Abstract
Mesenchymal stem cells (MSCs) are multipotent cells capable of differentiating into adipocytes, chondrocytes, or osteocytes. MSCs secrete an array of cytokines and express the LIFRβ (leukemia inhibitory factor receptor) chain on their surface. Mutations in the gene coding for LIFRβ lead to a syndrome with altered bone metabolism. LIFRβ is one of the signaling receptor chains for cardiotrophin-like cytokine (CLCF1), a neurotrophic factor known to modulate B and myeloid cell functions. We investigated its effect on MSCs induced to differentiate into osteocytes in vitro. Our results indicate that CLCF1 binds mouse MSCs, triggers STAT1 and -3 phosphorylation, inhibits the up-regulation of master genes involved in the control of osteogenesis, and markedly prevents osteoblast generation and mineralization. This suggests that CLCF1 could be a target for therapeutic intervention with agents such as cytokine traps or blocking mAbs in bone diseases such as osteoporosis.
Keywords: bone, cytokine, mesenchymal stem cells (MSCs), osteoblast, osteoporosis, cardiotrophin-like cytokine (CLCF1)
Introduction
CLCF12 was initially identified as a cytokine expressed by immune cells signaling through the LIFR (1, 2). It was later shown to require the soluble cytokine receptor–like protein CRLF1 as chaperone to be efficiently secreted and to be a ligand for the tripartite ciliary neurotrophic factor receptor (CNTFR) comprising CNTFRα and signaling chains gp130 and LIFRβ (3). Mutations inactivating the gene coding for LIFRβ are associated with severe, mostly lethal Stüve–Wiederman syndrome that comprises skeletal manifestations such as bent long bones, reduced bone volume, and osteoporosis (4–6). This indicates roles for cytokines signaling through LIFRβ in the control of bone mineralization and metabolism. Bone phenotypes were, however, not reported in CLCF1-deficient mice or patients with mutations in CLCF1, suggesting that the functions of this cytokine regarding osteogenesis are redundant or different from those of other cytokines signaling though LIFRβ (7–9). In support of the latter hypothesis, CLCF1 was shown to have modest inhibitory effects on osterix expression and mineralization in primary calvarial osteoblast cultures (10). Variations in CLCF1 levels have been recently associated with postmenopausal osteoporosis (11).
Osteogenesis is a complex multistep process, and it is likely that CLCF1 plays a role in cellular transitions. Differentiation of MSCs into osteoblasts can be induced in vitro (12). To further investigate the effect of CLCF1 on osteogenesis, we examined the capacity of CLCF1 to bind, activate JAK/STAT signaling, and regulate osteoblastic differentiation in mouse MSCs.
Results
CLCF1 binds and triggers signaling in MSCs
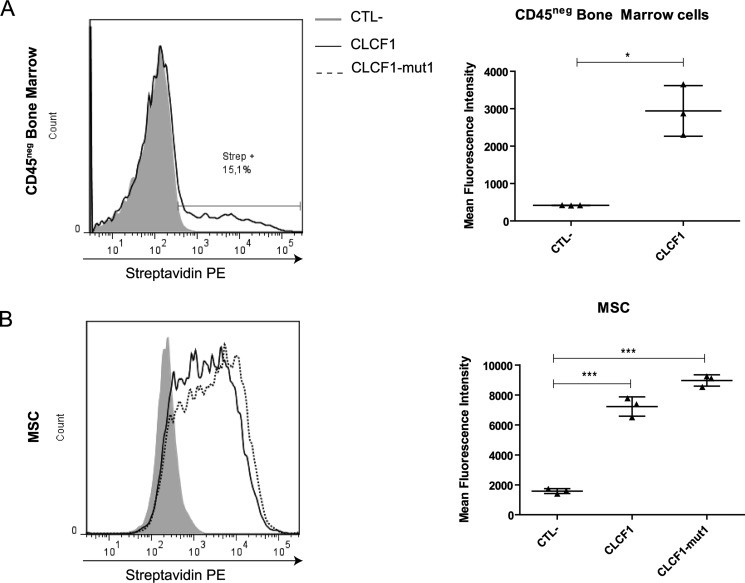
To assess whether CLCF1 has the potential to modulate MSC fate, we analyzed the binding of biotinylated CLCF1 to the nonhematopoietic (CD45−) mouse bone marrow cell fraction that comprises MSCs. Flow cytometry analysis of primary bone marrow cells incubated with biotinylated CLCF1 showed that 15–20% of the CD45− cells bind CLCF1 (Fig. 1A). To investigate the expression of CLCF1 receptors by MSCs, we expanded bone marrow cells under conditions favoring MSC growth to near homogeneity (≥99% CD45− and ≥99% Sca1+; Fig. S1). A distinct binding could be observed on a large fraction of the in vitro–expanded MSCs (Fig. 1B).
Figure 1.
CLCF1 binds to a CD45− population in mouse bone marrow and to MSCs. A, freshly isolated bone marrow cells (1 × 106 cells) were incubated with biotinylated CLCF1 (1 μg/ml) for 1 h and then stained with an allophycocyanin-conjugated anti-CD45.2 mAb and PE-conjugated streptavidin to detect the CLCF1 binding. Fluorescence was measured by flow cytometry. The gray-filled histogram and the black line show the PE-conjugated streptavidin control stain and the CLCF1 binding, respectively, both on the gated CD45neg population. The vertical dot plot shows the mean fluorescence intensity ± S.D. (error bars) of the CLCF1 binding compared with the control staining. Student's t test was used to assess statistical significance. *, p < 0.05; ***, p < 0.001 (n = 3 technical replicates). B, MSCs (1 × 106 cells) were incubated with biotinylated CLCF1 or CLCF1-mut1 (both at 1 μg/ml) for 1 h and then stained with a PE-conjugated streptavidin. Fluorescence was measured by flow cytometry. The gray-filled histogram represents the fluorescence of MSCs incubated with streptavidin alone. The solid and the dotted lines represent the fluorescence of MSCs incubated with CLCF1 + streptavidin and CLCF1-mut1 + streptavidin, respectively. The vertical dot plot shows the mean fluorescence intensity ± S.D. of the CLCF1 and the CLCF1-mut1 bindings compared with the control staining. Student's t test was used to assess statistical significance. *, p < 0.05; ***, p < 0.001 (n = 3 technical replicates).
