Abstract
Purpose of Review
Metabolic reprogramming is essential for the rapid proliferation of cancer cells and is thus recognized as a hallmark of cancer. In this review, we will discuss the etiologies and effects of metabolic reprogramming in colorectal cancer.
Recent Findings
Changes in cellular metabolism may precede the acquisition of driver mutations ultimately leading to colonocyte transformation. Oncogenic mutations and loss of tumor suppressor genes further reprogram CRC cells to upregulate glycolysis, glutaminolysis, one-carbon metabolism, and fatty acid synthesis. These metabolic changes are not uniform throughout tumors, as subpopulations of tumor cells may rely on different pathways to adapt to nutrient availability in the local tumor microenvironment. Finally, metabolic cross-communication between stromal cells, immune cells, and the gut microbiota enable CRC growth, invasion, and metastasis.
Summary
Altered cellular metabolism occurs in CRC at multiple levels, including in the cells that make up the bulk of CRC tumors, cancer stem cells, the tumor microenvironment, and host-microbiome interactions. This knowledge may inform the development of improved screening and therapeutics for CRC.
Keywords: Metabolism, Colorectal cancer, Warburg effect, Cancer stem cells, Tumor microenvironment, Microbiome
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed malignancy and the fourth leading cause of cancer-related deaths worldwide [1]. In the USA, over 50,000 people died of the disease in 2017 [2]. Additionally, although CRC incidence and mortality have decreased nationally overall due to advancements in screening and therapeutics, CRC incidence in younger adults (age <50 years) unexpectedly increased, including a rapid increase in distant-stage disease [2]. The current 5-year survival rate for patients with CRC ranges from 88.1% for cases diagnosed at stage I (23% of cases) to 12.6% for cases diagnosed at stage IV (20% of cases) [3].
Development of CRC is a gradual process defined by the accumulation of somatic mutations, chromosomal instability (CIN), microsatellite instability (MSI), and/or epigenetic alterations resulting in loss of tumor suppressor genes and activation of oncogenes [4]. Metabolic pathways are actively reprogrammed during this process, leading to increased glycolysis, glutaminolysis, and fatty acid (FA) synthesis. This metabolic reprogramming is now recognized as a hallmark of cancer [5, 6••]. In this review, we will discuss the etiologies and effects of altered cellular metabolism in CRC, including the role of tumor genetics and epigenetics, the tumor microenvironment (TME), and the microbiome. Understanding metabolic reprogramming and its consequences may contribute to improved CRC screening and treatment.
Metabolic Phenotypes in CRC
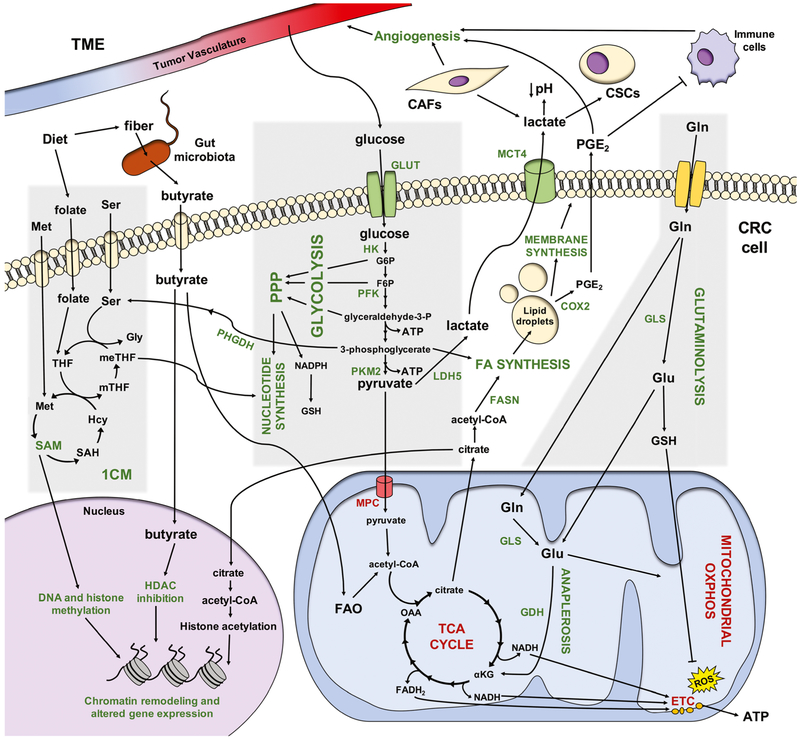
Metabolism is broadly defined as “the sum of biochemical processes in living organisms that either produce or consume energy” [7]. The primary source of cellular energy is glucose, a simple carbohydrate transported into cells by a family of transmembrane glucose transporters (GLUTs) [8]. In the cytoplasm, glucose is catabolized to pyruvate, generating two molecules of adenosine triphosphate (ATP), in a process known as glycolysis [8]. In the absence of oxygen, normal cells convert pyruvate to lactate, which is secreted from the cell. In contrast, in the presence of oxygen, pyruvate is transported into the mitochondria by the mitochondrial pyruvate carrier (MPC) and then converted to acetyl-CoA by pyruvate dehydrogenase (PDH) [8]. Acetyl-CoA, which can also be generated by the catabolism of fatty acids (FAs) and some amino acids, then enters the tricarboxylic acid (TCA) cycle, which reduces nicotinamide adenine dinucleotide (NAD+) and flavin adenine dinucleotide (FAD) to NADH and FADH2 [8]. These high-energy electron carriers are then used to create a proton gradient across the inner mitochondrial membrane that drives adenosine triphosphate (ATP) synthase in the electron transport chain (ETC). This process, known as oxidative phosphorylation (OXPHOS), results in an additional 32–36 molecules of ATP [8, 9]. Cancer cells, however, prioritize energy sources differently to support their rapid growth and proliferation (summarized in Fig. 1; abbreviations summarized in Table 1).
Fig. 1.
Overview of metabolic reprogramming in CRC. Metabolic processes and enzymes (emphasized in this review) that are upregulated in CRC cells with a Warburg phenotype are shown in green, while those that are downregulated in CRC are shown in red. Abbreviations: 1CM, one-carbon metabolism; αKG, α-ketoglutarate; ATP, adenosine triphosphate; CAF, cancer-associated fibroblast; COX2, cyclooxygenase-2; CSC, cancer stem cell; ETC, electron transport chain; F6P, fructose-6-phosphate; FA, fatty acid; FADH2, reduced flavin adenine dinucleotide; FAO, fatty acid oxidation; FASN, fatty acid synthase; G6P, glucose-6-phosphate; GDH, glutamate dehydrogenase; Gln, glutamine; GLS, glutaminase; Glu, glutamate; GLUT, glucose transporter; Gly, glycine; GSH, glutathione; Hcy, homocysteine; HDAC, histone deacetylase; HK, hexokinase; LDH5, lactate dehydrogenase5; MCT4, monocarboxylate transporter4; Met, methionine; meTHF, 5,10-methylene THF; MPC, mitochondrial pyruvate carrier; mTHF, 5-methyl THF; NADH, reduced nicotinamide adenine dinucleotide; NADPH, reduced nicotinamide adenine dinucleotide phosphate; OAA, oxaloacetate; OXPHOS, oxidative phosphorylation; PFK, phosphofructokinase; PGE2, prostaglandin E2; PHGDH, phosphoglycerate dehydrogenase; PKM2, pyruvate kinase isozyme M2; PPP, pentose phosphate pathway; SAH, S-adenosyl-L-homocysteine; SAM, S-adenosylmethionine; Ser, serine; TCA cycle, tricarboxylic acid cycle; THF, tetrahydrofolate
Table 1.
Abbreviations used in this review
| 1CM | One-carbon metabolism | LOH | Loss of heterozygosity |
| ACL | Adenosine triphosphate citrate lyase | MCT | Monocarboxylate transporter |
| αKG | α-ketoglutarate | Met | Methionine |
| AMPK | Adenosine monophosphate-activated protein kinase | meTHF | 5,10-methylene tetrahydrofolate |
| ATP | Adenosine triphosphate | MPC | Mitochondrial pyruvate carrier |
| CAF | Cancer-associated fibroblast | MSI | Microsatellite instability |
| CIMP | CpG island methylator phenotype | mTHF | 5-methyl tetrahydrofolate |
| CIN | Chromosomal instability | mTORC | Mammalian target of rapamycin complex |
| CKB | Creatine kinase brain-type | NAD+ | Nicotinamide adenine dinucleotide |
| COX2 | Cyclooxygenase-2 | NADH | Reduced nicotinamide adenine dinucleotide |
| CSC | Cancer stem cell | NADP+ | Nicotinamide adenine dinucleotide phosphate |
| DNMT | DNA methyltransferase | NADPH | Reduced nicotinamide adenine dinucleotide phosphate |
| EGF | Epidermal growth factor | OAA | Oxaloacetate |
| ETC | Electron transport chain | OXPHOS | Oxidative phosphorylation |
| F6P | Fructose-6-phosphate | PDGF | Platelet-derived growth factor |
| FA | Fatty acid | PDH | Pyruvate dehydrogenase |
| FAD | Flavin adenine dinucleotide | PDK | Pyruvate dehydrogenase kinase |
| FADH2 | Reduced flavin adenine dinucleotide | PFK | Phosphofructokinase |
| FAO | Fatty acid oxidation | PGE2 | Prostaglandin E2 |
| FASN | Fatty acid synthase | PHGDH | Phosphoglycerate dehydrogenase |
| FGF | Fibroblast growth factor | PI3K | Phosphatidylinositol 3-kinase |
| G6P | Glucose-6-phosphate | PIP2 | Phosphatidylinositol-4,5-bisphosphate |
| GDH | Glutamate dehydrogenase | PIP3 | Phosphatidylinositol-3,4,5-triphosphate |
| Gln | Glutamine | PKM2 | Pyruvate kinase isozyme M2 |
| GLS | Glutaminase | PPP | Pentose phosphate pathway |
| GLUT | Glucose transporter | ROS | Reactive oxygen species |
| GPT | Glutamate pyruvate transaminase | RTK | Receptor tyrosine kinase |
| GSH | Glutathione | SAH | S-adenosyl-L-homocysteine |
| Gly | Glycine | SAM | S-adenosylmethionine |
| HAT | Histone acetyltransferase | SCFA | Short-chain fatty acid |
| Hcy | Homocysteine | Ser | Serine |
| HDAC | Histone deacetylase | SREBP | Sterol regulatory element-binding protein |
| HIF | Hypoxia-inducible factor | TCA cycle | Tricarboxylic acid cycle |
| HK | Hexokinase | TGF-β | Transforming growth factor-β |
| HMT | Histone methyltransferase | THF | Tetrahydrofolate |
| IGF | Insulin-like growth factor | TME | Tumor microenvironment |
| LDH | Lactate dehydrogenase | VEGF | Vascular endothelial growth factor |
| LGR5 | Leucine-rich repeat-containing G protein-coupled receptor 5 |
The Warburg Effect
The unique metabolic phenotype of cancer cells was first observed by Otto Warburg in 1924 when he noted that, even in the presence of oxygen, cancer cells preferentially shift their metabolism toward glycolysis followed by lactic acid fermentation rather than OXPHOS [10]. This state of aerobic glycolysis is known as the “Warburg effect” [11, 12]. While OXPHOS produces more ATP per molecule of glucose, glycolysis produces ATP more rapidly. Additionally, increased glycolysis facilitates cellular utilization of glycolytic intermediates to synthesize macromolecules required to support rapid proliferation (reviewed in [13]). For instance, the glycolytic intermediates glucose-6-phosphate, fructose-6-phosphate, and glyceraldehyde-3-phosphate are converted to ribose-5-phosphate in the pentose phosphate pathway (PPP), which generates pentose sugars and ribose-5-phosphate to synthesize nucleotides required for DNA replication [13]. Glyceraldehyde-3-phosphate is also utilized to synthesize phospholipids to generate cell membranes, and 3-phosphoglycerate is a substrate for the synthesis of serine and glycine [13]. The decrease in OXPHOS is also thought to protect cells from toxic reactive oxygen species (ROS) and oxidative stress that would be generated during rapid proliferation [13]. The pyruvate that accumulates during aerobic glycolysis is reduced to lactate by lactate dehydrogenase 5 (LDH5) to regenerate NAD+ and then secreted from the cell by monocar-boxylate transporter 4 (MCT4) (reviewed in [14]).
