Abstract
Thymidylate synthase (TS), found in all organisms, is an essential enzyme responsible for the de novo synthesis of deoxythymidine monophosphate. The TS active sites of the protozoal parasite Cryptosporidium hominis and human are relatively conserved. Evaluation of antifolate compound 1 and its R-enantiomer 2 against both enzymes reveals divergent inhibitor selectivity and enzyme stereospecificity. To establish how C. hominis and human TS (ChTS and hTS) selectively discriminate 1 and 2, respectively, we determined crystal structures of ChTS complexed with 2 and hTS complexed with 1 or 2. Coupled with the previously determined structure of ChTS complexed with 1, we discuss a possible mechanism for enzyme stereospecificity and inhibitor selectivity.
Keywords: Enzyme stereospecificity, Inhibitor selectivity, Cryptosporidium hominis, Thymidylate synthase, Chiral recognition, X-Ray crystallography
Introduction
The ability of biological macromolecules to recognize chiral compounds is a fundamental feature of nature and is essential in life processes. Biological macromolecules have evolved to have chiral character and selectively bind to molecules with specific special configurations.[1] Preference for a single enantiomer is exemplified by natural amino acids and sugars composed of L-isomers and D-isomers, respectively.[2] Additionally, the high stereospecificity which occurs in biological interactions such as protein/ligand and enzyme/substrate, suggest that these effects are mediated through molecular recognition and strict chiral matching.[1, 3] The ability for biological systems to discriminate between enantiomers has implications for a diverse range of applications in fields such as environmental pollution, food flavoring and the pharmaceutical industry.[4–6]
While sharing identical chemical formula and atomic linkages, enantiomeric drugs are well known to display different pharmacological and toxicological effects.[5] For example S-thalidomide is a powerful teratogen, while its R-enantiomer was prescribed to pregnant women for lessening morning sickness.[1, 7] Additionally, L-propoxyphene is an antitussive agent, while D-propoxyphene is an analgesic.[8, 9] It is evident that stereoisomers have quite distinct biological activities. Through biochemical exploration of these processes we can determine potential benefits and risks of chiral compounds in humans.[1]
The essential enzyme thymidylate synthase (TS) has long been a chemotherapeutic target for cancer and infectious disease.[10, 11] TS, a highly conserved enzyme found in all organisms, is responsible for catalyzing the de novo synthesis of deoxythymidine monophosphate (dTMP) from deoxyuridine monophosphate (dUMP) and the cofactor 5,10-methylenetetrahydrofolate (CH2H4F), producing dihydrofolate in the process.[10, 12] Dihydrofolate reductase (DHFR) then utilizes dihydrofolate and the cofactor NADPH, to produce tetrahydrofolate and NADP+.[10, 13] In humans and most other species, both TS and DHFR are expressed as two separate enzymes.[14] In protozoal parasites, such as Cryptosporidium hominis (C. hominis), the two catalytic activities of TS and DHFR exist as one bifunctional enzyme.[14] Though highly conserved, the TS active site of both the C. hominis and human enzymes differ by only two residues, which reside in the binding region for the glutamate moiety of folate. In C. hominis TS (ChTS) these residues are S290 and A287, versus G83 and F80 in human TS (hTS), respectively.[10]
Previously we reported the 2.7 Å resolution X-ray crystal structure of ChTS complexed with the S-enantiomer of antifolate analog, (2-amino-4-oxo-4,7-dihydro-pyrrolo[2,3-d]pyrimidin-methyl-phenyl-L-glutamic acid), compound 1, along with the inhibitory activity of 1 for both ChTS and hTS, showing a preference for the parasitic enzyme.[11] (Compound 1 is referred to as 906 in several papers published by our lab.) These data suggest that the variant, non-conserved residues in the ChTS and hTS active site play an important role in determining the selectivity for compound 1. In the present study, we explore the biochemical and structural basis for selectivity by preparing the corresponding R-enantiomer, compound 2, and evaluating the inhibitory activity for each stereoisomer with the ChTS and hTS enzymes as well as determining the corresponding X-ray structures complexed with 1 or 2. We discuss the structural basis for inhibitor selectivity and enzyme chiral recognition for either S or R-enantiomer with each species of TS.
Materials and methods
Enzyme Preparation, inhibition assay, and analysis
ChTS-DHFR was expressed, purified and an estimation of protein concentration was determined as reported in previous study.[15] His6-human TS, wild type, (WT-hTS) was expressed, purified and an estimation of protein concentration was determined as previously described.[16] Δ7–29 human TS deletion mutant (mhTS) was expressed and purified as reported in previous studies with several modifications.[16–18] The supernatant was treated with ammonium sulfate to a final concentration of 60 % saturation and centrifuged at 15,000 RPM for 1 h at 4 °C. Pelleted protein was re-dissolved in lysis buffer and dialyzed overnight against the same buffer. The dialysate was passed through a DEAE sepharose Fast Flow column (GE Healthcare) and eluted with a gradient of potassium phosphate from 10 mM to 250 mM. Protein was passed through HiTrap phenyl HP column (GE Healthcare) equilibrated with buffer (20 mM Tris, pH 7.5, 0.1 mM EDTA, 0.5 mM TCEP, and 1 M ammonium sulfate) and eluted with a gradient of ammonium sulfate from 1 M to 250 mM. Protein was passed through Hitrap Blue HP column (GE Healthcare) equilibrated with buffer (20 mM Tris, pH 7.5, 0.1 mM EDTA, 0.5 mM TCEP, and 5 mM KCl) and eluted with gradient of KCl from 5 mM to 1 M. Finally, protein was loaded on a Superdex 200 Increase column (GE Healthcare), which was equilibrated and eluted with 10 mM potassium phosphate buffer, pH 7.5, and 0.5 mM TCEP. Protein concentration was estimated spectrophotometrically at 280 nm using an extinction coefficient of 43,890 M−1 cm−1.