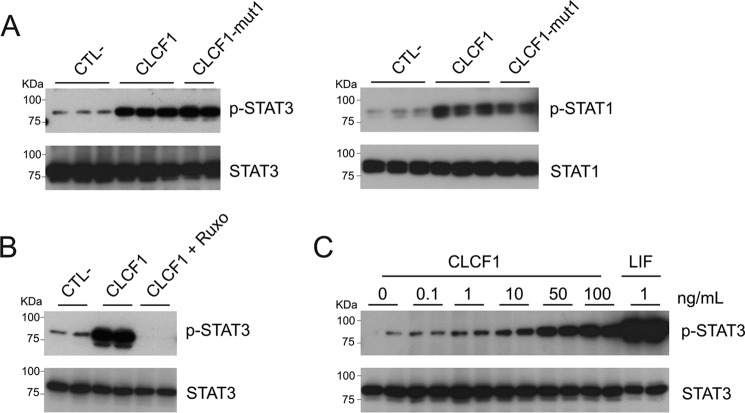
CLCF1 is a ligand for CNTFR (3) as well as for the multiligand receptors sortilin and SorLA (13, 14). MSCs express CNTFR and sortilin (15, 16). We therefore investigated whether CLCF1 could activate JAK/STAT signaling in MSCs. As CNTFR activation by CLCF1 induces STAT1 and STAT3 phosphorylation (3, 17), we focused our investigation on these two transcription factors. Up-regulation of both STAT1 and STAT3 tyrosine phosphorylation could be detected in response to CLCF1, and this up-regulation was inhibited by the JAK inhibitor ruxolitinib (Fig. 2, A–C). We compared CLCF1 with LIF, a cytokine that activates the signaling chains of the CNTFR (18, 19). Dose–response experiments indicate that CLCF1 is less potent than LIF in triggering STAT1 and STAT3 tyrosine phosphorylation (Fig. 2C). Whereas these results did not discriminate between the known CLCF1 receptors, they indicate that CLCF1 induces the activation of a prototypic cytokine JAK/STAT signaling in MSCs that could influence their differentiation.
Figure 2.
CLCF1 activates the phosphorylation of STAT1 and STAT3 in MSCs. A and B, MSCs were stimulated with CLCF1 (100 ng/ml), CLCF1-mut1 (100 ng/ml), or CLCF1 (100 ng/ml) with or without ruxolitinib (lane CLCF1 + Ruxo; 10 μm) for 15 min. Lysates were subjected to Western blot analysis using anti- phospho-STAT3 (p-STAT3), anti-STAT3, anti-phospho-STAT1 (p-STAT1), or anti-STAT1 mAb, respectively. Signals were revealed using horseradish peroxidase–labeled secondary antibody and chemiluminescence. C, MSCs were stimulated with 0, 0.1, 1, 10, 50, or 100 ng/ml CLCF1 or with 1 ng/ml LIF for 15 min. Lysates were analyzed for phospho-STAT3 and total STAT3 levels as described above.
To investigate whether CLCF1 activates MSCs via CNTFR, we used a derivative with site I inactivated by a W94A substitution (20) (CLCF1-mut1). Unlike WT CLCF1, CLCF1-mut1 does not bind or activate Ba/F3 transfectants expressing CNTFR (Fig. S2). We observed that CLCF1-mut1 could still bind MSCs (Fig. 1B) and induce STAT1 and STAT3 phosphorylation (Fig. 2A). To investigate whether CLCF1 binds MSCs via sortilin, we down-regulated sortilin expression in MSCs by RNAi. Whereas sortilin mRNA levels were reduced by 88% in MSCs transfected with sortilin-specific siRNA, no effect on the binding of CLCF1-mut1 could be observed (Fig. S3). Altogether, these results suggest that CLCF1 activates an alternative, so far elusive receptor on MSCs (21).
