Abstract
In the past half century, our version on cancer, from tumor initiation, growth, to metastasis, is dominated by genetic mutation. The importance of metabolism and epigenetics was not recognized until most recently. Extensive cell proliferation is one of the hallmarks of cancers. To support the energetic and anabolic demands of enhanced proliferation, tumors reprogram the pathways of nutrient procurement and metabolism. In this context, a new link between metabolic alterations and cancer progression has been unraveled over the last decade by the studies conducted in the area of cancer cell metabolism. Cancer cells are known to alter their metabolic profile during the course of tumorigenesis and metastasis thereby exhibiting a tightly regulated program of metabolic plasticity. Noteworthy, certain metabolic alteration are known to occur at the epigenetic level, thus making epigenetics and metabolism highly interwoven in a reciprocal manner. Metabolites that are generated during metabolic pathways, such as in glycolytic cycle and oxidative phosphorylation, serve as cofactors or substrates for the enzymatic reactions that catalyze the epigenetic modifications and transcriptional regulation. Several studies also indicate that the epigenome is sensitive to cellular metabolism. Since many of the metabolic alterations and consequently aberrated epigenetic regulation are common to a wide range of cancer types, they serve as promising targets for anti-cancer therapies. Here we discuss the latest findings in cancer cell metabolism, elucidating the major anabolic, catabolic and energetic demands required for sustaining cancer growth, and the influence of altered metabolism on epigenetics and vice versa.
A comprehensive research pertaining to metabolomic profiling and epigenome interactors/mediators in malignant neoplasias is imperative in deciphering the potential targets that can be exploited for the development of robust anti-cancer therapies.
Keywords: cancer cell metabolism, warburg effect, DNA and histone methylation, epigenetics
Introduction
The word metabolism is derived from the Greek word “mεtabolή (metabole)” that means “to change”. It defines a series of complex reactions that basically generate energy for maintaining life and its processes. It represents a fine balance between anabolism (i.e. building up) and catabolism (i.e. breakdown), resulting in the generation of energy rich biomolecule adenosine triphosphate (ATP). Metabolism is broadly classified as either oxidative or non-oxidative. Oxidative process is carried out in mitochondria whereas non-oxidative processes are completed in the cytoplasm. Collectively, metabolism serves three essential functions within the cells. First is ATP generation which is an absolute requirement for the energy consuming activities, second is the production of glycolytic intermediates, that are important for the anabolic reactions and lastly is the generation of metabolites that serves as a cofactor or substrates for several important enzymatic reactions that are implicated in epigenetic modification and gene regulation.
One of the most common phenotype that all cancer cells exhibit is the uncontrolled cell proliferation. To meet their rapid growth requirement, cancer cells undergo metabolic adaptations to sustain their survival under several stress conditions such as hypoxia and nutrient starvation. These metabolic adaptations renders altered metabolic activities/pathways in cancers cells relative to the normal cells. Such reprogrammed metabolism is considered as a hallmark of cancer, as several metabolic alterations are common across many other cancer cell types [1, 2]. Like normal cells, cancer cells too need to generate ATP, rely on metabolic intermediates or precursors for biosynthesis and most importantly to cope up with the oxidizing effects in a way to minimize the influence of reactive oxygen species (ROS). Accordingly, these altered metabolic and bioenergetic mechanisms, most notably the elevated biosynthesis and redox balance, are pivotal for the survival of cancer cells [3]. One can appreciate the fact that in spite of cancers being so genetically and histologically heterogenous and diverse, there are some common mechanisms that ultimately support the core functions of metabolism and redox balance in tumors [4]. Cancer cells are in constant interaction with their microenvironment [5] and metabolism in these cells are affected by both external and internal stimuli such as nutrient or oxygen supply and oncogenic signal transmissions, respectively. In recent years, metabolism in cancers have gained wide interest owing to the fact that they are closely associated with epigenetic regulation as it supplies metabolic intermediates which serve as cofactors for several important epigenetic enzymes. For instance, most of the chromatin modifying enzymes employ intermediary metabolites as cofactors and substrates for their respective reactions and it has been indicated that the epigenome is quite sensitive to the metabolic state of the cells [6–8]
Undoubtedly therefore, an altered metabolism in cancer cells is essential for distinct epigenetic programs that are likely to contribute to tumorigenesis, malignancy and the generation of cancer stem cells. In the current review, we provide a concise discussion of the major metabolic alterations that are exhibited in cancer cells to sustain their growth and the epigenetic control of cancer metabolism. Meanwhile, we discuss the effect of altered metabolism on the epigenetic landscape of cancer cells. Lastly, we briefly describe the potential therapeutic strategies that can exploit the metabolism and epigenetics in neoplasias.
Altered metabolism in cancers
Aerobic Glycolysis: the Warburg effect
Glycolysis is a metabolic pathway that involves the conversion of glucose into pyruvate. Adenosine triphosphate (ATP) and reduced nicotinamide adenine dinucleotide (NADH) are generated from the free energy released by this process. In 1920, Otto Heinrich Warburg observed that glycolysis was enhanced in cancer cells even in the presence of abundant oxygen. It was known that glycolysis normally increases under anaerobic conditions, however its increase in cancer cells in spite of surplus oxygen was quite a new phenomenon and was termed as aerobic glycolysis or the “Warburg effect” [9–11]. Thus, high rate of glucose catabolism into lactate represents the most ubiquitous metabolic phenotype observed across cancer cells, simultaneously resulting in the accumulation of lactate by-products in the tumor microenvironment [12]. In cancer cells, the Warburg effect is not much of an energy generating pathway, rather it serves as a mode for generating glycolytic & biosynthetic intermediates that acts as precursors to many other anabolic processes for the de novo synthesis of carbohydrates, nucleic acids, protein and fats thereby facilitating the survival and growth of cancer cells. These anabolic pathways majorly constitute the pentose phosphate pathway in cancer cells, hexamine biosynthesis pathway and serine-glycine pathway [13, 14]. Interestingly, cancer cells behave as metabolic parasites, owing to the fact that they procure nutrients from the host cells via activating several catabolic processes, among which aerobic glycolysis is most common [15, 16]. Sustained aerobic glycolysis has also been associated with the activation of oncogenes and loss of tumor suppressor genes, respectively, in certain cancers [17, 18].
