Abstract
HLA class I and KIR sequences were determined for Dogon, Fulani and Baka populations of western Africa, Mbuti of central Africa and Datooga, Iraqw and Hadza of eastern Africa. Study of 162 individuals identified 135 HLA class I alleles (41 HLA-A, 61 HLA-B and 33 HLA-C). Common to all populations are three HLA-C alleles (C1+C*07:01, C1+C*07:02 and C2+C*06:02), but no HLA-A or -B. Unexpectedly, no novel HLA class I was identified in these previously unstudied and anthropologically distinctive populations. In contrast, of 227 KIR detected, 22 are present in all seven populations and 28 are novel. A high diversity of HLA A-C-B haplotypes was observed. In six populations, most haplotypes are represented just once. But in the Hadza a majority of haplotypes occur more than once, with two having high frequencies and ten, intermediate frequencies. The centromeric part of the KIR locus exhibits an even balance between cenA and cenB in all seven populations. The telomeric part has an even balance of telA to telB balance in East Africa, but this changes across the continent to where telB is vestigial in West Africa. All four KIR ligands (A3/11, Bw4, C1 and C2) are present in six of the populations. HLA haplotypes of the Iraqw and Hadza encode two KIR ligands, whereas the other populations have an even balance between haplotypes encoding one and two KIR ligands. Individuals in these African populations have a mean of 6.8–8.4 different interactions between KIR and HLA class I, compared to 2.9–6.5 for non-Africans.
Introduction
HLA-A, -B and -C comprise the highly polymorphic MHC class I molecules of humans (1). They bind peptide antigens inside cells and carry them to the cell surface. There the complexes of peptide and MHC class I function as ligands for various receptors and co-receptors of human lymphocytes, notably NK cells and CD8 T cells (2). This pathway of antigen presentation enables killer lymphocytes to respond to cells and tissues compromised by virus infection, malignant transformation or other forms of cellular stress. By killing infected and transformed cells, the responding lymphocytes help prevent the spread of infection or cancer, facilitating disease elimination.
Whereas most nucleated human cells express HLA-A, -B and -C, either constitutively or cytokine induced, in the course of reproduction HLA-C, but not HLA-A or -B, is expressed by extravillous trophoblast cells of the fetus (3). Following embryo implantation, maternal uterine NK cells co-operate with fetal extravillous trophoblast cells in remodeling maternal arteries that will provide the placenta with blood for nourishing the fetus throughout pregnancy. In these ways HLA class I molecules contribute to two vital biological functions: reproduction and immune defense (4).
The structural variation that distinguishes the allotypes of HLA-A, -B and -C is germ line encoded and concentrated on residues of the α1 and α2 domains that bind the peptide or engage with NK or T cell receptors. Functionally distinctive HLA class I allotypes can differ by up to 47 amino acid substitutions (5). As a consequence, HLA class I allotypes differ in the peptide antigens they bind and the lymphocyte receptors that engage the peptide-HLA class I complexes.
The killer cell Immunoglobulin-like receptors (KIR) are a family of signaling NK cell receptors that recognize HLA class I and are principally expressed by NK cells (6, 7). Like HLA class I, KIR variation is germ-line encoded, in a gene family that exhibits haplotypic variation in gene number and gene content, combined with allotypic variation of each KIR gene (8). A given KIR gene encodes either an inhibitory receptor or an activating receptor. The inhibitory KIR are highly polymorphic and function to educate NK cells (9), enabling them to attack infected or malignant cells with compromised expression of HLA class I. Activating KIR exhibit considerably less polymorphism than inhibitory KIR and are now being seen to recognize complexes of HLA class I with conserved peptides of microbial pathogens (10, 11).
HLA-C is more recently evolved than HLA-A and -B. Only great apes have an HLA-C ortholog, whereas all apes and Old World monkeys have MHC-A and -B. A further distinction is that all HLA-C allotypes are recognized by inhibitory KIR, whereas that is only true for a minority of HLA-A and -B allotypes. Thus dimorphism at position 80 of HLA-C distinguishes the C1 epitope (N80) recognized by KIR2DL2/3 from the C2 epitope (K80) recognized by KIR2DL1. Activating KIR2DS1 and KIR2DS5 also recognize the C2 epitope. In contrast, KIR3DL1 recognizes the minority of HLA-A and -B epitopes that carry the Bw4 epitope and KIR3DL2 recognizes only HLA-A*03 and HLA-A*11. That KIR concentrate on HLA-C recognition points to the reproductive function of KIR interactions with HLA-C and KIR having been instrumental in the evolution of HLA-C (12, 13).
Because HLA genes are on chromosome 6 and the KIR genes are on chromosome 19, these cognate ligands and receptors segregate independently in human populations. Thus family members frequently have combinations of HLA class I ligands and KIR receptors that educate NK cells in different ways (9). A likely benefit of this situation is that infections are less readily transmitted from one family member to another, because any pathogen that adapts to the KIR-HLA combinations in one family member will not be adapted to their relatives having different combinations of KIR and HLA. This line of argument can also apply to pathogen transmission from one family to another or from one human population to another.
To determine how KIR and HLA class I coevolve in human populations we have studied these ligands and receptors in human populations that are anthropologically well defined. We found that the Yucpa, an indigenous population of South America, has an unusually high frequency of C1+HLA-C. Accompanying this abundance of C1 ligands, are two Yucpa-specific variants of KIR2DL3, the inhibitory C1 receptor (14). One variant is non-functional, the other has reduced affinity for C1. Thus the effect of increasing C1 frequency, was selection for less effective C1 receptors. In a complementary situation, the Khomani San of southern Africa have an unusually high frequency of C2+HLA-C. Accompanying this abundance of C2 ligands are two Khomani-San specific variants of KIR2DL1, the inhibitory C2 receptor. One variant is non-functional and the other has a substitution in the binding site that changes the receptor’s specificity from C2 to C1. Here the effect of increasing frequency of the C2 ligand is to decrease the frequency of C2 receptors and increase the frequency of C1 receptors (15).
Archaeological analysis of the fossil record, dating back from 195,000 – 300,000 years ago, points to Africa as the continent in which modern humans first emerged (16–19). Genetic evidence supports this model (16, 17). In particular, analysis of neutral genetic markers of mitochondrial DNA and nuclear DNA has shown that Africans have greater genetic diversity than the human populations of other continents (18–21). In contrast to neutral markers, much variation in highly polymorphic HLA class I and killer cell immunoglobulin-like receptors (KIR) is the product of natural selection (22, 23). Previous analyses of KIR and HLA class I diversity in Africans (Ghanaians), Asians (Japanese), Oceanians (Maori) and Amerindians (Yucpa) show similar trends to the neutral markers, with African populations having the most diversity. We therefore investigated HLA class I and KIR in seven anthropologically, well-defined populations representing eastern, central, and western sub-Saharan Africa (SSA), and for which little is known of their HLA and KIR.
Materials and Methods
Study populations
DNA samples from seven sub-Saharan African populations were genotyped for KIR and HLA class I genes at high resolution. These samples were collected in the context of studies looking at genetic markers other than HLA and KIR genes (24–27). Genomic DNA was isolated from saliva samples collected by buccal swab (Epicentre) from 13 Iraqw and 18 Datooga individuals, as described by Knight et al. 2003 (24). These two populations reside in northern Tanzania. A third Tanzanian population we studied is the Hadza, for which DNA samples from 52 individuals were collected in the Arusha district as described (25). The Baka (Gabon) and Mbuti (Democratic Republic of Congo) are pygmy populations; DNA samples were collected from 20 and 19 individuals, respectively (26, 27). We analyzed 29 family trios (both parents and one child) from Mali in West Africa. Genomic DNA was isolated from blood samples as described (28). The results from each child were used to inform the haplotype compositions of the parents, but were not included in calculating the frequencies of haplotypes and alleles. The Mali samples comprised Dogon (N=44) and Fulani (N=14).
We obtained HLA class I genotypes for 170 of the individuals typed. For 4 individuals, complete HLA-A, -B, and -C genotypes were not obtained because of technical failure.
HLA class I genotyping
The DNA samples from Iraqw, Datooga, Baka, Mbuti and Hadza individuals were genotyped for HLA-A, -B and -C at allele level, using bead-based SSOP hybridization detected with a Luminex-100 instrument (Luminex corp. Austin, TX). The assays were performed using LABType SSO reagents (One Lambda, Canoga Park, CA).
PCR amplification and pyrosequencing
High resolution KIR genotyping of DNA samples from Iraqw, Datooga, Baka, Mbuti and Hadza individuals was performed as described (23). Specific amplicons corresponding to individual KIR gene exons were made and subjected to pyrosequencing using a PSQ HS 96A instrument (Qiagen). Novel KIR alleles found in the African populations, were validated by generating an independent PCR amplicon, cloning each allele and sequencing several clones by Sanger methodology.