CLCF1 regulates the expression of transcription factors involved in the control of osteoblast differentiation
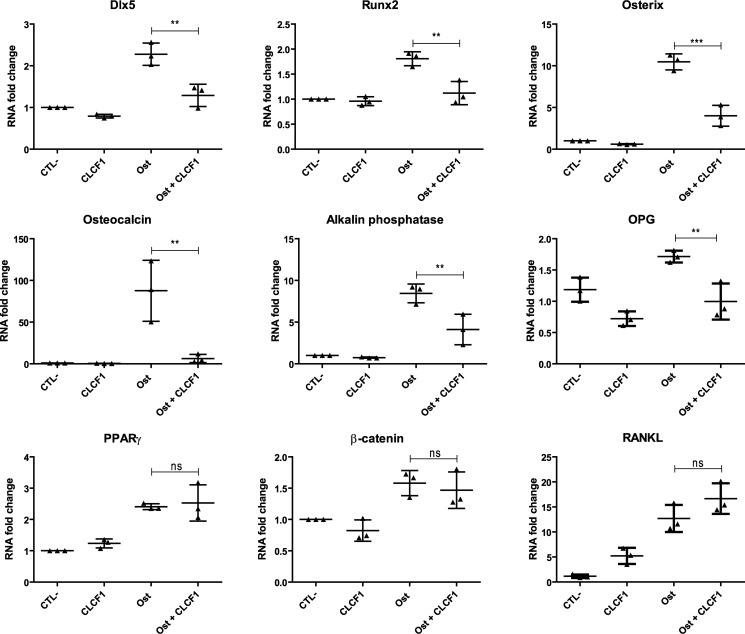
We next examined whether CLCF1 modulates the mRNA levels of transcription factors involved in osteoblast differentiation using quantitative RT-PCR. We compared MSC cultured under conditions inducing osteogenesis in the absence or presence of CLCF1 (Fig. 3). As expected, the mRNA levels of osterix, Runx2, and Dlx5, three transcription factors implicated in osteoblast differentiation (22–24), were strongly up-regulated in MSCs by the osteogenic culture medium (Fig. 3). The up-regulation of these transcription factors was markedly reduced in the presence of CLCF1 (Fig. 3). The observed effect was specific, as the level of the nuclear receptor PPARγ mRNA, involved in adipocyte differentiation (25) was unaffected by the presence of CLCF1 (Fig. 3). We also examined whether CLCF1 regulates the expression of mRNA encoding osteoclastogenesis-related factors. We observed that CLCF1 down-regulated the mRNA levels of osteoprotegrin (OPG), an osteoclastogenesis inhibitory factor (26), in MSCs cultured in either normal or osteogenic conditions. CLCF1 significantly up-regulated the osteoclast differentiation factor RANKL mRNA levels in MSCs expanded in normal medium but not in MSCs maintained in osteogenic conditions (Fig. 3).
Figure 3.
CLCF1 inhibits the induction of osteogenic specific genes in MSCs. MSCs were expanded in AMEM medium (CTL−), AMEM medium supplemented with CLCF1 (100 ng/ml) (CLCF1), osteogenic medium (Ost), or osteogenic medium supplemented with CLCF1 (100 ng/ml) (Ost + CLCF1) for 3 weeks. Dlx5, Runx2, osterix, osteocalcin, alkaline phosphatase, β-catenin, PPARγ, OPG, and RANKL mRNA expression was quantified by RT-qPCR. Results were normalized using the housekeeping gene HPRT mRNA levels. Vertical dot plots indicate mean mRNA -fold changes ± S.D. (error bars). Statistical significance was assessed using analysis of variance. **, p < 0.01; ***, p < 0.001; ns, not significant; n = 3 technical replicates.
CLCF1 inhibits the differentiation of MSCs into osteoblasts
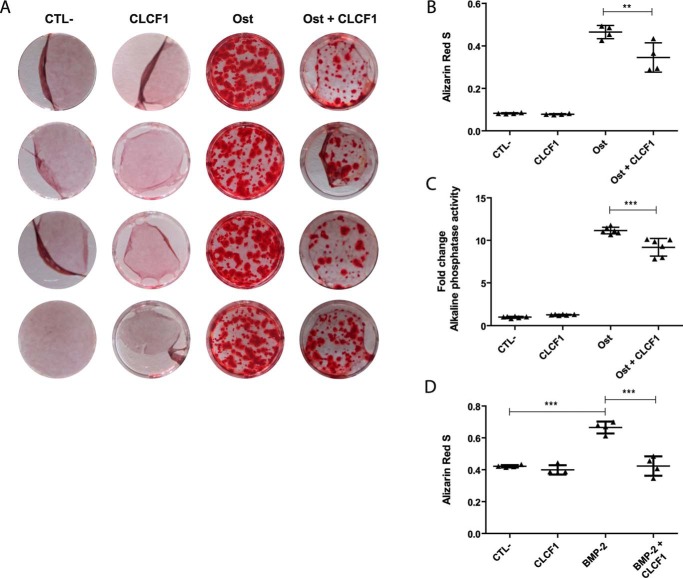
The up-regulation of the mRNA levels of the osteoblast markers alkaline phosphatase and osteocalcin was also reduced when the osteoblast differentiation was induced in the presence of CLCF1 (Fig. 3). To analyze whether this was associated with a reduction of mineralization, we quantified osteogenesis using alizarin red S staining (Fig. 4, A and B). A striking decrease of the staining was observed in MSC cultures in which osteoblast differentiation was induced in the presence of CLCF1. Alkaline phosphatase activity, a marker of osteogenic differentiation involved in bone mineralization (27), was also reduced, albeit less markedly than hydroxyapatite deposit formation assessed by alizarin red S staining (Fig. 4C). CLCF1 also down-regulated the hydroxyapatite deposit formation in MSCs induced to differentiate into osteoblasts using BMP-2 stimulation in additional experiments. (Fig. 4D).
Figure 4.
CLCF1 inhibits MSC differentiation into osteocytes. A, photographs of the alizarin red S–stained cells. MSCs were expanded in AMEM medium (CTL−), AMEM medium with CLCF1 (100 ng/ml) (CLCF1), osteogenic medium (Ost), or osteogenic medium supplemented with CLCF1 (100 ng/ml) (Ost + CLCF1) for 3 weeks. Calcium deposits were stained with alizarin red S. B, alizarin red S was extracted using cetylpyridinium chloride (10%), and the A570 nm was assessed using 1:5 diluted samples. Vertical dot plots represent mean A ± S.D. (error bars), n = 4 technical replicates. C, to quantify alkaline phosphatase activity, MSCs were lysed in 1% Triton X-100 and incubated with p-nitrophenyl phosphate for 10 min. The total protein concentration was used to normalize the values. Vertical dot plots represent mean alkaline phosphatase activity ± S.D., n = 6 technical replicates. Statistical significance was analyzed using analysis of variance. **, p < 0.01; ***, p < 0.001. D, MSCs were expanded in AMEM medium (CTL−), AMEM medium with CLCF1 (100 ng/ml) (CLCF1), BMP-2 osteogenic medium (BMP-2), or BMP-2 osteogenic medium supplemented with CLCF1 (100 ng/ml) (BMP-2 + CLCF1) for 3 weeks. Calcium deposits were stained with alizarin red S and extracted as in B, and the A570 nm was assessed using undiluted samples.
