Abstract
Two common genetic polymorphisms in the beta-1 adrenergic receptor (ADRB1 Ser49Gly [rs1801252] and Arg389Gly [rs1801253]) significantly affect receptor function in vitro. The objective of this study was to determine whether ADRB1 Ser49Gly and Arg389Gly are associated with recovery of left ventricular ejection fraction (LVEF) in patients with heart failure. Patients with heart failure and baseline LVEF ≤40% were genotyped (n=98), and retrospective chart review assessed the primary outcome of LVEF recovery to ≥40%. Un/adjusted logistic regression models revealed that Ser49Gly, but not Arg389Gly, was significantly associated with LVEF recovery in a dominant genetic model. The adjusted odds ratio for Ser49 was 8.2 (95% CI = 2.1 – 32.9; p = 0.003), and it was the strongest predictor of LVEF recovery among multiple clinical variables. In conclusion, patients with heart failure and reduced ejection fraction that are homozygous for ADRB1 Ser49 were significantly more likely to experience LVEF recovery than Gly49 carriers.
Keywords: beta-1 adrenergic receptor, genetics, polymorphism, beta-blocker, heart failure, left ventricular ejection fraction
Introduction
Heart failure (HF) is a prevalent health issue that currently affects approximately 6.5 million Americans, with that number expected to grow to over 8 million by 2030 [1]. While developments in medical therapy dramatically reduced heart failure mortality from 1979 to 2000, new evidence indicates that mortality improvements may be leveling off [1]. With one-year mortality still high at 29.6% and five-year mortality close to 50% [1], it is essential that improvements in HF outcomes do not stagnate. Left ventricular ejection fraction (LVEF) is a surrogate measure of clinical outcomes in patients with heart failure with reduced ejection fraction (HFrEF). Improvements in LVEF to near normal levels are associated with dramatically lower mortality rates than either persistent HFrEF or heart failure with preserved ejection fraction (HFpEF) (mortality 25% vs. 45% vs. 47%, respectively) [2]. Unfortunately, the percentage of HFrEF patients who achieve LVEF recovery is estimated to be only ≈20% [2,3]. Increasing the percentage of patients who recover LVEF has potential to dramatically improve HFrEF outcomes, but to achieve that goal we need a better understanding of the factors that predict LVEF recovery.
LVEF recovery can occur upon the removal of the inciting cardiac insult (e.g., hypertension or infection), but more often it occurs as the result of pharmacologic and non-pharmacologic therapies [4]. Effective non-pharmacologic therapy is cardiac resynchronization, and effective pharmacologic therapies are those that block neurohormonal activation (i.e., inhibitors of the sympathetic adrenergic and renin-angiotensin-aldosterone systems) [4].While many mechanisms contribute to HFrEF, it has long been known that excessive adrenergic stimulation is cardiotoxic [5]. Consistent with those effects, inhibition of the beta-1 adrenergic receptor using beta blocker therapy is strongly associated with left ventricular reverse remodeling and recovery in HFrEF patients [6,7]. However two common, non-synonymous genetic variants in the beta-1 adrenergic receptor (gene: ADRB1) have been characterized [8], and they dramatically affect beta-1 adrenergic receptor function in vitro: Ser49Gly (rs1801252) and Arg389Gly (rs1801253). The Gly49 allele increases agonist-promoted down-regulation of the receptor compared to the Ser49 allele [9], and the Gly389 allele decreases both basal and agonist-promoted beta-1 adrenergic receptor activity compared to the Arg389 allele [10]. Given that both Gly alleles decrease beta-1 adrenergic receptor expression and activity, our hypothesis was that HFrEF patients that are homozygous for Ser49 or Arg389 would benefit most from beta-blockade, and thus have improved LVEF recovery in response to optimal medical therapy. Therefore, the objective of this study was to determine whether ADRB1 Ser49Gly and/or Arg389Gly is/are associated with the recovery of LVEF in HFrEF patients treated in a HF specialty clinic.
Methods
Patient sample
This study was a retrospective chart review and genotyping of a convenience sample of 135 patients from the Heart Failure Clinic at the Ohio State University Wexner Medical Center in Columbus, Ohio, USA. Any patient with symptomatic American Heart Association/American College of Cardiology (AHA/ACC) stage C congestive heart failure [11] and willing to donate a blood sample for DNA analysis was enrolled into the overall study. The goal of the overall study was to investigate the effects of genetic variation on heart failure surrogate outcomes, regardless if the patient had HFrEF or HFpEF.Patients were enrolled from February 1999 to September 1999. Only patients with a documented baseline LVEF < 40%, known ADRB1 genotypes, and a documented follow-up LVEF at least 90 days since the baseline LVEF were included in this analysis. Previous work by our group identified five clinical variables significantly associated with higher rates of LVEF recovery: female sex, non-ischemic etiology, non-diabetics, increased systolic blood pressure, and decreased QRS duration [12]. Thus we collected as much of the data as possible for those five variables, plus the patients’ baseline age, self-reported race, body mass index (BMI), heart rate, serum creatinine, chronic kidney disease, atrial fibrillation, New York Heart Association (NYHA) functional class, left bundle branch block, and current vital status. If the patient did not have a diagnosis of chronic kidney disease anywhere in their electronic medical record at baseline, then it was recorded as stage 0. LVEFs were collected from echocardiograms performed as part of routine clinical care. The most extreme LVEFs for each patient were collected, i.e., the lowest LVEF in the electronic medical record was used as the baseline LVEF, and then the highest LVEF in the electronic medical record at least 90 days after the baseline LVEF was used as the follow-up LVEF. Clinical variables were obtained to the extent available through existing electronic medical records. The investigators that performed the retrospective chart review were blinded to the patients’ ADRB1 genotypes. Given that the patients were enrolled nearly 20 years ago, outpatient medication records were not available as part of the retrospective chart review. However the goal of the Heart Failure Clinic is to treat HFrEF patients with the optimal AHA/ACC guideline-recommended medical therapy, including beta-blockers and angiotensin converting enzyme inhibitors (or angiotensin receptor blockers), titrated to target doses [11]. All patients signed informed consent for the collection of DNA via a blood sample and tabulation of clinical variables from electronic medical records. This study was performed in accordance with the Institutional Review Board of The Ohio State University, which reviewed and approved this protocol.