Cloning and Sanger Sequencing of KIR alleles
KIR-PCR amplicons were sequenced directly in each direction using BigDye™ Terminator Chemistry v. 3.1 (Applied Biosystems, Foster City, CA) and an ABI 3730 Bioanalyzer (Applied Biosystems). When candidate new alleles were identified, they were cloned using the TOPO TA Cloning ® Kit (Invitrogen, Carlsbad, CA). Three or more clones for each candidate were sequenced, as recommended by the curators of the Immuno Polymorphism Database (IPD) (29). The sequences of validated new KIR alleles have been deposited in the IPD database (http://www.ebi.ac.uk/ipd/kir/). Their names and accession numbers are listed in Figure S1M.
Analysis of HLA and KIR genes in Mali family trios by Illumina Next Generation Sequencing
A pool of oligonucleotide capture probes was used to target the KIR genomic region and HLA genes for high throughput sequencing (30), with modifications to the library preparation as described (31). The fragments captured were subjected to paired-end sequencing using Illumina’s MiSeq instrument and V3 sequencing chemistry (Illumina Inc. San Diego, CA).
Processing and analysis of sequence data
The PING bioinformatics pipeline (30), was used for the assessment of KIR gene copy number and KIR allele content from the Next Generation Sequencing data. This pipeline has the capacity to provide information on new SNPs and new recombinant KIR alleles.
For the Baka and Mbuti samples, the data obtained from pyrosequencing, were supplemented with low-coverage (4x) whole genome sequencing data. To extract KIR-specific sequences, we used SAMtools 0.1.18 (32) to identify read pairs that mapped either within the KIR region (hg19 coordinates: 19:55,228,188–55,383,188) or to an unallocated chromosome 19 region (GenBank: GL000209.1) corresponding to an alternative KIR haplotype. The KIR alleles present were then determined using PING (30).
The Immuno Polymorphism Database (IPD/KIR database 2015 release) was used as the reference set of KIR alleles. HLA-A, -B and -C alleles were determined using the NGSengine 1.7.0 software (GenDX, Utrecht, the Netherlands) with the IMGT 3.18.0 combined reference set. These data were analyzed directly by the software, with no pre-filtering being needed for the HLA genes.
KIR haplotypes
Centromeric (cen) and telomeric (tel) KIR haplotypes were determined separately. For the Mali, family trios were used to determine the KIR haplotypes. For other populations, the cen and tel haplotypes were deduced from the patterns of linkage disequilibrium within each population. This was achieved by first determining the most common pairs of KIR alleles present at adjacent genes, then choosing the most likely combination for individual and iteration to define the entire cen or tel segment. The results obtained by this iterative approach were validated for the larger numbers of Dogon and Hadza individuals studied, because we could compare them to the results obtained using PHASE. A further test of their validity, was their comparison with the segregation of cen and tel KIR haplotypes in the Mali population.
Calculation of a population’s mean number of HLA-KIR interactions
For each individual in a cohort, the total number of different, functional interactions between their HLA and KIR was calculated as described (31). Only ligand-receptor pairs that have been defined by functional analysis were considered (33–40). These comprise Bw4+HLA-A and KIR3DL1 (33), Bw4+HLA-B and KIR3DL1 (33, 35), HLA-A*03 and KIR3DL2 (36), HLA-A*11 and KIR3DL2 (37), C2+HLA-C and KIR2DL1 (38), C1+ and C2+HLA-C and KIR2DL2 (38), C1+HLA-C and KIR2DL3 (38), C2+HLA-C and KIR2DS1 (38), HLA-C*16 and KIR2DS2 (40), HLA-A*11 and KIR2DS4 (34), a subset of HLA-C and KIR2DS4 (34), and C2+HLA-C and KIR2DS5 (only some KIR2DS5 allotypes) (39). In counting the interactions, homozygous ligands and receptors were counted as one and scores for each ligand-receptor pair could be 0, 1, 2 or 4.
Results
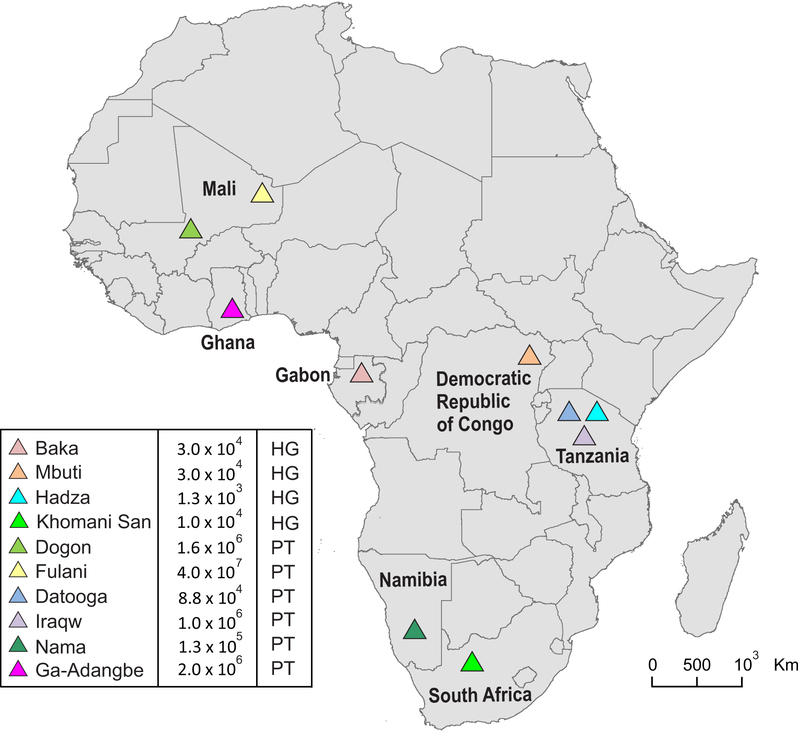
High-resolution genotyping was used to determine the allelic diversity of KIR and HLA class I genes in 180 individuals representing seven populations of sub-Saharan Africans (SSA). The study cohorts were small, varying between 13 and 52 individuals (Fig. S1A). Four of the populations; Dogon (Mali), Fulani (Mali), Datooga (Tanzania) and Iraqw (Tanzania) are pastoralists. The other three; Baka (Gabon), Mbuti (Democratic Republic of Congo) and Hadza (Tanzania) are hunter gatherers. The location of each of these populations is shown on the map in Figure 1. They include populations from eastern Africa, central Africa and western Africa. Also shown on the map are three SSA populations we previously characterized; the Ga-Adangbe (Ghana) (23), the Nama (Namibia) (31) and the Khomani San (South Africa) (31).
FIGURE 1. African populations studied.
This map of Africa shows the locations of the ten sub-Saharan African populations that have been allele-level genotyped for KIR and HLA class I. Previously studied were the Ga-Adangbe from Ghana in West Africa (23) and two KhoeSan hunter-gatherer (HG) populations, the Nama and the Khomani San, from southern Africa (15, 31). Of the seven populations, on which KIR and HLA class I are first reported here, the Baka and Mbuti Pygmies and the Hadza are hunter gatherers, and the Dogon, Fulani, Datooga and Iraqw are pastoralists (PT). The estimated size of each population is given in the key (https://en.wikipedia.org/wiki/Iraqw_people, https://en.wikipedia.org/wiki/Dogon_people, https://en.wikipedia.org/wiki/Mbuti_people, https://en.wikipedia.org/wiki/Datooga_people, https://en.wikipedia.org/wiki/Baka_people_(Cameroon_and_Gabon)), https://en.wikipedia.org/wiki/Hadza_people, https://en.wikipedia.org/wiki/Fula_people, https://en.wikipedia.org/wiki/Nama_people, https://en.wikipedia.org/wiki/Ga-Adangbe_people.