Discussion
We observed that CLCF1 binds MSCs and that CLCF1 promotes STAT1 and STAT3 tyrosine phosphorylation, indicating that these cells respond to this cytokine. We therefore investigated the effect of CLCF1 on the differentiation of MSCs into osteoblasts in vitro. A significant down-regulation of the expression of mRNA coding for osterix, Runx2, and Dlx5, the three transcription factors that regulate osteoblast differentiation (22–24), was detected in MSCs primed for osteogenesis in the presence of CLCF1. CLCF1 also inhibited the expression of osteocalcin and alkaline phosphatase mRNA, two markers of osteoblastic differentiation. We also observed that CLCF1 inhibited expression of OPG, an osteoclastogenesis inhibitory factor (26), by MSCs, indicating that CLCF1, besides modulating osteoblastogenesis, might have a complementary effect on bone metabolism by promoting osteoclast differentiation and therefore bone resorption. In line with these effects, CLCF1 markedly reduced mineralization and alkaline phosphatase activity. Our results are in accordance with previous reports regarding the effect of CLCF1 on osterix expression and mineralization in primary calvarial osteoblasts. The effects of CLCF1 on MSCs were more extensive than those reported (10) on calvarial osteoblast, as Runx2 and osteocalcin mRNA levels were also down-regulated in MSCs. Our observations further indicate that CLCF1, like CNTF, differs from LIF, cardiotrophin-1, oncostatin M, and neuropoietin, the other cytokines signaling through LIFRβ and gp130, in its effect on osteogenesis (28). Our observation that CLCF1 site I inactivation does not prevent MSC activation indicates that the recruited receptor does not comprise CNTFRα. We hypothesized that CLCF1 and CNTF may exert distinctive roles through sortilin, as both bind this alternative receptor (13) expressed by MSCs (15). Binding experiments with MSCs depleted from sortilin mRNA using siRNA transfection suggest that the CLCF1-binding receptor on MSCs is not sortilin either and remains to be identified, as is the one involved in the immunomodulatory roles of this cytokine (21). Nonetheless, CLCF1 binds MSCs, induces JAK/STAT signaling, and regulates MSC differentiation. As MSCs can be induced to generate several cell lineages besides osteoblasts, such as adipocytes, chondrocytes, myocytes, and neuron-like cells (29–32), it will be of interest to investigate whether these processes are also regulated by CLCF1.
CLCF1 requires CRLF1 as a chaperone for efficient secretion. The phenotype of CRLF1 knockout mice suggests that CLCF1 is required for hematopoietic stem cell maintenance (33), a role that could involve bone marrow stromal cells, such as MSCs. CLCF1 administration was shown to promote B cell expansion and myelopoiesis (2, 34, 35). Ligands of CNTFR are believed to have direct and indirect effects on hematopoietic cells, such as B cells (2, 34–36). Factors produced by bone marrow MSCs in response to CLCF1 could contribute to the indirect effect of CLCF1 on hematopoietic cells.
In conclusion, our results indicate that CLCF1 can influence MSC differentiation and confirm that it regulates osteogenesis in vitro, indicating a new facet of activities for this cytokine.
Experimental procedures
Isolation of BM-MSCs
All procedures conformed to the Canadian Council on Animal Care guidelines and were approved by the Animal Ethics Committee of the Université de Montréal (CDEA). Tibiae and femora of C57BL/6 female mice purchased from Charles River Laboratories were dissected and washed with PBS. Bone marrows were flushed using AMEM medium (Wisent Bioproducts, Saint-Jean-Baptiste, Quebec, Canada) containing 10% FBS, 4 mm l-glutamine, 1 IU/ml penicillin, and 100 μg/ml streptomycin. The recovered cells were incubated in a 100-mm Petri dish at 37 °C under a 5% CO2 atmosphere for 5 days. The MSC isolation and expansion protocol was adapted from Huang et al. (37). Briefly, nonadherent cells were washed away with PBS. Adherent cells were detached with trypsin/EDTA and split at a 1:3 ratio into 75-cm2 culture flasks to deplete the hematopoietic cell pool. Cells were passaged at confluence, and purity was assessed by flow cytometry using a FACS Canto II flow cytometer (BD Biosciences). For all experiments, MSCs at passage 7 and above were used.
CLCF1-binding assays
Freshly isolated bone marrow cells from C57BL/6 mice were depleted from red blood cells and incubated for 1 h on ice with biotinylated CLCF1 (38) (1 μg/ml) in PBS containing 1% BSA. Cells were washed and stained with phycoerythrin (PE)-conjugated Streptavidin (554061, BD Biosciences) and allophycocyanin-conjugated anti-mouse CD45.2 (17-0454-82, Thermo Fisher Scientific). The same protocol was used for the CLCF1 and the CLCF1-mut1 (W94A CLCF1 (38)) binding assays on the expanded MSCs without anti-CD45 staining. Fluorescence was detected and quantified by flow cytometry.
