahmad K, Henikoff S. Histone H3 variants specify modes of chromatin assembly. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16477–16484. doi: 10.1073/pnas.172403699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 49.Nandakumar V, Vaid M, Tollefsbol TO, Katiyar SK. Aberrant DNA hypermethylation patterns lead to transcriptional silencing of tumor suppressor genes in UVB-exposed skin and UVB-induced skin tumors of mice. Carcinogenesis. 2011;32:597–604. doi: 10.1093/carcin/bgq282. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Chen IP, Henning S, Faust A, Boukamp P, Volkmer B, Greinert R. UVA-induced epigenetic regulation of P16(INK4a) in human epidermal keratinocytes and skin tumor derived cells. Photochemical & photobiological sciences : Official journal of the European Photochemistry Association and the European Society for Photobiology. 2012;11:180–190. doi: 10.1039/c1pp05197k. [DOI] [PubMed] [Google Scholar]
- 51.Xu Y, Sun Y, Jiang X, Ayrapetov MK, Moskwa P, Yang S, Weinstock DM, Price BD. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. The Journal of cell biology. 2010;191:31–43. doi: 10.1083/jcb.201001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Polo SE, Roche D, Almouzni G. New histone incorporation marks sites of UV repair in human cells. Cell. 2006;127:481–493. doi: 10.1016/j.cell.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 53.Bodor DL, Rodriguez MG, Moreno N, Jansen LE. Analysis of protein turnover by quantitative SNAP-based pulse-chase imaging. Current protocols in cell biology. 2012;Chapter 8 doi: 10.1002/0471143030.cb0808s55. Unit8.8. [DOI] [PubMed] [Google Scholar]
- 54.Adam S, Dabin J, Bai SK, Polo SE. Imaging local deposition of newly synthesized histones in UVC-damaged chromatin. Methods in molecular biology (Clifton, N.J.) 2015;1288:337–347. doi: 10.1007/978-1-4939-2474-5_19. [DOI] [PubMed] [Google Scholar]
- 55.Latreille D, Bluy L, Benkirane M, Kiernan RE. Identification of histone 3 variant 2 interacting factors. Nucleic Acids Res. 2014;42:3542–3550. doi: 10.1093/nar/gkt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adam S, Polo SE, Almouzni G. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell. 2013;155:94–106. doi: 10.1016/j.cell.2013.08.029. [DOI] [PubMed] [Google Scholar]
- 57.Schwartz BE, Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 2005;19:804–814. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frey A, Listovsky T, Guilbaud G, Sarkies P, Sale JE. Histone H3.3 is required to maintain replication fork progression after UV damage. Current biology : CB. 2014;24:2195–2201. doi: 10.1016/j.cub.2014.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dinant C, Ampatziadis-Michailidis G, Lans H, Tresini M, Lagarou A, Grosbart M, Theil AF, van Cappellen WA, Kimura H, Bartek J, Fousteri M, Houtsmuller AB, Vermeulen W, Marteijn JA. Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Molecular cell. 2013;51:469–479. doi: 10.1016/j.molcel.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 60.Chervona Y, Hall MN, Arita A, Wu F, Sun H, Tseng HC, Ali E, Uddin MN, Liu X, Zoroddu MA, Gamble MV, Costa M. Associations between arsenic exposure and global posttranslational histone modifications among adults in Bangladesh, Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research. cosponsored by the American Society of Preventive Oncology. 2012;21:2252–2260. doi: 10.1158/1055-9965.EPI-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Vizcaya-Ruiz A, Barbier O, Ruiz-Ramos R, Cebrian ME. Biomarkers of oxidative stress and damage in human populations exposed to arsenic. Mutation research. 2009;674:85–92. doi: 10.1016/j.mrgentox.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 62.Brocato J, Costa M. Basic mechanics of DNA methylation and the unique landscape of the DNA methylome in metal-induced carcinogenesis. Critical reviews in toxicology. 2013;43:493–514. doi: 10.3109/10408444.2013.794769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boellmann F, Zhang L, Clewell HJ, Schroth GP, Kenyon EM, Andersen ME, Thomas RS. Genome-wide analysis of DNA methylation and gene expression changes in the mouse lung following subchronic arsenate exposure. Toxicological sciences : an official journal of the Society of Toxicology. 2010;117:404–417. doi: 10.1093/toxsci/kfq225. [DOI] [PubMed] [Google Scholar]
- 64.Zhou X, Sun H, Ellen TP, Chen H, Costa M. Arsenite alters global histone H3 methylation. Carcinogenesis. 2008;29:1831–1836. doi: 10.1093/carcin/bgn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Albig W, Doenecke D. The human histone gene cluster at the D6S105 locus. Human genetics. 1997;101:284–294. doi: 10.1007/s004390050630. [DOI] [PubMed] [Google Scholar]
