Abstract
Anaerobic bacteria inhabiting the human gastrointestinal tract have evolved various enzymes that modify host-derived steroids. The bacterial steroid-17,20-desmolase pathway cleaves the cortisol side chain, forming pro-androgens predicted to impact host physiology. Bacterial 20β-hydroxysteroid dehydrogenase (20β-HSDH) regulates cortisol side-chain cleavage by reducing the C-20 carboxyl group on cortisol, yielding 20β-dihydrocortisol. Recently, the gene encoding 20β-HSDH in Butyricicoccus desmolans ATCC 43058 was reported, and a nonredundant protein search yielded a candidate 20β-HSDH gene in Bifidobacterium adolescentis strain L2-32. B. adolescentis 20β-HSDH could regulate cortisol side-chain cleavage by limiting pro-androgen formation in bacteria such as Clostridium scindens and 21-dehydroxylation by Eggerthella lenta. Here, the putative B. adolescentis 20β-HSDH was cloned, overexpressed, and purified. 20β-HSDH activity was confirmed through whole-cell and pure enzymatic assays, and it is specific for cortisol. Next, we solved the structures of recombinant 20β-HSDH in both the apo- and holo-forms at 2.0–2.2 Å resolutions, revealing close overlap except for rearrangements near the active site. Interestingly, the structures contain a large, flexible N-terminal region that was investigated by gel-filtration chromatography and CD spectroscopy. This extended N terminus is important for protein stability because deletions of varying lengths caused structural changes and reduced enzymatic activity. A nonconserved extended N terminus was also observed in several short-chain dehydrogenase/reductase family members. B. adolescentis strains capable of 20β-HSDH activity could alter glucocorticoid metabolism in the gut and thereby serve as potential probiotics for the management of androgen-dependent diseases.
Keywords: microbiome; probiotic; androgen; glucocorticoid; oxidation–reduction (redox); 20β-hydroxysteroid dehydrogenase; Bifidobacteria; C-20 reduction; cortisol; Steroid-17,20-desmolase
Introduction
Hydroxysteroid dehydrogenases (HSDH)2 are pyridine nucleotide-dependent enzymes classified into three superfamilies, including aldo-keto reductase (AKR), long/medium-chain dehydrogenase/reductase (MDR), and short-chain dehydrogenase/reductase (SDR), which interconvert between enol and keto forms of hydroxyl groups on diverse small molecules (1, 2). The gut microbiome is exposed to a mixture of glucocorticoids and bile acids. Recent investigations identified bacterial strains of diverse taxa expressing stereo- and regio-specific HSDHs in the SDR and AKR families capable of oxidation and epimerization of bile acid hydroxyl groups at C-3, C-7, and C-12 (3–5). Cortisol and corticosterone are human steroid hormone glucocorticoids that can be metabolized by both host and bacterial enzymes (6). Cortisol is the major glucocorticoid in humans and is present in concentrations ranging from 5 to 20 μg/dl throughout the day (7). The various bacterial reactions altering steroid structure impact the host by generating potent signaling molecules capable of modulating host physiology.
Recent attention has been focused on the bacterial steroid-17,20-desmolase pathway (desABCD) converting cortisol to 11β-hydroxyandrostenedione (11β-OHAD) in Clostridium scindens and the zinc-dependent MDR family 20α-HSDH (desC), a predicted enzymatic regulator of the pathway (Fig. 1) (2, 8, 9). 11β-OHAD is a pro-androgen whose formation by gut bacteria (10) as well as urinary tract bacteria (9) may contribute to castration-resistant prostate cancer (11) and polycystic ovary syndrome (12). The bacterial steroid-17,20-desmolase pathway has also been reported in Clostridium cadavaris and Butyricicoccus desmolans (formerly Eubacterium desmolans) and is associated with NADH-dependent cortisol 20β-HSDH (desE) activity (9, 13). Previous studies also identified strains of Bifidobacterium adolescentis isolated from human and rat fecal dilutions expressing cortisol 20β-HSDH activity alone (14). Recently, the gene encoding gut bacterial NADH-dependent 20β-HSDH activity was reported in B. desmolans (9). Phylogenetic analysis identified numerous Bifidobacterium spp. strains, including strains of B. adolescentis, encoding putative cortisol 20β-HSDH (9).
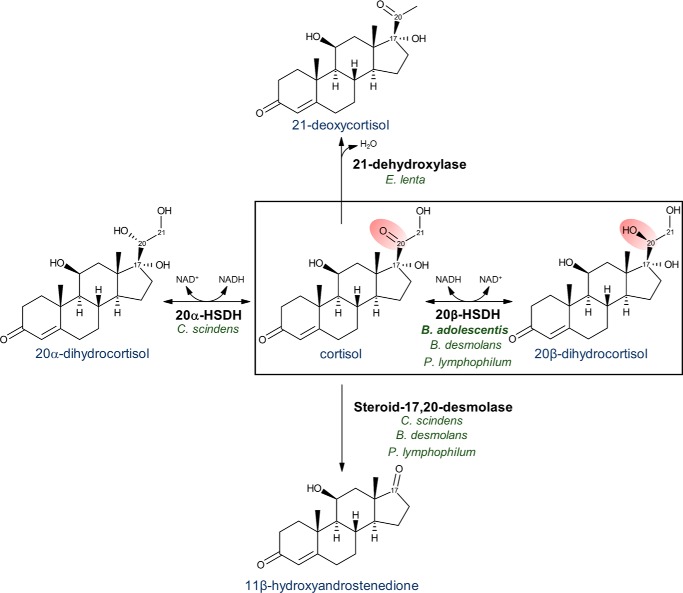
Figure 1.
Cortisol-metabolizing reactions by B. adolescentis, C. scindens, B. desmolans, P. lymphophilum, and E. lenta. Boxed reaction shows 20β-HSDH oxidizes the coenzyme NADH to transform cortisol into 20β-dihydrocortisol. Cortisol + NADH ⇌ 20β-dihydrocortisol + NAD+. This reaction reduces the double-bonded oxygen at C-20 (highlighted in red) on cortisol and is fully reversible.
Gut bacterial 20β-HSDH is a member of the SDR family of proteins (9). The SDR family constitutes one of the largest protein superfamilies distributed across all three domains of life (15), comprising functionally diverse members (16). SDR family enzymes include several Enzyme Commission (EC) classifications, including lyases and isomerases, along with pyridine nucleotide-dependent oxidoreductases, which constitute the majority of enzymes. Currently, in Uniprot there are 266,639 SDR entries, and members typically share only ∼20–30% amino acid residue identity in pairwise comparisons. Most SDR family members are 250–350 amino acids in length, with secondary structures consisting of a Rossmann fold domain containing 6–7 β-strands flanked by three α-helices (17). Members usually contain a conserved Gly-rich sequence critical in NAD(P)(H) binding, as well as a catalytic tetrad composed of Ser, Tyr, Lys, and Asn residues (15). The SDR family enzymes vary in substrate recognition, including sugars, dyes, prostaglandins, porphyrins, alcohols, and steroids (16). Consequently, substrate-binding residues are not highly conserved. Differentiating HSDHs from other functional classes of SDR family enzymes is important for identifying steroid biochemical pathways in gut metagenomes. At present, this task is impeded by a lack of primary amino acid sequences for several HSDHs whose activities have been reported in bacterial isolates (18), but also the need to determine residues important in differentiating HSDHs into functional categories (i.e. 7α-HSDH from 7β-HSDH and both from 20β-HSDH).
The first SDR family enzyme with HSDH activity crystallized was 3α,20β-HSDH from the soil bacterium Streptomyces hydrogenans (19). The 3α,20β-HSDH reversibly oxidizes the 3α-hydroxyl and 20β-hydroxyl groups of androgens and progestins. The S. hydrogenans 3α,20β-HSDH shares 31% amino acid sequence identity with the 20β-HSDH from B. adolescentis, which does not appear to function as a dual 3α-HSDH (9). Humans express glucocorticoid 20β-HSDH activity as well via carbonyl reductase 1 (CBR1) (20). 20β-Dihydrocortisol is detected in the urine of patients with Cushing's disease (21) and the plasma of hypertensive individuals (22). It is thus important to obtain a mechanistic understanding of gut bacterial 20β-HSDH because this enzyme is capable of generating 20β-dihydrocortisol and regulating both major microbial side-chain cleavages of cortisol: steroid-17,20-desmolase activity resulting in pro-androgens (6), and cortisol 21-dehydroxylation producing 21-deoxycortisol, a hypertensive compound (Fig. 1) (6, 23). By understanding the structure and function of 20β-HSDH, the ability of B. adolescentis strain L2-32 to shift glucocorticoid metabolism in the gut may be improved. 20β-HSDH activity could reduce formation and absorption of 11β-hydroxyandrogens and 21-deoxycortisol, which are implicated in chronic human disease.
Herein, we report the structures of NADH-dependent 20β-HSDH from B. adolescentis strain L2-32 at 2.2 Å for the apo-form and 2.0 Å for the binary complex with NADH. Through site-directed mutagenesis and binding-affinity studies, we have identified residues comprising the substrate-binding pocket and propose a catalytic mechanism for NADH-dependent C-20 reduction. A long, flexible N-terminal domain involved in overall 20β-HSDH stability was discovered, having widespread implications for other members of the diverse SDR family.
