Abstract
Background
The unfolded protein response (UPR) plays versatile roles in physiology and pathophysiology. Its connection to cell growth however remains elusive. Here, we sought to define the role of UPR in regulation of cardiomyocyte growth in the heart.
Methods
We used both gain- and loss-of-function approaches to genetically manipulate spliced X-box binding protein 1 (XBP1s), the most conserved signaling branch of the UPR, in the heart. In addition, primary cardiomyocyte culture was employed to address the role of XBP1s in cell growth in a cell-autonomous manner.
Results
We found that XBP1s expression is reduced in both human and rodent cardiac tissues under heart failure. Further, deficiency of XBP1s leads to decompensation and exacerbation of heart failure progression under pressure overload. On the other hand, cardiac-restricted overexpression of XBP1s prevents the development of cardiac dysfunction. Mechanistically, we found that XBP1s stimulates adaptive cardiac growth through activation of the mechanistic target of rapamycin (mTOR) signaling, which is mediated via FK506-binding protein 11 (FKBP11), a novel transcriptional target of XBP1s. Moreover, silencing of FKBP11 significantly diminishes XBP1s-induced mTOR activation and adaptive cell growth.
Conclusions
Our results reveal a critical role of the XBP1s-FKBP11-mTOR axis in coupling the UPR and cardiac cell growth regulation.
Keywords: XBP1s, unfolded protein response, heart failure, cardiac hypertrophy, mTOR, FKBP11
Introduction
Hypertension is one of the most important risk factors for cardiovascular disease.1 In response to high blood pressure, the heart manifests hypertrophic growth to ameliorate ventricular wall stress.2 This once adaptive process may decompensate and progress into heart failure. Despite extensive research, signaling pathways governing the transition are still elusive.
Approximately 40% of human proteins are predicted to be either transmembrane or secretory.3 The synthesis, folding, vesicular transportation, and localization of these proteins rely on proper function of the secretory pathway. The unfolded protein response (UPR) is a cellular adaptive process to cope with protein folding stress.4 Numerous studies have established that the UPR plays versatile roles during development and under physiological and pathophysiological conditions.
Accumulation of misfolded proteins in the secretory pathway stimulates three signaling branches of the UPR, PERK, IRE1/XBP1s, and ATF6, via distinct mechanisms.4 In so doing, the cell transiently slows down protein synthesis, upregulates chaperone production, and triggers ER-associated degradation to restore cellular homeostasis. Disturbance of the UPR contributes to etiology of many disorders, including diabetes, neurodegenerative disease, and cancer.5–7 Recent studies have highlighted the critical roles of the UPR in the heart.8 We have shown that overexpression of GRP78, a master regulator of the UPR, confers strong cardiac cardioprotection against ischemia/reperfusion (I/R) injury.9 ATF6 is activated by myocardial infarction, and overexpression of ATF6 protects the heart from ischemic damage.10 Additionally, the adaptive branch IRE1/XBP1s is potently and acutely stimulated by cardiac I/R.11 Cardiomyocyte-specific deletion of XBP1s leads to aggravation in cardiac dysfunction by I/R, suggesting that XBP1s plays a protective role.11 Despite these, role of the UPR and XBP1s in cardiac hypertrophic growth and heart failure progression remains to be fully elucidated.
Pathological cardiac remodeling involves metabolic derangements, calcium signaling alterations, hypoxia, and augmented demand on protein synthesis.12–14 Most, if not all, of these processes are potent inducers of the UPR. Additionally, the XBP1s pathway has been implicated in metabolic regulation, secretory protein synthesis, and phospholipid production.15 On the other hand, mechanistic target of rapamycin (mTOR) is a signaling nexus to orchestrate protein synthesis, lipid production, and cell growth.16 Accumulating evidence suggests that mTOR actively participates in hypertrophic growth in response to pressure overload.17 These findings suggest an intrinsic crosstalk between the UPR and mTOR signaling. Here, we set up to investigate the significance of XBP1s in regulating mTOR and the relevance in cardiac hypertrophic growth in response to pressure overload.
Methods
The data that support the findings of this study are available from the corresponding author on reasonable request. All mouse experiments were approved by the Institutional Animal Care and the Use Committee of University of Texas Southwestern Medical Center. Additional details of the experimental procedures are included in the online-only Data Supplement.
RNA isolation, reverse transcription, and quantitative PCR
Total RNA from cardiac tissues and NRVMs was isolated using an Aurum Total RNA Fatty and Fibrous Tissue kit (Bio-Rad, 732–6870) and a Quick-RNA Microprep kit (Zymo Research, R1051), respectively. A total of 250 ng RNA was used for reverse transcription (Bio-Rad, iScript, 1708891) and the cDNA was used for quantitative PCR to determine relative mRNA levels to 18s rRNA using a LightCycler machine (Roche) and the SYBR green reagent (Bimake, B21203). All primer sequences are listed in Supplemental Table 1.
Statistical Analysis
Data are expressed as means ± SEM. The Student’s t test (2-tailed) was used for comparison between two groups. The log rank test was conducted to calculate the statistical significance for survival curve. For comparison of more than two groups, two-way ANOVA was conducted, followed by Tukey’s test. A P < 0.05 was considered as statistically significant. Statistical analysis was done with Prism Graphpad 7.0.
Results
XBP1s expression is reduced in heart failure
We have shown that cardiac I/R triggers acute, potent induction of XBP1s, which is necessary and sufficient to protect the heart from reperfusion injury.11 However, role of XBP1s in cardiac hypertrophic growth and heart failure remains largely undefined.
We first sought to determine whether XBP1s expression is relevant in the pathophysiology of human heart failure. We collected human cardiac tissues from patients who underwent heart transplantation due to end-stage dilated cardiomyopathy (DCM) (Supplemental Table 2). Compared to normal donors, XBP1s mRNA level was significantly decreased in DCM patients by 60% (Figure 1A, P<0.05), indicating XBP1s might participate in the etiology of human heart failure. Consistent with the reduction of XBP1s, phosphorylation of IRE1α, a signature upstream event to drive XBP1s expression, was diminished in DCM hearts by 70% (Supplemental Figure 1A and 1B, P<0.01).
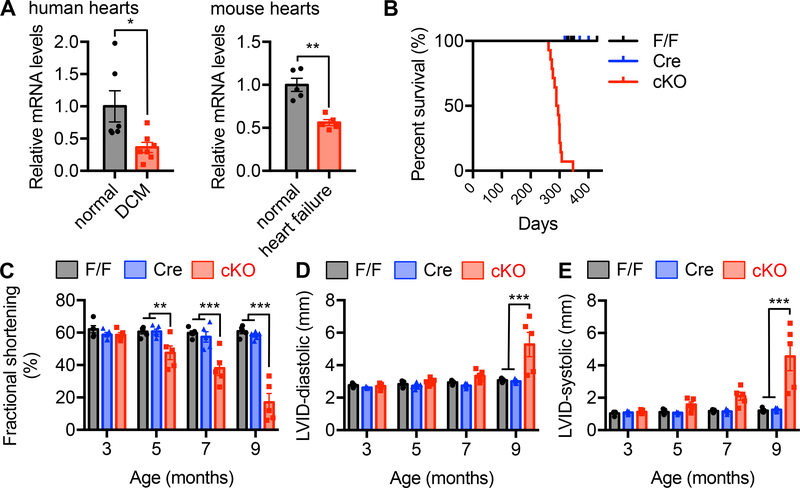
Figure 1. Cardiac-specific deficiency of XBP1s leads to cardiomyopathy in mice.
A. XBP1s expression was reduced in the hearts from human patients with dilated cardiomyopathy (DCM). N = 6 for normal controls; 7 for DCM. XBP1s expression was decreased in failing mouse hearts. Cardiac dilation and heart failure were triggered by thoracic aortic constriction (TAC). N = 5 for each group. Student’s t test was conducted.