Every HLA class I identified in the sub-Saharan Africans corresponds to a known allele
A total of 135 HLA class I alleles was identified in the 170 SSA individuals (Fig. S1B): 41 HLA-A alleles, 61 HLA-B alleles and 33 HLA-C alleles. Although the cohorts varied by up to four fold in size, the numbers of HLA class I alleles detected for each population were less variable, ranging from 34 to 55 (Fig. S1B). None of the HLA-A or HLA-B alleles is present in all seven populations, consistent with these genes having been subject to selection by fast evolving pathogens, such as RNA viruses (41). Contrasting with HLA-A and -B, three HLA-C alleles are common to all seven populations; these comprise C*06:02, C*07:01 and C*07:02. HLA-C*06:02 has the C2 epitope, whereas HLA-C*07:01 and C*07:02 have the C1 epitope (Fig. S1B). This difference between HLA-C and either HLA-A or -B is consistent with HLA-C polymorphism having evolved more slowly than HLA-A and -B polymorphism (42), a difference attributed to its distinctive role in reproduction (4). An unanticipated finding from this study is that all 135 of the HLA-A, -B and -C alleles present in these seven distinctive and unstudied SSA populations correspond to known alleles, for which the sequences were already in the HLA database (43). Thus knowledge of the breadth and depth of HLA class I in SSA populations is now seen to be more complete than previously appreciated. In the combined data set for the seven SSA populations (N=170): HLA-A*29:01, HLA-B*44:03 and HLA-C*16:01 are, respectively, the most common HLA-A, -B and -C alleles. HLA-A*29:01 and -B*44:03 reach highest frequency in the Hadza (21.6% and 37.5%, respectively), whereas HLA-C*16:01 reaches highest frequency in the Fulani (32.1%) (Fig. S1B).
High polymorphism and heterozygosity are characteristic features of the HLA-A, -B and -C genes in all seven SSA populations (Fig. 2A). Forty eight of the HLA class I alleles (36% of the total) are present in only one of the seven populations and within this dataset are candidate ‘population-specific’ alleles. Such alleles are present in all seven populations, but to varying degree (Fig. S1C); a majority of them have frequencies of less than 0.05, with the three most common having frequencies of 0.100 (HLA-A*66:03 in the Baka and HLA-B*44:01 in the Mbuti) or 0.105 (HLA-C*08:04 in the Baka) (Fig. S1C). Because of the small sample sizes, it is likely that some, of the putative ‘population-specific’ alleles are present in more than one of the populations.
FIGURE 2. HLA class I diversity in seven sub-Saharan African populations.
(A). Shown is the variation (k) and heterozygosity (H) for HLA class I alleles and haplotypes. (B) The frequency of HLA-A, -B and -C allotypes within each category of KIR ligand is shown for each population.
Extensive diversity of HLA class I haplotypes in sub-Saharan Africans
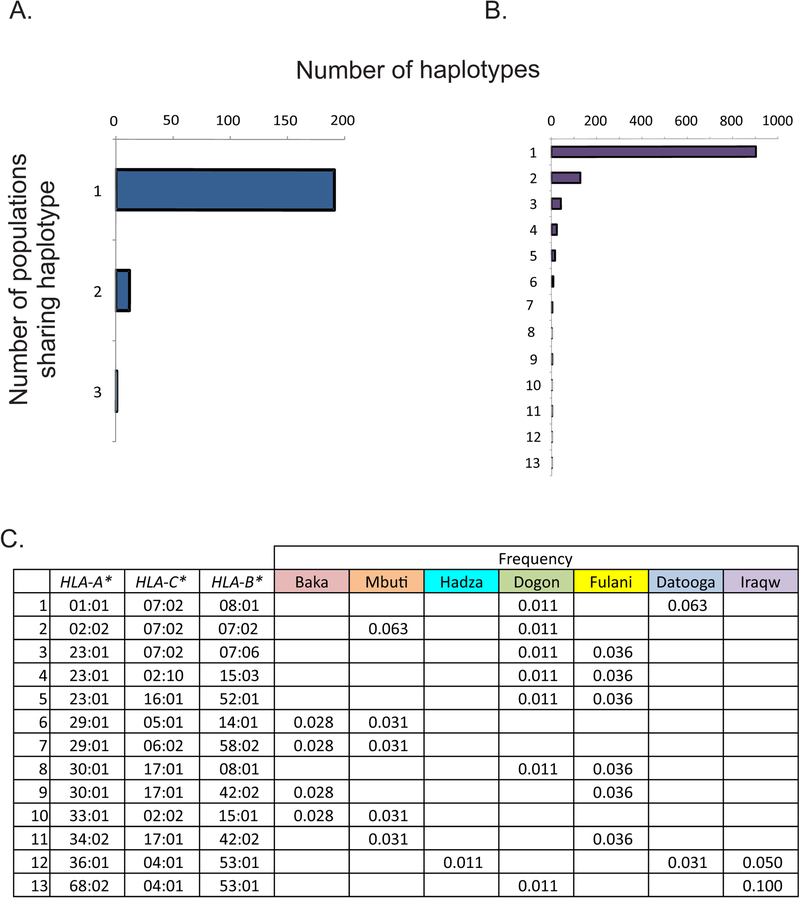
In the HLA region of the human genome, the order of the polymorphic HLA class I genes is HLA-A, HLA-C and HLA-B. In the seven SSA populations, we defined a total of 204 different HLA A-C-B haplotypes in the seven SSA populations. For short, these haplotypes will subsequently be called HLA haplotypes. Of the 204 haplotypes, 191 were observed in just one of the populations, twelve were observed in two populations and one was observed in three populations (Fig. 3A). No HLA haplotype was seen in more than three of the seven populations (Fig. 3C, S1D). This striking result shows how little sharing of HLA haplotypes there is among these anthropologically well-defined African populations. Expanding this analysis to include all African populations with defined HLA haplotypes (23 populations, 1134 HLA haplotypes) showed that 80% of the haplotypes were found only in one population. Most widespread was one haplotype present in 13 of the 23 populations (Fig. 3B). Haplotypes that are frequent in one population can be absent from the others. For example, the HLA-A*29:02, -C*16:01, -B*78:01 haplotype has high frequency in the Fulani (21.6%) but was not detected in the other six populations (Fig. S1D). Neither is this haplotype present in previously characterized SSA populations of western (23), and southern Africa (31). In contrast, this haplotype is present in 4% of African-Americans, an admixed population with genetic input from a variety of African populations (44).
FIGURE 3. Distribution of HLA class I haplotypes among seven sub-Saharan African populations.
(A). Shown are the numbers of HLA class I haplotypes observed in one, two or three of the seven populations. (B) The numbers of haplotypes present in one, two, three etcetera up to 13 are given for all African HLA class I haplotypes present in the database. (C) Shown are HLA class I haplotypes found in more than one population ordered by their HLA-A alleles.
In the Mbuti, Datooga and Iraqw cohorts, almost all HLA haplotypes were detected in only one individual, with three haplotypes being found in two individuals. Similarly, most Baka HLA haplotypes are represented once, with only six haplotypes being represented by two or three individuals (Fig. 4, S1E). In contrast, the spectra of Hadza HLA haplotype frequencies have lower complexity. Of 88 different HLA haplotypes, 25 are represented only once. But two haplotypes have high frequencies, being represented 13 or 14 times. At intermediate frequencies are ten HLA haplotypes, each observed in 2–8 individuals (Fig. 4, S1E). The less complex haplotype spectra observed in the Hadza supports the severe population bottleneck described by Henn et al for the population studied here (25). Different again is the Dogon, a population in which the majority of HLA haplotypes were detected in only one person. Among the minority haplotypes, one was represented twelve times, one six times, two four times, and six twice. Fulani HLA haplotypes exhibit a similar frequency spectrum to that of the Dogon, the other West African population (Fig. 4, S1E). Thus in six of the seven study populations, a majority (54–81%) of HLA haplotypes are represented once. Only in the Hadza are such singleton haplotypes a minority (28%) of the total number of HLA haplotypes.
FIGURE 4. HLA class I haplotype diversity in seven sub-Saharan African populations.
Shown for each SSA population, is the frequency distribution of all HLA class I haplotypes. Also given are the frequency distributions of HLA class I haplotypes in three non-African populations: North Americans (NAM), South East Asians (SEA) and West Asians (WAS). For each population the total number of haplotypes (2N) is indicated.
To assess if this frequency spectrum of HLA haplotypes is a more general phenomenon, we examined cohorts representing 25 other populations, for which the cohorts studied are of similar size to the seven SSA populations. Having no singleton HLA haplotypes is the Chihuahua Tarahumara population of Mexico (N=44). In contrast, 26–32% of HLA haplotypes are singletons in the Rukai, Yami, Puyuma, and Tsou populations of South East Asia. Singletons comprise 41–88% of the HLA haplotypes in the other 20 populations; they are particularly frequent, >80%, in populations of South African Mixed Ancestry, Tamil (South Asia), Mexican Mixed Ancestry and Kurds (West Asia) (Fig. 4, S1F). In conclusion, the diversity and distribution of HLA haplotypes we find in the seven SSA populations, are also characteristic of other human populations but not all.