TS activity was determined as reported in previous study.[15] Reaction conditions used to determine IC50 values for ChTS and WT-hTS TS were as previously described.[11] Data were fitted to an [Inhibitor] vs normalized response with variable slope equation using GraphPad Prism software (version 7.01).
Protein crystallization and structure determination
The purified ChTS-DHFR enzyme was co-crystallized with 2 by hanging drop vaporization as previously described.[11] The purified mhTS enzyme was co-crystallized with 1 by sitting drop vaporization with a protein:well solution ratio of 1:1 to screen crystallization conditions. Reservoir conditions for crystallization of 1 included 5 % (w/v) PEG 8000, 40 % (w/v) 2-methyl-2,4-pentanediol, and 100 mM sodium cacodylate, pH 6.5. mhTS enzyme was co-crystallized with 2 by sitting drop vaporization with a protein:well solution:seed solution ratio of 3:2:1 to screen crystallization conditions. Reservoir conditions for crystallization of 2 included 1.55 M sodium formate, and 0.1 mM sodium acetate, pH 5.0. Seed solution for crystallization of 2 consisted of 0.2 mM MgCl2, and 20 % (w/v) PEG 3350 with a dilution ratio 50:1 seed solution:seed stock. mhTS enzyme (approximately 13 mg/ml) was incubated with 20 mM Bis-Tris, pH 7.0, 2 mM DTT, 2 mM dUMP, and 5 mM of either 1 or 2 on ice for 1 h before setting up drops. Crystals grew in approximately 2 weeks at 22 °C.
Diffraction data for ChTS-DHFR with 2 and mhTS with 1 were collected on beam line 24-ID-E while mhTS with 2 was collected on beam line 24-ID-C through NE-CAT, using wavelength 0.979 Å. Data sets for best diffracting crystals were processed by XDS.[19] Phases were solved via molecular replacement using the program phaser.[20, 21] For ChTS-DHFR:2 a ChTS-DHFR structure in complex with four ligands (PDB ID 4Q0E)[22] was used as the search model for molecular replacement. For mhTS:2 and mhTS:1 an apo mhTS structure (PDB ID 1HZW)[17] was used as the search model for molecular replacement. The program COOT[23] was used for model building of the ligands into the electron density as well as residues 307–313 of mhTS. Structures were refined using Phenix.[24] The crystallographic statistics are summarized in Table 1. Iterative build simulated annealing omit σA- weighted 2mFo-Fc electron density maps were generated using Phenix Autobuild.[25]
Table:1.
Data collection and refinement statistics.
| mhTS: 1 | mhTS: 2 | ChTS-DHFR: 2 | |
|---|---|---|---|
| Data Collection | |||
| X-Ray Source | APS 24ID-E | APS 24ID-C | APS 24ID-E |
| Wavelength (Å) | 0.97915 | 0.979200 | 0.97915 |
| Space group | C 1 2 1 | P 1 21 1 | C 1 2 1 |
| Unite-cell parameters (Å) | a=154.96, | a=62.00, | a=212.61, |
| b=103.04, | b=124.81, | b=116.18, | |
| c=109.22 | c=80.36 | c=220.89 | |
| α=90,β=128.92, | α=90, β=92.09, | α=90, β=95.486, | |
| γ=90 | γ=90 | γ=90 | |
| Resolution range (Å) | 50.0–2.59 (2.75–2.59) | 50.0–2.88 (3.06–2.88) | 51.0–3.21 (3.41–3.21) |
| Completeness (%) | 98.7 (97.1) | 98.6 (94.5) | 98.2 (91.9) |
| Rsym (%) | 0.141 (1.16) | 0.227 (0.963) | 0.317 (1.73) |
| CC(1/2) (%) | 99.6 (80.7) | 98.4 (84.2) | 98.3 (61.6) |
| Avg. I/σ | 12.72 (1.44) | 6.69 (2.11) | 4.98 (1.06) |
| Redundancy | 6.6 (6.4) | 6.8 (6.7) | 6.9 (6.7) |
| Refinement Statisitics | |||
| No. Reflections (Unique Reflections) | 272583 (41155) | 184496 (27262) | 594193 (85851) |
| Rfree/Rwork | 0.2688/0.2402 | 0.2494/0.2131 | 0.2317/0.2179 |
| No. of Atoms | 9190 | 8992 | 20926 |
| Protein | 8957 | 8758 | 20266 |
| Ligands | 200 | 200 | 660 |
| Solvent | 33 | 34 | |
| R.M.S. deviation | |||
| Bond lengths (Å) | 0.002 | 0.002 | 0.003 |
| Bond angles (°) | 0.511 | 0.512 | 0.502 |
| Avg. B-factor | 67.98 | 51.35 | 90.53 |
| Protein | 67.97 | 49.99 | 88.62 |
| Ligand | 68.82 | 53.83 | 91.17 |
| Solvent | 62.34 | 37.00 | |
| Ramachandran Plot | |||
| Most favored region (%) | 95.83 | 94.86 | 93.97 |
| Allowed region (%) | 4.17 | 5.14 | 6.03 |
Values in parentheses are from highest resolution shell. One crystal was used for the data set.