To date, data suggest that most CRC cells demonstrate the “Warburg” metabolic phenotype. For example, primary and metastatic CRC tumors often exhibit higher glucose consumption than surrounding normal intestinal tissue, and this is taken advantage of in tumor imaging with 18F-deoxyglucose positron emission tomography (18FDG-PET) [15, 16]. In addition, CRC tumors exhibit increased intratumoral lactate [17, 18••]. Numerous genes and proteins important for glucose uptake and glycolysis are also upregulated in CRC, including hypoxia-inducible factor-1 α (HIF-1 α)[19], glucose transporter 1 (GLUT1) ([20–24]), hexokinase 1 (HK1) [24, 25], hexokinase 2 (HK2) [26], MCT4 [23, 27–29], pyruvate kinase M2 (PKM2) [24, 30, 31], lactate dehydrogenase A (LDHA, a monomer of LDH) [24, 32], and lactate dehydrogenase 5 (LDH5) [33]. Indeed, upregulation of many of the above genes has been correlated with tumor aggressiveness and poor prognosis in CRC [34].
Glutaminolysis
Glutamine dependency is another metabolic phenotype that supports Warburg metabolism and is common to many different cancer types (reviewed in [35]), including CRC [36]. Because glycolytic intermediates are diverted toward biosynthetic pathways, cancer cells utilize glutamine to replenish the TCA cycle in a process termed anaplerosis and, like glucose, synthesize macromolecules necessary for cell division and tumor growth [35]. First, glutamine is taken up by the cell and deaminated to glutamate by glutaminase 1 (GLS1). There, it is converted to α-ketoglutarate (αKG) by glutamate dehydrogenase (GDH), which can be employed to synthesize lipids and amino acids and generate ATP via the TCA cycle followed by OXPHOS [35]. These reactions also produce reduced nicotinamide adenine dinucleotide phosphate (NADPH), which is used to regenerate glutathione (GSH) [37]. GSH acts as an electron acceptor to neutralize ROS created by the increased metabolic flux in cancer cells (reviewed in [37]). GLS1 and GDH are both upregulated in CRC, and this correlates with tumor aggressiveness and poor patient prognosis [38–40].
Lipid Biosynthesis
Deregulated lipid metabolism is recognized as another metabolic phenotype in many different cancers (reviewed in [41, 42•]). Most normal cells preferentially use circulating FAs derived from dietary fat. In contrast, cancer cells not only increase uptake of extracellular lipids and lipoproteins but also upregulate de novo lipid biogenesis and cholesterol synthesis pathways, which produce lipid metabolites crucial to cell membranes, cell signaling, post-translational modification of proteins, neutralization of ROS, and energy storage [42•]. FAs are synthesized in the cytoplasm using citrate, which is converted to acetyl-CoA by cytoplasmic ATP citrate lyase (ACL) and malonyl-CoA, which is generated from acetyl-CoA by acetyl-CoA carboxylase [42•]. FA synthase (FASN) catalyzes the synthesis of palmitate, a 16-carbon saturated FA, by the repeated condensation of acetyl-CoA molecules with one molecule of malonyl-CoA [42•]. Palmitate provides the basic substrate for desaturation and elongation reactions that produce a large variety of FAs [42•]. Metabolic intermediates of these processes are substrates for the synthesis of cholesterol and phospholipids, which can be used to synthesize cell membranes and prostaglandins, key mediators of inflammation [42•]. Finally, surplus lipids are stored in complex organelles called lipid droplets and consumed to generate ATP via mitochondrial FA oxidation (FAO) in nutrient-poor environments (reviewed in [43]).
Lipogenic enzymes, including FASN, are upregulated in CRC (reviewed in [44]). Patients with stage III and IV CRC have higher circulating FASN levels than patients with stage I and II CRC [45••]. In CRC cell lines, high FASN expression upregulates both glycolysis and mitochondrial respiration and enables CRC cells to increase FAO in metabolic stress conditions [46]. Novel FASN inhibitors are being evaluated in phase I/II clinical trials for CRC treatment [47]. In addition to increased palmitate synthesis, CRC exhibits enhanced FA elongation [48]. Upregulation of other lipid metabolism genes, including PPARG and ACSL1, are correlated with poor prognosis in CRC [49, 50]. Silencing genes encoding sterol regulatory element-binding proteins (SREBPs) 1 and 2, which are transcription factors critical for lipogenesis, inhibited CRC growth in vitro and in xenograft models [51]. CRC cells also have high levels of lipid droplets, which serve as the site of prostaglandin E2 (PGE2) synthesis from arachidonic acid, a FA, by cyclooxygenase-2 (COX-2) [52, 53]. PGE2 is transported out of the cell, where it exerts both autocrine and paracrine effects by activating signaling pathways that control inflammation, proliferation, migration, apoptosis, and angiogenesis (reviewed in [54, 55]). Finally, lipid biogenesis pathways have been correlated with CRC epithelial-mesenchymal transition, invasion, and metastasis [56–59].
One-Carbon Metabolism
One-carbon metabolism (1CM) is also heavily utilized by many different cancer types, including CRC (reviewed in [60•, 61•, 62]). 1CM is essential for the increased nucleotide and FA synthesis required by highly proliferating cells and provides crucial substrates for chromatin remodeling [61•]. 1CM consists of three interconnected pathways that involve the transfer of a single carbon: the folate cycle, the methionine cycle, and the trans-sulfuration pathway [61•]. The amino acids serine, glycine, and threonine (converted to glycine) serve as carbon donors to initiate the cycle, converting tetrahydrofolate (THF, which is reduced from dietary folate) to 5,10-methylene THF (meTHF), which is further reduced to 5-methyl THF (mTHF) [61•]. mTHF donates a carbon to homocysteine, generating methionine. Adenylation of methionine generates S-adenosylmethionine (SAM), which serves as the methyl donor for histone methyltransferases (HMTs) and DNA methyltransferases (DNMTs) [61•]. SAM was recently found to be the most upregulated metabolite in CRC across all tumor stages [18••]. Additionally, polymorphisms in genes required for 1CM and dietary intake of folate have been shown to modify CRC risk [63, 64].
THF can also be converted to formyl-THF. Formyl-THF and meTHF donate carbon units to purine and thymidylate synthesis, and meTHF can be oxidized to support FA synthesis [61•]. These reactions also reduce NADP+ to NADPH. This NADPH, and glutathione (GSH) generated via the trans-sulfuration pathway, are important in other cellular redox reactions [61•]. Phosphoglycerate dehydrogenase (PHGDH), an enzyme that diverts 3-phosphoglycerate from glycolysis into the serine biosynthetic pathway to resupply 1CM, is upregulated in CRC tumors compared to adjacent normal tissue and is correlated with advanced TNM stage and tumor size [65]. This is particularly useful to cancer cells in the setting of glucose deprivation [66].
Oxidative Phosphorylation
Although the Warburg effect was initially thought to be due to defective mitochondrial OXPHOS, it is now thought that cancer cells, including CRC, use the TCA cycle (fueled by pyruvate, lactate, glutamine, and/or FAs) followed by OXPHOS to generate ATP, which is required for biosynthetic and housekeeping reactions (reviewed in [5, 67]). It is the downregulation of the MPC in CRC that results in increased aerobic glycolysis metabolism [68]. Additionally, there is intratumoral heterogeneity in metabolic phenotypes, with some cells relying more heavily on OXPHOS compared to others based on the local TME [69]. This will be discussed in detail later in this review (see “Intratumoral Heterogeneity”).
Metabolic Pathways Impact Epigenetics
Epigenetics is defined as changes in gene expression that are heritable during cell division and cannot be explained by alterations in primary DNA sequences [70]. Changes in metabolic flux impact epigenetics in both normal cells and cancer cells because metabolites serve as essential cofactors for chromatin remodeling enzymes responsible for epigenetic alterations (reviewed in [61•, 71]), which are abundant in CRC (reviewed in [70]). SAM, generated by the methionine cycle, serves as the methyl donor for HMTs and DNMTs [61•]. Thus, changes in intracellular SAM directly affect histone methylation associated with active gene transcription [72]. Increased methylation is common in CRC and is associated with poor survival rates [70]. Furthermore, aberrant DNA methylation has also been extensively demonstrated in CRC and occurs early in the adenoma to carcinoma sequence, as hypermethylation of CpG islands in gene promoters transcriptionally silence tumor suppressor genes (reviewed in [4, 54, 70]). The CpG island methylator phenotype (CIMP) pathway in CRC is characterized by particularly widespread altered promoter methylation [4].
On the other hand, CRC also exhibits global DNA hypomethylation (outside of CpG islands) [70]. Metabolites generated in the TCA cycle and ETC serve as cofactors for DNA and histone demethylation. αKG is required for activity of the TET family of DNA demethylases and the Jumonji C family of histone demethylases [61•]. The histone demethylation activity of lysine-specific demethylase 1 requires FAD, which is synthesized de novo from riboflavins and renewed through the oxidation of FADH2 in the ETC [61•].
Histone acetylation can also be influenced by metabolite production. Acetyl-CoA provides the acetyl group necessary for acetylation of lysine and other amino acid residues on histones [61•]. Nuclear acetyl-CoA is derived from citrate, which is generated in the mitochondria by the TCA cycle, transported out of the mitochondria, and converted back to oxaloacetate (OAA) and acetyl-CoA by nuclear ACL [73]. In the nucleus, acetyl-CoA serves as the substrate for histone acetyltransferases (HATs). Removal of acetyl groups by a class of histone deacetylases (HDACs) known as sirtuins (SIRTs) are dependent on NAD+ [61•]. Changes in metabolic flux shift the ratio between NAD+ and NADH and alter SIRT activity; increased glycolysis has been shown to decrease NAD+ availability, impairing SIRT-mediated histone deacetylation [61•].