Genotyping
Whole blood samples were stored at −80°C until the DNA was extracted using standard techniques [13]. ADRB1 Ser49Gly (rs1801252) genotype was determined using TaqMan® Genotyping Assay C___8898508_10 (ThermoFisher Scientific), and ADRB1 Arg389Gly (rs1801253) genotype was determined using TaqMan® Genotyping Assay C___8898494_10 (ThermoFisher Scientific). Several quality control measures were used to ensure accurate genotyping. Negative and positive controls were run with each 96-well plate. Negative controls used molecular grade water instead of a DNA sample, and the positive controls were DNA samples derived from the Centre d’Etude du Polymorphism Humain (CEPH) lymphoblastoid cell lines from the Coriell Institute for Medical Research (Camden, New Jersey, USA). Two samples of each genotype (A/A, A/G, and G/G for Ser49Gly and C/C, C/G, and G/G for Arg389Gly) were used as positive controls. After initial genotyping, a randomly selected 10% of patient samples were re-genotyped to confirm concordance. Investigators performing the genotyping were blinded to the clinical data collected via retrospective chart review.
Statistical analysis
Continuous baseline variables were described by the median ± interquartile range, and categorical baseline variables were described by counts and percentages. Baseline variables were described in all patients and stratified by the presence/absence of LVEF recovery, ADRB1 Ser49Gly genotype, and ADRB1 Arg389Gly genotype. LVEF recovery, the primary outcome, was defined as a documented follow-up LVEF ≥ 40% at least 90 days since the baseline LVEF. ADRB1 genotypes were analyzed using the dominant genetic model (i.e., Ser49 homozygotes versus Gly49 carriers and Arg389 homozygotes versus Gly389 carriers). The Mann-Whitney U test was used to compare continuous baseline variables between all strata, and the chi-square test (or when necessary, the Fisher’s exact test) was used to compare the categorical baseline variables between all strata. Unadjusted and adjusted logistic regression models were used to test the association of each ADRB1 genotype with the primary outcome of LVEF recovery. The primary model was adjusted for the five clinical variables previously identified by our group to be significantly associated with LVEF recovery [12]. Variables with >15% missing values had the missing values imputed as the mean of the available values. Imputation was necessary for three clinical covariates: QRS duration, ischemic etiology, and NYHA functional class were missing in 33%, 41%, and 35% of the sample at baseline (imputed as 117, 0.172, and 2.8 respectively). Ischemic/not ischemic was imputed as a numerical variable in the model. Specifically, for the patients in which non-ischemic and ischemic status could be verified, they were assigned values of 0 and 1, respectively. From those patients with known values, the mean was 0.172 (corresponding with a mean overall frequency of ischemic etiology = 17.2%). Therefore, for the patients with the unverifiable non/ischemic status, they were assigned values of 0.172, which would be numerically similar as the probability of having ischemic etiology in this patient sample. To ensure that imputation did not confound the results, we ran similarly adjusted models omitting the imputed variables as covariates. Our most conservative model included the five previously identified clinical covariates plus the patients’ age, self-reported race, and NYHA functional class. Hardy Weinberg equilibrium was tested using the chi-square test stratified by self-reported race. Statistical significance was defined as p < 0.05. Given the analytical sample size; the minor allele frequencies of ADRB1 Ser49Gly and Arg389Gly; and an a priori expected LVEF recovery rate of 25%; we estimated that we had 80% power to detect an odds ratio = 5 for Ser49Gly and an odds ratio = 4 for Arg389Gly for the primary outcome in univariate analysis. All statistical analyses were performed using SAS version 9.4 (Cary, NC).
Results
Of the 135 patients enrolled, 98 qualified for analysis. The most common reasons that patients were excluded from this analysis were that patients had a baseline LVEF > 40% (n = 23), or patients did not have a follow-up LVEF documented in their electronic medical record a least 90 days since the baseline LVEF (n = 13). Baseline characteristics overall and stratified by LVEF recovery are displayed in Table 1. A total of 36 (37%) patients had LVEF recovery to ≥ 40% Overall, the patients were primarily self-reported whites (80%; 19% self-reported blacks) and male (71%) with a median and interquartile range BMI of 29 ± 9 kg/m2 and heart rate 80 ± 13 bpm. The median age of the patients was 50 ± 17 years at baseline, with roughly half having diabetes (46%), 36% had atrial fibrillation, 37% had left bundle branch block, and 17% had ischemic etiology for their heart failure. The mean NYHA class was 2.8. The median baseline EF was 20 ± 11%, with the median and interquartile range for the change in LVEF being +2% ± 34%. The median and interquartile range for the length of time between the baseline and follow-up LVEFs was 4.4 ± 7.2 years, and the median follow-up LVEF value was 25% ± 36%. The only statistically significant differences between the patients that did and did not have LVEF recovery were female sex (p = 0.029) and ADRB1 Ser49Gly genotype (p = 0.002). Nearly twice as many women had LVEF recovery compared to men (54% vs. 30%, respectively). Approximately three times as many ADRB1 Ser49 homozygotes had LVEF recovery compared to ADRB1 Gly49 carriers (46% vs. 14%, respectively). Numerically more ADRB1 Gly389 carriers in the group recovered LVEF (61% vs 45%), but the difference was not statistically significant (p = 0.098). Consistent with the previous findings by our group [12], systolic blood pressure was numerically higher and QRS duration was numerically lower in patients with LVEF recovery, but those differences were not statistically significant.
Table 1.