Results
Evaluation of inhibitory activity
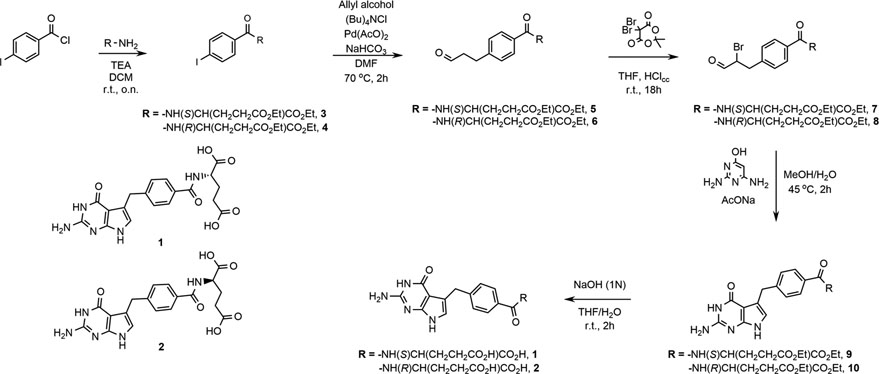
The synthesis of compounds 1 and 2 is depicted in Scheme 1. While the synthesis of 1 has been reported earlier,[11, 26, 27] the modified scheme reported here offers a simplified reaction route and improvements in yield from more readily accessible precursors. Compound 1 was previously evaluated for inhibitory activity against ChTS and WT-hTS where it demonstrated a 5-fold greater potency against ChTS (Table 2).[11] Here we similarly evaluated 2 inhibitory activity against both enzymes. The catalytic activity of ChTS was inhibited by 2 with an IC50 of 9.8 ± 0.6 μM, while WT-hTS was inhibited with and IC50 of 0.6 ± 0.2 μM. Compound 2 displays a 16-fold greater potency for the human form of the enzyme as compared to the parasite enzyme. The stereospecificity for S-enantiomer, 1, over R, 2, was 26-fold for the ChTS(Table 2). The preference is reversed for the hTS where 2, favored the R-enantiomer by 3-fold.
Scheme:1.
Table:2.
IC50(μcm) for ChTS and hTSa.
| Compound | ChTS IC50 | hTS IC50 | Fold-Selectivity |
|---|---|---|---|
| 1b | 0.38 ± 0.04 | 1.8 ± 0.5 | 5 |
| 2 | 9.8 ± 0.6 | 0.6 ± 0.2 | 16 |
| Fold-Stereospecificity | 26 | 3 |
SD from triplicate measurement.
Results from reference [11]
Overall structures of ChTS and hTS
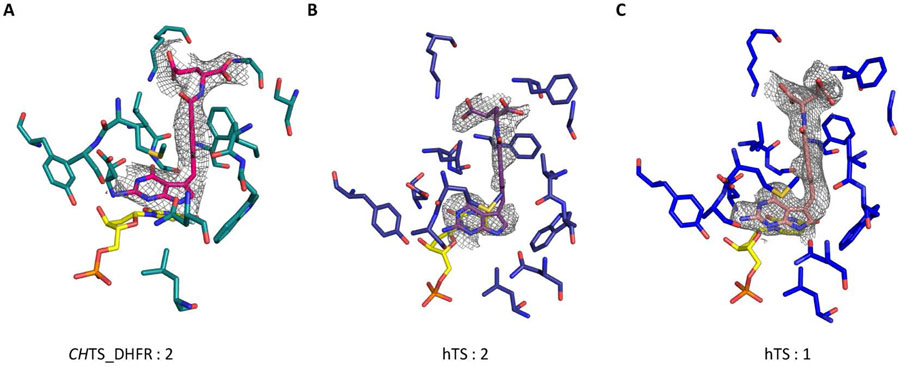
The crystal structure of ChTS-DHFR complexed with 2 (PDB ID: 6OJS) in addition to three other ligands of ChTS-DHFR, 5-fluorodeoxyuridine monophosphate (FdUMP), NADPH, and methotrexate (MTX), was determined with an asymmetric unit containing five protein molecules assembled in two homodimers and one monomer. The protein crystallized in a C2 space group with a similar crystal lattice as reported previously for ChTS-DHFR complexed with 1.[11]. All residues from 3–521 except for residues 181–192 were clearly defined in the electron density, allowing the ligand binding sites of the structure to be visualized (Fig. 1A and Fig. S1A). The Δ7–29 deletion mutant of hTS (mhTS) was utilized to obtain X-ray structures of mhTS complexed with either 1 or 2 in addition to the ligand deoxyuridine monophosphate (dUMP). mhTS complexed with 1 (PDB ID: 6OJV) crystallized in a C2 space group containing four protein molecules in the asymmetric unit assembled in one homodimer and two monomers. Where mhTS complexed with 2 (PDB ID: 6OJU) crystallized in a P21 space group containing four protein molecules in the asymmetric unit assembled as four monomers. Residues from 6–313 for both structures of mhTS were clearly defined in the electron density, allowing the ligand binding sites of the structure to be visualized (Fig. 1B and 1C, Fig. S1B and S1C). The best crystals yielded diffraction data to a resolution of 3.21 Å, 2.60 Å, and 2.88 Å for ChTS with 2 and hTS with 1 or 2, respectively.
Figure:1.
Omit σA- weighted 2mFo-Fc electron density maps of inhibitor compound. A. 2 complexed with ChTS B. 2 complexed with hTS C. 1 complexed with hTS. Corresponds to supplementary Fig S1. Omit σAweighted 2mFo-Fc electron density maps of inhibitor compound and TS active site.
