Abstract
Radioresistance poses a major challenge in the treatment of advanced rectal cancer. Therefore, understanding the detailed mechanisms of radioresistance may improve patient response to irradiation and the survival rate. To identify the novel targets that modulate the radiosensitivity of rectal cancer, we performed small RNA sequencing with human rectal cancer cell lines. Through bioinformatics analysis, we selected microRNA-310a (miR-130a) as a promising candidate to elucidate radioresistance. miR-130a was dramatically upregulated in radiosensitive rectal cancer cells and overexpression of miR-130a promotes rectal cancer cell radiosensitivity. Mechanically, miR-130a reversed the epithelial-mesenchymal transition phenotype of rectal cancer cells following inhibition of cell invasion upon irradiation. Moreover, miR-130a also inhibited the repair of irradiation-induced DNA damage followed by cell death. We identified that SOX4 was a direct target of miR-130a. Overexpression of SOX4 reversed the promotion activity of miR-130a on radiosensitivity. Together, our findings suggest that miR-130a functions as a radiosensitizer in rectal cancer and reveals a potential therapeutic target and preoperative prognostic marker for radiotherapy.
Introduction
Preoperative radiotherapy has been extensively used as a powerful standard treatment for selected rectal cancer patients to reduce the relative risk of a local failure and promote the patient survival [1], [2], [3]. However, tumor responses to radiotherapy are different and many display an intrinsic and therapy-induced resistance. Radioresistance leading to tumor recurrence and metastatic lesions with enhanced aggressiveness, and a consequent poor prognosis, is a major reason for treatment failure and shortening of patient survival [4]. Therefore, overcoming radioresistance and enhancing radiosensitivity to improve the radiation therapeutic response is urgently required. To solve this problem, it is necessary to understand the molecular mechanisms for radioresistance and discover the novel effectors to improve the efficacy of radiotherapy.
MicroRNAs (miRNAs) repress their target mRNAs by binding to the 3′-untranslated region (UTR) in a sequence-specific manner, causing cleavage of the target or inhibition of protein expression [5]. Accumulating evidence has suggested the involvement of miRNAs in the cellular response to ionizing radiation. For example, upregulation of miR-100 enhanced the radiosensitivity of colorectal cells to X-ray irradiation, which probably induced apoptosis and DNA double-strand breaks [6]. miR-205 was also suggested as a tumor radiosensitizer through inhibition of DNA damage repair by targeting ZEB1and Ubc13 [7]. However, expression of miR-95 promoted radiation resistance in a variety of cancer cells and recapitulated an aggressive phenotype following ionizing radiation through targeting sphingolipid phosphatase SGPP1 [8]. miR-622 was also highly detected in tumors of rectal cancer patients with nonregression after standard radiotherapy [9]. Although multiple miRNAs have been reported for regulation of radiation resistance through various mechanisms, other miRNAs still need to be identified for more efficient management of rectal cancer.
SOX4 is a member of the SOX (SRY-related HMG-box) transcription factor family, which is characterized as a highly conserved HMG-box, DNA-binding domain [10]. SOX4 activity has been reported to contribute to various cellular processes. For example, overexpression of SOX4 was correlated with increased expression of epithelial-mesenchymal transition (EMT) markers [11], [12] and enhancing β-catenin/TCF activity [13]. Increased SOX4 activity in many cancer types also contributes to cell survival and metastasis [14], [15]. Therefore, SOX4 is considered a potential oncogene based on its positive effect on tumor growth [16]. However, increasing evidence has also shown that SOX4 has a potential tumor suppressive activity. A previous study reported that forced expression of SOX4 strongly impaired cell viability and promoted apoptosis in bladder cancer cells [17]. SOX4 also inhibits the growth of glioblastoma cells by inducing cell cycle arrest and inhibiting cell growth [18]. Recent studies have demonstrated that SOX4 expression is regulated by a variety of miRNAs and most miRNAs are found to be down-regulated in many cancers that cause an increase of SOX4 [19], [20].
In this study, we aimed to identify and characterize the radiotherapy response-related miRNAs from rectal cancer cells. Based on sequencing and biochemical analysis, miR-130a was selected as a potential radiosensitizer for treatment of rectal cancer. Forced expression of miR-130a suppresses cell growth and colony formation of rectal cancer cells upon irradiation treatment. Mechanistically, miR-130a inhibited radiation-induced EMT phenotype, invasion and DNA damage repair by directly targeting SOX4.
Material and Methods
Cell Culture and Reagents
SNU70 and SNU1411 are human rectal cancer cell lines established from the malignant tumors of colorectal cancer patients and were purchased from the Korean Cell Line Bank, Korea. Two other rectal cancer cell lines (SW837 and SW1463) were obtained from the American Type Culture Collection. All these cell lines were authenticated with DNA fingerprinting and mycoplasma detection by suppliers. These rectal cell lines were maintained in RPMI 1640 (Welgene, Daegu, Korea) supplemented with 10% heat-inactivated FBS, 1% streptomycin/penicillin (Invitrogen, Carlsbad, CA) and 25 mM HEPES in a 5% CO2 atmosphere at 37 °C. Antibodies against p-ATM (5883) and γH2AX (9718S) were acquired from Cell Signaling Technology (Beverly, MA). Total ATM (sc-377,293), H2AX (sc-517,336), Mre11 (sc-135,992), Rad50 (sc-74,460) and SOX4 (sc-130,633) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The antibodies against NBS1 (ab23996) was obtained from Abcam (Eugene, CA) and β-actin (A5441) from Sigma-Aldrich (St. Louis, MO).
Generation of Overexpression Cell Lines
To generate the overexpression system, pCAG lentiviral vector (Myc-tagged SOX4) or pCDH-CMV-MCS-EF1-copGFP vector (System Biosciences, Palo Alto, CA) including pre-miR-130a DNA were transfected into Lenti293T packaging cells with packaging plasmids using a Lipofectamine 2000 reagent (Invitrogen). After 24 h of transfection, media was changed to complete media containing 10% FBS. Secreted lentiviruses were harvested after 48 h using 0.45 μm pore-size filters and stored in aliquots at −80 °C. Rectal cancer cells were infected with lentivirus three times with 8 μg/mL polybrene (Sigma-Aldrich). The infection efficiency was checked with western blotting and real time PCR.
Clonogenic Survival Assay
Cells were plated in 60 mm culture dishes and then exposed to various ranges of irradiation doses (3-9 Gy) using a 6-MV therapeutic linear accelerator (CLINAC EX, Varian). After irradiation, equal number of cells was plated in six well plate at clonogenic density (104 cells per well). Cells were incubated for 10–14 days until the colonies were large enough. Colonies were fixed in cold methanol then stained with crystal violet and positive colonies (>50 cells) were counted. Plating efficiency was calculated as the percentage of cells seeded that grew into colonies under a specific culture condition. Surviving fraction was calculated as the numbers of colonies counted divided by the number of cells plated with a correction for the plating efficiency.
