Abstract
Multiple sclerosis (MS) is a chronic inflammatory demyelinating and neurodegenerative disease of the central nervous system (CNS). The cause of MS is unknown, with no effective therapies available to halt the progressive neurological disability. Development of new and improvement of existing therapeutic strategies would therefore require a better understanding of MS pathogenesis, especially during the progressive phase of the disease. This can be achieved through development of biomarkers that can help to identify disease pathophysiology and monitor disease progression. Proteomics is a powerful and promising tool to accelerate biomarker detection and contribute to novel therapeutics. In this review, we provide an overview of how proteomic technology using CNS tissues and biofluids from MS patients has provided important clues to the pathogenesis of MS. We discuss current publications, pitfalls, as well as directions of future research involving proteomic approaches to understand the pathogenesis of MS.
Keywords: Multiple sclerosis, CNS, Proteomics, biomarkers
1. INTRODUCTION
1.1. Multiple sclerosis (MS)
MS is an autoimmune demyelinating and neurodegenerative disease of the central nervous system (CNS) and is the major cause of disability in young adults.[1, 2] The most common form of MS is relapsing-remitting MS (RRMS, 80–85%), which is characterized by relapses or acute attacks (new symptoms or worsening of existing symptoms) followed by periods of remission.[3] Episodes of relapses correlate with new inflammatory lesions as detected by Gadolinium (Gd)-enhancing regions with magnetic resonance imaging (MRI).[4] Pathologically, inflammatory demyelination is predominant in patients with RRMS with episodes of relapses and remissions. Most patients with RRMS eventually enter a stage known as secondary-progressive MS (SPMS) with irreversible neurological disability. Approximately 10–15% of patients experience a disease progression without RRMS called primary-progressive MS (PPMS),[5] with continuous neurological decline from onset that worsens overtime and eventually leads to permanent disability.[6, 7]
There are a number of anti-inflammatory therapies available for RRMS patients.[8, 9] These therapies, which are directed towards immune mechanisms, reduce the frequency and severity of MS attacks. However, these therapies are of little benefit to progressive MS patients. Significant effort has been made to understand the progressive disease course and pathologic characteristics of MS lesions. These include, but are not limited to, the roles of blood-brain barrier (BBB) disruption, oligodendrocyte death, reactive gliosis, and neuro-axonal loss. Several different techniques have been employed to investigate the diverse cellular processes that are affected during MS. These include genomics,[10] transcriptomics,[11, 12] epigenetics,[13-15] metabolomics,[16, 17] and pharmacogenomics[18, 19] comparing different stages of MS patients.
Similar to genomics, transcriptomics, and metabolomic profiling, proteomics has been developed as a large-scale, unbiased tool that has been and continues to be applied in the MS research field.[20, 21] This approach holds advantages over conventional genomics/transcriptomics tools, as it can identify post-translational modifications such as phosphorylation, glycosylation, and acetylation,[22, 23] which have been associated with MS pathogenesis.[24-26] In addition, proteomic-based approaches have been useful in identifying mutations associated with MS.[27, 28] Different proteomics approaches have been used in CNS tissues and biological fluids from MS patients to identify biomarkers and causative factors in disease pathogenesis. For example, using proteomic approaches (2D and LC-MS/MS), sera from SPMS patients revealed the presence of auto-antibodies against galectin-3[29] and the chloride-channel protein anoctamin 2 in MS cases.[30] Another portential marker, chitinase-3-like protein 1 (CHI3L1), has been observed in CSF samples from different MS types which was also identified by proteome analysis.[31, 32] This marker has been also obsereved other disease condtions and currently beiing trageted in pre-clincal study to treat severe asthma.[33, 34]
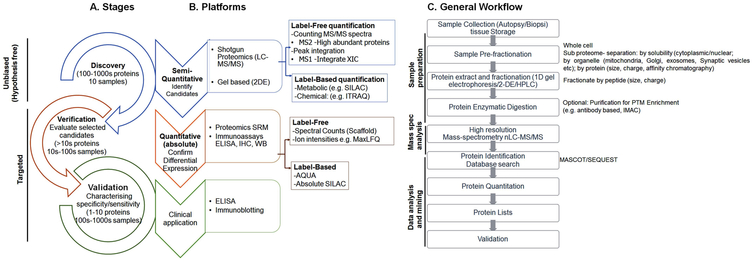
Over the past two decades, the extensive growth of mass spectrometry techniques towards higher resolution, accuracy, sensitivity, and scanning speed, as well as high through-put machinery, chromatographic techniques, and integrated software, have all been driving forces behind significant advancements in the field of proteomics.[21] Due to these developments, specific and sensitive information of the proteome (protein analysis and identification) of complex samples over a wide dynamic range is rapidly feasible.[35] This includes critical steps of sample collection, sample preparation, and liquid chromatography tandem mass spectrometry analysis (LC MS/MS), as well as bioinformatics-supported data analysis and validation studies (schematically represented in Figure 1). Proteins characterized by mass spectrometry analysis can be either intact proteins (top–down) or enzymatically-digested protein peptides (bottom–up), which is also known as shotgun proteomics.[36-38] In commonly-used bottom–up approaches, the digested peptide mixture is fractionated and separated by chromatography, ionized by electrospray ionization (ESI), and analyzed by a mass spectrometer.[39] Online application of nanoscale liquid chromatography techniques (compared to conventional LC-MS/MS) have further increased the potential to detect low levels of abundant proteins in limited samples.[40, 41] Another emerging class of proteomics technique is the protein microarray,[42] which is being used in combination with surface-enhanced laser desorption/ionization (SELDI) mass spectrometry to detect altered proteins.
Figure 1: Detailed workflow for quantitative proteomic experiments.
Panel A: Stages in the proteomic biomarker discovery process. For different stages, different mass spectrometric approaches can be used. A non-targeted approach is used normally for discovery/exploratory based-proteomic investigation.
Panel B: Different bottom-up proteomic approaches can be used to identify and (semi) quantify hundreds of proteins in complex mixtures. Labelling or label-free techniques can be used for protein quantification. 2D is another way of protein separation and to perform gel-based comparative analysis. Comparative proteomic analysis provides a list of differentially-abundant proteins. After discovery, stage markers can be filtered by validating in independent cohorts of patients using different platforms of absolute quantification such as ELISA or by using targeted/higher-specificity mass spectrometry. SRM can be used when the availability of antibodies with high specificity is lacking. The process “verification” provides verified markers. The verified small number protein markers can then be validated in clinical samples from MS patients.
Panel C shows a general work-flow for a proteomic experiment. These steps start with biological sample collection from well-characterized patients and include sample preparation and fractionation, separation, LC-MS, as well as protein identification and quantification and validation.
In this review, we provide an overview of the advanced proteomic analysis in biological/clinical samples from MS patients and how these results have shed light on the mechanisms underlying MS disease pathogenesis.