One mechanism that creates new HLA haplotypes is reciprocal recombination in the ~1,300 kb region that separates HLA-A from HLA-C. In the Mbuti, for example, the five haplotypes containing HLA-A*23:01 have different combinations of HLA-C and HLA-B alleles. Because the HLA-C and HLA-B genes are only separated by ~130 kb, the frequency of recombination between HLA-C and HLA-B is expected to be one tenth of that between HLA-A and HLA-C. Despite this constraint, we could identify 17 examples of recombination between HLA-B and HLA-C in the seven SSA populations (Fig. S1G). In the Mbuti two haplotypes have HLA-A*23:01 linked to HLA-C*02:02. One of these haplotypes HLA-B*15:01 and the other has HLA- B*15:02. This arrangement makes it highly likely that one of these haplotypes was derived from the other by reciprocal recombination between HLA-B and HLA-C. This difference could not have occurred through point mutation, because HLA-B*15:01 and HLA-B*15:02 differ by eight nucleotide substitutions, spread throughout exons 2 and 3, that cause five amino-acid substitutions in the α1 and α2 domains.
Hadza HLA haplotypes have a distinctive frequency spectrum
The Hadza have two high-frequency HLA haplotypes (A*23:01-C*07:01-B*41:02 and A*01:02-C*17:01-B*44:03) that account for 31% of all Hadza HLA haplotypes (Fig. S1E). That no HLA-A, -B or -C allele is shared by these two haplotypes points to them having complementary functions. For example, one haplotype encodes the C1 KIR ligand (carried by HLA-C*07:01) and the other encodes the C2 KIR ligand (carried by HLA-C*17:01). We tested Hardy-Weinberg equilibrium using three alleles; two were the high-frequency HLA haplotypes and the third was all other HLA haplotypes combined. The result of this test shows their frequencies are consistent with Hardy-Weinberg equilibrium. Consequently, the cohort of 44 Hadza includes one C*07:01 homozygote, one C*17:01 homozygote and two C*07:01/C*17:01 heterozygotes. At moderate frequency in the Hadza are three other HLA haplotypes that together account for a further 20% of the HLA haplotypes (Fig. S1E).
The Dogon has one common HLA haplotype, which is of similar frequency to the two common Hadza haplotypes. The one feature this Dogon haplotype (A*30:01-C*17:01-B*42:01) shares with a common Hadza haplotype is HLA-C*17:01, an allele specific to SSA populations that encodes a structurally divergent HLA-C allotype (Fig. S1E).
The HLA haplotypes of the seven SSA populations were compared to those of previously studied SSA populations. These include a Ghanaian West African population (23), KhoeSan populations of southern Africa (31), as well as populations for which HLA haplotype data are deposited in the Allele Frequency Net Database (www.allelefrequencies.net/) (44, 45). Of 204 HLA haplotypes present in the seven SSA populations reported here, 28% of them are present in other SSA populations. Among these other SSA populations, the most common haplotype is HLA-A*30:01 -C*17:01 -B*42:01 (Fig. S1H). With a frequency of 13%, it is the most common HLA haplotype of the Dogon. Of the 60 different HLA class I haplotypes identified in the Dogon only 26 are shared with the other SSA populations (Fig. 5A, S1I). Most similar to the Dogon, sharing 13 haplotypes, are the Ghanaians. In contrast, the Hadza share only one haplotype (HLA-A*36:01 -C*04:01 -B*53:01) with the other six sub-Saharan African populations reported here (Fig. 3C).
FIGURE 5. HLA class I diversity in Africans and other populations.
(A). Heat map showing frequencies of 231 HLA class I haplotypes common to African populations. In the color key on the right, red denotes the highest and blue the lowest frequency haplotype.
(B). Shown for each population are the number of HLA-A, HLA-B and HLA-C alleles, as well as the total number of HLA-A, -B and -C alleles, present in the seven sub-Saharan African populations (SSA), Yucpa (South America: SA) (14), Maori and Polynesians (Oceana: OCE) (47), Japanese (East Asia: EA) (48), Europeans (45), Khomani San (31), Nama (31) and Ghanaians (23). For each population the total number of HLA-A, -B and -C alleles is given.
In summary, 73% of the HLA class I haplotypes in the seven SSA populations are specific to their respective population and not observed in other SSA populations. Among the seven populations, the Hadza has the most distinctive repertoire and diversity of HLA class I haplotypes.
Comparison of sub-Saharan African HLA class I diversity to that in other populations
Comparison of the HLA class I diversity in populations worldwide shows the KhoeSan of southern Africa have the highest diversity (31) and Yucpa South Amerindians have the least (Fig. 5B) (46). Among the seven SSA populations, the Dogon has the highest number of different HLA class I alleles (k= 55). The Datooga has a comparable number of alleles (k=53), although the cohort (N=16) is much smaller that of the Dogon (N=44). Unlike most populations, in which there are more different HLA-B alleles than HLA-A alleles (14, 23, 31, 47, 48), the Iraqw, Hadza and Mbuti have a greater number of HLA-A alleles than HLA-B alleles (Fig. 5B).
Six of seven sub-Saharan African populations have the four HLA epitopes recognized by KIR
The four HLA class I epitopes recognized by KIR are the A3/11 epitope carried by HLA-A3 and HLA-A11, the Bw4 epitope carried by subsets of HLA-A and -B allotypes, and the C1 and C2 epitopes carried by subsets of HLA-C allotypes. These epitopes are also called KIR ligands. Six of the seven SSA populations have all four KIR ligands, whereas the Iraqw have Bw4, C1 and C2 but lack A3/11. That we failed to detect the A3/11 epitope in the Iraqw could be due to its absence from this population, or to inadequate sampling, because only ten Iraqw individuals were studied (Fig. 2B). Intriguing is the abundance in Iraqw of Bw4, which is carried by 25% of HLA-A allotypes and 82% of HLA-B allotypes. Consequently, >70% of Iraqw individuals have two or more HLA-A and -B allotypes bearing the Bw4 epitope (Fig. S1J). In the seven SSA populations, the frequency of A3/11 ranges from 0–14% and that of Bw4 ranges from 8.3–82% (Fig. 2B). Each Hadza individual has one or more Bw4+HLA-A/B allotypes and >50% of individuals have two Bw4+HLA-A/B allotypes (Fig. S1J). The largest contributor to this elevated frequency is B*44:03, possibly a consequence of selective pressure due to disease (25). Worldwide, the Bw4+HLA-B allotype reaches its highest frequencies in the Hadza (69%) and the Iraqw (82%). Third in this hierarchy is the Ga-Adangbe, another SSA population, with a Bw4+HLA-B frequency of 53%. Baka combine the lowest frequency of Bw4+ HLA-A/B allotypes (39%) with a relatively high frequency, 12.5%, of HLA-A allotypes having the A3/11 epitope (Fig. 2B).
Worldwide analysis identified five populations that completely lack the A3/11 epitope (Fig. S1K). They are all indigenous Amerindian populations: the Chimila, Yucpa and Bari of South America, and the Chihuahua Tarahumara and NaDene of North America. Characterizing these populations is an abundance of the Bw4 epitope. With 58% Bw4+HLA-A and 48% Bw4+HLA-B, the Chimila has the highest Bw4 frequency. In contrast, the Yucpa has the lowest Bw4 frequency: 17% Bw4+HLA-A and 11% Bw4+HLA-B. The trend in Yucpa, for Bw4+HLA-A to dominate Bw4+HLA-B, increases in the Bari, who have 72% Bw4+HLA-A and 6% Bw4+HLA-B, and is highest in the Paiwan of South East Asia who have 86% Bw4+HLA-A and 26% Bw4+HLA-B (Fig. S1K).
African KIR diversity varies with geographical location
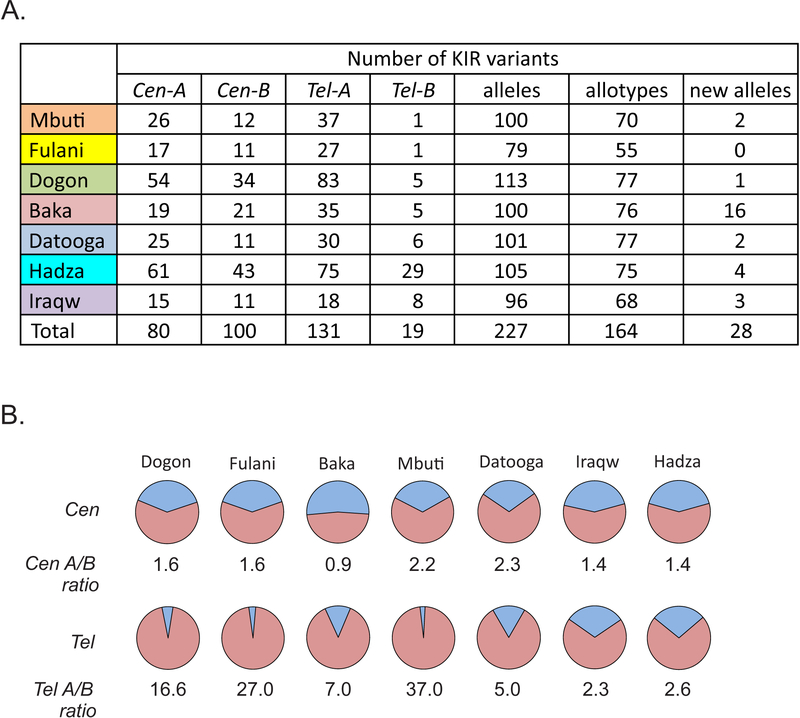
In the seven SSA populations, we detected 227 KIR alleles that encode 164 KIR allotypes (Fig. 6A). The individual populations have 79–113 KIR alleles that encode 55–77 KIR allotypes. Common to the seven populations are 22 KIR alleles encoding 17 KIR allotypes. These comprise 9.7% of the total KIR. For KIR3DL1, 3DL2 and 3DL3, the most polymorphic KIR genes, only seven alleles (three 3DL3, two 3DL1 and two 3DL2 alleles) are present in all populations, showing how each population has a different KIR repertoire (Fig. S1L).