Small RNA Sequencing and Bioinformatics Analysis
A small RNA library was generated using a commercial preparation kit according to the manufacturer's instructions (Illumina, San Diego, CA). Total RNA was isolated on a 15% TBE-urea PAGE gel (Invitrogen) and 18 to 30 bp band was eluted for small RNA. An SRA 5′ and SRA 3′ adaptors were ligated to the small RNA with T4 RNA ligase. The small RNA was processed to single-stranded cDNA using SuperScript II kit (Invitrogen) and SRA RT primers. Then, cDNA was amplified using Illumina's PCR primer set. The amplified cDNA was precipitated, resuspended, and sequenced. A small RNA library was sequenced using an Illumina 1G Genome Analyzer according to the manufacturer's instructions. Analysis of the sequencing data was performed using mirTools with default parameters.
Luciferase Reporter Assay
The 3′UTR of SOX4 gene which contains putative miR-130a binding site was amplified and cloned into pMIR-REPORT™ Luciferase vector. SOX4 3′UTR mutant construct was generated by which miR-130a putative binding site was mutated by replacing the UUGCACUU sequence with AACGUGAU sequence.
NBS1 promoter region located 1.1 kb upstream of the transcription start site which contains one putative SOX4 responsive element (SRE) at approximate positions −746 to −740 was cloned into the pGL3B reporter vector. A mutated version of NBS1 promoter was also generated by replacing wild-type SRE (AACAAAG) into mutant sequence (AATGGAG). All mutant constructs were generated using EZ change Site-Directed Mutagenesis Kit following the manufacturer's instructions (Enzynomics, Daejeon, Korea). β-galactosidase was used as a control to normalize the transfection efficiency. At 48 h after transfection, cells were washed twice in PBS and lysed for luciferase and β-galactosidase assay using Dual Luciferase Reporter Assay system (Promega, Madison, WI). All assays were performed in triplicate and data expressed as the mean of three independent experiments.
RNA Isolation and Real-Time PCR
Total RNA was extracted with TRIzol reagent (Invitrogen) and used for first-strand cDNA synthesis using PrimeScript II RT reagent kit (Takara, Madison, WI). SYBR-green Premix Ex-Tag II (Takara) was used for the quantification of SOX4 and EMT markers with a real time quantitative PCR with Prism 7900HT sequence detection system (Applied Biosystems, Foster City, CA). The primer sequences for this study were listed in Table S1. The data was normalized with cyclophilin as control. Quantification of the mature form of miRNAs were performed using the Taqman MicroRNA Assay kit (Applied Biosystems) according to the manufacturer's protocol and RNU6B was used as the internal control. TaqMan probe sequences for this study were listed in Table S2. Data were analyzed using the comparative Ct method. The experiments were performed in triplicates and expressed as the mean ± S.D.
Western Blotting Assay
Proteins were extracted with a lysis buffer (25 mM HEPES (pH 7.5), 150 mM NaCl, 1% Triton X-100, 10% glycerol, 5 mM EDTA and a protease inhibitor cocktail) for 30 min and clarified by centrifugation at 13,200×g for 15 min at 4 °C. The protein concentrations were measured by the BCA method (Pierce, Rockford, IL). For immunoblotting, equal amounts of protein lysates were separated by SDS–polyacrylamide gel electrophoresis, followed by blotting onto the polyvinylidene difluoride membrane. Membranes were blocked for non-specific antibody binding with a blocking solution for 1 h. Next, membranes were incubated with specific primary antibodies in 3% BSA in TBST overnight with gentle agitation, followed by peroxidase-conjugated secondary antibodies. Immunoreactive proteins were checked with the chemiluminescence method according to the manufacturer's protocol (Pierce).
Matrigel Invasion Assay
Invasive ability of radioresistant cells was determined using commercial matrigel transwell chambers (BD Bioscience, San Jose, CA). Briefly, cells were plated in 60 mm culture dishes and then exposed to 6 Gy dose of irradiation. After irradiation, 5×103 cells/500 μl in free serum media were transferred to the pre-coated Matrigel upper chamber (24-well insert, 8 μm pore size, BD Bioscience) and 750 μl complete media was added to the lower chamber for providing chemoattractant and preventing dehydration. After 48 h of incubation, non-invaded cells were removed from upper wells with cotton tips and invaded cells were fixed in cold methanol then stained with crystal violet. Excess dye was washed away with tap water and invaded cells were photographed (40×) in five independent fields for each well.
Immunofluorescence
Cells were seeded on chamber slides overnight and fixed with 4% (v/v) paraformaldehyde for 20 min at room temperature, followed by permeabilization with 0.5% TritonX-100 in phosphate-buffered saline for 30 min. Next, cells were blocked for non-specific binding with 5% goat serum in Tween-20 and PBS for 1 h. Then, cells were stained with specific primary antibodies overnight at 4 °C and subsequently with Alexa Fluor 488-conjugated goat anti-mouse secondary antibody or Alexa Fluor 594-conjugated goat anti-rabbit secondary antibody (Invitrogen). Cell nuclei were stained with 4`,6-diamidino-2-phenylindole (DAPI). After staining, fluorescence images were acquired using an LSM700 confocal microscope (Carl Zeiss, Thornwood, NY).
Tumor Xenograft Experiments
BALB/c nu/nu mice (6 weeks old, 16~18 g, Charles River Laboratories, Yokohama, Japan) were maintained according to Institutional Animal Care and Use Committee-approved protocols of the Lee Gil Ya Cancer and Diabetes Institute, Gachon University (Incheon, Korea). SNU70RR, miR-130a overexpressing SNU70RR, or miR-130a/SOX4 overexpressing SNU70RR cells (1 × 106) were mixed in a ratio (1:1) with matrigel (BD Bioscience) and the mixtures were subcutaneously injected into the left flanks of mice. When the xenograft tumor size grew approximately to 150 mm3, single dose (15 Gy) of irradiation was delivered to mice. During irradiation, unanesthetized mice were manually fixed inside the irradiation shield. Tumor volume was determined every 3 days by caliper measurements and calculated using the modified ellipse formula (volume = length × width2/2). After tumor size reached at 1500 mm3, the mice were scarified and tumors were excised for further analysis.
Statistical Analysis
Results from at least three separate experiments were expressed as mean ± SD. Two tails Student's t test was conducted for comparisons between control and treatment groups. Differences between or among samples were considered as significant at P < .05.
