Abstract
Thiol-based redox regulation via ferredoxin-thioredoxin (Trx) reductase/Trx controls various functions in chloroplasts in response to light/dark changes. Trx is a key factor of this regulatory system, and five Trx subtypes, including 10 isoforms, have been identified as chloroplast-localized forms in Arabidopsis thaliana. These subtypes display distinct target selectivity, and, consequently, they form a complicated redox regulation network in chloroplasts. In this study, we developed a FRET-based sensor protein by combining CFP, YFP, and the N-terminal region of CP12, a redox-sensitive regulatory and Trx-targeted protein in chloroplasts. This sensor protein enabled us to monitor the redox change of chloroplast thioredoxin in vivo, and we therefore designated this protein “change in redox state of Trx” (CROST). Using CP12 isoforms, we successfully prepared two types of CROST sensors that displayed different affinities for two major chloroplast Trx isoforms (f-type and m-type). These sensor proteins helped unravel the real-time redox dynamics of Trx molecules in chloroplasts during the light/dark transition.
Keywords: biosensor, chloroplast, fluorescence, FRET), redox regulation, thioredoxin
Introduction
To survive under fluctuating environmental conditions, green plants have evolved various systems for regulating metabolism. The light condition is an important environmental factor for plant cell metabolism. Indeed, important biological functions, such as carbon dioxide fixation in plants, are adequately regulated under varying light conditions. For this purpose, the thiol-based redox regulation system in chloroplasts is a key regulation system for controlling chloroplast metabolism under fluctuating light conditions. In this regulation system, reducing power produced by water splitting in the light-driven electron transport chain is transferred to thioredoxin (Trx)2 via ferredoxin and ferredoxin-Trx reductase. Trx is a ubiquitous protein possessing a characteristic WCGPC motif as an active center. Via reduction and oxidation of two Cys residues in the motif and the dithiol-disulfide exchange reaction with target proteins, Trx regulates the activities of target proteins by reducing/oxidizing the disulfide bond on their molecular surfaces (1). Therefore, detection of Trx activity in vivo is important for understanding the states of the redox regulation system and various Trx-regulated enzymes in chloroplasts such as ATP synthase (2), four enzymes involved the Calvin–Benson cycle (3), the malate valve (4), glucose 6-phosphate dehydrogenase for the oxidative pentose phosphate pathway (4, 5), and the enzymes involved in chlorophyll biosynthesis (6).
Genome sequencing of the model plant Arabidopsis thaliana revealed that 10 Trx isoforms, classified into five subtypes (f, m, x, y, and z), are localized in chloroplasts. These five subtypes of Trx exhibit different target selectivities and form a complicate redox network, although these Trx isoforms maintain a common structural motif named Trx-fold (7). The target selectivity of Trx isoforms might be determined by their different molecular characteristics, such as the surface charges of the protein molecule (8) and their midpoint redox potentials (9). However, the details of the target discrimination mechanism of Trx are unclear (10). Recent studies revealed that some proteins possessing Trx-like motifs also function in the redox network in chloroplasts. For example, NADPH-Trx reductase C (NTRC), which has both NTR and Trx domains in one molecule, can reduce several Trx target proteins, such as 2-Cys proxiredoxin (2-CysPrx), peroxiredoxin Q, and the Mg-chelatase I subunit, using reducing power provided by NADPH (11). In addition, we recently reported the physiological significance of TrxL2, which has a WCRKC sequence in its active center motif, in the chloroplast redox network. TrxL2 has a higher midpoint redox potential than Trx, and it can oxidize Trx target proteins using oxidation power derived from 2-CysPrx and reactive oxygen species (12). We then clarified that the TrxL2–2-CysPrx pathway is the oxidation pathway for redox-regulated enzymes in chloroplasts in the dark. This important finding reminded us of the significance of the reduction/oxidation balance of the whole redox network in chloroplasts as a metabolism-regulatory system for adapting to fluctuating environmental conditions.
To date, Trx activities in vivo have been assessed by visualizing the redox states of target proteins using SDS-PAGE mobility shift analysis following chemical modification of free thiol groups (13–15). However, this method requires disruption of the desired cells or organelles. To monitor the in vivo redox state, roGFP, the most well-known redox sensor protein, was developed by introducing two cysteine residues at positions Ser147 and Gln204 in the WT GFP or the GFP S65T mutant, and these Cys residues act as switches to detect the redox state as a redox-dependent change in fluorescence (16). roGFP has been widely used for nondestructive observation in cells. However, roGFP cannot react directly with Trx or small redox molecules such as GSH and hydrogen peroxide. Instead, roGFP was reduced and oxidized by glutaredoxin (Grx), a Trx superfamily protein containing the CPYC motif in its active center (17). Although several redox sensor proteins, such as Oba-Q (18), rxYFP (19), rxRFP (20), and Re-Q (21), have been developed, these sensor proteins also cannot detect the redox change in Trx molecules directly. For this purpose, TrxRFP was recently developed as a genetically encoded sensor by fusing rxRFP and human Trx1 (22). This Trx1 was reduced by human Trx reductase and oxidized by human Trx peroxidases in a similar manner as native Trx1. In this system, the redox states of rxRFP and Trx1 in the fused TrxRFP molecule are in equilibrium, and the redox state of Trx1 can be observed by measuring the change in the fluorescence intensity of rxRFP. However, TrxRFP overexpression in cells might affect the intracellular redox states because TrxRFP functions similarly as Trx in vivo. In addition, the signal obtained from TrxRFP illustrates the redox status of Trx1 in the fused molecule but not that of other endogenous Trx molecules. Therefore, it is difficult to directly apply TrxRFP to the observation of Trx molecules in photosynthetic organisms.