Metabolic Changes May Precede Somatic Mutations
The majority of CRC develops progressively from normal mucosa to adenoma to carcinoma, which occurs by several major pathways [74]. The most common mechanism is the CIN pathway characterized by widespread loss of heterozygosity (LOH) in tumor suppressor genes and numerous chromosomal copy number variations and structural abnormalities [75]. Other mechanisms include the MSI pathway, caused by defects in the DNA mismatch repair system, and the CIMP pathway, in which the promoter sequences of tumor suppressor genes are hypermethylated (reviewed in [76]).
Findings from numerous studies in both mice and humans support the theory that metabolic and epigenetic alterations throughout the colon promote genomic instability that leads to neoplastic transformation, known as “field carcinogenesis” or “condemned mucosa” (reviewed in [77, 78]). These alterations have been shown to exist in grossly and even histologically “normal” colon tissue and likely develop due to a combination of exogenous factors including diet and obesity and endogenous factors including genetic CRC risk, preceding and promoting focal neoplastic transformation due to LOH [77].
In accordance with the field carcinogenesis model, morphologically normal colon tissue in mice with genetic susceptibility to CRC exhibits increased lactate [79] and transcriptomic changes associated with multiple metabolic pathways, including glucose metabolism and insulin signaling [80, 81]. Cruz et al. also recently showed that tissue from rectal biopsies of grossly normal tissue obtained from patients with one or more pre-cancerous colonic lesions has significant increases in the expression of HIF1A, GLUT1, PKM2, and LDHA, indicating upregulated glycolysis [82]. They also found increased expression of mitochondrial genes that promote mitochondrial fission and fusion, uncoupling between proton flow and ATP synthase activity, and mitochondrial copy number, indicating altered OXPHOS [82].
Effects of CRC Driver Mutations on Cellular Metabolism
Mutations in genes that functionally contribute to CRC development or progression are considered “driver” mutations rather than “passenger” mutations that occur as a byproduct of CRC development [83]. While metabolic flux changes were initially believed to be passive “side effects” of oncogenesis, it is now thought that metabolic pathways are actively reprogrammed by oncogenes, providing a selective advantage as tumors evolve to adjust to nutrient availability in the TME [5, 7, 13].
The WNT Signaling Pathway
Ninety-three percent of sporadic CRC tumors exhibit hyperactive WNT signaling, of which 80% is caused by biallelic inactivation of the adenomatous polyposis coli (APC) tumor suppressor gene or activating mutations in β-catenin (CTNNB1) [84]. WNT signaling is critical in embryonic development and homeostasis of the adult gastrointestinal tract (reviewed in [85]). β-catenin, the major effector of the canonical WNT signaling pathway, translocates to the nucleus and associates with TCF/LEF transcription factors to activate expression of target genes that control cell fate specification, stem cell function, proliferation, and migration [85]. Pate and colleagues recently established a direct link between WNT signaling and increased glycolysis by identifying pyruvate dehydrogenase kinase 1 (PDK1) as a novel WNT target gene [86••]. PDK1 phosphorylates pyruvate dehydrogenase (PDH), inhibiting pyruvate conversion to acetyl-CoA and subsequent flux to OXPHOS [87]. They also asserted that MCT1 may be another novel WNT target gene due to occupation of its promoter by TCF4 and its downregulation in xenograft tumors after expression of dominant negative TCF/LEF [86••] The significance of MCT1 will be discussed later in this review (see “The Tumor Microenvironment”).
Another transcriptional target of the WNT signaling pathway is MYC, which controls glycolysis, nucleotide synthesis, lipid synthesis, and mitochondrial bioenergetics in various cancers (reviewed in [88]). However, in addition to WNT signaling, MYC deregulation can be a consequence of aberrant transcription factor activity, PI3K signaling, receptor tyrosine kinases (RTKs), hormones, and growth factors [89]. In 2017, the Soga group utilized a combined transcriptomics and metabolomics approach to evaluate paired normal and tumor tissues obtained from 275 patients with CRC [18••]. The group found consistent overexpression of MYC in all cancer stages regardless of mutations in APC, CTNNB1, or other genes involved in CRC carcinogenesis. MYC overexpression correlated with expression changes in 231 unique metabolic genes; those involved in purine and pyrimidine synthesis, glycolysis, the PPP, FA synthesis, 1CM, and histone methylation were upregulated, while those in the TCA cycle and FAO were downregulated [18••]. The corresponding changes in metabolite levels occurred at the premalignant adenoma stage and remained similar throughout the adenoma-carcinoma sequence, indicating that global metabolic reprogramming was driven by MYC independent of WNT hyperactivation or other driver mutations in CRC [18••].
The PI3K Signaling Pathway
Phosphatidylinositol 3-kinases (PI3Ks) are a family of intracellular signal transduction enzymes critical for the response of normal cells to growth factors, including platelet-derived growth factor (PDGF), epidermal growth factor (EGF), insulin-like growth factor (IGF), and insulin (reviewed in [90, 91]). When these growth factors bind their respective RTKs and G protein-coupled receptors, phosphatidylinositol 3-kinase α (PI3Kα) phosphorylates phosphatidylinositol-4,5-bisphosphate (PIP2), converting it to phosphatidylinositol-3,4,5-triphosphate (PIP3) [90]. This second messenger activates numerous effectors—most importantly, the serine/threonine kinases AKT and mammalian target of rapamycin (mTOR)—that alter cell-cycle progression and activate the metabolic program necessary to meet the requirements for the growth and synthesis of new cells [90]. The mTORC1 complex, which consists of mTOR, Raptor, and mLST8, has numerous phosphorylation targets and is a key metabolic regulator upon which many different signaling pathways converge, including the RAS pathway, which will be discussed next (reviewed in [92]). In general, mTORC1 acts as a sensor of cellular energy status, directing metabolic flux toward anabolism (when active) or catabolism (when inactive) in response to nutrient and oxygen availability [92]. mTORC1 activates SREBP to induce transcription of genes required for de novo lipid synthesis and increase flux through the PPP to generate nucleotide precursors [92]. mTORC1 also increases the translation of HIF-1 α, a transcription factor that drives angiogenesis and the expression of genes critical for glycolysis, including GLUT1 and phosphofructokinase (PFK) [93]. Activated mTORC1 also suppresses protein catabolism and autophagy, the lysosomal degradation of cytoplasmic proteins, organelles, and lipids to promote survival during nutrient deprivation [92].
Mutations leading to hyperactive PI3K signaling occur in the majority of CRC tumors [84]. The PIK3CA gene, which encodes the p110α catalytic subunit of PI3Kα, is one of the most frequently mutated genes in human cancers [94], including approximately 30% of CRC [84, 95]. Other common alterations in the PI3K pathway include activating mutations in IGF2 and IRS2 (which codes for a protein linking the IGF-1 receptor to PI3K) and inactivation of the genes encoding the PI3K inhibitors PTEN and PIK3R1 [84]. In various cell types (nonspecific to CRC), the PI3K pathway has been shown to assist in GLUT1 translocation to the membrane [96, 97] and reduce endocytosis of GLUT1 and GLUT4 [98]. AKT increases glucose metabolism by phosphorylating HK2 [99], PFKFB2 (which allosterically regulates PFK-1) [100], and mTOR [101].
Recent work by Hao and colleagues demonstrated that oncogenic PIK3CA mutations also affect glutamine dependency in CRC by upregulating glutamate pyruvate transaminase 2 (GPT2), which converts glutamate to αKG [102••]. As stated previously, this replenishes the TCA cycle, generating ATP and intermediates for lipid and amino acid synthesis. Interestingly, this relationship between oncogenic PI3Kα and GPT2 is AKT-independent; PI3Kα activates RSK2 kinase, which phosphorylates ATF4, preventing its ubiquitin-mediated degradation and allowing it to activate GPT2 [102••].
RTK-RAS Signaling
The RTK-RAS-MAPK pathway is an additional pro-proliferative signaling cascade that is integrated with the PI3K pathway, as the two pathways are activated by many of the same growth factors, can activate mTORC1, and either cross-activate or cross-inhibit each other depending on the balance of extracellular and intracellular signals (reviewed in [103]). The RTK-RAS pathway is activated when a ligand binds to a cell-surface RTK (i.e., EGFR), leading to the activation of RAS. RAS recruits and activates RAF, which activates the MEK/ERK cascade leading to the activation of transcription factors critical for cell survival and proliferation [103].
The RTK-RAS-MAPK pathway is altered in the majority of CRC tumors [84]. Specifically, mutations in KRAS that confer constitutive activity are present in 43% of non-hypermutated tumors and 30% of hypermutated tumors [84]. Mutations in BRAF, the most common of which is the V600E mutation, exhibit a pattern of mutual exclusivity with mutations in KRAS (and its family member, NRAS), and are present in only 3% of non-hypermutated CRCs, but a substantial 47% of hypermutated tumors [84]. These oncogenic mutations in KRAS and BRAF have been strongly correlated with metabolic dysregulation in CRC [104]. The CRC cell lines DLD-1, which has an oncogenic mutation in KRAS, and RKO, which has an oncogenic mutation in BRAF, display increased GLUT1 expression and a Warburg phenotype [105, 106]. Analysis of isogenic DLD-1 and RKO CRC cell lines expressing KRASG13D or BRAFv600E mutations identified significant changes in metabolic proteins involved in glycolysis, the nonoxidative PPP, glutamine metabolism, and the phosphoserine biosynthetic pathway, as well as increases in glucose uptake and lactate production, compared to those expressing wild-type KRAS or BRAF [107••]. However, according to Hao et al., isogenic HCT116 and DLD-1 clones with either wild-type or mutant KRAS knockout do not exhibit differential glutamine dependency [102••]. Oncogenic KRAS has also been shown to decrease ROS production and increase mitochondrial OXPHOS efficiency by activating mitochondrial phospholipid synthesis via HIF-1α and HIF-2α transcriptional programs [108].
Finally, two groups have recently demonstrated that KRASG12v and BRAFV600E mutations affects metabolic pathways differently in CRC [109, 110]. A proteomics and metabolomics approach by Fritsche-Guenther and colleagues showed that glycolytic flux was upregulated in Caco-2-KRASG12v cells but decreased in Caco-2-BRAFV600E cells as compared to Caco-2 controls at physiologic glucose levels [110]. Mechanistically, they found that this was because activation of mTOR by BRAFV600E was uncoupled from control of mTORC activity by AMPK, which is known to be a cellular sensor of glucose availability (reviewed in [111]). Inhibition of mTOR decreased cell viability in cells with either mutation, a finding corroborated by many other groups [112–114], indicating that mTOR is a potential therapeutic target in BRAFV600E-driven CRC [110].
p53 Signaling
The main function of the p53 tumor suppressor is to mediate cell-cycle arrest, apoptosis, and senescence in response to cellular stress. The role of TP53 loss in tumorigenesis is evident as TP53 is the most commonly mutated gene in human cancer (36.1% of cancers in 20 different tissues) [115]. Aberrant p53 signaling, usually caused by biallelic TP53 inactivation, occurs in 64% of non-hypermutated sporadic CRC cases and 47% of hypermutated cases [84]. p53 signaling regulates glycolysis and OXPHOS both directly and indirectly (reviewed in [116–118]). In various cell types, wild-type p53 suppresses glycolysis by repressing transcription of GLUT1 [119, 120], GLUT3 (via repression of the NF-κB pathway) [121], GLUT4 [119], and GLUT12 [120]. p53 also inhibits the expression of mitochondrial PDH kinase 2 (PDK2), which negatively regulates PDH, leading to increased conversion of pyruvate to acetyl-CoA [122], and activates expression of PARK2, which augments the expression of the PDH subunit-encoding gene PDHA1 [123]. Conversely, p53 promotes mitochondrial OXPHOS (reviewed in [124]) by maintaining the mitochondrial genome [125–127] and inducing the expression of cytochrome c oxidase and genes required for its synthesis [128, 129]. It follows that loss of TP53 in CRC would promote glycolysis and prevent OXPHOS. Dominant negative p53 (as opposed to p53 loss) also contributes to CRC pathogenesis (reviewed in [130]). For example, dominant negative p53 directly stimulates glycolysis by promoting GLUT1 translocation to the plasma membrane and by activating HK-2 (reviewed in [118]).