Results
miR-130a is Down-regulated in Radioresistant Rectal Cancer Cells
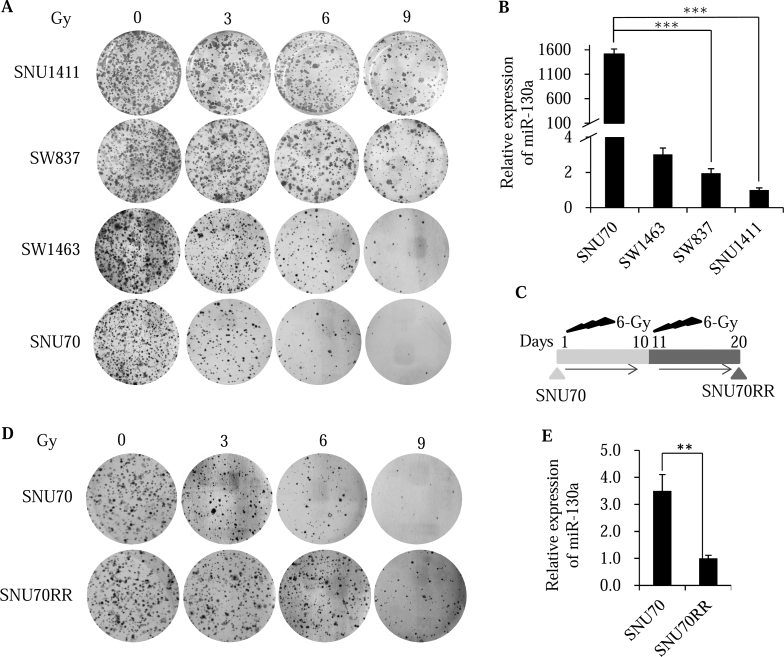
To determine the sensitivity of rectal cell lines to irradiation, colony formation assay was performed using four rectal cancer cells (SNU70, SNU1411, SW837 and SW1463) after treatment with various types of single dose radiation. The radiosensitivity of the four rectal cell lines was, in descending order, SNU70, SW1463, SW837 and SNU1411 (Figure 1A and S1A). Because the persistent presence of γH2AX foci revealed that delayed DNA damage repair and was associated with radiosensitivity [21], the expression of γH2AX foci was confirmed with immunofluorescence staining after irradiation. Consistently, γH2AX foci appeared prominently in SNU70 cells and was less expressed in SNU1411 and SW837 cells after irradiation (Figure S1B).
Figure 1.
miR-130a is down-regulated in radioresistant rectal cancer cells. (A) Rectal cancer cells were seeded into six well plates and irradiated with indicated doses. After 14 days, the cells were fixed and stained with crystal violet. The images were captured and showed colony growth in four rectal cancer cells. (B) Relative miR-130a expression in different rectal cancer cells was evaluated with real time PCR analysis. Small nuclear RNA (RNU6B) was used as the internal control. (C) Schematic diagram showing the generation of a radioresistant subline (SNU70RR) from its parental SNU70. SNU70 cells were treated one time with 6 Gy of irradiation followed by 10 days of incubation. Surviving cells were pooled and selected with 6 Gy of irradiation one more time. Ten days after the second irradiation, radioresistant SNU70RR cells were used for further experiments. (D) SNU70 and SNU70RR were seeded into six well plates and irradiated with the indicated doses. After 14 days, the cells were fixed and stained with crystal violet. The images were captured and showed colony growth. (E) Expression of miR-130a in SNU70 and SNU70RR cells was compared with real time PCR analysis. Small nuclear RNA (RNU6B) was used as the internal control. The data were presented with the mean ± S.D. of three independent experiments. (⁎⁎P < .01, ⁎⁎⁎P < .001).
To identify the candidate miRNAs involved in regulating radiosensitivity, we performed small RNA sequencing to profile the miRNA expression between four rectal cancer cell lines. Twenty-two miRNAs were differentially expressed, 14 upregulated and 8 down-regulated miRNAs, by at least 2 fold in two radiosensitive cell lines compared with radioresistant cells (Table S3). Among candidate miRNAs, we focused on miR-130a which is the most overexpressed miRNA in the radiosensitive SNU70 cell compared with resistant cells. Using miRNA specific real time PCR, we confirmed that miR-130a expression was markedly decreased in SNU1411 cells compared with SNU70 cells consistent with sequencing data (Figure 1B).
To examine the role of miR-130a on radioresistance in rectal cancer cells, we established a radioresistant rectal cancer cell line from the most radiosensitive SNU70 cell (SNU70RR) by repeating radiation and selection (Figure 1C). The colony formation (Figure 1D) and survival fractions (Figure S1C) in SNU70RR cells were significantly higher compared with parental SNU70 cells. Consistent with radioresistance, miR-130a expression was markedly down-regulated in SNU70RR cells (Figure 1E). Taken together, these data suggest that miR-130a may be involved in the radioresistance of rectal cancer cells.
miR-130a Enhances the Radiosensitivity of Rectal Cancer Cells
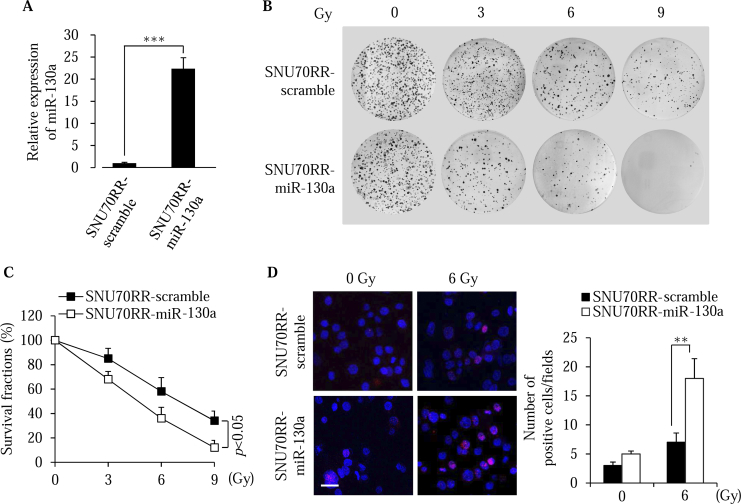
To clarify the role of miR-130a in radioresistance of rectal cancer cells, we overexpressed miR-130a in established radioresistant SNU70RR and the most resistant SNU1411 cells. Overexpression of miR-130a in SNU70RR and SNU1411 was confirmed by real time PCR analysis (Figure 2A and S2A). Expression of miR-130a in radioresistant cells (SNU70RR) suppressed colony formation followed by reducing survival fractions after irradiation (Figure 2, B and C). Colony formations of SNU1411-miR-130a cells were also significantly suppressed compared with SNU1411-scramble cells (Figure S2, B and C). Additionally, γH2AX foci were sustained for a longer time in SNU70RR-miR-130a, but almost disappeared in radioresistant parental cells (Figure 2D). These data suggest that miR-130a sensitizes the rectal cancer cells in response to radiation by inhibiting the repair of radiation-induced DNA damage followed by cell death.