2. APPLICATION OF PROTEOMICS TO IDENTIFY BIOMARKERS FOR AND PATHOGENESIS OF MS
In the current study, we conducted a literature review on MS biomarkers using the words “multiple sclerosis”, “cerebrospinal fluid”, “Plasma or serum”, “urine”, “saliva”, “CNS tissue,” and “proteomics” to search the PubMed database from 2013 to 2019. The results from the identified studies are discussed in detail below.
2.1. Proteomic studies using circulatory body fluids
a). Cerebrospinal fluid (CSF)
CSF is mainly produced by the choroid plexus in the cerebral ventricles and is absorbed in arachnoid granulations. CSF is in close contact with MS lesions in the CNS (periventricular white-matter lesions, grey-matter lesions, and spinal cord lesions),[43] making it a pathologically-relevant fluid for MS-related biomarkers.[44] Changes in CSF can be reflective of neuroinflammation, metabolic damage, BBB disruption, and/or brain tissues damage (axonal, neuronal, astroglial and oligodendrocyte death), which may be useful in predicting the progression of clincally-isoalated syndrome (CIS) into clinically-definitive MS.[45] Moreover, disease-associated changes in protein expression, post-translational modifications, and/or protein turnover within the CNS may mirror corresponding changes in CSF protein content.[26, 46] Other advantages of using CSF include its availability from a living MS patient before disease symptoms appear, as well as the ability to perform longitudinal follow-up measures during the MS disease course, which may be helpful in identifying useful biomarkers and discovering important contributions to disease pathogenesis.[47] Over the past five years, several proteomic research studies have been conducted in CSF samples from MS patients to identify biomarkers related to disease pathogenesis, clinical symptoms, treatment efficacy, dysregulated pathways and many other aspects of this disease discussed below (Table 1).
Table 1.
CSF biomarker proteomic studies in MS
| CSF studies | Number of patients | Main finding | Method | Ref | |
|---|---|---|---|---|---|
| 1 | Diagnostic autoantibody peptide markers | 29 MS and 30 healthy controls | Five complementarity determining regions specific to MS | Label free LC-MS/MS | [28] |
| 2 | Diagnosis, prognosis and disease course | 6 RRMS patients and 18 neurological disease controls Validation set: 13 early-MS, 13 RRMS and 13 neurological controls | Secretogranin-1 to be increased in early-MS patients compared to RRMS and neurological controls | iTRAQ, SRM | [31] |
| 3 | Diagnostic and prognostic biomarkers MS | Discovery- 28 RRMS, control- 28 Verification- 40 CIS, 38 RRMS, 16 PMS and 29 control | CHI3L1, CHI3L2 | TMT-LC-MS/MS, ELISA | [32] |
| 4 | Early MS markers | First-attack MS patients compared to RRMS | CSF proteins in first-attack patients were differentially enriched for gray matter components | 2D-LC-MS/MS | [44] |
| 5 | First attack of suspected MS compared to controls | 47 CIS patients and 45 control | Ig kappa chain C region, amyloid-like protein 1 | Label-free LC-MS/MS (LTQ-Orbitrap), ELISA | [45] |
| 6 | Autoantibody repertoire in paired serum and CSF samples from RRMS, PPMS, and control | 10 cases each | Epstein-Barr virus protein EBNA1 homologous to the N-terminal part of human crystallin alpha-B | High-density peptide microarrays | [46] |
| 7 | Diagnostic markers (RRMS) | 21 RRMS and 21 ONDs controls | Oligo-myelin glycoprotein and noelin glycosylation status | TMT label and LC-MS/MS | [48] |
| 8 | Prognostic biomarkers for identifying patients with a high risk of developing RR MS | 11 MS and 15 healthy control | (Ig) γ-1 chain C region, Ig heavy chain V-III region BRO and Ig κ chain C | Nanoscale LC coupled with tandem MS | [52] |
| 9 | Prediction of CIS-MS conversion | CIS-MS to RRMS (n = 23) and non RRMS (n = 19) | HOXB3 | Label-free LC-MS/MS, ELISA | [53] |
| 10 | MS clinical courses | 24 CIS, 16 RRMS, 11 PMS | Secretogranin II, Protein 7B2 upregulated in RRMS Fibrinogen, Fibrinopeptide-A downregulated in CIS Tymosin β4- CIS vs RRMS patients | MALDI-TOF | [56] |
| 11 | Prognostic MS markers | 39 children with initial ADS, 18 of whom were diagnosed with MS and 21 of whom had a monophasic disease course | Increased abundance of CNS gray matter-related proteins | Label free LC-MS/MS | [58] |
| 12 | Treatment markers MS patients before and after Natalizumab treatment | Discovery set-17 RRMS (before and after Natalizumab treatment) | Ig µ chain C region, haptoglobin, Chitinase-3-like protein 1 | Label free LC-MS/MS, SRM, ELISA | [60] |
Studies using proteomic analysis of CSF have identified several MS disease-specific proteins.[44, 45] In one such study, seven biomarker candidates, namely CHI3L1, secretogranin-1 (Scg1), cerebellin-1, neuroserpin, cell surface glycoprotein MUC18, testican-2, and glutamate receptor 4 were idenitified using isobaric tags for relative abundance and quantitation (iTRAQ) labeling and Orbitrap MS analysis of CSF samples from MS patients.[31] In the followup validation study from independent MS patients, intracellular calcium binding protein Sg1 showed increased protein levels in early-MS patients compared to RRMS. Recently, pathways and processes affected in MS were analyzed using proteome and deglycoproteome analysis, which detected 484 proteins and 180 deglycopeptides that are significantly changed in RRMS patients. These proteins belonged to biological categories such as inflammation, extracellular matrix organization, cell adhesion, immune response, and neuron development.[48] In another study that investigated biomarkers segregating CIS from progession to confirmed MS, twenty-two differentially-abundant CSF proteins were identified.[32] Of these proteins, CHI3L1 was proposed to be useful for differentiating CIS cases from RRMS/progressive MS (PMS) patients. Interestingly, this protein has also been identified and reported in other studies in MS patients and many other neurodegenerative diseases,[49-51] and was found to be correlated with the number of Gd-enhancing T2 lesions observed in brain MRI scans performed at baseline and during disability progression at follow-up. CHI3L1, therefore, may be useful as a prognostic biomarker to predict the conversion of CIS into definite MS and disability progression for future studies. Further, comparing CSF proteome profiles from 11 MS cases and 15 control subjects, Pavelek et al. (2016)[52] found nine significantly decreased and seventeen significantly increased proteins in the CSF of MS patients compared to controls. Among these, immunoglobulin (Ig) γ−1 chain C region, Ig heavy chain V-III region BRO and Ig κ chain C region were previously linked to RRMS and CIS,[45] providing rationale for their use as prognostic biomarkers for identifying patients with a high risk of developing RRMS. Similarly, LC-MS/MS analysis of 42 CIS cases resulted in the identification of 637 proteins, out of which 132 were significantly dysregulated.[53] Pathway analysis revealed enrichement of three main pathways: 1) inflammatory response, 2) cellular growth, and 3) tissue proliferation. Validation studies using ELISA confirmed increased HOXB3 levels in the CSF of CIS pateints, augmenting its possible use as a biomarker to predict CIS-MS conversion future studies. Furthermore, CSF from MS patients are now being targetted to find out neurlogical markers specific to a clincal sympotms. In a recent study, authors campared CSF protemoe profile of MS patients showing fatigue symptoms to MS patients without fatigue as well healthy control.[54] Using a shotgun proteomics and label-free quantitative proteomics approach, authors found 591 CSF proteins and majority of downregulated proteins were associated with synaptic plasticity, energy homeostasis and other pathways. They further valiadted proteomics findings using western blot and found decreased levels of protein kinase C-binding protein NELL2, neural cell adhesion molecule L1-like protein, and reelin in MS patients with fatigue.[54]
The presence of immunoglobulins in CSF and intrathecal lymphocytes supports an IgG-mediated pathogenesis. Based on these reports, various studies have explored immunoglobulin types and autoantibodies in CSF samples. In one study, Hecker et al. (2016)[46] screened 3991 peptides in CSF samples from RRMS and PPMS patients and found 54 peptides specific to MS patients compared with controls (p values <0.05). The study also detected the highest signals for a peptide mapping to Epstein-Barr virus protein EBNA1 (amino acids 392–411), and homologous to the N-terminal part of human crystallin alpha-B. In a separate study involving MS patient CSF samples,[28] mutated sequences in IgG complementarity-determining region (CDR) between MS types, but not in healthy controls, was detected. These results exemplifies the utility of proteomic based approaches in identifying key pathogenic mechanisms underlying MS pathogenesis through study of the CSF.