Given the fact that cancer cells use the catabolism of glucose by glycolysis as its major energy generating mechanism, glycolysis serves as an early attractive target for cancer therapy. Perhaps, many tumors display a significant increase in glucose uptake when compared to their normal adjacent tissue counterparts [19]. In this context, monocarboxylate transporters (MCTs) facilitate the transport of lactate out of the cells and are essential for sustaining the elevated rates of glucose catabolism in cancer cells. Hence MCTs are also considered as candidate targets for cancer therapy [20].
The Pentose Phosphate Pathway
A metabolic pathway parallel to glycolysis is the pentose phosphate pathway, also referred as the phosphogluconate pathway and the hexose monophosphate shunt. It’s an anabolic pathway that generates NADPH, 5-carbon sugars i.e. the pentoses and ribose 5-phosphate. While ribose sugars serve as the precursor for nucleotide biosynthesis, NADPH serves as a reducing agent for the macromolecules synthesis. This pathway is essential in assisting the cancer cells to meet their anabolic demands and survive the oxidative and nutritional stress.
The PI3K Pathway
Constitutive activation of PI3K pathway has been frequently displayed in cancer cells. Activation of PI3K further results in the activation of AKT, which is a pro-survival kinase. AKT has been known as the major regulator of glucose uptake and enhances glucose metabolism via glycolysis and PPP [3, 21]. More-over, it drives the Warburg effect in cancer cells by stimulating the expression of glucose transporters and direct phosphorylation of Hexokinase 2 (HK2) and Phosphofructokinase (PFK) enzymes, hence subjecting glucose to the glycolic pathway [22].
Reactive Oxygen Species: The Redox Balance
Cellular metabolism generates several toxic by-products as well. Among them, reactive oxygen species (ROS) are the major ones that comprise H2O2, superoxide O2 and hydroxyl radical OH [23]. The main metabolic source of ROS is from mitochondrial oxidative phosphorylation. ROS have a pronounced damaging effect on proteins, lipids and nucleotides at elevated levels [24]. Cancer cells have adopted a unique mechanism for the detoxification of ROS, the glutathione (GSH) oxidation–reduction coupled to NADPH reduction–oxidation. GSH and TRX, the two proteins that act in neutralizing the ROS-mediated cellular damage and toxicity, are in fact synthesized through mechanism where NADPH serves as the main precursors [25]. Noteworthy, elevated levels of NADPH have been a common feature in cancer cells that helps them to combat excessive ROS as well as to assist in their survival by supporting anabolic pathways. Interestingly an emerging concept of redox balance in cancer cells has been recently proposed, which explains the loop between tumor initiation, metabolic activity and cancer maintenance. With the onset of tumorigenesis which is accompanied by rapid cell proliferation, the metabolic activity of cancer cells is dramatically increased. With an elevated metabolic rate, an excessive production of ROS happens, which further activates signal transduction pathways thereby supporting the growth, survival and metabolism of cancer cells [26].
Glutamine, c-Myc and p53
Glutamine is the most rapidly utilized amino acid in cancer cells [27, 28] and in vitro assays show that glutamine is required in high levels for the survival of cancer cells in culture medium [29]. Glutamine is able to provide acetyl-CoA as a precursor of the biosynthesis of macromolecules that support the growth of tumor cells, especially under those conditions where the oxidation of pyruvate to acetyl-CoA is undermined either by the deterioration of electron transport chain or due to hypoxic environment [30, 31]. Cells harboring activating RAS mutations have been found to have elevated glutamine uptake thereby leading to GSH production to cope up with the oxidative stress [32]. Myc also regulates glutamine utilization by cancer cells. In this context, it facilitates the transportation of glutamine as well as down-regulates the expression of microRNAs such as miR23a/b that are involved in the downregulation of the expression of glutaminase [33, 34]. Hence c-Myc being itself an oncogene, upregulates the expression of glucose and glutamine transporters [33, 35]. In fact, MYC was the first gene associated with glycolysis regulation in aerobic cells via the direct stimulation of LDHA [36, 37]. Other potent genes such as tumor suppressor p53 is known to inhibit glycolysis and shunts the glucose into the pentose phosphate pathway. This is mediated by TIGAR, a transcriptional target gene of p53 [38]. Therefore, glycolysis is favored by the loss of p53; whereas p53 facilitates efficient mitochondrial oxidative phosphorylation [39, 40].
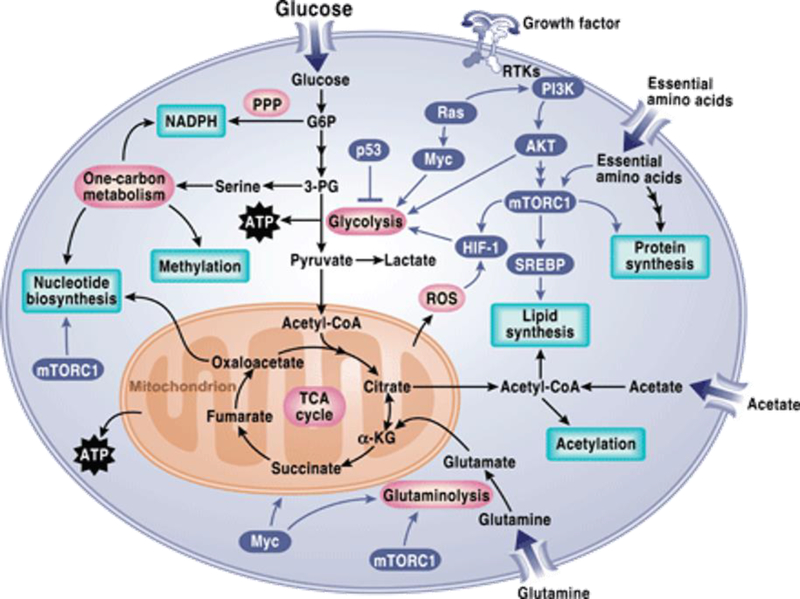
While tumor cells are in constant interaction with their micro-environment, the conditions within the tumor micro-environment significantly influence the cancer cell metabolism. Due to extensive cell proliferation and rapid anabolic demands to sustain their growth and survival, cancer cells often encounter stress conditions such as nutrient starvation and poor oxygen supply. To overcome these limitations, cancer cells have adopted several nutrient scavenging tactics. Restricted supply of nutrients occurs primarily due to the poor perfusion stemming from poor vascularization and elevated interstitial pressure. Pancreatic ductal adenocarcinoma (PDAC), is one such example [41, 42]. In order to maintain their viability in limited nutrient and oxygen supply conditions, cancer cells have evolved mechanisms that sustain their ATP/ADP ratio. One such robust mechanism that prevails under the conditions of nutrient starvation is “self-eating” or “autophagy” [43]. Autophagy process generates several breakdown products that serve as critical raw materials for the building blocks of cellular macromolecules. This biosynthesis helps sustain energy production under the conditions of stress and nutrient deprivation [44] [45]. In NSCLC cells with oncogenic Kras or B Raf fueling the tumors, autophagy has been found to sustain mitochondrial function by providing intracellular supply of glutamine [44, 46]. Signaling cascades that regulates cancer metabolism has been summarized in figure 1.