However, some actions of wild-type p53 seem paradoxical in the context of widespread TP53 mutations and Warburg metabolism in cancer cells. Wild-type p53 induces transcription of TIGAR, which shunts glycolytic intermediates into the pentose phosphate pathway by hydrolyzing fructose-2,6-bisphosphate, an allosteric activator of PFK-1 [131]. This generates GSH to mitigate ROS production. Another transcriptional target of p53 is glutaminase 2 (GLS2), which converts glutamine to glutamate for entry into the TCA cycle [132, 133]. While this may drive oxidative metabolism, it could also support anaplerosis in cancer cells. Thus, it appears that the actions of p53 are context-dependent [118], and mechanisms of p53 metabolic reprogramming in CRC need to be investigated further.
Intratumoral Heterogeneity
While tumor genetics were once studied in aggregate, we have come to appreciate the complex intratumoral heterogeneity resulting in distinct tumor cell populations, including cancer stem cells (CSCs) and neoplastic stromal cells recruited by tumor cells [6, 134]. Further contributing to tumor heterogeneity, tumor cells exhibit metabolic plasticity that enables them to adapt to environmental factors affected by the local TME, such as oxygenation, pH, and nutrient availability [135•].
Cancer Stem Cells
The CSC hypothesis suggests that tumor cells exist in a hierarchy rather than uniform, clonal populations [135•]. CSCs constitute a small population of tumor cells having stem cell properties, such as self-renewal, the potential for proliferation and multilineage differentiation, and the ability to initiate tumor formation (reviewed in [135•, 136]). CSCs may generate the bulk of tumor cells and have also been implicated as drivers of metastasis and therapeutic resistance [135•]. CSCs in CRC have been identified by numerous different markers, including CD144, CD133, CD24, CD29, CD26, CD166, CD326, leucine-rich repeat-containing G protein-coupled receptor 5(LGR5), BMI1, ALDH1, and type III deiodinase [136]. Two different, but not necessarily mutually exclusive, models of CSC origin have been proposed in CRC: the transformation of normal adult stem cells and the reprogramming of cancer cells resulting in the acquisition of stem cell properties [135•]. While the acquisition of stem cell properties may be due to genetic changes in these cells, the “metabostemness” theory posits that stem cell properties are dependent on cell metabolism and its epigenetic and transcriptional regulation of gene expression [61•, 135•, 137]. However, it is unclear whether CSCs in CRC exhibit a consistent metabolic phenotype. These discrepancies may be affected by differences in methods, models, cellular environment, and the marker by which the CSCs were identified.
Several distinct populations of intestinal stem cells, identified by location and distinct markers, have been identified in the adult small and large intestine (reviewed in [138, 139]). The most thoroughly studied stem cells in the normal adult intestine are the crypt base columnar (CBC) stem cells that express LGR5, a membrane receptor for R-spondin that enhances WNT signaling [140]. Although most intestinal epithelial cancers occur in the large intestine (colon) in humans, the role of LGR5+ stem cells has been most thoroughly studied in the small intestine using mouse models and ex vivo stem cell culture. LGR5+ stem cells have been shown to generate all cell lineages of the small intestinal epithelium by lineage tracing in vivo [141, 142], and single-sorted LGR5+ cells generate three-dimensional organoids containing all differentiated cell types when cultured ex vivo [143]. The role of LGR5+ cells as the potential cell-of-origin in CRC was illuminated when LGR5+ stem cell-specific deletion of Apc caused the formation and sustained growth of small intestinal adenomas; in contrast, Apc deletion in transit amplifying cells led to initiation of microadenomas whose growth rapidly stalled [144]. Work by Rodríguez-Coleman et al. showed that Paneth cells, which form the CBC niche, rely on glycolysis and secrete lactate, which is then taken up by LGR5+ CBCs to fuel high levels of OXPHOS [145•]. This mitochondrial OXPHOS generated ROS that activated the p38 MAPK pathway, inducing crypt differentiation [145•]. These findings were consistent with studies indicating that upregulation of OXPHOS enhanced self-renewal and longevity in Drosophila intestinal stem cells [146] and that mitochondrial dysfunction in murine intestinal crypts resulted in loss of stemness and proliferation [147]. However, as pointed out by commentary written by Roper and Yilmaz, it is unclear whether these metabolic phenotypes are applicable to stem cells in the colon, which does not contain Paneth cells [148]. A study by a different group showed that LGR5-GFP stem cells isolated from murine colon by fluorescence activated cell sorting (FACS) exhibited a Warburg phenotype [149]. Additionally, Pate and colleagues recently determined that intestinal stem cells, which are highest in WNT tone, exhibit increased aerobic glycolysis compared to more differentiated cells [86••]. Finally, Schell et al. demonstrated that intestinal stem cells downregulate the MPC to reduce pyruvate flux into the mitochondria, and MPC expression increases following differentiation in ex vivo cultures derived from both small intestine and colon [150, 151]. CD133+ CSCs isolated from CRC cell lines and patients exhibit a metabolic signature characterized by high levels of enzymes and metabolites involved in both glycolysis and the TCA cycle, as well as cysteine and methionine metabolism [152]. Using Raman spectroscopy, Trinato et al. found that CSCs characterized by CD133+ status and high WNT/β-catenin activity exhibited more lipid droplets than both normal cells and CD133− CRC cells [153]. Of note, these studies need to be carefully evaluated in light of recent findings that FACS introduces oxidative stress that causes metabolic changes in cells [154].
The Tumor Microenvironment
The CRC TME consists of extracellular matrix, cancer-associated fibroblasts (CAFs), immune cells (including mast cells, neutrophils, eosinophils, basophils, tumor-associated macrophages, dendritic cells, myeloid-derived suppressor cells, natural killer cells, B lymphocytes, and T lymphocytes), adipocytes, endothelial cells, pericytes, and platelets (reviewed in [134]). To support their uncontrolled proliferation, invasion, and metastasis, tumor cells alter transcriptional and metabolic programs in these other cell populations through the exchange of growth factors, cytokines, and metabolites [155•]. These species include PDGF, transforming growth factor-β (TGF-β), interleukins 4 and 6 (IL-4 and IL-6), IGF-2, fibroblast growth factor-2 (FGF-2), PGE2, and ROS [134]. Cancer cells have also been shown to metabolically reprogram stromal cells at sites of metastasis (reviewed in [69]).
CAFs are one such stromal cell type co-opted by tumor cells to support their growth; in fact, CAFs have been described as “architects of tumor pathogenesis” [156]. CAFs can be reprogrammed toward a glycolytic phenotype in breast and head/neck carcinomas in a process called “reverse Warburg” metabolism, so-named because of its implication that aerobic glycolysis can take place in the tumor-associated stroma and not only epithelial tumor cells (reviewed in [69, 135•, 157, 158]). Here, microenvironmental cues from the tumor cells, particularly OXPHOS-generated ROS, induce a Warburg effect in fibroblasts, which then release lactate, pyruvate, glutamine, and ketone bodies (metabolic byproducts of FAO) via upregulation of MCT4 and other transporters [159]. These energy-rich metabolic byproducts are taken up by carcinoma cells via MCT1 and used to replenish the TCA cycle to support OXPHOS [159]. While this has not been investigated extensively in CRC, MCT1 is upregulated in tumor cells [23, 27]. Additionally, lactate produced by glycolytic CRC cells can fuel oxidative metabolism in oxygenated tumor cells [160]. Of note, CAFs also produce hepatocyte growth factor, EGF, IGF-1 and IGF-2, FGF-2 and FGF-7, PGE2, vascular endothelial growth factor (VEGF), stromal cell-derived factor, interleukins, and even miRNAs that activate metabolic reprogramming in tumor cells [134].
Adipocytes are also co-opted to fuel CRC growth. Wen et al. recently showed that adipocytes from colon cancer patients released abundant FAs that were taken up, catabolized via autophagy to replenish the TCA cycle and OXPHOS, and stored as lipid droplets by co-cultured SW480 and DLD1 CRC cells and CRC tumor organoids [161•]. These FAs enabled the tumor cells to survive nutrient deprivation [161•]. This may serve as one mechanism by which obesity increases CRC risk and decreases survival in CRC patients [162–165]. Increased uptake of FAs by cancer cells also contributes to cancer cachexia, massive weight loss in the setting of cancer characterized by release of FAs into the plasma (reviewed in [157]).
We also now know that the polarization and/or activation of immune cells is dependent on metabolic reprogramming (reviewed in [166•]). Lactate has been shown to cause macrophages to adopt anti-inflammatory transcription profiles [166•]. Additionally, some cancers generate “oncometabolites” that actively suppress the anti-tumor activity of immune cells in the TME [167]. In CRC, commensal gut microbiota, and immunologic responses to the microbiota, also shape the colonic immune environment [168]. Thus, the metabolic reprogramming of immune cells in CRC cells needs to be further investigated.
It is important to note that, while many of the studies explored in this review characterize the metabolic behavior of CRC using cell lines grown in an abundance of glucose and/or glutamine, the local TME varies within tumors, leading to metabolic compartmentalization [155•]. Because the proliferation of tumor cells is limited by access to oxygen and glucose, tumors recruit their own network of blood vessels via angiogenesis. VEGF and PGE2 are both pro-angiogenic factors released by epithelial CRC cells and tumor-associated stromal cells [155•]. However, the tumor-associated vasculature is heterogeneous and leaky; thus, it does not deliver oxygen and/or nutrients to all tumor cells efficiently or equally. Tumor cells adapt to this changing and/or limited nutrient availability by preferentially utilizing different metabolic programs [155•, 160]. Metabolic plasticity is also essential for metastasis, in which tumor cells extravasate and colonize a completely different TME (reviewed in [69]). Loo et al. demonstrated that CRC cells release creatine kinase brain-type (CKB), which phosphorylates liver-derived creatine (a metabolite of glycine and arginine) to phosphocreatine extracellularly [169•]. The phosphocreatine is imported into metastatic CRC cells using the SLCA8 transporter and used as a high-energy phosphate donor to produce ATP, bypassing OXPHOS and allowing the cancer cells to survive hypoxia. Inhibition of this pathway by miRNAs or a small-molecule inhibitor suppressed metastatic colonization in the liver [169•].