Baseline characteristics in all patients and stratified by LVEF recovery status
| Overall (n=98) | LVEF Recovered (n=36) 36.7% | LVEF Not recovered (n=62) 63.3% | *p | |
|---|---|---|---|---|
| Age (years) | 49.5 ± 17.0 | 47.0 ± 13.0 | 50.0 ± 18.0 | 0.463 |
| Female sex | 28 (28.6%) | 15 (41.7%) | 13 (21.0%) | 0.029 |
| Self-reported whites | 78 (79.6%) | 29 (80.6%) | 49 (79.0%) | 1.000 |
| Self-reported blacks | 19 (19.4%) | 7 (19.4%) | 12 (19.4%) | |
| Ischemic Etiology | 10 (17.2%) | 3 (13.6%) | 7 (19.4%) | 0.727 |
| Diabetes | 45 (46.4%) | 19 (52.8%) | 26 (42.6%) | 0.333 |
| Atrial fibrillation | 35 (35.7%) | 15 (41.7%) | 20 (32.3%) | 0.349 |
| †Stage of chronic kidney disease | 1.0 ± 1.5 | 1.0 ± 1.3 | 1.0 ± 1.6 | 0.873 |
| Serum creatinine (mg/dL) | 1.1 ± 0.4 | 1.0 ± 0.4 | 1.1 ± 0.4 | 0.435 |
| BMI (kg/m2) | 29.0 ± 9.2 | 29.7 ± 10.5 | 28.1 ± 10.3 | 0.511 |
| SBP (mmHg) | 118.0 ± 20.0 | 120.0 ± 22.0 | 116.6 ± 19.0 | 0.084 |
| Heart Rate (bpm) | 80.0 ± 13.0 | 80.0 ± 13.3 | 80.0 ± 15.0 | 0.944 |
| QRS Duration (ms) | 110.0 ± 48.0 | 98.0 ± 26.0 | 114.0 ± 46.0 | 0.139 |
| †NYHA functional class | 2.8 ± 0.9 | 2.5 ± 0.9 | 3.0 ± 0.8 | 0.063 |
| Left bundle branch block | 35 (36.5%) | 11 (30.6%) | 24 (40.0%) | 0.352 |
| Baseline LVEF (%) | 20 ± 11 | 20 ± 9 | 20 ± 11 | 0.953 |
| Follow-up LVEF (%) | 25 ± 36 | 55 ± 8 | 20 ± 12 | <.0001 |
| Change in LVEF (%) | 2 ± 34 | 35 ± 13 | -3 ± 9 | <.0001 |
| Time between baseline and follow-up LVEFs (years) | 4.4 ± 7.2 | 4.1 ± 5.3 | 4.5 ± 8.0 | 0.565 |
| ADRB1 Gly49 carriers | 29 (29.6%) | 4 (11.1%) | 25 (40.3%) | 0.002 |
| ADRB1 Gly389 carriers | 46 (48.9%) | 21 (60.0%) | 25 (42.4%) | 0.098 |
| Deaths | 53 (54.1%) | 20 (55.6%) | 33 (53.2%) | 0.823 |
ADRB1 = gene for the beta-1 adrenergic receptor; BMI = body mass index; Gly = glycine; LVEF = left ventricular ejection fraction; NYHA = New York Heart Association functional class; SBP = systolic blood pressure; Ser = serine
p-value for patients with LVEF recovery to ≥ 40% versus patients with follow-up EF < 40%. Bolded p-values indicate statistical significance
Reported as mean ± sd instead of median ± interquartile range because the medians were identical and thus did not reveal subtle differences in the values
Genotyping results of the patient sample for ADRB1 Ser49Gly (rs1801252) revealed 69 A/A homozygotes, 27 A/G heterozygotes and 2 G/G homozygotes; hence 29 (30%) were ADRB1 Gly49 carriers. The genotyping call rate was 100%, and the genotype distributions were consistent with Hardy-Weinberg equilibrium within the self-reported white and black race groups (p = 0.998 and p = 0.414, respectively). The frequency of the Gly49 allele in this patient sample was 0.16 for the self-reported whites (n = 78) and 0.16 for the self-reported blacks (n = 19). These values are similar to previously reported allele frequencies for individuals with European (0.12) and African (0.17) ancestry residing in the United States [14]. Genotyping results of the patient sample for ADRB1 Arg389Gly (rs1801253) revealed 48 A/A homozygotes, 37 A/G heterozygotes and 9 G/G homozygotes; hence n = 46 (49%) were ADRB1 Gly389 carriers. The genotyping call rate for Arg389Gly was 96%, and the genotype distributions were consistent with Hardy-Weinberg equilibrium within the self-reported white and black race groups (p = 0.178 and p = 0.120, respectively). The frequency of the Gly389 allele in this patient sample was 0.28 for the self-reported whites (n = 74) and 0.37 for the self-reported blacks (n = 19). These values are similar to previously reported allele frequencies for individuals with European (0.33) and African (0.37) ancestry residing in the United States. [14] There was 100% concordance between the observed and reported genotypes for the positive controls and the 10% of samples that were randomly re-genotyped for both ADRB1 polymorphisms. These results support accurate genotyping.
Baseline characteristics overall and stratified by ADRB1 Ser49Gly genotype are displayed in Supplementary Table 1. The only statistically significant differences between the two genotype groups were the follow-up LVEF (35% in the ADRB1 Ser49 homozygotes vs. 20% in the ADRB1 Gly49 carriers; p = 0.002); change in LVEF (+6% in the ADRB1 Ser49 homozygotes vs. −3% in the ADRB1 Gly49 carriers; p = 0.004); and the percentage of patients with LVEF recovery to > 40% (46% in the ADRB1 Ser49 homozygotes vs. 14% in the ADRB1 Gly49 carriers p = 0.002). The difference in NYHA class at baseline was nearly significant between the Ser49 homozygotes and Gly49 carriers (2.7 vs. 3.1, respectively; p = 0.051). Notably, baseline LVEF, demographics, comorbidities, and vital signs were similar between ADRB1 Ser49 homozygotes and ADRB1 Gly49 carriers. Baseline characteristics overall and stratified by ADRB1 Arg389Gly genotype are displayed in Supplementary Table 2. The only statistically significant difference between ADRB1 Arg389 homozygotes and ADRB1 Gly389 carriers was the duration of time between the baseline and follow-up LVEF measurements. The duration of time was significantly longer in the ADRB1 Arg389 homozygotes than in the ADRB1 Gly389 carriers (median 6.8 vs. 2.6 years, respectively; p = 0.002).
Unadjusted and adjusted logistic regression models for ADRB1 Ser49Gly are displayed in Table 2. Univariate logistic regression found that patients who were homozygous for Ser49 were more than 5 times more likely to recover their LVEF to ≥ 40% than those who were carriers of Gly49 (odds ratio = 5.4; 95% confidence interval = [1.7 - 17.2]; p= 0.004). Regardless of the number or types of clinical covariates used to adjust the logistic regression models, Ser49Gly was significantly associated with LVEF recovery. The most conservative model adjusted for eight baseline clinical characteristics: patients’ age, self-reported race, sex, systolic blood pressure, QRS duration, diabetes, ischemic etiology, and NYHA class. Despite this rigorous covariate adjustment, ADRB1 Ser49 homozygous genotype was an independent, statistically significant, and strong factor associated with LVEF recovery (adjusted odds ratio = 8.2; CI= [2.1, 32.9]; p= 0.003). The results were similar even when only patients with complete data for non/ischemic etiology were included (total n = 54). Table 3 shows the results for all variables in the most conservatively adjusted multivariable model. ADRB1 Ser49 homozygous genotype and female sex remained statistically significant, and Ser49Gly was by far the strongest predictor of LVEF recovery. Supplemental Table 3 shows a similar model, but it only includes the patients that had complete data for non/ischemic etiology.