Figure 1. Signaling pathways regulating metabolism in cancer cells.
Network of several major anabolic and catabolic pathways results in energy production as well as generates protein, lipids and nucleotide biosynthesis through glycolysis, oxidative phosphorylation and pentose phosphate pathway. These metabolic pathways exert control over signaling via regulating reactive oxygen species (ROS), acetylation, and methylation. [PPP, pentose phosphate pathway; G6P, glucose-6-phosphate; 3-PG, 3-phosphoglycerate; ATP, adenosine 5´-triphosphate; mTORC1, mTOR complex 1; α-KG, α-ketoglutarate; RTK, receptor tyrosine kinase.]
Image is adapted from [91].
Crosstalk between metabolism and epigenetics in cancer
Epigenetics and Cancer metabolism
Cancer metabolism is known to affect the epigenetic landscape of the cells by at least three different cellular mechanism. First and foremost is the reprogramming of the metabolic pathways. Such a reprogramming results in the alteration of the metabolite levels. Some of the metabolites are important co-factors or substrates of the critical enzymes for epigenetic modifications. Second mechanism pertains to the nuclear production of metabolites by the metabolic enzymes that are translocated to the nucleus. Lastly, is the production of oncometabolites that regulate the activities of several potent epigenetic enzymes. An accumulation of oncometabolites in cancer cells is a very important factor that drives tumor progression.
Epigenetic regulation of gene expression is known to occur at the DNA, histone and RNA level. In this context, DNA methylation, histone methylation, acetylation, ubiquitination, phosphorylation, and microRNA-dependent gene silencing have been well-characterized [47]. Aberrant DNA methylation has been associated with pathological gene expressions in a variety of human cancers [48]. Global DNA hypomethylation and site-specific CpG promoter hypermethylation are the most common epigenetic alterations observed in cancers.
Metabolites and cofactors play an essential role in mediating the activities of several epigenetic associated enzymes. Examples of such metabolites generated via metabolic process includes, S-adenosyl methionine (SAM) produced from one carbon cycle, a-ketoglutarate and flavin adenine dinucleotide (FAD) from the TCA cycle, acetyl-CoA generated from glycolysis and NAD+ generated from the conjunction of glycolysis and oxidative phosphorylation.
Metabolic control of DNA/histone methylation
S-adenosylmethionine (SAM), which is synthesized from methionine and ATP serves as the universal methyl donor in mammals. Methylation of DNA is mediated by DNA methyltransferases (DNMT) that use SAM as methyl donor. Methylation of histones too require SAM where the methylation occurs at the lysine or arginine residue and the reaction is catalyzed by histone methyltransferases (HMT) [49]. Therefore, the abundance or availability of SAM can directly affect the methylation status of DNA and histone proteins. Methylation reactions involves the transfer of a methyl group from SAM that results in the generation of Sadenosylhomocysteine (SAH) which is recycled back in the methionine cycle. Interestingly, SAH is inhibitory to methyltransferases. Thus, the cellular SAM/SAH ratio is an important indicator of the “methylation potential” of a cell and serves as a potent determinant in the regulation of chromatin methylation [50].
On the contrary, reversal of DNA and histone methylation events are brought by the demethylase activities of DNA and histone demethylases. There are two groups of histone demethylases: lysine specific demethylase family, LSD 1 and LSD2, and JmjC-containing family proteins, both of which are flavin adenine dinucleotide (FAD)-dependent oxidase enzymes. The JmjC family demethylases are ferrous ion-dependnet oxygenases using a-ketoglutarate (aKG) as a co-factor for the enzymatic activation [51, 52]. Notably, the catalytic co-factors FAD and αKG are generated as TCA cycle-intermediary metabolites. Similarly, the activity of DNA demethylase TET family proteins also requires FAD and αKG. In contrast, other TCA cycle metabolites, including succinate and fumarate, are serving as antagonists for the JmjC family demethylases [53]. Hence the TCA metabolic pathway in mitochondria can directly contribute to the overall epigenetic regulation of the genome.
Acetylation of histones is yet another important epigenetic mechanism that contributes to several chromatin dependent process such as DNA replication, damage & repair, transcriptional activation, cell cycle and gene regulation. Two class of enzymes are involved in the dynamic regulation of histone acetylation. These are the histone acetyltransferases (HAT) and histone deacetylases (HDAC) [54]. One of the key metabolites essential for energy production through oxidative phosphorylation and aids anabolic process is acetyl-CoA [55, 56]. Interestingly, acetyl-CoA acts as a substrate for HATs, where HATs transfer the acetyl moiety of acetyl-CoA to the lysine residues of histones. This reaction is majorly associated with transcriptional activation [57, 58] and is critical for rapidly proliferating cells, such as cancer cells. Therefore, variations in the level of cellular acetyl-CoA tightly influences the HATs-mediated histone acetylation. Studies in yeast [58, 59] and mammalian cells [60] indicated that glycolytic activities dynamically regulate the cellular acetyl-CoA levels which correspondingly modulates the HATs mediated acetylation of histone proteins.
Similarly, reversal of histone acetylation is carried by a group of enzymes known as HDACs. They remove the acetyl group from the lysine residues on histone. This results in the tight wrapping of DNA by histones, hence functioning opposite to that of HATs. Acetylation levels within the cell is attributed towards the fine balance between the activities of HAT and HDAC. Different cellular metabolites antagonize the activities of HDACs. Among them butyrate is a potent antagonist that inhibits the activities of HDACs I, II and IV [61]. Butyrate is produced by colonial bacteria that basically ferments the dietary fibers. Cancer metabolism such as increased production of lactic acid by glycolysis contributes to the generation of local acidic pH within the tumor microenvironment. This condition has been known to promote histone deacetylation [62] and interestingly favor an aggressive, pro-metastatic phenotype of the cancer cells [63]. Also, low histone acetylation levels are correlated with poorer prognosis of cancer patients [64]. Another metabolite NAD+ serves as a catalytic cofactor for HDAC III (Sirtuins)-mediated histone deacetylation reactions. The summarized concept has been illustrated in figure 2.