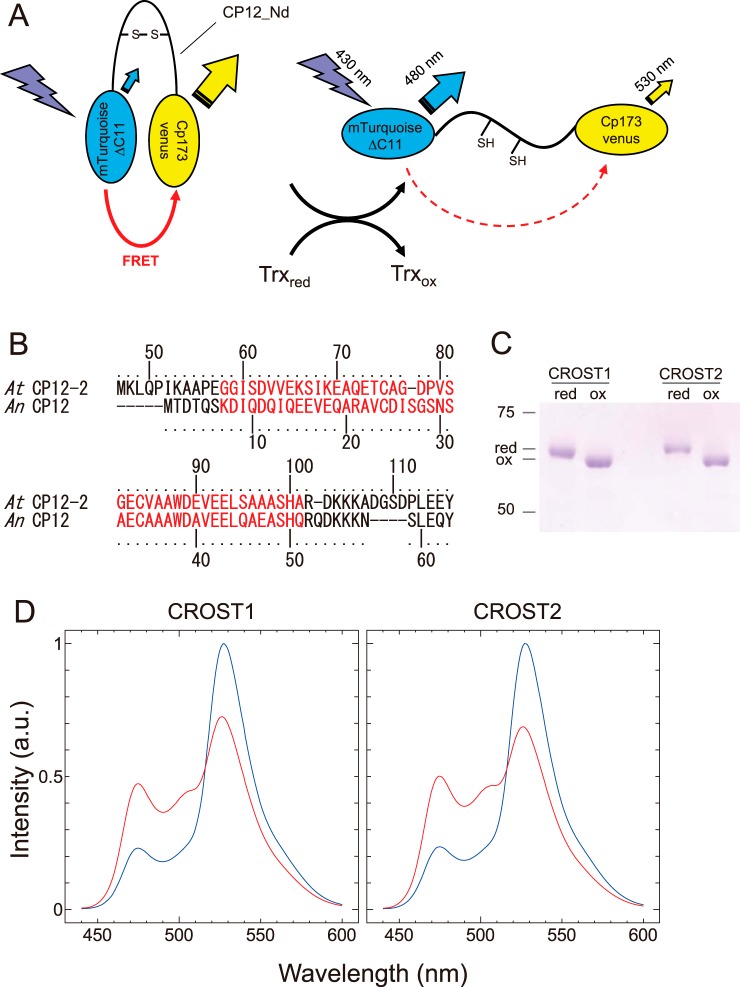
In this study, we report newly designed sensor probes to detect the redox changes of Trx by combining CFP, YFP, and a partial sequence of CP12 (Fig. 1A). CP12 is a Trx target protein conserved among photosynthetic organisms that forms a supercomplex with glyceraldehyde-3-phosphate dehydrogenase and phosphoribulokinase and suppresses their activities in the dark or under limited light or stress conditions (23). CP12 in chloroplasts has two cysteine pairs that can form disulfide bonds at the N- and C-terminal sides. Trx-f can reduce these disulfide bonds and induce a conformational change of CP12 (24). We therefore applied the partial sequence of CP12 to our Trx sensor. We then fused the N-terminal domain of CP12 with mTurquoise (CFP) and cp173-mVenus (YFP). When the fused protein interacted with the reduced form of Trx, the protein exhibited a remarkable change in fluorescence based on FRET. In addition, we succeeded in conferring Trx specificity to the sensor protein using CP12 from a different origin.
Figure 1.
The molecular design of CROST sensors. A, schematics of CROST sensors. Mutants of CFP (mTurquoise featuring a deletion of 11 C-terminal amino acids) and YFP (cp173Venus) were connected by the N-terminal domain of CP12 derived from A. thaliana or Anabaena sp. PCC7120. B, amino acid sequences of CP12. The N-terminal domains used for CROST sensors are highlighted in red. C, oxidation (ox) and reduction (red) of CROST proteins were confirmed by nonreducing SDS-PAGE following the labeling with 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonate. D, redox-dependent fluorescence spectral changes of CROST proteins. Fluorescence emission spectra of oxidized (blue lines) and reduced (red lines) CROST proteins at 25 °C in 50 mm Tris-HCl (pH 7.5), 50 mm NaCl, and 1 mm EDTA were measured with excitation at 430 nm. CROST proteins were oxidized during the purification process and, therefore, reduced in the presence of 100 μm DTTred and 0.2 μΜ AtTrx-f1 before the measurement. a.u., arbitrary unit.
Results and discussion
Development of probes for redox changes of Trx
CP12 of A. thaliana (CP12-2) has two pairs of cysteines (Cys75-Cys84 and Cys117-Cys126) that form disulfide bonds under oxidizing conditions, and this protein is considered the target of Trx-f in vivo (24). Although the molecular structure of CP12 was unsolved, a large conformational change of the molecule caused by changes of the redox status of these cysteine pairs was expected. We therefore applied the partial sequence of the N-terminal domain of CP12-2 from Gly58 to Ala101, including the Cys75-Cys84 pair, which was designated as AtCP12_Nd, to prepare the sensor protein (Fig. 1B, red). As a donor protein for FRET, mTurquoiseΔ11, a variant of CFP lacking the C-terminal 11 amino acids, was adopted, and cp173-mVenus, a circular permutation mutant of YFP having the amino acid at number 173 as its N terminus, was used as the acceptor protein (Fig. 1A). These two GFP derivatives were connected by AtCP12_Nd, and the redox changes of cysteines in the CP12 region were then confirmed using the mobility shift on SDS-PAGE following the labeling of free thiols on the proteins with 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonate (13) (Fig. 1C). This new probe nicely displayed high FRET efficiency in the oxidized form, and a remarkable decrease of the signal was noted for the reduced form (Fig. 1D). We then designated this sensor protein change in redox state of thioredoxin 1 (CROST1). Next we prepared another sensor protein using CP12 from the cyanobacterium Anabaena sp. PCC7120, as the major Trx in cyanobacteria is Trx-m, and they do not possess Trx-f. A part of CP12 from Anabaena, Lys7-Gln51 (Fig. 1B, red), was fused to mTurquoiseΔ11 at the N terminus and cp173-mVenus at the C terminus. This protein also exhibited a change in fluorescence caused by the redox change of the CP12 molecule (Fig. 1D), and it was designated CROST2.