Results
B. adolescentis expresses 20β-HSDH activity
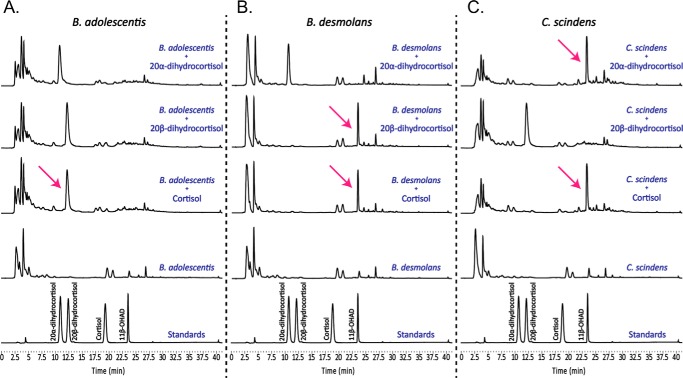
The 20β-HSDH sequence from B. desmolans ATCC 43058 was used to query the nonredundant protein sequences database (BLASTP) (9). An annotated SDR family oxidoreductase (WP_003810233.1) from B. adolescentis was found with a sequence identity of 59%. This protein was selected for further study because it has the potential to be of great pharmaceutical interest, as strains of B. adolescentis have potential probiotic function, i.e. capable of conferring beneficial effects on the host. 20β-HSDH catalyzes the conversion of cortisol to 20β-dihydrocortisol in the boxed reaction depicted in Fig. 1, and to determine whether B. adolescentis strain L2-32 possesses 20β-HSDH activity, whole-cell extracts were screened for the activity at 48 h. The result, as shown in Fig. 2, was net conversion of cortisol to 20β-dihydrocortisol. As controls, B. desmolans ATCC 43058 and C. scindens ATCC 35704 were both shown to metabolize cortisol, as well as 20β-dihydrocortisol and 20α-dihydrocortisol, respectively. 20α-Dihydrocortisol is not metabolized by B. adolescentis strain L2-32 or B. desmolans ATCC 43058, both of which possess desE, a gene previously shown to encode NAD(H)-dependent 20β-HSDH in B. desmolans and Bifidobacterium scardovii (9). 20β-Dihydrocortisol is not a metabolite for C. scindens ATCC 35704, which was previously shown to harbor the desC gene, expressing NAD(H)-dependent 20α-HSDH (8). The desAB genes, encoding steroid-17,20-desmolase, are present in both C. scindens ATCC 35704 and B. desmolans ATCC 43058. As expected, net conversions of 20α-dihydrocortisol to 11β-OHAD and 20β-dihydrocortisol to 11β-OHAD were observed in cultures of C. scindens ATCC 35704 and B. desmolans ATCC 43058, respectively. In contrast, B. adolescentis strain L2-32 lacks the desAB genes, and incubation with cortisol resulted in net conversion to 20β-dihydrocortisol under anaerobic conditions.
Figure 2.
Whole-cell assays of B. adolescentis strain L2-32, B. desmolans ATCC 43058, and C. scindens ATCC 35704. A, B. adolescentis completely converts cortisol into 20β-dihydrocortisol. 20α-Dihydrocortisol is not metabolized. In a highly reductive environment and without pressure from a steroid-17,20-desmolase, B. adolescentis does not convert 20β-dihydrocortisol into cortisol. B, B. desmolans completely converts cortisol into 11β-OHAD and 20β-dihydrocortisol into 11β-OHAD. 20α-Dihydrocortisol is not metabolized. C, C. scindens completely converts cortisol into 11β-OHAD and 20α-dihydrocortisol into 11β-OHAD. 20β-Dihydrocortisol is not metabolized. Standards are reused in each panel for ease of comparison to reaction products above.
Biochemical characterization of recombinant 20β-HSDH
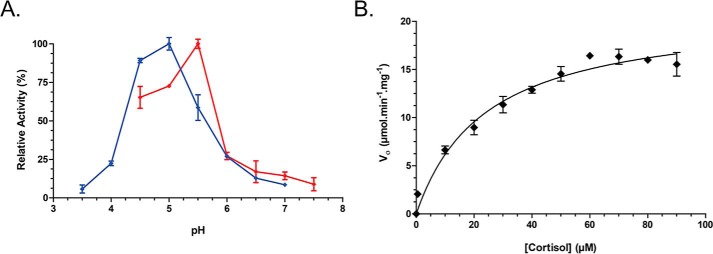
To determine whether the SDR family oxidoreductase identified in B. adolescentis (WP_003810233.1) encodes a 20β-HSDH (DesE), the gene was cloned into the pET-28a(+) expression vector. The N-terminal His6-tagged recombinant protein was overexpressed in Escherichia coli BL21(DE3)RIPL and purified via metal-affinity chromatography as a single band on SDS-PAGE, and the molecular mass was similar to the theoretical calculated subunit molecular mass of 31.7 kDa (Fig. 3A). The native molecular mass of the recombinant 20β-HSDH (r20β-HSDH) protein was determined by gel-filtration chromatography with reference to protein standards. r20β-HSDH displayed an elution volume of 13.9 ml, corresponding to a molecular mass of 130.5 ± 2.8 kDa (Fig. 3B). Thus, the gel-filtration data, together with the subunit molecular mass determined by SDS-PAGE, suggest that r20β-HSDH forms a homotetrameric quaternary structure in solution. A tetrameric structure was also seen in the structural data, and the proposed tetramer is shown in Fig. 5B. The purified r20β-HSDH exhibited a pH optimum of 5.0 in the reductive direction, and in the oxidative direction the optimum was pH 5.5 (Fig. 4A).
Figure 3.
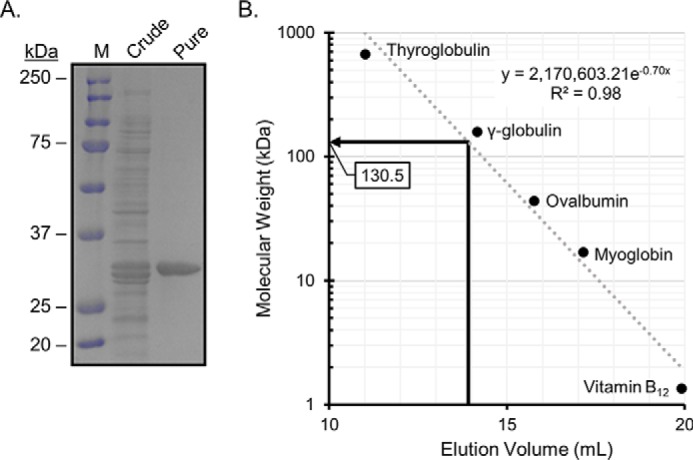
Purified 20β-HSDH from B. adolescentis, strain L2-32. A, SDS-polyacrylamide gel of crude and purified 20β-HSDH showing a subunit size of 32 ± 0.12 kDa. Lane M, molecular mass markers. B, native molecular size analysis of purified 20β-HSDH via gel-filtration chromatography.
Figure 5.
Structural characterization of 20β-HSDH. A, apo structure of 20β-HSDH showing the SDR-characteristic Rossmann fold consisting of seven central β-strands (β3–β2–β1–β4–β5–β6–β7) flanked by six α-helices. Right panel depicts the extended N terminus with no electron density before residue 17 and truncation sites. B, proposed tetramer of 20β-HSDH based on crystallographic arrangement. C, NADH-binding pocket showing five side chains and three peptidyl backbone interactions. D, surface representation of the NADH-binding pocket highlighting residues that make van der Waals contacts to NADH (yellow). The Gly-rich region is highlighted in pink and also makes van der Waals contacts to NADH.
Figure 4.
pH optimum and Michaelis-Menten saturation curve of WT 20β-HSDH. A, pH optimization of 20β-HSDH in the reductive (blue) and oxidative (red) direction. B, saturation curve showing the reduction of cortisol by WT 20β-HSDH.
The Michaelis-Menten saturation curve was determined from the primary reaction of cortisol reduction performed by r20β-HSDH (Fig. 4B). The saturation curve was used to estimate the kinetic constants that are reported in Table 1. The purified r20β-HSDH has a Km of 24.07 μm when cortisol is the substrate. The Vmax, kcat, and kcat/Km values for cortisol were determined to be 21.08 μmol·min−1·mg−1, 668.3 min−1, and 27.77 μm−1·min−1, respectively. Substrate-specificity studies revealed that cortisol was the primary substrate utilized by 20β-HSDH (Table 2). Hydroxyl groups on rings C and D are important for substrate specificity by r20β-HSDH. We observed an ∼34% reduction in relative activity with 11-deoxycortisol and an ∼61% reduction in activity with corticosterone, which lacks a 17-hydroxyl group. Reduction of the 3-oxo-Δ4 to 3α-, 5β-H (tetrahydrocortisol) results in an ∼56% reduction in activity relative to cortisol. It was previously determined that recombinant steroid-17,20-desmolase from C. scindens ATCC 35704 also showed significant reduction in side-chain cleavage activity with tetrahydrocortisol (8) but not allotetrahydrocortisol (3α-,5α-H).3 Consistent with the whole-cell conversion data from B. adolescentis strain L2-32 (Fig. 2A), the oxidative direction with 20β-dihydrocortisol and NAD+ has only 0.23% of the relative activity compared with cortisol and NADH. As expected, 20α-dihydrocortisol did not serve as a substrate, nor did we detect formation of 20α-derivatives with purified recombinant r20β-HSDH in the presence of NADH. NADP(H) was tested, but we did not observe activity with this cofactor.