Figure 2.
miR-130a enhances the sensitivity to irradiation of rectal cancer cells. (A) Relative expression of miR-130a was compared in SNU70RR cells with miRNA-specific real time PCR. The data were normalized with the RNU6B gene as the internal control. (B) Representative images of colony formation in SNU70RR-scramble and SNU70RR-miR-130a cells after exposure to different irradiation doses. Irradiated cells were maintained for 14 days to form the colonies and stained with crystal violet. (C) The graph shows survival fractions of SNU70RR-scramble and SNU70RR-miR-130a cells were calculated from colony formation in (B). (D) Expression of γH2AX was detected in SNU70RR-miR-130a cells after irradiation treatment for 24 h with immunofluorescence analysis. The bar graph shows the average number of γH2AX positive cells per field. Scale bar: 20 μm. The data were presented with the mean ± S.D. of three independent experiments. (⁎⁎P < .01, ⁎⁎⁎P < .001).
miR-130a Suppresses the EMT Process and Invasion of Rectal Cancer Cells
Recent studies have found that EMT phenotypic expression and cancer stem cell phenotypes promote the radioresistance of various human cancer cells [22], [23], [24]. Interestingly, SNU70RR-scramble cells looked elongated, spindle-like mesenchymal shape, whereas the morphology of miR-130a overexpressing cells became rounded and had a cuboidal-like epithelial shape (Figure S3A). Thus, we hypothesized that overexpression of miR-130a might induce a reversal of the EMT process in radioresistant cells. Consistent with morphological changes, differential expression of epithelial cell markers (E-cadherin and ZO-1) or the mesenchymal cell marker (N-cadherin) was detected with confocal microscopy in SNU70RR-miR-130a cells (Figure S3B). A change in EMT markers by miR-130a was also confirmed with real time PCR and western blotting (Figure S3, C and D). The same phenomenon that occurs with EMT gene expression was also detected with SNU1411-miR-130a cells (Figure S3, E and F).
Cell invasion is a characteristic feature of EMT and metastasis [25], [26]. In keeping with enhancing the EMT process, irradiation can also promote cancer invasion and metastasis in various types of cancer. Therefore, we determined the effect of miR-130a on the invasion ability of radioresistant rectal cancer cells by performing transwell assay. As expected, cell invasion of miR-130a overexpressing cells was significantly lower than with control cell (Figure S3G). These results suggest that miR-130a sensitizes rectal cancer cells to irradiation by suppressing the EMT process and invasion ability.
SOX4 is a Direct Target of miR-130a
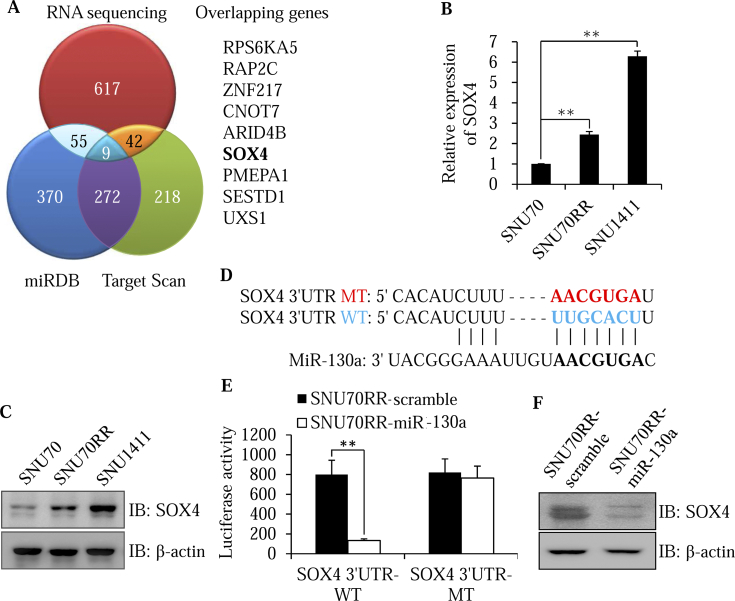
To identify the potential targets involved in radiosensitivity of miR-130a, we performed three-pronged approaches, including two bioinformatics algorithms (miRDB and Target Scan) in combination with RNA sequencing profiling generated from SNU70 and SNU1411 cells (Figure 3A). Among nine overlapping theoretical miR-130a target genes, SOX4 stood out as the best candidate owing to its ability as a master regulator of the EMT process [12] as well as being an inducer of tumor invasion and chemo-radioresistance in human cancer [27], [28]. To confirm the transcriptional regulation of miR-130a on expression of SOX4, we examined the expression of SOX4 in different rectal cancer cells. In contrast with level of miR-130a, SOX4 was down-regulated in radiosensitive SNU70 cells and upregulated in radioresistant SNU70RR and SNU1411 cells at both mRNA and protein levels (Figure 3, B and C). To determine whether miR-130a directly targets SOX4, wild-type or mutant 3′UTR fragments of SOX4 containing the miR-130a binding site were subcloned into luciferase reporter plasmid (Figure 3D). The luciferase activity of SOX4–3′UTR-WT was significantly decreased in miR-130a overexpressing SNU70RR cells compared with scramble cells (Figure 3E). However, inhibitory activity of miR-130a disappeared in the mutant reporter construct. Moreover, immunoblotting analysis indicated that overexpression of miR-130a significantly decreased the endogenous SOX4 protein (Figure 3F). Taken together, these results suggest that SOX4 is a direct target of miR-130a.
Figure 3.
SOX4 is a direct target of miR-130a. (A) Venn diagram showing the overlap between predicted target genes of miR-130a from miRDB, Target Scan and RNA sequencing profiling generated from SNU70 and SNU1411 cells. The nine overlapping genes were presented. (B and C) SOX4 expression levels in SNU70, SNU70RR and SW1411 cells were measured with real time PCR (B) and western blotting (C). The data were normalized with cyclophilin as the internal control for real time PCR. (D) Schematic illustration for a potential miR-130a binding site of the SOX4 3′UTR. The mutant sequence (MT) of the SOX4 3′UTR was presented by comparing it with the wild-type sequence (WT). (E) Luciferase assay was performed after transfection with the wild-type or mutant construct in SNU70RR cells. After incubation for 48 h, relative luciferase activity was determined with β-galactosidase as a normalization control. Data are presented as the mean ± S.D. of three independent experiments. (F) The level of SOX4 was measured in SNU70RR-miR-130a cells with western blotting. β-actin was used as the loading control. (⁎⁎P < .01).
Restoration of SOX4 Reverses the Suppressive Effects of miR-130a on the EMT Process and Invasion of Radioresistant Cells
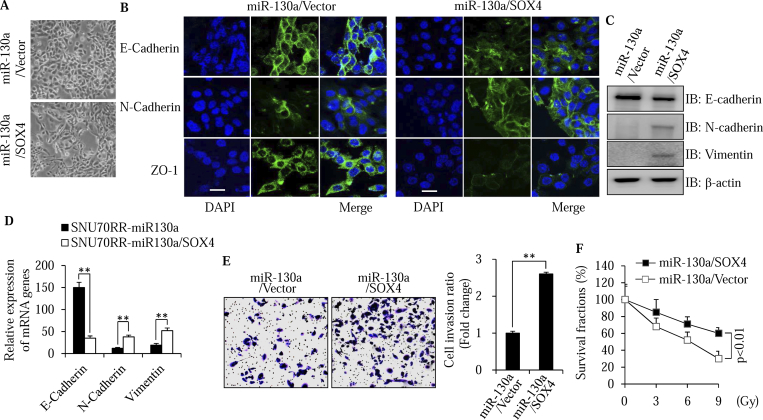
Based on the function of SOX4 as a master regulator of EMT, we considered whether the suppressive effects of miR-130a on the EMT process and invasion ability of radioresistant cells are dependent on its regulation of SOX4. A rescue experiment was performed with a SOX4 construct without a miR-130a binding 3′UTR sequence (Figure S4A). Indeed, rescue of SOX4 induced the morphologic change of mesenchymal cells to an epithelial phenotype in miR-130a overexpressing cells (Fig. 4A). Reversal of the EMT process was also confirmed with immunofluorescence staining (Figure 4B), western blotting (Figure 4C) and real time PCR analysis (Figure 4D) for EMT markers. In addition, ectopic expression of SOX4 reversed the invasion suppressive ability of miR-130a (Figure 4E). We next considered whether restoration of SOX4 could reverse the radiosensitivity induced by miR-130a. Clonogenic survival assay indicated that restoration of SOX4 increased the survival fractions of SNU70RR- miR-130a cells upon irradiation treatment (Figure 4F and S4B). Taken together, these data suggest that SOX4 can reverse the radiosensitivity induced by miR-130a through enhancement of the EMT process.