B. Cardiac-specific knockout of XBP1s caused early mortality. XBP1s deletion was achieved by crossing XBP1F/F with the αMHC-Cre mouse models. All conditional knockout (cKO) mice died before 350 days old, while the controls did not show mortality. N = 14 for F/F; 6 for αMHC-Cre; 14 for cKO. The log rank test was conducted. P<0.001 between F/F and cKO, and Cre and cKO, respectively.
C. XBP1s cKO mice showed gradual deterioration of cardiac function. Fractional shortening (%) was determined by echocardiography with conscious animals. N = 5 per group.
D. XBP1s deficient mice displayed an increase in left ventricular inner diameter (LVID) at diastole. N = 5 for each group.
E. XBP1s cKO showed elevated left ventricular diameter at systole. N = 5 for each group. Two-way ANOVA was conducted with the selection of repeated measures, followed by Bonferroni’s multiple comparisons test. *, P<0.05; **, P<0.01; ***, P<0.001.
We went on to investigate whether XBP1s was reduced in a mouse model of heart failure that exhibits similar clinical features as human patients. We subjected adult mice to thoracic aortic constriction (TAC) to trigger dilated cardiomyopathy from pressure overload. This surgical procedure leads to profound pathological hypertrophic remodeling, cardiac dysfunction, heart failure, and dilation.18 We showed that, like the findings from DCM patients, cardiac XBP1s expression was strongly suppressed by 40% in heart failure rodents (Figure 1A, P<0.01). These data together suggest that XBP1s expression is downregulated in end-stage heart failure, implying that XBP1s reduction may be part of the pathological progression.
Deficiency of XBP1s in cardiomyocytes leads to early mortality
Next, we set up to investigate whether XBP1s plays a role in maintaining cardiac homeostasis. We took advantage of the cardiomyocyte-specific XBP1 knockout mouse model since whole body deletion causes defects in cardiogenesis and embryonic lethality.19 We crossed XBP1F/F animals to the cardiomyocyte-restricted Cre transgenic mouse model (αMHC-Cre). In the conditional knockout (cKO) animals, both full-length and spliced XBP1 were eliminated in cardiomyocytes. Although emerging evidence suggests a role of full-length XBP1 in smooth muscle cells,20 the contribution of XBP1 in pathological cardiac remodeling remains to be fully illustrated. The cKO mice did not show detectable differences during birth, weaning, and maturation (data not shown).
The cKO mice had a shorter lifespan and all cKO animals died around 300 days old (Figure 1B, P<0.001 between cKO and single transgenic mice by the log rank test). To determine whether this early mortality was due to dysfunction of the heart, we evaluated cardiac performance. The cKO mice progressively lost their cardiac contractility, as shown by reduction of fractional shortening (FS) at 5 (20% decrease, P<0.01), 7 (40% decrease, P<0.001), and 9 (70% decrease, P<0.001) months of age (Figure 1C), while heart rate remained unchanged (Supplemental Figure 1C and 1D). This decrease of FS stemmed from systolic dysfunction (Figure 1D and 1E, P<0.001 between cKO and single transgenic mice at 9 months old). Collectively, these results indicate that deficiency of XBP1s in cardiomyocytes leads to systolic cardiac dysfunction, highlighting a critical role of XBP1s in maintaining cardiac homeostasis.
XBP1s deficiency exacerbates heart failure progression by pressure overload
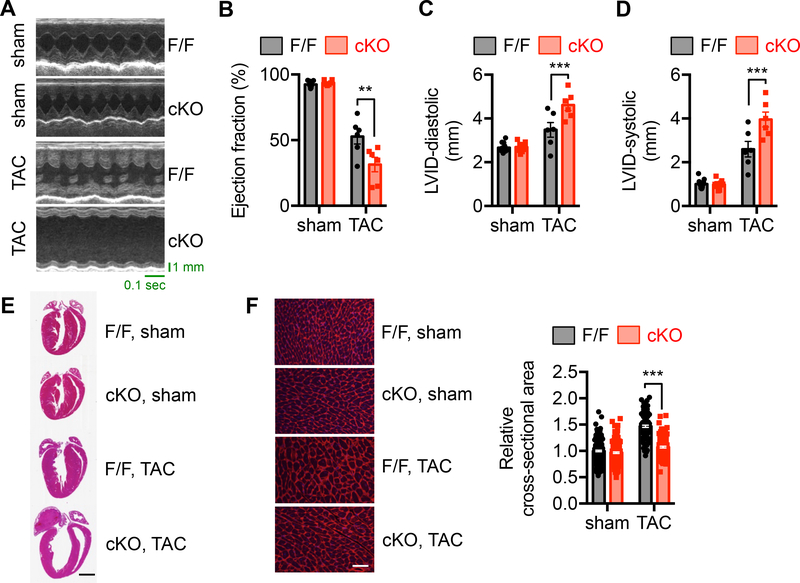
After establishing a critical role of XBP1s in cardiac homeostasis, we next sought to assess XBP1s function under pressure overload, a common pathological condition in hypertensive patients. TAC in control animals led to decline in cardiac function, as indicated by decreases in ejection fraction (EF) (Figure 2A and 2B, P<0.01 between F/F and cKO after TAC), FS (Supplemental Figure 2A), and systolic performance (Figure 2C and 2D). Importantly, elimination of XBP1s from cardiomyocytes significantly exacerbated this response. Moreover, ventricular wall thickness was reduced by XBP1s deletion (Supplemental Figure 2B through 2D, P<0.05 between F/F and cKO after TAC), which was also evident at the histological level (Figure 2E) and by quantification of cardiomyocyte size (Figure 2F, 30% decrease between F/F and cKO after TAC, P<0.001). Further, elevation of relative wet lung weight by twofold indicates that lack of XBP1s in cardiomyocytes causes not only cardiac dysfunction but also heart failure (Supplemental Figure 2E, P<0.001). These data suggest XBP1s is required for the heart to mount an adaptive growth response in response to pressure overload.
Figure 2. XBP1s deficiency exacerbates heart failure progression by pressure overload.
A. Control and cKO animals of 8 weeks old were used for sham or TAC surgery. Cardiac function was determined 2 weeks later. Representative echocardiography images of control and cKO animals after sham or TAC surgery are shown.
B. XBP1s deficiency in the heart led to deterioration of cardiac function and accelerated progression to cardiomyopathy, as revealed by a decrease in ejection fraction (%). N = 6–9.
C. Diastolic LVID was significantly elevated in the cKO mice after TAC. N = 6–13.
D. LVID at systole was increased by XBP1s deficiency in the heart. N = 6–13.
E. Representative cardiac images showed deficiency of ventricular growth in the cKO mice after TAC. Scale bar: 2 mm.
F. Wheat germ agglutinin (WGA) staining was performed to visualize cardiac myocytes (left). Scale bar: 50 μm. Quantification is shown at the right. N = 209 for sham/FF; 188 for sham/cKO; 115 for TAC/FF; 143 for TAC/cKO. Two-way ANOVA analysis was conducted, followed by Tukey’s test. **, P<0.01; ***, P<0.001.
Decompensation of cardiac growth under pressure overload represents a major mechanism for ventricular wall thinning, cardiac dilation, and heart failure. Since XBP1s might play a role in stimulating cardiac growth and its level was reduced in heart failure, we asked the question whether restoration of XBP1s in cardiomyocytes could enhance adaptive cardiomyocyte growth and prevent cardiac dysfunction.