CSF analysis has also been utilized to find biomarkers related to the neurodegenerative phase of MS.[55] Liguori et al. (2014)[56] carried out CSF analysis on 24 CIS, 16 RRMS, and 11 PMS patients using a matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) approach. Results from this study showed significant upregulation of Secretogranin II and Protein 7B2, with concomitant downregulation of fibrinogen and Fibrinopeptide A in CIS compared to PMS patients. In addition, they also found that Thymosin β4 levels can be used to separate CIS and RRMS patients. Interestingly, this protein has been reported to promote oligodendrogenesis in the demyelinating CNS by activating the epidermal growth factor receptor (EGFR) signaling pathway.[57] The speific role of Thymosin β4 in MS pathogenesis requires further investigation. Proteomic approaches to CSF protein identification have been used in children with acquired CNS demyelinating syndrome (ADS) to differentiate monophasic MS from the group progressing to definite MS.[58] The results of this study showed that CSF from definite MS cases had increased CNS gray matter-related proteins, whereas children with a monophasic disease course had increased abundance of innate immunity-related proteins.
Proteomic approaches have also been used effectively to determine the effects of MS therapies.[59, 60] A report from Stoop et al. (2013)[60] evaluted the effects of natalizumab treatment (before and after) in CSF samples of MS pateints using electrospray Orbitrap MS/MS and found a number of candidate biomarkers that were significantly different before and after treatment. Among these, three proteins (Ig µ chain C region, haptoglobin, and CHI3L1) were further validated using in new sample sets. All three of these proteins were found to be significantly lower in the CSF of MS patients after 1 year of natalizumab treatment suggesting prognostic value of these markers towards disease modifying therapies.
In addition, CSF proteomic studies have been useful for identifying novel pathways and post-traslational protein modifications in MS. For example, a quantitative and deglycoproteome analysis of CSF from RRMS cases resulted in more than 2000 proteins and 1700 deglycopeptides being identified,[48] of which 484 proteins and 180 deglycopeptides were significantly changed between RRMS and control samples. These identified proteins were found to be associated with inflammation, extracellular matrix organization, cell adhesion, immune response, and neuron development.
In recent years, CSF-derived extracellular vesicles (EVs), such as exosomes and macrovesicles, have generated considerable interest as many CNS cell specific proteins have been found to be enriched in EVs proteome[61, 62] Interestingly, many of these EV proteins have been also found to be linked with RNA in disease and cellular stress and different neurodegenerative diseases.[63, 64] A quantitative proteomic analysis of exosomes isolated from the CSF of patients with neuromyelitis optica (NMO) identified proteins signatures (e.g. glial fibrillary acidic protein (GFAP) and fibronectin) that can be useful in differentiating NMO from MS and idiopathic longitudinally-extensive transverse myelitis.[65] These studies demonstrated EV enrichment in CSF derived from MS patients and allowed detailed analysis of protein profiles that may offer opportunities to identify novel biomarkers and therapeutic approaches in CNS inflammatory diseases.
b). Blood Serum/Plasma
CSF is collected by an invasive lumbar puncture procedure that is mainly performed for clinical indications. The clinical usefulness of CSF biomarkers is restricted, particularly due to the invasiveness of the procedure and the risks associated with repeated lumbar puncture. On the other hand, blood-based markers are more important tools for clinical use, as blood is routinely collected as a clinical specimen. Several emerging (serum and plasma) biomarkers have been identified for MS using proteomic analysis, which have been summarized in Table 2.
Table 2:
Blood Serum/plasma proteomic studies in MS
| Serum studies | Number of patients |
Main finding | Method | Ref | |
|---|---|---|---|---|---|
| 1 | Biomarker for SPMS | immortalized human brain microvascular endothelial cellsSera 11 SPMS and 12 RRMS | Galectin-3 is a possible immunological target molecule of the pathogenic auto-antibodies | 2D-LC-MS/MS, Western blot | [29] |
| 2 | Marker for MS patient’s characterization | 2,169 plasma samples from MS cases | Anoctamin 2 identified as an autoimmune target in MS | Protein microarrays, immunofluorescence | [30] |
| 3 | Protein biomarkers of brain atrophy in SPMS | 140 SPMS patients, longitudinal serum samples - baseline, 6, 12 and 24 months | Free hemoglobin in the serum was associated with the rate of brain atrophy SPMS | SELDI-TOF, ELISA | [66] |
| 4 | Biomarkers of disease progression | 14 serum samples (n = 7 from each MS phenotype) benign and aggressive cases of MS | 11 proteins classified samples into the two disease groups inflammation, opsonization, and complement activation | iTRAQ-MALDI-TOF/TOF | [67] |
| 5 | Predicting clinical relapse of MS | 54 MS and 55 healthy controls | Autophagy-related gene16L2 | MALDI-TOF, immunoblotting and real-time PCR | [68] |
| 6 | Diagnostic and prognostic markers (pathology related) | 18 RRMS in relapsing (Group I) or remitting phase (pooled)and 7 controls | Vitamin D-binding protein, Apolipoprotein A-IV levels of oxidative stress elevated with the disease progression | 2D-MALDI TOF, Redox proteomics | [69] |
In a longitudinal study combining SELDI-TOF-Mass spectroscopy and MRI in 475 serum samples from 140 SPMS patients, Lewin et al. (2016)[66] reported a significant correlation between the rate of brain atrophy and a rise in the concentration of proteins at 15.1 kDa and 15.9 kDa in the serum. They further validated free serum hemoglobin and lactate hydrogenase by ELISA. Based on these findings, the authors concluded that in PMS, low-grade chronic intravascular hemolysis may be a potential source of the iron whose deposition along blood vessels in MS plaques can contribute to neurodegeneration and consequent brain atrophy. Further, iTRAQ analysis of serum proteome profiles from 14 RRMS patients (aggressive and benign) identified 108 proteins belonging to inflammation, opsonization, and complement activation.[67] Many of the proteins from this list have been proposed to act as biomarkers to monitor MS progression and treatment. In a similar study, Yin et al. (2014)[68] analyzed serum proteome profiles from MS patients (n=54) and healthy controls (n=55) using bead fractionation/MALDI-TOF analyses, and identified 11 significantly different peptides between the two groups. The authors further validated their proteome results by applying independent techniques like immunoblotting and real-time PCR.