Table 1.
Steady-state kinetic parameters of 20β-HSDH and active-site mutants
| Protein | Vmax | Km | kcat | Relative kcat | kcat/Km | Relative kcat/Km |
|---|---|---|---|---|---|---|
| μmol·min−1·mg−1 | μm | min−1 | % | μm−1·min−1 | % | |
| Wildtype | 21.08 ± 1.13a | 24.07 ± 3.80 | 668.30 ± 50.54 | 100 | 27.77 ± 3.65 | 100 |
| S181A | b | – | – | – | – | – |
| S183A | 4.70 ± 0.10 | 9.49 ± 2.60 | 148.70 ± 11.24 | 22 | 15.68 ± 2.06 | 56 |
a Values represent the means ± S.D. based on three or more replications.
b – means no activity was detected.
Table 2.
Substrate specificity of 20β-HSDH
| Steroid | Trivial name | Coenzyme | Relative activity |
|---|---|---|---|
| % | |||
| 4-Pregnen-11β,17,21-triol-3,20-dione | Cortisol | NADH | 100.00 ± 0.77a |
| 4-Pregnen-17,21-diol-3,20-dione | 11-Deoxycortisol | NADH | 66.03 ± 0.75 |
| 4-Pregnen-11β,21-diol-3,20-dione | Corticosterone | NADH | 39.32 ± 0.25 |
| 5β-Pregnan-3α,11β,17,21-tetrol-20-one | Tetrahydrocortisol | NADH | 44.29 ± 0.29 |
| 4-Pregnen-11β,17,20β,21-tetrol-3-one | 20β-Dihydrocortisol | NAD+ | 0.23 ± 0.04 |
| 4-Pregnen-17,20β-diol-3-one | 17α,20β-Dihydroxyprogesterone | NAD+ | 0.09 ± 0.05 |
| 4-Pregnen-11β,20β,21-triol-3-one | 20β-Dihydrocorticosterone | NAD+ | –b |
| 4-Pregnen-17,20α-diol-3-one | 17α,20α-Dihydroxyprogesterone | NAD+ | – |
| 4-Pregnen-11β,17,20α,21-tetrol-3-dione | 20α-Dihydrocortisol | NAD+ | – |
a Values represent the means ± S.D. from three or more replications.
b – means no activity was detected.
Overall crystallographic structure of 20β-HSDH
The apo crystal structure of r20β-HSDH was solved using molecular replacement with the structure of an SDR from Mycobacterium marinum (PDB code 3R1I) and refined at a resolution of 2.2 Å (Rw = 0.1960 and Rf = 0.2310) (Table 3). The tertiary structure contains motifs characteristic of the SDR superfamily, including a Rossmann fold consisting of seven central β-strands (β3–β2–β1–β4–β5–β6–β7) flanked by six α-helices (Fig. 5A, left). The structure shows two monomers in the asymmetric unit that align with an RMSD of 0.339 Å. The largest differences between the monomers are the 2 Å shift in the α4 helix near the β4–α4 loop, the length of the β4, β5, and β6 strands, and the α6 helix. These differences are likely due to the flexibility of the adjacent loops that could not be modeled in either monomer and may play a role in substrate binding. The monomer with the most unambiguous density in this region is shown in Fig. 5A. In addition to the Rossman fold, r20β-HSDH contains a partial α-helical N terminus of ∼40 residues that extends outward from the core protein structure and interacts with other monomers when the protein is in its tetramer formation (Fig. 5, A, right, and B).
Table 3.
Crystallographic data collection and refinement statistics
| Structure | Apo | NADH-bound |
|---|---|---|
| PDB code | 6M9U | 6OW4 |
| Resolution range (Å) | 50.69–2.2 (2.279–2.2) | 103.71–2 (2.045–2) |
| Space group | I 4 2 2 | C 1 2 1 |
| Unit cell | 168.371 168.371 126.961 90 90 90 | 165.078 134.71 96.396 90 100.15 90 |
| Total no. of reflections | 338,597 (33468) | 353,360 (16341) |
| No. of unique reflections | 46,287 (4574) | 73,226 (3662) |
| Multiplicity | 7.3 (7.3) | 4.8 (4.5) |
| Completeness (%) | 99.57 (99.72) | 54.3 (8.7) |
| Mean I/σ (I) | 9.52 (1.66) | 6.3 (1.7) |
| Wilson B-factor | 41.11 | 16.22 |
| R-merge | 0.140 (1.715) | 0.147 (0.806) |
| R-meas | 0.151 (1.845) | 0.178 (0.976) |
| R-pim | 0.055 (0.673) | 0.072 (0.412) |
| CC1/2 | 0.998 (0.492) | 0.995 (0.670) |
| Reflections used in refinement | 46,144 (4561) | 69,583 (174) |
| Reflections used for R-free | 1997 (198) | 3673 (9) |
| R-work | 0.196 (0.381) | 0.187 (0.279) |
| R-free | 0.231 (0.410) | 0.238 (0.146) |
| No. of nonhydrogen atoms | 4192 | 17,642 |
| Macromolecules | 3853 | 16,839 |
| Ligands | 179 | 352 |
| Solvent | 160 | 451 |
| Protein residues | 501 | 2227 |
| RMSD (bonds) | 0.011 | 0.013 |
| RMSD (angles) | 1.46 | 1.64 |
| Ramachandran favored (%) | 96.70 | 94.43 |
| Ramachandran allowed (%) | 3.09 | 5.44 |
| Ramachandran outliers (%) | 0.21 | 0.14 |
| Rotamer outliers (%) | 3.24 | 0.80 |
| Clashscore | 6.46 | 2.37 |
| Average B-factor | 59.99 | 26.73 |
| Macromolecules | 58.89 | 26.85 |
| Ligands | 84.00 | 25.91 |
| Solvent | 59.50 | 23.01 |
We attempted to crystallize the ternary complex of NADH and cortisol bound to the inactive S181A mutant of r20β-HSDH (Table 1 and Fig. S1). Crystals diffracted to 2.0 Å (Rw = 0.187 and Rf = 0.238) and show NADH clearly bound in the active site, but no density for cortisol is seen (Fig. S2, A and B). These crystals were of the C121 space group, with eight monomers in the asymmetric unit (Table 3). The monomers showed little variation and aligned with RMSD values ranging from 0.14 to 0.24 Å. The largest difference between monomers is the helical character of the N-terminal extension and a 1 Å shift in a flexible loop (residues 235–245) close to where cortisol is predicted to bind (Fig. S2C). This loop is near the α6 helix and was not resolved in the apo structure. The inability to fully resolve this loop in all monomers suggests flexibility when cortisol is not bound.
The NADH was bound within a deep pocket and makes five side-chain and three peptidyl backbone hydrogen-bonding or electrostatic interactions (Fig. 5, B–D). The carboxylic acid of Asp-101 is positioned 2.9 Å away from the adenine nucleotide, whereas the carboxylic acid of Asp-72 is positioned 2.5 and 2.8 Å from the O2 and O3 hydroxyls of the adenine ribose, respectively. The side-chain hydroxyl of Ser-235 is 2.5 Å from the α-phosphate group as well as within 3.3 Å of the carboxamide N of the nicotinamide. The Nϵ of Lys-204 coordinates the O2 and O3 (3.3 and 3.1 Å) of the nicotinamide ribose, and Tyr-200, part of the catalytic tetrad, is directed toward the N of the pyridine ring and the adjacent ribose (3.4 and 2.6 Å). Additionally, the O3 of the nicotinamide ribose interacts with the peptidic N of Gly-130 (3.5 Å) and peptidic O of Asn-128 (2.7 Å), whereas the nicotinamide carboxamide is positioned 2.9 and 3.0 Å from Val-233 (Fig. 5C). van der Waals interactions are seen between NADH and Cys-100, Val-102, the Asn-128 side chain, Val-131, Thr-179, Gly-231, Ile-237, and Phe-238 (yellow in Fig. 5D) and, most notably, Gly-48, -51, and -52, which fall within the Gly-rich region characteristic of SDR family enzymes (pink in Fig. 5D).
The overall structure and NADH-binding pocket of r20β-HSDH are similar to the NADH-bound structure of 3α,20β-HSDH from S. hydrogenans, which is the only structure of an SDR enzyme with known 20β-HSDH activity (Fig. S2D). The two structures align with an RMSD ∼1 Å for 975 atoms. Significant differences are seen in the nicotinamide position and the enzyme's predicted substrate-binding site, especially around the 20β-HSDH α6-helix, likely allowing for the 3α-HSDH activity seen in the S. hydrogenans enzyme (Fig. S2D). Despite these changes, most of the protein–cofactor interactions are conserved. Notable differences are Arg-16 that interacts with the bis-phosphate in the 3α,20β-HSDH is replaced by a glycine in our structure. In addition, Thr-187 in 3α,20β-HSDH is replaced by Ser-235 in our structure and is too far (>3.5 Å) from the cofactor to make a meaningful interaction. Except for Arg-16 in 3α,20β-HSDH, the Gly-rich region characteristic of SDR enzymes provides a similar docking site for the adenine side of NADH (GAAGGLG in 20β-HSDH and GGARGLG in 3α,20β-HSDH), and the catalytic tetrad is completely conserved (Fig. S2D).