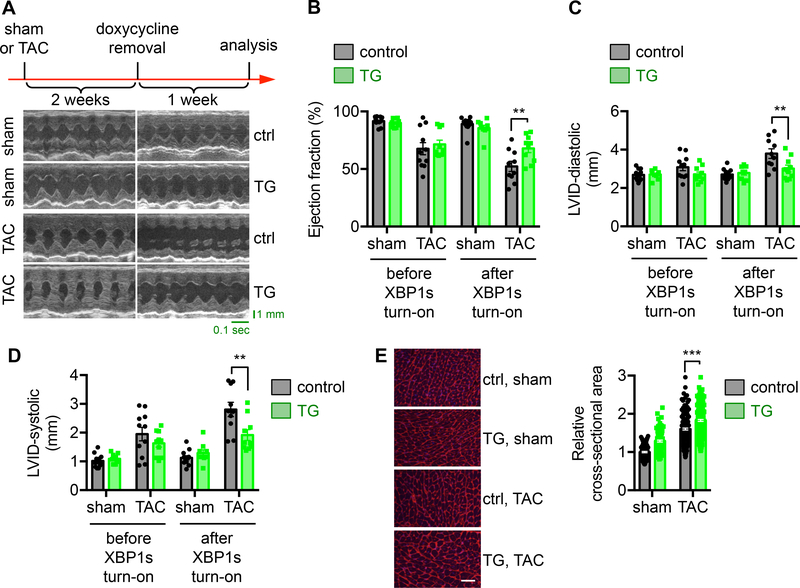
Here, we took advantage of an inducible overexpression system (Supplemental Figure 3A). We crossed the TRE-XBP1s transgenic mouse11 with the αMHC-tTA animal. The transgene XBP1s is under the control of 7 tetracycline responsive elements (TRE). In the double “tet-off” transgenic mice, transcriptional factor tTA is expressed only in cardiomyocytes, which is dissociated from TRE in the presence of doxycycline. Upon removal of doxycycline, tTA undergoes transformational change, binds TRE, and stimulates transgene XBP1s expression. This mouse model confers specificity, inducibility, and reproducibility. We included doxycycline in the drinking water during breeding, pregnancy, and weaning. We then switched to the regular drinking water for a week. XBP1s was only induced in the double transgenic hearts upon doxycycline removal (Supplemental Figure 3B and 3C).
We first conducted TAC to trigger cardiac hypertrophic growth and heart failure progression (Figure 3A). Two week later, doxycycline was withdrawn from drinking water to trigger XBP1s overexpression in the transgenic mice when the heart did not show detectable dysfunction (Figure 3B through 3D) and endogenous cardiac XBP1s expression began to decline by 50% in the TAC group (Supplemental Figure 3D, P<0.05). XBP1s protein level in the transgenic mice was significantly upregulated, rescuing the cardiac XBP1s expression from TAC-induced decrease (Supplemental Figure 3E, P<0.001). We assayed cardiac function after another week. Importantly, we found that TAC led to cardiomyopathy at this time in the control mice, which was significantly alleviated by XBP1s expression in the transgenic animals. This was associated with improved systolic performance (Figure 3B through 3D, and Supplemental Figure 3F, P<0.01 between control and TG groups after XBP1s turn-on), restoration of ventricular wall thickness (Supplemental Figure 3G), and prevention of heart failure. Moreover, cardiomyocyte size was significantly increased (Figure 3E, P<0.001 between control and TG mice after TAC), consistent with the notion that XBP1s promotes adaptive cardiac cell growth. Taken together, these results indicate that XBP1s restoration in cardiomyocytes confers adaptive growth and protects from dilation and heart failure in response to pressure overload.
Figure 3. Overexpression of XBP1s in the heart improves cardiac function in response to pressure overload.
A. XBP1s expression was turned on after 2 weeks of TAC, before the onset of cardiac dysfunction (indicated as doxycycline removal). After another week, control animals (single TRE-XBP1s or αMHC-tTA transgenics alone under water without doxycycline) displayed cardiomyopathy and cardiac dysfunction, while transgenic mice (TRE-XBP1s and αMHC-tTA double transgenics under water without doxycycline) showed significant improvements in cardiac function, as evidenced by representative echocardiographic images.
B. Ejection fraction (%) was significantly increased by XBP1s overexpression. N = 9–13.
C. LVID at diastole was reduced in the XBP1s transgenic mice after TAC. N = 9–13.
D. Systolic LVID in the transgenic mice was improved. N = 9–13.
E. WGA staining was conducted (left). Scale bar: 50 μm. Quantification of the relative cross-sectional area shows significant upregulation of cardiomyocyte size by XBP1s overexpression (right). N = 128 for sham/control; 104 for sham/TG; 198 for TAC/control; 226 for TAC/TG. Two-way ANOVA analysis was conducted, followed by Tukey’s test. **, P<0.01; ***, P<0.001.
XBP1s is required for cardiomyocyte growth in response to hypertrophic stimuli
We showed that deficiency of XBP1s in cardiomyocytes diminishes adaptive cardiac hypertrophy while forced overexpression of XBP1s improves cardiac function under pressure overload. These data strongly imply an important role of XBP1s in cardiac myocyte growth.
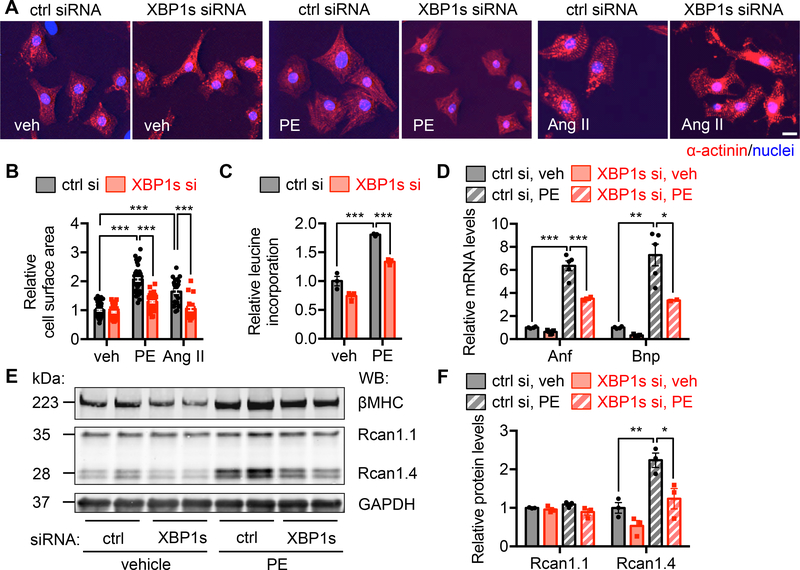
To directly address the role of XBP1s in cardiomyocyte growth in a cell-autonomous manner, we turned to cultured neonatal rat ventricular myocytes (NRVMs), isolated from 1–2 days old Sprague-Dawley rats. Phenylephrine (PE) is a α1-adrenergic receptor agonist, which is commonly used to stimulate cardiomyocyte hypertrophic growth.21 We found that PE treatment (50 μM) in NRVMs led to significant upregulation of XBP1s at the protein level (Supplemental Figure 4A), indicating a potential role of XBP1s in cardiomyocyte growth. Next, we first silenced XBP1s by siRNA transfection (Supplemental Figure 4B and 4C). Immunofluorescence staining showed that PE increased cardiomyocyte size, which was severely diminished by XBP1s silencing (Figure 4A and 4B). Additionally, we used Angiotensin II as an alternative hypertrophic stimulus. A similar response was discovered (Figure 4A and 4B). We next included a trace amount of radiolabeled 3H-leucine in the culture medium during PE treatment and harvested NRVMs to detect leucine incorporation. We found that PE increased protein synthesis, which was reduced by XBP1s knockdown (Figure 4C). At the molecular level, PE induction of Anf and Bnp, markers of cardiomyocyte hypertrophy, was strongly suppressed by XBP1s silencing (Figure 4D). Rcan1 is a target of the calcineurin/NFAT pathway.22 The splicing variant Rcan1.4 is directly related to hypertrophic growth, while Rcan1.1 remains constant as an internal control. Consistently, we found that Rcan1.4 was elevated by PE treatment, and XBP1s knockdown led to a diminished response while Rcan1.1 protein level was not affected (Figure 4E and 4F). Importantly, we evaluated and confirmed the role of XBP1s in cardiomyocyte hypertrophic growth by using another independent XBP1s siRNA (Supplemental Figure 4D through 4H). In aggregate, these data support that XBP1s is required for hypertrophic growth in cardiac myocytes.