Western blot analysis
MSCs were plated at 5 × 104 cells/cm2 overnight and serum-starved for 24 h. MSCs were then stimulated for 15 min with LIF (PeproTech), recombinant murine CLCF1 (100 ng/ml), or recombinant CLCF1-mut1 (100 ng/ml). CLCF1 and CLCF1-mut1 were produced, purified, and tested as described previously (38). When included during the stimulation, ruxolitinib (Selleckchem, Houston, TX) was used at 10 μm. Cells were washed with ice-cold PBS and lysed in radioimmune precipitation assay lysis buffer containing protease (cOmpleteTM, Millipore-Sigma) and phosphatase inhibitors (Thermo Fisher Scientific). Proteins were subjected to PAGE and electrotransferred to polyvinylpyrrolidone blotting membranes. Membranes were sequentially incubated with rabbit antibodies specific for phospho-STAT1 (7649S, Cell Signaling Technology), phospho-STAT3 (4904S, Cell Signaling Technology), STAT1 (9172, Cell Signaling Technology), or STAT3 (9131L, Cell Signaling Technology) and with horseradish peroxidase–conjugated anti-rabbit IgG (R&D Systems). Signals were revealed by chemiluminescence.
Osteogenic differentiation
Once MSCs reached 90% confluence, the osteogenic induction medium was added and replaced every 3–4 days for 3 weeks. The MSCs were differentiated according to Stagg et al. (39). The osteogenic medium consisted of the AMEM culture medium described above supplemented with 0.1 μm dexamethasone, 20 mm β-glycerophosphate, and 200 μm l-ascorbic acid 2-phosphate. The BMP-2 osteogenic medium consisted of the AMEM medium described above supplemented with 100 ng/ml BMP-2 (PeproTech), dexamethasone (10 nm), β-glycerophosphate (5 mm), and l-ascorbic acid 2-phosphate (170 μm) (40, 41). Control wells contained nonsupplemented AMEM medium. CLCF1 was used at a concentration of 100 ng/ml.
Alizarin red S staining and quantification
MSCs incubated for 3 weeks in control or osteogenic medium were washed three times and fixed for 1 h in PBS, 4% formaldehyde. The fixed cells were then washed three times with water, stained in 40 mm alizarin red S (Sigma-Aldrich), pH 4.1–4.3, for 20 min, and washed with water. Staining was analyzed by microscopy. Photos of the wells were taken using a Fujifilm FinePix F770EXR camera. For mineralization quantification, cells were gently shaken at room temperature in 10% cetylpyridinium chloride, 10 mm sodium phosphate, pH 7.0, to extract the red dye (42). The absorbance (A570 nm) of the extracts diluted 1:5 was assessed using a Viktor2 microplate reader (PerkinElmer Life Sciences, Woodbridge, Canada).
Alkaline phosphatase activity
MSCs incubated for 3 weeks in control or osteogenic medium were washed with PBS and lysed in 1% Triton X-100, 10 mm Tris-HCl, pH 7.4. The cell lysates were scraped and subjected to three freeze-thaw cycles. Cell lysate aliquots were incubated at a 5:100 ratio with p-nitrophenyl phosphate liquid substrate (N7653, Sigma-Aldrich). The reactions were stopped using 25 μl of 3 m NaOH, and A405 nm was measured. Protein concentration of cell lysates was quantified using a bicinchoninic acid protein assay kit (Thermo Fisher Scientific).
Quantitative PCR (qPCR)
MSCs incubated for 3 weeks in control or osteogenic medium were detached, and total RNA was isolated using TRIzolTM (Thermo Fisher Scientific). RNA was further purified using RNeasy Mini Kit columns (Qiagen). RNA concentrations were measured using a NanoDrop spectrophotometer (Thermo Fisher Scientific). RNA integrity was assessed using a BioAnalyzer (Agilent Technologies). A high-capacity cDNA reverse transcription kit (Applied Biosystems) was used to generate the cDNA using aliquots of 1 μg of RNA as template. Levels of specific mRNA were quantified with a QuantStudioTM 7 Flex Real-Time PCR System. Results were normalized using the HPRT mRNA levels as an endogenous control that was not regulated by CLCF1. Primer pairs used are indicated in Table 1.
Table 1.
Forward and reverse primers used for RT-qPCR
| Gene | Forward sequence | Reverse sequence |
|---|---|---|
| Alpl | cggatcctgaccaaaaacc | tcatgatgtccgtggtcaat |
| Runx2 | gcccaggcgtatttcaga | tgcctggctcttcttactgag |
| Sp7 | agcaccaatggactcctctc | gggtgggtagtcatttgcat |
| Dlx5 | agcccctaccaccagtacg | gctccgccacttctttctc |
| Bglap2 | agactccggcgctacctt | ctcgtcacaagcagggttaag |
| Ctnnb1 | ccaatggcttggaatgaga | gggatcatcctggcgata |
| Hprt | tcctcctcagaccgctttt | cctggttcatcatcgctaatc |
| Sort1 | cggatatcacgacgactcag | gagcctcagggagtgtagga |
| Gapdh | tgtccgtcgtggatctgac | cctgcttcaccaccttcttg |
Author contributions
J.-F. G., S. P., and S. N. conceptualization; S. N., S. P., V. L., and F. R. formal analysis; J.-F. G., S. N. and M. S. writing-review and editing; J.-F. G. supervision and writing of original draft.