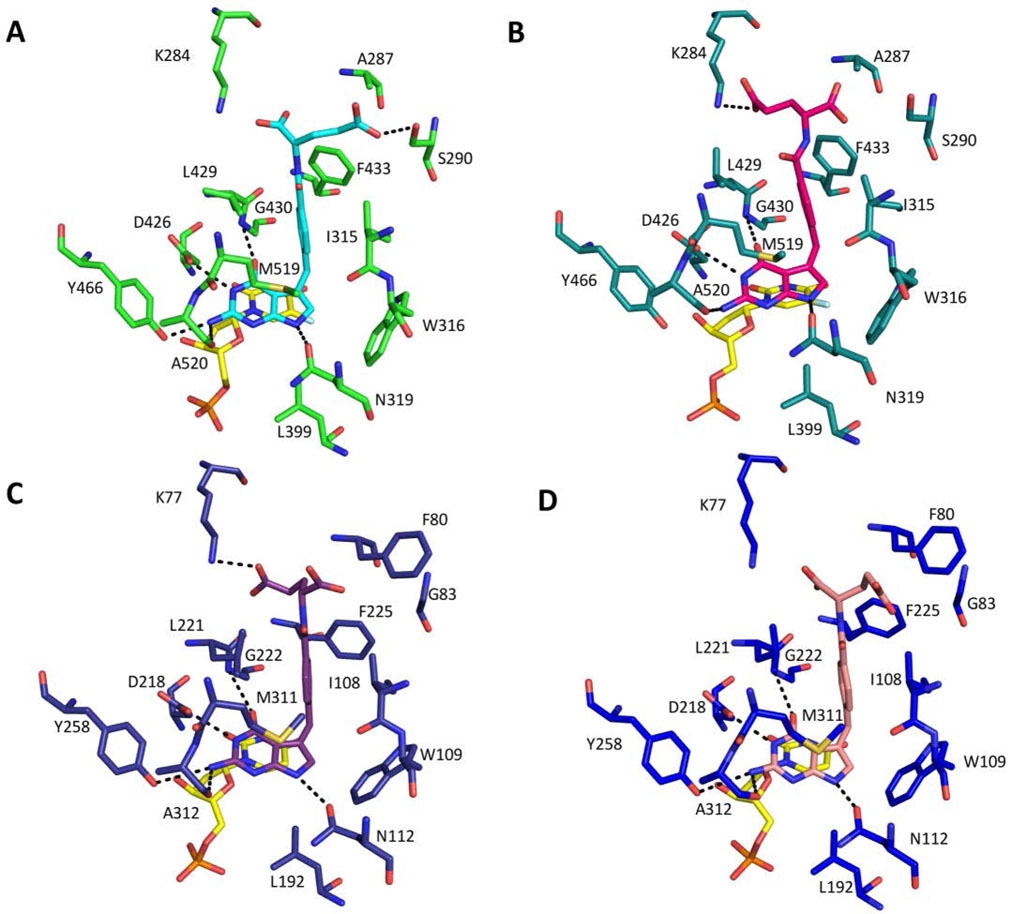
The five protein molecules of the ChTS structure with 2 asymmetric subunit can be superimposed with an average root-mean-square-deviation (RMSD) of 0.41 Å for 508 Cα atoms. Compound 2 binds into the pocket where the folate substrate, CH2H4F, would be and adopts a similar conformation in all chains with the greatest variation in the molecule occurring within the glutamate moiety (Fig. S2A). The structure of chain A was used as representative of the five chains in the asymmetric unit. The ChTS active site and 2 form similar interactions to those previously reported for 1 (Fig. 2A).[11] As observed previously, the ChTS active site predominantly consists of hydrophobic residues, A287, I315, W316, L399, L429, G430, F433, Y466, and M519. Compound 2 binds close to FdUMP with the pyrimidine ring of FdUMP forming a stacking interaction with the pyrrolo[2,3-d]pyrimidine of 2. The pyrrolo[2,3-d]pyrimidine of 2 forms several Van der Waals and hydrophobic interactions with residues W316, L399 and Y466. Residues I315, L429, F433, and M519 form hydrophobic interactions with the phenyl moiety of 2, while the glutamate moiety interacts with the non-conserved residue A287. Four hydrogen bonds are formed between the pyrrolo[2,3-d]pyrimidine of 2 and the TS active site. The carbonyl O of D426 hydrogen bonds with N3 and the carbonyl O of N319 hydrogen bonds with N7. The amino backbone of G430 hydrogen bonds with the 4-oxo group and the backbone carbonyl O of A520 hydrogen bonds with the 2-amino group. Additionally, the γ-carboxylate of the glutamate moiety is within hydrogen bonding distance of residue K284 amino group. Importantly, unlike the structure of ChTS complexed with 1, compound 2 does not form hydrogen bonds with S290, or Y466 (Fig. 2B).
Figure:2.
Active site residues of ChTS (green tones) and hTS (blue tones) A. ChTS complexed with 1 (cyan) and FdUMP (yellow). B ChTS complexed with 2 (magenta) and FdUMP (yellow). C. hTS complexed with 1 (purple) and dUMP (yellow). D. hTS complexed with 1 (pink) and dUMP (yellow). Corresponds to supplementary Fig S3. Active site residues which form hydrogen bonding interactions.
The four protein molecules in the asymmetric subunit, from mhTS structure with 1 or 2, can be superimposed with an average RMSD of 0.45 Å and 0.37 Å for 291 Cα atoms, respectively. Both enantiomers bind into the CH2H4F pocket and adopt a similar conformation in all chains with the greatest variation in the molecule occurring within the glutamyl moiety. Additionally, there is a small shift for the pyrrolo[2,3-d]pyrimidine moiety for both enantiomers in chain D as compared to the other chains in the asymmetric subunit (Fig. S2B and S2C). Chain A from each crystal structure of mhTS was used representative of the four chains in the asymmetric unit. As was observed for ChTS, the mhTS active site predominantly consists of hydrophobic residues, F80, I108, W109, L192, L221, G222, F225, Y258, and M311. Both 1 and 2 binds close to dUMP in the mhTS active site, with the pyrimidine ring of dUMP forming a stacking interaction with the pyrrolo[2,3-d]pyrimidine to both stereoisomers. Several Van der Waals and hydrophobic interactions are observed between the pyrrolo[2,3-d]pyrimidine and residues W109, L192 and Y258. Residues I108, L221, F225, and M311 form hydrophobic interactions with the phenyl moiety, while the glutamate moiety of both stereoisomers hydrophobically interacts with the non-conserved residue F80. Five hydrogen bonds are formed between the pyrrolo[2,3-d]pyrimidine of both enantiomers and the TS active site. The carbonyl O of D218 hydrogen bonds with N3 and the carbonyl O of N112 hydrogen bonds with N7. The amino backbone of G222 hydrogen bonds with the 4-oxo group and the hydroxyl group of Y258 and the backbone carbonyl O of A312 hydrogen bonds with the 2-amino group. Additionally, γ-carboxylate O of the glutamate moiety in 2 is within hydrogen bonding distance of residue K77 amino group (Fig. 2C) and 2D).