Characterization of CROST sensors in vitro
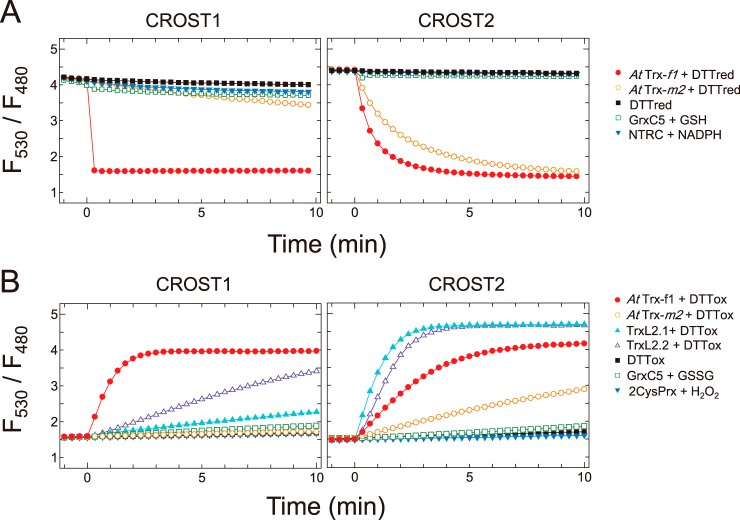
We digitalized the FRET signal changes of the sensor proteins by calculating the ratio of fluorescence intensity at 530 and 480 nm (Fig. 2). Initially, reduction of the oxidized form of CROST1 was examined in the presence of various reductants. When Trx-f1 from A. thaliana was applied together with 100 μm of the reduced form of DTT (DTTred) at time 0, the signal rapidly decreased from 4.1 to 1.5 (Fig. 2A, left panel, closed circles), indicating that the full reduction of CROST1 occurs within 20 s. By contrast, CROST1 was not reduced when the reduced form Trx-m2 from A. thaliana was applied (Fig. 2A, left panel, open circles). In addition, CROST1 was hardly reduced by 100 μm DTTred (Fig. 2A, closed squares), glutaredoxin C5 (GrxC5) from A. thaliana and 2 mm GSH (Fig. 2A, open squares) or by NTRC and 100 μm NADPH (Fig. 2A, closed reverse triangles). These results clearly indicate that CROST1 is a specific sensor for efficiently detecting the redox status of Trx-f. Similarly, reduction of CROST2 was not observed when DTTred, GrxC5 or NTRC was used as a reductant (Fig. 2A, right panel). However, CROST2 was reduced by Trx-m2 in addition to Trx-f1. This result suggests that CROST2 is a redox sensor that has a lower specificity to Trx isoforms. (Fig. 2A, right panel).
Figure 2.
Reduction and oxidation of CROST proteins. A, reduction of CROST1 (left) and CROST2 (right) by reductants. Fluorescence intensity ratios (530/480 nm) of 0.1 μm oxidized CROST proteins at 25 °C in 50 mm Tris-HCl (pH 7.5), 50 mm NaCl, and 1 mm EDTA were plotted before and after addition of reductants at time 0 as follows: closed circles, 0.2 μm AtTrx-f1 + 100 μm DTTred; open circles, 0.2 μm AtTrx-m2 + 100 μm DTTred; closed squares, 100 μm DTTred; open squares, 0.2 μm AtGrxC5 + 2 mm GSH; closed inverted triangles, 0.2 μm AtNTRC + 100 μm NADPH. B, oxidation of CROST1 (left) and CROST2 (right) by oxidants. Fluorescence intensity ratios of 0.1 μm reduced CROST proteins were plotted before and after addition of reductants at time 0 as follows: closed circles, 0.2 μm AtTrx-f1 + 50 mm DTTox; open circles, 0.2 μm AtTrx-m2 + 50 mm DTTox; closed triangles, 1 μm AtTrxL2.1 + 50 mm DTTox; open triangles, 1 μm AtTrxL2.2 + 50 mm DTTox; closed squares, 50 mm DTTox; open squares, 0.2 μm AtGrxC5 + 1 mm GSSG; closed inverted triangles, 0.2 μm At2CysPrx + 0.1 mm H2O2. In the presence of 0.2 μm AtTrx-f1 and 0.5 mm DTTred, 10 μm CROST sensors were reduced for 30 min at 25 °C before measurements and diluted to a useful concentration.
Because we recently revealed a new protein functioning in the chloroplast redox network that inactivates thiol-regulated enzymes in the dark via oxidation (12), we examined the oxidation process of CROST proteins. Oxidation of the reduced form CROST1 was observed when the protein was incubated with Trx-f1 and the oxidized form of DTT (DTTox), whereas the newly assigned chloroplast oxidation factors (TrxL2 proteins) oxidized CROST1 much slower than Trx-f1 (Fig. 2B, left panel). DTTox, GrxC5 plus GSSG, or 2-CysPrx together with H2O2 did not oxidize CROST1. In contrast, CROST2 was efficiently oxidized by TrxL2.1 and TrxL2.2 (12). These results indicate that CROST proteins can reflect changes in the redox states of Trx-f or Trx-m in the solution to their FRET signals, whereas the chemical redox molecules do not affect the redox status of CROST proteins.
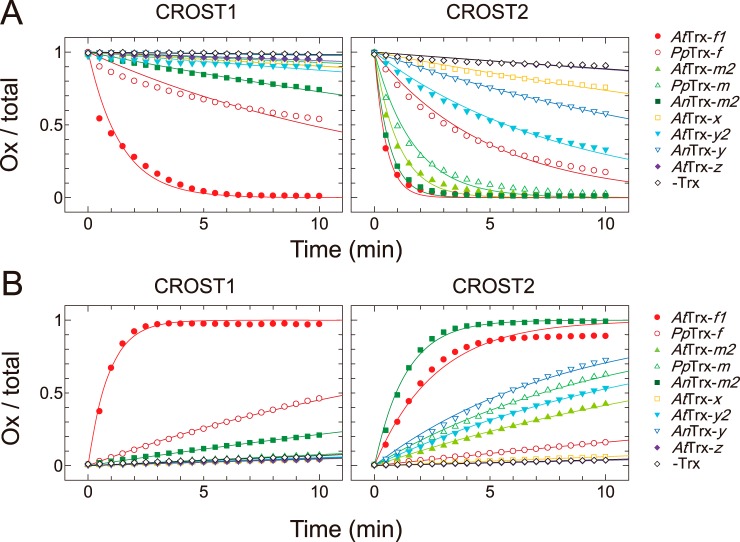
The Trx isoform specificities of CROST sensors were then examined using nine Trx isoforms derived from A. thaliana, Physcomitrella patens, and Anabaena sp. PCC7120 (A. 7120). Because the reaction rate of CROST1 with AtTrx-f1 was sufficiently fast, as shown in Fig. 2A, a Trx isoform concentration of 30 nm was used together with 100 μm DTTred in this study. Among the Trx isoforms examined, AtTrx-f1 and PpTrx-f could efficiently reduce CROST1 (Fig. 3A, left), and the rate constants were obtained (Fig. 4A). We then examined the Trx isoform specificity of CROST1 in the oxidation process. When 0.1 μm CROST1 was incubated with 1 μm Trx isoforms together with 50 mm DTTox, slow oxidation was observed. Again, AtTrx-f1 and PpTrx-f could oxidize CROST1 under these conditions (Figs. 3B, left panel, and 4B). Although slow reduction and oxidation of CROST1 were observed when AnTrx-m1 was used (Fig. 3, A and B, left), the rate constants were much smaller than those of f-type isoforms (Fig. 4, A and B). These results indicate that CROST1 is an f-type Trx-specific sensor that is reduced or oxidized via the redox equilibrium with f-type Trx.