Figure 4.
SOX4 reverses the suppressive effects of miR-130a on the EMT process and invasion of radioresistant cells. (A) Morphological changes in SOX4-overexpressing cells were compared with control cells with bright-phase microscopy. The images were captured with 200× magnification. (B-D) Expression of EMT markers including E-cadherin, N-cadherin, vimentin and ZO-1, was detected with immunofluorescence confocal microscopy (B), western blotting (C) and real time PCR (D). The data were normalized with cyclophilin as the internal control for real time PCR. Scale bar: 20 μm. (E) Representative images showing the invasive activity of SNU70RR-miR-130a/SOX4 cells were captured after 48 h of irradiation treatment. The invaded cells were stained with crystal violet. The graph shows the fold difference of invasive cells compared with control cells from five different fields of three independent experiments. (F) Rectal cancer cells were seeded into six well plates and irradiated with indicated doses. After 14 days, the cells were fixed and stained with crystal violet. The images were captured and showed colony growth in four rectal cancer cells. Percentage of survived cells was calculated by comparing with untreated control. (⁎⁎P < .01).
SOX4 Activates ATM-Mediated DNA Damage Repair through Induction of NBS1
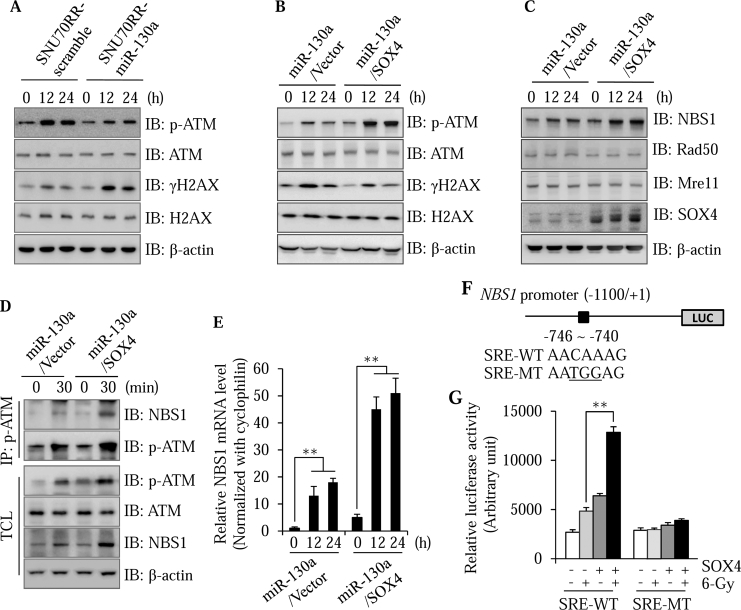
As shown in Figure 2D, miR-130a overexpressing cells were less effective at repairing DNA damage compared with scramble cells. Overexpression of miR-130a induced accumulation of γH2AX foci, which were considered a marker for unrepaired damaged DNA, in the nucleus after irradiation treatment. As ATM is a critical regulator of the DNA repair process, we speculated that miR-130a might delay the repair of DNA damage through suppression of ATM signaling. To test this, we examined phosphorylated ATM which is indicative of ATM activation in SNU70RR-miR-130a upon irradiation treatment. Overexpression of miR-130a significantly suppressed the phosphorylation of ATM compared with SNU70RR parental cells (Figure 5A). In contrast with the inhibition of ATM signaling, γH2AX was markedly increased in miR-130a overexpressing cells (Figure 5A). These results indicated that miR-130a has a suppressive activity on ATM signaling in response to irradiation.
Figure 5.
SOX4 activates ATM signaling in response to irradiation through transcriptional regulation of NBS1. (A) To determine the effect of miR-130a on irradiation-induced DNA damage, SNU70RR cells were irradiated with a 6 Gy dose and collected at 12 or 24 h after treatment. The protein lysates were isolated and analyzed for expression of p-ATM, total ATM, γH2AX and total H2AX by western blotting. (B) After 12 or 24 h of irradiation, miR-130a or SOX4 cells were harvested and analyzed with western blotting for p-ATM, total ATM, γH2AX and total H2AX. β-actin was used as the loading control. (C) SNU70RR-miR-130a and SNU70RR-miR-130a/SOX4 cells were delivered with 6 Gy single dose of irradiation. After 12 or 24 h of treatment, the expression of NBS1, Rad50, Mre11 and SOX4 was determined with western blotting. β-actin was used as the loading control. (D) At 30 min after irradiation, cells were lysed to immunoprecipitate the p-ATM protein with specific antibody. Interaction of NBS1 was detected with western blotting. Total cell lysates (TCL) were analyzed as the normalization control. (E) The relative expression level of NBS1 was measured with real time PCR in SOX4 cell after irradiation. The data were normalized with cyclophilin as the internal control and represents one of at least three repeated experiments. (F) Schematic diagram showing the −1100/+1 region of the human NBS1 promoter luciferase construct. The locations of the putative SRE and point mutations are indicated. (G) Cells were cotransfected with the SRE wild-type or mutant reporter construct and β-galactosidase. Luciferase activity was determined after 48 h of transfection. Data represent the mean ± S.D. of three independent experiments and show the normalized ratio of luciferase activity to β-galactosidase activity. (⁎⁎P < .01).
We next aimed to determine whether miR-130a suppresses ATM activation through down-regulation of SOX4. Expectedly, restoration of SOX4 in SNU70RR-miR-130a cells reversed the activation of ATM signaling accompanied with less accumulation of γH2AX (Figure 5B). In response to DNA damage, the MRN complex (Mre11, Rad50 and Nbs1 proteins) is required for ATM activation [29]. The interaction between the MRN complex and ATM facilitates the activation of ATM and enhances the affinity of p-ATM for its substrates [30]. To investigate the mechanistic role of SOX4 on ATM-dependent DNA damage repair, we assessed the expression of MRN components in the presence of SOX4. As shown in Figure 5C, restoration of SOX4 markedly enhanced the expression of NBS1 by irradiation treatment. In contrast, expression of SOX4 did not affect the expression of other MRN components. To confirm that SOX4 enhances the interaction of NBS1 and p-ATM upon irradiation treatment, we performed an immunoprecipitation assay. The interaction of NBS1 and p-ATM was significantly increased after irradiation in keeping with increased p-ATM levels in miR-130a/SOX4 cells (Figure 5D). This suggests that SOX4 increases the activation of ATM signaling through an increase in NBS1 expression and facilitation of the interaction between NBS1 and p-ATM.