FIGURE 6. KIR diversity in seven sub Saharan African populations.
(A). Shown are the number of cenA, cenB, telA and telB haplotypes identified in each population. Also shown are the numbers of different KIR alleles, KIR allotypes and new KIR alleles characterized in this study.
(B). Shown are the frequencies of the centromeric (cenA) and telomeric (telA) KIR A haplotypes (red) and the centromeric (cenB) and telomeric (telB) KIR B haplotypes (blue) in each population ordered by their A/B haplotype ratio.
Specific to the seven SSA populations are 28 new KIR alleles, each specific to one of the populations (Fig. 6A). Sixteen of these KIR are Baka-specific, whereas the other populations have between 0 and 4 new alleles. The Baka are a population of African pygmies, who are genetically highly diverged from the non-pygmy populations of Africans (49, 50). Thus the abundance of new KIR alleles in the Baka is likely to be a reflection of this divergence. Defining these new alleles are 33 nucleotide substitutions, ten of which are silent and 23 are coding changes (Fig. S1M). Fifteen of the new alleles were detected as single observations, and 13 were present in two or more individuals. Most common is KIR2DL5B*020, which is present in 12 Hadza. Single nucleotide substitutions define 22 of the new alleles, six are defined by two or more substitutions. Defining KIR2DL5B*020 is a substitution that terminates translation at residue 283 in the cytoplasmic domain (Fig. S1M). Although this shortens the cytoplasmic tail, the ITIM motif is retained.
Three other new KIR alleles have changes that are predicted to alter KIR function. Distinguishing Baka-specific KIR2DL4*031 is a substitution that terminates translation at position 279 in an ITIM motif of the cytoplasmic tail. The second new allele, also Baka-specific, is KIR3DL1*081N, for which a nucleotide deletion alters the reading frame in codon 85 and causes early termination. The third new allele, Hadza-specific KIR3DL3*059, has a substitution in codon 86, which is known to prevent cell-surface expression of KIR3DL1*004 (51). For the other 16 other new KIR alleles that have coding changes, the substitutions are at sites that are not associated with functional change (Fig. S1M).
For each population the KIR alleles were assembled into haplotypes (see Materials and Methods). A total of 180 centromeric and 148 telomeric KIR haplotypes were defined (Fig. 6A and S1N–O). Each of the seven populations has a relatively even balance of cenA and cenB haplotypes, in which the cenA/B ratio ranges from 0.9 to 2.3 (Fig. 6B). In contrast, the ratio of telA to telB haplotypes varies greatly, from 2.3 to 37, and correlates partly with geographical location (Fig. 1). In the West and Central African populations, Dogon, Fulani and Mbuti have higher telA/B ratios of 16.6–37; and the East African populations of Hadza, Datooga and Iraqw have lower telA/B ratios of 2.3–5.0 (Fig. 6B). Exceptional is the Baka, who have a low-intermediate telA/B ratio (7.0). Overall we observe a gradient from east to west across sub-Saharan Africa in which the cenA/B ratio remains relatively constant, whereas the telA/B ratio progressively increases to the point where telA is dominant in West African populations and telB is rare. This is consistent with balancing selection operating on the cen region, with the tel region being subject to directional selection. In all seven populations the frequencies of cenA and cenB, and of telA and telB, are consistent with Hardy-Weinberg equilibrium.
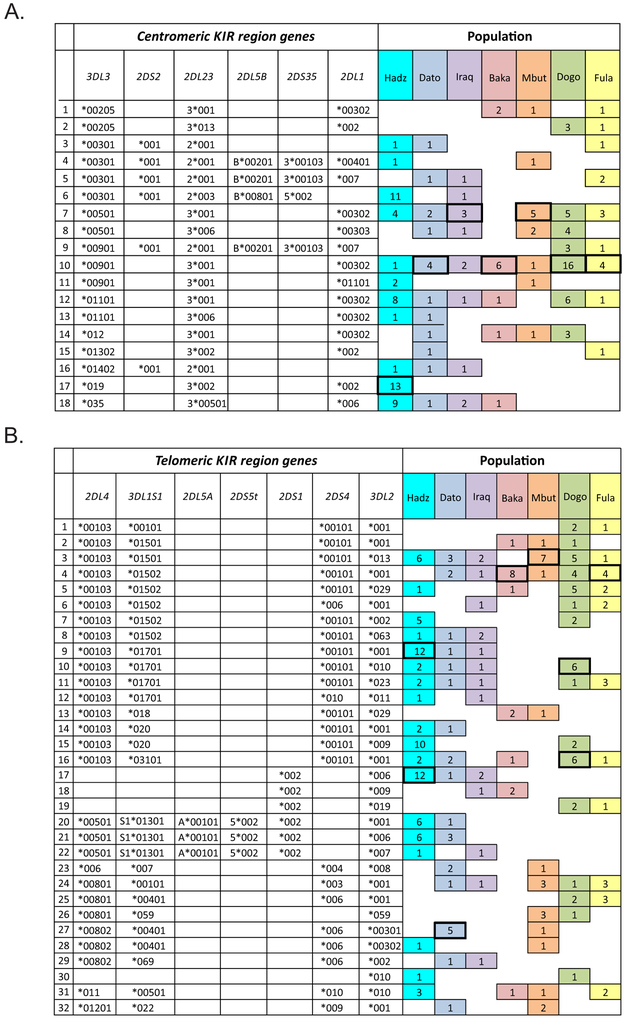
Common to all the sub-Saharan populations is the centromeric haplotype, 3DL3*00901–2DL3*001–2DL1*00302. It is also the most frequent centromeric haplotype in four of the populations: Datooga, Baka, Dogon and Fulani. This haplotype is common to populations in eastern, central and western Africa (Fig. 7A). The dominant centromeric haplotype in the Hadza is 3DL3*019–2DL3*002–2DL1*002. Distinguishing this haplotype is 3DL3*019, which is absent from the other six SSA populations, but present in Europeans at a frequency of 0.5%. The second most frequent centromeric haplotype of the Hadza is 3DL3*00301–2DS2*001–2DL2*00301–2DL5B*00801–2DS5*002, which lacks the KIR2DL1 gene. This haplotype is also present in one Iraqw individual (Fig. 7A).
FIGURE 7. KIR haplotypes segregating in seven populations of sub-Saharan Africans.
Shown on the left are 18 centromeric KIR haplotypes (A) and 32 telomeric KIR haplotypes (B). Included are the most common KIR haplotypes present in each population and all of the KIR haplotypes present in two or more populations. Shown on the right are the distributions of the 50 KIR haplotypes among the seven SSA populations (colored boxes) and the occurrence of each haplotype in each population (the number in each box). The boxes corresponding to the most frequent centromeric and telomeric KIR haplotypes have bold outlines.
No telA or telB haplotype is common to all seven SSA populations. Two common telB haplotypes are present in the three east African populations: Hadza (11.5% frequency), Iraqw and Datooga (Fig. 7B). Haplotype 2DS1*002–3DL2*006 has a deletion of the KIR2DL4-KIR2DS5 module and is common in KhoeSan (31). Haplotype 2DL4*00103–3DL1*01701–2DS4*00101–3DL2*001 has 3DL1*01701, an African-specific allele (23).
Diversity of interaction between HLA class I and KIR in sub-Saharan Africans
For the seven SSA populations, we determined the number of KIR ligands encoded by each HLA haplotype. For each population we then determined the distribution of haplotypes encoding one, two or three KIR ligands. The analysis was performed in two ways: one weighted each haplotype according to its population frequency, the other gave each distinctive haplotype an equal weight. The two methods gave similar results (Fig. S1P).