Figure 3.
Reduction and oxidation of CROST proteins by five Trx subtypes. A, reduction levels of CROST1 (left) and CROST2 (right) at 25 °C in 50 mm Tris-HCl (pH 7.5), 50 mm NaCl, and 1 mm EDTA were plotted. At time 0, each Trx subtype was added at a final concentration of 30 nm for CROST1 and 500 nm for CROST2 with 100 μm DTTred: closed circles, AtTrx-f1; open circles, PpTrx-f; closed triangles, AtTrx-m2; open triangles, PpTrx-m; closed squares, AnTrx-m2; open squares, AtTrx-x; closed inverted triangles, AtTrx-y2; open inverted triangles, AnTrx-y; closed diamonds, AtTrx-z; open diamonds, without Trx. B, oxidation levels of CROST1 (left) and CROST2 (right) were plotted under the conditions described for A. Trx (1 μm) was used in the presence of 50 mm DTTox. The symbols follow those used in A. CROST sensors (10 μm) were reduced in the presence of 0.2 μm AtTrx-f1 and 0.5 mm DTTred for 30 min at 25 °C before measurements and diluted to a useful concentration.
Figure 4.
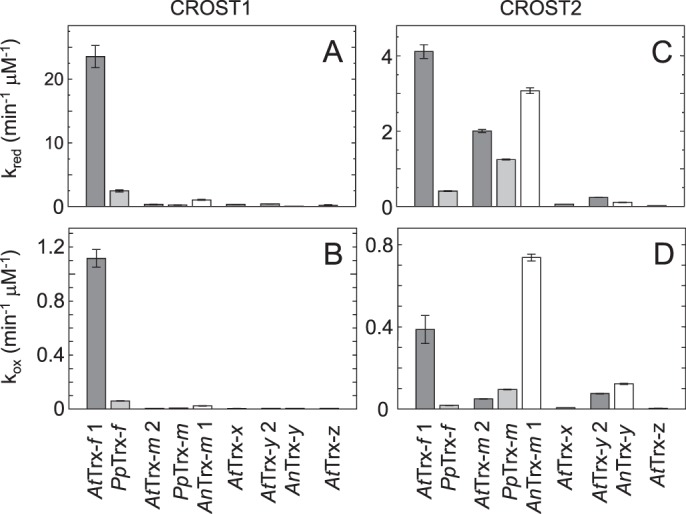
Rate constants of CROST reduction and oxidation. A and B, reaction rate constants for CROST1 were determined via fitting with the first-order equation (see “Experimental procedures”) of the reduction process data shown in Fig. 3A, left (A), and the oxidation process data shown in Fig. 3B, left (B). C and D, reaction rate constants for CROST2 were determined via fitting with the first-order equation (see “Experimental procedures”) of the reduction process data shown in Fig. 3A, right (C), and the oxidation process data shown in Fig. 3B, right (D).
To determine the specificity of CROST2, 500 nm Trx isoforms were used because the apparent reduction rates of CROST2 were much slower than those of CROST1 (Fig. 2). Based on the rate constants calculated from the reduction time course data (Fig. 3A, right), CROST2 could detect changes in the redox states of AtTrx-f, PpTrx-f, AtTrx-m2, PpTrx-m, and AnTrx-m1 (Fig. 4C). When the oxidation process was examined, AnTrx-m1 was more effective than AtTrx-f1 (Figs. 3B, right, and 4D), and this must be attributable to the use of A. 7120–origin CP12 to prepare CROST2. Hence, CROST2 must be applicable to indicate the redox states of m-type Trx in cells lacking f-type Trx, such as cyanobacteria.
Although there was a large difference in the reduction rate constants of CROST1 (0.40 s−1 μm−1) and CROST2 (0.068 s−1 μm−1) using AtTrx-f1, the reduction rate constant of CROST2 for AtTrx-f1 was equivalent to that of chloroplast F1-ATPase for spinach-derived Trx-f (0.057 s−1 μm−1) (25). Therefore, CROST2 must have a certain reactivity with Trx even under physiological conditions. The rate constant of the reduction process of CROST1 for AtTrx-f1 was 21-fold larger than that for the oxidized process (Fig. 4, A and B), suggesting that the oxidation of AtCP12-2 used by Trx-f1 to reduce CROST1 does not occur efficiently in vivo. Conversely, the rate constant of the reduction process of CROST2 for AnTrx-m1 was only 4.2-fold larger than that for the oxidation process (Fig. 4, C and D), suggesting that the N-terminal domain of AnCP12 used for CROST2 must be easily oxidized by AnTrx-m1 in A. 7120 cells.
We then measured the midpoint redox potentials of CROST1 and CROST2 to reveal the cause of the difference of reduction rate constants of AtTrx-f1. Consequently, a midpoint redox potential of −266 mV was obtained for CROST1 at pH 7.5 (Fig. 5A, left). This value was considerably larger than that of AtTrx-f1 (−321 mV) (9) and similar to that of TrxL2.1 (−258 mV) (12). In contrast, CROST2 displayed a more negative midpoint redox potential of −296 mV (Fig. 5A). Because the rate of electron transfer between two redox proteins is influenced by the difference of their midpoint redox potentials, the observed difference at ∼30 mV between CROST1 and CROST2 must be sufficient to explain the difference of the reduction rate constants observed in Fig. 4. The lower midpoint redox potential of CROST2 may explain the difficulty of various Trx isoforms in reducing this protein.