To understand the detailed mechanism for SOX4-induced NBS1 upregulation, we first checked the level of NBS1 mRNA using real time PCR. As shown in Figure 5E, NBS1 mRNA was significantly upregulated upon irradiation treatment in the presence of SOX4, suggesting that NBS1 is regulated at the transcriptional level by SOX4. To further determine whether SOX4 directly regulates the expression of NBS1, 1.1 kb of human NBS promoter was cloned into luciferase reporter vector. This region contains one putative SRE located between −746 and− 740 (Figure 5F). With forced expression of SOX4, NBS1 promoter activity was increased about threefold after irradiation treatment (Figure 5G). Induction of luciferase activity completely disappeared because of the mutated SRE region. These data suggest that SOX4 directly regulates the expression of NBS1 at the transcriptional level.
miR-130a Radiosensitizes In Vivo Rectal Cancer in a Xenograft Model
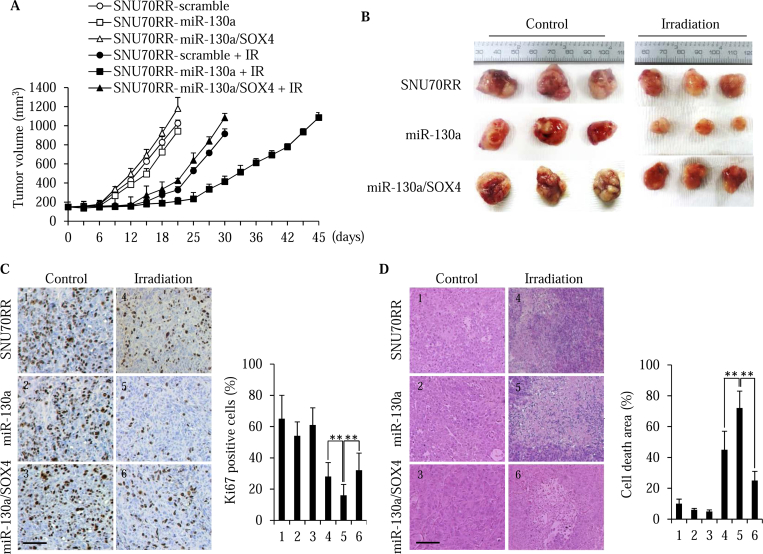
To further confirm the effects of miR-130a overexpression on tumor growth and radiation responses, we generated in vivo xenograft tumors in athymic nude mice using SNU70RR-scramble, SNU70RR-miR-130a and SNU70RR-miR-130a/SOX4 cells. When the tumor size reached 150 mm3 in volume, a single dose of 15 Gy radiation was delivered locally to the mice. After irradiation treatment, mice bearing SNU70RR-miR-130a tumors were more radiosensitive compared with mice with SNU70RR-scramble tumors (Figure 6, A and B). In contrast, restoration of SOX4 completely suppressed the radiosensitivity induced by miR-130a. Consistent with a delay of the tumor growth rate, overexpression of miR-130a reduced the proliferation of cancer cells compared with scramble or SOX4 restored cells (Figure 6C). Additionally, histopathological examination indicated that a significant increase of cell death was observed in SNU70RR-miR-130a tumors and was reversed by expression of SOX4 (Figure 6D). Taken together, these data suggest that miR-130a radiosensitizes rectal cancer in vivo and SOX4 is a master regulator of radioresistance.
Figure 6.
miR-130a overcomes the radioresistance of in vivo rectal cancer in a xenograft model. (A) The growth curves of SNU70RR-control, miR-130a and miR-130a/SOX4 xenograft tumors without or with irradiation (IR) treatment. Mice were locally delivered a single 15 Gy dose of irradiation. After the tumor size reached 1500 mm3, the mice were scarified. The tumor size was measured with caliper every 3 days (n = 6 mice per group). (B) Representative images of tumors after sacrifice. The tumors from the miR-130a expressing cells were markedly smaller after irradiation compared with other cells. (C) Left, immunohistochemical staining for Ki-67 for SNU70RR-scramble, SNU70RR-miR-130a and SNU70RR-miR-130a/SOX4 xenograft tumors. Right, a graph showing the percentage of Ki-67 positive nuclei. (D) Left, representative image of H&E staining for SNU70RR-scramble, SNU70RR-miR-130a and SNU70RR-miR-130a/SOX4 xenograft tumors with the area of cell death. Right, a graph showing the percentage of cell death areas in the tumors. The images were captured and analyzed from at least five different fields. The scale bar represents 50 μm. (⁎⁎P < .01).
Discussion
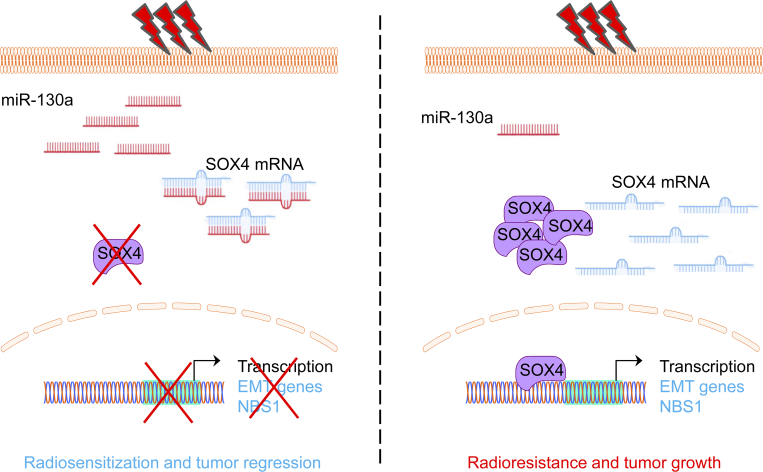
Recently, accumulating evidence indicates that dysregulated expression of miRNAs is correlated with radioresistance in various human cancers [31], [32]. In this study, we compared the radiosensitivity of four rectal cancer cell lines and analyzed the miRNA expression profiles using small RNA sequencing. In this study, we first demonstrate that miR-130a dramatically enhances the radiosensitivity of rectal cancer cells in both in vitro and in vivo models. Functionally, miR-130a reverses the EMT process and suppresses ATM-dependent DNA damage repair signaling upon irradiation by targeting of SOX4 (Figure 7). When the level of miR-130a is low, rectal cancer cells have a high level of SOX4 to enhance the transcription of EMT-related and NBS1 genes, resulting in radioresistance and tumor growth.
Figure 7.
Schematic diagram showing the radiosensitizer activity of miR-130a by targeting SOX4. Radiosensitive rectal cancer cells have a high expression of miR-130a to target SOX4 and suppress transcription of the EMT-related genes and NBS1. In contrast, radioresistant rectal cancer cells shows the low expression of miR-130a leading to high level of SOX4. As a result, SOX4 promotes radioresistance by inducing the EMT process and activating the DNA repair process. In conclusion, miR-130a functions as a radiosensitizer in rectal cancer cells and can be used as a preoperative prognostic biomarker for radiotherapy.