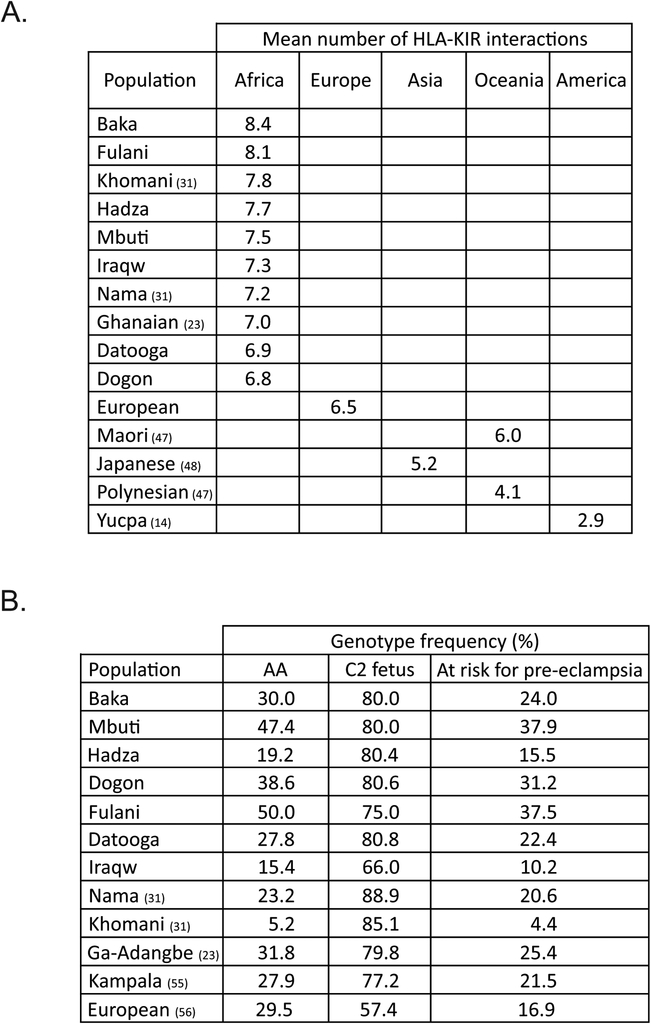
Although each population exhibited a different frequency distribution of HLA haplotypes (Fig. 8), they clearly formed two groups. For the first group, (Baka, Mbuti, Dogon, Fulani and Datooga) HLA haplotypes encoding one KIR ligand and HLA haplotypes encoding two KIR ligands have comparable frequency, whereas haplotypes encoding three KIR ligands are less frequent (Fig. 8). For the second group, comprising Iraqw and Hadza, the majority of HLA haplotypes encode two KIR ligands. We found no significant difference between the ligand distributions in the Iraqw and the Hadza, as assessed by χ2 analysis. In the seven SSA populations, the mean number of HLA-KIR interactions per individual varied from 6.8–8.4, higher values than the 2.9–6.5 interactions seen in populations outside of Africa (Fig. 9A).
FIGURE 8. HLA class I allele frequency spectra for seven sub Saharan African populations.
Venn diagrams (pie charts) give the frequencies of HLA-A, -B and -C alleles for each population. ‘n’ denotes the number of different alleles for each gene. The frequencies of allotypes having epitopes recognized by KIR are color-coded: yellow, the A3/11 epitope; green, the Bw4 epitope; red, the C1 epitope and blue, the C2 epitope. The column on the right gives the relative frequencies of HLA class I haplotypes, encoding one (yellow), two (orange) or three (red) KIR ligands.
FIGURE 9. Diversity in the functional potential of KIR-HLA interactions in human populations.
(A). For sub-Saharan Africans (23, 31) and other populations (14, 47, 48) the combined KIR and HLA class I genotype was used to define, for each individual, the total number of different pairs of interacting KIR and HLA class I ligands. Averaging the number of such interactions over the population gives the ‘Mean number of HLA-KIR interactions’.
(B). Pregnancies in which the mother is homozygous for C1 and KIR A, and the fetus expresses C2, are at risk for pre-eclampsia. Shown are the predicted frequencies of such pregnancies in sub-Saharan African populations and Europeans (23, 31, 55, 56).
Discussion
Sequence analysis of HLA-A, -B and -C was first applied to the common alleles of Europeans and subsequently to populations of other continents, including Africans. This led to the discovery of numerous HLA class I alleles that are restricted in their distribution to particular populations or groups of populations. It became a pattern that new alleles would be discovered when high-resolution molecular analysis was applied to non-Caucasoid populations. For example, the indigenous populations of the Americas, proved a particularly rich source of new HLA class I alleles (52). With this background, our expectation was that some novel HLA class I alleles would emerge from analysis of the anthropologically and geographically diverse set of seven SSA populations studied here. It was therefore striking that no new HLA class I variants emerged from this study. In contrast, 28 new KIR were found among a total of 227 KIR identified in the seven populations. More than half of the new KIR, 16, are from the Baka population of Gabon. From this and previous studies we can estimate a range for the numbers of KIR and HLA class I alleles that are present in anthropologically well-defined human populations. Having least diversity is the Yucpa of South America, who have 19 HLA class I and 29 KIR alleles (14). Having most diversity is the Nama of southern Africa, who have 97 HLA class I and 202 KIR alleles (31).
A significant finding to emerge from this investigation is the high diversity of HLA A-C-B haplotypes in the SSA populations. Most of the haplotypes are present in just one individual, and few haplotypes have higher frequencies. In this regard the Hadza are exceptional because they have two haplotypes at comparably high frequency, which account for 5% (2 of 37) of the haplotypes, as well as 27% (10 of 37) of the haplotypes that have intermediate frequencies. Extending this form of analysis to other populations, showed how high HLA A-C-B haplotype diversity is not specific to SSA populations, but a general phenomenon worldwide. Among the 25 populations analyzed 41–88% of the HLA A-C-B haplotypes were present in one member of the study cohort. Exceptional are four South Asian populations (Rukai, Yami, Puyuma and Tsou) in which such singleton haplotypes are a minority, and one North American population, Mexico Chihuahua Tarahumara, in which they are not found at all.
The more frequent centomeric KIR haplotypes are present in all seven SSA populations. One exception is the Hadza’s most frequent centromeric KIR haplotype, which is specific to this population. In contrast, the Hadza’s most frequent telomeric KIR haplotype, 2DS1*002-3DL2*006, has moderate frequency in the Iraqw and highest frequency in the KhoeSan. This haplotype is specific to Africans (31). Telomeric KIR haplotype 2DL4*00501–3DS1*001301-2DL5A*00101-2DS5*002-3DL2*006 is present in Hadza, Datooga and differs from the common European haplotype, 2DL4*00501-3DS1*001301-2DL5A*00101-2DS3*002-3DL2*007, at KIR2DS3 and KIR3DL2.
In the SSA populations the ratio of the KIR A/B haplotypes also correlates with geographical location. While the ratio of centromeric haplotypes stays relatively constant across Africa, the telA/B ratio progressively increases from east to west, with the exception of Baka, and is lowest in the Tanzanian populations. This difference is likely the result of varying selective pressures. Disease correlation studies indicate that telA correlates with greater resistance to pathogens (8), whereas telB correlates with reproductive success (53). Possible candidates in western Africa that selected for telA and against telB include endemic pathogens, such as malaria, or any of the many rapidly evolving viruses that continually evolve to thwart the human immune response (54).
Comparing the HLA class I haplotypes present in the seven SSA populations with other African population haplotypes in the database revealed very little sharing among these populations pointing again to their unique HLA class I diversity. Comparing this diversity with populations worldwide shows the highest diversity in the KhoeSan from southern Africa and the lowest in the Yucpa. Yucpa also lack allotypes carrying the A3/11 epitope and has the lowest frequency of Bw4+ allotypes. In the SSA populations where A3/11 is absent there is an abundance of Bw4+ epitopes. Consequently populations with the lowest frequency of Bw4+ allotypes have a relative high frequency of A3/11+ allotypes. This is not always the case in other cohorts. Populations with a low frequency of Bw4+ allotypes do not necessarily have a high frequency of A3/11+ allotypes.
The distribution of KIR ligands is similar in most of the SSA populations (Dogon, Fulani, Baka, Mbuti and Datooga). In these populations a majority of HLA haplotypes encode one or two KIR ligands and a minority of haplotypes encode three KIR ligands. A somewhat different distribution is seen in Hadza and Iraqw for which the majority of HLA haplotypes encode two KIR ligands and the minority comprises roughly equal numbers of HLA haplotypes encoding one or three KIR ligands.