Figure 4. XBP1s is required for cardiac myocyte growth in response to hypertrophic stimuli.
A. Immunofluorescence staining for α-actinin. XBP1s was silenced by siRNA transfection and the cells were treated by phenylephrine (PE, 50 μM) or Angiotensin II (Ang II, 1 μM) for 48 hrs. Scale bar: 20 μm.
B. Cardiomyocyte surface area quantification of A. PE or Ang II treatment increased cardiomyocyte size, which was blunted by XBP1s silencing. N = 56 for veh/ctrl si; 57 for veh/XBP1s si; 49 for PE/ctrl si; 50 for PE/XBP1s si; 36 for Ang II/ctrl si; 28 for Ang II/XBP1s si.
C. XBP1s knockdown attenuated PE-induced growth as revealed by a 3H-leucine incorporation assay. N = 3 per group.
D. Induction of hypertrophic markers by PE was diminished by XBP1s knockdown, as assessed by quantitative RT-PCR. N = 4 for ctrl si; 5 for XBP1s si.
E. Immunoblotting showed that the induction of Rcan1.4 by PE was blunted by XBP1s silencing. In contrast, Rcan1.1 expression was not affected. GAPDH was used as a loading control.
F. Quantification of E. N = 3 for each group. Two-way ANOVA analysis was performed, followed by Tukey’s test. *, P<0.05; **, P<0.01; ***, P<0.001.
XBP1s expression is sufficient to stimulate cardiomyocyte hypertrophic growth
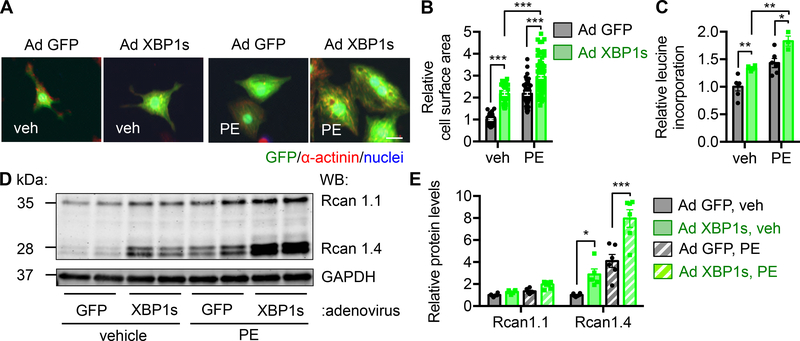
We showed that XBP1s is indispensible for cardiac cell growth. Next we asked the questions whether XBP1s is sufficient to drive cardiomyocyte growth. We induced XBP1s expression in NRVMs by adenovirus infection (Supplemental Figure 5A). We found that overexpression of XBP1s increased cardiac myocyte size and further potentiated PE-induced hypertrophic growth (Figure 5A and 5B), which was confirmed by elevated protein synthesis (Figure 5C). Consistently, expression of XBP1s combined with PE led to a higher level of Bnp than PE treatment alone (Supplemental Figure 5B). The elevation of Rcan1.4 at the protein level further corroborated these findings (Figure 5D and 5E). Taken together, these data suggest that XBP1s expression in cardiac myocytes is sufficient to drive cell growth.
Figure 5. XBP1s expression is sufficient to stimulate cardiomyocyte hypertrophic growth.
A. XBP1s overexpression in cardiomyocytes led to an increase in cell size. NRVMs were infected by adenovirus expressing control GFP or XBP1s, followed by PE treatment. Note that the XBP1s-expressing virus is bi-cistronic for GFP and XBP1s, and GFP positivity indicates infection by either GFP-only control or XBP1s-GFP virus. Scale bar: 20 μm.
B. Quantification of A. N = 34 for veh/Ad GFP; 33 for veh/Ad XBP1s; 51 for PE/Ad GFP; 107 for PE/Ad XBP1s.
C. Protein synthesis was elevated by XBP1s overexpression as revealed by 3H-leucine incorporation. N = 3 for PE/Ad XBP1s. N = 6 for the other groups.
D. XBP1s overexpression potentiated PE-induced upregulation of Rcan1.4 at the protein level. As a control, Rcan1.1 did not show a similar trend.
E. Quantification of D. N = 5 for Rcan1.4/Ad XBP1s. N = 6 for the other groups. Two-way ANOVA analysis was conducted, followed by Tukey’s test. *, P<0.05; **, P<0.01; ***, P<0.001.
XBP1s stimulates the mTOR signaling
We set out to determine the mechanism by which XBP1s, a signaling branch of the UPR, induced cell growth. Although a stimulatory role of XBP1s in anabolism has been discovered in liver, muscle, and adipose tissues,23 direct links to cell growth are still missing. Accumulating evidence suggests that the mechanistic target of rapamycin (mTOR) signaling plays a pivotal role in the heart in response to physiological or pathophysiological cues.17 mTOR is a master regulator of cell growth and a hub for numerous signaling pathways.24 These findings indicate a direct connection between XBP1s and mTOR.
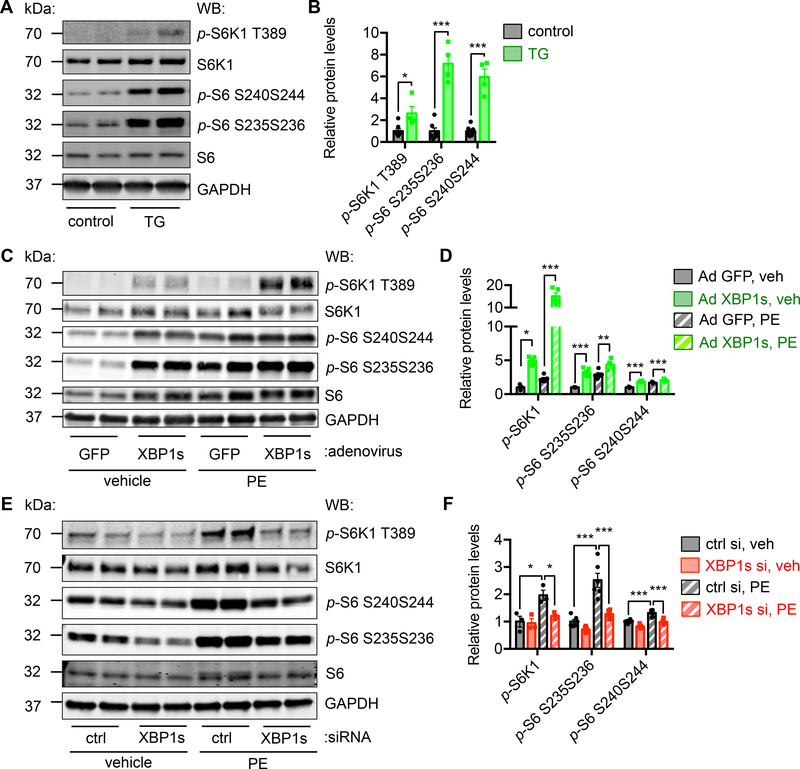
In the XBP1s transgenic hearts, we found mTOR signaling was strongly upregulated, as revealed by increases in the phosphorylation of downstream targets S6K1, S6, and 4EBP1 (Figure 6A and 6B, and Supplemental Figure 6A, 2-fold P<0.05 for p-S6K1/S6K1 and 6-fold P<0.001 for p-S6/S6 by Student’s t test). This was not a phenomenon exclusive to cardiomyocytes. Inducible overexpression of XBP1s in hepatocytes in liver25 and adipocytes in fat26 led to similar activation of mTOR (Supplemental Figure 6B and 6C).