Interestingly, findings of serum proteome profiles in a pre-symptomatic MS patient cohort (n=100) showed a different protein profile compared to the control group,[69] which indicates that proteomic changes can be detected prior to disease onset. Moreover, while looking for BBB impairment in SPMS patients, Nishihara et al. (2017)[29] identified autoantibodies against galectin-3 in 10 SPMS sera samples compare to other neurological disease samples. Functional studies showed that galectin-3 is possibly involved in regulating intercellular adhesion molecule-1 (ICAM-1) and phospho-nuclear factor-kappa (NFκ) B p65 expression in brain microvascular endothelial cells (BMECs), hence upregulated auto-antibodies again galectin-3 induce BBB breakdown in SPMS patients.
Serum proteome profiles of MS patients have been used to study differentially-expressed and oxidatively-modified low-abundant serum proteins by combining proteome and redox proteome analysis.[70] In one such study, the authors identified increased levels of ceruloplasmin, antithrombin III, clusterin, apolipoprotein E, and complement C3 in MS patients compared with healthy controls, whereas redox proteomics analysis showed a progressive trend of vitamin D-binding protein oxidation from remission to relapse. Recently, whole-proteome peptide microarrays have been used to profile different proteins in a given sample. In one such study using this method, a peptide array hosting 1.74 million 12-mer peptides with a six amino acid lateral shift was applied in order to identify autoantibodies proteome present in plasma and serum sample of MS patients.[71] The preliminary findings from this study were further used to verify IgG reactivity with a larger sample set (n = 448) using bead-based peptide microarrays and confirmed the presence of antibodies against different proteins like YBX2, JUNB, MAFA, etc. Moreover, this study also reported many proteins (MUC16, STK19, PRMT3, etc.) that can be used to differentiate CIS from SPMS.
In line with these findings, 1063 MS patients and 1106 healthy control plasma samples were used to determine IgG reactivity against 384 antigens, which yielded 196 unique proteins.[30] One such peptide fragment, anoctamin 2 (ANO2), showed the greatest difference between MS and control samples, which was also validated for its cellular expression in human brain tissue. These findings suggest that variations in serum/plasma protein contents can be used to distinct different MS disease stages as well as differentiate from control subjects.
2.2. Proteomic studies using urine from MS patients
Urine is an attractive biofluid that can be collected non-invasively and has an enriched metabolic profile. Greater fluctuations could occur in urine than in blood, reflecting changes in the human body. Previous reports have shown that neopterin and nitric oxide metabolites could act as markers of disease activity in MS.[72] While urine has not been suggested as the best choice, especially for CNS biomarker research, altered metabolites in urine from MS patients have been found to correlate with alterations in energy and fatty acid metabolism, mitochondrial activity, and gut microbiota.[73] There are two reported MS and related studies performed with urine samples over the past five years that have used proteomic approaches (Table 3). In the first study, using label-free MC-MS techniques, the authors profiled longitudinal changes in proteome in a cohort of pregnant MS patients (n=31) and pregnant controls (n=8) at third trimester and first post-partum intervals.[74] In this study, the authors identified 402 proteins, of which trefoil factor 3 and lysosomal-associated membrane protein 2 have been found to play role in the innate immune system. Another three proteins with a significantly decreased ratio were plasma glutamate carboxypeptidase, Ig mu chain C region, and osteoclast-associated immune-like receptor. In a second study, Neilson et al. compared urine proteomes of NMO (n=32) to MS patients (n=46) using quantitative LC-MS/MS after trypsin digestion and iTRAQ labelling and identified 1112 different proteins.[75] In the future, larger clinical studies are required to verify and expand upon these results, as urine biomarkers could be the most convenient for clinical application if proven to be reliable.
Table 3.
Urine proteomic studies in MS
| Urine studies | Number of patients |
Main finding | Method | Ref | |
|---|---|---|---|---|---|
| 1 | Pregnancy-related MS markers | 31 pregnant MS patients and 8 pregnant controls Longitudinal study | Pregnancy-related peptides were significantly elevated in MS compared with controls | Label free LC-MS/MS | [74] |
| 2 | Biomarkers differentiating NMO/NMO spectrum disorders from MS | NMO/NMO-SD patients (n = 32), MS (n = 46)Healthy subjects (n = 31) | Ig-light chains Increased in NMO/NMO-SD. | iTRAQ labellingLC-MS/MS | [75] |
2.3. Proteomic studies using saliva in MS patients
Another body fluid that can be non-invasively collected is saliva, which can easily be repeatedly collected and stored. In addition to having secreted proteins from salivary glands, saliva also contains proteins from the gingival cervicular fluid, oral microflora, and also plasmatic proteins. Diagnostic tests on saliva are common and cost-effective, particularly for patients who need to monitor their hormone levels or the effectiveness of ongoing therapies.[75] The human saliva proteome has been profiled from healthy and disease cases. Cataloging experimental reports from various laboratories, a total of 2290 proteins have been identified in human saliva.[77] Of these, nearly 40% have been suggested to be candidate markers for diseases such as cancer, cardiovascular disease, and stroke. The identified protein classes belong to amylases, proline-rich proteins, statherin, histatin, mucin, and cystatins.[78] However, there are not many studies available from MS patients. Recently, using a top-down proteomic approach, Manconi et al. (2018)[79] compared the salivary proteomes and peptidomes of MS patients to those of healthy controls and showed significant differences between salivary proteomic profiles. Proteins identified in saliva of MS patients belonged to inflammation and immune response, which is a typical MS pathology. Overall, saliva presents a potential option to be used for MS diagnostic purposes as well as for monitoring treatment responses. Hence, future studies should be planned for larger populations covering different spectra of MS disease stages, as these findings could be useful to understand disease initiation, progression, and pathogenesis.