Although the apo structure aligns well with the NADH-bound form (RMSD 0.49–0.56 Å across the NADH-bound monomers), significant rearrangements occur near the active site to accommodate NADH binding (Fig. S2E). The β3 to α3 loop shifts up to 1.4 Å, allowing Asp-101 to interact with the adenine ring. Most striking is the loss of the discrete secondary structure toward the end of β4 and β5 allowing the nicotinamide ribose to sit in the space occupied by the 5th β-sheet in the apo structure. Along with the shortening of β4 and β5, the α4 and α5 helices shift ∼2 and 3 Å, respectively. Movement of α5 allows a 3.8 Å shift of Tyr-200, which then interacts with the nicotinamide and the adjacent ribose. Neither the loops between β4 and α4 nor between β5 and α5 were resolved in the apo structure; however, these loops were well-defined in the NADH-bound structure and may help form the binding site for cortisol. Most significantly, the shortening of β4 allows the loop connecting to α5 to swing toward the α6 helix and C terminus of the protein. This movement elongates the binding cleft and positions Ser-181 (Ala in our NADH-bound structure) for potential interaction with cortisol.
Catalytic residues and predicted binding of NADH and cortisol
The conserved catalytic tetrad (NSYK), characteristic of the SDR superfamily, is conserved in 20β-HSDH (green in Fig. S3). Mutation of the putative catalytic serine (Ser-181) and tyrosine (Tyr-200) to an alanine completely abolished activity based on both spectrophotometric assay and TLC separation of overnight reaction products (Table 1 and Fig. S1A). Wildtype (WT) r20β-HSDH was incubated with cortisol, forming 20β-dihydrocortisol. 11β-OHAD showed no observable conversion, confirming B. adolescentis 20β-HSDH does not have 3α-HSDH activity (Fig. S1A). To determine whether cortisol binds to the S181A mutant, we performed isothermal titration calorimetry (ITC) in the presence of NADH (2 mm) or a combination of NADH + cortisol (1 mm) (Fig. S1B). NADH bound tightly, with a Kd of 28.97 μm. However, cortisol was shown to bind with much less affinity, suggesting that Ser-181 is important for cortisol binding and explaining the absence of cortisol binding in attempts to obtain the ternary complex. The S183A mutant retained 56% activity relative to the WT. The Y200A mutant showed no activity, and ITC experiments with 2 mm NADH suggest loss of binding, confirming the importance of Tyr-200 in hydrogen bonding to the nicotinamide nitrogen. Comparison of circular dichroism (CD) spectra between WT and 20β-HSDH mutants indicate these alanine substitutions did not significantly affect secondary structural elements (Fig. S1C).
Extended N terminus is important for secondary and quaternary structures
In both the apo and holo structures of r20β-HSDH, we were not able to fully resolve the N terminus (Fig. 5A, right). In the most complete monomers found in the NADH-bound structure, there was no electron density for the first 10 residues, suggesting a flexible N terminus. Other SDR family proteins whose structures have been solved, and which contain a large, extended N-terminal region, include a quinuclidinone reductase (RrQR) from Rhodotorula rubra (24) and an S-specific carbonyl reductase from Candida parapsilosis (25). There was no electron density for the first six amino acid residues of RrQR, indicating a flexible region. An extended structure was observed from residues 7–19 and observed hydrophobic stacking between Pro-10 and Tyr-229 of the protomer indicating possible quaternary stability (24). The S-specific carbonyl reductase from C. parapsilosis is another example of a pyridine nucleotide-dependent SDR member that contains an extended N-terminal peptide. Comparison between WT and the N-terminally truncated mutant form (Asp-31) showed no observable change in catalytic activity, and only a marginal decrease in tetrameric stability in the truncated form (25). To assess the magnitude of the extended N-terminal abundance throughout the SDR family, a representative full-sequence alignment was produced (Fig. S4). The B. adolescentis 20β-HSDH sequence and 555 sequences annotated as SDR family with predicted HSDH function and bacterial in origin were obtained from the UniProt database. Within these predicted bacterial HSDHs, about 10% (39/556) have an extended N terminus of greater than or equal to 10 amino acid residues. The extended N-terminal regions were not conserved throughout the alignment. Based on this subset of SDR sequences along with the functionally different quinuclidinone reductase and carbonyl reductase, a significant portion of SDR family members likely have an extended N terminus.
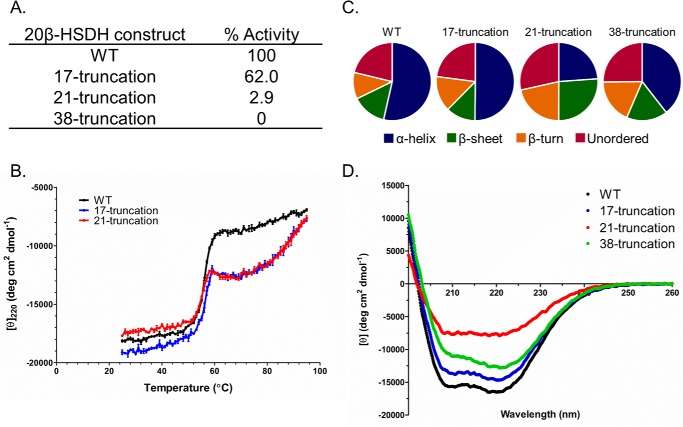
The functional role, if any, of this N-terminal region of 20β-HSDH and the nearest homologs are understudied. We generated the N-terminal truncations of varying lengths (r20β-HSDHΔ1–17, r20β-HSDHΔ1–21, and r20β-HSDHΔ1–38) to determine the effect of truncation on oligomerization, monomeric secondary structure, and enzymatic activity. The locations of these truncations are indicated on the structure in Fig. 5A, right, although there is no density for any residues before 17 in the structure. The purified r20β-HSDH and mutants were separated by gel-filtration chromatography, resulting in a calculated mass of 130.5 ± 2.8 kDa (subunit 31.69 kDa) for r20β-HSDH–WT, 123.2 ± 5.8 (subunit 29.9 kDa) for r20β-HSDHΔ1–17, 95.1 ± 8.6 (subunit 29.4 kDa) for r20β-HSDHΔ1–21, and 43.4 ± 5.4 (subunit 28.7 kDa) for r20β-HSDHΔ1–38 (Fig. S5). r20β-HSDHΔ1–17 is predicted to be tetrameric much like r20β-HSDH–WT, which was supported by the crystal structure (Fig. 5B). However, r20β-HSDHΔ1–21 and r20β-HSDHΔ1–38 were estimated to be trimeric and a mix of dimeric or monomeric forms, respectively.
We next determined the effect of extended N-terminal truncation on enzyme activity and thermal stability. Enzymatic activities of truncated derivatives were compared relative to the WT (Fig. 6A). r20β-HSDHΔ1–17 retained 62% activity when cortisol was the substrate and NADH co-substrate relative to r20β-HSDH–WT. Temperature-dependent CD of r20β-HSDHΔ1–17 resulted in a transition temperature (ΔTm) of 55.70 ± 0.099 °C (Fig. 6B). r20β-HSDHΔ1–21 retained only 2.9% activity relative to r20β-HSDH–WT with a ΔTm of 54.7 ± 0.052 °C. r20β-HSDHΔ1–38 lost complete enzyme activity, and the CD of r20β-HSDH–WT versus r20β-HSDHΔ1–38 resulted in a shift in transition temperature from 56.16 ± 0.10 to 43.3 ± 0.11 °C. These results indicate an important role of the 38-residue extended N terminus in protein stability as deletion of the first 21 residues effectively abolished enzyme activity with a 1.45 °C shift in Tm.
Figure 6.
CD analysis and relative activity of WT and truncated 20β-HSDH. A, relative activity based on spectrophotometric assay. B, thermal stability of WT, 17-, and 21-truncated 20β-HSDH by temperature-dependent CD. C, percentage secondary structure calculated with DichroWeb according to D and CD spectra of WT and truncated proteins.
CD in the far-UV region was performed to assess the impact of N-terminal truncation on secondary protein structure (Fig. 6, C and D). CD spectra were analyzed using DichroWeb, producing relative composition of α-helices, β-sheets, β-turns, and unordered regions. The relative amount of α-helix was reduced in each of the truncated proteins compared with r20β-HSDH–WT. r20β-HSDHΔ1–21 composition was most different with ∼10% increase in β-sheet and β-turn, and 30% decrease in α-helix compared with WT. The percent of unordered regions was largely stable across all truncated proteins (21.3–28.3%). These results indicate an important role of the extended N terminus in overall protein stability illustrated by changes in secondary structure, depletion of enzyme activity, and loss of oligomerization (Fig. S6).
Discussion
Bacterial 20β-HSDH is an NADH-dependent oxidoreductase belonging to the SDR family and is expressed by some strains of B. adolescentis (9). The gene encoding NAD(H)-dependent 20β-HSDH from B. adolescentis strain L2-32 was cloned, overexpressed, and structurally characterized in the apo and NADH-bound forms. The 20β-HSDH crystal structure had a tetrameric configuration with the catalytic tetrad and coenzyme-binding domains characteristic of the SDR family (Fig. 5) (15). Ser-181 is predicted to stabilize cortisol, and mutation to an alanine results in loss of substrate binding. The enzymatic reaction is proposed to initiate as a proton transfer from Ser-181 to the C-20 carbonyl of cortisol. Next, a hydride is predicted to be transferred to C-20 from NADH. From there, a subsequent proton relay likely occurs that follows the order: NADH → Ser-181, Tyr-200 → 2′OH ribose, and bulk solvent → Tyr-200. Determination of the structure of the ternary complex will be critical to confirm the catalytic mechanism and cortisol binding. Multiple attempts to crystallize with both NADH and cortisol bound at varying concentrations were unsuccessful. A possible avenue for future experiments is to use NAD+ and 20β-dihydrocortisol rather than NADH and cortisol. Alternatively, a cortisol derivative such as carbenoxolone could be utilized. Carbenoxolone was bound in the S. hydrogenans 3α,20β-HSDH structure, although this substrate seemed to inhibit NADH binding as well (26).