Discussion
Stereospecificity of ChTS
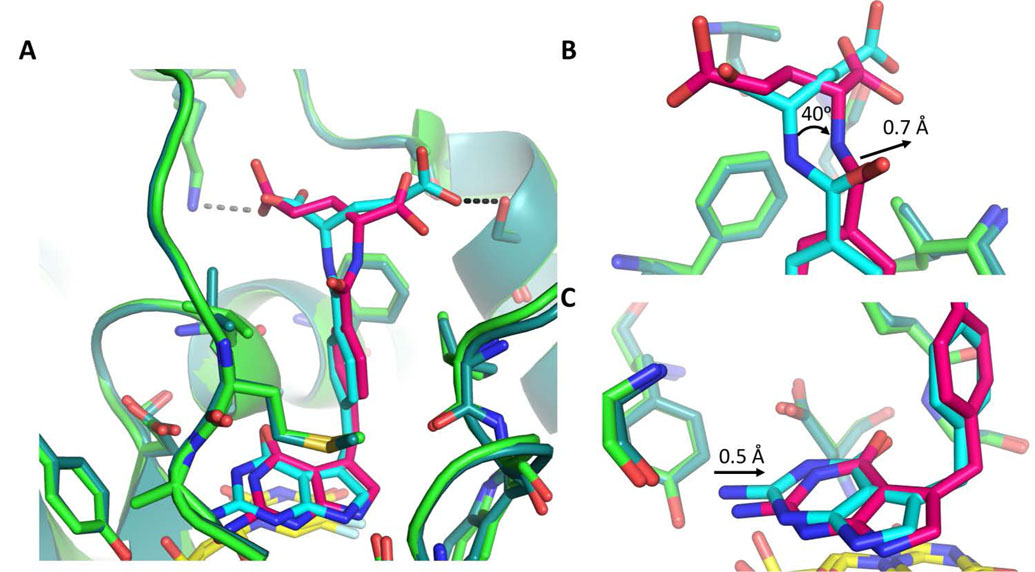
Structurally the ChTS active site with either 1 or 2 bound is similar; however, there are several key differences (Fig. 3A). First, in examining active site residues, there is a ~ 120° clockwise rotation of the L429 side chain in the structure for 2 as compared to the previously published structure of 1.[11] However, because the structure for 2 was solved at a lower resolution, 3.21 Å as compared to 2.78 Å for the structure of 1, the positioning of the L429 side chain may not be reliable.
Figure:3.
Comparison of ChTS bound to 1 (light green/cyan) and ChTS bound 2 (dark green/magenta) with FdUMP (yellow). A. Comparison of 1 and 2 bound in ChTS active site. ChTS stabilizes 1 with a hydrogen bonding interaction with residue S290 (Black dotted line). ChTS hydrogen bonding interaction between 2 and residue K284 may induce conformational strain (gray dotted line). B. Zoom in view of glutamate moiety of 1 and 2. C. Zoom in view of the pyrrolo[2,3-d]pyrimidine of 1 and 2.
Secondly, comparing both 1 and 2 bound in ChTS reveals that 2 conformationally strained. The pyrrolo[2,3-d]pyrimidine and phenyl moieties for both compounds are similarly placed within the CH2H4F binding pocket. However, the position of the γ-carboxylate of the glutamate moiety of 1 versus 2 is different. In the previously solved ChTS with 1, the γ-carboxylate formed a hydrogen bond with S280.[11] In the case of ChTS with 2, the γ-carboxylate extends to hydrogen bond with K284 (see Fig. 2A versus 2B), which in turn induces a rotation of the amino linker region by ~ 40°, in addition to ~ 0.7 Å shift of the amino linker towards M519 (Fig. 3B). These conformational changes appear to propagate a ~ 0.5 Å shift of the pyrrolo[2,3-d]pyrimidine moiety of 2 out of hydrogen bonding distance with Y466 (Fig. 3C). Compound 1, which is not under similar conformational strain is able to maintain hydrogen bonding distance with Y466.