2.4. Proteomic studies using CNS tissue from MS brain
Available brain tissues in MS research have been confined to those involving biopsy and autopsy tissues. As all biofluids do not have a direct connection to disease etiology, and animal models only mimic certain aspects of the disease, the CNS represents a critical avenue for MS research to explore demyelination, axonal degeneration, perivascular inflammation, and gliosis. There are various reports available for proteome profiling in animal models of MS, but we found only four studies conducting human CNS tissue proteome analysis (Table 4). In one of the earliest studies on MS tissue, the authors profiled proteomes of three major lesion types - acute, chronic active, and chronic inactive demyelinating lesions - from six MS cases (two acute, one chronic, one progressive, one secondary-progressive, and one chronic progressive).[80] In this study, laser-capture microdissection (LCM) and LC-MS/MS proteomic analysis approaches were implemented and resulted in 158, 416, and 236 proteins unique to acute plaque, chronic active, and chronic plaques, respectively. However, one drawback of this study was that the authors compared their findings in MS lesions with healthy brain tissue, not with normal-appearing white matter (NAWM) from MS patient brain samples.
Table 4.
CNS proteomic studies in MS
| Study (Brain) | Number of patients | Main finding | Method | Ref | |
|---|---|---|---|---|---|
| 1 | Pathogenesis | Autopsied brain samples of two female MS patients (acute MS) Immune response study: Serum from 20 additional MS patients, and seven control patients |
Mutated forms ofProteolipid Protein 1, mutant-specific immuneresponse | SDS-PAGEnLC-MS/MSPeptide microarrays, NGS | [27] |
| 2 | Remyelination promoting proteins | Postmortem MS tissue | EphrinB3 target for therapies to promote remyelination in demyelinating disease | SDS-PAGE nLC-MS/MSLCM, in vitro/ in vivo study | [81] |
| 3 | Pathogenesis and/or remyelination Markers | Discovery set- 2 Postmortem brain blocks SPMS Validation-12PMS SPMS/PPMS |
Thymosin β-4 in partially remyelinated lesions | H&E, LFB, MALDI-IMS, LC-MS/MS, IHC | [82] |
| 4 | Neuroprotection | Five postmortem MS cortical tissueand four control brains | Hemoglobin β subunit (Hbb) interacting proteins (ATP synthase, histones, and a histone lysine demethylase) | LC-MS/MS, neuronal cultures, IHC | [85] |
A more recent study using LCM and LC-MS/MS in human MS brain tissue reported the presence of EphrinB3, an oligodendrocyte differentiation inhibitor, in demyelinated white-matter lesions (WMLs).[81] In another study, the authors employed MALDI-MS to profile peptides and proteins expressed in NAWM, grey matter and MS brain lesions with different extents of remyelination.[82] One important finding from this study was that lesions with low remyelination had compounds of molecular weight smaller than 5,300Da, whereas completely remyelinated lesions had molecular weights of more than 15,200Da. Furthermore, they also identified demyelinated lesion rim-associated proteins like thymosin β4 in 12 MS samples by LC-MS/MS. This protein has been reported to play a role in neurite extension, reducing inflammation, and regulating remyelination by prompting p38-mitogen activated pathway.[83, 84] Interestingly, CSF levels of this protein have been used to discriminate CIS from RRMS patients.[56] Furthermore, LC-MS/MS analysis of MS cortical proteins has revealed hemoglobin interaction proteins (ATP synthase, histones, and histone lysine demethylase).[85] We are also currently profiling proteins in LCM-collected WMLs and comparing them to surrounding NAWM using Quadrupole/Orbitrap MS proteomics approach. In the future, it is imperative that the involvement of individual cell types from CNS tissue should be explored for differential proteins and possibly for any disease-specific modifications.
3. Future challenges and directions
Over the years, the capability of mass spectrometry approaches to quantify nearly-complete proteomes from large biological samples from patients has dramatically changed the MS field. It has allowed unbiased screens and unforeseen discoveries. However, comprehensive proteome measurements have been limited to biological samples (CNS tissue, CSF, blood, etc.) composed of many different types of cells, indicating only the average cell population. When biological samples contain heterogeneous cells, this average cell population may not be represent changes in any specific cell types. Understanding single-cell function is important to elucidate protein expression in distinct cell populations and to understand MS disease progression. Many existing approaches are now being applied to single cells.
Single-cell isolation is typically accomplished using micromanipulation, microfluidics, or fluorescence-activated cell sorting (FACS).[86] In addition, novel techniques like LCM are also emerging to isolate single cells with even greater accuracy and specificity.[87] The benefit of LCM compared to other procedures of cell isolation such as FACS is that the positional information of cells is retained without dissociation of tissues and no live cells with genetic labels are needed. Previously, in RNA studies, LCM has been applied for topographical mapping of brain regions and neuronal populations in other neurological diseases,[88-90] and thus it offers a great opportunity to explore the involvement of specific CNS cell types in MS disease pathogenesis. In a recent report, authors have outlined specific technological improvements and strategies for single-cell proteomics that can increase sensitivity and throughput of single-cell mass spectrometry.[91] In addition to increasing the sensitivity throughput of single-cell proteomics, the above two technological advances can be applied to different proteomic techniques to identify and quantify the number of proteins per cell.
When choosing an optimal workflow to study cell proteomic profiles in MS disease, it is important to consider that proteins cannot be amplified (produced in small quantities by each cell), thus protein analysis at the single-cell level will need further development to complete the work flow as well as improvements in the sensitivity of analytical technologies. Furthermore, approaches and mass spectrometry technologies such as sample handling need to be developed in order to minimize sample loss. Proteomic analysis of single cells based on new mass spectrometry technologies such as the CyTOF are beginning to emerge.[92] CyTOF uses antibodies with metal labels as detection surrogates to measure proteins. Further, single-cell proteomics can be combined with single-cell genomics. Such knowledge of complicated biological cellular networks will allow the discovery of new ways to diagnose and treat MS.
4. Conclusion
Proteomic analysis of CNS tissue and body fluids is a powerful technique to identify biomarkers and therapeutic candidates. Over the past several years, advancements in the field of proteomic research have been instrumental in identifying new biomarkers in MS disease like CHI3L1, Tymosin β4, SCGs etc. However, validation and application of these markers is yet to be conducted in new cases using different techniques. Despite this, proteomic analysis application in the field of MS is still lagging behind than other omics techniques, although many studies have reported correlations with several potential candidates. One of the main reasons hindering application of this technique in detecting biomarkers of disease progression or pathogenesis is the complexity of the disease itself. MS affects both the CNS and immune system, making it extremely difficult to detect underlying pathogenic mechanisms. It is therefore expected that MS will not have a single biomarker, but rather a combination of several candidates that reflect the pathogenesis to its full extent. Designing of experiments that target each of the components of the disease is needed. In addition, modern techniques of cell separation and proteomic advancements must be used to determine the cellular targets that are indicative of each of these disease components. Moreover, there is an urgent need for development of biomarkers that reflect the repair mechanisms in MS. As we enter into the new era of MS therapeutics directed at repair, these markers can be used for validation of therapeutic candidates.
Acknowledgements
The authors would like to thank Dr. Christopher Nelson for editorial assistance. The authors are supported by funding of the NIH, NINDS (NS096148) and the National Multiple Sclerosis Society, USA (RG 5298) to RD.