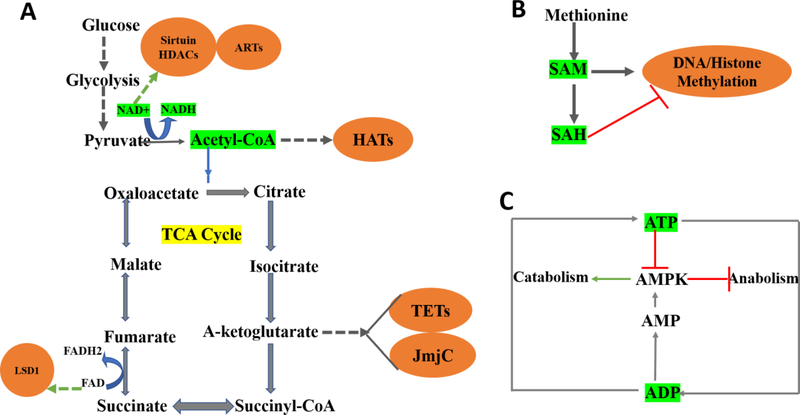
Figure 2. Metabolites serves as cellular rheostats and regulate the epigenetic processes.
Variations in the concentration of several metabolites that act as either substrate or cofactors for key epigenetic enzymes, influence chromatin modification; also, by a feedback mechanism that dynamically regulates the entire process. (A) The tricarboxylic acid (TCA) generates metabolites that link energy pathways with epigenetic chromatin modifications, highlighted in orange. Glycolysis generates acetyl-CoA that feeds itself into the TCA cycle further and serves as a substrate for histone acetyltransferases (HATs). Pyruvate to acetyl-CoA conversion generates nicotinamide adenine dinucleotide (NAD+), which is needed by the sirtuin histone deacetylases (HDACs; histone deacetylation) and ADP-ribosyltransferases (ARTs). α-Ketoglutarate and Flavin adenine dinucleotide (FAD) serve as cofactors for DNA ten-eleven translocations (TETs) and histone demethylases (Jumonji C domain containing JmjC), LSD1. (B) Product metabolite SAdenosyl methionine (SAM) generated via one carbon cycle acts as a methyl donor to the histonemodifying enzymes, histone methyltransferases (HMT) & DNA methyltransferases (DNMT) thereby facilitating histone and DNA methylation. On the contrary, S-adenosylhomocysteine (SAH) negatively regulates this process, indicating that SAM/SAH ratio is essential in regulating DNA and histone methylation. (C) Cellular ATP/ADP ratio has a physiological role where conversion of adenosine triphosphate (ATP) to adenosine diphosphate (ADP) aids in anabolic process. Catabolism, however, relies on ADP to ATP conversion where activation of AMPactivated protein kinase (AMPK) is critical in regulating this balance. Therefore, within the cellular environment, NAD+ /NADH, Acetyl-CoA/Co-A, SAM/SAH, ATP/ADP ratio act as sensory signals (highlighted in green) governing the various epigenetic process.
Genetic and epigenetic alteration of metabolic enzymes in cancer
Several studies indicated that mutations in the metabolic enzymes subjects the cells to tumorigenesis [7, 65]. Such mutations facilitate the generation of oncometabolites that ultimately influence the epigenetic regulation of DNA and histone. Mutations were frequently observed on the NADP+-dependent isocitrate dehydrogenase (cytosolic IDH1 and mitochondrial IDH2), succinate dehydrogenase (SDH) and fumarate hydratase (FH). Inactivating mutations in these metabolic enzymes results in the stacking up of 2-hydroxyglutarate, succinate, and fumarate, respectively [66]. These mutations are of oncogenic nature where both succinate and fumarate inhibit the enzymatic activities of the TET and Jmj-C family of proteins [53]. Cancer cells carrying IDH1/IDH2 mutations display hypermethylation of DNA and histones. Gliomas and blood cancers frequently exhibit oncogenic mutations in IDH1 and IDH2 [67, 68]. Nicotinamide N-methyltransferase (NNMT) is yet another metabolic enzyme catalyzing the transfer of methyl moiety from SAM to nicotinamide thereby catabolizing SAM to 1-Methyl Nicotinamide (1MNA), is aberrantly expressed in several cancers and is linked with enhanced migratory and invasive behaviors. Cancer cells overexpressing NNMT have been shown to exhibit alterations in their SAM and histone methylation levels, along with the procurement of more aggressive phenotype [50]. The reaction catalyzed by NNMT hampers the SAM-mediated DNA and histone methylation process. The entire process has been summarized in figure 3.
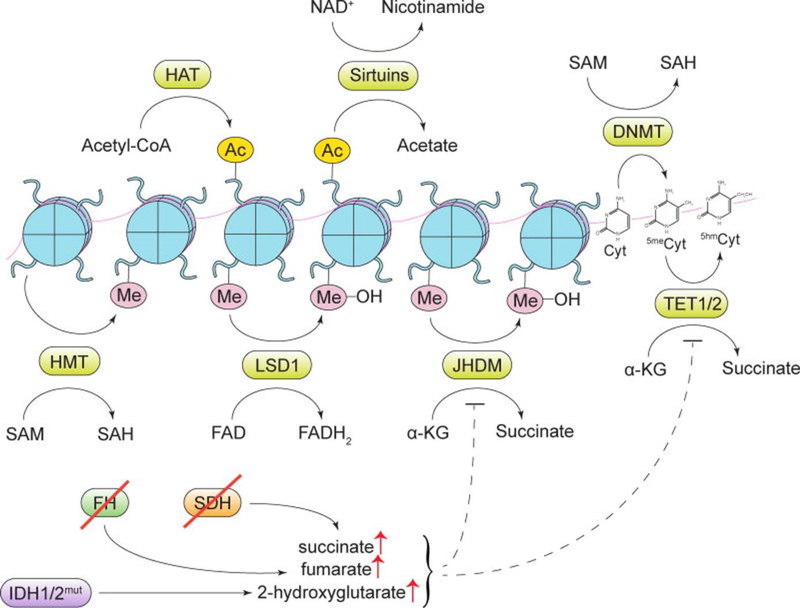
Figure 3. Cellular metabolites contribute to gene regulation and their fluctuating levels affect the epigenetic process.