Figure 6. XBP1s stimulates mTOR signaling in the heart.
A. Cardiomyocyte-specific overexpression of XBP1s increased mTOR signaling in the heart. XBP1s expression in the heart was turned on for 2 weeks by removal of doxycycline from drinking water. Cardiac tissues were subjected to Western blotting to detect the mTOR signaling. GAPDH was used as loading control.
B. Quantification of A. Normalization was done with phosphorylation levels to respective total proteins. Student’s t test was conducted. N = 4–7.
C. XBP1s overexpression in vitro augmented mTOR activity. NRVMs were infected by adenovirus expressing either GFP or XBP1s. PE treatment was then conducted for 24 hrs.
D. Quantification of C. N = 5 for each group.
E. XBP1s knockdown decreased the mTOR activity. XBP1s expression was reduced by siRNA transfection in NRVMs and PE was incubated for 24 hrs. GAPDH was used as a loading control.
F. Quantification of E. N = 3–6. Two-way ANOVA analysis was conducted, followed by Tukey’s test. *, P<0.05; **, P<0.01; ***, P<0.001.
Next, we asked whether mTOR induction in XBP1s transgenic mice increased cardiac cell size. We induced XBP1s expression by doxycycline withdrawal, followed by rapamycin administration (I.P., daily, 2 mg/kg body weight). The hearts were harvested after a week. We found that XBP1s induction led to an increase in cardiomyocyte size by 60% (P<0.001 between control and TG mice), which was suppressed by rapamycin (Supplemental Figure 6D). Consistently, XBP1s-triggered activation of mTOR was inhibited by rapamycin (Supplemental Figure 6E), highlighting a critical role of mTOR in mediating the pro-growth response of XBP1s. On the other hand, conditional knockout of XBP1s in cardiomyocytes caused a decrease in mTOR signaling at the age of 5 months old (Supplemental Figure 7A), which was associated with reduction in cardiomyocyte size by 10% (Supplemental Figure 7B, P<0.05) and the emerging of cardiac dysfunction (Figure 1C).
To further evaluate whether the induction of mTOR pathway was a cell-autonomous phenomenon, we overexpressed XBP1s in NRVMs. Consistently, the mTOR pathway was stimulated (Supplemental Figure 8A). These increases were sensitive to mTOR inhibition by rapamycin or torin-1 (Supplemental Figure 8B). Moreover, XBP1s-mediated activation of mTOR was further increased by PE treatment (Figure 6C and 6D). In contrast, knockdown of XBP1s significantly diminished PE-induced activation of mTOR (Figure 6E and 6F). Another independent siRNA against XBP1s further confirmed the necessity of XBP1s in mTOR activation by PE (Supplemental Figure 8C). Collectively, these results suggest that XBP1s directly stimulates the mTOR signaling.
FKBP11 is a direct target of XBP1s
Our findings indicate that XBP1s couples cell growth with mTOR stimulation. To further dissect the underlying mechanism, we mined microarray data from XBP1s transgenics. We found that FK506 binding protein 11 (FKBP11) was among the top 10 most upregulated transcripts by XBP1s.25 FKBP11 belongs to a family of FK506 binding proteins that have prolyl isomerase activity.27 Many members of the FKBP family have been implicated in the regulation of mTOR.28
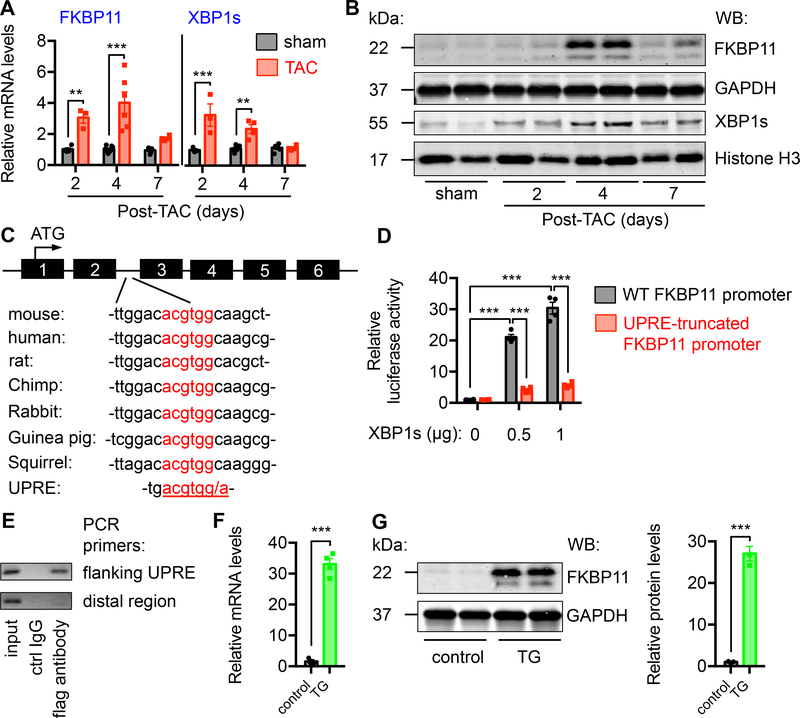
We first evaluated the relevance of FKBP11 in the heart during hypertrophic growth in response to pressure overload. We subjected wild type mice to TAC. We harvested hearts at different time. Pressure overload led to cardiac hypertrophic growth, as shown by a progressive increase in the ratios of heart weight/body weight21 and gene expression of hypertrophic markers (Supplemental Figure 9A). We found FKBP11 mRNA was strongly and acutely induced by TAC (Figure 7A, P<0.01 for day 2 and P<0.001 for day 4 after TAC), which coincided with elevation of XBP1s and other downstream targets of XBP1s (Figure 7A, and Supplemental Figure 9B). Immunoblotting showed consistent augmentation (Figure 7B). The induction of XBP1s by pressure overload displayed an acute pattern (Figure 7B, and Supplemental Figure 9C), and the protein level of XBP1s was reduced in the heart under the failing condition (Supplemental Figure 9D). Both mRNA and protein levels of FKBP11 manifested a similar pattern as XBP1s during hypertrophic growth, suggesting a causal link.
Figure 7. FKBP11 is a direct target of XBP1s.
A. FKBP11 expression was acutely induced by pressure overload in the heart. TAC was conducted in wild type animals. Hearts were harvested at different time points post surgery. Relative FKBP11 mRNA level was determined by quantitative RT-PCR. Note that XBP1s expression was also upregulated by TAC. Two-way ANOVA analysis was conducted, followed by Tukey’s test. N = 3–8.
B. FKBP11 protein level was augmented by TAC in the heart as shown by immunoblotting, along with induction of XBP1s. GAPDH and Histone H3 were used as loading controls for whole cell lysates and nuclear extracts, respectively.
C. A conserved region in the second intron of FKBP11 genome, which resembles the consensus sequence of XBP1s-binding site UPRE (unfolded protein response element). The translational initiation site is in the first exon of FKBP11 genomic DNA.
D. XBP1s expression stimulated FKBP11 promoter activity in a dose-dependent manner. The conserved region of FKBP11 promoter was engineered into a luciferase reporter construct. Co-transfection of a XBP1s-expressing plasmid led to an increase in luciferase activity. Note that a truncated mutant of FKBP11 promoter lacking the UPRE region did not show significant promoter activity. N = 4 per group. Two-way ANOVA analysis was performed, followed by Tukey’s test.
E. Chromatin immunoprecipitation (ChIP) assay was conducted to determine the binding of XBP1s to the FKBP11 promoter. NRVMs were infected by adenovirus expressing flag-tagged XBP1s. ChIP was performed with either control mouse IgG or anti-flag antibody. PCR was conducted using primers spanning the UPRE site. Primers from the distal region were used as a negative control.
F. The mRNA level of FKBP11 was upregulated in the XBP1s transgenic hearts, as determined by quantitative RT-PCR. Student’s t test was performed. N = 3–4.