Abbreviations:
- BBB
Blood brain barrier
- CDR
Complementarity-determining region
- CIS
Clinically isolated syndrome
- CNS
Central nervous system
- CSF
Cerebrospinal fluid
- ELISA
Enzyme-Linked immunosorbent assay
- ESI
Electrospray ionization
- FACS
Fluorescence-activated cell sorting
- ITRAQ
Isobaric tags for relative abundance and quantitation
- LCM
Laser-capture microdissection
- LC-MS/MS
Liquid chromatography tandem mass spectrometry analysis
- MALDI
Matrix-assisted laser desorption/ionization
- MRI
Magnetic resonance imaging
- MS
Multiple sclerosis
- NAWM
Normal appearing white matter
- SDS-PAGE
One-dimensional gel electrophoresis
- PMS
Progressive multiple sclerosis
- PPMS
Primary progressive multiple sclerosis
- RRMS
Relapsing remitting multiple sclerosis
- SELDI
Surface-enhanced laser desorption/ionization
- SPMS
Secondary progressive multiple sclerosis
- SRM
Selected reaction monitoring
- TOF
Time-of-flight
- 2D
Two-dimensional polyacrylamide gel electrophoresis
- IgG
Immunoglobulin G
- WMLs
White matter lesions
Footnotes
Conflict of interest: None reported.
References:
- [1].Lassmann H, Cold Spring Harb Perspect Med. 2018, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].G. B. D. N. D. C. Group, Lancet Neurol. 2017, 16, 877.28931491 [Google Scholar]
- [3].Compston A, Coles A, Lancet. 2008, 372, 1502. [DOI] [PubMed] [Google Scholar]
- [4].Lublin FD, Reingold SC, Neurology. 1996, 46, 907. [DOI] [PubMed] [Google Scholar]
- [5].Miller DH, Leary SM, Lancet Neurol. 2007, 6, 903. [DOI] [PubMed] [Google Scholar]
- [6].Criste G, Trapp B, Dutta R, Handb Clin Neurol. 2014, 122, 101. [DOI] [PubMed] [Google Scholar]
- [7].Schirmer L, Antel JP, Bruck W, Stadelmann C, Brain Pathol. 2011, 21, 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Garg N, Smith TW, Brain Behav. 2015, 5, e00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jones JL, Coles AJ, Experimental Neurology. 2010, 225, 34. [DOI] [PubMed] [Google Scholar]
- [10].Oksenberg JR, Baranzini SE, Sawcer S, Hauser SL, Nat Rev Genet. 2008, 9, 516. [DOI] [PubMed] [Google Scholar]
- [11].Dutta R, Chomyk AM, Chang A, Ribaudo MV, Deckard SA, Doud MK, Edberg DD, Bai B, Li M, Baranzini SE, Fox RJ, Staugaitis SM, Macklin WB, Trapp BD, Ann Neurol. 2013, 73, 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gerrard B, Singh V, Babenko O, Gauthier I, Wee Yong V, Kovalchuk I, Luczak A, Metz GAS, Neuroscience. 2017, 359, 299. [DOI] [PubMed] [Google Scholar]
- [13].Chomyk AM, Volsko C, Tripathi A, Deckard SA, Trapp BD, Fox RJ, Dutta R, Sci Rep. 2017, 7, 8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Keller A, Leidinger P, Steinmeyer F, Stahler C, Franke A, Hemmrich-Stanisak G, Kappel A, Wright I, Dorr J, Paul F, Diem R, Tocariu-Krick B, Meder B, Backes C, Meese E, Ruprecht K, Mult Scler. 2014, 20, 295. [DOI] [PubMed] [Google Scholar]
- [15].KhorshidAhmad T, Acosta C, Cortes C, Lakowski TM, Gangadaran S, Namaka M, Mol Neurobiol. 2016, 53, 1092. [DOI] [PubMed] [Google Scholar]
- [16].Stoessel D, Stellmann JP, Willing A, Behrens B, Rosenkranz SC, Hodecker SC, Sturner KH, Reinhardt S, Fleischer S, Deuschle C, Maetzler W, Berg D, Heesen C, Walther D, Schauer N, Friese MA, Pless O, Front Hum Neurosci. 2018, 12, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Reinke SN, Broadhurst DL, Sykes BD, Baker GB, Catz I, Warren KG, Power C, Mult Scler. 2014, 20, 1396. [DOI] [PubMed] [Google Scholar]
- [18].Grossman I, Knappertz V, Laifenfeld D, Ross C, Zeskind B, Kolitz S, Ladkani D, Hayardeny L, Loupe P, Laufer R, Hayden M, Prog Neurobiol. 2017, 152, 114. [DOI] [PubMed] [Google Scholar]
- [19].Comabella M, Vandenbroeck K, Curr Neurol Neurosci Rep. 2011, 11, 484. [DOI] [PubMed] [Google Scholar]
- [20].Singh V, Hintzen RQ, Luider TM, Stoop MP, J Neuroimmunol. 2012, 248, 40. [DOI] [PubMed] [Google Scholar]
- [21].Ghaste M, Mistrik R, Shulaev V, Int J Mol Sci. 2016, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wuhrer M, Selman MH, McDonnell LA, Kumpfel T, Derfuss T, Khademi M, Olsson T, Hohlfeld R, Meinl E, Krumbholz M, J Neuroinflammation. 2015, 12, 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Villen J, Gygi SP, Nat Protoc. 2008, 3, 1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Harauz G, Proteomics. 2017, 17. [DOI] [PubMed] [Google Scholar]
- [25].Lillico R, Zhou T, Khorshid Ahmad T, Stesco N, Gozda K, Truong J, Kong J, Lakowski TM, Namaka M, J Proteome Res. 2018, 17, 55. [DOI] [PubMed] [Google Scholar]
- [26].Ren RJ, Dammer EB, Wang G, Seyfried NT, Levey AI, Transl Neurodegener. 2014, 3, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Qendro V, Bugos GA, Lundgren DH, Glynn J, Han MH, Han DK, Proteomics. 2017, 17. [DOI] [PubMed] [Google Scholar]
- [28].Singh V, Stoop MP, Stingl C, Luitwieler RL, Dekker LJ, van Duijn MM, Kreft KL, Luider TM, Hintzen RQ, Mol Cell Proteomics. 2013, 12, 3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nishihara H, Shimizu F, Kitagawa T, Yamanaka N, Akada J, Kuramitsu Y, Sano Y, Takeshita Y, Maeda T, Abe M, Koga M, Nakamura K, Kanda T, Mult Scler. 2017, 23, 382. [DOI] [PubMed] [Google Scholar]