Addition or removal of epigenetic marks by several key epigenetic enzymes is dependent on metabolites that act as substrates or cofactors of these enzymes. [HAT, histone acetyltransferase enzymes; Ac, an acetyl mark; SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine; DNMT, DNA methyltransferase enzymes; HMT, histone methyltransferase enzymes; Me, a methyl mark; LSD1, lysine-specific histone demethylase 1; JHDM, Jumonji domain-containing histone demethylase enzymes; Cyt, cytosine; 5meCyt, 5-methylcytosine; 5hmCyt, 5-hydroxymethylcytosine; TET1/2, ten-eleven translocation methylcytosine dioxygenase 1/2; α-KG, α-ketoglutarate; SDH, succinate dehydrogenase; FH, fumarate hydratase; IDH1/2, isocitrate dehydrogenase 1/2]. Image is adapted from[92]
Apart from genetic mutations, epigenetic events also contribute to the alteration of metabolic enzymes in cancer. Examples of such metabolic enzymes include Hexokinase isoform 2 (HK2) and Fructose-1,6-bisphosphatase (FBP1). Epigenetic events such as hypomethylation of the promoter is responsible for the upregulation of HK2 in glioblastoma and hepatic carcinoma [69, 70]. Promoter methylation results in the silencing of FBP1 in the cancers of stomach, liver and colon tissues [71]. While increased HK2 levels facilitates enhanced glycolytic flux, gluconeogenesis is regulated by FBP1 in cancer cells.
Metabolism also affects the genetic process that are essential in the regulation of cell growth, survival, differentiation and overall homeostasis. Metabolic enzymes have roles in DNA based processes that includes gene expression, DNA replication and DNA damage response. Enzymes involved in acetyl-CoA synthesis, such as ACSS2, PDC, ACLY; other enzymes like pyruvate kinase, 6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4 (PFKFB4), α-ketoglutarate dehydrogenase complex and fumarase etc. are few examples [72]. Metabolic status of cancer cells too have an influence on the genetic events related to DNA folding and DNA-damage-repair pathways, eventually affecting the genomic stability of the cells. In view of this, several metabolic factors are known to affect the DNA damage and repair mechanisms. For example, methyl- and acetyl-group donors generated via different metabolic pathways have an impact on DNA folding and double strand break repair. Perhaps, limitations in the amount of acetyl-group donors has been known to distort the usual DNA organization, further affecting the DNA folding that is critical for the DNA double stand break repair process [73]. Other process linked to DNA repair and DNA replication are also affected by the availability of the metabolic products such as glutamine, aspartate and nucleotide intermediates in the cell’s microenvironment. Increased glycolysis exhibited by cancer cells has been found to confer radio-resistance, where such increase in the glycolytic pathway has augmented the repair of the radiation induced DNA damage pathways particularly the non-homologous end joining (NHEJ) and homologous recombination (HR) pathways of DNA double strand break repair [74]. One of the key side pathways of glycolysis, the pentose phosphate pathway (PPP) generates metabolic intermediates which are essential component for the nucleotide and protein synthesis, thereby affecting the replication of DNA in the proliferating cells. Metabolic enzyme such as phosphoinositide 3-kinase, is an important one where its activity is essentially required for the synthesis of nucleotides [75]. Noteworthy, apart from metabolism affecting the genetic landscape of cancer cells, defects in the metabolic pathways also results in a plethora of pathological conditions collectively called as single gene disorders or inborn errors of metabolism. This can be attributed to many faulty events falling within the realms of metabolism such as enzyme deficiency, accumulation of toxic metabolic intermediates, deregulated metabolic signaling pathways etc. Metabolic ataxias such as Hartnup disease, hyperammonemia, maple syrup urine disease, mitochondrial disorders, pediatric neuropathy, metabolic encephalopathy, Alpers disease etc are few examples depicting the crosstalk between metabolism and genetics. Metabolic enzyme deficiency disorders itself accounts for a number of diseases affecting the genes and its regulation and expression [76]. Deficiencies of important metabolic enzymes and components includes but not limited to glucose-6-phosphatase (G6Pase), fructose-1,6-bisphosphatase (FBPase), phosphoenolpyruvate carboxykinase (PEPCK), pyruvate dehydrogenase complex (PDHC), succinate dehydrogenase (SDH), fumarate hydratase (FH), glucose-6-phosphate dehydrogenase, ribose-5-phosphate isomerase (RPI), and transaldolase (TALDO).
Targeting metabolism for cancer therapy
Several investigations and studies over the past decade in the field of cancer metabolism has undoubtedly reflected the dire need of metabolic supply for cancer cells in order to sustain their growth and progression. This becomes the rationale for new drug discovery and cancer therapeutic approach where selectively starving cancer cells of their unquenchable metabolic demands can potentially inhibit the proliferation and growth of cancer cells. However, the choice of drugs targeting the cancer cells must be carefully determined such that they should be relatively nontoxic to the normal cells. Perhaps a better and comprehensive understanding of the metabolic pathways implicated in cancer cell metabolism is imperative towards the designing of effective drugs that are detrimental to cancer cells while rendering the normal cells relatively safe.
Recent advancements in the area of cancer drug discovery have spotlighted on the inhibitors of several potent metabolic pathways that are harnessed by cancer cells [77, 78]. Inhibiting the activity of some key metabolic enzymes holds great potential. However, because of their physiological role in the normal cells, targeting such enzymes can be conventionally toxic [79]. Genetic ablation of the metabolic enzyme LDH-A, that catalyzes the conversion of pyruvate to lactate; has been shown to delay the progression of myeloid leukemia [80]. Interestingly, LDH-A is the first metabolic target of MYC oncogene and pharmacological inhibition of LDH-A demonstrated a dwindling effect of MYC driven tumors in the xenograft models [81]. Another important mechanism of eliciting nutrient deprivation in cancers, is the blocking of those transport channels that aids the transportation of nutrients within the cells. Since the rate limiting step in the glycolysis process is the uptake of glucose by cancer cells; therefore, targeting glucose transporters (GLUT) is a well-justified approach towards cancer drug discovery of small molecules or inhibitors performing the task. Cytochalasin B and distinct tyrosine kinase inhibitors are one such examples of GLUT inhibiters, where they hold great promise in the treatment of neoplasias [82]. Hence targeting metabolic pathways implicated in cancer cell survival and progression, such as glycolysis, mitochondrial metabolism, glutamate metabolism, and autophagy, offers effective strategies for novel drug discovery scheme targeting the respective entities. For instance, BPTES [bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl) ethyl sulfide], an inhibitor of glutaminase activity is being tested for anticancer effects [83]. Mitochondrial metabolism is crucial for the survival of healthy as well as cancer cells. Interestingly, with the unraveling of anticancer effects of metformin, an anti-diabetic drug has further focused on mitochondria-mediated metabolic pathways to be exploited as key target pathways for cancer therapy [84]. In this context, inhibiting mitochondrial complex I by the drug biguanide phenformin has displayed anticancer characteristics also [85].