Table 2.
Unadjusted and adjusted logistic regression models for the association of ADRB1 Ser49 homozygous status with recovery of LVEF to ≥ 40%.
| Logistic Regression Model | n | OR (for Ser49 homozygotes) |
95% CI | *p |
|---|---|---|---|---|
| Univariate | 98 | 5.4 | 1.7 – 17.2 | 0.004 |
| Adjusted for age, sex, and race | 97 | 6.8 | 2.0 – 23.4 | 0.002 |
| Adjusted for sex, SBP, diabetes | 89 | 6.9 | 1.9 – 25.0 | 0.003 |
| Adjusted for age, sex, SBP, diabetes, QRS duration,a and ischemic etiologya | 89 | 8.4 | 2.2 – 32.2 | 0.002 |
| Adjusted for age, sex, race, SBP and diabetes | 89 | 6.9 | 1.9 – 25.2 | 0.004 |
| Adjusted for age, race, sex, SBP, diabetes, QRS duration,a NYHA class,a and ischemic etiologya | 89 | 8.2 | 2.1 – 32.9 | 0.003 |
| Adjusted for age, race, sex, SBP, diabetes, QRS duration,a NYHA class,a and ischemic etiologyb | 54 | 10.9 | 1.3 – 89.5 | 0.026 |
ADRB1 = gene for the beta-1 adrenergic receptor; CI = confidence interval; Gly = glycine; LVEF = left ventricular ejection fraction; NYHA = New York Heart Association; OR = odds ratio; SBP = systolic blood pressure; Ser = serine
Mean values were used to impute missing data for n = 32 missing QRS duration, n = 40 missing ischemic etiology, and n = 34 missing NYHA functional class.
This model did not include patients that had non/ischemic etiology imputed. It only included patients in which non/ischemic etiology data was available.
p-value for ADRB1 Ser49 homozygotes versus Gly49 carriers within the specified model. Bolded p-values indicate statistical significance
Table 3.
Full logistic regression model including all clinical covariates and ADRB1 Ser49 homozygous status for the primary outcome of LVEF recovery to ≥ 40%.
| Variable | OR | 95% CI | *p |
|---|---|---|---|
| ADRB1 Ser49 homozygotes | 8.17 | 2.07 – 32.22 | 0.003 |
| Age | 1.00 | 0.96 – 1.04 | 0.991 |
| Female sex | 4.07 | 1.27 – 13.03 | 0.018 |
| African-American race | 1.01 | 0.29 – 3.51 | 0.983 |
| Systolic blood pressure | 1.03 | 0.99 – 1.06 | 0.074 |
| Non-diabetic | 0.84 | 0.28 – 2.51 | 0.757 |
| aQRS duration | 0.98 | 0.96 – 1.01 | 0.122 |
| aIschemic etiology | 0.93 | 0.17 – 5.10 | 0.933 |
| aNYHA functional class | 0.69 | 0.34 – 1.39 | 0.296 |
ADRB1 = gene for the beta-1 adrenergic receptor; CI = confidence interval; Gly = glycine; LVEF = left ventricular ejection fraction; OR = odds ratio; SBP = systolic blood pressure; Ser = serine
Mean values were used to impute missing data for n = 32 missing QRS duration, n = 40 missing ischemic etiology, and n = 34 missing NYHA functional class.
p-value is for type III analysis of effects; bolded values indicate statistical significance (p < 0.05)
Unadjusted and adjusted logistic regression models for ADRB1 Arg389Gly are displayed in Table 4. None of the models for ADRB1 Arg389Gly were statistically significant for an association with LVEF recovery (all p > 0.05). The model only adjusted for patient age, sex, and race trended toward statistical significance (odds ratio = 0.42; 95% confidence interval = 0.17 - 1.1; p = 0.063). However the estimated odds ratio for that model and all other models for Arg389Gly were in the opposite direction than what would be expected based on the previous literature [10]. The distributions for the change in LVEF in patients stratified by ADRB1 Ser49Gly and Arg389Gly genotypes are displayed in Figures 1 and 2. The distribution for Ser49Gly more clearly shows a shift toward improved LVEF recovery for Ser49 homozygotes than the distribution for Arg389Gly.
Table 4.
Unadjusted and adjusted logistic regression models for the association of ADRB1 Arg389 homozygous status with recovery of LVEF to ≥ 40%.
| Logistic Regression Model | n | OR (for Arg389 homozygotes) |
95% CI | *p |
|---|---|---|---|---|
| Univariate | 94 | 0.49 | 0.21 - 1.2 | 0.101 |
| Adjusted for age, sex, and race | 93 | 0.42 | 0.17 - 1.1 | 0.063 |
| Adjusted for sex, SBP, diabetes | 86 | 0.49 | 0.19 - 1.3 | 0.142 |
| Adjusted for age, sex, SBP, diabetes, QRS duration, and ischemic etiologya | 86 | 0.55 | 0.20 - 1.5 | 0.230 |
| Adjusted for age, sex, race, SBP and diabetes | 85 | 0.48 | 0.18 - 1.3 | 0.148 |
| Adjusted for age, race, sex, SBP, diabetes, QRS duration, NYHA class, and ischemic etiologya | 85 | 0.55 | 0.19 - 1.5 | 0.251 |
| Adjusted for age, race, sex, SBP, diabetes, QRS duration,a NYHA class,a and ischemic etiologyb | 51 | 0.55 | 0.13 – 2.2 | 0.404 |
ADRB1 = gene for the beta-1 adrenergic receptor; Arg = Arginine; CI = confidence interval; Gly = glycine; LVEF = left ventricular ejection fraction; OR = odds ratio; SBP = systolic blood pressure
Mean values were used to impute missing data for n = 31 missing QRS duration, n = 39 missing ischemic etiology, and n = 34 missing NYHA functional class.
This model did not include patients that had non/ischemic etiology imputed. It only included patients in which non/ischemic etiology data was available.
p-value for ADRB1 Arg389Gly genotype within the specified model. Bolded p-values indicate statistical significance
Fig.1.