- [30].Ayoglu B, Mitsios N, Kockum I, Khademi M, Zandian A, Sjoberg R, Forsstrom B, Bredenberg J, Lima Bomfim I, Holmgren E, Gronlund H, Guerreiro-Cacais AO, Abdelmagid N, Uhlen M, Waterboer T, Alfredsson L, Mulder J, Schwenk JM, Olsson T, Nilsson P, Proc Natl Acad Sci U S A. 2016, 113, 2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kroksveen AC, Jaffe JD, Aasebo E, Barsnes H, Bjorlykke Y, Franciotta D, Keshishian H, Myhr KM, Opsahl JA, van Pesch V, Teunissen CE, Torkildsen O, Ulvik RJ, Vethe H, Carr SA, Berven FS, Proteomics. 2015, 15, 3361. [DOI] [PubMed] [Google Scholar]
- [32].Hinsinger G, Galeotti N, Nabholz N, Urbach S, Rigau V, Demattei C, Lehmann S, Camu W, Labauge P, Castelnovo G, Brassat D, Loussouarn D, Salou M, Laplaud D, Casez O, Bockaert J, Marin P, Thouvenot E, Mult Scler. 2015, 21, 1251. [DOI] [PubMed] [Google Scholar]
- [33].Vaananen T, Vuolteenaho K, Kautiainen H, Nieminen R, Mottonen T, Hannonen P, Korpela M, Kauppi MJ, Laiho K, Kaipiainen-Seppanen O, Luosujarvi R, Uusitalo T, Uutela T, Leirisalo-Repo M, Moilanen E, Group NE-RS, PLoS One. 2017, 12, e0183294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang J, Sheng Z, Yang W, Cai Y, Cell Physiol Biochem. 2016, 38, 461. [DOI] [PubMed] [Google Scholar]
- [35].Lapek JD Jr., Greninger P, Morris R, Amzallag A, Pruteanu-Malinici I, Benes CH, Haas W, Nat Biotechnol. 2017, 35, 983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Karas M, Hillenkamp F, Anal Chem. 1988, 60, 2299. [DOI] [PubMed] [Google Scholar]
- [37].Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM, Science. 1989, 246, 64. [DOI] [PubMed] [Google Scholar]
- [38].Wu CC, MacCoss MJ, Curr Opin Mol Ther. 2002, 4, 242. [PubMed] [Google Scholar]
- [39].Stoop MP, Dekker LJ, Titulaer MK, Burgers PC, Sillevis Smitt PA, Luider TM, Hintzen RQ, Proteomics. 2008, 8, 1576. [DOI] [PubMed] [Google Scholar]
- [40].Stoop MP, Dekker LJ, Titulaer MK, Lamers RJ, Burgers PC, Sillevis Smitt PA, van Gool AJ, Luider TM, Hintzen RQ, J Proteome Res. 2009, 8, 1404. [DOI] [PubMed] [Google Scholar]
- [41].Jackson DH, Banks RE, Proteomics Clin Appl. 2010, 4, 250. [DOI] [PubMed] [Google Scholar]
- [42].Aguilar-Mahecha A, Kuzyk MA, Domanski D, Borchers CH, Basik M, PLoS One. 2012, 7, e38290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Brandle SM, Obermeier B, Senel M, Bruder J, Mentele R, Khademi M, Olsson T, Tumani H, Kristoferitsch W, Lottspeich F, Wekerle H, Hohlfeld R, Dornmair K, Proc Natl Acad Sci U S A. 2016, 113, 7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Schutzer SE, Angel TE, Liu T, Schepmoes AA, Xie F, Bergquist J, Vecsei L, Zadori D, Camp DG 2nd, Holland BK, Smith RD, Coyle PK, PLoS One. 2013, 8, e66117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Stoop MP, Runia TF, Stingl C, van der Vuurst de Vries RM, Luider TM, Hintzen RQ, Proteomics Clin Appl. 2017, 11. [DOI] [PubMed] [Google Scholar]
- [46].Hecker M, Fitzner B, Wendt M, Lorenz P, Flechtner K, Steinbeck F, Schroder I, Thiesen HJ, Zettl UK, Mol Cell Proteomics. 2016, 15, 1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Perga S, Giuliano Albo A, Lis K, Minari N, Falvo S, Marnetto F, Caldano M, Reviglione R, Berchialla P, Capobianco MA, Malentacchi M, Corpillo D, Bertolotto A, PLoS One. 2015, 10, e0129291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kroksveen AC, Guldbrandsen A, Vaudel M, Lereim RR, Barsnes H, Myhr KM, Torkildsen O, Berven FS, J Proteome Res. 2017, 16, 179. [DOI] [PubMed] [Google Scholar]
- [49].Comabella M, Fernandez M, Martin R, Rivera-Vallve S, Borras E, Chiva C, Julia E, Rovira A, Canto E, Alvarez-Cermeno JC, Villar LM, Tintore M, Montalban X, Brain. 2010, 133, 1082. [DOI] [PubMed] [Google Scholar]
- [50].Llorens F, Thune K, Tahir W, Kanata E, Diaz-Lucena D, Xanthopoulos K, Kovatsi E, Pleschka C, Garcia-Esparcia P, Schmitz M, Ozbay D, Correia S, Correia A, Milosevic I, Andreoletti O, Fernandez-Borges N, Vorberg IM, Glatzel M, Sklaviadis T, Torres JM, Krasemann S, Sanchez-Valle R, Ferrer I, Zerr I, Mol Neurodegener. 2017, 12, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Thompson AG, Gray E, Thezenas ML, Charles PD, Evetts S, Hu MT, Talbot K, Fischer R, Kessler BM, Turner MR, Ann Neurol. 2018, 83, 258. [DOI] [PubMed] [Google Scholar]
- [52].Pavelek Z, Vysata O, Tambor V, Pimkova K, Vu DL, Kuca K, Stourac P, Valis M, Biomed Rep. 2016, 5, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Timirci-Kahraman O, Karaaslan Z, Tuzun E, Kurtuncu M, Baykal AT, Gunduz T, Tuzuner MB, Akgun E, Gurel B, Eraksoy M, Kucukali CI, Acta Neurol Belg. 2018. [DOI] [PubMed] [Google Scholar]
- [54].Valko PO, Roschitzki B, Faigle W, Grossmann J, Panse C, Biro P, Dambach M, Spahn DR, Weller M, Martin R, Baumann CR, J Sleep Res. 2018, e12721. [DOI] [PubMed] [Google Scholar]
- [55].van Luijn MM, van Meurs M, Stoop MP, Verbraak E, Wierenga-Wolf AF, Melief MJ, Kreft KL, Verdijk RM, t Hart BA, Luider TM, Laman JD, Hintzen RQ, J Neuropathol Exp Neurol. 2016, 75, 86. [DOI] [PubMed] [Google Scholar]
- [56].Liguori M, Qualtieri A, Tortorella C, Direnzo V, Bagala A, Mastrapasqua M, Spadafora P, Trojano M, PLoS One. 2014, 9, e103984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Zhang J, Zhang ZG, Li Y, Lu M, Zhang Y, Elias SB, Chopp M, Neurobiol Dis. 2016, 88, 85. [DOI] [PubMed] [Google Scholar]
- [58].Singh V, van Pelt ED, Stoop MP, Stingl C, Ketelslegers IA, Neuteboom RF, Catsman-Berrevoets CE, Luider TM, Hintzen RQ, Neurol Neuroimmunol Neuroinflamm. 2015, 2, e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Stoop MP, Rosenling T, Attali A, Meesters RJ, Stingl C, Dekker LJ, van Aken H, Suidgeest E, Hintzen RQ, Tuinstra T, van Gool A, Luider TM, Bischoff R, J Proteome Res. 2012, 11, 4315. [DOI] [PubMed] [Google Scholar]