G. The protein level of FKBP11 was significantly increased in the XBP1s transgenic hearts, as revealed by immunoblotting. GAPDH was used as a loading control. N = 3 for each group. Student’s t test was conducted. **, P<0.01; ***, P<0.001.
We then surveyed the FKBP11 promoter. We identified a conserved stretch of nucleotides that resemble the unfolded protein response element (UPRE), a classical binding site of XBP1s (Figure 7C). We engineered this region into a luciferase reporter. When this reporter construct was co-transfected with a XBP1s-expressing plasmid, FKBP11 promoter activity was induced by XBP1s in a dose-dependent manner (Figure 7D). More importantly, when the binding site was truncated, XBP1s-mediated upregulation of luciferase activity was diminished. Further chromatin immunoprecipitation assay confirmed the binding of XBP1s to FKBP11 promoter (Figure 7E). These results collectively suggest that FKBP11 may be a direct transcriptional target of XBP1s.
We found that FKBP11 mRNA was significantly increased in the XBP1s transgenic hearts (Figure 7F, P<0.001), which was confirmed at the protein level (Figure 7G, P<0.001). Further, overexpression of XBP1s in NRVMs led to upregulation of FKBP11 mRNA (Supplemental Figure 9E) and protein (Supplemental Figure 9F) levels. In addition, the increase in FKBP11 by XBP1s was observed in XBP1s transgenic liver25 (Supplemental Figure 9G) and XBP1s transgenic fat tissues26 (Supplemental Figure 9H). Collectively, these data provide convincing evidence that FKBP11 is a direct target of XBP1s.
FKBP11 is required for XBP1s-mediacted mTOR activation
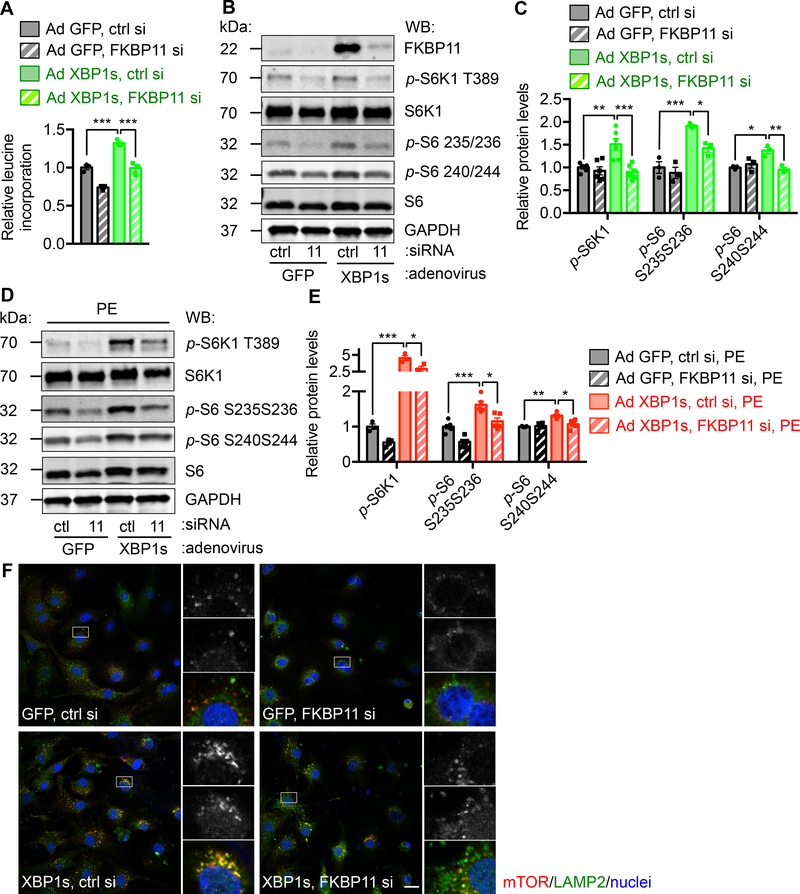
We have shown that XBP1s leads to strong activation of mTOR signaling and consequent cell growth. We next asked whether FKBP11 might be involved in the pro-growth effect of XBP1s. In NRVMs, FKBP11 was reduced by siRNA transfection (Supplemental Figure 10A). Adenovirus-mediated overexpression of XBP1s was then conducted, which increased protein synthesis as assessed by 3H-leucine incorporation (Figure 8A). Knockdown of FKBP11 significantly diminished this response. Consistently, cardiomyocyte growth that was induced by overexpression of XBP1s showed significant reduction by FKBP11 silencing (Supplemental Figure 10B and 10C). Importantly, upregulation of the mTOR signaling by XBP1s was inhibited by FKBP11 silencing (Figure 8B and 8C, and Supplemental Figure 10D). XBP1s overexpression led to increases in hypertrophic markers, which was further augmented by PE (Supplemental Figure 10E). However, FKBP11 silencing significantly diminished this response, as shown by suppression of Bnp and Rcan1.4. Importantly, mTOR signaling was inhibited (Figure 8D and 8E, and Supplemental Figure 10F). The contribution of FKBP11 to XBP1s-mediated cell growth and mTOR activation was further validated with another independent siRNA against FKBP11 (Supplemental Figure 11A and 11B). We went on to dissect the mechanism by which the XBP1s/FKBP11 axis regulates mTOR. By confocal immunofluorescence staining, we showed that the colocalization of mTOR and LAMP2, a lysosomal marker, was enhanced by XBP1s overexpression (Figure 8F). This interaction was however significantly reduced by FKBP11 silencing, highlighting a potential activation mechanism by XBP1s as increasing mTOR translocation to lysosomes.
Figure. 8. FKBP11 is required for XBP1s-mediated mTOR activation.
A. FKBP11 was required for XBP1s-induced hypertrophic growth. NRVMs were infected by adenovirus expressing either GFP or XBP1s. FKBP11 was silenced by siRNA transfection. A 3H-leucine incorporation assay was conducted to assess cell growth. N = 3 for each group.
B. FKBP11 knockdown led to a decrease in XBP1s-induced mTOR activation. NRVMs were first infected by adenovirus to overexpression control GFP or XBP1s. FKBP11 siRNA was then used to transfect NRVMs and cell lysates were extracted for immunoblotting. GAPDH was used as a loading control.
C. Induction of mTOR activity by XBP1s was significantly inhibited by FKBP11 silencing as shown by quantification of B. N = 3–6.
D. Silencing of FKBP11 led to a decrease in PE and XBP1s-induced mTOR activation. NRVMs were first infected by adenovirus expressing GFP or XBP1s. FKBP11-specific siRNA was used to transfect the cells. PE treatment was conducted for 24 hrs and immunoblotting was performed to examine the mTOR signaling.
E. Quantification of D. N = 3–6.
F. Overexpression of XBP1s in NRVMs increased colocalization of mTOR and LAMP2, a lysosomal marker. FKBP11 knockdown significantly diminished this effect. Scale bar: 20 μm. Two-way ANOVA analysis was performed, followed by Tukey’s test. *, P<0.05; **, P<0.01; ***, P<0.001.
We next probed the role of FKBP11 in XBP1s-mediated cardiomyocyte growth using a gain-of-function approach. XBP1s silencing led to a significant decrease in cardiomyocyte growth, as revealed by reduction in leucine incorporation and cardiomyocyte size (Supplemental Figure 12A through 12C). Adenovirus-mediated overexpression of FKBP11 in NRVMs significantly rescued this growth defect. XBP1s cKO in the heart exacerbated cardiac response to pressure overload (Figure 2A through 2D), and caused defective cardiomyocyte growth (Figure 2E and 2F). To examine whether overexpression of FKBP11 might rescue cardiomyocyte growth in vivo, we generated adeno-associated virus to express FKBP11 using the AAV9 system. After TAC, we injected AAV9 FKBP11 virus along with AAV9 GFP controls to the animals. FKBP11 expression was confirmed after 2 weeks (Supplemental Figure 12D), which led to an increase in cardiomyocyte size in the cKO mice by 30% (Supplemental Figure 12E, P<0.001). Echocardiography showed a significant improvement in cardiac function by FKBP11 overexpression (Supplemental Figure 12F and 12G). Collectively, these data suggest that FKBP11, a direct target of XBP1s, is required for XBP1s-mediated mTOR activation and cardiac cell growth.