Contributions From Gut Microbiota
Rapid improvement of high-throughput screening technologies has advanced our understanding of the role of the gastrointestinal microbiota in human health and disease. The gastrointestinal microbiota is composed of at least 1000 identified bacterial species [170]. The most common species belong to the phyla Firmicutes, Actinobacteria, and Bacteroidetes, while Lactobacillae, Streptococci, and Enterobacteria are present in smaller numbers [171]. The “microbiome” consists of approximately 3.3 million non-redundant genes encoded by the microbiota, 150 times the number of genes in the human genome [170, 172]. The microbiome is highly enriched for genes involved in the metabolism of starch, sugars, short-chain FAs (SCFAs) and the synthesis of essential amino acids and vitamins [173].
Dysbiosis has been associated with many risk factors of CRC, including obesity, diabetes, inflammatory bowel disease, and with CRC itself (reviewed in [168]). A number of studies have shown intestinal dysbiosis in patients with pre-cancerous adenomas and overt CRC, implicating the microbiome in tumor initiation and progression (reviewed in [174, 175]). Studies have shown that decreases in protective microbes and increases in harmful microbes can lead to chronic inflammation and CRC pathogenesis [175]. CRC has been associated with increased Fusobacterium and Prevotella and decreased butyrate-producing bacteria [174]. Furthermore, different molecular subtypes of CRC have been associated with enrichment of distinct microbial populations [176].
In general, the gut microbiota broadly affects cancer susceptibility through several mechanisms involving metabolism, including harvesting otherwise inaccessible substrates from the diet, metabolizing xenobiotics, and producing metabolites that cause chronic inflammation [177, 178]. The impact of this “chemical communication” between gut bacteria and colonic epithelial cells using specific metabolites on CRC tumorigenesis was recently reviewed by Bhutia et al. [179]. Here, we will focus on the role of butyrate, a SCFA that has been particularly implicated as a link between the gut microbiome, diet, metabolism, and CRC.
While some dietary butyrate is available from bovine milk products [180], most butyrate is produced in the colon by intestinal microbiota as a fermentation product of polysaccharides, including dietary fiber and resistant starch (reviewed in [181]). Butyrate-producing species predominantly belong to the Firmicutes phylum, but also include Actinobacteria, Bacteroidetes, Fusobacteria, and others [182–185]. The microbiota of CRC patients has been shown to include fewer butyrate-producing bacteria compared to patients without CRC [186, 187], and fecal butyrate levels are inversely correlated with CRC tumor size [188, 189].
The “butyrate paradox” refers to the differential effects of butyrate on CRC cells compared to normal differentiated colonocytes (reviewed in [190]). Butyrate supplies over 70% of the energy required by colonocytes [191]. By oxidizing butyrate, colonocytes rely less on glucose oxidation and can divert pyruvate and glutamine to other pathways. Butyrate is metabolized in the mitochondrial β-oxidation pathway to acetyl-CoA, which enters the TCA cycle. As stated previously, nuclear ACL converts citrate generated in the TCA cycle back to acetyl-CoA, the substrate for FA synthesis and histone acetylation by HATs [73]. Thus, increased butyrate uptake by normal colonocytes not only provides cellular energy but also leads to histone acetylation in an ACL-dependent mechanism [192••]. While administration of butyrate either has no significant effect on, or stimulates the growth of, normal colonocytes, it inhibits the growth and induces differentiation of CRC cell lines [193, 194]. This is a direct effect of Warburg metabolism, in which decreased usage of OXPHOS reduces butyrate flux into the TCA cycle, causing butyrate to accumulate intracellularly and in the nucleus, where it functions as an HDAC inhibitor (HDACi) [192••]. In support of this, while the ACL-dependent mechanism predominates in CRC cell lines treated with 0.5 mM butyrate, the HDACi mechanism predominates in CRC cells treated with 2–5 mM butyrate. Although both mechanisms result in increased histone acetylation, ACL-dependent target genes are predominantly pro-proliferative, while HDACi-dependent target genes are predominantly pro-apoptotic [192••]. This established a mechanism by which aberrant cell metabolism causes an epigenetic shift that contributes to tumorigenesis. A recent study by Li et al. demonstrated an additional mechanism to explain the effect of butyrate on CRC cell lines, in which butyrate directly binds to and inhibits PKM2 [195•], a less active isoform of PK that is highly expressed in anabolic cells, including cancer cells, to direct glycolytic intermediates away from the TCA cycle (reviewed in [196]). Finally, fiber-rich diets and the subsequent increase in butyrate have also been shown to attenuate risk factors of CRC, including inflammation, obesity, and insulin resistance (reviewed in [181]). Of note, butyrate is also a potent inhibitor of intestinal stem cell proliferation; however, they are located deep in colonic crypts and are thus protected from this effect by the metabolism of butyrate by differentiated colonocytes closer to the luminal surface [197].
Conclusions
CRC is widespread and confers high morbidity and mortality. Thus, there is great need for improved CRC screening and therapeutics. While colonoscopy is the gold standard method for detecting pre-cancerous lesions and CRC, it is costly and invasive. Thus, metabolic profiling of intestinal mucosa may one day augment colonoscopy-based screening approaches. Numerous studies in mice and in humans have attempted to classify metabolites found in the blood, stool, and urine that may be useful in detecting pre-cancerous lesions and/or overt CRC. Additionally, routine clinical evaluation of patients with overt CRC remains based on tumor size, histology, involvement of lymph nodes, and tumor genetics. Assessing the metabolic profile of CRC tumors in patients may one day be a component of a patients’ staging evaluation.
In this review, we discussed the etiologies and effects of altered cellular metabolism in CRC, including in cells that make up the bulk of CRC tumors, CSCs, stromal and immune cells in the TME, and the gut microbiota. It is clear that metabolic dysregulation is by no means an isolated hallmark of cancer—it contributes to, and likely drives, other hallmarks of cancer including genomic instability and mutational burden, sustained proliferative signaling, tumor-promoting inflammation, activation of invasion and metastasis, induction of angiogenesis, avoiding immune destruction, and resisting cell death.
While many studies have characterized the metabolic phenotypes of CRC cell lines or CRC tumors in aggregate (including the upregulation of glycolytic enzymes, increased glutaminolysis, and increased FA synthesis), we still have much to learn about the metabolic programs active in individual CRC cells. For example, it is still unclear whether CSCs in CRC adopt a predominantly Warburg metabolic phenotype or whether they utilize OXPHOS using substrates generated by a “reverse Warburg” mechanism. Finally, while the metabolic reprogramming of CAFs and tumor-associated immune cells has been well-characterized in other cancers, the cross-talk between epithelial CRC cells and their immunologic niche needs to be further explored.
Acknowledgements
This work was supported by National Institutes of Health grants R01DK099204 to C.S.W., 1F30DK120149 and T32GM007347 to R.E.B., 5F32 DK108492 to S.P.S., P30DK058404 (Vanderbilt Digestive Disease Research Center); and an Office of Medical Research, Department of Veterans Affairs (Merit Review Grants) 1I01 BX001426 to C.S.W. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Conflict of Interest The authors declare they have no conflict of interest.
Human and Animal Rights and Informed Consent This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–91. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–93. [DOI] [PubMed] [Google Scholar]
- 3.Cronin KA, Lake AJ, Scott S, Sherman RL, Noone A-M, Howlader N, et al. Annual report to the nation on the status ofcancer, part I: national cancer statistics. Cancer. 2018;124:2785–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell NIH Public Access; 2012;21:297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.••.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. [DOI] [PubMed] [Google Scholar]; This commentary by Hanahan and Weinberg updated their original “Hallmarks of Cancer” to include metabolic reprogramming.
- 7.DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: what do metabolic outliers teach us? Cell. 2012;148:1132–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leninger A, Nelson D, Cox M. Lehninger principles of biochemistry. 7th ed. New York: Worth Publishers; 2017. [Google Scholar]
- 9.Rich PR. The molecular machinery of Keilin’s respiratory chain. Biochem Soc Trans. 2003;31:1095–105. [DOI] [PubMed] [Google Scholar]
- 10.The metabolism of tumours: investigations from the Kaiser Wilhelm Institute for Biology, Berlin-Dahlem. J Am Med Assoc. 1931;96:1982. [Google Scholar]
- 11.Warburg O On the origin of cancer cells. Science. 1956;123:309–14. [DOI] [PubMed] [Google Scholar]
- 12.Weinhouse S, Warburg O, Burk D, Shade AL. On respiratory impairment in cancer cells. Science. 1956;124:267–269. [DOI] [PubMed] [Google Scholar]
- 13.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330:1340–4. [DOI] [PubMed] [Google Scholar]
- 14.Fisel P, Schaeffeler E, Schwab M. Clinical and functional relevance of the monocarboxylate transporter family in disease pathophysiology and drug therapy. Clin Transl Sci. 2018;11:352–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maffione AM, Lopci E, Bluemel C, Giammarile F, Herrmann K, Rubello D. Diagnostic accuracy and impact on management of 18F-FDG PET and PET/CT in colorectal liver metastasis: a meta-analysis and systematic review. Eur J Nucl Med Mol Imaging. 2015;42:152–63. [DOI] [PubMed] [Google Scholar]
- 16.Chung JK, Lee YJ, Kim C, Choi SR, Kim M, Lee K, et al. Mechanisms related to [18F]fluorodeoxyglucose uptake of human colon cancers transplanted in nude mice. J Nucl Med. 1999;40:339–46. [PubMed] [Google Scholar]
- 17.Jiménez B, Mirnezami R, Kinross J, Cloarec O, Keun HC, Holmes E, et al. 1 H HR-MAS NMR spectroscopy of tumor-induced local metabolic “field-effects” enables colorectal cancer staging and prognostication. J Proteome Res. 2013;12:959–68. [DOI] [PubMed] [Google Scholar]
- 18.••.Satoh K, Yachida S, Sugimoto M, Oshima M, Nakagawa T, Akamoto S, et al. Global metabolic reprogramming of colorectal cancer occurs at adenoma stage and is induced by MYC. Proc Natl Acad Sci U S A. 2017;114:E7697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study showed that metabolic reprogramming occurs in response to MYC hyperactivation and does not correlate with mutations in APC, CTNNB1, or other genes involved in colorectal carcinogenesis.