Supplementary Material
Acknowledgments
We thank the Institute for Research in Immunology and Cancer genomic platform for the RT-qPCR. We thank Dr. Moutih Rafei, Yun Cui, and Jamila Abusara (Départment de Pharmacologie et Physiologie, Université de Montréal) for kind help with MSC cultures and for the initial donation of MSCs. We thank Dr. Armelle LeCampion (Flow Cytometry Core Facility, Université de Montréal) for support with flow cytometry.
This work was supported by Canadian Institutes of Health Research (CIHR) Grants MOP-57832 and PTJ 159654 (to J.-F. G.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3.
- CLCF1
- cardiotrophin-like cytokine factor-1
- CNTF
- ciliary neurotrophic factor
- CNTFR
- CNTF receptor
- LIF
- leukemia inhibitory factor
- LIFR
- LIF receptor
- MSC
- mesenchymal stem cell
- PE
- phycoerythrin
- PPAR
- peroxisome proliferator-activated receptor
- OPG
- osteoprotegrin
- STAT
- signal transducers and activators of transcription
- qPCR
- quantitative PCR.
References
- 1. Shi Y., Wang W., Yourey P. A., Gohari S., Zukauskas D., Zhang J., Ruben S., and Alderson R. F. (1999) Computational EST database analysis identifies a novel member of the neuropoietic cytokine family. Biochem. Biophys. Res. Commun. 262, 132–138 10.1006/bbrc.1999.1181 [DOI] [PubMed] [Google Scholar]
- 2. Senaldi G., Varnum B. C., Sarmiento U., Starnes C., Lile J., Scully S., Guo J., Elliott G., McNinch J., Shaklee C. L., Freeman D., Manu F., Simonet W. S., Boone T., and Chang M. S. (1999) Novel neurotrophin-1/B cell-stimulating factor-3: a cytokine of the IL-6 family. Proc. Natl. Acad. Sci. U.S.A. 96, 11458–11463 10.1073/pnas.96.20.11458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Elson G. C., Lelièvre E., Guillet C., Chevalier S., Plun-Favreau H., Froger J., Suard I., de Coignac A. B., Delneste Y., Bonnefoy J. Y., Gauchat J. F., and Gascan H. (2000) CLF associates with CLC to form a functional heteromeric ligand for the CNTF receptor complex. Nat. Neurosci. 3, 867–872 10.1038/78765 [DOI] [PubMed] [Google Scholar]
- 4. Dagoneau N., Scheffer D., Huber C., Al-Gazali L. I., Di Rocco M., Godard A., Martinovic J., Raas-Rothschild A., Sigaudy S., Unger S., Nicole S., Fontaine B., Taupin J. L., Moreau J. F., Superti-Furga A., et al. (2004) Null leukemia inhibitory factor receptor (LIFR) mutations in Stuve-Wiedemann/Schwartz-Jampel type 2 syndrome. Am. J. Hum. Genet. 74, 298–305 10.1086/381715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gaspar I. M., Saldanha T., Cabral P., Vilhena M. M., Tuna M., Costa C., Dagoneau N., Daire V. C., and Hennekam R. C. (2008) Long-term follow-up in Stuve-Wiedemann syndrome: a clinical report. Am. J. Med. Genet. 146A, 1748–1753 10.1002/ajmg.a.32325 [DOI] [PubMed] [Google Scholar]
- 6. Jung C., Dagoneau N., Baujat G., Le Merrer M., David A., Di Rocco M., Hamel B., Mégarbané A., Superti-Furga A., Unger S., Munnich A., and Cormier-Daire V. (2010) Stuve-Wiedemann syndrome: long-term follow-up and genetic heterogeneity. Clin. Genet. 77, 266–272 10.1111/j.1399-0004.2009.01314.x [DOI] [PubMed] [Google Scholar]
- 7. Rousseau F., Gauchat J. F., McLeod J. G., Chevalier S., Guillet C., Guilhot F., Cognet I., Froger J., Hahn A. F., Knappskog P. M., Gascan H., and Boman H. (2006) Inactivation of cardiotrophin-like cytokine, a second ligand for ciliary neurotrophic factor receptor, leads to cold-induced sweating syndrome in a patient. Proc. Natl. Acad. Sci. U.S.A. 103, 10068–10073 10.1073/pnas.0509598103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zou X., Bolon B., Pretorius J. K., Kurahara C., McCabe J., Christiansen K. A., Sun N., Duryea D., Foreman O., Senaldi G., Itano A. A., and Siu G. (2009) Neonatal death in mice lacking cardiotrophin-like cytokine is associated with multifocal neuronal hypoplasia. Vet. Pathol. 46, 514–519 10.1354/vp.08-VP-0239-B-BC [DOI] [PubMed] [Google Scholar]
- 9. Hahn A. F., Waaler P. E., Kvistad P. H., Bamforth J. S., Miles J. H., McLeod J. G., Knappskog P. M., and Boman H. (2010) Cold-induced sweating syndrome: CISS1 and CISS2: manifestations from infancy to adulthood. Four new cases. J. Neurol. Sci. 293, 68–75 10.1016/j.jns.2010.02.028 [DOI] [PubMed] [Google Scholar]
- 10. McGregor N. E., Poulton I. J., Walker E. C., Pompolo S., Quinn J. M., Martin T. J., and Sims N. A. (2010) Ciliary neurotrophic factor inhibits bone formation and plays a sex-specific role in bone growth and remodeling. Calcif. Tissue Int. 86, 261–270 10.1007/s00223-010-9337-4 [DOI] [PubMed] [Google Scholar]