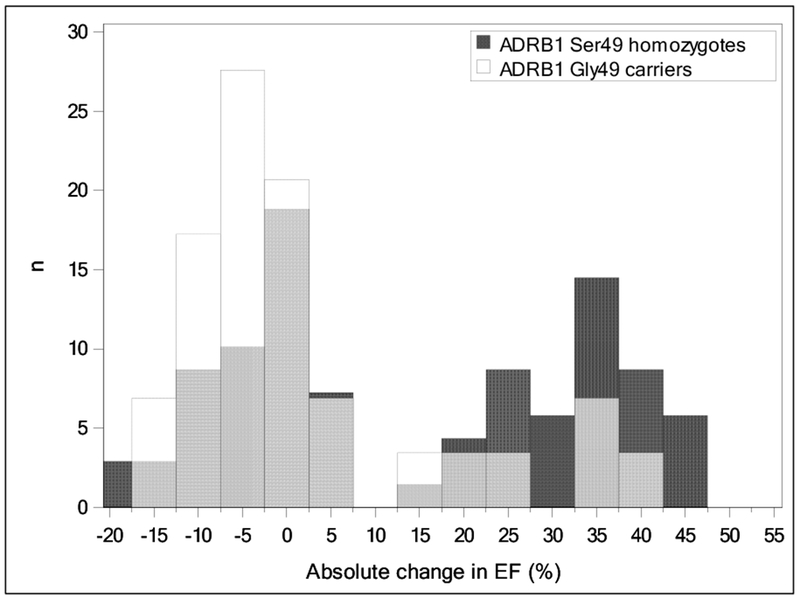
Overlaid histograms of absolute change in ejection fraction (EF) for ADRB1 Ser49 homozygotes (dark gray columns) vs Gly49 carriers (white columns). Light gray columns indicate overlapping data.
Fig.2.
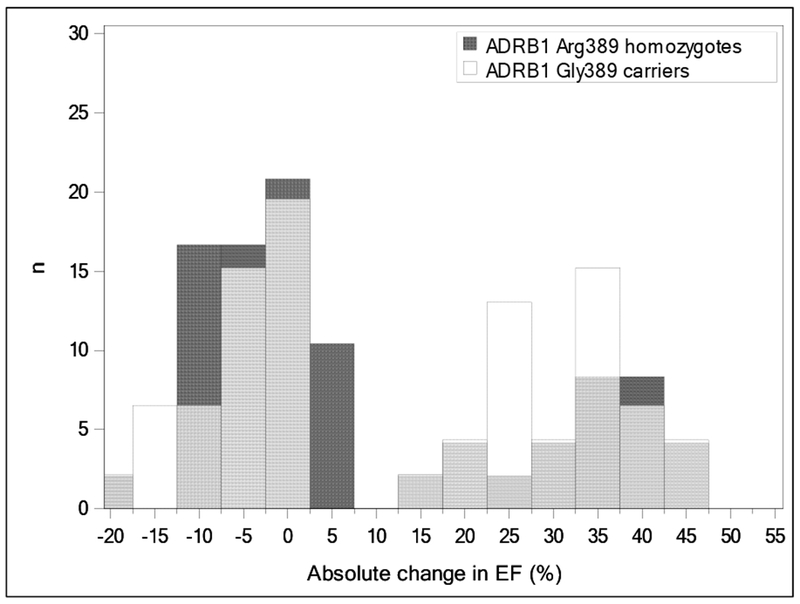
Overlaid histograms of absolute change in ejection fraction (EF) for ADRB1 Arg389 homozygotes (dark gray columns) vs Arg389 carriers (white columns). Light gray columns indicate overlapping data.
Discussion
The aim of the present study was to investigate the association between ADRB1 Ser49Gly and Arg389Gly with recovery of LVEF in patients with HFrEF treated in a HF specialty clinic. Homozygous ADRB1 Ser49, but not Arg389, was significantly associated with LVEF recovery. The association of ADRB1 Ser49Gly with LVEF recovery was independent of rigorous adjustment for multiple clinical variables, including those previously shown to be associated with recovery of ventricular function [12], and it was by far the strongest predictor of LVEF recovery among all variables assessed. Patients homozygous for the major allele Ser49 had significantly higher rates of LVEF recovery than Gly49 carriers. Our findings are consistent with prior in vitro data and studies assessing survival outcomes from beta-blocker treatment in patients with heart failure [15,16].
Cell line data has shown that beta-1 adrenergic receptors with the Gly49 allele undergo increased down-regulation when exposed to long-term stimulation compared to receptors with Ser49 [9]. This result is supported by another cell line study by Levin et al, which found increased catecholamine-induced desensitization in cells carrying the Gly49 allele [17]. As excessive adrenergic stimulation has been associated with cardiotoxicity [5], down-regulation of the beta-1 adrenergic receptor may be cardioprotective. This supports the hypothesis that Gly49 is a beneficial adaptation in patients with HF, and that patients without the Gly49 allele (Ser49 homozygotes) may benefit more from treatment with beta-blockers; a treatment expected to have been implemented in a HF specialty clinic.
Four previous studies did not find a significant association of ADRB1 Ser49Gly with ventricular remodeling responses to beta-blockers [18,19,20,21]. The major difference between our study and those four previous studies is the duration of follow-up. The median length of time between the baseline and follow-up LVEF measurements in our study was 4.4 years, whereas the longest length of follow-up in any of the previous studies was 1.6 years; with most of the studies having following up of several months. Despite using only a 90 day minimum between the baseline and follow-up LVEFs in our study, our results may still represent persistent LVEF recovery because of the long duration of time observed between the extreme LVEF measurements. Specifically, we selected the lowest documented LVEF as the baseline LVEF, and the highest documented LVEF at least 90 days since the baseline LVEF as the follow-up LVEF. By selecting the highest LVEF documented, we collected the maximum observed effect that resulted from a myriad of different therapeutic strategies used to treat HFrEF in a HF specialty clinic. Therefore the effects of Ser49Gly on LVEF recovery may be long-term effects and have gone undetected in previous studies. This would explain why the results of our study are consistent with the results of survival outcome studies, which also had a few years of follow-up [15, 16].
Four previous studies found that ADRB1 Arg389Gly was significantly associated with ventricular remodeling responses to beta-blockers [18,21,22,23], but consistent with our study, three previous studies also did not find a significant association of ADRB1 Arg389Gly [19,24,25]. Therefore the true association of ADRB1 Arg389Gly with left ventricular remodeling responses to beta-blockers is not clear. All of these studies, including our own, were small (each n < 500). Left ventricular remodeling is only a surrogate for clinical outcomes in patients with HFrEF, and thus the reasons for the discordant results between these ventricular remodeling studies and clinical outcomes studies discussed below are not clear. The effects of these genetic polymorphisms on survival responses to beta-blockers could be through additional mechanisms than just ventricular remodeling. For example, the beta-1 adrenergic receptor is not only expressed in the heart, but also in the kidney, where it affects the release of renin. The renin-angiotensin-aldosterone system is a critical system in the neurohormonal pathophysiology of heart failure.