- 19.Furlan D, Sahnane N, Carnevali I, Cerutti R, Uccella S, Bertolini V, et al. Up-regulation and stabilization of HIF-1alpha in colorectal carcinomas. Surg Oncol. 2007;16(Suppl 1):S25–7. [DOI] [PubMed] [Google Scholar]
- 20.Haber RS, Rathan A, Weiser KR, Pritsker A, Itzkowitz SH, Bodian C, et al. GLUT1 glucose transporter expression in colorectal carcinoma. Cancer. 1998;83:34–40. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Ye C, Chen C, Xiong H, Xie B, Zhou J, et al. Glucose transporter GLUT1 expression and clinical outcome in solid tumors: a systematic review and meta-analysis. Oncotarget. 2017;8:16875–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng W, Cui G, Tang C-W, Zhang X-L, Dai C, Xu Y-Q, et al. Role of glucose metabolism related gene GLUT1 in the occurrence and prognosis of colorectal cancer. Oncotarget. 2017;8:56850–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martins SF, Amorim R, Viana-Pereira M, Pinheiro C, Costa RFA, Silva P, et al. Significance of glycolytic metabolism-related protein expression in colorectal cancer, lymph node and hepatic metastasis. BMC Cancer. 2016;16:535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graziano F, Ruzzo A, Giacomini E, Ricciardi T, Aprile G, Loupakis F, et al. Glycolysis gene expression analysis and selective metabolic advantage in the clinical progression of colorectal cancer. Pharmacogenomics. 2017;17:258–64. [DOI] [PubMed] [Google Scholar]
- 25.He X, Lin X, Cai M, Zheng X, Lian L, Fan D, et al. Overexpression of hexokinase 1 as a poor prognosticator in human colorectal cancer. Tumor Biol. 2016;37:3887–95. [DOI] [PubMed] [Google Scholar]
- 26.Ho N, Coomber BL. Hexokinase II expression is correlated with colorectal cancer prognosis. Cancer Treat Commun. 2016;6:11–6. [Google Scholar]
- 27.Pinheiro C, Longatto-Filho A, Scapulatempo C, Ferreira L, Martins S, Pellerin L, et al. Increased expression of monocarboxylate transporters 1, 2, and 4 in colorectal carcinomas. Virchows Arch. 2008;452:139–46. [DOI] [PubMed] [Google Scholar]
- 28.Nakayama Y, Torigoe T, Inoue Y, Minagawa N, Izumi H, Kohno K, et al. Prognostic significance of monocarboxylate transporter 4 expression in patients with colorectal cancer. Exp TherMed 2012;3:25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HK, Lee I, Bang H, Kim HC, Lee WY, Yun SH, et al. MCT4 expression is a potential therapeutic target in colorectal cancer with peritoneal carcinomatosis. Mol Cancer Ther. 2018;17:838–48. [DOI] [PubMed] [Google Scholar]
- 30.Hardt PD, Mazurek S, Toepler M, Schlierbach P, Bretzel RG, Eigenbrodt E, et al. Faecal tumour M2 pyruvate kinase: a new, sensitive screening tool for colorectal cancer. Br J Cancer. 2004;91:980–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uppara M, Adaba F, Askari A, Clark S, Hanna G, Athanasiou T, et al. A systematic review and meta-analysis of the diagnostic accuracy of pyruvate kinase M2 isoenzymatic assay in diagnosing colorectal cancer. World J Surg Oncol. 2015;13:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Wang H, Liu A, Fang C, Hao J, Wang Z. Lactate dehydrogenase A negatively regulated by miRNAs promotes aerobic glycolysis and is increased in colorectal cancer. Oncotarget. 2015;6:19456–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koukourakis MI, Giatromanolaki A, Simopoulos C, Polychronidis A, Sivridis E. Lactate dehydrogenase 5 (LDH5) relates to up-regulated hypoxia inducible factor pathway and metastasis in colorectal cancer. Clin Exp Metastasis. 2005;22:25–30. [DOI] [PubMed] [Google Scholar]
- 34.Fang S, Fang X. Advances in glucose metabolism research in colorectal cancer. Biomed Rep. 2016;5:289–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li T, Le A. Glutamine metabolism in cancer. Adv Exp Med Biol. 2018;1063:13–32. [DOI] [PubMed] [Google Scholar]
- 36.Miyo M, Konno M, Nishida N, Sueda T, Noguchi K, Matsui H, et al. Metabolic adaptation to nutritional stress in human colorectal cancer. Sci Rep. 2016;6:38415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alberghina L, Gaglio D. Redox control of glutamine utilization in cancer. Cell Death Dis. 2014;5:e1561–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu G, Zhu J, Yu M, Cai C, Zhou Y, Yu M, et al. Glutamate dehydrogenase is a novel prognostic marker and predicts metastases in colorectal cancer patients. J Transl Med. 2015;13:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang F, Zhang Q, Ma H, Lv Q, Zhang T. Expression of glutaminase is upregulated in colorectal cancer and of clinical significance. Int J Clin Exp Pathol. 2014;7:1093–100. [PMC free article] [PubMed] [Google Scholar]
- 40.Song Z, Wei B, Lu C, Li P, Chen L. Glutaminase sustains cell survival via the regulation of glycolysis and glutaminolysis in colorectal cancer. Oncol Lett. 2017;14:3117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baenke F, Peck B, Miess H, Schulze A. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis Model Mech. 2013;6:1353–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.•.Beloribi-Djefaflia S, Vasseur S, Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5:e189. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review discusses the many roles of lipid metabolism in cancer.
- 43.Cohen S Lipid droplets as organelles. Int Rev Cell Mol Biol. 2018;337:83–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Notarnicola M, Messa C, Caruso MG. A significant role of lipogenic enzymes in colorectal cancer. Anticancer Res. 2012;32:2585–90. [PubMed] [Google Scholar]
- 45.••.Notarnicola M, Tutino V, Calvani M, Lorusso D, Guerra V, Caruso MG. Serum levels of fatty acid synthase in colorectal cancer patients are associated with tumor stage. J Gastrointest Cancer. 2012;43:508–11. [DOI] [PubMed] [Google Scholar]; This paper describes the use of FASN inhibitors in patient-derived CRC xenograft tumor models.
- 46.Zaytseva YY, Harris JW, Mitov MI, Kim JT, Butterfield DA, Lee EY, et al. Increased expression of fatty acid synthase provides a survival advantage to colorectal cancer cells via upregulation of cellular respiration. Oncotarget. 2015;6:18891–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zaytseva YY, Rychahou PG, Le A-T, Scott TL, Flight RM, Kim JT, et al. Preclinical evaluation of novel fatty acid synthase inhibitors in primary colorectal cancer cells and a patient-derived xenograft model of colorectal cancer. Oncotarget. 2018;9:24787–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karger S, Mika A, Kobiela J, Czumaj A, Chmielewski M, Stepnowski P, et al. Hyper-elongation in colorectal cancer tissue-cerotic acid is a potential novel serum metabolic marker of colorectal malignancies. Cell Physiol Biochem. 2017;41:722–30. [DOI] [PubMed] [Google Scholar]
- 49.da Costa AC, Filho PRS, Júnior SA, de Oliveira FF, Begnami MD, de Lima VCC, et al. Prognostic value of factors associated with hypoxia and lipid metabolism in patients with colorectal cancer. Appl Cancer Res. 2017;37:44. [Google Scholar]
- 50.Fernández LP, Ramos-Ruiz R, Herranz J, Martín-Hernández R, Vargas T, Mendiola M, et al. The transcriptional and mutational landscapes of lipid metabolism-related genes in colon cancer. Oncotarget. 2018;9:5919–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wen Y-A, Xiong X, Zaytseva YY, Napier DL, Vallee E, Li AT, et al. Downregulation of SREBP inhibits tumor growth and initiation by altering cellular metabolism in colon cancer. Cell Death Dis. 2018;9:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mutoh M, Komiya M, Teraoka N, Ueno T, Takahashi M, Kitahashi T, et al. Overexpression of low-density lipoprotein receptor and lipid accumulation in intestinal polyps in Min mice. Int J Cancer. 2009;125:2505–10. [DOI] [PubMed] [Google Scholar]
- 53.Accioly MT, Pacheco P, Maya-Monteiro CM, Carrossini N, Robbs BK, Oliveira SS, et al. Lipid bodies are reservoirs of cyclooxygenase-2 and sites of prostaglandin-E2 synthesis in colon cancer cells. Cancer Res Am Assoc Cancer Res. 2008;68:1732–40. [DOI] [PubMed] [Google Scholar]
- 54.Hagland HR, Berg M, Jolma IW, Carlsen A, Søreide K. Molecular pathways and cellular metabolism in colorectal cancer. Dig Surg. 2013;30:12–25. [DOI] [PubMed] [Google Scholar]
- 55.Wang D, Dubois RN. Prostaglandins and cancer. Gut. 2006;55:115–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sánchez-Martínez R, Cruz-Gil S, de Cedrón MG, Álvarez-Fernández M, Vargas T, Molina S, et al. A link between lipid metabolism and epithelial-mesenchymal transition provides a target for colon cancer therapy. Oncotarget. 2015;6:38719–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cruz-Gil S, Sanchez-Martinez R, Gomez de Cedron M, Martin-Hernandez R, Vargas T, Molina S, et al. Targeting the lipid metabolic axis ACSL/SCD in colorectal cancer progression by therapeutic miRNAs: miR-19b-1 role. J Lipid Res. 2018;59:14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zaytseva YY, Rychahou PG, Gulhati P, Elliott VA, Mustain WC, O’Connor K, et al. Inhibition of fatty acid synthase attenuates CD44-associated signaling and reduces metastasis in colorectal cancer. Cancer Res. 2012;72:1504–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang H, Xi Q, Wu G. Fatty acid synthase regulates invasion and metastasis of colorectal cancer via Wnt signaling pathway. Cancer Med. 2016;5:1599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.•.Newman AC, Maddocks ODK. One-carbon metabolism in cancer. Br J Cancer. 2017;116:1499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review provides a broad overview of 1CM in cancer.
- 61.•.Ryall JG, Cliff T, Dalton S, Sartorelli V. Metabolic reprogramming of stem cell epigenetics. Cell Stem Cell. 2015;17:651–62. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review discusses the mechanisms by which metabolic intermediates contribute to epigenetic reprogramming.