Translating metabolic alterations into therapeutic opportunities has potential outcomes dealing with cancer therapy related research and development processes. Though it offers great platform where oncogenic metabolic alterations can be selectively targeted; however one cannot fully ignore the challenges accompanying such research paradigms [86]. Tumor heterogeneity and existence of a dynamic range of metabolic profile of cancer cells within the same cancer type are major roadblocks that allows a cancer cell to escape the deleterious effect of metabolic inhibitors. Safety of metabolic inhibitors in patients is also of great concern, e.g. drugs targeting glycolytic pathways can counter affect the otherwise healthy brain cells that are high in glycolytic activity [87]. Perhaps, while selecting metabolic inhibitors, having information regarding patient’s metabolic data is also indispensable. Regarding therapeutics targeting epigenetic processes, success has been so far achieved in hematological malignancies. However challenges are still prevalent in targeting solid tumors. Also replication status of the cells remain one of the important prerequisite for the DNMT inhibitors, that essentially require tumor cells to be in replicating mode for successful incorporation into DNA. Hence non-replicating populations tend to be resistant to such agents. Stem cells within the cancer, are one such example of cell population that are difficult to target. Complexity and heterogeneity of the cancer cell epigenome, dynamics repertoire of the chemotherapy resistance cancer cells, pharmacokinetic half-life of the drugs, and biological differences in gene regulation between solid and hematological tumors are few of the challenges that needs to be addressed when dealing with epigenetic modifiers as cancer therapies. Comprehensive analysis of the relationship between epigenetic marks in germline DNA and tumor DNA can offer many modalities that can be exploited for developing better therapies targeting the aberrant epigenetic events [88].
Perspectives
It is now evident, supported by a large body of research over the past decade that metabolic reprogramming does contributes to the epigenetic alteration in cancers. Oncogenic insults accompanied with tumor microenvironment exerts selective pressure that renders the cancer cells to adopt altered metabolism, which in turn supports the energy and metabolic demands of these cells thereby facilitating tumor growth and maintenance. Such a metabolic switch not only supplies the cancer cells with enhanced nutrient synthesis precursors, but also maintains an adequately balanced redox potential that is the core of cancer cell survival and sustainability.
Therefore, exploiting cancer metabolism has been the focus of cancer therapy where antifolates served as the first among such targeted anti-cancer therapies. Though the field has advanced with new developments, some of the aspects pertaining to metabolic reprogramming in cancers remains poorly understood. For example, it is still elusive to fully decipher the critical downstream factors that mediate the neoplastic functions of some oncometabolites in cancer cells. Heterogeneity of tumors is yet another challenge, where inconsistent metabolic phenotypes are observed, that too when evaluated across single solid tumors in human subjects [89]. Experimental conditions, composition of growth media utilized for culturing the cancer cells etc. plays a critical role in modulating the dynamics of metabolic alterations. Therefore, one has to be extremely cautious in interpreting the metabolic influx data generated via cell culture systems and juxtaposing it to clinical data. Perhaps, differences exist between the artificially in vitro cultured cell models and in vivo observations made regarding tumor cell metabolic susceptibilities [90].
Individual cellular compartments within a cell needs to be fully profiled when studying metabolism in cancer and its related epigenetic effects in tumorigenesis. A plethora of metabolic enzymes have been discovered in cell’s nucleus, however their individual contribution in regulating the epigenetic landscape still needs to be well defined. Since multiple cellular compartments such as nucleus, cytoplasm and mitochondria are involved in harboring several metabolic events in cancer, it is essential to learn how the metabolites transport/oscillate from one compartment to another. Additionally, more robust experimental approach needs to be adopted in overcoming the technical challenges faced while measuring the metabolites in distinct cellular domain. This will facilitate in determining which epigenetic regulatory metabolic enzymes were catalytically active in which of the cellular compartments and what are the signals that trigger their nuclear localization. Establishment of better computational or analytical strategies and comprehensive metabolomic profiling of not only cancer cells, but also other cells of the tumor microenvironment such as lymphocytes, cancer associated fibroblasts, endothelial cells, would further facilitate a better understanding of the metabolic reprogramming in cancers. Better assessment of metabolism in human tumors and mice models, along with connecting the missing links between obesity, diabetes and cancer is surely warranted. Knowledge gained in these areas along with connecting the loop between oncogenic signaling, metabolism and chromatin organization, would assist in effective designing of the targeted therapies in cancer that utilize metabolism and epigenetic regulation as the key players.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hanahan D and Weinberg RA, Hallmarks of cancer: the next generation. Cell, 2011. 144(5): p. 646–74. [DOI] [PubMed] [Google Scholar]
- 2.Pavlova NN and Thompson CB, The Emerging Hallmarks of Cancer Metabolism. Cell Metab, 2016. 23(1): p. 27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward PS and Thompson CB, Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell, 2012. 21(3): p. 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantor JR and Sabatini DM, Cancer cell metabolism: one hallmark, many faces. Cancer Discov, 2012. 2(10): p. 881–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hui L and Chen Y, Tumor microenvironment: Sanctuary of the devil. Cancer Lett, 2015. 368(1): p. 7–13. [DOI] [PubMed] [Google Scholar]
- 6.Lu C and Thompson CB, Metabolic regulation of epigenetics. Cell Metab, 2012. 16(1): p. 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaelin WG Jr. and McKnight SL, Influence of metabolism on epigenetics and disease. Cell, 2013. 153(1): p. 56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaochar S and Tu BP, Gatekeepers of Chromatin: Small Metabolites Elicit Big Changes in Gene Expression. Trends in biochemical sciences, 2012. 37(11): p. 477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warburg O, Wind F, and Negelein E, THE METABOLISM OF TUMORS IN THE BODY. The Journal of General Physiology, 1927. 8(6): p. 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warburg O, On respiratory impairment in cancer cells. Science, 1956. 124(3215): p. 269–70. [PubMed] [Google Scholar]