Discussion
Under conditions of elevated workload, cardiac myocytes respond by increases in cell volume and size.29 To maintain cardiac function, this hypertrophic growth requires a homonymous interplay of various pathways, including metabolism, protein turnover, organelle biogenesis, and membrane production.13, 30 Here, we provide evidence that mTOR, a master anabolism regulator, and XBP1s, the most conserved branch of the UPR, are intimately coupled. At both in vitro and in vivo levels, XBP1s directly stimulates mTOR. Loss of XBP1s in cardiomyocytes leads to functional impairments of the heart and early mortality. Mechanistically, we show that XBP1s directly upregulates FKBP11 at the transcriptional level, which greatly contributes to XBP1s-mediated mTOR activation and cell growth. Taken together, our results uncover a previously unappreciated link between mTOR, the UPR, and cell growth in the heart in response to pressure overload.
UPR and cardiac hypertrophic growth
The UPR is an anciently conserved adaptive process for cell to deal with protein folding stress.31 Emerging evidence strongly suggests that the UPR participates in the pathogenesis of various heart diseases.32 Studies from many groups, including ours, show that cardiac ischemia/reperfusion (I/R) leads to acute activation of ATF6 and XBP1s, which is cardioprotective.10, 11
Role of the UPR in cardiac hypertrophic growth is less defined. Earlier studies show that an UPR downstream effector CHOP is induced by cardiac hypertrophy.33 However, the stimulation profile of the UPR during hypertrophic growth and the functional significance remain to be fully addressed. Cardiac growth is a dynamic, complex process, involving interplays of multiple pathways, above and beyond simple enlargement of cardiac cells.13 It consists of de novo protein synthesis, membrane expansion, and organelle biogenesis. We show here that XBP1s is induced as early as 2 days post pressure overload. Consistently, Lynch et al. found that ATF6 is induced at 48 hrs after TAC.34 More recently, Glembotski and colleagues show that pressure overload in the heart stimulates ATF6 expression, which confers an adaptive growth response through direct upregulation of RheB.35 This acute, dynamic induction also occurs for autophagy, another cellular housekeeping process.30 In aggregate, these findings suggest that acute, adaptive responses may be elevated at the early phase of pressure overload, and dysregulation in these processes may contribute to pathological cardiac remodeling and heart failure.
Activation of XBP1s and mTOR in cardiac hypertrophy
Emerging evidence suggests that XBP1s is a versatile player in metabolism.23 XBP1s is upregulated in adipose tissues,36 liver,25 and heart (data not shown) by metabolic challenges. Insulin treatment stimulates XBP1s.37, 38 On the other hand, we have shown that XBP1s increases glucose assimilation by transcriptionally activating multiple genes of glucose metabolic pathways.11, 25 In liver25 and heart (data not shown), XBP1s is acutely and potently induced by fasting/refeeding, a condition of cell growth. Moreover, Glimcher and colleagues have shown that XBP1s stimulates a subset of genes involved in hepatic lipogenesis.39 Taken together, these data strongly implicate XBP1s as a positive regulator of cell growth and anabolism.
mTOR is a signaling nexus of nutrient sensing and metabolic control.40 Insulin signaling leads to activation of Akt, which triggers the phosphorylation of TSC1/2 and activation of mTORC1.41 On the other hand, mTOR is an indispensible component of the insulin pathway, involved in protein synthesis, lipogenesis, and cell growth. The aforementioned connections between XBP1s, insulin signaling, and mTOR strongly point to a model in which XBP1s is directly linked to mTOR. Previous studies have suggested that mTOR activation may lead to UPR activation due to augmented demand of protein synthesis and folding. Here, our findings on coupling of XBP1s and mTOR provide a missing piece to explain a direct stimulation signal from XBP1s to mTOR, which may form a feed-forward circle to ensure correct protein folding and cellular homeostasis. Although the XBP1s/mTOR axis presented here is implicated in cardiac hypertrophic growth, the same mechanism may exist in other contexts of cell growth under more generalized conditions.
Conclusions and Perspectives
The heart manifests hypertrophic growth in response to pressure overload. This adaptive reaction may decompensate to heart failure. A better understanding of underlying mechanisms holds great promises to arrest disease progression. Here, we show that cardiac hypertrophic growth is accompanied by acute activation of XBP1s. Further, we provide evidence that XBP1s directly stimulates the mTOR signaling, which may be one of the mediators contributing to XBP1s-related adaptive growth. Our findings underscore the importance of the XBP1s/mTOR axis in cardiomyocyte growth and cardiac functional maintenance.
Supplementary Material
Clinical Perspective.
What Is New?
The unfolded protein response (UPR) is reduced in heart failure.
The adaptive branch of the UPR, spliced X-box binding protein 1 (XBP1s), induces cardiac growth in response to pressure overload.
XBP1s directly activates the pro-growth mTOR signaling through transcriptionally upregulating FK-506 binding protein 11.
What Are the Clinical Implications?
Heart failure is a leading cause of death worldwide with hypertension as one of the most important risk factors.
Pathological cardiac remodeling by hypertension involves numerous signaling pathways.
The adaptive UPR confers significant cardioprotection against the development of heart failure.
Acknowledgements
We thank the Molecular Pathology Core (John Shelton) for help with histology. We thank Dr. Ann-Hwee Lee (Regeneron Pharmaceuticals, Inc.) for providing the XBP1F/F mouse model. X. Wang and Z.V.W conceived and designed the study. X. Wang performed most experiments with the help from Y.D. (luciferase assay and liver and adipose tissue transgenic mouse models), G.Z. (mouse colony maintenance and echocardiography), C.L. (ChIP assay), G.D. (FKBP11 knockdown), H.I.M. (animal surgeries), D.H.T. (cell culture), X.L. (NRVM isolation), D.S.J. (human studies), D.L.L. (echocardiography), A.F. (promoter assay), and X. Wei (human studies). X. Wang and Z.V.W. wrote the manuscript with the help of L.X., T.G.G., P.E.S., and X.J.
Sources of Funding
This work was supported by grants from American Heart Association (14SDG18440002 to ZVW) and NIH (R01-HL137723 to ZVW.
Footnotes
Disclosures
None.