- 62.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martinelli M, Scapoli L, Mattei G, Ugolini G, Montroni I, Zattoni D, et al. A candidate gene study of one-carbon metabolism pathway genes and colorectal cancer risk. Br J Nutr. 2013;109:984–9. [DOI] [PubMed] [Google Scholar]
- 64.Liu AY, Scherer D, Poole E, Potter JD, Curtin K, Makar K, et al. Gene-diet-interactions in folate-mediated one-carbon metabolism modify colon cancer risk. Mol Nutr Food Res. 2013;57:721–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jia X-Q, Zhang S, Zhu H-J, Wang W, Zhu J-H, Wang X-D, et al. Increased expression of PHGDH and prognostic significance in colorectal cancer. Transl Oncol. 2016;9:191–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma L, Tao Y, Duran A, Llado V, Galvez A, Barger JF, et al. Control of nutrient stress-induced metabolic reprogramming by PKCζin tumorigenesis. Cell. 2013;152:599–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng J Energy metabolism of cancer: glycolysis versus oxidative phosphorylation (review). Oncol Lett. 2012;4:1151–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schell JC, Olson KA, Jiang L, Hawkins AJ, Van Vranken JG, Xie J, et al. A role for the mitochondrial pyruvate carrier as a repressor of the Warburg effect and colon cancer cell growth. Mol Cell. 2014;56:400–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lehuede C, Dupuy F, Rabinovitch R, Jones RG, Siegel PM. Metabolic plasticity as a determinant of tumor growth and metastasis. Cancer Res. 2016;76:5201–8. [DOI] [PubMed] [Google Scholar]
- 70.Puccini A, Berger MD, Naseem M, Tokunaga R, Battaglin F, Cao S, et al. Colorectal cancer: epigenetic alterations and their clinical implications. Biochim Biophys Acta. 1868;2017:439–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim J-A, Yeom YI. Metabolic signaling to epigenetic alterations in cancer. Biomol Ther (Seoul). Korean Soc Appl Pharm. 2018;26:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zee BM, Levin RS, Xu B, LeRoy G, Wingreen NS, Garcia BA. In vivo residue-specific histone methylation dynamics. J Biol Chem. 2010;285:3341–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. [DOI] [PubMed] [Google Scholar]
- 75.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–7. [DOI] [PubMed] [Google Scholar]
- 76.Hisamuddin IM, Yang VW. Molecular genetics of colorectal cancer: an overview. Curr Colorectal Cancer Rep. 2006;2:53–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luo Y, Yu M, Grady WM. Field cancerization in the colon: a role for aberrant DNA methylation? Gastroenterol Rep. 2014;2:16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Patel A, Tripathi G, Gopalakrishnan K, Williams N, Arasaradnam RP. Field cancerisation in colorectal cancer: a new frontier or pastures past? World J Gastroenterol. 2015;21:3763–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Backshall A, Alferez D, Teichert F, Wilson ID, Wilkinson RW, Goodlad RA, et al. Detection of metabolic alterations in non-tumor gastrointestinal tissue of the Apc Min/+ mouse by 1 H MAS NMR spectroscopy. J Proteome Res. 2009;8:1423–30. [DOI] [PubMed] [Google Scholar]
- 80.Leclerc D, Lévesque N, Cao Y, Deng L, Wu Q, Powell J, et al. Genes with aberrant expression in murine preneoplastic intestine show epigenetic and expression changes in normal mucosa of colon cancer patients. Cancer Prev Res (Phila). 2013;6:1171–81. [DOI] [PubMed] [Google Scholar]
- 81.Rao CV, Sanghera S, Zhang Y, Biddick L, Reddy A, Lightfoot S, et al. Systemic chromosome instability resulted in colonic transcriptomic changes in metabolic, proliferation, and stem cell regulators in Sgo1−/+ mice. Cancer Res NIH. 2016;76:630–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dela Cruz M, Ledbetter S, Chowdhury S, Tiwari AK, Momi N, Wali RK, et al. Metabolic reprogramming of the premalignant colonic mucosa is an early event in carcinogenesis. Oncotarget. 2017;8:20543–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rao CV, Yamada HY. Genomic instability and colon carcinogenesis: from the perspective of genes. Front Oncol. 2013;3:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Network TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–205. [DOI] [PubMed] [Google Scholar]
- 86.••.Pate KT, Stringari C, Sprowl-Tanio S, Wang K, TeSlaa T, Hoverter NP, et al. Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer. Embo J. 2014;33:1454–73. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report was the first to establish a direct metabolic transcriptional target of WNT signaling.
- 87.Roche TE, Baker JC, Yan X, Hiromasa Y, Gong X, Peng T, et al. Distinct regulatory properties of pyruvate dehydrogenase kinase and phosphatase isoforms. Prog Nucleic Acid Res Mol Biol. 2001;70:33–75. [DOI] [PubMed] [Google Scholar]
- 88.Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med. 2013;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV MYC, metabolism, and cancer. Cancer Discov. 2015;5:1024–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K pathway in human disease. Cell. 2017;170:605–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu M, Grady WM. Therapeutic targeting of the phosphatidylinositol 3-kinase signaling pathway: novel targeted therapies and advances in the treatment of colorectal cancer. Therap Adv Gastroenterol. 2012;5:319–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell.. 2017;168:960–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. [DOI] [PubMed] [Google Scholar]
- 96.Bentley J, Itchayanan D, Barnes K, McIntosh E, Tang X, Downes CP, et al. Interleukin-3-mediated cell survival signals include phosphatidylinositol 3-kinase-dependent translocation of the glucose transporter GLUT1 to the cell surface. J Biol Chem. 2003;278:39337–48. [DOI] [PubMed] [Google Scholar]
- 97.Rathmell JC, Fox CJ, Plas DR, Hammerman PS, Cinalli RM, Thompson CB. Akt-directed glucose metabolism can prevent Bax conformation change and promote growth factor-independent survival. Mol Cell Biol. 2003;23:7315–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Waldhart AN, Dykstra H, Peck AS, Boguslawski EA, Madaj ZB, Wen J, et al. Phosphorylation of TXNIP by AKT mediates acute influx of glucose in response to insulin. Cell Rep. 2017;19:2005–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Roberts DJ, Tan-Sah VP, Smith JM, Miyamoto S. Akt phosphorylates HK-II at Thr-473 and increases mitochondrial HK-II association to protect cardiomyocytes. J Biol Chem. 2013;288:23798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Deprez J, Vertommen D, Alessi DR, Hue L, Rider MH. Phosphorylation and activation of heart 6-phosphofructo-2-kinase by protein kinase B and other protein kinases of the insulin signaling cascades. J Biol Chem. 1997;272:17269–75. [DOI] [PubMed] [Google Scholar]
- 101.Dibble CC, Cantley LC. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 2015;25:545–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.••.Hao Y, Samuels Y, Li Q, Krokowski D, Guan B-J, Wang C, et al. Oncogenic PIK3CA mutations reprogram glutamine metabolism in colorectal cancer. Nat Commun. 2016;7:11971. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report showed that PIK3CA mutations can reprogram CRC metabolism, increasing glutamine dependency.
- 103.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 2011;36:320–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yun J, Mullarky E, Lu C, Bosch KN, Kavalier A, Rivera K, et al. Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science. 2015;350:1391–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.••.Hutton JE, Wang X, Zimmerman LJ, Slebos RJC, Trenary IA, Young JD, et al. Oncogenic KRAS and BRAF drive metabolic reprogramming in colorectal cancer. Mol Cell Proteomics. 2016;15:2924–38. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper used a unique strategy to identify changes in levels of metabolic enzymes driven by oncogenic mutations in KRAS and BRAF.
- 108.Chun SY, Johnson C, Washburn JG, Cruz-Correa MR, Dang DT, Dang LH. Oncogenic KRAS modulates mitochondrial metabolism in human colon cancer cells by inducing HIF-1α and HIF-2α target genes. Mol Cancer. 2010;9:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sun Y, Liu Z, Zou X, Lan Y, Sun X, Wang X, et al. Mechanisms underlying 3-bromopyruvate-induced cell death in colon cancer. J Bioenerg Biomembr. 2015;47:319–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fritsche-Guenther R, Zasada C, Mastrobuoni G, Royla N, Rainer R, Roßner F, et al. Alterations of mTOR signaling impact metabolic stress resistance in colorectal carcinomas with BRAF and KRAS mutations. Sci Rep. 2018;8:9204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lin S-C, Hardie DG. AMPK: sensing glucose as well as cellular energy status. Cell Metab. 2018;27:299–313. [DOI] [PubMed] [Google Scholar]
- 112.Bhagwat SV, Gokhale PC, Crew AP, Cooke A, Yao Y, Mantis C, et al. Preclinical characterization of OSI-027, a potent and selective inhibitor of mTORC1 and mTORC2: distinct from rapamycin. Mol Cancer Ther. 2011;10:1394–406. [DOI] [PubMed] [Google Scholar]
- 113.Blaser B, Waselle L, Dormond-Meuwly A, Dufour M, Roulin D, Demartines N, et al. Antitumor activities of ATP-competitive inhibitors of mTOR in colon cancer cells. BMC Cancer. 2012;12:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Faes S, Duval AP, Planche A, Uldry E, Santoro T, Pythoud C, et al. Acidic tumor microenvironment abrogates the efficacy of mTORC1 inhibitors. Mol Cancer. 2016;15:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349:1483–9. [DOI] [PubMed] [Google Scholar]
- 116.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. [DOI] [PubMed] [Google Scholar]
- 117.Napoli M, Flores ER. The p53 family orchestrates the regulation of metabolism: physiological regulation and implications for cancer therapy. Br J Cancer. 2017;116:149–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Itahana Y, Itahana K. Emerging roles of p53 family members in glucose metabolism. Int J Mol Sci. 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–33. [DOI] [PubMed] [Google Scholar]
- 120.Zawacka-Pankau J, Grinkevich VV, Hünten S, Nikulenkov F, Gluch A, Li H, et al. Inhibition of glycolytic enzymes mediated by pharmacologically activated p53: targeting Warburg effect to fight cancer. J Biol Chem. 2011;286:41600–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kawauchi K, Araki K, Tobiume K, Tanaka N. Loss of p53 enhances catalytic activity of IKKbeta through O-linked beta-N-acetyl glucosamine modification. Proc Natl Acad Sci U S A. 2009;106:3431–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Contractor T, Harris CR. p53 negatively regulates transcription of the pyruvate dehydrogenase kinase Pdk2. Cancer Res. 2012;72:560–7. [DOI] [PubMed] [Google Scholar]
- 123.Zhang C, Lin M, Wu R, Wang X, Yang B, Levine AJ, et al. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci U S A. 2011;108:16259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kamp WM, Wang P-Y, Hwang PM. TP53 mutation, mitochondria and cancer. Curr Opin Genet Dev. 2016;38:16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kulawiec M, Ayyasamy V, Singh KK. p53 regulates mtDNA copy number and mitocheckpoint pathway J Carcinog. 2009;8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lebedeva MA, Eaton JS, Shadel GS. Loss of p53 causes mitochondrial DNA depletion and altered mitochondrial reactive oxygen species homeostasis. Biochim Biophys Acta. 2009;1787:328–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bourdon A, Minai L, Serre V, Jais J-P, Sarzi E, Aubert S, et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet. 2007;39:776–80. [DOI] [PubMed] [Google Scholar]
- 128.Okamura S, Ng CC, Koyama K, Takei Y, Arakawa H, Monden M, et al. Identification of seven genes regulated by wild-type p53 in a colon cancer cell line carrying a well-controlled wild-type p53 expression system. Oncol Res. 1999;11:281–5. [PubMed] [Google Scholar]
- 129.Matoba S, Kang J-G, Patino WD, Wragg A, Boehm M, Gavrilova O, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–3. [DOI] [PubMed] [Google Scholar]
- 130.Li X-L, Zhou J, Chen Z-R, Chng W-J. P53 mutations in colorectal cancer—molecular pathogenesis and pharmacological reactivation. World J Gastroenterol. 2015;21:84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bensaad K, Tsuruta A, Selak MA, Vidal MNC, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–20. [DOI] [PubMed] [Google Scholar]
- 132.Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci U S A. 2010;107:7455–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A. 2010;107:7461–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Colangelo T, Polcaro G, Muccillo L, D’Agostino G, Rosato V, Ziccardi P, et al. Friend or foe?: the tumour microenvironment dilemma in colorectal cancer. Biochim Biophys Acta Rev Cancer. 2017;1867:1–18. [DOI] [PubMed] [Google Scholar]
- 135.•.De Francesco EM, Sotgia F, Lisanti MP. Cancer stem cells (CSCs): metabolic strategies for their identification and eradication. Biochem J. 2018;475:1611–34. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review provides a summary of CSC metabolism in many different cancer types.