Figure 5.
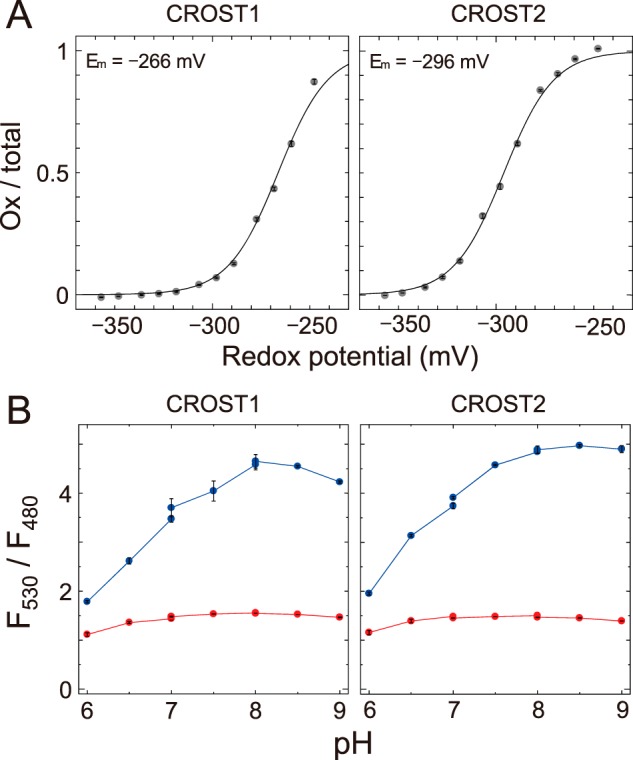
Redox potential and pH dependence of CROST sensors. A, the oxidation levels of CROST1 (left) and CROST2 (right) were quantified as the ratios of the oxidized forms to the total and plotted against the redox potential of DTT buffer (pH 7.5). Data were fitted to the Nernst equation for two electrons exchanged (y = 1/(1 + exp(0.078 (x − Em)))). The midpoint redox potential value shown in the figure is the mean ± S.D. (four independent experiments using two different protein preparations). B, fluorescence ratios (F530/F480) of recombinant CROST1 (left) and CROST2 (right) under different pH levels were measured for their oxidized (blue lines) and reduced (red lines) forms. The buffers MES (for pH 6–7), PIPES (for pH 7–8), and HEPES (for pH 8–9) were used.
The disadvantages of CROST sensor proteins are their remarkable pH dependences and small fluorescent signal changes under low pH conditions (Fig. 5B). The observed pH dependencies may be attributable to the pH dependence of the YFP chromophore. Under low-pH conditions, the YFP chromophore of cp173-mVenus is not ionized, and, therefore, it cannot function as a FRET acceptor. However, CROST sensors fortunately had large dynamic ranges at a pH of ∼7–8 (Fig. 5B), which is equivalent to the pH in various organelles, and the F530/F480 values under the oxidized conditions were more than double compared with those under the reduced condition. These sensors should therefore be useful for monitoring the intracellular redox states of Trx molecules.
Visualization of the redox states of Trx in chloroplasts
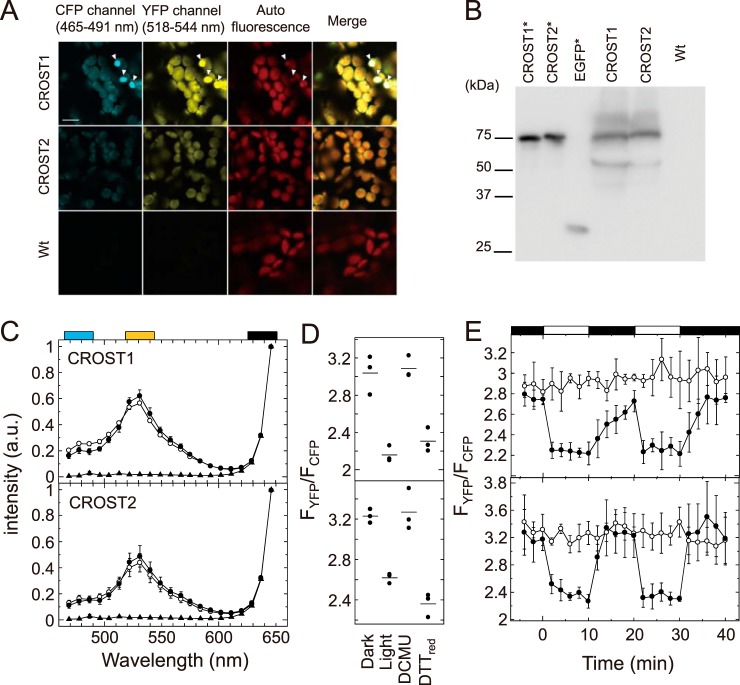
To observe the redox status of Trx in living cells, we expressed CROST1 and CROST2 in A. thaliana chloroplasts and observed the fluorescence of the sensor proteins using an LSM780 confocal fluorescence microscope (Zeiss) (Fig. 6A). Because of the unexpected appearance of bright spots, which were obviously differentiated from the fluorescence signals from the cytosol or chloroplasts and might be ascribed to the aggregation of sensor proteins (Fig. 6A, arrowheads), we calculated the fluorescence intensities in the image by removing these bright spots. In addition, we confirmed the expression of CROST proteins in leaves by immunoblot analysis (Fig. 6B). Degradation of a small part of CROST sensors was observed. The emerged degraded fluorescent proteins may affect the fluorescence intensity ratios of YFP (518–544 nm) and CFP (465–491 nm) channels (FYFP/FCFP) in measurements, especially when free CFP appeared in cells. The autofluorescence of chlorophyll is also a possible disturbance factor for the in situ measurement. Fluorescence spectra from A. thaliana cells expressing CROST sensors were acquired using the λ mode, and we confirmed that the effects of degradation of the sensors and the autofluorescence of chlorophyll in chloroplasts was weak enough and did not influence the fluorescence signals of CROST sensors (Fig. 6C). Leaves of WT plants were used as a control. By changing the light intensity, we could observe a slight decrease in signal intensity at 530 nm and an increase in the second peak at 480 nm (Fig. 6C). FYFP/FCFP ratios were then calculated from these spectra (Fig. 6D). As controls, FYFP/FCFP ratios were determined in the presence of 20 mm DTTred or 20 μm 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU). Finally, we observed the change in FYFP/FCFP during the mutual light/dark changes using CROST1 (Fig. 6E, top, closed circles) and CROST2 (Fig. 6E, bottom, closed circles) introduced into the chloroplasts of A. thaliana. When the plant leaves were illuminated, both CROST sensors immediately shifted to the reduced state, implying that Trx proteins in chloroplasts, mainly Trx-f, were reduced. When the light source was removed, FYFP/FCFP gradually increased, indicating that Trx-f was slowly oxidized in chloroplasts. The signal decay of CROST1 was obviously slower than that of CROST2. This difference might be ascribed to the difference of the midpoint redox potentials of these two sensors. CROST1 had a higher redox potential than CROST2 (Fig. 5A). In addition, the reactivity of CROST2 was not restricted to f-type Trx, but it was sufficient against Trx proteins and TrxL2. This property might explain the rapid oxidation of CROST2 even under the light/dark transition (Figs. 2 and 3). In contrast, we could not detect any changes in FYFP/FCFP when 20 μm DCMU was applied (Fig. 6E, open circles). Thus, we succeeded in observing the real-time redox dynamics of Trx molecules in chloroplasts caused by photosynthetic electron transport according to the light/dark transition.