- 11.Vander Heiden MG, Cantley LC, and Thompson CB, Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science, 2009. 324(5930): p. 1029–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koppenol WH, Bounds PL, and Dang CV, Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer, 2011. 11(5): p. 325–37. [DOI] [PubMed] [Google Scholar]
- 13.Lunt SY and Vander Heiden MG, Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol, 2011. 27: p. 441–64. [DOI] [PubMed] [Google Scholar]
- 14.Hosios AM, et al. , Amino Acids Rather than Glucose Account for the Majority of Cell Mass in Proliferating Mammalian Cells. Dev Cell, 2016. 36(5): p. 540–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Outschoorn UE, et al. , Energy transfer in “parasitic” cancer metabolism: Mitochondria are the powerhouse and Achilles’ heel of tumor cells. Cell Cycle, 2011. 10(24): p. 4208–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pavlides S, et al. , The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle, 2009. 8(23): p. 3984–4001. [DOI] [PubMed] [Google Scholar]
- 17.Levine AJ and Puzio-Kuter AM, The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science, 2010. 330(6009): p. 1340–4. [DOI] [PubMed] [Google Scholar]
- 18.Cairns RA, Harris IS, and Mak TW, Regulation of cancer cell metabolism. Nat Rev Cancer, 2011. 11(2): p. 85–95. [DOI] [PubMed] [Google Scholar]
- 19.Vander Heiden MG, Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov, 2011. 10(9): p. 671–84. [DOI] [PubMed] [Google Scholar]
- 20.Doherty JR and Cleveland JL, Targeting lactate metabolism for cancer therapeutics. The Journal of Clinical Investigation, 2013. 123(9): p. 3685–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elstrom RL, et al. , Akt stimulates aerobic glycolysis in cancer cells. Cancer Res, 2004. 64(11): p. 3892–9. [DOI] [PubMed] [Google Scholar]
- 22.Robey RB and Hay N, Is Akt the “Warburg kinase”?-Akt-energy metabolism interactions and oncogenesis. Semin Cancer Biol, 2009. 19(1): p. 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finkel T, Signal transduction by reactive oxygen species. J Cell Biol, 2011. 194(1): p. 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sabharwal SS and Schumacker PT, Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles’ heel? Nat Rev Cancer, 2014. 14(11): p. 709–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nathan C and Ding A, SnapShot: Reactive Oxygen Intermediates (ROI). Cell, 2010. 140(6): p. 951–951.e2. [DOI] [PubMed] [Google Scholar]
- 26.Chandel NS and Tuveson DA, The promise and perils of antioxidants for cancer patients. N Engl J Med, 2014. 371(2): p. 177–8. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Pavlova NN, and Thompson CB, Cancer cell metabolism: the essential role of the nonessential amino acid, glutamine. The EMBO Journal, 2017. 36(10): p. 1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.COLES N and JOHNSTONE R, Glutamine metabolism in Ehrlich ascites-carcinoma cells. Biochemical Journal, 1962. 83(2): p. 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eagle H, Nutrition needs of mammalian cells in tissue culture. Science, 1955. 122(3168): p. 501–14. [DOI] [PubMed] [Google Scholar]
- 30.Mullen AR, et al. , Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature, 2011. 481(7381): p. 385–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wise DR, et al. , Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of αketoglutarate to citrate to support cell growth and viability. Proceedings of the National Academy of Sciences of the United States of America, 2011. 108(49): p. 19611–19616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trachootham D, et al. , Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer Cell, 2006. 10(3): p. 241–52. [DOI] [PubMed] [Google Scholar]
- 33.Wise DR, et al. , Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A, 2008. 105(48): p. 18782–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao P, et al. , c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature, 2009. 458(7239): p. 762–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osthus RC, et al. , Deregulation of glucose transporter 1 and glycolytic gene expression by cMyc. J Biol Chem, 2000. 275(29): p. 21797–800. [DOI] [PubMed] [Google Scholar]
- 36.Shim H, et al. , c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A, 1997. 94(13): p. 6658–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dang CV, et al. , The c-Myc target gene network. Semin Cancer Biol, 2006. 16(4): p. 253–64. [DOI] [PubMed] [Google Scholar]
- 38.Bensaad K, et al. , TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell, 2006. 126(1): p. 107–20. [DOI] [PubMed] [Google Scholar]
- 39.Matoba S, et al. , p53 regulates mitochondrial respiration. Science, 2006. 312(5780): p. 1650–3. [DOI] [PubMed] [Google Scholar]
- 40.Wang PY, Zhuang J, and Hwang PM, p53: exercise capacity and metabolism. Curr Opin Oncol, 2012. 24(1): p. 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koong AC, et al. , Pancreatic tumors show high levels of hypoxia. Int J Radiat Oncol Biol Phys, 2000. 48(4): p. 919–22. [DOI] [PubMed] [Google Scholar]
- 42.Neesse A, et al. , Stromal biology and therapy in pancreatic cancer. Gut, 2011. 60(6): p. 861–8. [DOI] [PubMed] [Google Scholar]
- 43.Rabinowitz JD and White E, Autophagy and metabolism. Science, 2010. 330(6009): p. 1344–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo JY, et al. , Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev, 2011. 25(5): p. 460–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang ZJ, et al. , The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther, 2011. 10(9): p. 1533–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strohecker AM and White E, Autophagy promotes Braf (V600E)-driven lung tumorigenesis by preserving mitochondrial metabolism. Autophagy, 2014. 10(2): p. 384–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Margueron R and Reinberg D, Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet, 2010. 11(4): p. 285–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Easwaran H, Tsai HC, and Baylin SB, Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell, 2014. 54(5): p. 716–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Varier RA and Timmers HT, Histone lysine methylation and demethylation pathways in cancer. Biochim Biophys Acta, 2011. 1815(1): p. 75–89. [DOI] [PubMed] [Google Scholar]
- 50.Ulanovskaya OA, Zuhl AM, and Cravatt BF, NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat Chem Biol, 2013. 9(5): p. 300–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klose RJ, Kallin EM, and Zhang Y, JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet, 2006. 7(9): p. 715–27. [DOI] [PubMed] [Google Scholar]
- 52.Shi YJ, et al. , Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell, 2005. 19(6): p. 857–64. [DOI] [PubMed] [Google Scholar]