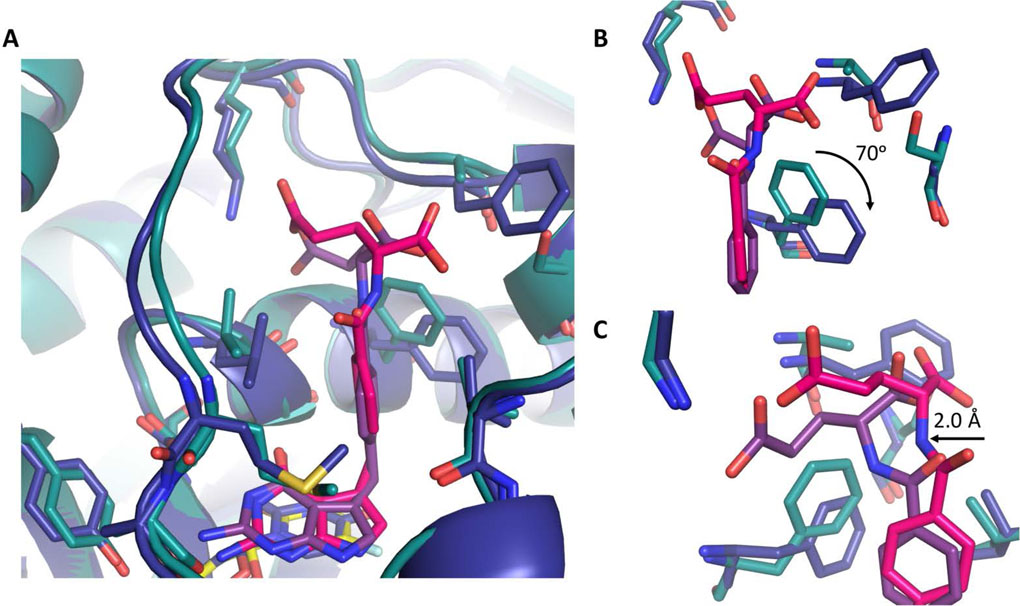
A further comparison of 2 in both ChTS and hTS active sites reveals hTS residue F225 is rotated ~70° clockwise, as compared to its corresponding ChTS residue F433 (Fig. 4A and 4B). All other residues within both active sites remain structurally unchanged. The rotation of F225 provides further access for the glutamate moiety of 2 to shift between ~ 2.0 Å into hTS active site to form a hydrogen bond with residue K77 (Fig. 4C). On the other hand, F433 of ChTS , remains stationary upon binding of either 1 or 2. The inflexibility of F433 appears to force the glutamate moiety to sample a strained conformation while forming a hydrogen bond with K284, giving rise to the observed conformational strain in the 2 amino linker region.
Figure:4.
Comparison of ChTS (green) bound to 2 (magenta) and FdUMP (yellow) with hTS (blue) bound to 2 (purple) and dUMP (yellow). A. Comparison of 2 bound in ChTS and hTS active site. B. Zoom in view hTS residue F225 position versus ChTS residue F433 upon binding of 2. C. Zoom in view of glutamate moiety of 2 interactions with both ChTS and hTS. Rotation of hTS residue F225 provides greater access allowing 2 to shift deeper into active site.
A combination of factors appears to account for the 26-fold stereospecificity of ChTS for the S-enantiomer, 1. ChTS destabilizes the binding of the R-enantiomer, 2, by inducing conformational strain in the compound, through a more rigid active site that does not accommodate the position of the glutamate moiety. Compound 1 would not be subject to similar strain due to the positioning of its glutamate moiety. Additionally, ChTS further stabilizes 1 by structurally ordering the glutamate moiety through the formation of a hydrogen bond with S290.[11]
Stereospecificity of hTS
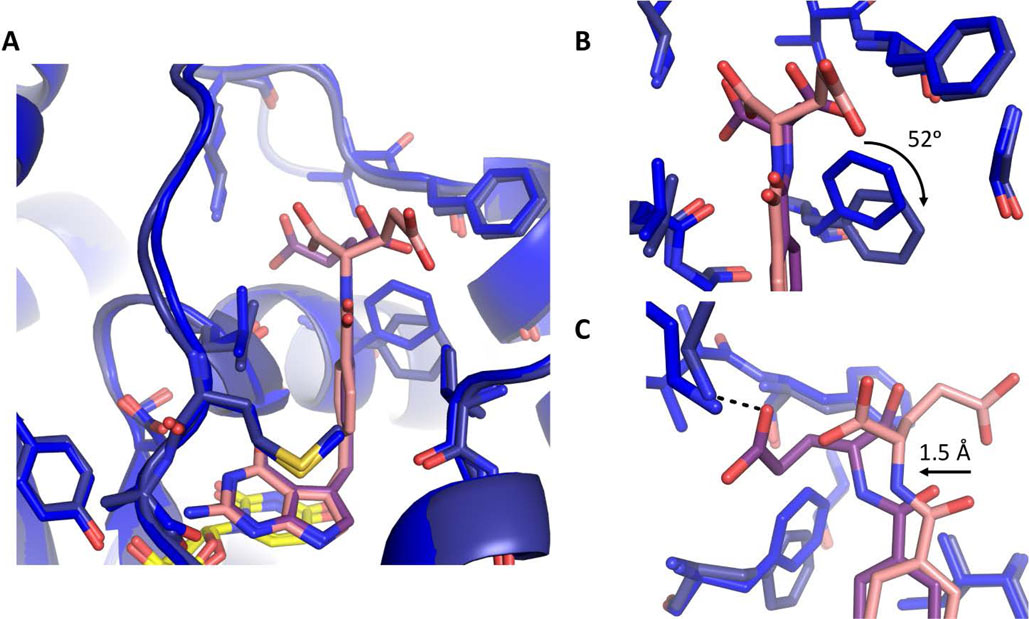
Similarly, we conducted a structural comparison of 1 and 2 bound in hTS active site to determine the mechanism for hTS stereospecificity (Fig. 5A). Both 1 and 2 form similar hydrophobic and hydrogen bonding interactions with hTS active site. However, 2 can form an additional hydrogen bond between its glutamate moiety γ-carboxylate and residue K77. The glutamate moiety of 1 does not form hydrogen bonding interactions with hTS active site, rather the γ-carboxylate group interacts with solvent. Further comparison of the two hTS structures, reveals that F225 undergoes a ~52° clockwise rotation when 2 is bound (Fig. 5B). This rotation provides greater space for the glutamate moiety of 2, allowing this region to shift ~ 1.5 Å towards F225, thus allowing the glutamate moiety to sample a less strained conformation while forming a hydrogen bond with K77 (Fig. 5C).
Figure:5.
Comparison of hTS bound to 1 (blue/pink) and hTS bound to 2 (navy/purple) with dUMP (yellow). A. Comparison of 1 and 2 bound in hTS active site. B. Zoom in view hTS residue F225 position upon binding 1 or 2. C. Zoom in view of glutamate moiety of 1 and 2 interactions with hTS. hTS stabilizes the glutamate moiety of 2 through a hydrogen bonding interaction with K77(black dotted line).