Short-chain dehydrogenase/reductase family enzymes are extremely diverse, currently spanning 266,639 entries in the Uniprot database. SDR family proteins recognize diverse substrates, from steroids to sugars, alcohols, and xenobiotics (16). At present, there are 475 SDR family structure entries in PDB and about 216 of those are HSDHs. To determine how widespread this extended N terminus may be, a full sequence alignment was created composed of bacterial HSDHs in the SDR family from the UniProt database (Fig. S4). A total of 555 predicted HSDH sequences were compiled, along with the B. adolescentis 20β-HSDH sequence, and 39 of them exhibited an extended N terminus with an unconserved amino acid sequence. Many of these 39 sequences were only annotated based on inferred homology, except for 053547_MYCTU and 3BHDP_RUMGV. The probable SDR dehydrogenase/reductase and the possible 17β-hydroxysteroid dehydrogenase, 053547_MYCTU, were first identified from the complete genome sequence of Mycobacterium tuberculosis strain H37Rv, although the function has not been experimentally determined (27). Notably, 3BHDP_RUMGV in Ruminococcus gnavus ATCC 29149 was functionally studied and shown to have bile acid 3β-HSDH activity by TLC and GC coupled to MS (5). Unfortunately, neither of these proteins have been crystallized and can corroborate the extended N termini demonstrated by sequence alignment. Nonetheless, extended N-terminal regions seem to be frequent (∼10%) across putative bacterial HSDHs and are likely prevalent throughout the entire SDR family, as well. Takeshita et al. (24) noted the flexible N terminus in a quinuclidinone reductase, but it was not studied in depth (PDB code 4O0L, Fig. S3). Zhang et al. (35) investigated a carbonyl reductase extended N terminus in more detail and concluded that it conferred some stability to the oligomer; however it did not impact enzymatic activity (PDB code 3CTM, Fig. S3). This contrasts the extensive analysis performed in this study on the B. adolescentis 20β-HSDH N terminus, which displays complete abolition of activity when fully truncated, as well as marked changes in oligomerization and secondary structure. Fig. S6 represents a potential model for the effect of N-terminal truncation on r20β-HSDH structure/function where truncation of the first 17 amino acid residues resulted in 40% loss of activity relative to WT while retaining a tetrameric structure. Truncation of the next four residues (r20β-HSDHΔ1–21) resulted in a precipitous drop in enzymatic activity to 2% of WT and exhibited a trimeric configuration. Finally, the full truncation of the 38-residue extended N terminus resulted in complete loss of activity and a mixed dimeric and monomeric form. Future studies could utilize targeted site-directed mutagenesis to determine which residues along the flexible N terminus, specifically between residues 17 and 21, are essential for stability. The role of the N-terminal region in 20β-HSDH stability and activity could have broad impacts across proteins of diverse functional classes within the vast SDR family.
The corticosteroids β-cortol and β-cortolone have been identified in human urine samples (28, 29). β-Cortol is formed from tetrahydrocortisol, and β-cortolone is produced from tetrahydrocortisone, both by the proposed action of a human 20β-HSDH (28). Recent work identified at least one enzyme, CBR1, with NADPH-dependent 20β-HSDH activity (20). It was determined that 20β-dihydrocortisol displayed weak glucocorticoid receptor agonism, indicating that 20β-dihydrocortisol formation by gut bacteria may have important physiological effects on the host. In zebrafish, Danio rerio, the gene encoding 20β-HSDH is distinct from that in mammals (30). Zebrafish 20β-HSDH type 2 catalyzes the conversion of cortisone to 20β-hydroxycortisone, which is important in zebrafish stress response through inactivation and excretion of both endogenous and exogenous cortisol (30). Importantly, bacterial 20β-HSDH enzymes are becoming more well-studied, as evidenced by previous work in B. desmolans (9) and the current study in B. adolescentis. Further work will be needed to determine the contribution of gut bacteria to formation of 20β-derivatives of human glucocorticoids.
B. adolescentis is among the first bacteria to colonize the gastrointestinal tract of breast-fed infants; the species was first isolated from the stool of a healthy 2-year-old infant in 1996 (31, 32). The presence of B. adolescentis has been associated with numerous health benefits; however, the mechanisms by which this species and other bifidobacteria confer these benefits on the host have yet to be fully understood. There are currently 31 known species of bifidobacteria (33) with few among a limited selection of commonly used probiotic organisms (34). B. adolescentis strain L2-32 encoding a 20β-HSDH has the potential to be a novel therapeutic probiotic based on the important regulatory role of 20β-HSDH in the steroid-17,20-desmolase pathway (9).
Bacterial steroid-17,20-desmolase (DesAB) converts cortisol into 11β-OHAD (Fig. 1) (8). This compound is pro-androgenic due to the ability of both host (35) and microbial (6) enzymes to biotransform it into 11-ketotestosterone (11-KT). The androgen 11-KT has been shown previously to activate androgen receptor (AR) with similar effectiveness to testosterone itself (11). AR is a ligand-dependent transcription factor that regulates genes for cell proliferation and death (36). This recently discovered pro-androgenic microbe-mediated cortisol metabolism could directly impact androgen-dependent diseases, such as castration-resistant prostate cancer and polycystic ovary syndrome (12).
Bacterial 20β-HSDH can be thought of as acting as an enzymatic “switch” by converting cortisol to 20β-dihydrocortisol, either regulating bacterial cortisol 21-dehydroxylation or effectively blocking the steroid-17,20-desmolase pathway (Fig. 1) (9). B. desmolans and a urinary isolate, Propionimicrobium lymphophilum, have both 20β-HSDH and steroid-17,20-desmolase activity (9), whereas C. scindens expresses steroid-17,20-desmolase and 20α-HSDH (8). B. adolescentis encodes only 20β-HSDH; therefore, B. adolescentis could be a potential inter-species modulator in gut environments with steroid-17,20-desmolase and 20α-HSDH activities. Another competing cortisol biotransformation is 21-dehydroxylase activity in Eggerthella lenta (37). E. lenta lacks both 20α-HSDH and 20β-HSDH, and modification of the cortisol/corticosterone side chain blocks 21-dehydroxylation (38). 21-Dehydroxylation of cortisol produces 21-deoxycortisol, a potent inhibitor of the host kidney 11β-hydroxysteroid dehydrogenase-2 (11β-HSD2) enzyme (23). Inhibition of 11β-HSD2 was shown to cause severe hypertension in humans with an inactivating mutation in this gene (6). Thus, a B. adolescentis probiotic with the 20β-HSDH “switch” could alleviate the inhibitory effects of 21-deoxycortisol by shifting cortisol metabolism toward 20β-dihydrocortisol. The B. adolescentis 20β-HSDH is about 400 times more active in the reductive direction with cortisol as substrate, the preferred direction for a probiotic that seeks to reduce androgen excess or concentrations of 21-deoxycortisol. However, the solved crystal structure supports targeted mutagenesis of binding residues to optimize activity in either direction. Future in vivo investigations are needed to assess the efficacy and safety of 20β-HSDH–encoding B. adolescentis strain L2-32 as a probiotic organism.
Materials and methods
Bacterial strains, enzymes, reagents, and materials
B. adolescentis, strain L2-32, was purchased through BEI Resources (catalog no. HM-633). Turbo Competent E. coli DH5α (high efficiency) cells were purchased from New England Biolabs (Ipswich, MA). BL21-CodonPlus (DE3)-RIPL–competent E. coli cells were purchased from Agilent Technologies (catalog no. 230280). Phusion High-Fidelity DNA polymerases, restriction endonucleases, and T4 DNA ligase were purchased from New England Biolabs. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was purchased from Gold Biotechnology. TALON His-tag purification resin was purchased from Takara Bio USA, Inc. Corning Spin-X UF concentrators with a 10-kDa MWCO were purchased from MilliporeSigma. Steroids were purchased from Steraloids. All other materials were purchased from Thermo Fisher Scientific and MilliporeSigma.
Whole-cell steroid conversion assay
B. adolescentis strain L2-32, B. desmolans ATCC 43058, and C. scindens ATCC 35704 were cultivated in anaerobic brain heart infusion (BHI) broth. Steroids were dissolved in methanol and added to sterile anaerobic BHI medium at a concentration of 50 μm. The samples were then seeded separately with 0.1 ml of each bacterial culture and incubated at 37 °C for 48 h. After incubation, the products were extracted by vortexing the samples with 2 volumes of ethyl acetate for 3 min followed by recovery of the organic phase. The organic phase was then evaporated under nitrogen gas. The remaining residues were dissolved in 100 μl of methanol and analyzed using a high-performance LC (HPLC) system (Shimadzu, Japan) equipped with a C18 analytical column (Capcell Pak c18, Shiseido, Japan). The mobile phase contained acetonitrile/water with 0.01% formic acid, and the flow rate was maintained at 0.2 ml·min−1. A DAD detector was used at a wavelength of 254 nm. Peak retention times and peak areas of samples were compared with standard steroid molecules.