Figure 6.
Visualization of intracellular Trx activities. A, fluorescence images of transgenic plant leaves expressing CROST1 and CROST2 and of WT leaves were obtained using an Ar 458-nm laser for excitation. Images from 465 to 491 nm for CFP and from 518 to 544 nm for YFP were taken. Chlorophyll autofluorescence from 624 to 651 nm was also assessed, and merged images were created (scale bar = 10 μm). B, immunoblot analysis of the expression of CROST1 and CROST2 in leaves using an antibody against GFP. Recombinant CROST1, CROST2, and EGFP were used as controls (asterisks). C, emission spectra of CROST1 (top) and CROST2 (bottom) expressed in chloroplasts were measured using a confocal microscopy under dark (closed circles) and light (50 μmol photons m−2 s−1, open circles) conditions. Emission spectra of WT A. thaliana cells were measured as a control (closed triangles). The obtained spectra were standardized at the chlorophyll autofluorescence region (black box) using the least square method. Cyan and yellow boxes show the region used for observation of CFP and YFP, respectively. a.u., arbitrary unit. D, fluorescence intensity ratios of CROST1 (top) and CROST2 (bottom) in the dark and under light (50 μmol photons m−2 s−1) conditions were calculated and plotted. Fluorescence intensity ratios in the presence of 20 mm DTTred and 20 μm DCMU were determined as well. The results of three independent experiments are shown. Bars show the average. E, fluorescence intensity ratio changes of CROST1 (top) and CROST2 (bottom) under mutual light (50 μmol photons m−2 s−1)/dark conditions in the absence (closed circles) and presence (open circles) of 20 μm DCMU were calculated at every 2 min and plotted. The light period is indicated by white rectangles above the graph. Results obtained from three independent experiments are shown with error bars.
Experimental procedures
Gene construction
The complementary DNAs of mTurquoiseΔ11, cp173m-Venus, the N-terminal domains of AtCP12-2 (At3g56690) and AnCP12 (pfam02672) were amplified by PCR and fused using hot fusion methods (26) (Fig. 1B) with the pET-23a vector to express the protein with a C-terminal His6 tag. Complementary DNA fragments encoding the mature protein-coding regions, which were predicted by TargetP (27), of AtTrx-f1 (At3g02730), AtTrx-m2 (At4g03520), AtTrx-x (At1g50320), AtTrx-y2 (At1g43560), AtTrx-z (At3g06730), PpTrx-f (PHPAT.004G014000.1), and PpTrx-m (PHPAT.005G085400.1) were cloned into the pET-23a vector (Novagen) to express the protein with a C-terminal His6 tag. Other expression plasmids were prepared as in our earlier studies (9, 28, 29).
Protein expression and purification
Escherichia coli strains BL21(DE3) and BL21(DE3) pLysS were transformed with each expression plasmid. BL21(DE3) pLysS was used to express A. thaliana-derived Trx, and BL21(DE3) was used for P. patens-derived Trx and CROST sensors. Transformed E. coli was then cultured at 37 °C. Expression of the desired protein was induced using 0.5 mm isopropyl β-d-thiogalactopyranoside, followed by further cultivation for 16 h at 21 °C. Collected cells were disrupted and ultracentrifuged (125,000 × g for 40 min) to obtain supernatant containing the desired protein for subsequent purification. His-tagged A. thaliana–derived Trxs and P. patens–derived Trx and CROST sensors were purified via Ni-NTA affinity chromatography using Ni-NTA–agarose (Qiagen) resin. Ni-NTA–agarose resin was washed in 25 mm Tris-HCl (pH 8.0) containing 20 mm imidazole and eluted using the same buffer containing 250 mm imidazole. Elusions including A. thaliana–derived Trxs were applied to Superdex 75 Increase 10/300 GL (GE Healthcare) columns and purified. Eluates including P. patens–derived Trx were dialyzed and concentrated. Eluates containing CROST sensors were applied to a Toyopearl Butyl-650 (Tosoh, Tokyo, Japan) column and eluted using 20 mm Tris-HCl (pH 8.0) with an ammonium sulfate reverse gradient from 30% to 0%. Peak fractions eluted from the column were collected. Eluates containing CROST sensors were incubated in the presence of 0.5 mm diamide for 10 min at 25 °C, dialyzed, and concentrated. Other proteins that were partially purified by Ni-NTA–agarose chromatography were then purified according to the methods described in our earlier studies (9, 28, 29). The purity of the recombinant proteins used was examined by SDS-PAGE (Fig. S1).