- 136.Hatano Y, Fukuda S, Hisamatsu K, Hirata A, Hara A, Tomita H. Multifaceted interpretation of colon cancer stem cells. Int J Mol Sci. 2017;18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Menendez JA, Alarcón T. Metabostemness: a new cancer hall-mark. Front Oncol. 2014;4:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Barker N, van de Wetering M, Clevers H. The intestinal stem cell Genes Dev. Cold Spring Harbor Laboratory Press; 2008;22:1856–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Umar S Intestinal stem cells. Curr Gastroenterol Rep. 2010;12:340–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.De Lau W, Barker N, Low TY, Koo B-K, Li VSW, Teunissen H, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476. [DOI] [PubMed] [Google Scholar]
- 141.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. [DOI] [PubMed] [Google Scholar]
- 142.Zhu L, Gibson P, Currle DS, Tong Y, Richardson RJ, Bayazitov IT, et al. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature. 2009;457:603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5. [DOI] [PubMed] [Google Scholar]
- 144.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–11. [DOI] [PubMed] [Google Scholar]
- 145.•.Rodríguez-Colman MJ, Schewe M, Meerlo M, Stigter E, Gerrits J, Pras-Raves M, et al. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature. 2017;543:424–7. [DOI] [PubMed] [Google Scholar]; This report identified metabolic synergy between CBCs and Paneth cells in the intestinal stem cell niche.
- 146.Rera M, Bahadorani S, Cho J, Koehler CL, Ulgherait M, Hur JH, et al. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab. 2011;14:623–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Berger E, Rath E, Yuan D, Waldschmitt N, Khaloian S, Allgäuer M, et al. Mitochondrial function controls intestinal epithelial stemness and proliferation. Nat Commun. 2016;7:13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Roper J, Yilmaz ÖH. Metabolic teamwork in the stem cell niche. Cell Metab. 2017;25:993–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fan Y-Y, Davidson LA, Callaway ES, Wright GA, Safe S, Chapkin RS. A bioassay to measure energy metabolism in mouse colonic crypts, organoids, and sorted stem cells. Am J Physiol Liver. 2015;309:G1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Schell JC, Wisidagama DR, Bensard C, Zhao H, Wei P, Tanner J, et al. Control of intestinal stem cell function and proliferation by mitochondrial pyruvate metabolism. Nat Cell Biol. 2017;19:1027–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wei P, Dove KK, Bensard C, Schell JC, Rutter J. The force is strong with this one: metabolism (over)powers stem cell fate. Trends Cell Biol. 2018;28:551–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen K-Y, Liu X, Bu P, Lin C-S, Rakhilin N, Locasale JW, et al. A metabolic signature of colon cancer initiating cells. Conf Proc Annu Int Conf IEEE Eng Med Biol Soc 2014;2014:4759–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tirinato L, Liberale C, Di Franco S, Candeloro P, Benfante A, La Rocca R, et al. Lipid droplets: a new player in colorectal cancer stem cells unveiled by spectroscopic imaging. Stem Cells. 2015;33:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Llufrio EM, Wang L, Naser FJ, Patti GJ. Sorting cells alters their redox state and cellular metabolome. Redox Biol. 2018;16:381–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.•.Reina-Campos M, Moscat J, Diaz-Meco M. Metabolism shapes the tumor microenvironment. Curr Opin Cell Biol. 2017;48:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review describes the metabolic interactions between cells in the TME.
- 156.Marsh T, Pietras K, McAllister SS. Fibroblasts as architects of cancer pathogenesis. Biochim Biophys Acta - Mol Basis Dis. 2013;1832:1070–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Martinez-Outschoorn UE, Lisanti MP, Sotgia F. Catabolic cancer-associated fibroblasts transfer energy and biomass to anabolic cancer cells, fueling tumor growth. Semin Cancer Biol. 2014;25:47–60. [DOI] [PubMed] [Google Scholar]
- 158.Avagliano A, Granato G, Ruocco MR, Romano V, Belviso I, Carfora A, et al. Metabolic reprogramming of cancer associated fibroblasts: the slavery of stromal fibroblasts. Biomed Res Int. 2018;2018:6075403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Fu Y, Liu S, Yin S, Niu W, Xiong W, Tan M, et al. The reverse Warburg effect is likely to be an Achilles’ heel of cancer that can be exploited for cancer therapy. Oncotarget. 2017;8:57813–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sonveaux P, Végran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest. 2008;118:3930–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.•.Wen Y-A, Xing X, Harris JW, Zaytseva YY, Mitov MI, Napier DL, et al. Adipocytes activate mitochondrial fatty acid oxidation and autophagy to promote tumor growth in colon cancer. Cell Death Dis. 2017;8:e2593. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrated a direct role of tumor-associated adipocytes in CRC metabolism.
- 162.Hong S, Cai Q, Chen D, Zhu W, Huang W, Li Z. Abdominal obesity and the risk of colorectal adenoma. Eur J Cancer Prev. 2012;21:523–31. [DOI] [PubMed] [Google Scholar]
- 163.Dai Z, Xu Y-C, Niu L. Obesity and colorectal cancer risk: a meta-analysis of cohort studies. World J Gastroenterol. 2007;13:4199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ning Y, Wang L, Giovannucci EL. A quantitative analysis of body mass index and colorectal cancer: findings from 56 observational studies. Obes Rev. 2010;11:19–30. [DOI] [PubMed] [Google Scholar]
- 165.Sinicrope FA, Foster NR, Yothers G, Benson A, Seitz JF, Labianca R, et al. Body mass index at diagnosis and survival among colon cancer patients enrolled in clinical trials of adjuvant chemotherapy. Cancer. 2013;119:1528–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.•.Hobson-Gutierrez SA, Carmona-Fontaine C. The metabolic axis of macrophage and immune cell polarization. Dis Model Mech. 2018;11. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review describes how immune cell polarization is triggered by intracellular metabolic reprogramming.
- 167.Bunse L, Pusch S, Bunse T, Sahm F, Sanghvi K, Friedrich M, et al. Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nat Med. 2018;24:1192–203. [DOI] [PubMed] [Google Scholar]
- 168.Parekh PJ, Balart LA, Johnson DA. The influence of the gut microbiome on obesity, metabolic syndrome and gastrointestinal disease. Clin Transl Gastroenterol. 2015;6:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.•.Loo JM, Scherl A, Nguyen A, Man FY, Weinberg E, Zeng Z, et al. Extracellular metabolic energetics can promote cancer progression. Cell. 2015;160:393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrated a novel mechanism by which CRC cells adapt to nutrient availability in the metastatic niche.
- 170.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Riaz Rajoka MS, Shi J, Mehwish HM, Zhu J, Li Q, Shao D, et al. Interaction between diet composition and gut microbiota and its impact on gastrointestinal tract health. Food Sci Hum Wellness. 2017;6:121–30. [Google Scholar]
- 172.Dusko ES. Metagenomics of the intestinal microbiota: potential applications. Gastroentérologie Clin Biol. 2010;34:S23–8. [DOI] [PubMed] [Google Scholar]
- 173.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dulal S, Keku TO. Gut microbiome and colorectal adenomas. Cancer J. 2014;20:225–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Keku TO, Dulal S, Deveaux A, Jovov B, Han X. The gastrointestinal microbiota and colorectal cancer. Am J Physiol Gastrointest Liver. 2015;308:G351–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Purcell RV, Visnovska M, Biggs PJ, Schmeier S, Frizelle FA. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci Rep. 2017;7:11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hullar MAJ, Burnett-Hartman AN, Lampe JW. Gut microbes, diet, and cancer. Cancer Treat Res. 2014:377–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sheflin AM, Whitney AK, Weir TL. Cancer-promoting effects of microbial dysbiosis. Curr Oncol Rep. 2014;16:406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bhutia YD, Ogura J, Sivaprakasam S, Ganapathy V. Gut microbiome and colon cancer: role of bacterial metabolites and their molecular targets in the host. Curr Colorectal Cancer Rep. 2017;13:111–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Parodi PW. Fatty acid composition of Australian butter and milk fats. Aust J Dairy Technol. 1970;25:200–205. [Google Scholar]
- 181.McNabney S, Henagan T. Short chain fatty acids in the colon and peripheral tissues: a focus on butyrate, colon cancer, obesity and insulin resistance. Nutrients. 2017;9:1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wong JMW, de Souza R, Kendall CWC, Emam A, Jenkins DJA. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40:235–43. [DOI] [PubMed] [Google Scholar]
- 183.Vital M, Gao J, Rizzo M, Harrison T, Tiedje JM. Diet is a major factor governing the fecal butyrate-producing community structure across Mammalia, Aves and Reptilia. ISME J. 2015;9:832–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio. 2014;5:e00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. [DOI] [PubMed] [Google Scholar]
- 186.Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol. 2008;23:1298–303. [DOI] [PubMed] [Google Scholar]
- 188.Boutron-Ruault M-C, Marteau P, Lavergne-Slove A, Myara A, Gerhardt M-F, Franchisseur C, et al. Effects of a 3-mo consumption of short-chain fructo-oligosaccharides on parameters of colorectal carcinogenesis in patients with or without small or large colorectal adenomas. Nutr Cancer. 2005;53:160–8. [DOI] [PubMed] [Google Scholar]
- 189.Monleón D, Morales JM, Barrasa A, López JA, Vázquez C, Celda B. Metabolite profiling of fecal water extracts from human colorectal cancer. NMR Biomed 2009;22:342–8. [DOI] [PubMed] [Google Scholar]
- 190.Lupton JR. Microbial degradation products influence colon cancer risk: the butyrate controversy. J Nutr. 2004;134:479–82. [DOI] [PubMed] [Google Scholar]
- 191.Roediger WE. Utilization of nutrients by isolated epithelial cells of the rat colon. Gastroenterology. 1982;83:424–9. [PubMed] [Google Scholar]
- 192.••.Donohoe DR, Collins LB, Wali A, Bigler R, Sun W, Bultman SJ. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol Cell. 2012;48:612–26. [DOI] [PMC free article] [PubMed] [Google Scholar]; This seminal paper showed that the “butyrate paradox” is due to the differences in metabolic programming between normal enterocytes and CRC cells.
- 193.Velcich A, Palumbo L, Jarry A, Laboisse C, Racevskis J, Augenlicht L. Patterns of expression of lineage-specific markers during the in vitro-induced differentiation of HT29 colon carcinoma cells. Cell Growth Differ. 1995;6:749–57. [PubMed] [Google Scholar]
- 194.Archer SY, Meng S, Shei A, Hodin RA. p21(WAF1) is required for butyrate-mediated growth inhibition of human colon cancer cells. Proc Natl Acad Sci U S A. 1998;95:6791–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.•.Li Q, Cao L, Tian Y, Zhang P, Ding C, Lu W, et al. Butyrate suppresses the proliferation of colorectal cancer cells via targeting pyruvate kinase M2 and metabolic reprogramming. Mol Cell Proteomics. 2018;17:1531–45. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report identified an HDAC-independent mechanism by which butyrate inhibits the growth of CRC cells.
- 196.Dong G, Mao Q, Xia W, Xu Y, Wang J, Xu L, et al. PKM2 and cancer: the function of PKM2 beyond glycolysis. Oncol Lett. 2016;11:1980–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kaiko GE, Ryu SH, Koues OI, Collins PL, Solnica-Krezel L, Pearce EJ, et al. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell. 2016;165:1708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]