- 66.Brocato J, Fang L, Chervona Y, Chen D, Kiok K, Sun H, Tseng HC, Xu D, Shamy M, Jin C, Costa M. Arsenic induces polyadenylation of canonical histone mRNA by down-regulating stem-loop-binding protein gene expression. The Journal of biological chemistry. 2014;289:31751–31764. doi: 10.1074/jbc.M114.591883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brocato J, Chen D, Liu J, Fang L, Jin C, Costa M. A Potential New Mechanism of Arsenic Carcinogenesis: Depletion of Stem-Loop Binding Protein and Increase in Polyadenylated Canonical Histone H3.1 mRNA. Biological trace element research. 2015;166:72–81. doi: 10.1007/s12011-015-0296-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeitlin SG, Baker NM, Chapados BR, Soutoglou E, Wang JY, Berns MW, Cleveland DW. Double-strand DNA breaks recruit the centromeric histone CENP-A. Proc Natl Acad Sci U S A. 2009;106:15762–15767. doi: 10.1073/pnas.0908233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Helfricht A, Wiegant WW, Thijssen PE, Vertegaal AC, Luijsterburg MS, van Attikum H. Remodeling and spacing factor 1 (RSF1) deposits centromere proteins at DNA double-strand breaks to promote non-homologous end-joining. Cell cycle (Georgetown, Tex.) 2013;12:3070–3082. doi: 10.4161/cc.26033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pessina F, Lowndes NF. The RSF1 histone-remodelling factor facilitates DNA double-strand break repair by recruiting centromeric and Fanconi Anaemia proteins. PLoS biology. 2014;12:e1001856. doi: 10.1371/journal.pbio.1001856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K. Active establishment of centromeric CENP-A chromatin by RSF complex. The Journal of cell biology. 2009;185:397–407. doi: 10.1083/jcb.200903088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kato T, Sato N, Hayama S, Yamabuki T, Ito T, Miyamoto M, Kondo S, Nakamura Y, Daigo Y. Activation of Holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer research. 2007;67:8544–8553. doi: 10.1158/0008-5472.CAN-07-1307. [DOI] [PubMed] [Google Scholar]
- 73.Kar S, Deb M, Sengupta D, Shilpi A, Parbin S, Torrisani J, Pradhan S, Patra S. An insight into the various regulatory mechanisms modulating human DNA methyltransferase 1 stability and function. Epigenetics. 2012;7:994–1007. doi: 10.4161/epi.21568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci U S A. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ejlassi-Lassallette A, Mocquard E, Arnaud MC, Thiriet C. H4 replication-dependent diacetylation and Hat1 promote S-phase chromatin assembly in vivo. Molecular biology of the cell. 2011;22:245–255. doi: 10.1091/mbc.E10-07-0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nagarajan P, Ge Z, Sirbu B, Doughty C, Agudelo Garcia PA, Schlederer M, Annunziato AT, Cortez D, Kenner L, Parthun MR. Histone acetyl transferase 1 is essential for mammalian development, genome stability, and the processing of newly synthesized histones H3 and H4. PLoS genetics. 2013;9:e1003518. doi: 10.1371/journal.pgen.1003518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 78.Campos EI, Fillingham J, Li G, Zheng H, Voigt P, Kuo WH, Seepany H, Gao Z, Day LA, Greenblatt JF, Reinberg D. The program for processing newly synthesized histones H3.1 and H4. Nature structural & molecular biology. 2010;17:1343–1351. doi: 10.1038/nsmb.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ejlassi-Lassallette A, Thiriet C. Replication-coupled chromatin assembly of newly synthesized histones: distinct functions for the histone tail domains. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2012;90:14–21. doi: 10.1139/o11-044. [DOI] [PubMed] [Google Scholar]
- 80.Zhang H, Han J, Kang B, Burgess R, Zhang Z. Human histone acetyltransferase 1 protein preferentially acetylates H4 histone molecules in H3.1-H4 over H3.3-H4. The Journal of biological chemistry. 2012;287:6573–6581. doi: 10.1074/jbc.M111.312637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fang L, Chen D, Yu C, Li H, Brocato J, Huang L, Jin C. Mechanisms Underlying Acrolein-Mediated Inhibition of Chromatin Assembly. Molecular and cellular biology. 2016;36:2995–3008. doi: 10.1128/MCB.00448-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen D, Fang L, Li H, Tang MS, Jin C. Cigarette smoke component acrolein modulates chromatin assembly by inhibiting histone acetylation. The Journal of biological chemistry. 2013;288:21678–21687. doi: 10.1074/jbc.M113.476630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen D, Fang L, Mei S, Li H, Xu X, Des Marais TL, Lu K, Liu XS, Jin C. Regulation of Chromatin Assembly and Cell Transformation by Formaldehyde Exposure in Human Cells. Environmental health perspectives. 2017;125:097019. doi: 10.1289/EHP1275. [DOI] [PMC free article] [PubMed] [Google Scholar]