Our findings are consistent with previous studies investigating clinical outcome responses to beta-blockers in patients with heart failure (i.e., survival benefit). In retrospective (n = 184) and prospective (n = 190) studies of Swedish patients with idiopathic dilated cardiomyopathy, patients homozygous for Ser49 gained more survival benefit from beta-blockers than patients carrying Gly49 [26,27]. In a prospective, multicenter heart failure patient registry in the U.S. (total n = 715), beta-blocker use was associated with reduced mortality in the overall study population (adjusted HR = 0.72, 95% CI 0.56 - 0.91, p = 0.004), and Ser49 homozygotes (adjusted HR = 0.56, 95% CI 0.42 - 0.75, p < 0.001), but not Gly49 carriers (adjusted HR = 1.28, 95% CI 0.80 - 2.04, p = 0.31) [16]. In a subsequent and independent prospective heart failure registry at a single center in the U.S. (n = 822), only Ser49 homozygotes had a significant reduction in the risk for mortality from beta-blockers (adjusted HR = 0.41, p = 0.022 in Ser49 homozygotes; adjusted HR = 0.88, p = 0.81 in Gly49 carriers). All of the patients in these studies were treated with FDA-approved beta-blockers.
The survival data for ADRB1 Arg389Gly also supports our findings, in that Arg389Gly may not have a significant effect on beta-blocker response, albeit for FDA-approved beta-blockers. Two prospective heart failure registries [28,29] and a genetic analysis of a landmark beta-blocker randomized controlled trial [30] did not find a significant association of Arg389Gly with survival benefit from FDA-approved beta-blockers. The strongest evidence to support an effect of Arg389Gly on beta-blocker survival benefit comes from a genetic analysis (n = 1,040) of the Beta-Blocker Evaluation of Survival Trial (BEST); the landmark clinical trial assessing the effectiveness of bucindolol on mortality in patients with heart failure [10,31]. Patients homozygous for Arg389 had a statistically significant improvement in survival compared with placebo (HR = 0.62; p = 0.03), whereas Gly389 carriers did not (HR = 0.90; p = 0.57). Bucindolol has pharmacologic effects that are unique compared to FDA-approved beta-blockers, in that it causes exaggerated sympatholysis (i.e., decreases in norepinephrine levels) [10]. Therefore bucindolol may be affected by ADRB1 Ser49Gly and Arg389Gly in ways that are unique from FDA-approved beta-blockers without sympatholytic effects.
This study has several limitations. Our study was small (n = 98 total). Only n = 29 patients were Gly49 carriers, and only n = 4 of the Gly49 carriers experienced the primary outcome of LVEF recovery. Therefore the results could be due to chance. However the low p-value for LVEF recovery within the Gly49 carriers (p = 0.003) indicates that our results are unlikely due to chance, and more likely represent a significantly lower LVEF recovery rate in the Gly49 carriers than in Ser49 homozygotes. Despite the small sample size, the long duration of follow-up between LVEF measurements (4.4 years) still provides a large number of patient-years, especially compared to most previous studies in which the length of follow-up was only several months [18,19,20,21]. Our study was underpowered to detect the significance of the many previously identified clinical predictors of LVEF recovery and mortality rates [12,32]. However, it is worth noting that our most conservative model identified sex as being an independent predictor of LVEF recovery in addition to Ser49Gly. QRS duration and systolic blood pressure did not reach, but were trending toward, statistical significance (p-values of 0.017, 0.125, and 0.070 respectively). Conversely, our study did not identify an association for diabetes, ischemic etiology, atrial fibrillation, or left bundle branch block. Subjects in this study were selected as a convenience sample from a single site, creating the possibility of selection bias and non-generalizability of results to outside sites. In addition, DNA collection occurred nearly 20 years ago, complicating data collection from electronic medical records. This was particularly evident with ischemic etiology, QRS interval duration data, and NYHA class, which were missing many values. Thus our models either excluded those variables or relied on imputation of missing data, which could incorporate bias. Variables of potential importance that we were unable to retrieve from the electronic medical records were glomerular filtration rate, right ventricular ejection fraction, and duration of heart failure.
Outpatient medication records were not available within the time frame of the study, and thus we were not able to verify that patients were treated with optimal medical therapy. We can only assume that because the patients were managed in a heart failure specialty clinic at a major academic medical center that they were treated with (or at least attempted to be treated with) beta-blockers effective for HFrEF and at target doses. However given that the patients were enrolled in 1999, and the landmark beta-blocker clinical trials were published around that same time, we cannot assume widespread beta-blocker use had occurred yet. However it may be reasonable to assume that beta-blocker treatment was similar between the Ser49 homozygotes and the Gly49 carriers. First, the providers were unaware of the patients’ Ser49Gly genotypes. Second, patient characteristics that would limit beta-blocker dose titration were not significantly different between the genotype groups at baseline. Specifically, mean systolic blood pressure, heart rate, age, and BMI were all similar, if not identical, between the Ser49 homozygotes and Gly49 carriers.
Conclusion
ADRB1 Ser49Gly, but not Arg389Gly, was significantly associated with recovery of LVEF in patients with HFrEF. HFrEF patients that were homozygous for Ser49 had significantly more LVEF recovery than Gly49 carriers. ADRB1 Ser49 homozygous status was by far the strongest predictor of LVEF recovery among several clinical variables assessed. These data suggest that genetic factors may be the strongest determinant of recovery of ventricular performance beyond recognized clinical variables. The findings point to the need for further research into genetic determinants of recovery of ventricular function, which may constitute therapeutic targets that will promote normalization of ventricular function in an increasing proportion of patients with HFrEF.
Supplementary Material
Clinical Relevance.
These findings from a small study at a single site need to be replicated in a larger study of multiple sites to be clinically relevant. The potential clinical relevance is that a patient’s ADRB1 Ser49Gly (rs1801252) genotype may be a predictor of whether or not the patient with HFrEF will have recovery of their LVEF to ≥ 40%. This genotype was the strongest predictor of LVEF recovery, even compared to multiple clinical variables. Thus, if validated, HFrEF patients that are ADRB1 Gly49 carriers may need additional monitoring or therapies compared to Ser49 homozygotes.