Fluorescence spectroscopy and calculation of the reduced and oxidized ratio
Fluorescence emission spectra were measured using an FP-8500 fluorescence spectrophotometer (Jasco, Tokyo, Japan). An excitation wavelength of 430 nm was used to excite CROST sensors. Emission fluorescence intensities from CROST sensors at 480 (F480) and 530 nm (F530) were measured, and the fluorescence ratio (Fr) was calculated using Equation 1:
| (Eq. 1) |
To establish fully reduced or oxidized conditions, CROST sensors were incubated with 100 μm DTTred or 50 mm DTTox in the presence of 1 μm AtTrx-f1 for 10 min at 25 °C, and Fr values were obtained as Fred and Fox. The oxidation ratio, Ox/total, was then calculated using Equation 2:
| (Eq. 2) |
Determination of rate constants
To eliminate the influence of direct reduction by DTT, we calculated the rate constant from Ox/total data in the presence of Trx isoforms after subtracting Ox/total data in the presence of only DTT at each time. Then the resulting Ox/total value at the indicated time was plotted and fitted using Equation 3 to determine the rate constant kred and Equation 4 to determine kox:
| (Eq. 3) |
| (Eq. 4) |
[Trx] is the concentration of Trx.
Determination of the midpoint redox potential of CROST sensors
CROST sensor proteins (0.1 μm) were incubated in 25 mm Tris-HCl (pH 7.5), 50 mm DTTox, various concentrations of DTTred (1 μm to 50 mm), and 1 μm AtTrx-f1 for 3 h at 25 °C. The midpoint redox potential values of CROST sensors were calculated by fitting the titration data of the reduction level of proteins to the Nernst equation. A value of −357 mV was used as a midpoint redox potential of DTT at pH 7.5 (9).
Expression and observation in chloroplasts of A. thaliana
The expression plasmids pRI201/PtCROST1 and pRI201/PtCROST2 were prepared by appending the recA DNA recombination family protein leader sequence to the 5′ end of the CROST1 and CROST2 coding genes cloned in the expression vector pRI201-AN (Takara). The A. thaliana plants were transformed with the expression plasmids using Agrobacterium tumefaciens and then grown at 22 °C under 16 h light/8 h dark conditions. For the observation, AxioObserverZ1 with LSM780 (Carl Zeiss) was used with an Ar 458-nm laser (25 milliwatt). The laser power was held at less than 0.1% to avoid activating the photosystem. A multi-channel GaAsP detector was used in the photon-counting and λ modes. The intensity of the fluorescence image was then digitalized using ImageJ image analysis software.
Author contributions
K. S. and T. H. conceptualization; K. S. data curation; K. S., Y. Y., and N. F. investigation; K. S. and T. H. writing-original draft; N. F. and Y. F. visualization; Y. F. resources; Y. F. and S. M. methodology; K. Y. and T. H. supervision; K. Y. and S. M. writing-review and editing; T. H. validation.
Supplementary Material
Acknowledgments
We thank the Center for Biological Resources and Informatics, Suzukakedai Design, and the Manufacturing Division of the Tokyo Institute of Technology for technical support.
This work was supported by MEXT-KAKENHI Grant 16H06556 (to T. H.) and the Dynamic Alliance for Open Innovation Bridging Human, Environment, and Materials. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Fig. S1.
- Trx
- thioredoxin
- NTRC
- NADPH-Trx reductase C
- 2-CysPrx
- 2-Cys proxiredoxin
- Grx
- glutaredoxin
- CFP
- cyan fluorescent protein
- CROST
- change in redox state of Trx
- DCMU
- 3-(3,4-dichlorophenyl)-1,1-dimethylurea
- Ni-NTA
- nickel-nitrilotriacetic acid
- Fr
- fluorescence ratio
- DTTox
- oxidized form of DTT
- DTTred
- reduced form of DTT.
References
- 1. Buchanan B. B. (1980) Role of light in the regulation of chloroplast enzymes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 31, 341–374 10.1146/annurev.pp.31.060180.002013 [DOI] [Google Scholar]
- 2. Mills J. D., and Mitchell P. (1982) Modulation of coupling factor ATPase activity in intact chloroplasts: reversal of thiol modulation in the dark. Biochim. Biophys. Acta 679, 75–83 10.1016/0005-2728(82)90257-2 [DOI] [Google Scholar]
- 3. Buchanan B. B. (1991) Regulation of CO2 assimilation in oxygenic photosynthesis: the ferredoxin/thioredoxin system. Perspective on its discovery, present status, and future development. Arch. Biochem. Biophys. 288, 1–9 10.1016/0003-9861(91)90157-E [DOI] [PubMed] [Google Scholar]
- 4. Scheibe R., and Anderson L. E. (1981) Dark modulation of NADP-dependent malate dehydrogenase and glucose-6-phosphate dehydrogenase in the chloroplast. Biochim. Biophys. Acta 636, 58–64 10.1016/0005-2728(81)90075-X [DOI] [PubMed] [Google Scholar]
- 5. Wenderoth I., Scheibe R., and von Schaewen A. (1997) Identification of the cysteine residues involved in redox modification of plant plastidic glucose-6-phosphate dehydrogenase. J. Biol. Chem. 272, 26985–26990 10.1074/jbc.272.43.26985 [DOI] [PubMed] [Google Scholar]
- 6. Ikegami A., Yoshimura N., Motohashi K., Takahashi S., Romano P. G., Hisabori T., Takamiya K., and Masuda T. (2007) The CHLI1 subunit of Arabidopsis thaliana magnesium chelatase is a target protein of the chloroplast thioredoxin. J. Biol. Chem. 282, 19282–19291 10.1074/jbc.M703324200 [DOI] [PubMed] [Google Scholar]
- 7. Katti S. K., LeMaster D. M., and Eklund H. (1990) Crystal structure of thioredoxin from Escherichia coli at 1.68 A resolution. J. Mol. Biol. 212, 167–184 10.1016/0022-2836(90)90313-B [DOI] [PubMed] [Google Scholar]
- 8. Geck M. K., Larimer F. W., and Hartman F. C. (1996) Identification of residues of spinach thioredoxin f that influence interactions with target enzymes. J. Biol. Chem. 271, 24736–24740 10.1074/jbc.271.40.24736 [DOI] [PubMed] [Google Scholar]