- 53.Xiao M, et al. , Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev, 2012. 26(12): p. 1326–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shahbazian MD and Grunstein M, Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem, 2007. 76: p. 75–100. [DOI] [PubMed] [Google Scholar]
- 55.Wellen KE and Thompson CB, A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol, 2012. 13(4): p. 270–6. [DOI] [PubMed] [Google Scholar]
- 56.Pietrocola F, et al. , Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab, 2015. 21(6): p. 805–21. [DOI] [PubMed] [Google Scholar]
- 57.Racey LA and Byvoet P, Histone acetyltransferase in chromatin: Evidence for in vitro enzymatic transfer of acetate from acetyl-coenzyme A to histones. Experimental Cell Research, 1971. 64(2): p. 366–370. [DOI] [PubMed] [Google Scholar]
- 58.Cai L, et al. , Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell, 2011. 42(4): p. 426–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Friis RM, et al. , A glycolytic burst drives glucose induction of global histone acetylation by picNuA4 and SAGA. Nucleic Acids Res, 2009. 37(12): p. 3969–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee JV, et al. , Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab, 2014. 20(2): p. 306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Candido EP, Reeves R, and Davie JR, Sodium butyrate inhibits histone deacetylation in cultured cells. Cell, 1978. 14(1): p. 105–13. [DOI] [PubMed] [Google Scholar]
- 62.McBrian MA, et al. , Histone acetylation regulates intracellular pH. Mol Cell, 2013. 49(2): p. 310–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joyce JA and Pollard JW, Microenvironmental regulation of metastasis. Nat Rev Cancer, 2009. 9(4): p. 239–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kurdistani SK, Histone modifications in cancer biology and prognosis. Prog Drug Res, 2011. 67: p. 91–106. [DOI] [PubMed] [Google Scholar]
- 65.Soga T, Cancer metabolism: key players in metabolic reprogramming. Cancer Sci, 2013. 104(3): p. 275–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dang L, et al. , Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature, 2009. 462(7274): p. 739–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parsons DW, et al. , An integrated genomic analysis of human glioblastoma multiforme. Science, 2008. 321(5897): p. 1807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ward PS, et al. , The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell, 2010. 17(3): p. 225–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wolf A, et al. , Developmental profile and regulation of the glycolytic enzyme hexokinase 2 in normal brain and glioblastoma multiforme. Neurobiol Dis, 2011. 44(1): p. 84–91. [DOI] [PubMed] [Google Scholar]
- 70.Goel A, Mathupala SP, and Pedersen PL, Glucose metabolism in cancer. Evidence that demethylation events play a role in activating type II hexokinase gene expression. J Biol Chem, 2003. 278(17): p. 15333–40. [DOI] [PubMed] [Google Scholar]
- 71.Chen M, et al. , Promoter hypermethylation mediated downregulation of FBP1 in human hepatocellular carcinoma and colon cancer. PLoS One, 2011. 6(10): p. e25564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li X, et al. , Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nature Reviews Molecular Cell Biology, 2018. 19(9): p. 563–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sivanand S, et al. , Nuclear Acetyl-CoA Production by ACLY Promotes Homologous Recombination. Mol Cell, 2017. 67(2): p. 252–265.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bhatt AN, et al. , Transient elevation of glycolysis confers radio-resistance by facilitating DNA repair in cells. BMC cancer, 2015. 15: p. 335–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Juvekar A, et al. , Phosphoinositide 3-kinase inhibitors induce DNA damage through nucleoside depletion. Proceedings of the National Academy of Sciences, 2016. 113(30): p. E4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chaturvedi S, et al. , Human Metabolic Enzymes Deficiency: A Genetic Mutation Based Approach. Scientifica, 2016. 2016: p. 9828672–9828672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hamanaka RB and Chandel NS, Targeting glucose metabolism for cancer therapy. J Exp Med, 2012. 209(2): p. 211–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rodriguez-Enriquez S, et al. , Canonical and new generation anticancer drugs also target energy metabolism. Arch Toxicol, 2014. 88(7): p. 1327–50. [DOI] [PubMed] [Google Scholar]
- 79.Erez A and DeBerardinis RJ, Metabolic dysregulation in monogenic disorders and cancer finding method in madness. Nat Rev Cancer, 2015. 15(7): p. 440–8. [DOI] [PubMed] [Google Scholar]
- 80.Wang YH, et al. , Cell-state-specific metabolic dependency in hematopoiesis and leukemogenesis. Cell, 2014. 158(6): p. 1309–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fantin VR, St-Pierre J, and Leder P, Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell, 2006. 9(6): p. 425–34. [DOI] [PubMed] [Google Scholar]
- 82.Bailey KM, et al. , Targeting the metabolic microenvironment of tumors. Adv Pharmacol, 2012. 65: p. 63–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xiang Y, et al. , Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. J Clin Invest, 2015. 125(6): p. 2293–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weinberg SE and Chandel NS, Targeting mitochondria metabolism for cancer therapy. Nat Chem Biol, 2015. 11(1): p. 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Birsoy K, et al. , Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature, 2014. 508(7494): p. 108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Muir A, Danai LV, and Vander Heiden MG, Microenvironmental regulation of cancer cell metabolism: implications for experimental design and translational studies. Dis Model Mech, 2018. 11(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yamaguchi R and Perkins G, Challenges in targeting cancer metabolism for cancer therapy. EMBO reports, 2012. 13(12): p. 1034–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Graham JS, Kaye SB, and Brown R, The promises and pitfalls of epigenetic therapies in solid tumours. European Journal of Cancer, 2009. 45(7): p. 1129–1136. [DOI] [PubMed] [Google Scholar]
- 89.Hensley CT, et al. , Metabolic Heterogeneity in Human Lung Tumors. Cell, 2016. 164(4): p. 68194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Davidson SM, et al. , Environment Impacts the Metabolic Dependencies of Ras-Driven NonSmall Cell Lung Cancer. Cell Metab, 2016. 23(3): p. 517–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.DeBerardinis RJ and Chandel NS, Fundamentals of cancer metabolism. Science Advances, 2016. 2(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pavlova NN and Thompson CB, The Emerging Hallmarks of Cancer Metabolism. Cell metabolism, 2016. 23(1): p. 27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]