Sources of Funding
J.A.L. was supported by a Post-Doctoral Fellowship from the American Heart Association (14POST20100054), the NIH student loan repayment program (L30 HL110279), and a New Investigator Award from the American Association of Colleges of Pharmacy. J.D.E. was supported by the Michigan Institute for Clinical and Health Research (MICHR) Summer Immersion Program (grant UL1TR002240 from the National Center for Advancing Translational Sciences [NCATS]).
Abbreviations
- ADRB1
gene for the beta-1 adrenergic receptor
- AHA
American Heart Association
- ACC
American College of Cardiology
- Arg
arginine
- BMI
body mass index
- CEPH
Centre d’Etude du Polymorphism Humain lymphoblastoid cell lines
- CI
confidence interval
- FDA
United States Food and Drug Administration
- Gly
glycine
- HF
heart failure
- HFpEF
heart failure with preserved ejection fraction
- HFrEF
heart failure with reduced ejection fraction
- LVEF
left ventricular ejection fraction
- NYHA
New York Heart Association
- OR
odds ratio
- SBP
systolic blood pressure
- Ser
serine
Footnotes
Dedication
In sorrow, we would like to dedicate this work to our friend and co-author, Joseph P. Kitzmiller. Dr. Kitzmiller passed away on October 3, 2018; shortly before our submission of this paper.
Disclosures
None
Human Subjects/Informed Consent Statement
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000. Informed consent was obtained from all patients for being included in the study.
Animal Studies
No animal studies were carried out by the authors for this article.
References
- 1.Benjamin EJ, Virani SS, Callaway CW, Chamberlain AM, Chang AR, Cheng S, et al. (2018). Heart Disease and Stroke Statistics-2018 Update: A Report From the American Heart Association (review-article). Circulation, 137(12), e67–e492. doi: 10.1161/CIR.0000000000000558. [DOI] [PubMed] [Google Scholar]
- 2.Lupón J, Díez-López C, de Antonio M, Domingo M, Zamora E, Moliner P, et al. (2017). Recovered heart failure with reduced ejection fraction and outcomes: a prospective study. European Journal of Heart Failure. 19(12), 1615–1623. doi: 10.1002/ejhf.824. [DOI] [PubMed] [Google Scholar]
- 3.Teeter WA, Thibodeau JT, Rao K, Brickner ME, Toto KH, Nelson LL, et al. (2012). The natural history of new-onset heart failure with a severely depressed left ventricular ejection fraction: Implications for timing of implantable cardioverter-defibrillator implantation. American Heart Journal, 164(3), 358–364. doi: 10.1016/j.ahj.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Merlo M, Caiffa T, Gobbo M, Adamo L, & Sinagra G (2018). Reverse remodeling in Dilated Cardiomyopathy: Insights and future perspectives. International journal of cardiology. Heart & vasculature, 18, 52–57. doi: 10.1016/j.ijcha.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mann D, Kent R, Parsons B, & Cooper G 85 (1992) Adrenergic Effects on the Biology of the Adult Mammalian Cardiocyte. Circulation, 85(2),790–804. doi: 10.1161/01.CIR.85.2.790 [DOI] [PubMed] [Google Scholar]
- 6.MERIT-HF Study Group. (1999). Effect of metoprolol CR/XL in chronic heart failure: Metoprolol CR/XL Randomised Intervention Trial in-Congestive Heart Failure (MERIT-HF). The Lancet, 353(9169), 2001–2007. doi: 10.1016/S0140-6736(99)04440-2. [DOI] [PubMed] [Google Scholar]
- 7.Hernandez AF, Hammill BG, O’Connor CM, Schulman KA, Curtis LH, & Fonarow GC (2009). Clinical effectiveness of beta-blockers in heart failure: findings from the OPTIMIZE-HF (Organized Program to Initiate Lifesaving Treatment in Hospitalized Patients with Heart Failure) Registry. Journal of the American College of Cardiology, 53(2), 184. doi: 10.1016/j.jacc.2008.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Podlowski S, Wenzel K, Luther HP, Muller J, Bramlage P, Baumann G, et al. (2000). β1-Adrenoceptor gene variations: a role in idiopathic dilated cardiomyopathy? Journal of Molecular Medicine, 78(2), 87–93. doi: 10.1007/s001090000080. [DOI] [PubMed] [Google Scholar]
- 9.Rathz DA, Brown KM, Kramer LA, & Liggett SB (2002). Amino Acid 49 Polymorphisms of the Human [beta] 1-Adrenergic Receptor Affect Agonist-Promoted Trafficking. Journal of Cardiovascular Pharmacology. 39(2), 155–60. [DOI] [PubMed] [Google Scholar]
- 10.Liggett SB, Mialet-Perez J, Thaneemit-Chen S, Weber SA, Greene SM, Hodne D, et al. (2006). A polymorphism within a conserved beta1-adrenergic receptor motif alters cardiac function and beta-blocker response in human heart failure. Proceedings of the National Academy of Sciences, 103(30), 11288–11293. doi: 10.1073/pnas.0509937103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yancy CW, Jessup M, Bozkurt B, Butler J, Casey DE, Drazner MH, et al. (2013). 2013 ACCF/AHA Guideline for the Management of Heart Failure: A Report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Journal of the American College of Cardiology. 62(16), e147–e239. doi: 10.1016/j.jacc.2013.05.019 [DOI] [PubMed] [Google Scholar]
- 12.Binkley PF, Lesinski A, Ferguson JP, Hatton PS, Yamokoski L, Hardikar S, et al. (2008). Recovery of normal ventricular function in patients with dilated cardiomyopathy: Predictors of an increasingly prevalent clinical event. American Heart Journal, 155(1), 69–74. doi: 10.1016/j.ahj.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Miller SA, Dykes DD, & Polesky HF (1988). A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic acids research, 16(3), 1215–1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.1000 Genomes Project, C. (2015). A global reference for human genetic variation (Article). Nature, 526, 68. doi: 10.1038/nature15393 https://www.nature.com/articles/nature15393#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanfear DE, Peterson EL, Zeld N, Wells K, Sabbah HN, & Williams K (2015). Beta Blocker Survival Benefit in Heart Failure is Associated with ADRB1 Ser49Gly Genotype. Journal of Cardiac Failure, 21(8, Supplement), S50. doi: 10.1016/j.cardfail.2015.06.169. [DOI] [Google Scholar]