Cloning and formation of the 20β-HSDH expression vector
Genomic DNA was extracted and purified from B. adolescentis, strain L2-32, using the FastDNA spin kit from MP Biomedicals. The gene encoding 20β-HSDH (WP_003810233.1) was amplified using Phusion High-Fidelity DNA polymerase and the following primers: 5′-ATATATcatatgATGGCAGTGAATCATCGCAGATTCCA-3′ and 5′-ATATATctcgagTTAGAAGACGCTTAGCCGCCATCTAC-3′. The restriction endonuclease sites for NdeI and XhoI are shown in lowercase, respectively. The PCR product was purified and double-digested with NdeI and XhoI restriction endonucleases. The digested PCR product was purified and cloned into the pET-28a(+) (Novagen, Madison, WI) vector using T4 DNA ligase to create the expression construct. The pET-28a(+) vector used had previously been re-engineered by replacing the kanamycin resistance gene with the ampicillin resistance gene. The resulting construct was transformed into Turbo Competent E. coli DH5α (high efficiency) cells by heat-shock at 43 °C for 35 s and outgrown for 1 h in Super Optimal Broth with Catabolite Repression (SOC) (Ipswich, MA) before growing on LB agar plates supplemented with ampicillin (100 μg/ml) for 16 h at 37 °C. Single colonies from the LB agar plates were inoculated into 5 ml of LB medium containing ampicillin (100 μg/ml) and grown overnight at 37 °C at 220 rpm. The cells were then centrifuged at 4000 × g for 10 min at 4 °C, and plasmids were extracted and purified from the pellets using the QIAprep Spin miniprep kit. Both strands of the plasmid DNA were sequenced for confirmation of the insert and absence of mutations using T7 promoter and T7 terminator primers (W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana-Champaign). Once overexpressed, the expected protein should include an N-terminus His6-tag and a thrombin cleavage site.
Site-directed mutagenesis
Plasmids containing the 20β-HSDH gene construct were mutated using the QuikChange Lightning site-directed mutagenesis kit. Primers containing the desired mutations were designed as follows: 5′-CTGATCTTCACAGCCgcgCTGTCCGACACAA-3′ and 5′-TTGTGTCCGGACAGcgcGGCTGTGAAGATCAG-3′ for serine 181 to alanine (S181A); and 5′-TCTTCACAGCCTCCCTGgcgGGACACAACGCGAACTA-3′ and 5′-TATTCGCGTGTGTCCcgcCAGGGAGGCTGTGAAGA-3′ for serine 183 to alanine (S183A). Mutated bases are shown in lowercase. The resulting mutated constructs were transformed into Turbo Competent E. coli DH5α (high efficiency) cells by heat shock at 43 °C for 35 s and outgrown for 1 h in Super Optimal Broth with Catabolite Repression (SOC) (New England Biolabs, Ipswich, MA) before growing on LB agar plates supplemented with ampicillin (100 μg/ml) for 16 h at 37 °C. Single colonies from the LB agar plates were inoculated into 5 ml of LB medium containing ampicillin (100 μg/ml) and grown overnight at 37 °C at 220 rpm. The cells were then centrifuged at 4000 × g for 10 min at 4 °C, and plasmids were extracted and purified from the pellets using the QIAprep Spin miniprep kit. Mutations were confirmed by sequencing of the mutated plasmid using T7 promoter and T7 terminator primers (W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana-Champaign).
Overexpression and purification of WT and mutant 20β-HSDH proteins
For protein expression, recombinant plasmids extracted from the E. coli DH5α cells were transformed into BL21-CodonPlus (DE3)-RIPL Competent E. coli cells by heat shock at 43 °C for 35 s and outgrown for 1 h in Super Optimal Broth with Catabolite Repression (SOC) (New England Biolabs, Ipswich, MA) before growing on LB agar plates supplemented with ampicillin (100 μg/ml) and chloramphenicol (50 μg/ml). After 16 h of growth at 37 °C, five colonies were picked to inoculate 10 ml of LB medium containing ampicillin (100 μg/ml) and chloramphenicol (50 μg/ml) and were grown for 6 h at 37 °C with shaking at 220 rpm. The turbid pre-culture was then added to 1 liter of LB medium containing the same antibiotics and shaken at 37 °C at 220 rpm. At an A600 of 0.6, the culture was induced with 0.25 mm IPTG, after which the culture was incubated at 16 °C at 220 rpm for 16 h. Cells were pelleted by centrifugation at 4000 × g for 30 min at 4 °C. The pellets were resuspended in 25 ml of buffer containing 50 mm Tris-Cl and 300 mm NaCl at pH 7.9. The resuspended cells were homogenized using an EmulsiFlex C-3 homogenizer by passing the samples through the homogenizer four times, and the cell debris was removed by centrifugation at 20,000 × g for 30 min at 4 °C. The recombinant protein in the soluble lysate was then purified using metal chelate chromatography followed by removal of the His6 tag by use of thrombin. The recombinant protein was further purified using anion-exchange chromatography to remove the excess thrombin. The resulting purified protein was analyzed using SDS-PAGE.
TLC
Reaction mixtures were made using 50 μm substrate, 150 μm cofactor, and 5 nm enzyme in 150 mm NaCl, 50 mm Tris-Cl buffer at pH 7.8. They were incubated overnight at room temperature and extracted by vortexing 2 volumes of ethyl acetate twice. The organic layer was recovered and evaporated under nitrogen gas. The products were dissolved in 30 μl of methanol and spotted on a TLC plate (silica gel IB2-F flexible TLC sheet, 20 × 20 cm, 250-μm analytical layer; Avantor Performance Materials, LLC, PA). The steroids were separated with an isooctane/ethyl acetate/acetic acid (5:25:0.2, v/v) mobile phase and visualized by spraying with 50% sulfuric acid in ethanol and heating for 10 min at 100 °C.
Gel-filtration chromatography
Gel-filtration chromatography was performed on a Superose 6 10/300 GL analytical column (GE Healthcare) attached to an ÄKTAxpress chromatography system (GE Healthcare) at 4 °C. The purified protein was concentrated to 10 mg/ml and loaded onto the analytical column equilibrated with 50 mm Tris-Cl and 150 mm NaCl at pH 7.5 and was eluted at a flow rate of 0.3 ml/min. The native molecular mass of the 20β-HSDH proteins were determined by comparing elution volume with that of Gel Filtration Standard proteins (Bio-Rad): thyroglobulin, γ-globulin, ovalbumin, myoglobin, vitamin B12.
Optimal pH determination
The 20β-HSDH pH optimum was measured spectrophotometrically by monitoring the reductive biotransformation of cortisol to 20β-dihydrocortisol as well as the reverse oxidative biotransformation of 20β-dihydrocortisol to cortisol. For the reductive direction, measurements were taken by analyzing the decrease of NADH at 340 nm. For the oxidative direction, measurements were taken by analyzing the increase of NADH at 340 nm. The buffers used in the pH profiling contained 50 mm buffering agent and 150 mm NaCl. Buffering agents used were as follows: sodium acetate trihydrate (pH 3.5–5.5), sodium phosphate monobasic monohydrate (pH 6.5–7.5), Tris (pH 8.0–9.0), glycine (pH 10–11). For the reductive reaction, 50 nm enzyme was added to 50 μm cortisol and 150 μm NADH in 200 μl of buffer. For the oxidative reaction, 50 nm enzyme was added to 50 μm 20β-dihydrocortisol and 150 μm NAD+ in 200 μl of buffer. For each reaction, the decrease or increase in NADH in the reaction mixture was measured continuously for 5 min, and the data were used in calculating the initial velocities.
Enzyme activity assays and determination of kinetic parameters
r20β-HSDH WT and mutant activities were measured spectrophotometrically by monitoring the reductive biotransformation of cortisol to 20β-dihydrocortisol as well as the reverse oxidative biotransformation of 20β-dihydrocortisol to cortisol by measuring the levels of NADH at 340 nm. The standard reaction mixture contained 50 mm sodium acetate trihydrate buffer (pH 5.0), 150 μm cofactor (NADH), 10 nm enzyme and a varied substrate concentration between 5 and 90 μm. GraphPad Prism was used to plot data, create Michaelis-Menten saturation curves, and Lineweaver-Burk plots and to calculate kinetic parameters.
Substrate-specificity assay
A saturating concentration of 50 μm substrate, as determined by the enzyme activity analyses, was used as the concentration for analyzing various other steroid substrates and their activities. Similar to the enzyme activity assay, 20β-HSDH activity was measured spectrophotometrically by monitoring the biotransformation of substrate by measuring the levels of NADH at 340 nm. The standard reaction mixtures contained sodium acetate trihydrate buffer (pH 5.0 or pH 5.5), 150 μm cofactor (NADH or NAD+), 50 nm enzyme, and 50 μm substrate.
CD spectroscopy
Determination of CD spectra for 20β-HSDH WT, its site-directed mutants, and the truncated derivatives was carried out using a J-815 CD spectropolarimeter (Jasco, Tokyo, Japan). Protein samples were prepared at a concentration of 0.2 mg/ml in 10 mm KH2PO4 buffer (pH 7.9). For the measurements, a quartz cell with a path length of 0.1 cm was used. CD scans were carried out at 25 °C from 190 to 260 nm at a speed of 50 nm/min with a 0.1-nm wavelength pitch, with five accumulations. Data files were analyzed on the DICHROWEB on-line server (http://www.cryst.ac.uk/cdweb/html/home.html)4 (39) using the CDSSTR algorithm with reference set 4, which is optimized for analysis of data recorded in the range of 190 to 240 nm. Mean residue ellipticity was calculated using millidegrees recorded, molecular weight, number of amino acids, and concentration of protein.