- 11. Ge J. R., Xie L. H., Chen J., Li S. Q., Xu H. J., Lai Y. L., Qiu L. L., and Ni C. B. (2018) Liuwei Dihuang pill treats postmenopausal osteoporosis with shen (kidney) yin deficiency via Janus kinase/signal transducer and activator of transcription signal pathway by up-regulating cardiotrophin-like cytokine factor 1 expression. Chin. J. Integr. Med. 24, 415–422 10.1007/s11655-016-2744-2 [DOI] [PubMed] [Google Scholar]
- 12. Cheng S. L., Yang J. W., Rifas L., Zhang S. F., and Avioli L. V. (1994) Differentiation of human bone marrow osteogenic stromal cells in vitro: induction of the osteoblast phenotype by dexamethasone. Endocrinology 134, 277–286 10.1210/endo.134.1.8275945 [DOI] [PubMed] [Google Scholar]
- 13. Larsen J. V., Hansen M., Møller B., Madsen P., Scheller J., Nielsen M., and Petersen C. M. (2010) Sortilin facilitates signaling of ciliary neurotrophic factor and related helical type 1 cytokines targeting the gp130/leukemia inhibitory factor receptor β heterodimer. Mol. Cell. Biol. 30, 4175–4187 10.1128/MCB.00274-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Larsen J. V., Kristensen A. M., Pallesen L. T., Bauer J., Vægter C. B., Nielsen M. S., Madsen P., and Petersen C. M. (2016) Cytokine-like factor 1, an essential facilitator of cardiotrophin-like cytokine:ciliary neurotrophic factor receptor α signaling and sorLA-mediated turnover. Mol. Cell. Biol. 36, 1272–1286 10.1128/MCB.00917-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maeda S., Nobukuni T., Shimo-Onoda K., Hayashi K., Yone K., Komiya S., and Inoue I. (2002) Sortilin is upregulated during osteoblastic differentiation of mesenchymal stem cells and promotes extracellular matrix mineralization. J. Cell. Physiol. 193, 73–79 10.1002/jcp.10151 [DOI] [PubMed] [Google Scholar]
- 16. Djouad F., Bony C., Häupl T., Uzé G., Lahlou N., Louis-Plence P., Apparailly F., Canovas F., Rème T., Sany J., Jorgensen C., and Noël D. (2005) Transcriptional profiles discriminate bone marrow-derived and synovium-derived mesenchymal stem cells. Arthritis Res. Ther. 7, R1304–R1315 10.1186/ar1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lelièvre E., Plun-Favreau H., Chevalier S., Froger J., Guillet C., Elson G. C., Gauchat J. F., and Gascan H. (2001) Signaling pathways recruited by the cardiotrophin-like cytokine/cytokine-like factor-1 composite cytokine: specific requirement of the membrane-bound form of ciliary neurotrophic factor receptor α component. J. Biol. Chem. 276, 22476–22484 10.1074/jbc.M101681200 [DOI] [PubMed] [Google Scholar]
- 18. Davis S., Aldrich T. H., Stahl N., Pan L., Taga T., Kishimoto T., Ip N. Y., and Yancopoulos G. D. (1993) LIFR β and gp130 as heterodimerizing signal transducers of the tripartite CNTF receptor. Science 260, 1805–1808 10.1126/science.8390097 [DOI] [PubMed] [Google Scholar]
- 19. Ip N. Y., Nye S. H., Boulton T. G., Davis S., Taga T., Li Y., Birren S. J., Yasukawa K., Kishimoto T., and Anderson D. J. (1992) CNTF and LIF act on neuronal cells via shared signaling pathways that involve the IL-6 signal transducing receptor component gp130. Cell 69, 1121–1132 10.1016/0092-8674(92)90634-O [DOI] [PubMed] [Google Scholar]
- 20. Perret D., Guillet C., Elson G., Froger J., Plun-Favreau H., Rousseau F., Chabbert M., Gauchat J. F., and Gascan H. (2004) Two different contact sites are recruited by cardiotrophin-like cytokine (CLC) to generate the CLC/CLF and CLC/sCNTFRα composite cytokines. J. Biol. Chem. 279, 43961–43970 10.1074/jbc.M407686200 [DOI] [PubMed] [Google Scholar]
- 21. Tormo A. J., Letellier M. C., Lissilaa R., Batraville L. A., Sharma M., Ferlin W., Elson G., Crabé S., and Gauchat J. F. (2012) The cytokines cardiotrophin-like cytokine/cytokine-like factor-1 (CLC/CLF) and ciliary neurotrophic factor (CNTF) differ in their receptor specificities. Cytokine 60, 653–660 10.1016/j.cyto.2012.08.014 [DOI] [PubMed] [Google Scholar]
- 22. Heo J. S., Lee S. G., and Kim H. O. (2017) Distal-less homeobox 5 is a master regulator of the osteogenesis of human mesenchymal stem cells. Int. J. Mol. Med. 40, 1486–1494 10.3892/ijmm.2017.3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Otto F., Thornell A. P., Crompton T., Denzel A., Gilmour K. C., Rosewell I. R., Stamp G. W., Beddington R. S., Mundlos S., Olsen B. R., Selby P. B., and Owen M. J. (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89, 765–771 10.1016/S0092-8674(00)80259-7 [DOI] [PubMed] [Google Scholar]
- 24. Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J. M., Behringer R. R., and de Crombrugghe B. (2002) The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108, 17–29 10.1016/S0092-8674(01)00622-5 [DOI] [PubMed] [Google Scholar]
- 25. Hegele R. A., Cao H., Frankowski C., Mathews S. T., and Leff T. (2002) PPARG F388L, a transactivation-deficient mutant, in familial partial lipodystrophy. Diabetes 51, 3586–3590 10.2337/diabetes.51.12.3586 [DOI] [PubMed] [Google Scholar]