- 16.Talameh J, Garrand A, Ghali J, Oren RM, Dunlap S, Bakel AV, et al. (2012). Beta-1 adrenergic receptor genotype Ser49Gly is associated with beta-blocker survival benefit in patients with heart failure. Journal of the American College of Cardiology, 59(13, Supplement), E861. doi: 10.1016/S0735-1097(12)60862-6. [DOI] [Google Scholar]
- 17.Levin MC, Marullo S, Muntaner O, Andersson B, & Magnusson Y (2002). The Myocardium-protective Gly-49 Variant of the β1-Adrenergic Receptor Exhibits Constitutive Activity and Increased Desensitization and Down-regulation. Journal of Biological Chemistry, 277(34), 30429–30435. doi: 10.1074/jbc.M200681200. [DOI] [PubMed] [Google Scholar]
- 18.Terra SG, Hamilton KK, Pauly DF, Lee CR, Patterson JH, Adams KF, et al. (2005). Beta1-adrenergic receptor polymorphisms and left ventricular remodeling changes in response to beta-blocker therapy. Pharmacogenetics and genomics, 15(4), 227. [DOI] [PubMed] [Google Scholar]
- 19.de Groote P, Helbecque N, Lamblin N, Hermant X, Mc Fadden E, Foucher-Hossein C, et al. (2005). Association between beta-1 and beta-2 adrenergic receptor gene polymorphisms and the response to beta-blockade in patients with stable congestive heart failure. Pharmacogenetics and Genomics, 15(3), 137–142. doi: 10.1097/01213011-200503000-00001. [DOI] [PubMed] [Google Scholar]
- 20.Nonen S, Okamoto H, Fujio Y, Takemoto Y, Yoshiyama M, Hamaguchi T, et al. (2008). Polymorphisms of norepinephrine transporter and adrenergic receptor α1D are associated with the response to β-blockers in dilated cardiomyopathy. The Pharmacogenomics Journal, 8(1), 78–84. doi: 10.1038/sj.tpj.6500450. [DOI] [PubMed] [Google Scholar]
- 21.Chen L, Meyers D, Javorsky G, Burstow D, Lolekha P, Lucas M, et al. (2007). Arg389Gly-beta1-adrenergic receptors determine improvement in left ventricular systolic function in nonischemic cardiomyopathy patients with heart failure after chronic treatment with carvedilol. Pharmacogenetics and genomics, 17(11), 941. doi: 10.1097/FPC.0b013e3282ef7354 [DOI] [PubMed] [Google Scholar]
- 22.Perez JM, Rathz DA, Petrashevskaya NN, Hahn HS, Wagoner LE, Schwartz A, et al. (2003). [beta]1-adrenergic receptor polymorphisms confer differential function and predisposition to heart failure. Nature Medicine, 9(10), 1300. doi: 10.1038/nm930. [DOI] [PubMed] [Google Scholar]
- 23.Luo M, Bi Y, & Xu YX (2007). Effects of metoprolol on beta1 adrenergic receptor polymorphism and receptor density in urban Chinese patients with heart failure. Chin Med J (Engl), 120(19), 1720–3. [PubMed] [Google Scholar]
- 24.Hu H, Jui H-Y, Hu F-C, Chen Y-H, Lai L-P, & Lee C-M (2007). Predictors of Therapeutic Response to Beta-blockers in Patients with Heart Failure in Taiwan. Journal of the Formosan Medical Association, 106(8), 641–648. doi: 10.1016/S0929-6646(08)60021-2. [DOI] [PubMed] [Google Scholar]
- 25.Metra M, Covolo L, Pezzali N, Zaca V, Bugatti S, Carlo L, et al. (2010). Role of Beta-Adrenergic Receptor Gene Polymorphisms in the Long-Term Effects of Beta-Blockade with Carvedilol in Patients with Chronic Heart Failure. Cardiovascular Drugs and Therapy, 24(1), 49–60. doi: 10.1007/s10557-010-6220-5. [DOI] [PubMed] [Google Scholar]
- 26.Borjesson M, Magnusson Y, Hjalmarson A, Andersson B (2000). A novel polymorphism in the gene coding for the beta1-adrenergic receptor associated with survival in patients with heart failure. European Heart Journal, 21(22), 1853–1858. doi: 10.1053/euhj.1999.1994. [DOI] [PubMed] [Google Scholar]
- 27.Magnusson Y, Levin MC, Eggertsen R, Nystrom E, Mobini R, Schaufelberger M, et al. (2005). Ser49Gly of beta1-adrenergic receptor is associated with effective beta-blocker dose in dilated cardiomyopathy. Clinical pharmacology and therapeutics, 78(3), 221 10.1016/j.clpt.2005.06.004 [DOI] [PubMed] [Google Scholar]
- 28.Cresci S, Kelly RJ, Cappola TP, Diwan A, Dries D, Kardia SL, et al. (2009). Clinical and Genetic Modifiers of Long-Term Survival in Heart Failure. Journal of the American College of Cardiology, 54(5), 432–444. doi: 10.1016/j.jacc.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sehnert AJ, Daniels SE, Elashoff M, Wingrove JA, Burrow CR, Horne B, et al. (2008). Lack of Association Between Adrenergic Receptor Genotypes and Survival in Heart Failure Patients Treated With Carvedilol or Metoprolol. Journal of the American College of Cardiology, 52(8), 644–651. doi: 10.1016/j.jacc.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 30.White HL, Boer RA, Maqbool A, Greenwood D, Veldhuisen DJ, Cuthbert R, et al. (2003). An evaluation of the beta-1 adrenergic receptor Arg389Gly polymorphism in individuals with heart failure: a MERIT-HF sub-study. European Journal of Heart Failure, 5(4), 463–468. doi: 10.1016/S1388-9842(03)00044-8. [DOI] [PubMed] [Google Scholar]
- 31.The Beta-Blocker Evaluation of Survival Trial Investigators. (2001). A Trial of the Beta-Blocker Bucindolol in Patients with Advanced Chronic Heart Failure. New England Journal of Medicine, 344(22), 1659–1667. doi: 10.1056/NEJM200105313442202. [DOI] [PubMed] [Google Scholar]
- 32.Zou C.-h., Zhang J, Zhang Y.-h., Wei B.-q., Wu X.-f., Zhou Q, et al. (2014). Frequency and Predictors of Normalization of Left Ventricular Ejection Fraction in Recent-Onset Nonischemic Cardiomyopathy. The American Journal of Cardiology, 113(10), 1705–1710. doi: 10.1016/j.amjcard.2014.02.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