- 9. Yoshida K., Hara S., and Hisabori T. (2015) Thioredoxin selectivity for thiol-based redox regulation of target proteins in chloroplasts. J. Biol. Chem. 290, 19540 10.1074/jbc.A115.647545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yoshida K., and Hisabori T. (2018) Determining the rate-limiting step for light-responsive redox regulation in chloroplasts. Antioxidants 7, E153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoshida K., and Hisabori T. (2016) Two distinct redox cascades cooperatively regulate chloroplast functions and sustain plant viability. Proc. Natl. Acad. Sci. U.S.A. 113, E3967–E3976 10.1073/pnas.1604101113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoshida K., Hara A., Sugiura K., Fukaya Y., and Hisabori T. (2018) Thioredoxin-like2/2-Cys peroxiredoxin redox cascade supports oxidative thiol modulation in chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 115, E8296–E8304 10.1073/pnas.1808284115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kobayashi T., Kishigami S., Sone M., Inokuchi H., Mogi T., and Ito K. (1997) Respiratory chain is required to maintain oxidized states of the DsbA-DsbB disulfide bond formation system in aerobically growing Escherichia coli cells. Proc. Natl. Acad. Sci. U.S.A. 94, 11857–11862 10.1073/pnas.94.22.11857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Motohashi K., Kondoh A., Stumpp M. T., and Hisabori T. (2001) Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc. Natl. Acad. Sci. U.S.A. 98, 11224–11229 10.1073/pnas.191282098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yoshida K., Matsuoka Y., Hara S., Konno H., and Hisabori T. (2014) Distinct redox behaviors of chloroplast thiol enzymes and their relationships with photosynthetic electron transport in Arabidopsis thaliana. Plant Cell Physiol. 55, 1415–1425 10.1093/pcp/pcu066 [DOI] [PubMed] [Google Scholar]
- 16. Hanson G. T., Aggeler R., Oglesbee D., Cannon M., Capaldi R. A., Tsien R. Y., and Remington S. J. (2004) Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 279, 13044–13053 10.1074/jbc.M312846200 [DOI] [PubMed] [Google Scholar]
- 17. Meyer A. J., Brach T., Marty L., Kreye S., Rouhier N., Jacquot J. P., and Hell R. (2007) Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J. 52, 973–986 10.1111/j.1365-313X.2007.03280.x [DOI] [PubMed] [Google Scholar]
- 18. Sugiura K., Nagai T., Nakano M., Ichinose H., Nakabayashi T., Ohta N., and Hisabori T. (2015) Redox sensor proteins for highly sensitive direct imaging of intracellular redox state. Biochem. Biophys. Res. Commun. 457, 242–248 10.1016/j.bbrc.2014.12.095 [DOI] [PubMed] [Google Scholar]
- 19. Ostergaard H., Henriksen A., Hansen F. G., and Winther J. R. (2001) Shedding light on disulfide bond formation: engineering a redox switch in green fluorescent protein. EMBO J. 20, 5853–5862 10.1093/emboj/20.21.5853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fan Y., Chen Z., and Ai H. W. (2015) Monitoring redox dynamics in living cells with a redox-sensitive red fluorescent protein. Anal. Chem. 87, 2802–2810 10.1021/ac5041988 [DOI] [PubMed] [Google Scholar]
- 21. Sugiura K., Tanaka H., Kurisu G., Wakabayashi K. I., and Hisabori T. (2019) Multicolor redox sensor proteins can visualize redox changes in various compartments of the living cell. Biochim. Biophys. Acta Gen. Subj. 1863, 1098–1107 10.1016/j.bbagen.2019.01.016 [DOI] [PubMed] [Google Scholar]
- 22. Fan Y., Makar M., Wang M. X., and Ai H. W. (2017) Monitoring thioredoxin redox with a genetically encoded red fluorescent biosensor. Nat. Chem. Biol. 13, 1045–1052 10.1038/nchembio.2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wedel N., Soll J., and Paap B. K. (1997) CP12 provides a new mode of light regulation of Calvin cycle activity in higher plants. Proc. Natl. Acad. Sci. U.S.A. 94, 10479–10484 10.1073/pnas.94.19.10479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marri L., Zaffagnini M., Collin V., Issakidis-Bourguet E., Lemaire S. D., Pupillo P., Sparla F., Miginiac-Maslow M., and Trost P. (2009) Prompt and easy activation by specific thioredoxins of Calvin cycle enzymes of Arabidopsis thaliana associated in the GAPDH/CP12/PRK supramolecular complex. Mol. Plant 2, 259–269 10.1093/mp/ssn061 [DOI] [PubMed] [Google Scholar]
- 25. Schwarz O., Schürmann P., and Strotmann H. (1997) Kinetics and thioredoxin specificity of thiol modulation of the chloroplast H+-ATPase. J. Biol. Chem. 272, 16924–16927 10.1074/jbc.272.27.16924 [DOI] [PubMed] [Google Scholar]
- 26. Fu C., Donovan W. P., Shikapwashya-Hasser O., Ye X., and Cole R. H. (2014) Hot fusion: an efficient method to clone multiple DNA fragments as well as inverted repeats without ligase. PLoS ONE 9, e115318 10.1371/journal.pone.0115318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emanuelsson O., Nielsen H., Brunak S., and von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016 10.1006/jmbi.2000.3903 [DOI] [PubMed] [Google Scholar]
- 28. Mihara S., Yoshida K., Higo A., and Hisabori T. (2017) Functional Significance of NADPH-thioredoxin reductase C in the antioxidant defense system of Cyanobacterium anabaena sp. PCC 7120. Plant Cell Physiol. 58, 86–94 [DOI] [PubMed] [Google Scholar]
- 29. Mihara S., Wakao H., Yoshida K., Higo A., Sugiura K., Tsuchiya A., Nomata J., Wakabayashi K. I., and Hisabori T. (2018) Thioredoxin regulates G6PDH activity by changing redox states of OpcA in the nitrogen-fixing cyanobacterium Anabaena sp. PCC 7120. Biochem. J. 475, 1091–1105 10.1042/BCJ20170869 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.