- [60].Stoop MP, Singh V, Stingl C, Martin R, Khademi M, Olsson T, Hintzen RQ, Luider TM, J Proteome Res. 2013, 12, 1101. [DOI] [PubMed] [Google Scholar]
- [61].Thompson AG, Gray E, Mager I, Fischer R, Thezenas ML, Charles PD, Talbot K, El Andaloussi S, Kessler BM, Wood M, Turner MR, Proteomics. 2018, 18, e1800257. [DOI] [PubMed] [Google Scholar]
- [62].Chiasserini D, van Weering JR, Piersma SR, Pham TV, Malekzadeh A, Teunissen CE, de Wit H, Jimenez CR, J Proteomics. 2014, 106, 191. [DOI] [PubMed] [Google Scholar]
- [63].Kadota T, Yoshioka Y, Fujita Y, Kuwano K, Ochiya T, Semin Cell Dev Biol. 2017, 67, 39. [DOI] [PubMed] [Google Scholar]
- [64].Quek C, Hill AF, Biochem Biophys Res Commun. 2017, 483, 1178. [DOI] [PubMed] [Google Scholar]
- [65].Lee J, McKinney KQ, Pavlopoulos AJ, Han MH, Kim SH, Kim HJ, Hwang S, Clin Chim Acta. 2016, 462, 118. [DOI] [PubMed] [Google Scholar]
- [66].Lewin A, Hamilton S, Witkover A, Langford P, Nicholas R, Chataway J, Bangham CRM, Wellcome Open Res. 2016, 1, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Tremlett H, Dai DL, Hollander Z, Kapanen A, Aziz T, Wilson-McManus JE, Tebbutt SJ, Borchers CH, Oger J, Cohen Freue GV, J Proteomics. 2015, 118, 2. [DOI] [PubMed] [Google Scholar]
- [68].Yin L, Liu J, Dong H, Xu E, Qiao Y, Wang L, Zhang L, Jia J, Li L, Geng X, Neurosci Lett. 2014, 562, 34. [DOI] [PubMed] [Google Scholar]
- [69].Wallin MT, Oh U, Nyalwidhe J, Semmes J, Kislinger T, Coffman P, Kurtzke JF, Jacobson S, Eur J Neurol. 2015, 22, 591. [DOI] [PubMed] [Google Scholar]
- [70].Fiorini A, Koudriavtseva T, Bucaj E, Coccia R, Foppoli C, Giorgi A, Schinina ME, Di Domenico F, De Marco F, Perluigi M, PLoS One. 2013, 8, e65184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zandian A, Forsstrom B, Haggmark-Manberg A, Schwenk JM, Uhlen M, Nilsson P, Ayoglu B, J Proteome Res. 2017, 16, 1300. [DOI] [PubMed] [Google Scholar]
- [72].Rejdak K, Leary SM, Petzold A, Thompson AJ, Miller DH, Giovannoni G, Mult Scler. 2010, 16, 1066. [DOI] [PubMed] [Google Scholar]
- [73].Gebregiworgis T, Nielsen HH, Massilamany C, Gangaplara A, Reddy J, Illes Z, Powers R, J Proteome Res. 2016, 15, 659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Singh V, Stingl C, Stoop MP, Zeneyedpour L, Neuteboom RF, Smitt PS, Hintzen RQ, Luider TM, J. Proteome Res. 2015, 14, 2065. [DOI] [PubMed] [Google Scholar]
- [75].Nielsen HH, Beck HC, Kristensen LP, Burton M, Csepany T, Simo M, Dioszeghy P, Sejbaek T, Grebing M, Heegaard NH, Illes Z, PLoS One. 2015, 10, e0139659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Ardito F, Perrone D, Cocchi R, Lo Russo L, L. A DE, Giannatempo G, Lo Muzio L, Oncol Lett. 2016, 11, 1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Loo JA, Yan W, Ramachandran P, Wong DT, J Dent Res. 2010, 89, 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Oppenheim FG, Salih E, Siqueira WL, Zhang W, Helmerhorst EJ, Ann N Y Acad Sci. 2007, 1098, 22. [DOI] [PubMed] [Google Scholar]
- [79].Manconi B, Liori B, Cabras T, Vincenzoni F, Iavarone F, Lorefice L, Cocco E, Castagnola M, Messana I, Olianas A, J Proteomics. 2018, 187, 212. [DOI] [PubMed] [Google Scholar]
- [80].Han MH, Hwang SI, Roy DB, Lundgren DH, Price JV, Ousman SS, Fernald GH, Gerlitz B, Robinson WH, Baranzini SE, Grinnell BW, Raine CS, Sobel RA, Han DK, Steinman L, Nature. 2008, 451, 1076. [DOI] [PubMed] [Google Scholar]
- [81].Syed YA, Zhao C, Mahad D, Mobius W, Altmann F, Foss F, Gonzalez GA, Senturk A, Acker-Palmer A, Lubec G, Lilley K, Franklin RJM, Nave KA, Kotter MRN, Acta Neuropathol. 2016, 131, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Maccarrone G, Nischwitz S, Deininger SO, Hornung J, Konig FB, Stadelmann C, Turck CW, Weber F, J Chromatogr B Analyt Technol Biomed Life Sci. 2017, 1047, 131. [DOI] [PubMed] [Google Scholar]
- [83].Vartiainen N, Pyykonen I, Hokfelt T, Koistinaho J, Neuroreport. 1996, 7, 1613. [DOI] [PubMed] [Google Scholar]
- [84].Santra M, Chopp M, Zhang ZG, Lu M, Santra S, Nalani A, Santra S, Morris DC, Glia. 2012, 60, 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Brown N, Alkhayer K, Clements R, Singhal N, Gregory R, Azzam S, Li S, Freeman E, McDonough J, J Mol Neurosci. 2016, 59, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Hu P, Zhang W, Xin H, Deng G, Front Cell Dev Biol. 2016, 4, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Datta S, Malhotra L, Dickerson R, Chaffee S, Sen CK, Roy S, Histol Histopathol. 2015, 30, 1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Garrido-Gil P, Fernandez-Rodriguez P, Rodriguez-Pallares J, Labandeira-Garcia JL, Histochem Cell Biol. 2017, 148, 299. [DOI] [PubMed] [Google Scholar]
- [89].Tagliafierro L, Bonawitz K, Glenn OC, Chiba-Falek O, Front Mol Neurosci. 2016, 9, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Mauney SA, Woo TW, Sonntag KC, Methods Mol Biol. 2018, 1723, 203. [DOI] [PubMed] [Google Scholar]
- [91].Specht H, Slavov N, J Proteome Res. 2018, 17, 2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Kay AW, Strauss-Albee DM, Blish CA, Methods Mol Biol. 2016, 1441, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]