- 26. Udagawa N., Takahashi N., Yasuda H., Mizuno A., Itoh K., Ueno Y., Shinki T., Gillespie M. T., Martin T. J., Higashio K., and Suda T. (2000) Osteoprotegerin produced by osteoblasts is an important regulator in osteoclast development and function. Endocrinology 141, 3478–3484 10.1210/endo.141.9.7634 [DOI] [PubMed] [Google Scholar]
- 27. Siffert R. S. (1951) The role of alkaline phosphatase in osteogenesis. J. Exp. Med. 93, 415–426 10.1084/jem.93.5.415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pasquin S., Sharma M., and Gauchat J. F. (2016) Cytokines of the LIF/CNTF family and metabolism. Cytokine 82, 122–124 10.1016/j.cyto.2015.12.019 [DOI] [PubMed] [Google Scholar]
- 29. Kuznetsov S. A., Krebsbach P. H., Satomura K., Kerr J., Riminucci M., Benayahu D., and Robey P. G. (1997) Single-colony derived strains of human marrow stromal fibroblasts form bone after transplantation in vivo. J. Bone Miner. Res. 12, 1335–1347 10.1359/jbmr.1997.12.9.1335 [DOI] [PubMed] [Google Scholar]
- 30. Johnstone B., Hering T. M., Caplan A. I., Goldberg V. M., and Yoo J. U. (1998) In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp. Cell Res. 238, 265–272 10.1006/excr.1997.3858 [DOI] [PubMed] [Google Scholar]
- 31. Wakitani S., Saito T., and Caplan A. I. (1995) Myogenic cells derived from rat bone marrow mesenchymal stem cells exposed to 5-azacytidine. Muscle Nerve 18, 1417–1426 10.1002/mus.880181212 [DOI] [PubMed] [Google Scholar]
- 32. Woodbury D., Schwarz E. J., Prockop D. J., and Black I. B. (2000) Adult rat and human bone marrow stromal cells differentiate into neurons. J. Neurosci. Res. 61, 364–370 [DOI] [PubMed] [Google Scholar]
- 33. Alexander W. S., Rakar S., Robb L., Farley A., Willson T. A., Zhang J. G., Hartley L., Kikuchi Y., Kojima T., Nomura H., Hasegawa M., Maeda M., Fabri L., Jachno K., Nash A., et al. (1999) Suckling defect in mice lacking the soluble haemopoietin receptor NR6. Curr. Biol. 9, 605–608 10.1016/S0960-9822(99)80266-8 [DOI] [PubMed] [Google Scholar]
- 34. Senaldi G., Stolina M., Guo J., Faggioni R., McCabe S., Kaufman S. A., Van G., Xu W., Fletcher F. A., Boone T., Chang M. S., Sarmiento U., and Cattley R. C. (2002) Regulatory effects of novel neurotrophin-1/b cell-stimulating factor-3 (cardiotrophin-like cytokine) on B cell function. J. Immunol. 168, 5690–5698 10.4049/jimmunol.168.11.5690 [DOI] [PubMed] [Google Scholar]
- 35. Pasquin S., Laplante V., Kouadri S., Milasan A., Mayer G., Tormo A. J., Savin V., Sharma M., Martel C., and Gauchat J. F. (2018) Cardiotrophin-like cytokine increases macrophage-foam cell transition. J. Immunol. 201, 2462–2471 10.4049/jimmunol.1800733 [DOI] [PubMed] [Google Scholar]
- 36. Askmyr M., White K. E., Jovic T., King H. A., Quach J. M., Maluenda A. C., Baker E. K., Smeets M. F., Walkley C. R., and Purton L. E. (2015) Ciliary neurotrophic factor has intrinsic and extrinsic roles in regulating B cell differentiation and bone structure. Sci. Rep. 5, 15529 10.1038/srep15529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang S., Xu L., Sun Y., Wu T., Wang K., and Li G. (2015) An improved protocol for isolation and culture of mesenchymal stem cells from mouse bone marrow. J. Orthop. Translat. 3, 26–33 10.1016/j.jot.2014.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pasquin S., Chehboun S., Dejda A., Meliani Y., Savin V., Warner G. J., Bosse R., Tormo A., Mayer G., Sharma M., Sapieha P., Martel C., and Gauchat J.-F. (2018) Effect of human very low-density lipoproteins on cardiotrophin-like cytokine factor 1 (CLCF1) activity. Sci. Rep. 8, 3990 10.1038/s41598-018-22400-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stagg J., Pommey S., Eliopoulos N., and Galipeau J. (2006) Interferon-γ-stimulated marrow stromal cells: a new type of nonhematopoietic antigen-presenting cell. Blood 107, 2570–2577 10.1182/blood-2005-07-2793 [DOI] [PubMed] [Google Scholar]
- 40. Chen C., Uludağ H., Wang Z., and Jiang H. (2012) Noggin suppression decreases BMP-2-induced osteogenesis of human bone marrow-derived mesenchymal stem cells in vitro. J. Cell. Biochem. 113, 3672–3680 10.1002/jcb.24240 [DOI] [PubMed] [Google Scholar]
- 41. Honda Y., Ding X., Mussano F., Wiberg A., Ho C.-M., and Nishimura I. (2013) Guiding the osteogenic fate of mouse and human mesenchymal stem cells through feedback system control. Scientific Reports 3, 3420 10.1038/srep03420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gregory C. A., Gunn W. G., Peister A., and Prockop D. J. (2004) An alizarin red-based assay of mineralization by adherent cells in culture: comparison with cetylpyridinium chloride extraction. Anal. Biochem. 329, 77–84 10.1016/j.ab.2004.02.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.