The Hadza has the most distinctive diversity of HLA class I, sharing only one haplotype with the other populations in this study. The two high frequency haplotypes do not have any HLA alleles in common and likely have complementary functions. The presence of these high frequency haplotypes in the Hadza could reflect their benefits in the immune response to infection and/or the reproductive success of individuals carrying these haplotypes.
As an assessment of the functional effect of KIR-HLA interactions, we compared the risk of pre-eclampsia among the human populations for which both KIR and HLA class I genotypes were known for each individual. Pregnancies in which the fetus expresses C2 and the mother is homozygous for C1 and KIR cenA are at risk for pre-eclampsia (53). This correlation suggests that inhibitory interaction between fetal C2 and maternal KIR2DL1 contributes to preeclampsia. This effect can be offset if the mother expresses an activating C2 receptor, which in Europeans is KIR2DS1*002 and in Africans is the subset of KIR2DS5 allotypes that recognize C2 (39, 55). In particular, 2DS5*006 is significantly associated with protection of Ugandan women from pre-eclampsia (39). Among the seven populations studied here, the Mbuti and Fulani are predicted to have the highest risk of pre-eclampsia, whereas the Iraqw are predicted to have the lowest risk (Fig. 9B). To explore what might provide protection for these populations, we calculated the frequencies of their KIR-AB and KIR-BB genotypes and grouped them according to the presence of the C2 ligand, as well as the activating, KIR2DS1 and KIR2DS5, and inhibitory, 2DL1, C2 receptors. The analysis suggests that protection in the Iraqw could come from KIR2DS1, which has high frequency (46.2%), whereas protection in the Baka could come from KIR2DS5 which is present in 45% of individuals having either an AB or BB KIR genotype. In the Hadza both KIR2DS1 and KIR2DS5 could provide protection (Fig. S1Q).
On starting this project we expected to discover new HLA-A, -B and -C coding sequence alleles that are specific to one or more sub-Saharan African populations. Instead, we found no single allele that had not been seen before. Because Africans are genetically the most diverse of human population groups, and the least well studied, our results raise the possibility that most if not all of HLA-A, -B and -C coding sequences are now known. In the context of hematopoietic cell transplantation for genetic and malignant diseases, the next challenge is to characterize polymorphisms in non-coding regions of HLA genes that influence the expression and function of their protein products. The KIR genes also have a genetic variation and polymorphism that bears comparison to that of the HLA genes. In this study 28 previously unknown KIR coding sequence alleles were defined, showing how further exploration will be needed to map the full extent of human KIR diversity.
Supplementary Material
Key points.
High levels of KIR and HLA diversity are present in all seven African populations.
Although 28 novel KIR alleles were detected, all HLA class I alleles are familiar.
KIRtelA and telB are balanced in East Africa, but KIRtelA dominates in West Africa.
Acknowledgments
This study supported by NIH grants AI090905 and AI17892 to P.P.
References
- 1.Trowsdale J, and Knight JC. 2013. Major histocompatibility complex genomics and human disease. Annu. Rev. Genomics Hum. Genet 14: 301–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cruz FM, Colbert JD, Merino E, Kriegsman BA, and Rock KL. 2017. The biology and underlying mechanisms of cross-presentation of exogenous antigens on MHC-I molecules. Annu. Rev. Immunol 35: 149–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.King A, Boocock C, Sharkey AM, Gardner L, Beretta A, Siccardi AG, and Loke YW. 1996. Evidence for the expression of HLAA-C class I mRNA and protein by human first trimester trophoblast. J. Immunol 156: 2068–2076. [PubMed] [Google Scholar]
- 4.Parham P, and Moffett A. 2013. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat. Rev. Immunol 13: 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson J, Guethlein LA, Cereb N, Yang SY, Norman PJ, Marsh SGE, and Parham P. 2017. Distinguishing functional polymorphism from random variation in the sequences of >10,000 HLA-A, -B and -C alleles. PLoS Genet. 13: e1006862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long EO 2002. Tumor cell recognition by natural killer cells. Semin. Cancer Biol 12: 57–61. [DOI] [PubMed] [Google Scholar]
- 7.Campbell KS, and Purdy AK. 2011. Structure/function of human killer cell immunoglobulin-like receptors: lessons from polymorphisms, evolution, crystal structures and mutations. Immunology 132: 315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bashirova AA, Martin MP, McVicar DW, and Carrington M. 2006. The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annu. Rev. Genomics Hum. Genet 7: 277–300. [DOI] [PubMed] [Google Scholar]
- 9.Boudreau JE, and Hsu KC. 2018. Natural killer cell education and the response to infection and cancer therapy: stay tuned. Trends Immunol 39: 222–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naiyer MM, Cassidy SA, Magri A, Cowton V, Chen K, Mansour S, Kranidioti H, Mbirbindi B, Rettman P, Harris S, Fanning LJ, Mulder A, Claas FHJ, Davidson AD, Patel AH, Purbhoo MA, and Khakoo SI. 2017. KIR2DS2 recognizes conserved peptides derived from viral helicases in the context of HLA-C. Sci. Immunol 2: eaal5296. [DOI] [PubMed] [Google Scholar]
- 11.Sim MJ, Malaker SA, Khan A, Stowell JM, Shabanowitz J, Peterson ME, Rajagopalan S, Hunt DF, Altmann DM, Long EO, and Boyton RJ. 2017. Canonical and cross-reactive binding of NK cell inhibitory receptors to HLA-C allotypes is dictated by peptides bound to HLA-C. Front. Immunol 8: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guethlein LA, Norman PJ, Hilton HG, and Parham P. 2015. Co-evolution of MHC class I and variable NK cell receptors in placental mammals. Immunol. Rev 267: 259–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parham P, and Guethlein LA. 2018. Genetics of natural killer cells in human health, disease, and survival. Annu. Rev. Immunol 36: 519–548. [DOI] [PubMed] [Google Scholar]
- 14.Gendzekhadze K, Norman PJ, Abi-Rached L, Graef T, Moesta AK, Layrisse Z, and Parham P. 2009. Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc. Natl. Acad. Sci. U.S.A 106: 18692–18697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilton HG, Norman PJ, Nemat-Gorgani N, Goyos A, Hollenbach JA, Henn BM, Gignoux CR, Guethlein LA, and Parham P. 2015. Loss and gain of natural killer cell receptor function in an African hunter-gatherer population. PLoS Genet 11: e1005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lahr MM, and Foley RA. 1998. Towards a theory of modern human origins: geography, demography, and diversity in recent human evolution. Am. J. Phys. Anthropol Suppl 27: 137–176. [DOI] [PubMed] [Google Scholar]
- 17.Tishkoff SA, and Williams SM. 2002. Genetic analysis of African populations: human evolution and complex disease. Nat. Rev. Genet 3: 611–621. [DOI] [PubMed] [Google Scholar]
- 18.Chen YS, Torroni A, Excoffier L, Santachiara-Benerecetti AS, and Wallace DC. 1995. Analysis of mtDNA variation in African populations reveals the most ancient of all human continent-specific haplogroups. Am. J. Hum. Genet 57: 133–149. [PMC free article] [PubMed] [Google Scholar]
- 19.Ingman M, Kaessmann H, Paabo S, and Gyllensten U. 2000. Mitochondrial genome variation and the origin of modern humans. Nature 408: 708–713. [DOI] [PubMed] [Google Scholar]
- 20.Jorde LB, Watkins WS, Bamshad MJ, Dixon ME, Ricker CE, Seielstad MT, and Batzer MA. 2000. The distribution of human genetic diversity: a comparison of mitochondrial, autosomal, and Y-chromosome data. Am. J. Hum. Genet 66: 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watson E, Forster P, Richards M, and Bandelt HJ. 1997. Mitochondrial footprints of human expansions in Africa. Am. J. Hum. Genet 61: 691–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, Bruno D, Carrington CV, Chandanayingyong D, Chang YH, Crespi C, Saruhan-Direskeneli G, Fraser PA, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Mila J, Park MH, Pitchappan RM, Ramdath DD, Shiau MY, Stephens HA, Struik S, Verity DH, Vaughan RW, Tyan D, Davis RW, Riley EM, Ronaghi M, and Parham P. 2007. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat. Genet 39: 1092–1099. [DOI] [PubMed] [Google Scholar]