References
- 1.Benjamin EJ, Virani SS, Callaway CW, Chamberlain AM, Chang AR, Cheng S, Chiuve SE, Cushman M, Delling FN, Deo R, de Ferranti SD, Ferguson JF, Fornage M, Gillespie C, Isasi CR, Jimenez MC, Jordan LC, Judd SE, Lackland D, Lichtman JH, Lisabeth L, Liu S, Longenecker CT, Lutsey PL, Mackey JS, Matchar DB, Matsushita K, Mussolino ME, Nasir K, O’Flaherty M, Palaniappan LP, Pandey A, Pandey DK, Reeves MJ, Ritchey MD, Rodriguez CJ, Roth GA, Rosamond WD, Sampson UKA, Satou GM, Shah SH, Spartano NL, Tirschwell DL, Tsao CW, Voeks JH, Willey JZ, Wilkins JT, Wu JH, Alger HM, Wong SS and Muntner P. Heart Disease and Stroke Statistics-2018 Update: A Report From the American Heart Association. Circulation. 2018;137:e67–e492. [DOI] [PubMed] [Google Scholar]
- 2.Drazner MH. The progression of hypertensive heart disease. Circulation. 2011;123:327–334. [DOI] [PubMed] [Google Scholar]
- 3.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J and Ponten F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 4.Walter P and Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. [DOI] [PubMed] [Google Scholar]
- 5.Hetz C and Mollereau B. Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat Rev Neurosci. 2014;15:233–249. [DOI] [PubMed] [Google Scholar]
- 6.Back SH and Kaufman RJ. Endoplasmic reticulum stress and type 2 diabetes. Annu Rev Biochem. 2012;81:767–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang M and Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat Rev Cancer. 2014;14:581–597. [DOI] [PubMed] [Google Scholar]
- 8.Glembotski CC. The role of the unfolded protein response in the heart. J Mol Cell Cardiol. 2008;44:453–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bi X, Zhang G, Wang X, Nguyen C, May HI, Li X, Al-Hashimi AA, Austin RC, Gillette TG, Fu G, Wang ZV and Hill JA. Endoplasmic Reticulum Chaperone GRP78 Protects Heart From Ischemia/Reperfusion Injury Through Akt Activation. Circ Res. 2018;122:1545–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin JK, Blackwood EA, Azizi K, Thuerauf DJ, Fahem AG, Hofmann C, Kaufman RJ, Doroudgar S and Glembotski CC. ATF6 Decreases Myocardial Ischemia/Reperfusion Damage and Links ER Stress and Oxidative Stress Signaling Pathways in the Heart. Circ Res. 2017;120:862–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang ZV, Deng Y, Gao N, Pedrozo Z, Li DL, Morales CR, Criollo A, Luo X, Tan W, Jiang N, Lehrman MA, Rothermel BA, Lee AH, Lavandero S, Mammen PP, Ferdous A, Gillette TG, Scherer PE and Hill JA. Spliced X-box binding protein 1 couples the unfolded protein response to hexosamine biosynthetic pathway. Cell. 2014;156:1179–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leong HS, Brownsey RW, Kulpa JE and Allard MF. Glycolysis and pyruvate oxidation in cardiac hypertrophy--why so unbalanced? Comp Biochem Physiol A Mol Integr Physiol. 2003;135:499–513. [DOI] [PubMed] [Google Scholar]
- 13.Heineke J and Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006;7:589–600. [DOI] [PubMed] [Google Scholar]
- 14.Frey N, McKinsey TA and Olson EN. Decoding calcium signals involved in cardiac growth and function. Nat Med. 2000;6:1221–1227. [DOI] [PubMed] [Google Scholar]
- 15.Glimcher LH. XBP1: the last two decades. Ann Rheum Dis. 2010;69 Suppl 1:i67–i71. [DOI] [PubMed] [Google Scholar]
- 16.Saxton RA and Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168:960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sciarretta S, Volpe M and Sadoshima J. Mammalian target of rapamycin signaling in cardiac physiology and disease. Circ Res. 2014;114:549–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rothermel BA, Berenji K, Tannous P, Kutschke W, Dey A, Nolan B, Yoo KD, Demetroulis E, Gimbel M, Cabuay B, Karimi M and Hill JA. Differential activation of stress-response signaling in load-induced cardiac hypertrophy and failure. Physiol Genomics. 2005;23:18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masaki T, Yoshida M and Noguchi S. Targeted disruption of CRE-binding factor TREB5 gene leads to cellular necrosis in cardiac myocytes at the embryonic stage. Biochem Biophys Res Commun. 1999;261:350–356. [DOI] [PubMed] [Google Scholar]
- 20.Zhao G, Fu Y, Cai Z, Yu F, Gong Z, Dai R, Hu Y, Zeng L, Xu Q and Kong W. Unspliced XBP1 Confers VSMC Homeostasis and Prevents Aortic Aneurysm Formation via FoxO4 Interaction. Circ Res. 2017;121:1331–1345. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Zhang Y, Ding G, May HI, Xu J, Gillette TG, Wang H and Wang ZV. Temporal dynamics of cardiac hypertrophic growth in response to pressure overload. Am J Physiol Heart Circ Physiol. 2017;313:H1119–h1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vega RB, Rothermel BA, Weinheimer CJ, Kovacs A, Naseem RH, Bassel-Duby R, Williams RS and Olson EN. Dual roles of modulatory calcineurin-interacting protein 1 in cardiac hypertrophy. Proc Natl Acad Sci U S A. 2003;100:669–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J and Ozcan U. Unfolded protein response signaling and metabolic diseases. J Biol Chem. 2014;289:1203–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Efeyan A, Comb WC and Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517:302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng Y, Wang ZV, Tao C, Gao N, Holland WL, Ferdous A, Repa JJ, Liang G, Ye J, Lehrman MA, Hill JA, Horton JD and Scherer PE. The Xbp1s/GalE axis links ER stress to postprandial hepatic metabolism. J Clin Invest. 2013;123:455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng Y, Wang ZV, Gordillo R, Zhu Y, Ali A, Zhang C, Wang X, Shao M, Zhang Z, Iyengar P, Gupta RK, Horton JD, Hill JA and Scherer PE. Adipocyte Xbp1s overexpression drives uridine production and reduces obesity. Mol Metab. 2018;11:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonner JM and Boulianne GL. Diverse structures, functions and uses of FK506 binding proteins. Cell Signal. 2017;38:97–105. [DOI] [PubMed] [Google Scholar]
- 28.Hausch F, Kozany C, Theodoropoulou M and Fabian AK. FKBPs and the Akt/mTOR pathway. Cell Cycle. 2013;12:2366–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill JA and Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–1380. [DOI] [PubMed] [Google Scholar]
- 30.Wang ZV, Rothermel BA and Hill JA. Autophagy in hypertensive heart disease. J Biol Chem. 2010;285:8509–8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ron D and Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. [DOI] [PubMed] [Google Scholar]
- 32.Glembotski CC. Endoplasmic reticulum stress in the heart. Circ Res. 2007;101:975–984. [DOI] [PubMed] [Google Scholar]
- 33.Okada K, Minamino T, Tsukamoto Y, Liao Y, Tsukamoto O, Takashima S, Hirata A, Fujita M, Nagamachi Y, Nakatani T, Yutani C, Ozawa K, Ogawa S, Tomoike H, Hori M and Kitakaze M. Prolonged endoplasmic reticulum stress in hypertrophic and failing heart after aortic constriction: possible contribution of endoplasmic reticulum stress to cardiac myocyte apoptosis. Circulation. 2004;110:705–712. [DOI] [PubMed] [Google Scholar]
- 34.Lynch JM, Maillet M, Vanhoutte D, Schloemer A, Sargent MA, Blair NS, Lynch KA, Okada T, Aronow BJ, Osinska H, Prywes R, Lorenz JN, Mori K, Lawler J, Robbins J and Molkentin JD. A thrombospondin-dependent pathway for a protective ER stress response. Cell. 2012;149:1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blackwood EA, Hofmann C, Santo Domingo M, Bilal AS, Sarakki A, Stauffer W, Arrieta A, Thuerauf DJ, Kolkhorst FW, Muller OJ, Jakobi T, Dieterich C, Katus HA, Doroudgar S and Glembotski CC. ATF6 Regulates Cardiac Hypertrophy by Transcriptional Induction of the mTORC1 Activator, Rheb. Circ Res. 2019;124:79–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH and Hotamisligil GS. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. [DOI] [PubMed] [Google Scholar]
- 37.Park SW, Zhou Y, Lee J, Lu A, Sun C, Chung J, Ueki K and Ozcan U. The regulatory subunits of PI3K, p85alpha and p85beta, interact with XBP-1 and increase its nuclear translocation. Nat Med. 2010;16:429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winnay JN, Boucher J, Mori MA, Ueki K and Kahn CR. A regulatory subunit of phosphoinositide 3-kinase increases the nuclear accumulation of X-box-binding protein-1 to modulate the unfolded protein response. Nat Med. 2010;16:438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee AH, Scapa EF, Cohen DE and Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laplante M and Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang J and Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.