Thermal denaturation studies were performed by monitoring CD (millidegrees) at 220.5 nm from 25 to 95 °C at a rate of 1 °C/min. Each protein was run in triplicate with 0.2 mg/ml protein and 10 mm KH2PO4 buffer at pH 7.9.
Isothermal titration calorimetry
ITC analysis was performed with a VP-ITC microcalorimeter and 1.94-ml cell volume from MicroCal, Inc. The enzymes were dialyzed with 150 mm NaCl, 50 mm Tris buffer at pH 7.8. The pyridine nucleotide (NADH) was dissolved in this buffer, and cortisol was dissolved in methanol. 50 μm protein was injected with 28 successive 10-μl aliquots of 2 mm NADH at 300-s intervals. The protein and 2 mm NADH were then injected similarly with 1 mm cortisol. The data were fitted to a nonlinear regression model using a single binding site with the MicroCal Origin software. The thermodynamic parameters were calculated using the Gibbs free energy equation (ΔG = ΔH − TΔS) and ΔG = −RTln(Ka).
Crystallization
WT 20β-HSDH crystals were obtained using the PEGRx HT kit from Hampton-Research. Purified 20β-HSDH protein was concentrated between a range of 15 and 20 mg/ml. Sitting drops were prepared consisting of 30 μl of well-condition and 300 nl of protein mixed with 300 nl of well-condition. Crystallization screening was performed in 96-well trays using Art Robbins PHOENIX robot. Plates were placed in 4 °C and single crystals formed after 2 days using condition 69 of PEGRx HT containing 20% v/v 2-propanol, 0.1 m MES monohydrate (pH 6.0), and 20% w/v PEG monomethyl ether 2000. The crystals continued to grow up to day 7. No further optimization was necessary. The crystals were overlaid with paraffin oil to prevent evaporation and 20% ethylene glycol as a cryoprotectant. Crystals were picked and immediately flash-cooled in liquid nitrogen.
For the NADH-bound structure, mutant S181A 20β-HSDH–purified recombinant protein was incubated with 2.5 mm NADH and 0.25 mm or 0.5 mm cortisol for 2 h at 4 °C. Crystals were grown in condition 86 of the Hampton PEG/Ion screen containing 0.05 m citric acid, 0.05 m Bis-tris propane (pH 5.0), and 16% w/v PEG 3350. The condition was then optimized in hanging-drop format using 18–20% w/v PEG 3350. S181A crystals were frozen using the same process outlined for the WT 20β-HSDH.
X-ray data collection, processing, structure determination, and refinement
X-ray data were collected on the ID-D beamline at the Life Sciences Collaborative Access Team of the Advanced Photon Source at Argonne National Laboratory. Data for the apo structure were processed and scaled in HKL2000 (40). The crystal structure of the short-chain type dehydrogenase/reductase from M. marinum (PDB code 3R1I) was selected using the MORDA pipeline; Molrep and Refmac from CCP4 are used in this automation to test and refine initial models (41). The final molecular replacement model was rebuilt in Autobuild of the Phenix package (42). For the NADH-bound structure, data were processed in Autopro applying an anisotropy correction in Staraniso (43, 44). Because of the highly anisotropic data, the completeness for this structure appears low when calculated conventionally (isotropic) but the Staraniso program utilizes ellipsoidal completeness to determine the resolution cutoff. Ellipsoidal completeness for these data are high, 92.1% overall and 63.3% for the outer shell. Additionally, the electron density maps are well-defined as shown in Fig. S2, A and B.
The apo structure was used as the molecular replacement model in Phaser-MR of the Phenix package (45). Each structure was refined via multiple rounds of manual model building in Coot and refinement with Phenix.refine (46, 47). The X-ray processing and refinement statistics are presented in Table 3.
Protein Data Bank accession code
The apo 20β-HSDH structure can be found in the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) under the accession number 6M9U. The NADH-bound structure can be found under the accession number 6OW4.
Multiple sequence alignment
The SDR family sequences were aligned using Clustal Omega (1.2.4) (https://www.ebi.ac.uk/Tools/msa/clustalo/) (48).4 The secondary structural elements were rendered using the ESPript 3.0 web server (49).
Author contributions
H. L. D., R. M. P., and S. M. M. data curation; H. L. D., R. M. P., and N. M. K. software; H. L. D., R. M. P., S. M. M., and Z. W. formal analysis; H. L. D. and R. M. P. validation; H. L. D., R. M. P., and S. M. M. investigation; H. L. D., R. M. P., and S. M. M. visualization; H. L. D., R. M. P., S. M. M., S. D., I. C., N. M. K., and J. M. R. methodology; H. L. D., S. M. M., J. M. R., and Z. W. formal analysis; H. L. D. project administration; H. L. D., R. M. P., S. D., I. C., N. M. K., and J. M. R. writing-review and editing; S. D. and J. M. R. conceptualization; N. M. K., Z. W., I. C., and J. M. R. resources; S. D., N. M. K., and J. M. R. supervision; J. M. R. funding acquisition.
Supplementary Material
Acknowledgments
The following reagent was obtained through BEI Resources, NIAID, National Institutes of Health, as part of the Human Microbiome Project: Bifidobacterium adolescentis, strain L2-32, HM-633.
This work was supported by new faculty Start-up Grant Hatch ILLU-538-916 through the Department of Animal Sciences, University of Illinois at Urbana-Champaign (to J. M. R.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S6.
The atomic coordinates and structure factors (codes 6M9U and 6OW4) have been deposited in the Protein Data Bank (http://wwpdb.org/).
L. Ly, S. Devendran, and J. M. Ridlon, unpublished data.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- HSDH
- hydroxysteroid dehydrogenase
- AKR
- aldo-keto reductase
- MDR
- long/medium chain dehydrogenase/reductase
- SDR
- short-chain dehydrogenase/reductase
- 11β-OHAD
- 11β-hydroxyandrostenedione
- PDB
- Protein Data Bank
- Bis-tris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- RrQR
- quinuclidinone reductase
- 11-KT
- 11-ketotestosterone
- AR
- androgen receptor
- r20β-HSDH
- recombinant 20β-HSDH
- RMSD
- root mean square deviation
- 11-KT
- 11-ketotestosterone
- IPTG
- isopropyl β-d-1-thiogalactopyranoside
- BHI
- brain heart infusion
- ITC
- isothermal titration calorimetry
- CD
- circular dichroism.