There are several factors which contribute to a 3-fold increase in stereospecificity of hTS for 2. hTS stabilizes the binding of 2 by structurally ordering glutamate moiety through a hydrogen bond with K77, whereas the glutamate moiety of 1 is less ordered as it interacts with solvent. Additionally, the rotation of F225 affords greater space for glutamate moiety of 2, relieving conformational strain while hydrogen bonded to K77.
Inhibitor selectivity
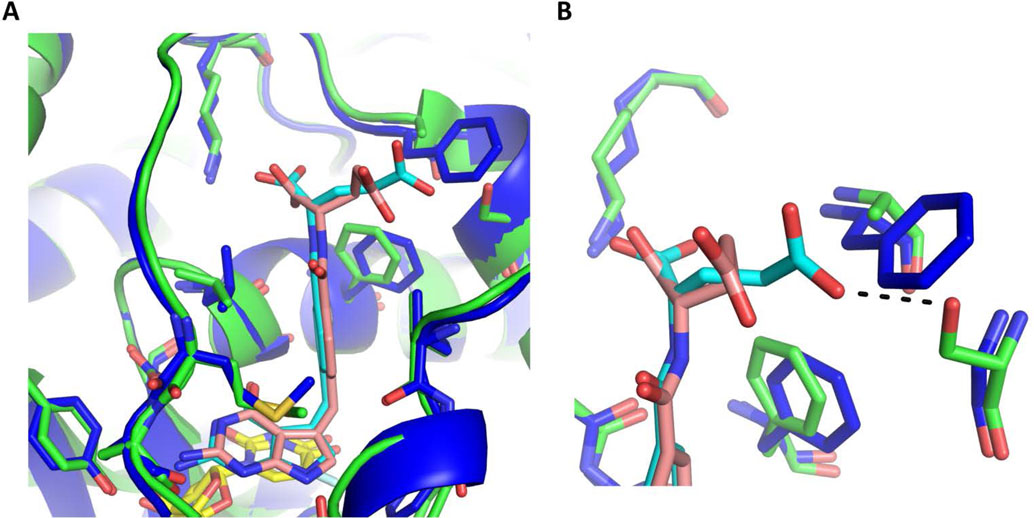
Previously we reported 1 inhibits ChTS with 5-fold greater selectivity than hTS.[11] A comparison of 1 bound to both ChTS and hTS active sites, reveals that 1 forms similar hydrophobic and hydrogen bonding interactions in both active sites, with one exception. Compound 1 forms an additional hydrogen bond between the glutamate γ-carboxylate and ChTS residue S290 (Fig. 6A and 6B). This hydrogen bond stabilizes 1 by structurally ordering its glutamate moiety within the ChTS active site. However, when 1 is bound in hTS the glutamate moiety freely samples different conformations as it interacts with solvent (Fig. 6B).
Figure:6.
Comparison of ChTS (green) bound to 1 (cyan) and FdUMP (yellow) with hTS (blue) bound to 1 (pink) and dUMP (yellow). A. Comparison of 1 bound in ChTS and hTS active site. B. Zoom in view of glutamate moiety of 1 interactions with ChTS and hTS. ChTS stabilizes the glutamate moiety of 1 through a hydrogen bonding interaction with ChTS residue S290 (black dotted line).
We reported here that 2 inhibits hTS with 16-fold greater selectivity. As described earlier, a comparison of 2 in both active sites, revealed hTS residue F225 rotates ~70° clockwise, as compared to its corresponding ChTS residue F433. The rotation of F225 provides greater access allowing the glutamate moiety to shift ~ 2.0 Å deeper into the active site, and sample a less strained conformation while forming a hydrogen bond with K77 (Fig. 4C). On the other hand, F433 does not rotate to afford greater access, forcing the glutamate moiety to sample a strained conformation while forming a hydrogen bond with K284, giving rise to conformational strain at 2 amino linker region. This would suggest the selectivity of 2 is determined by the degree of conformational strain induced by the active site.
In summary, the results presented here demonstrate a structural basis for ChTS and hTS stereospecificity as well as inhibitor selectivity. We showed that the enzyme stereospecificity is determined in part by the rigidity of the active site, while at the same time stabilizing the preferred inhibitor glutamate moiety through hydrogen bonding interactions. Additionally, inhibitor selectivity is determined by the ability of these enzymes to provide greater structural stability to the inhibitor, whether through the stabilization of the γ-carboxylate of the glutamate moiety in 1 by ChTS or through the relief of conformational strain in 2 by hTS. Taken together, this information is useful for further structure-guided design efforts to develop more selective ChTS inhibitors.
Supplementary Material
Acknowledgements
This work is supported by the National Institutes of Health under Award Numbers (AI083146) to K.S.A., and GM32136 to W.L.J. and Training Grant (5T32AI007404-23) to D.J.C.