Based on small RNA sequencing data, a number of miRNAs differentially regulated in rectal cancer cells and most of these were reported to be involved in the radioresistance of cancer cells. For instance, miR-16-5p has been reported to regulate the cellular radiosensitivity of human prostate cancer cells. Overexpression of miR-16-5p enhances the radiosensitivity of cancer cells through suppression of cell proliferation, reduction of cell viability and induction of cell cycle arrest [33]. The miR-15a/15b/16 family was also shown to enhance the radiosensitivity of breast cancer cells by influencing G2/M checkpoint proteins [34]. Another study has also shown that let-7 g suppresses the expression of NF-κB1 and enhances the efficiency of radiotherapy for lung cancer treatment [35]. These studies suggested that miRNAs identified with small RNA sequencing could be potential candidates for overcoming of radioresistance in rectal cancer.
Recently, miR-130a has emerged as a tumor suppressor miRNA that suppresses cancer cell proliferation, invasion and migration by targeting oncogenic proteins. For instance, miR-130a suppresses the tumor growth, migration and EMT phenotype of glioma cells by directly targeting HMGB2 [36]. MiR-130a also inhibited the cell cycle progression and proliferation upon irradiation treatment by targeting c-myc [37]. miR-130a is down-regulated in variety of human cancers, which is consistent with the tumor suppressive activity [38], [39]. Moreover, miR-130a was significantly down-regulated in nonresponders compared with responders during chemo-radiation therapy for locally advanced rectal cancer [40]. Based on these results, miR-130a is a critical regulator for radioresistance of rectal cancer cells and can be a potential preoperative biomarker for radiotherapy to select responders. In contrast, miR-130a also involved in tumor growth and poorer prognosis as oncogene for colorectal cancer [41], [42], [43]. To solve these discrepancies for double-edged sword's activity of miR-130a, further study will be required using in vivo model and patient's sample.
A recent study has demonstrated that EMT could affect the therapeutic resistance in breast cancer [41] and can be a target of miRNAs to overcome radioresistance. Previous studies suggested that SOX4 is a master regulator of EMT [12] and upregulation of SOX4 is associated with colorectal cancer progression [42]. In addition to regulation of the EMT process, SOX4 was also induced by DNA damage in an ATM/ATK-dependent manner [43]. In this study, we also confirmed that SOX4 is a key regulator for the expression of EMT-related genes and NBS1. Therefore, SOX4 may be used as a prognostic biomarker for radiotherapy nonresponders and for screening of therapeutics to overcome radioresistance.
Author Contributions
Huyen Trang Ha Thi, Hye-Youn Kim and Suntaek Hong designed the study. Huyen Trang Ha Thi, Hye-Youn Kim and Young-Mi Kim performed the experiments and analyzed the data. Huyen Trang Ha Thi and Suntaek Hong wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
Funding: This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (2016R1A2B2008007 to S.H. and 2017R1D1A1B03030485 to H.T.H.T.).
Conflicts of interest: The authors declare no conflicts of interest.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2019.07.005.
Appendix A. Supplementary Data
Supplementary material
References
- 1.Wang X, Zheng B, Lu X, Bai R, Feng L, Wang Q, Zhao Y, He S. Preoperative short-course radiotherapy and long-course radiochemotherapy for locally advanced rectal cancer: Meta-analysis with trial sequential analysis of long-term survival data. PLoS One. 2018;13 doi: 10.1371/journal.pone.0200142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cedermark B, Dahlberg M, Glimelius B, Pahlman L, Rutqvist LE, Wilking N. Improved survival with preoperative radiotherapy in resectable rectal cancer. N Engl J Med. 1997;336:980–987. doi: 10.1056/NEJM199704033361402. [DOI] [PubMed] [Google Scholar]
- 3.Glimelius B, Gronberg H, Jarhult J, Wallgren A, Cavallin-Stahl E. A systematic overview of radiation therapy effects in rectal cancer. Acta Oncol. 2003;42:476–492. doi: 10.1080/02841860310012301. [DOI] [PubMed] [Google Scholar]
- 4.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang XD, Xu XH, Zhang SY, Wu Y, Xing CG, Ru G, Xu HT, Cao JP. Role of miR-100 in the radioresistance of colorectal cancer cells. Am J Cancer Res. 2015;5:545–559. [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang P, Wang L, Rodriguez-Aguayo C, Yuan Y, Debeb BG, Chen D, Sun Y, You MJ, Liu Y, Dean DC. miR-205 acts as a tumour radiosensitizer by targeting ZEB1 and Ubc13. Nat Commun. 2014;5:5671. doi: 10.1038/ncomms6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang X, Taeb S, Jahangiri S, Emmenegger U, Tran E, Bruce J, Mesci A, Korpela E, Vesprini D, Wong CS. miRNA-95 mediates radioresistance in tumors by targeting the sphingolipid phosphatase SGPP1. Cancer Res. 2013;73:6972–6986. doi: 10.1158/0008-5472.CAN-13-1657. [DOI] [PubMed] [Google Scholar]
- 9.Ma W, Yu J, Qi X, Liang L, Zhang Y, Ding Y, Lin X, Li G, Ding Y. Radiation-induced microRNA-622 causes radioresistance in colorectal cancer cells by down-regulating Rb. Oncotarget. 2015;6:15984–15994. doi: 10.18632/oncotarget.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van de Wetering M, Oosterwegel M, van Norren K, Clevers H. Sox-4, an Sry-like HMG box protein, is a transcriptional activator in lymphocytes. EMBO J. 1993;12:3847–3854. doi: 10.1002/j.1460-2075.1993.tb06063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Liang Q, Lei Y, Yao M, Li L, Gao X, Feng J, Zhang Y, Gao H, Liu DX. SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res. 2012;72:4597–4608. doi: 10.1158/0008-5472.CAN-12-1045. [DOI] [PubMed] [Google Scholar]
- 12.Tiwari N, Tiwari VK, Waldmeier L, Balwierz PJ, Arnold P, Pachkov M, Meyer-Schaller N, Schubeler D, van Nimwegen E, Christofori G. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 2013;23:768–783. doi: 10.1016/j.ccr.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 13.Sinner D, Kordich JJ, Spence JR, Opoka R, Rankin S, Lin SC, Jonatan D, Zorn AM, Wells JM. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol. 2007;27:7802–7815. doi: 10.1128/MCB.02179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao YL, Sun YM, Chau GY, Chau YP, Lai TC, Wang JL, Horng JT, Hsiao M, Tsou AP. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008;27:5578–5589. doi: 10.1038/onc.2008.168. [DOI] [PubMed] [Google Scholar]