References
- 1. Penning T. M. (1997) Molecular endocrinology of hydroxysteroid dehydrogenases. Endocr. Rev. 18, 281–305 10.1210/edrv.18.3.0302 [DOI] [PubMed] [Google Scholar]
- 2. Ridlon J. M., Ikegawa S., Alves J. M., Zhou B., Kobayashi A., Iida T., Mitamura K., Tanabe G., Serrano M., De Guzman A., Cooper P., Buck G. A., and Hylemon P. B. (2013) Clostridium scindens: a human gut microbe with a high potential to convert glucocorticoids into androgens. J. Lipid Res. 54, 2437–2449 10.1194/jlr.M038869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ridlon J. M., Harris S. C., Bhowmik S., Kang D. J., and Hylemon P. B. (2016) Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7, 22–39 10.1080/19490976.2015.1127483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mythen S. M., Devendran S., Méndez-García C., Cann I., and Ridlon J. M. (2018) Targeted synthesis and characterization of a gene cluster encoding NAD(P)H-dependent 3α-, 3β-, and 12α-hydroxysteroid dehydrogenases from Eggerthella CAG:298, a gut metagenomic sequence. Appl. Environ. Microbiol. 84, e02475–17 10.1128/AEM.02475-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Devlin A. S., and Fischbach M. A. (2015) A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat. Chem. Biol. 11, 685–690 10.1038/nchembio.1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morris D. J., and Ridlon J. M. (2017) Glucocorticoids and gut bacteria: “The GALF Hypothesis” in the metagenomic era. Steroids 125, 1–13 10.1016/j.steroids.2017.06.002 [DOI] [PubMed] [Google Scholar]
- 7. Krieger D. T., Allen W., Rizzo F., and Krieger H. P. (1971) Characterization of the normal temporal pattern of plasma corticosteroid levels. J. Clin. Endocrinol. Metab. 32, 266–284 10.1210/jcem-32-2-266 [DOI] [PubMed] [Google Scholar]
- 8. Devendran S., Mythen S. M., and Ridlon J. M. (2018) The desA and desB genes from Clostridium scindens ATCC 35704 encode steroid-17,20-desmolase. J. Lipid Res. 59, 1005–1014 10.1194/jlr.M083949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Devendran S., Méndez-García C., and Ridlon J. M. (2017) Identification and characterization of a 20β-HSDH from the anaerobic gut bacterium Butyricicoccus desmolans ATCC 43058. J. Lipid Res. 58, 916–925 10.1194/jlr.M074914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bokkenheuser V. D., Morris G. N., Ritchie A. E., Holdeman L. V., and Winter J. (1984) Biosynthesis of androgen from cortisol by a species of Clostridium recovered from human fecal flora. J. Infect. Dis. 149, 489–494 10.1093/infdis/149.4.489 [DOI] [PubMed] [Google Scholar]
- 11. Pretorius E., Africander D. J., Vlok M., Perkins M. S., Quanson J., and Storbeck K.-H. (2016) 11-Ketodihydrotestosterone in castration-resistant prostate cancer: potent androgens which can no longer be ignored. PLoS ONE 11, e0159867 10.1371/journal.pone.0159867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pretorius E., Arlt W., and Storbeck K.-H. (2017) A new dawn for androgens: novel lessons from 11-oxygenated C19 steroids. Mol. Cell. Endocrinol. 441, 76–85 10.1016/j.mce.2016.08.014 [DOI] [PubMed] [Google Scholar]
- 13. Bokkenheuser V. D., Winter J., Morris G. N., and Locascio S. (1986) Steroid desmolase synthesis by Eubacterium desmolans and Clostridium cadavaris. Appl. Environ. Microbiol. 52, 1153–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Winter J., Cerone-McLernon A., O'Rourke S., Ponticorvo L., and Bokkenheuser V. D. (1982) Formation of 20β-dihydrosteroids by anaerobic bacteria. J. Steroid Biochem. 17, 661–667 10.1016/0022-4731(82)90568-4 [DOI] [PubMed] [Google Scholar]
- 15. Filling C., Berndt K. D., Benach J., Knapp S., Prozorovski T., Nordling E., Ladenstein R., Jörnvall H., and Oppermann U. (2002) Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J. Biol. Chem. 277, 25677–25684 10.1074/jbc.M202160200 [DOI] [PubMed] [Google Scholar]
- 16. Kallberg Y., Oppermann U., Jörnvall H., and Persson B. (2002) Short-chain dehydrogenases/reductases (SDRs). Coenzyme-based functional assignments in completed genomes. Eur. J. Biochem. 269, 4409–4417 10.1046/j.1432-1033.2002.03130.x [DOI] [PubMed] [Google Scholar]
- 17. Rossmann M. G., Ford G. C., Watson H. C., and Banaszak L. J. (1972) Molecular symmetry of glyceraldehyde-3-phosphate dehydrogenase. J. Mol. Biol. 64, 237–245 10.1016/0022-2836(72)90332-4 [DOI] [PubMed] [Google Scholar]
- 18. Edenharder R., and Schneider J. (1985) 12β-Dehydrogenation of bile acids by Clostridium paraputrificum, C. tertium, and C. difficile and epimerization at carbon-12 of deoxycholic acid by cocultivation with 12α-dehydrogenating Eubacterium lentum. Appl. Environ. Microbiol. 49, 964–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ghosh D., Weeks C. M., Grochulski P., Duax W. L., Erman M., Rimsay R. L., and Orr J. C. (1991) Three-dimensional structure of holo 3α,20β-hydroxysteroid dehydrogenase: a member of a short-chain dehydrogenase family. Proc. Natl. Acad. Sci. U.S.A. 88, 10064–10068 10.1073/pnas.88.22.10064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morgan R. A., Beck K. R., Nixon M., Homer N. Z. M., Crawford A. A., Melchers D., Houtman R., Meijer O. C., Stomby A., Anderson A. J., Upreti R., Stimson R. H., Olsson T., Michoel T., Cohain A., et al. (2017) Carbonyl reductase 1 catalyzes 20β-reduction of glucocorticoids, modulating receptor activation and metabolic complications of obesity. Sci. Rep. 7, 1–11 10.1038/s41598-016-0028-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schöneshofer M., Weber B., and Nigam S. (1983) Increased urinary excretion of free 20α- and 20β-dihydrocortisol in a hypercortisolemic but hypocortisoluric patient with Cushing's disease. Clin. Chem. 29, 385–389 [PubMed] [Google Scholar]
- 22. Kornel L., Miyabo S., Saito Z., Cha R.-W., and Wu F.-T. (1975) Corticosteroids in human blood. VIII. Cortisol metabolites in plasma of normotensive subjects and patients with essential hypertension. J. Clin. Endocrinol. Metab. 40, 949–958 10.1210/jcem-40-6-949 [DOI] [PubMed] [Google Scholar]
- 23. Latif S. A., Pardo H. A., Hardy M. P., and Morris D. J. (2005) Endogenous selective inhibitors of 11β-hydroxysteroid dehydrogenase isoforms 1 and 2 of adrenal origin. Mol. Cell. Endocrinol. 243, 43–50 10.1016/j.mce.2005.08.006 [DOI] [PubMed] [Google Scholar]
- 24. Takeshita D., Kataoka M., Miyakawa T., Miyazono K., Kumashiro S., Nagai T., Urano N., Uzura A., Nagata K., Shimizu S., and Tanokura M. (2014) Structural basis of stereospecific reduction by quinuclidinone reductase. AMB Express 4, 6 10.1186/2191-0855-4-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang R., Zhu G., Zhang W., Cao S., Ou X., Li X., Bartlam M., Xu Y., Zhang X. C., and Rao Z. (2008) Crystal structure of a carbonyl reductase from Candida parapsilosis with anti-Prelog stereospecificity. Protein Sci. 17, 1412–1423 10.1110/ps.035089.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ghosh D., Erman M., Wawrzak Z., Duax W. L., and Pangborn W. (1994) Mechanism of inhibition of 3α,20β-hydroxysteroid dehydrogenase by a licorice-derived steroidal inhibitor. Structure 2, 973–980 10.1016/S0969-2126(94)00099-9 [DOI] [PubMed] [Google Scholar]
- 27. Cole S. T, Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E. 3rd., Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 10.1038/31159 [DOI] [PubMed] [Google Scholar]
- 28. Cho H. J., Kim J. D., Lee W. Y., Chung B. C., and Choi M. H. (2009) Quantitative metabolic profiling of 21 endogenous corticosteroids in urine by liquid chromatography-triple quadrupole-mass spectrometry. Anal. Chim. Acta 632, 101–108 10.1016/j.aca.2008.10.059 [DOI] [PubMed] [Google Scholar]
- 29. Romanoff L. P., Parent C., Rodriguez R. M., and Pincus G. (1959) Urinary excretion of β-cortolone (3α,17α,20β,21-tetrahydroxypregnane-11-one) in young and elderly men and women. J. Clin. Endocrinol. Metab. 19, 819–826 10.1210/jcem-19-7-819 [DOI] [PubMed] [Google Scholar]
- 30. Tokarz J., Norton W., Möller G., Hrabé de Angelis M., and Adamski J. (2013) Zebrafish 20β-hydroxysteroid dehydrogenase type 2 is important for glucocorticoid catabolism in stress response. PLoS ONE 8, e54851 10.1371/journal.pone.0054851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yoshioka H., Fujita K., Sakata H., Murono K., and Iseki K. (1991) Development of the normal intestinal flora and clinical significance in infants and children. Bifidobacteria Microflora 10, 11–17 10.12938/bifidus1982.10.1_11 [DOI] [Google Scholar]
- 32. Tissier H. (1900) Recherche Sur La Flore Intestinale Des Nourissons (Etat normal et pathologique). Ph.D. thesis, Univ of Paris, Paris, France [Google Scholar]
- 33. Lee J.-H., and O'Sullivan D. J. (2010) Genomic insights into Bifidobacteria. Microbiol. Mol. Biol. Rev. 74, 378–416 10.1128/MMBR.00004-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O'Toole P. W., Marchesi J. R., and Hill C. (2017) Next-generation probiotics: the spectrum from probiotics to live biotherapeutics. Nat. Microbiol. 2, 17057 10.1038/nmicrobiol.2017.57 [DOI] [PubMed] [Google Scholar]
- 35. Storbeck K. H., Bloem L. M., Africander D., Schloms L., Swart P., and Swart A. C. (2013) 11β-Hydroxydihydrotestosterone and 11-ketodihydrotestosterone, novel C19 steroids with androgenic activity: a putative role in castration resistant prostate cancer? Mol. Cell. Endocrinol. 377, 135–146 10.1016/j.mce.2013.07.006 [DOI] [PubMed] [Google Scholar]
- 36. D'Errico I., and Moschetta A. (2008) Nuclear receptors, intestinal architecture and colon cancer: an intriguing link. Cell. Mol. Life Sci. 65, 1523–1543 10.1007/s00018-008-7552-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Feighner S. D., Bokkenheuser V. D., Winter J., and Hylemon P. B. (1979) Characterization of a C21 neutral steroid hormone transforming enzyme, 21-dehydroxylase, in crude cell extracts of Eubacterium lentum. Biochim. Biophys. Acta 574, 154–163 10.1016/0005-2760(79)90094-8 [DOI] [PubMed] [Google Scholar]
- 38. Feighner S. D., and Hylemon P. B. (1980) Characterization of a corticosteroid 21-dehydroxylase from the intestinal anaerobic bacterium, Eubacterium lentum. J. Lipid Res. 21, 585–593 [PubMed] [Google Scholar]
- 39. Whitmore L., and Wallace B.A. (2008) Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 89, 392–400 [DOI] [PubMed] [Google Scholar]
- 40. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 41. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., Womack T., and Bricogne G. (2011) Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 10.1107/S0907444911007773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tickle I. J., Flensburg C., Keller P., Paciorek W., Sharff A., Vonrhein C., Bricogne G. (2018) STARANISO (http://staraniso.globalphasing.org/cgi-bin/staraniso.cgi) Global Phasing Ltd., Cambridge, UK [Google Scholar]
- 45. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y. M., Buso N., and Lopez R. (2015) The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43, W580–W584 10.1093/nar/gkv279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.