References
- [1].Peng W, Ding F, Enantioselective recognition of an isomeric ligand by a biomolecule: mechanistic insights into static and dynamic enantiomeric behavior and structural flexibility, Mol Biosyst, 13 (2017) 2226–2234. [DOI] [PubMed] [Google Scholar]
- [2].Tsuchiya H, Mizogami M, Discrimination of Stereoisomers by Their Enantioselective Interactions with Chiral Cholesterol-Containing Membranes, Molecules, 23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sundaresan V, Abrol R, Biological chiral recognition: the substrate’s perspective, Chirality, 17 Suppl (2005) S30–39. [DOI] [PubMed] [Google Scholar]
- [4].Lopes P, Kataky R, Chiral interactions of the drug propranolol and alpha1-acid-glycoprotein at a micro liquid-liquid interface, Anal Chem, 84 (2012) 2299–2304. [DOI] [PubMed] [Google Scholar]
- [5].Tsuchiya H, Mizogami M, The membrane interaction of drugs as one of mechanisms for their enantioselective effects, Med Hypotheses, 79 (2012) 65–67. [DOI] [PubMed] [Google Scholar]
- [6].Lammerhofer M, Chiral recognition by enantioselective liquid chromatography: mechanisms and modern chiral stationary phases, J Chromatogr A, 1217 (2010) 814–856. [DOI] [PubMed] [Google Scholar]
- [7].Cassina M, Cagnoli GA, Zuccarello D, Di Gianantonio E, Clementi M, Human teratogens and genetic phenocopies. Understanding pathogenesis through human genes mutation, Eur J Med Genet, 60 (2017) 22–31. [DOI] [PubMed] [Google Scholar]
- [8].Zhang J, Zhou F, Niu F, Lu M, Wu X, Sun J, Wang G, Stereoselective regulations of P-glycoprotein by ginsenoside Rh2 epimers and the potential mechanisms from the view of pharmacokinetics, PLoS One, 7 (2012) e35768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wainer IW, Three-dimensional view of pharmacology, Am J Hosp Pharm, 49 (1992) S4–8. [PubMed] [Google Scholar]
- [10].Martucci WE, Vargo MA, Anderson KS, Explaining an unusually fast parasitic enzyme: folate tail-binding residues dictate substrate positioning and catalysis in Cryptosporidium hominis thymidylate synthase, Biochemistry, 47 (2008) 8902–8911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kumar VP, Cisneros JA, Frey KM, Castellanos-Gonzalez A, Wang Y, Gangjee A, White AC Jr., Jorgensen WL, Anderson KS, Structural studies provide clues for analog design of specific inhibitors of Cryptosporidium hominis thymidylate synthase-dihydrofolate reductase, Bioorg Med Chem Lett, 24 (2014) 4158–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Carreras CW, Santi DV, The catalytic mechanism and structure of thymidylate synthase, Annu Rev Biochem, 64 (1995) 721–762. [DOI] [PubMed] [Google Scholar]
- [13].Benkovic SJ, Hammes-Schiffer S, A perspective on enzyme catalysis, Science, 301 (2003) 1196–1202. [DOI] [PubMed] [Google Scholar]
- [14].Kumar VP, Frey KM, Wang Y, Jain HK, Gangjee A, Anderson KS, Substituted pyrrolo[2,3-d]pyrimidines as Cryptosporidium hominis thymidylate synthase inhibitors, Bioorg Med Chem Lett, 23 (2013) 5426–5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Atreya CE, Anderson KS, Kinetic characterization of bifunctional thymidylate synthase-dihydrofolate reductase (TS-DHFR) from Cryptosporidium hominis: a paradigm shift for ts activity and channeling behavior, J Biol Chem, 279 (2004) 18314–18322. [DOI] [PubMed] [Google Scholar]
- [16].Pedersen-Lane J, Maley GF, Chu E, Maley F, High-level expression of human thymidylate synthase, Protein Expr Purif, 10 (1997) 256–262. [DOI] [PubMed] [Google Scholar]
- [17].Almog R, Waddling CA, Maley F, Maley GF, Van Roey P, Crystal structure of a deletion mutant of human thymidylate synthase Delta (7–29) and its ternary complex with Tomudex and dUMP, Protein Sci, 10 (2001) 988–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sayre PH, Finer-Moore JS, Fritz TA, Biermann D, Gates SB, MacKellar WC, Patel VF, Stroud RM, Multi-targeted antifolates aimed at avoiding drug resistance form covalent closed inhibitory complexes with human and Escherichia coli thymidylate synthases, J Mol Biol, 313 (2001) 813–829. [DOI] [PubMed] [Google Scholar]
- [19].Kabsch W, Xds, Acta Crystallogr D Biol Crystallogr, 66 (2010) 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].McCoy AJ, Solving structures of protein complexes by molecular replacement with Phaser, Acta Crystallogr D Biol Crystallogr, 63 (2007) 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ, Phaser crystallographic software, J Appl Crystallogr, 40 (2007) 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].O’Neil RH, Lilien RH, Donald BR, Stroud RM, Anderson AC, Phylogenetic classification of protozoa based on the structure of the linker domain in the bifunctional enzyme, dihydrofolate reductase-thymidylate synthase, J Biol Chem, 278 (2003) 52980–52987. [DOI] [PubMed] [Google Scholar]
- [23].Emsley P, Cowtan K, Coot: model-building tools for molecular graphics, Acta Crystallogr D Biol Crystallogr, 60 (2004) 2126–2132. [DOI] [PubMed] [Google Scholar]
- [24].Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH, PHENIX: a comprehensive Python-based system for macromolecular structure solution, Acta Crystallogr D Biol Crystallogr, 66 (2010) 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Adams PD, Read RJ, Zwart PH, Hung LW, Iterative-build OMIT maps: map improvement by iterative model building and refinement without model bias, Acta Crystallogr D Biol Crystallogr, 64 (2008) 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Aso K, Imai Y, Yukishige K, Ootsu K, Akimoto H, Pyrrolo[2,3-d]pyrimidine thymidylate synthase inhibitors: design and synthesis of one-carbon bridge derivatives, Chem Pharm Bull(Tokyo), 49 (2001) 1280–1287. [DOI] [PubMed] [Google Scholar]
- [27].Wang Y, Cherian C, Orr S, Mitchell-Ryan S, Hou Z, Raghavan S, Matherly LH, Gangjee A, Tumor-targeting with novel non-benzoyl 6-substituted straight chain pyrrolo[2,3-d]pyrimidine antifolates via cellular uptake by folate receptor alpha and inhibition of de novo purine nucleotide biosynthesis, J Med Chem, 56 (2013) 8684–8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.