- 23.Norman PJ, Hollenbach JA, Nemat-Gorgani N, Guethlein LA, Hilton HG, Pando MJ, Koram KA, Riley EM, Abi-Rached L, and Parham P. 2013. Co-evolution of human leukocyte antigen (HLA) class I ligands with killer-cell immunoglobulin-like receptors (KIR) in a genetically diverse population of sub-Saharan Africans. PLoS Genet. 9: e1003938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knight A, Underhill PA, Mortensen HM, Zhivotovsky LA, Lin AA, Henn BM, Louis D, Ruhlen M, and Mountain JL. 2003. African Y chromosome and mtDNA divergence provides insight into the history of click languages. Curr. Biol 13: 464–473. [DOI] [PubMed] [Google Scholar]
- 25.Henn BM, Gignoux CR, Jobin M, Granka JM, Macpherson JM, Kidd JM, Rodriguez-Botigue L, Ramachandran S, Hon L, Brisbin A, Lin AA, Underhill PA, Comas D, Kidd KK, Norman PJ, Parham P, Bustamante CD, Mountain JL, and Feldman MW. 2011. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc. Natl. Acad. Sci. U.S.A 108: 5154–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, Chen Z, Chu J, Carcassi C, Contu L, Du R, Excoffier L, Ferrara GB, Friedlaender JS, Groot H, Gurwitz D, Jenkins T, Herrera RJ, Huang X, Kidd J, Kidd KK, Langaney A, Lin AA, Mehdi SQ, Parham P, Piazza A, Pistillo MP, Qian Y, Shu Q, Xu J, Zhu S, Weber JL, Greely HT, Feldman MW, Thomas G, Dausset J, and Cavalli-Sforza LL. 2002. A human genome diversity cell line panel. Science 296: 261–262. [DOI] [PubMed] [Google Scholar]
- 27.Patin E, Siddle KJ, Laval G, Quach H, Harmant C, Becker N, Froment A, Regnault B, Lemee L, Gravel S, Hombert JM, Van der Veen L, Dominy NJ, Perry GH, Barreiro LB, Verdu P, Heyer E, and Quintana-Murci L. 2014. The impact of agricultural emergence on the genetic history of African rainforest hunter-gatherers and agriculturalists. Nat commun. 5: 3163. [DOI] [PubMed] [Google Scholar]
- 28.Ba A, Beley S, Chiaroni J, Bailly P, and Silvy M. 2015. RH diversity in Mali: characterization of a new haplotype RHD*DIVa/RHCE*ceTI(D2). Transfusion 55: 1423–1431. [DOI] [PubMed] [Google Scholar]
- 29.Robinson J, Halliwell JA, McWilliam H, Lopez R, and Marsh SG. 2013. IPD--the Immuno Polymorphism Database. Nucleic Acids Res. 41: D1234–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norman PJ, Hollenbach JA, Nemat-Gorgani N, Marin WM, Norberg SJ, Ashouri E, Jayaraman J, Wroblewski EE, Trowsdale J, Rajalingam R, Oksenberg JR, Chiaroni J, Guethlein LA, Traherne JA, Ronaghi M, and Parham P. 2016. Defining KIR and HLA class I genotypes at highest resolution via high-throughput sequencing. Am. J. Hum. Genet 99: 375–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nemat-Gorgani N, Hilton HG, Henn BM, Lin M, Gignoux CR, Myrick JW, Werely CJ, Granka JM, Moller M, Hoal EG, Yawata M, Yawata N, Boelen L, Asquith B, Parham P, and Norman PJ. 2018. Different selected mechanisms attenuated the inhibitory interaction of KIR2DL1 with C2(+) HLA-C in two indigenous human populations in southern Africa. J. Immunol 200: 2640–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and S. Genome Project Data Processing. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foley BA, De Santis D, Van Beelen E, Lathbury LJ, Christiansen FT, and Witt CS. 2008. The reactivity of Bw4+ HLA-B and HLA-A alleles with KIR3DL1: implications for patient and donor suitability for haploidentical stem cell transplantations. Blood 112: 435–443. [DOI] [PubMed] [Google Scholar]
- 34.Graef T, Moesta AK, Norman PJ, Abi-Rached L, Vago L, Older Aguilar AM, Gleimer M, Hammond JA, Guethlein LA, Bushnell DA, Robinson PJ, and Parham P. 2009. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J. Exp. Med 206: 2557–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gumperz JE, Litwin V, Phillips JH, Lanier LL, and Parham P. 1995. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J. Exp. Med 181: 1133–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dohring C, Scheidegger D, Samaridis J, Cella M, and Colonna M. 1996. A human killer inhibitory receptor specific for HLA-A1,2. J. Immunol 156: 3098–3101. [PubMed] [Google Scholar]
- 37.Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, Rowland-Jones S, and Braud VM. 2004. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur. J. Immunol 34: 1673–1679. [DOI] [PubMed] [Google Scholar]
- 38.Hilton HG, Guethlein LA, Goyos A, Nemat-Gorgani N, Bushnell DA, Norman PJ, and Parham P. 2015. Polymorphic HLA-C receptors balance the functional characteristics of KIR haplotypes. J. Immunol 195: 3160–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blokhuis JH, Hilton HG, Guethlein LA, Norman PJ, Nemat-Gorgani N, Nakimuli A, Chazara O, Moffett A, and Parham P. 2017. KIR2DS5 allotypes that recognize the C2 epitope of HLA-C are common among Africans and absent from Europeans. Immun. Inflamm. Dis 5: 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moesta AK, Graef T, Abi-Rached L, Older Aguilar AM, Guethlein LA, and Parham P. 2010. Humans differ from other hominids in lacking an activating NK cell receptor that recognizes the C1 epitope of MHC class I. J. Immunol 185: 4233–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin MP, and Carrington M. 2013. Immunogenetics of HIV disease. Immunol. Rev 254: 245–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buhler S, Nunes JM, and Sanchez-Mazas A. 2016. HLA class I molecular variation and peptide-binding properties suggest a model of joint divergent asymmetric selection. Immunogenetics 68: 401–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, and Marsh SG. 2015. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 43: D423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gonzalez-Galarza FF, Takeshita LY, Santos EJ, Kempson F, Maia MH, da Silva AL, Teles e Silva AL, Ghattaoraya GS, Alfirevic A, Jones AR, and Middleton D. 2015. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 43: D784–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gonzalez-Galarza FF, McCabe A, Melo Dos Santos EJ, Takeshita L, Ghattaoraya G, Jones AR, and Middleton D. 2018. Allele Frequency Net Database. Methods. Mol. Biol 1802: 49–62. [DOI] [PubMed] [Google Scholar]
- 46.Gendzekhadze K, Norman PJ, Abi-Rached L, Layrisse Z, and Parham P. 2006. High KIR diversity in Amerindians is maintained using few gene-content haplotypes. Immunogenetics 58: 474–480. [DOI] [PubMed] [Google Scholar]
- 47.Nemat-Gorgani N, Edinur HA, Hollenbach JA, Traherne JA, Dunn PP, Chambers GK, Parham P, and Norman PJ. 2014. KIR diversity in Maori and Polynesians: populations in which HLA-B is not a significant KIR ligand. Immunogenetics 66: 597–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, and Parham P. 2006. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J. Exp. Med 203: 633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verdu P 2016. African Pygmies. Curr. Biol 26: R12–14. [DOI] [PubMed] [Google Scholar]
- 50.Shriner D, Tekola-Ayele F, Adeyemo A, and Rotimi CN. 2018. Genetic ancestry of Hadza and Sandawe peoples reveals ancient population structure in Africa. Genome Biol. Evol 10: 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pando MJ, Gardiner CM, Gleimer M, McQueen KL, and Parham P. 2003. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J. Immunol 171: 6640–6649. [DOI] [PubMed] [Google Scholar]
- 52.Parham P, Arnett KL, Adams EJ, Little AM, Tees K, Barber LD, Marsh SG, Ohta T, Markow T, and Petzl-Erler ML. 1997. Episodic evolution and turnover of HLA-B in the indigenous human populations of the Americas. Tissue antigens 50: 219–232. [DOI] [PubMed] [Google Scholar]
- 53.Hiby SE, Walker JJ, O’Shaughnessy K M, Redman CW, Carrington M, Trowsdale J, and Moffett A. 2004. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J. Exp. Med 200: 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rotimi CN, Bentley AR, Doumatey AP, Chen G, Shriner D, and Adeyemo A. 2017. The genomic landscape of African populations in health and disease. Hum. Mol. Genet 26: R225–R236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakimuli A, Chazara O, Byamugisha J, Elliott AM, Kaleebu P, Mirembe F, and Moffett A. 2014. Pregnancy, parturition and preeclampsia in women of African ancestry. Am. J. Obstet. Gynecol 210: 510–520 e511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vierra-Green C, Roe D, Hou L, Hurley CK, Rajalingam R, Reed E, Lebedeva T, Yu N, Stewart M, Noreen H, Hollenbach JA, Guethlein LA, Wang T, Spellman S, and Maiers M. 2012. Allele-level haplotype frequencies and pairwise linkage disequilibrium for 14 KIR loci in 506 European-American individuals. PLoS ONE 7: e47491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.