- 15.Sun R, Jiang B, Qi H, Zhang X, Yang J, Duan J, Li Y, Li G. SOX4 contributes to the progression of cervical cancer and the resistance to the chemotherapeutic drug through ABCG2. Cell Death Dis. 2015;6:e1990. doi: 10.1038/cddis.2015.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu P, Ramachandran S, Ali Seyed M, Scharer CD, Laycock N, Dalton WB, Williams H, Karanam S, Datta MW, Jaye DL. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res. 2006;66:4011–4019. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- 17.Aaboe M, Birkenkamp-Demtroder K, Wiuf C, Sorensen FB, Thykjaer T, Sauter G, Jensen KM, Dyrskjot L, Orntoft T. SOX4 expression in bladder carcinoma: clinical aspects and in vitro functional characterization. Cancer Res. 2006;66:3434–3442. doi: 10.1158/0008-5472.CAN-05-3456. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Jiang H, Shao J, Mao R, Liu J, Ma Y, Fang X, Zhao N, Zheng S, Lin B. SOX4 inhibits GBM cell growth and induces G0/G1 cell cycle arrest through Akt-p53 axis. BMC Neurol. 2014;14:207. doi: 10.1186/s12883-014-0207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin Y, Zhao M, Xie Q, Zhang H, Wang Q, Ma Q. MicroRNA-338-3p functions as tumor suppressor in breast cancer by targeting SOX4. Int J Oncol. 2015;47:1594–1602. doi: 10.3892/ijo.2015.3114. [DOI] [PubMed] [Google Scholar]
- 20.Chen B, Liu J, Qu J, Song Y, Li Y, Pan S (2017). MicroRNA-25 suppresses proliferation, migration, and invasion of osteosarcoma by targeting SOX4 Tumour Biol 39, 1010428317703841. [DOI] [PubMed]
- 21.Banath JP, Macphail SH, Olive PL. Radiation sensitivity, H2AX phosphorylation, and kinetics of repair of DNA strand breaks in irradiated cervical cancer cell lines. Cancer Res. 2004;64:7144–7149. doi: 10.1158/0008-5472.CAN-04-1433. [DOI] [PubMed] [Google Scholar]
- 22.Su H, Jin X, Zhang X, Zhao L, Lin B, Li L, Fei Z, Shen L, Fang Y, Pan H. FH535 increases the radiosensitivity and reverses epithelial-to-mesenchymal transition of radioresistant esophageal cancer cell line KYSE-150R. J Transl Med. 2015;13:104. doi: 10.1186/s12967-015-0464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H, Luo H, Jiang Z, Yue J, Hou Q, Xie R, Wu S. Fractionated irradiation-induced EMT-like phenotype conferred radioresistance in esophageal squamous cell carcinoma. J Radiat Res. 2016;57:370–380. doi: 10.1093/jrr/rrw030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Theys J, Jutten B, Habets R, Paesmans K, Groot AJ, Lambin P, Wouters BG, Lammering G, Vooijs M. E-Cadherin loss associated with EMT promotes radioresistance in human tumor cells. Radiother Oncol. 2011;99:392–397. doi: 10.1016/j.radonc.2011.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 26.Christofori G. New signals from the invasive front. Nature. 2006;441:444–450. doi: 10.1038/nature04872. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Zeng S, Jiang X, Lai D, Su Z (2017). SOX4 induces tumor invasion by targeting EMT-related pathway in prostate cancer Tumour Biol 39, 1010428317694539. [DOI] [PubMed]
- 28.Yoon TM, Kim SA, Cho WS, Lee DH, Lee JK, Park YL, Lee KH, Lee JH, Kweon SS, Chung IJ. SOX4 expression is associated with treatment failure and chemoradioresistance in oral squamous cell carcinoma. BMC Cancer. 2015;15:888. doi: 10.1186/s12885-015-1875-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paull TT, Lee JH. The Mre11/Rad50/Nbs1 complex and its role as a DNA double-strand break sensor for ATM. Cell Cycle. 2005;4:737–740. doi: 10.4161/cc.4.6.1715. [DOI] [PubMed] [Google Scholar]
- 31.Wang N, Tan HY, Feng YG, Zhang C, Chen F, Feng Y. microRNA-23a in human cancer: its roles, mechanisms and therapeutic relevance. Cancers (Basel) 2018;11 doi: 10.3390/cancers11010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ni J, Bucci J, Chang L, Malouf D, Graham P, Li Y. Targeting microRNAs in prostate cancer radiotherapy. Theranostics. 2017;7:3243–3259. doi: 10.7150/thno.19934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F, Mao A, Tang J, Zhang Q, Yan J, Wang Y, Di C, Gan L, Sun C, Zhang H. microRNA-16-5p enhances radiosensitivity through modulating Cyclin D1/E1-pRb-E2F1 pathway in prostate cancer cells. J Cell Physiol. 2019;234:13182–13190. doi: 10.1002/jcp.27989. [DOI] [PubMed] [Google Scholar]
- 34.Mei Z, Su T, Ye J, Yang C, Zhang S, Xie C. The miR-15 family enhances the radiosensitivity of breast cancer cells by targeting G2 checkpoints. Radiat Res. 2015;183:196–207. doi: 10.1667/RR13784.1. [DOI] [PubMed] [Google Scholar]
- 35.Arora H, Qureshi R, Jin S, Park AK, Park WY (2011). miR-9 and let-7g enhance the sensitivity to ionizing radiation by suppression of NFkappaB1 Exp Mol Med43, 298-304. [DOI] [PMC free article] [PubMed]
- 36.Tang C, Yang Z, Chen D, Xie Q, Peng T, Wu J, Qi S. Downregulation of miR-130a promotes cell growth and epithelial to mesenchymal transition by activating HMGB2 in glioma. Int J Biochem Cell Biol. 2017;93:25–31. doi: 10.1016/j.biocel.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Challagundla KB, Sun XX, Zhang Q, Dai MS. MicroRNA-130a associates with ribosomal protein L11 to suppress c-Myc expression in response to UV irradiation. Oncotarget. 2015;6:1101–1114. doi: 10.18632/oncotarget.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Acunzo M, Visone R, Romano G, Veronese A, Lovat F, Palmieri D, Bottoni A, Garofalo M, Gasparini P, Condorelli G. miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene. 2012;31:634–642. doi: 10.1038/onc.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boll K, Reiche K, Kasack K, Morbt N, Kretzschmar AK, Tomm JM, Verhaegh G, Schalken J, von Bergen M, Horn F. MiR-130a, miR-203 and miR-205 jointly repress key oncogenic pathways and are downregulated in prostate carcinoma. Oncogene. 2013;32:277–285. doi: 10.1038/onc.2012.55. [DOI] [PubMed] [Google Scholar]
- 40.Lim SH-S, Ip E, Chua W, Ng W, Henderson C, Shin J-S, Harris BHL, Barberis A, Cowley M, Souza PLD. Serum microRNA expression during neoadjuvant chemoradiation for rectal cancer. J Clin Oncol. 2017;35 [Google Scholar]
- 41.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang B, Li Y, Tan F, Xiao Z. Increased expression of SOX4 is associated with colorectal cancer progression. Tumour Biol. 2016;37:9131–9137. doi: 10.1007/s13277-015-4756-5. [DOI] [PubMed] [Google Scholar]
- 43.Pan X, Zhao J, Zhang WN, Li HY, Mu R, Zhou T, Zhang HY, Gong WL, Yu M, Man JH. Induction of SOX4 by DNA damage is critical for p53 stabilization and function. Proc Natl Acad Sci U S A. 2009;106:3788–3793. doi: 10.1073/pnas.0810147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material